Abstract
A target with therapeutic potential, lysine-specific demethylase 1A (KDM1A) is a regulator of gene expression whose tower domain is a protein–protein interaction motif. This domain facilitates the interaction of KDM1A with coregulators and multiprotein complexes that direct its activity to nucleosomes. We describe the design and characterization of a chimeric ‘towerless’ KDM1A, termed nΔ150 KDM1AΔTower KDM1B chimera (chKDM1AΔTower), which incorporates a region from the paralog lysine-specific demethylase 1B (KDM1B). This chimera copurifies with FAD and displays demethylase activity, but fails to bind the partner protein corepressor of the RE1-silencing transcription factor (CoREST). We conclude that KDM1A catalysis can be decoupled from tower-dependent interactions, lending chKDM1AΔTower useful for dissecting molecular contributions to KDM1A function.
Keywords: KDM1A/LSD1, Tower domain, Chimera, Deletion mutant, Enzyme engineering, CoREST
1. Introduction
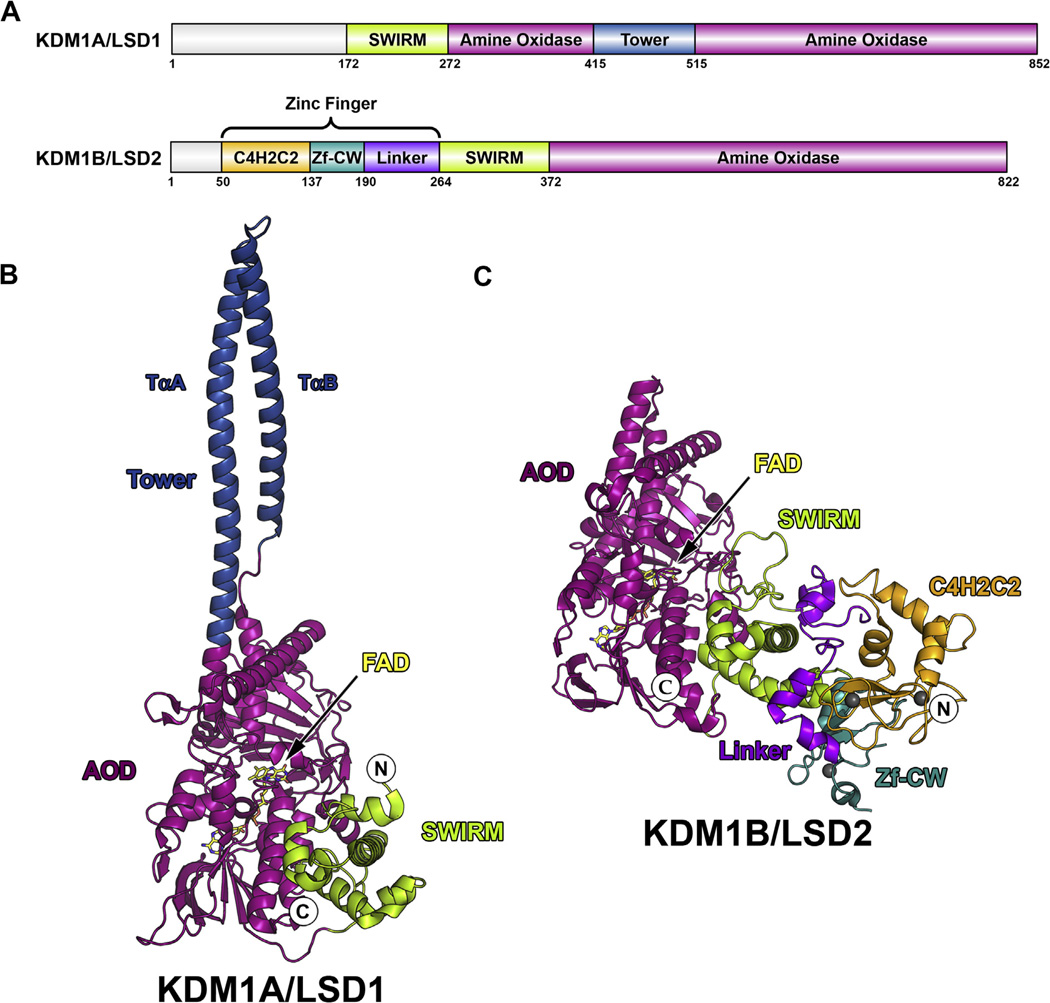
In 2004, lysine-specific demethylase 1A (KDM1A also known as LSD1/BHC110/AOF2/KIAA0601/p110b) became the first histone demethylase to be isolated and characterized [1] and has subsequently been identified as a potential therapeutic target [2–4]. KDM1A is a flavin-dependent amine oxidase that demethylates mono- and dimethylated lysine residues at positions 4 and 9 on histone H3 (H3K4me1/2 and H3K9me1/2), leading to gene activation and repression, respectively [1,5,6]. This 852 amino acid (aa) polypeptide is composed of three structured domains (Fig. 1A). The catalytic amine oxidase domain (AOD) houses a single, non-covalently bound flavin adenine dinucleotide (FAD) cofactor required for catalysis while the N-terminal SWI3p, Rsc8p, and Moira (SWIRM) domain is commonly found in chromatin-associating proteins (Fig. 1B) [7–10]. The ‘tower’ domain is a nearly 100 aa AOD insert that forms an approximately 90 Å long antiparallel coiled–coil from two α-helices (termed TαA and TαB) linked by a tight turn (Fig. 1B) [7,8]. Unprecedented among related amine oxidases [1], the tower domain serves as a protein interaction motif and facilitates KDM1A incorporation into multiprotein regulatory complexes that dictate its cellular function [11].
Fig. 1.
Domain maps and structural overview of KDM1A and KDM1B. (A) Domain maps of KDM1A and KDM1B. SWIRM domains are shown in green, AODs are shown in magenta, tower domain is shown in blue, C4H2C2 domain is shown in orange, Zf-CW domain is shown in cyan, and linker is shown in purple. Structural overview of (B) KDM1A (PDB 2IW5) and (C) KDM1B (PDB 4HSU). Coloring scheme follows that of domain map above, FAD cofactor is shown in yellow, and N and C-termini are labeled. Zinc ions in KDM1B structure are shown as gray spheres.
One of the best-studied tower domain interaction partners is the corepressor of the RE1-silencing transcription factor (CoREST/RCOR1/KIAA0071), which facilitates demethylation of nucleosomal substrates by bridging KDM1A to chromatin through its DNA-binding SANT domains [8,12–17]. Additionally, CoREST orchestrates the inclusion of enzymes such as KDM1A and histone deacetylases as catalytic subunits within modular multiprotein complexes [18,19]. KDM1A activity is further governed by other homologous proteins that bind the tower domain and perform similar functions in different molecular contexts, such as the CoREST paralogs RCOR2 and RCOR3 and the MTA proteins [17,20–22]. Therefore, one of the most significant challenges is understanding the impact of these tower-dependent interactions on KDM1A specificity, catalysis, and positioning within chromatin.
In order to evaluate these tower-dependent interactions independent of demethylase activity, a KDM1A tower domain deletion mutant is highly desired. Indeed, several groups have generated such mutants, but were limited by a lack of enzymatic activity [7,8,21,23]. These studies suggest that either the tower domain is inherently required for KDM1A catalysis or that the methods chosen to bridge the tower region were not conducive to proper active site folding. Interestingly the recent characterization of the KDM1A paralog KDM1B provides a unique opportunity to investigate this dichotomy [24]. KDM1B lacks a tower domain (Fig. 1A) but conserves the AOD architecture, sharing a 2.0 Å RMSD compared to KDM1A, despite a modest sequence similarity (<25%) (Fig. 1B and C). As this paralog overcomes exclusion of the tower domain in a manner that preserves a KDM1A-like active site conformation, we sought to use it as a template for engineering a chimeric KDM1A enzyme.
Here we report the rational design and characterization of a KDM1A tower domain deletion chimera. KDM1A and KDM1B sequence and structural alignments suggested that KDM1B residues V494–L531 could replace the KDM1A tower domain (residues T389–R524) with minimal active site disturbance. This construct, termed chKDM1AΔTower, copurified from Escherichia coli cellular lysates with a stoichiometric equivalent of FAD. As expected, chKDM1AΔTower failed to bind CoREST. However, unlike previous tower deletion mutants, our chimera exhibits kinetic parameters nearly identical to those of unaltered KDM1A and KDM1B, suggesting that the tower domain is not required for catalytic activity. The chKDM1AΔTower chimera therefore decouples tower-dependent protein interactions from catalysis and provides a tool to assess the effects of KDM1A mistargeting and orphanization.
2. Materials and methods
2.1. Reagents and materials
Clones of genes encoding Homo sapiens nΔ150 KDM1A (UniProtKB accession No. O60341) and full-length KDM1B (UniProtKB accession No. Q8NB78) were codon optimized for E. coli by GenScript (Piscataway, NJ) and subcloned into pET-15b (Novagen) with NdeI and XhoI (New England Biolabs; NEB). The pDB-HisGST vector was obtained from the DNASU Plasmid Repository. Buffer salts were obtained from Sigma, EMD Millipore, and JT Baker. Tween 20 was obtained from AMRESCO. Protein purification was conducted using an ÄKTA FPLC (Amersham Biosciences).
2.2. Alignment of KDM1A and KDM1B and generation of a chimera model
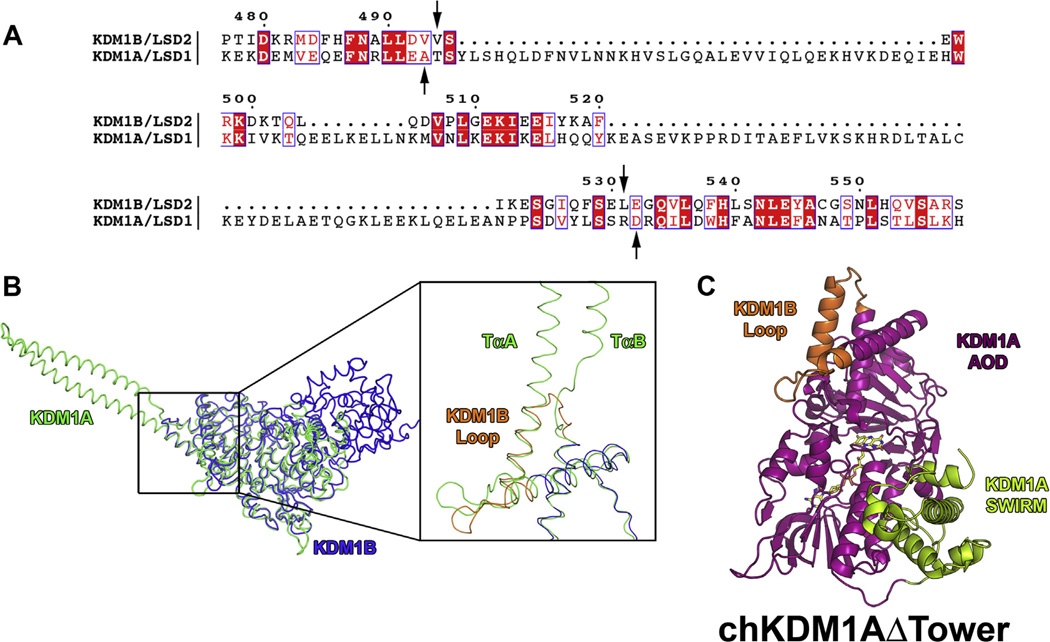
Primary amino acid sequences of KDM1A and KDM1B were aligned using Clustal Omega [25]. A sequence alignment figure (Fig. 2A) was generated utilizing ESPript 3.0 [26]. Structural alignment of PDB files 2IW5 and 4HSU (KDM1A [13] and KDM1B [27], respectively) was conducted with PyMOL [28] (Fig. 2B). To generate a composite model of the chimera, the headers of PDB files 2IW5 and 4HSU were deleted and coordinates superimposed using Coot [29]. KDM1A tower domain residues T389–R524 were replaced with KDM1B residues V494–L531. The resultant chain was renumbered and coordinates of the chimera composite model exported into PyMOL (Fig. 2C). All-atom contacts were validated with MolProbity [30].
Fig. 2.
Sequence and structural alignment of KDM1A and KDM1B from H. sapiens and structural model of chKDM1AΔTower. Only residues 171–852 of KDM1A were used for alignment as per Karytinos et al. (A) Sequence alignment of KDM1A and KDM1B. Numbering is based on the primary amino acid sequence of KDM1B. Residues that are invariant in the two enzymes are highlighted in red and conservative mutations are indicated by red font. Arrows indicate break and splice points chosen for the chimera. (B) Structural alignment of KDM1A and KDM1B. KDM1A is shown in green and KDM1B is shown in blue. Inset shows ~90° rotation and close up of the aligned structures with TαA and TαB of the tower domain denoted and the KDM1B loop shown in orange. (C) Structural model of chKDM1AΔTower. AOD is shown in magenta, SWIRM domain is shown in green, and KDM1B loop is shown in orange.
2.3. Cloning of chKDM1AΔTower
Joining KDM1A fragments L151–A388 and D525–M852 to KDM1B residues V494–L531 formed the chimera sequence. A previously described pET-15b vector containing 6 × His nΔ150 KDM1A (residues 151–852) [31] was used as a template for construction of the chimera. This entire vector was amplified with exclusion of the KDM1A tower domain (residues T389–R524) using primers that incorporated SalI and KpnI restriction sites and PFU Turbo DNA polymerase (Agilent) under the following conditions: an initial denaturation step for 2 min at 95 °C, 30 cycles of denaturation for 30 s at 95 °C, annealing for 30 s over a gradient from 54 to 65 °C, elongation for 8 min at 72 °C, and a final elongation step for 10 min at 72 °C. The KDM1B insert (residues V494–L531) was amplified with complementary restriction sites under similar conditions, but with an elongation of 1 min at 72 °C. The resulting insert and vector were digested and ligated utilizing a Quick Ligation Kit (NEB). Restriction sites were removed using a Q5 Site-Directed Mutagenesis Kit (NEB) under the following conditions: an initial denaturation step for 1 min at 98 °C, 30 cycles of denaturation for 30 s at 98 °C, annealing for 30 s over a gradient from 59 to 70 °C, elongation for 4 min at 72 °C, and a final elongation step for 2 min at 72 °C. For primers, see Table S1.
2.4. Expression and purification of KDM1A and chKDM1AΔTower
Expression and purification of wild type nΔ150 KDM1A was conducted as previously described [15,31]. The 6 × His-tagged nΔ150 KDM1AΔTower KDM1B chimera in pET-15b (i.e. chKDM1AΔTower) was expressed in BL21 Star (DE3) E. coli (Invitrogen) at 15 °C. chKDM1AΔTower was purified under similar conditions to wild type nΔ150 KDM1A with minor modifications. Approximately 4.5 mg of chKDM1AΔTower per liter of culture were obtained at >95% purity. For more details, please see the Supplementary Material.
2.5. Cofactor analysis of chKDM1AΔTower
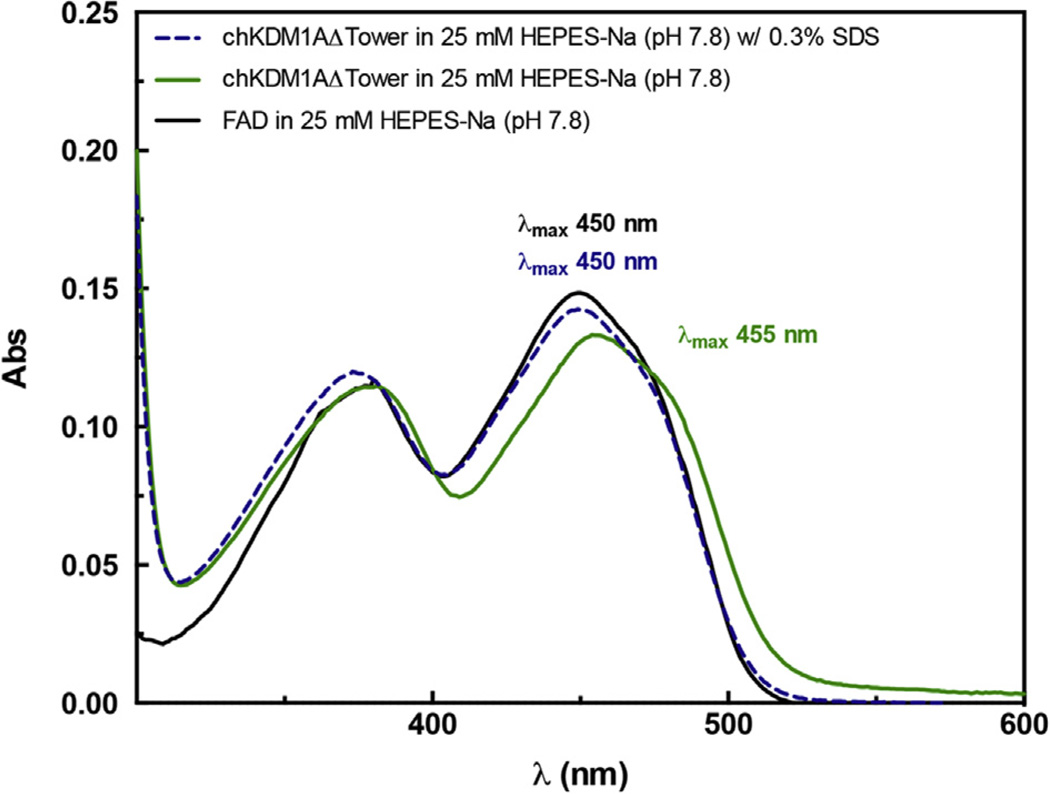
The method of Aliverti et al. was employed to determine the FAD molar extinction coefficient [32]. chKDM1AΔTower in gel filtration buffer at 1.5–2.0 mg/mL was incubated with SDS to a final concentration of 0.3% (w/v) and monitored until the UV trace stabilized. 15 µM FAD disodium salt hydrate (Sigma) in gel filtration buffer was used as a standard. The chimera was calculated to copurify with FAD in 1.1:1 ratio (FAD:protein) and to have a molar extinction coefficient of 10350 M−1 cm−1 at 455 nm using ε450 = 11300 M−1 cm−1 for free FAD (FADfree). This value was routinely used to determine protein concentration.
2.6. Expression and purification of His-GST-CoREST-C and His-GST
The His-GST-CoREST-C construct was expressed in BL21 (DE3) E. coli (Novagen) at 19 °C. Protein was purified by immobilized metal affinity chromatography (IMAC) and ion exchange chromatography (IEC). Approximately 2.0 mg protein per liter of culture were obtained at >75% purity. The His-GST construct was expressed and purified in a similar manner. For more details, see the Supplementary Material.
2.7. Steady-state demethylase assay
A peptide corresponding to the N-terminal 21 amino acids of histone H3 with a dimethylated K4 residue (H3K4me21–21) was prepared as previously described [31,33]. A continuous, fluorescence-based, steady-state kinetic assay was employed as previously described with slight modifications [31,33]. An HRP-coupled (1 U/mL of HRP) assay monitored enzymatic peroxide production in 50 mM Tris–HCl (pH 7.85) with 0.01% (w/v) CHAPS using Amplex Red (50 µM) as the fluorogenic electron acceptor. All measurements were performed in a final volume of 60 µL at 25 °C in 96-well format (Corning 3693). Reactions were initiated with the addition of enzyme (0.35 µM final concentration). The product, resorufin, was monitored with 535 nm excitation and 597 nm emission wavelengths. Data were analyzed using GraphPad Prism 6.0 (GraphPad Software, San Diego, CA). All data sets were corrected for background and zero by subtraction of the no enzyme control and the initial time point, respectively. Initial velocities within the 10% product conversion limit were fit to the Henri–Michaelis–Menten equation with non-linear regression analysis.
2.8. His-GST-CoREST-C pull-down interaction assay
A total of 15 µg of purified His-GST-CoREST-C (residues 286–482) or 9 µg His-GST were incubated with 12 µg nΔ150 KDM1A or 10 µg chKDM1AΔTower at 4 °C for 16 h in 100 µL of binding buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (pH 7.4), 10 mM DTT, and 0.05% (v/v) Tween 20) containing 20 µL of Glutathione Sepharose 4 Fast Flow beads (GE Healthcare). Flow-through (supernatant) was removed and beads were washed 3 times with 400 µL of binding buffer, resuspended in 80 µL of 2× SDS sample buffer, and denatured for 2 min at 100 °C. Input, beads (bound), and flow-through were analyzed by SDS–PAGE (4–20% gradient, Bio-Rad) and visualized with Coomassie Brilliant Blue.
3. Results
3.1. Design of the chKDM1AΔTower chimera from KDM1A and KDM1B alignments
To rationally design a tower domain deletion mutant of KDM1A, we looked to its ‘towerless’ paralog KDM1B, which performs identical chemical reactions but interacts with a non-identical set of biomolecules. We therefore used sequence and structural alignments to define a sequence from KDM1B that could replace the tower domain of KDM1A and maintain active site geometry. As predicted, a large gap in the sequence alignment of KDM1A and KDM1B reflects the KDM1A tower domain with areas of considerable sequence conservation on either side (Fig. 2A). We should note that a small internal area of apparent homology is likely an artifact of shared α-helical secondary structure. Subsequent structural alignment and analysis indicated that while the region C-terminal to the tower domain is highly similar between the two enzymes, the corresponding N-terminal region in KDM1B contains a solvent exposed loop not conserved in KDM1A (Fig. 2B). In order to maintain the interhelical contacts in this region (e.g. KDM1B W497 with K512 and I516 with V493), we extended the tower replacement sequence to include this entire loop and also supplanted a portion of the KDM1A α-helix that contacts the base of the tower domain (termed αCox or Sα1) [7,8]. The final chKDM1AΔTower design incorporated these KDM1B elements into KDM1A, with a final sequence of KDM1A fragments L151–A388 and D525–M852 linked by KDM1B fragment V494–L531 (Fig. S1).
Next, a preliminary evaluation of this design was performed by generating a composite structural model of chKDM1AΔTower using Coot (Fig. 2C). Subsequent visual inspection of the model in PyMOL revealed no gross structural abnormalities. Additionally, analysis of all-atom contacts with MolProbity [30] supported this observation, as no steric clashes in the replaced tower region were visible. We therefore predicted that replacement of the KDM1A tower with the KDM1B loop would result in a properly folded, active enzyme.
3.2. Construction, expression, and purification of the chKDM1AΔTower chimera
We next sought to generate the chKDM1AΔTower chimera in which the KDM1B loop was inserted into the KDM1A sequence in place of the tower domain using restriction digestion cloning. After insertion, restriction sites were removed to yield a ‘seamless’ chimera that contained only residues from the primary amino acid sequences of KDM1A and KMD1B (Fig. S2). The chimeric clone was then overexpressed in E. coli under nearly identical conditions as reported for wild type KDM1A [15,31] resulting in a visible band at the expected molecular weight of approximately 70kDa (Fig. S3A). Purification of chKDM1AΔTower from E. coli cellular lysates using a series of chromatographic separations resulted in a >95% homogenous sample as assessed by SDS–PAGE (Fig. S3B).
Our purified chKDM1AΔTower solution appears yellow in color (Fig. S3C), suggesting it retains the essential FAD cofactor as previously observed for KDM1A and KDM1B. Indeed, a UV–visible spectrum of the chKDM1AΔTower produced characteristic flavin cofactor absorbance maxima at 380 and 455 nm (Fig. 3) [34]. As compared to an authentic FAD standard, the molar extinction coefficient (FADbound) of the chKDM1AΔTower construct was determined to be 10350 M−1 cm−1 at 455 nm [32,34] (Fig. 3). Furthermore, full cofactor occupancy of chKDM1AΔTower was noted, as a stoichiometric amount of FAD was released upon SDS treatment. This FAD cofactor was also released into solution as a function of heat or TCA denaturation (data not shown). These data therefore suggest that, like KDM1A and KDM1B, the FAD cofactor of the chimera is non-covalently bound and potentially poised to initiate catalytic demethylation.
Fig. 3.
chKDM1AΔTower copurifies with a non-covalently bound FAD cofactor. chKDM1AΔTower was evaluated by UV–Vis and has absorption maxima at 380 and 455 nm (solid green line). The chimera was denatured by treatment with SDS, releasing the cofactor (dashed blue line). The spectrum of the denatured sample directly overlays with a FAD standard (solid black line).
3.3. chKDM1AΔTower exhibits similar demethylase activity as KDM1A and KDM1B
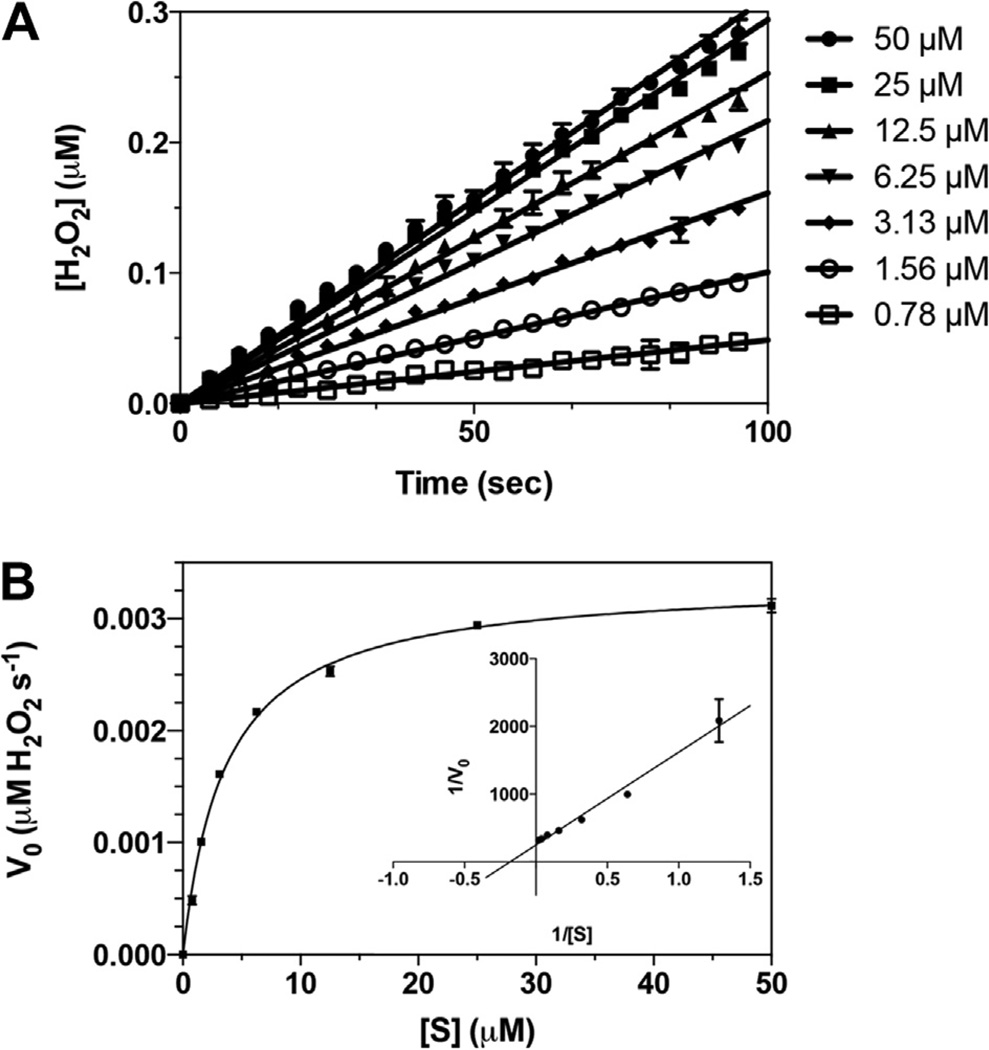
As the chKDM1AΔTower-bound flavin chromophore is suggestive of properly assembled catalytic machinery, we next tested for bona fide enzymatic activity. Steady-state kinetic parameters of chKDM1AΔTower were evaluated by monitoring enzymatic production of H2O2 using a peptide substrate. As shown in Fig. 4, chKDM1AΔTower exhibited well-behaved linear velocities and demethylated H3K4me21–21 with a of 3.21 ± 0.16 µM, a of 0.57 ± 0.01 min−1, and a catalytic efficiency of 0.18 ± 0.01 µM−1 min−1. These values compare very favorably to that of wild type KDM1A and KDM1B enzymes (Table 1). In fact, the of chKDM1AΔTower was identical to that of KDM1B, and only approximately 10-fold lower than that of KDM1A [24,31]. Interestingly, values of both and of the chimera were situated neatly near those of the two enzymes (Table 1). These data suggest that chKDM1AΔTower preserves elements from both KDM1A and KDM1B as anticipated, and clearly maintains integrity of the amine oxidase active site. Thus, by grafting the KDM1B loop onto KDM1A, we generated a tower domain deletion mutant that retains wild type-like demethylase activity.
Fig. 4.
chKDM1AΔTower is an active enzyme. (A) Representative linear fit of initial rates from substrate titration against chKDM1AΔTower. Plots are within 10% product conversion. Concentrations indicated are that of the H3K4me21–21 peptide substrate. (B) Representative initial velocity curve of the catalytic activity of chKDM1AΔTower. Data are fit to the Henri–Michaelis–Menten equation. Inset is a reciprocal plot, 1/V0 (s µM−1 H2O2) vs. 1/[S] (µM−1), to illustrate linear nature of data.
Table 1.
Kinetic Parameters of the Catalytic Activity of chKDM1AΔTower with the First 21 Amino Acids of H3 Dimethylated at K4 (H3K4me21–21).
3.4. chKDM1AΔTower does not interact with KDM1A binding partner CoREST
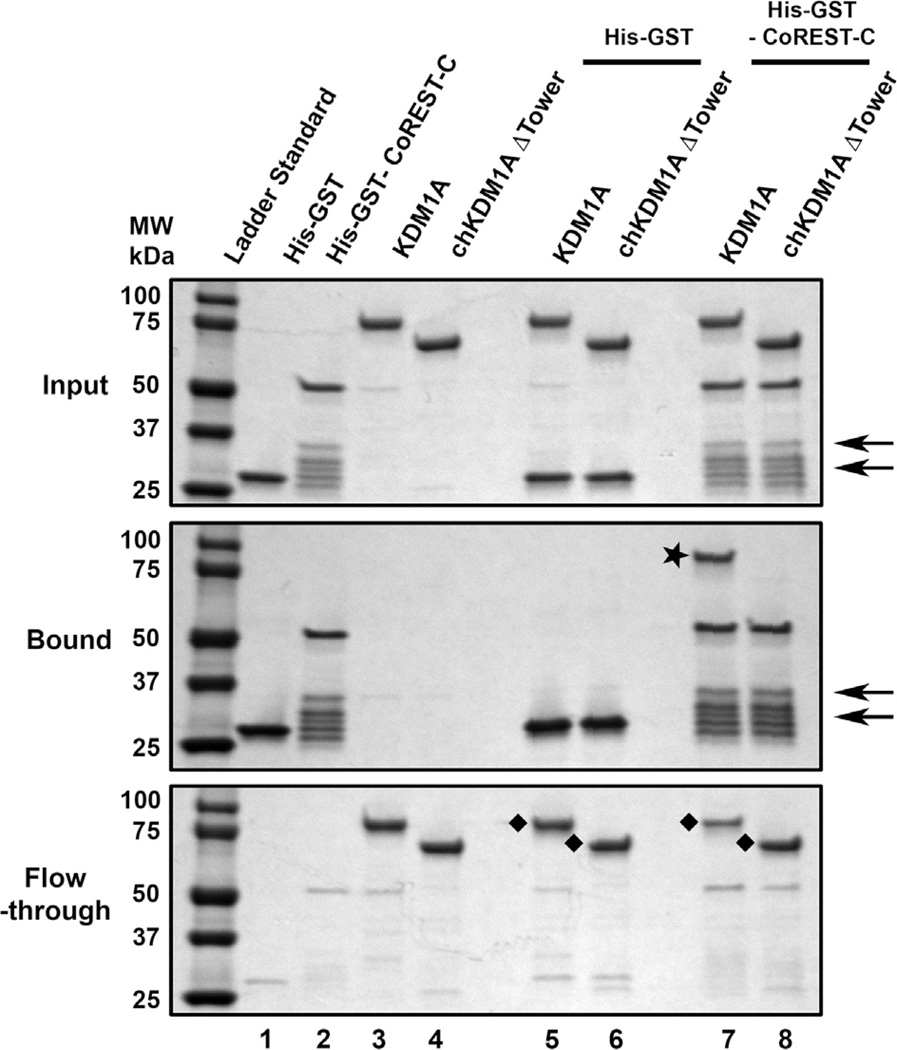
Having achieved one of our goals in engineering an active KDM1A tower deletion mutant, we next determined if a known tower domain interacting protein was precluded from binding the chimera. The tower domain of wild type KDM1A is a well-known protein recruitment motif for CoREST [8,12,13]. We therefore hypothesized that this interaction would be abrogated in the chimera. A GST pull-down assay previously used to evaluate CoREST binding to wild type KDM1A was employed to assess if chKDM1AΔTower interacts with the C-terminal region of CoREST (CoREST-C; residues 286–482) [12]. Immobilized His-GST-CoREST-C or His-GST were incubated with either wild type KDM1A or chKDM1AΔTower, extensively washed, and the input, flow-through, and bound proteins visualized by SDS–PAGE. As previously reported, His-GST-CoREST-C significantly bound wild type KDM1A as compared to the GST control. Conversely, immobilized His-GST-CoREST-C showed no enrichment for chKDM1AΔTower compared to the control (Fig. 5). These data are in agreement with previous reports that KDM1A interaction with the C-terminal fragment of CoREST in vitro is mediated mainly through the tower domain and does not receive significant contributions from the AOD and SWIRM domains [8,13]. Hence our chimera effectively abrogates association with CoREST, and presumably any other protein that relies predominantly on the tower domain for binding.
Fig. 5.
Unlike wild type KDM1A, chKDM1AΔTower is incapable of interacting with CoREST. Comparable wild type and deletion mutant of KDM1A (chKDM1AΔTower) were incubated with glutathione Sepharose beads in the presence or absence of His-GST-CoREST-C or His-GST (negative control). The input (top), bound (middle), and flow-through (bottom) proteins were analyzed by SDS–PAGE. The star (⋆) in middle indicates bound KDM1A (lane 7) and the diamonds (◆) in bottom indicate the unbound KDM1A (lanes 5 and 7) and chKDM1AΔTower (lanes 6 and 8) after incubation with His-GST-CoREST-C or His-GST. The arrows indicate major truncation products of the His-GST-CoREST-C protein.
4. Discussion
There is currently a remarkable lack of tools for studying the influence of tower-dependent interactions on KDM1A recruitment into modular multiprotein complexes, substrate specificity, and chromatin localization. To address this issue, we here report what is, to the best of our knowledge, the first KDM1A tower deletion enzyme that retains catalytic activity comparable to wild type KDM1A and KDM1B. Using sequence and structure-driven design principles, we have rationally selected boundaries for excision of theKDM1Atower domain and replacement with a bridging segment from KMD1B. This chimera, termed chKDM1AΔTower, copurifies with a stoichiometric equivalent of FAD and exhibits catalytic demethylase activity commensurate with its parent enzymes. Additionally, our in vitro pull-down studies demonstrate that the absence of a tower domain in chKDM1AΔTower precludes the tower-dependent binding interaction observed between CoREST and KDM1A, effectively uncoupling catalysis from tower-dependent interactions.
Our rational design demonstrates that the tower domain is not required for catalytic demethylation, thereby overcoming the catalytic inactivity of previously reported KDM1A tower deletion mutants. We suspect the catalytic deficiencies of these mutants may be attributed to subtle changes in active site geometry that alter substrate positioning [35]. However, it appears that insertion of the bridging KDM1B sequence instead mimics the native span between KDM1A residues A388 and D525 and facilitates folding of the chimera into a catalytically competent conformation. Although the catalytic efficiency of chKDM1AΔTower varies only minimally from that of KDM1A, we cannot rule out the possibility that our design may yet be catalytically suboptimal.
By combining catalytic activity with the lack of a tower domain, our chimera provides a much-needed probe for dissecting the influence of tower-dependent protein interactions on KDM1A function. KDM1A and KDM1B are believed to assemble in a modular fashion into larger, multivalent complexes that dictate their intracellular function [11]. As the tower domain is responsible for a large number of KDM1A interactions, chKDM1AΔTower provides an ideal tool for determining the effects of removing KDM1A from the control of its interaction partners. We predict this will likely result in mistargeting of the enzyme on chromatin and alteration of downstream genetic programming. For example, we suspect that chKDM1AΔTower will no longer localize with CoREST in their coregulated promoter regions, thereby functionally disrupting the transcriptional programs for this subset of genes. Beyond KDM1A, this strategy of splicing related regions between functionally related proteins may prove to be an invaluable tool for investigating domains for which standard domain deletion proves intractable. This strategy is specifically useful when investigating enzymes that exist as subunits of multimeric complexes.
Supplementary Material
Acknowledgments
We thank members of the McCafferty laboratory for their thoughtful insight during the preparation of this manuscript. This work was kindly supported by the U.S. DoD CDMRP Grant W81XWH-13-1-0400 (to D.G.M.) and by NIH Predoctoral Training Grants and T32-GM008487-19 in Structural Biology and Biophysics (to J.M.B.) and T32-GM007105-39 in Pharmacological Sciences (to J.E.L.).
Abbreviations
- AOD
amine oxidase domain
- chKDM1AΔTower
nΔ150 KDM1AΔTower KDM1B chimera
- CoREST
corepressor of the RE1-silencing transcription factor
- KDM1A
lysine-specific demethylase 1A
- KDM1B
lysine-specific demethylase 1B
- SWIRM
SWI3p, Rsc8p, and Moira
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.febslet.2015.07.028.
Author Contributions: J.B. and D.M. conceived this study; J.B., C.P., and D.M. planned experiments; J.B., A.M., C.P., J.L., and F.H. executed experiments and gathered data; J.B. and D.M. analyzed data; J.B., J.L., and D.M. wrote and edited the manuscript; D.M. supervised all aspects of this study.
References
- 1.Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Friso S, Carvajal CA, Fardella CE, Olivieri O. Epigenetics and arterial hypertension: the challenge of emerging evidence. Transl. Res. 2015;165:154–165. doi: 10.1016/j.trsl.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Kristie TM. Dynamic modulation of HSV chromatin drives initiation of infection and provides targets for epigenetic therapies. Virology. 2015;479–480:555–561. doi: 10.1016/j.virol.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat. Rev. Drug Discovery. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 5.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 6.Laurent B, Ruitu L, Murn J, Hempel K, Ferrao R, Xiang Y, Liu S, Garcia BA, Wu H, Wu F, Steen H, Shi Y. A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol. Cell. 2015;57:957–970. doi: 10.1016/j.molcel.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stavropoulos P, Blobel G, Hoelz A. Crystal structure and mechanism of human lysine-specific demethylase-1. Nat. Struct. Mol. Biol. 2006;13:626–632. doi: 10.1038/nsmb1113. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Yang Y, Wang F, Wan K, Yamane K, Zhang Y, Lei M. Crystal structure of human histone lysine-specific demethylase 1 (LSD1) Proc. Natl. Acad. Sci. U.S.A. 2006;103:13956–13961. doi: 10.1073/pnas.0606381103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Da G, Lenkart J, Zhao K, Shiekhattar R, Cairns BR, Marmorstein R. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2057–2062. doi: 10.1073/pnas.0510949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aravind L, Iyer LM. The SWIRM domain: a conserved module found in chromosomal proteins points to novel chromatin-modifying activities. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0039. Research0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burg JM, Link JE, Morgan BS, Heller FJ, Hargrove AE, McCafferty DG. KDM1 class flavin-dependent protein lysine demethylases. Biopolymers. 2015;104:213–246. doi: 10.1002/bip.22643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Yang M, Gocke CB, Luo X, Borek D, Tomchick DR, Machius M, Otwinowski Z, Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 15.Hwang S, Schmitt AA, Luteran AE, Toone EJ, McCafferty DG. Thermodynamic characterization of the binding interaction between the histone demethylase LSD1/KDM1 and CoREST. Biochemistry. 2011;50:546–557. doi: 10.1021/bi101776t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim SA, Chatterjee N, Jennings MJ, Bartholomew B, Tan S. Extranucleosomal DNA enhances the activity of the LSD1/CoREST histone demethylase complex. Nucleic Acids Res. 2015;43:4868–4880. doi: 10.1093/nar/gkv388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pilotto S, Speranzini V, Tortorici M, Durand D, Fish A, Valente S, Forneris F, Mai A, Sixma TK, Vachette P, Mattevi A. Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2752–2757. doi: 10.1073/pnas.1419468112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. U.S.A. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Upadhyay G, Chowdhury AH, Vaidyanathan B, Kim D, Saleque S. Antagonistic actions of Rcor proteins regulate LSD1 activity and cellular differentiation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:8071–8076. doi: 10.1073/pnas.1404292111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, Li R, Li Y, Zhang Y, Li Q, Yi X, Shang Y. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- 22.Barrios AP, Gomez AV, Saez JE, Ciossani G, Toffolo E, Battaglioli E, Mattevi A, Andres ME. Differential properties of transcriptional complexes formed by the CoREST family. Mol. Cell. Biol. 2014;34:2760–2770. doi: 10.1128/MCB.00083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi YI, Evers BM, Zhou BP. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A. A novel mammalian flavin-dependent histone demethylase. J. Biol. Chem. 2009;284:17775–17782. doi: 10.1074/jbc.M109.003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson DG, Higgins DG. () Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F, Yang H, Dong Z, Fang J, Wang P, Zhu T, Gong W, Fang R, Shi YG, Li Z, Xu Y. Structural insight into substrate recognition by histone demethylase LSD2/KDM1b. Cell Res. 2013;23:306–309. doi: 10.1038/cr.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schrödinger: LLC; [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaweska H, Henderson Pozzi M, Schmidt DM, McCafferty DG, Fitzpatrick PF. Use of pH and kinetic isotope effects to establish chemistry as rate-limiting in oxidation of a peptide substrate by LSD1. Biochemistry. 2009;48:5440–5445. doi: 10.1021/bi900499w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aliverti A, Curti B, Vanoni MA. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol. Biol. 1999;131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt DM, McCafferty DG. Trans-2-phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46:4408–4416. doi: 10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 34.Macheroux P. UV–visible spectroscopy as a tool to study flavoproteins. Methods Mol. Biol. 1999;131:1–7. doi: 10.1385/1-59259-266-X:1. [DOI] [PubMed] [Google Scholar]
- 35.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem. Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.