Abstract
The Ecteinascidin family comprises a number of biologically active compounds, containing two to three tetrahydroisoquinoline subunits. Although isolated from marine tunicates, these compounds share a common pentacyclic core with several antimicrobial compounds found in terrestrial bacteria. Among the tetrahydroisoquinoline natural products, Ecteinascidin 743 (Et-743) stands out as the most potent antitumor antibiotics that it is recently approved for treatment of a number of soft tissue sarcomas. In this article, we will review the backgrounds, the mechanism of action, the biosynthesis, and the synthetic studies of Et-743. Also, the development of Et-743 as an antitumor drug is discussed.
1. Isolation, Structure and Biological Activity of Et-743
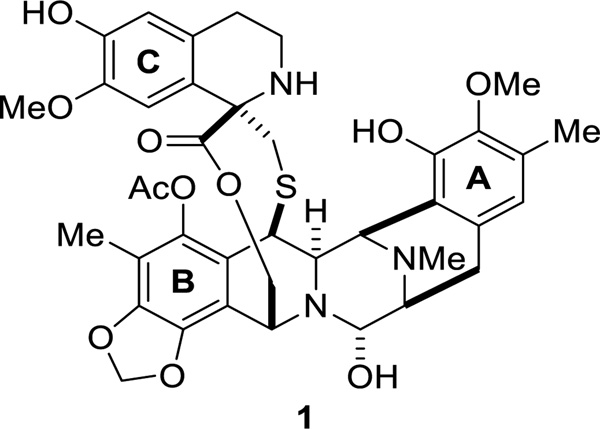
Ecteinascidin 743 (Et-743, 1) is a marine natural product possessing remarkable biological activity against several tumor cell lines.1 It was first isolated in 1990 from a Caribbean tunicate extract Ecteinascidia turbinata,2 and was later identified to be produced by the bacterial symbiont Candidatus Endoecteinascidia frumentensis.3–5 In 1992, Sakai et al. confirmed the structure of Et-743 via the X-ray crystal structure of the derived N12-oxide.6 Et-743 contains three tetrahydroisoquinoline (THIQ) units A, B, and C (Figure 1). The connection between the fully functionalized THIQ units A and B forms a rigid pentacyclic core. Unit C is connected to the pentacycle through a 10-membered macrolactone. Unit A and B are believed to promote the alkylation of DNA through hydrogen bonding with the target DNA. Unit C interacts with DNA-binding proteins, resulting in Et-743’s antitumor activity.7, 8
Figure 1.
Structure of Et-743.
Among the Et family members, Et-743 and Et-729 show the most potent antitumor activity on several carcinoma cell types (Table 1).9 The major emphasis has been on Et-743 because it was the most abundant compound from the tunicate extract.2
Table 1.
Bioactivities of the Ets.
| IC50 (ng/mL) | IC50 (µg/mL) | µg/dics | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L1210 | P388 | A549 | HT29 | MEL28 | CV-1 | PS | DNA | RNA | DNAp | RNAp | Bs | |
| Et-743 | 5.0 | 0.2 | 0.2 | 0.5 | 5.0 | 1.0 | >1 | 0.1 | 0.03 | 2.0 | 0.1 | 0.02 |
| Et-729 | <1.0 | 0.2 | 0.2 | 0.5 | 5.0 | 2.5 | >1 | 0.2 | 0.02 | 1.5 | 0.05 | 0.08 |
| Et-815 | 25 | 2.5 | 5.0 | 5.0 | 5.0 | >1 | 0.1 | 5.0 | 0.75 | |||
| Et-759B | 5.0 | 5.0 | 5.0 | 10 | 25 | >1 | 0.7 | 0.5 | >1 | 3.90 | ||
| Et-745B | 25 | 5.0 | 10 | 10 | 25 | >1 | 0.5 | 3.0 | ||||
| Et-759C | 1.0 | 2.5 | 2.5 | 2.5 | 2.5 | >1 | 0.5 | >5 | 0.1 | |||
| Et-745 | 10 | 20 | 25 | 50 | 50 | >1 | 0.3 | 5.0 | 6.50 | |||
| Et-731 | 100 | 100 | 100 | 200 | 200 | >1 | 6.20 | |||||
| Et-736 | 0.5 | 1.0 | 2.5 | 2.5 | 2.5 | 0.5 | 0.4 | 0.1 | 0.5 | 0.38 | ||
| Et-722 | 1.0 | 1.0 | 2.0 | 2.0 | 5.0 | 0.9 | 0.4 | 0.1 | >1 | 0.5 | 0.70 | |
| Et-594 | 10 | 20 | 25 | 25 | 25 | 0.8 | 0.5 | 0.5 | 1.0 | 0.37 | ||
| Et-597 | 2.0 | 2.0 | 2.0 | 2.0 | 2.5 | 0.7 | 0.08 | 0.01 | 0.25 | 0.14 | ||
| Et-583 | 10 | 10 | 10 | 5.0 | 25 | 1.0 | 1.0 | 0.4 | 0.5 | 0.74 | ||
L1210 = murine lymphocytic leukemia cells; P388 = murine lymphoblastic cells; A549 = human lung carcinoma; HT29 = human colon carcinoma; MEL28 = human melanoma; CV-1 = monkey kidney cells; PS = protein synthesis inhibition; DNA = DNA synthesis inhibition; RNA = RNA synthesis inhibition; DNAp = DNA polymerase inhibition; RNAp = RNA polymerase inhibition; Bs = Bacillus subtilis.
Et-743 has an effect on cell proliferation against a wide range of cancer subtypes at low concentration.10, 11 Izbicka et al. illustrated that a continuous exposure of the tumor to Et-743 resulted in a significant improvement of the antitumor activity in vitro (Table 2).10 Their results also revealed that breast, lung, melanoma and ovarian cancers are the most suitable targets for Et-743.
Table 2.
Antitumor activity of Et-743 against human tumor colonies after one-hour exposure (1HR) versus continuous exposure (CE).10
| Tumor type | 0.1 nM | 1.0 nM | 10 nM | 100 nM | 1000 nM | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1HR | CE | 1HR | CE | 1HR | CE | 1HR | CE | 1HR | CE | |
| Ovary | 0% | 0% | 11% | 22% | 20% | 45% | 0% | 58% | 0% | 67% |
| Breast | 0% | 0% | 0% | 0% | 0% | 40% | 20% | 79% | 40% | 100% |
| Non-small cell lung | 0% | 0% | 0% | 0% | 0% | 50% | _ | 69% | _ | 85% |
| Colon | 0% | 0% | 0% | 0% | 0% | 50% | _ | 43% | _ | 71% |
| Melanoma | _ | _ | _ | _ | _ | 71% | _ | 88% | _ | 86% |
| Kidney | 0% | 0% | 0% | 0% | 0% | 43% | 0% | 50% | 0% | 67% |
| Sarcoma | 0% | 0% | 0% | 0% | 100% | 75% | _ | 67% | _ | 67% |
| Corpus uteri | _ | _ | _ | _ | _ | 67% | _ | 67% | _ | 67% |
| Peritoneum | 0% | 0% | 0% | 0% | 50% | 100% | _ | _ | _ | |
| Head/neck | 0% | 0% | 0% | 0% | 0% | 100% | _ | _ | _ | |
| Lymphoma | _ | _ | _ | _ | _ | 0% | _ | 0% | _ | 0% |
| Pancreas | _ | _ | _ | _ | _ | 100% | _ | 100% | _ | 100% |
| Mesothelioma | _ | _ | _ | _ | _ | 100% | _ | 100% | _ | 100% |
| Prostate | _ | _ | _ | _ | _ | 0% | _ | 0% | _ | 0% |
| Stomach | _ | _ | _ | _ | _ | 0% | _ | 0% | _ | 100% |
2. Mechanism of action of Et-743
At micromolar concentration, Et-743 was found to inhibit a number of DNA-binding proteins, including NF-Y, TATA binding protein, E2F, and SRF/TCF.12 It also activates the formation of topoisomerase I-mediated cleavage complexes, and perturbs microtubule arrangement at this high range of concentration.13–15 These bioactivities were observed at suprapharmacological concentrations of Et-743, rendering these auxiliary effects of Et-743.
Interestingly, the incubation of NF-Y with Et-743 prior to the addition of DNA results in inhibition at a significantly reduced drug level (at 10 µM range). This result suggested that transcription factor NF-Y is the target of Et-743, not the CCAAT box on which NF-Y acts. Since NF-Y interacts with several essential genes involving DNA metabolism and cell-cycle regulators,16 the inhibition of this protein can cause a delay in S-phase progression and accumulation of cells in G2/M phase, which is in agreement with the observation of Simoens and coworkers.17 The inhibition of transcription factor NF-Y, however, still occurs at high concentration of Et-743, three times higher than the pharmacological concentration when pre-incubating NF-Y with Et-743.
The study on Et-743-induced suppression of the multidrug resistant gene MDR1 on the human colon carcinoma cell line SW620 by Jin et al. supports transcription factor NF-Y as one of Et-743’s target in its mechanism of action.18 In contrast to the in vitro study, the inhibition of MDR1 promoter activation occurs at 50 nM; hence this activity may follow a mechanism unrelated to DNA alkylation. Possibly, at its pharmacological concentration, Et-743 interferes with the binding of NF-Y to other co-activators, such as PCAF, which in turn regulates the expression of several critical genes.
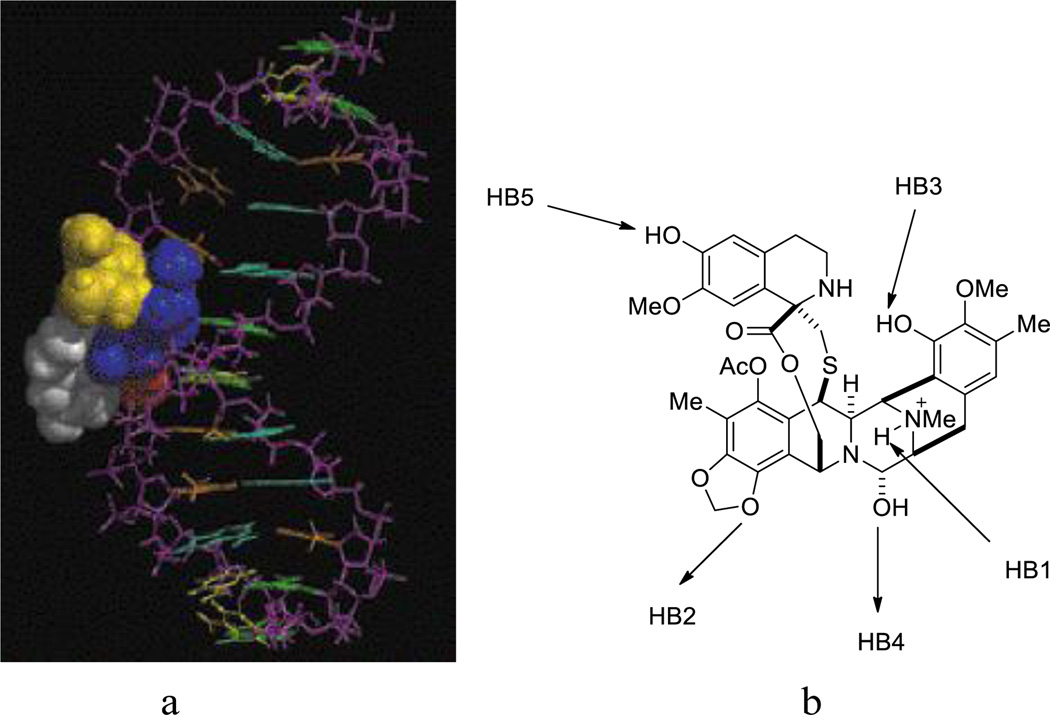
To date, the most widely accepted mechanism of action of Et-743 involves double-strand breaks (DBSs) induced by the transcription-coupled nucleotide excision repair (TC-NER) system following the binding of Et-743 to the minor groove of DNA (Figure 2a). Et-743 alkylates DNA at the N2 of a guanine, forming a DNA adduct.7, 19, 20 The presence of the adduct widens the minor groove of the target DNA, and bends DNA toward the major groove.21, 22 This event is reversible, favoring 5’-CGG, TGG, GGC, AGC sequences, in which the middle bold G is the site of alkylation.23 Computer-based dynamic simulation studies illustrated that these sequences optimize the hydrogen-bonding network to both strands of the DNA and orient the carbinolamine of Et-743 on top of the center guanine (Figure 2b). Seaman and Hurley proposed that the above-favored sequences optimize the coordination of hydrogen bond 1 (HB1) and HB2. The combination of HB1, HB2, and HB3 positions and stabilizes HB4 at the site of alkylation.24 The protonated amine (N12) also acts as a catalyst in the generation of the active iminium ion (N2), thus promoting the alkylation.20 Despite numerous thermodynamically favored target sequences, the efficiency of DNA alkylation by Et-743 is low, suggesting that other factors can contribute to the alkylation process.19 Possibly, the protein–DNA complex increases the sequence selectivity of Et-743.25
Figure 2.
a. Et-743-DNA complex. b. Hydrogen-bond network of Et-743 to DNA. The arrows represent the direction of the hydrogen bonds from a donor to an acceptor.
MDR1 promoter contains two GC-rich sequences, thus they are a target for Et-743 alkylation. This event leads to the suppression of P-glycoprotein (P-gp) expression and was found to reverse the sensitivity of P-gp/MDR1 overexpressing cells (KB-8-5 and KB-C-2) to doxorubicin and vincristine, the two known substrates of P-gp/MDR1.26 Interestingly, Et-743 activity is not subdued at the P-gp levels in human tumor cells, but only at high expression of P-gp as seen in LLC-PK1 pig kidney and Madine-Darby Canine kidney cells.27 This discovery suggested that the use of Et-743 in combination with other antitumor drugs may be beneficial in tumor models resistant to conventional treatments.
DNA adducts formed with Et-743 was found to block gene expressions by causing RNA polymerase II arrest during transcription and competing with certain transcription factors, such as Sp1, for the binding site on DNA.28 These events happen preferentially at the binding site of the fusion proteins FUS-CHOP and EWS-FLI1, leading to the displacement of these oncogenic transcription factors from the promoter.29, 30 FUS-CHOP and EWS-FLI1 were known for their role in tumor initiation, specifically myxoid/round cell liposarcoma type I and II by FUS-CHOP and Ewing sarcoma by EWS-FLI1, resulting in the tumor selectivity of Et-743 on these sarcoma.31–33
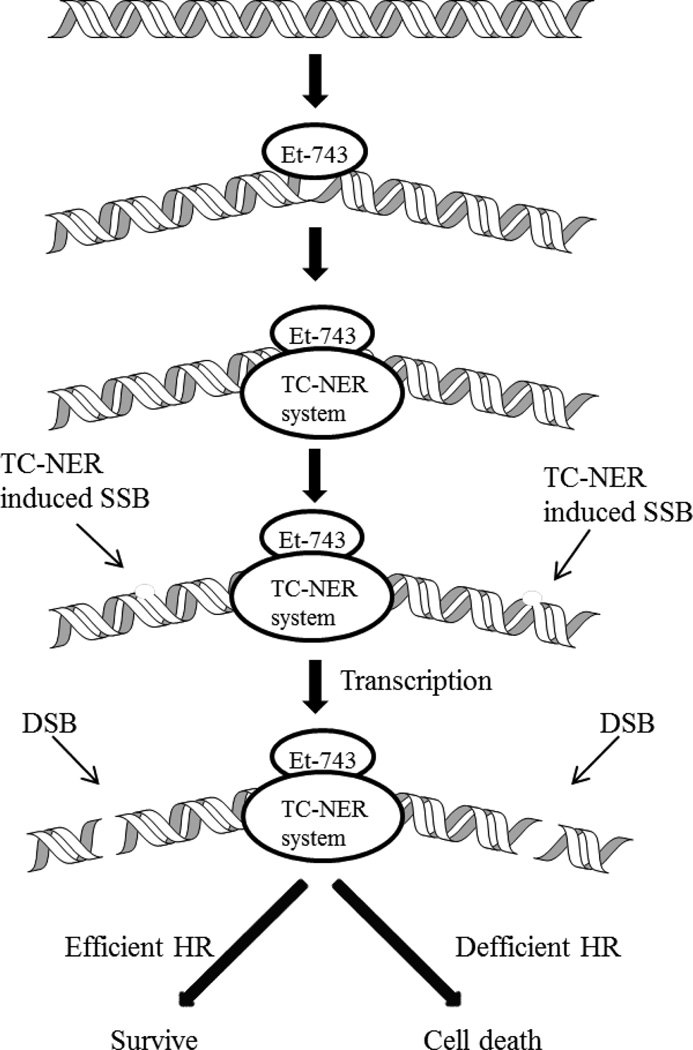
While other known DNA alkylating agents, such as mitomycin C and cisplatin, requires a deficient nucleotide excision repair (NER) system to efficiently exert their biological activity, Et-743 works the best in the presence of a proficient NER system. Cells deficient in NER genes (XPG, XPA, XPD, or XPF) are 2- to 8-fold less sensitive to Et-743, and XPG-deficient cells are more resistant to the drug than other strains.34–36 Moreover, cells deficient in XPC (the gene responsible for recognition of damages in global genome NER (GG-NER)) showed no change in sensitivity to Et-743, elucidating that the toxicity of Et-743 is independent on the GG-NER but is involved the TC-NER machinery.34 Upon formation of a DNA adduct, TC-NER is recruited to the lesion by the use of the XPA gene product (Figure 3). XPD will then unwind the damaged region, and XPF and XPG gene products will induce the single-strand break (SSB) on the opposite strand to the alkylation.28 An Et-743-induced adduct traps the XPG-DNA complex, generating irreversible SSBs,37 which are transformed into double-strand breaks (DSBs) during transcription.35, 38 Homologous recombination (HR) repair can recognize and repair the DSBs induced by Et-743; hence cells lacking HR repair (common in human tumors) are significantly more sensitive to Et-743.38, 39
Figure 3.
Mode of action of Et-743, dependent on DNA replication.
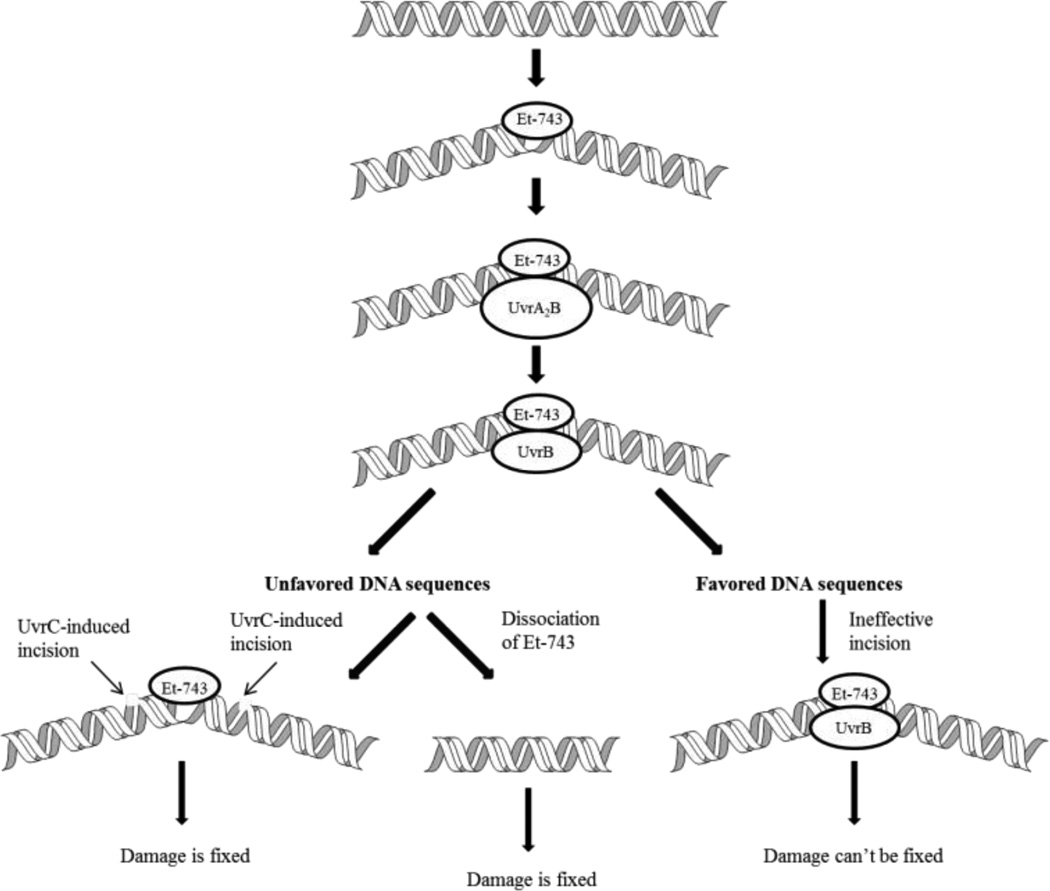
Studies of Zewail-Foote et al. on the less complicated bacterial nuclease UvrABC system demonstrated that the incision event induced by Et-743-DNA adduct is more efficient at the less favored sequence 5’-AGT.40 Based on this result, they proposed a different mechanism of how Et-743 induces apoptosis (Figure 4). The UvrA protein of the UvrABC machinery recognizes the Et-743-DNA lesion and initiates the repair mechanism by forming a dimer with another UvrA protein, then a heterotrimer UvrA2B. This event creates a six-base pair bubble around the alkylated guanine, bending DNA toward its major groove, and resulting in the dissociation of the UvrA2. UvrC is recruited to the UvrB-DNA complex to incise at both ends of the DNA containing the alkylated guanine, thus the lesion can be fixed. Et-743 can also dissociate from the bubble before the incision happens depending on the stability of the Et-743-DNA adduct, which also leads to cell survival. When the repair mechanism occurs at the favored DNA sequences, the incision is not as effective or does not happen at all, generating a toxic lesion and cell death.
Figure 4.
Incision effectiveness at unfavored DNA sequences versus favored DNA sequences.
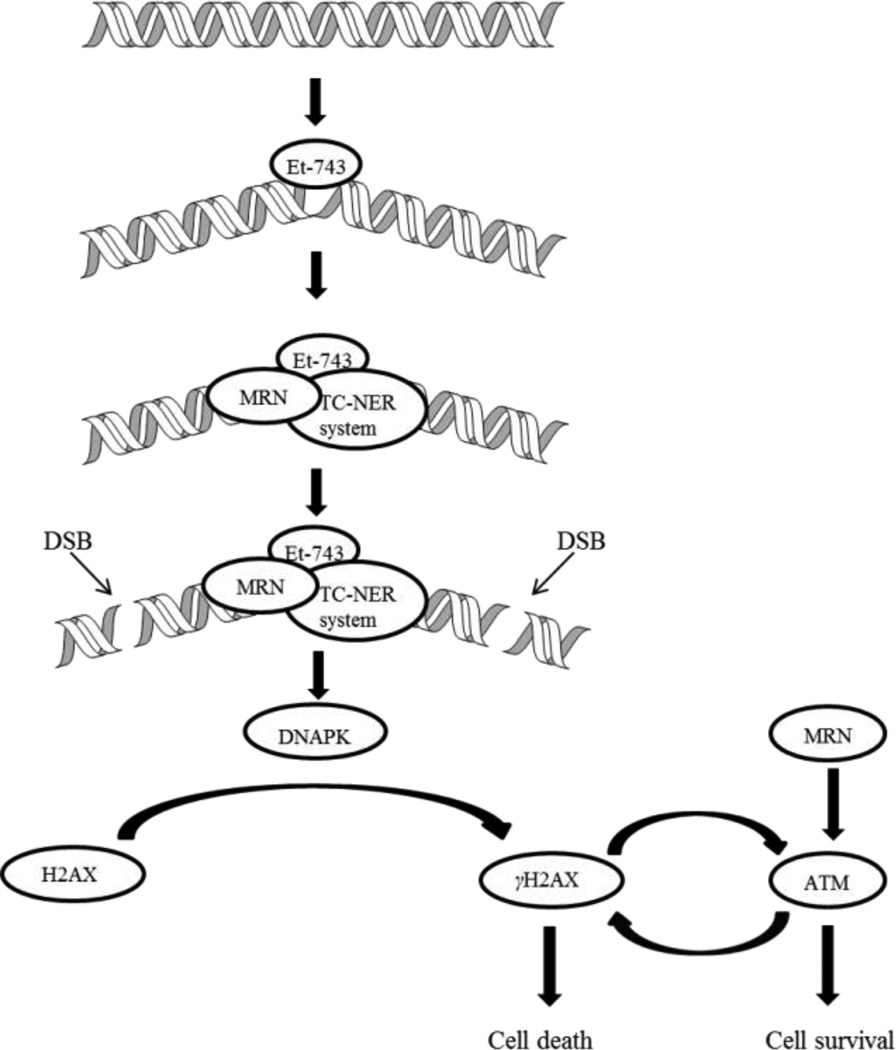
More recently, Guirouilh-Barbat et al. discovered a direct DSB pathway mediated by the TC-NER and Mre11-Rad50-Nbs1 (MRN) complex, independent of replication.41 The authors proposed that the TC-NER-mediated SSBs induced by Et-743 recruits the MRN complex, causing DSBs (Figure 5). DNA-dependent protein kinase (DNAPK) recognizes these DSBs and phosphorylates serine 139 of H2AX protein (γH2AX) around DSB sites, causing apoptosis. MRN and γH2AX also activates the ataxia telangiectasia-mutated (ATM) gene, which in turn will phosphorylate more H2AX, thus intensifying the cell death signal. Besides amplifying the effect of Et-743, MRN and ATM are associated with DNA-damage repair as well via activation of DNA-damage checkpoints, which can result in cell survival.42
Figure 5.
Et-743-induced DSBs mediated by TC-NER and MRN independent of replication.
A part of Et-743’s biological activity was associated with its ability to modulate the tumor microenvironment as a rapid reduction of blood vessels and mononuclear phagocytes, especially tumor-associated macrophages (TAM), was observed around the tumor.43 The reduction of macrophages was resulted from the activation of enzyme caspase-8, which initiates the extrinsic apoptotic pathway in leukocytes. The binding of decoy R3 onto TRAIL receptor inhibits this apoptotic pathway in neutrophils and T cells, and hence their resistance to Et-743.
These studies reveal that the detailed interactions of Et-743 in cells are enormously complex and much may yet be discovered regarding how this drug reacts at the biochemical and cellular levels.
3. Biosynthesis of the Ecteinascidins
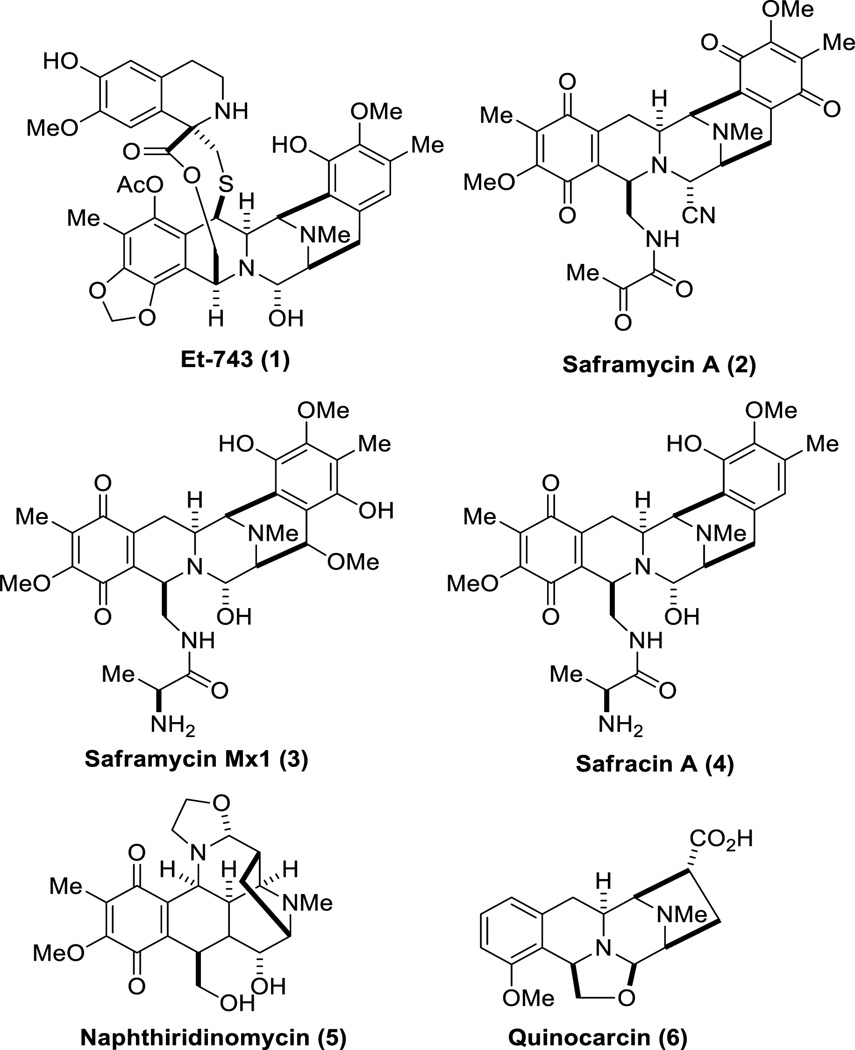
The biosynthetic studies of the ecteinascidins face several obstacles due to the fact that the producing bacterial symbiont has yet to be successfully cultured making it extremely difficult to obtain high quality samples of DNA. In addition, the ecteinascidins and their biosynthetic precursors are present in trace amounts in the tunicate extract. Despite these intrinsic challenges, information about the biosynthesis of these alkaloids has been partially obtained, owing largely to the structural similarity between the ecteinascidins, specifically Et-743, and other THIQ family members, including saframycin B (2),44 saframycin Mx1 (3),45 safracin A (4),46 naphthyridinomycin (5),47 and quinocarcin (6) (Figure 6).48, 49
Figure 6.
Structural similarity between Et-743 (1) and saframycin B (2), saframycin Mx1 (3), safracin A (4), naphthiridinomycin (5) and quinocarcin (6).
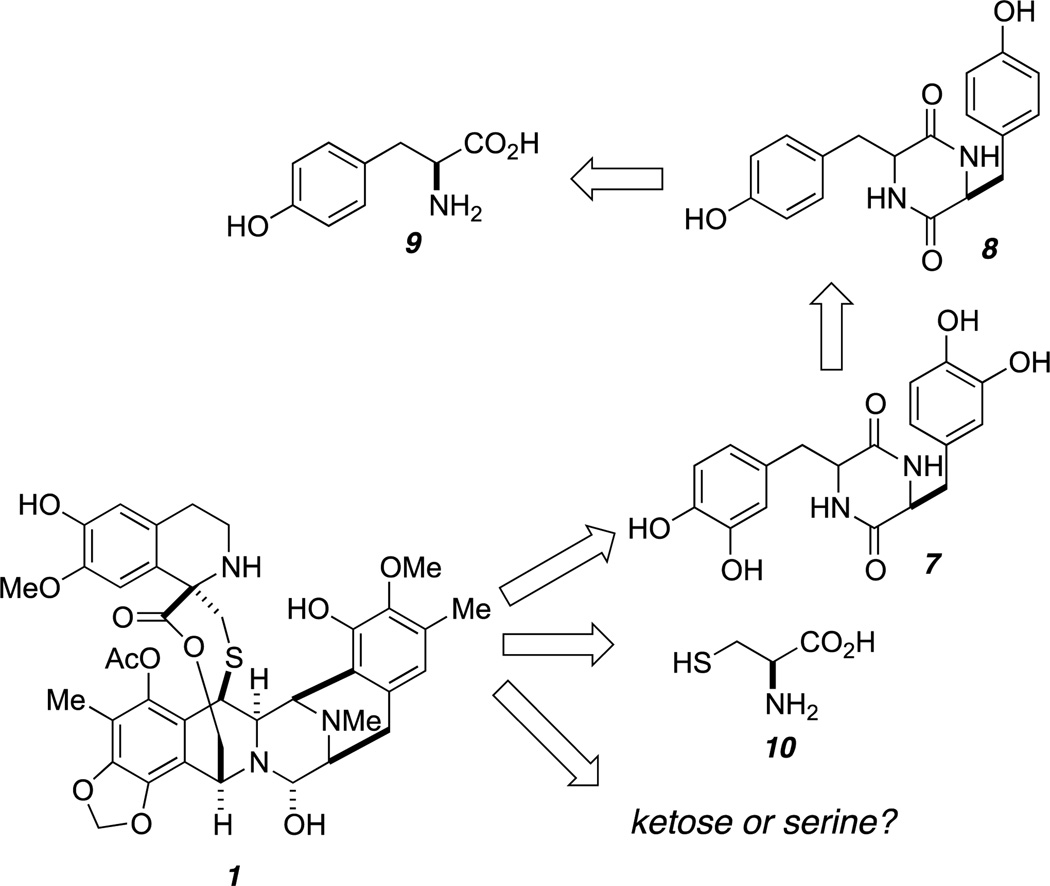
An early study from Kerr’s research group revealed that tyrosine and cysteine are associated with the biosynthesis of Et-743.50 Serine was originally proposed to make up the α-hydroxyethyl moiety (C1-C22) of Et-743 (Figure 7). However, there was no strong evidence to support this hypothesis. Studies by Peng et al. elucidated the role of ketoses in the biosynthesis of naphthyridinomycin and quinocarcin, in which the ketose phosphates are converted to α-hydroxyethyl-thiamine pyrophosphate prior to their incorporation into these natural products.51 Et-743 shares a similar α-hydroxyethyl moiety with these two bacterial metabolites; hence its structure may also arise from a ketose as well.
Figure 7.
The origin of Et-743.
The addition of 14C-labeled diketopiperazines (DKPs) 7 and 8 to the cell-free extract of E. tubinata afforded radiolabeled Et-743 (Figure 7). These two DKPs were found to incorporate into Et-743 at a significantly higher level than 14C-labeled tyrosine (9), suggesting that 8 could be the first committed precursor in Et-743 biosynthesis, and that oxidation of 8 to generate 7 could happen before its incorporation into the pentacyclic core of Et-743.52
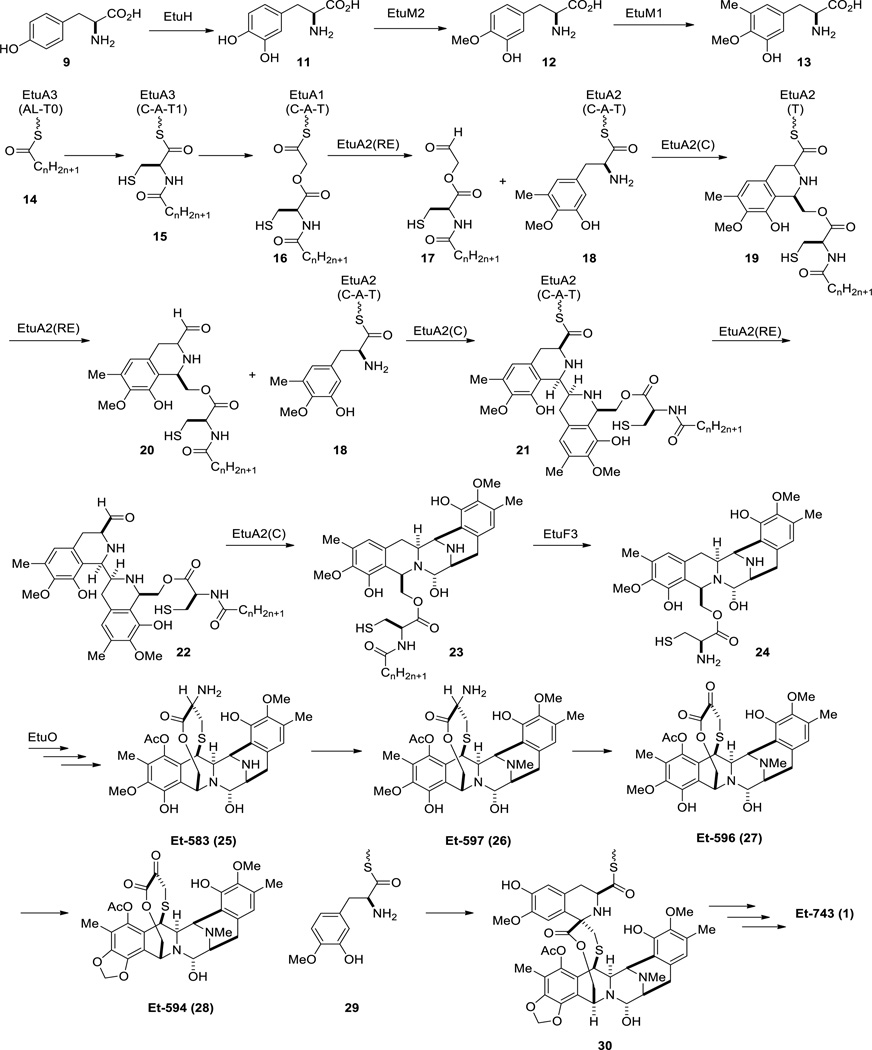
The challenge of culturing the tunicate bacterial symbiont Candidatus Endoecteinascidia frumentensis has been an obstacle to the discovery of the gene cluster responsible for Et-743 biosynthesis. To gain insights of how Et-743 is produced in the Nature, Rath and coworkers utilized metagenomic sequencing methodology on the tunicate consortium, followed by genetic comparison between the resulting DNA pools with those of the known bacterial THIQ biosynthetic genes, specifically of saframycin A (Sfm), saframycin Mx1 (Saf) and safracin (Sac).53–55 Their studies resulted in the identification of the gene cluster encoding the enzymes associated with the biosynthesis of Et-743 (Etu) (Scheme 1).3
Scheme 1.
Proposed biosynthetic pathway of Et-743 (1).
EtuH is a homologue of SfmD, a tyrosine hydroxylase, functioning to hydroxylate the C3 of a tyrosine. EtuM1, a homologue of SacF and SfmM2, and EtuM2, a homologue of SacC and SfmM3, are believed to C- and O-methylate 3-hydroxytyrosine, respectively. The combination of EtuH, EtuM1 and EtuM2 was proposed to produce tyrosine derivative 13 as previously observed in saframycin and safracin biosynthetic pathways (Scheme 1).56
The non-ribosomal peptide synthetase (NRPS) responsible for the biosynthesis of the ecteinascidins contains three modules, EtuA1, EtuA2 and EtuA3 (Figure 8). The common starter module AL-T is located in EtuA3, where a long-chain fatty acid is activated as a thioester 14 (Scheme 1). The EtuA3 domain was predicted to incorporate cysteine to the fatty acid, followed by the elongation of the chain at the EtuA1 domain with a glycolic acid to provide 16. Unlike conventional NRPSs, in which all the components are incorporated collinearly, the biosynthesis of the THIQ family was found to be noncollinear.57, 58 The C domain of SfmC was found to act as a Pictet-Spenglerase to form the mono- and bis-THIQ intermediates in the biosynthesis of saframycin A.59 The role of the C domain of the EtuA2 module may catalyze a similar transformation in the biosynthesis of the ecteinascidins. Hence, after the reductive release of 16, aldehyde 17 is condensed with the primary amine of 18 followed by ring closure of the resulting imine via Pictet-Spengler reaction to yield the first THIQ intermediate 19. The sequence is repeated to insert a second tyrosine derivative 13, followed by reductive release of the resulting bis-THIQ intermediate to generate aldehyde 22. Internal condensation of the aldehyde with the secondary amine gives the pentacyclic core 23, which will undergo hydrolysis of the long chain N-acyl moiety to provide free amine 24 under catalysis of EtuF3 (a homologue of penicillin acylase).60 EtuO is a homologue of SfmO2 and SacJ, FAD-dependent monooxygenases, and is proposed to act as a catalyst for the insertion of the last hydroxyl group on the pentacyclic core of Et-743.55 There is no reference or evidence of the last few steps in Et-743 biosynthesis. However, the presence of other isolated ecteinascidins can serve as hints of how pentacycle 24 can be converted to Et-743. Pentacycle 24 can be transformed into Et-583 (25), which is then methylated at the secondary amine to give Et-597 (26). Subsequent transamination of Et-597 may happen to provide Et-596 (27), which will incorporate the dioxymethylene to yield Et-594 (28). Condensation of Et-594 with 3-hydroxy-4-methoxytyrosine (29), followed by the Pictet-Spengler reaction of the resulting imine, gives rise to intermediate 30, which can be converted to Et-743 via an unknown transformation. Although more work will have to be done before a complete biosynthetic pathway for the ecteinascidins can be mapped, the work of Rath and coworkers provided the first molecular genetic basis of how the ecteinascidins are made in Nature.
Figure 8.
Gene cluster encoding the biosynthesis of Et-743. A: NRPS; D: DNA processing; F: fatty acid biosynthesis; H: hydrolase; M: methyltransferases; N: amidotransferases; O: monooxygenase; P: pyruvate cassette; R: regulatory enzymes; T: drug transporter; U: unknown function.
4. Synthetic Studies of Et-743
Due to its scarcity, being derived from a marine origin, Et-743 has garnered substantial interest from the synthetic community which has enabled its introduction into clinical use. While the field of THIQ synthesis is quite mature, the complex molecular structure of Et-743, relative to other members of the family, have rendered it a particularly alluring and relevant synthetic target. Currently, Et-743 is manufactured commercially by a semi-synthetic process from cyanosafracin B and the enabling synthetic technology to convert cyanosafracin B into commercial grade Et-743 relied on the pioneering synthetic studies reported from the Corey laboratory.61
4-1. Corey’s total synthesis of Et-743
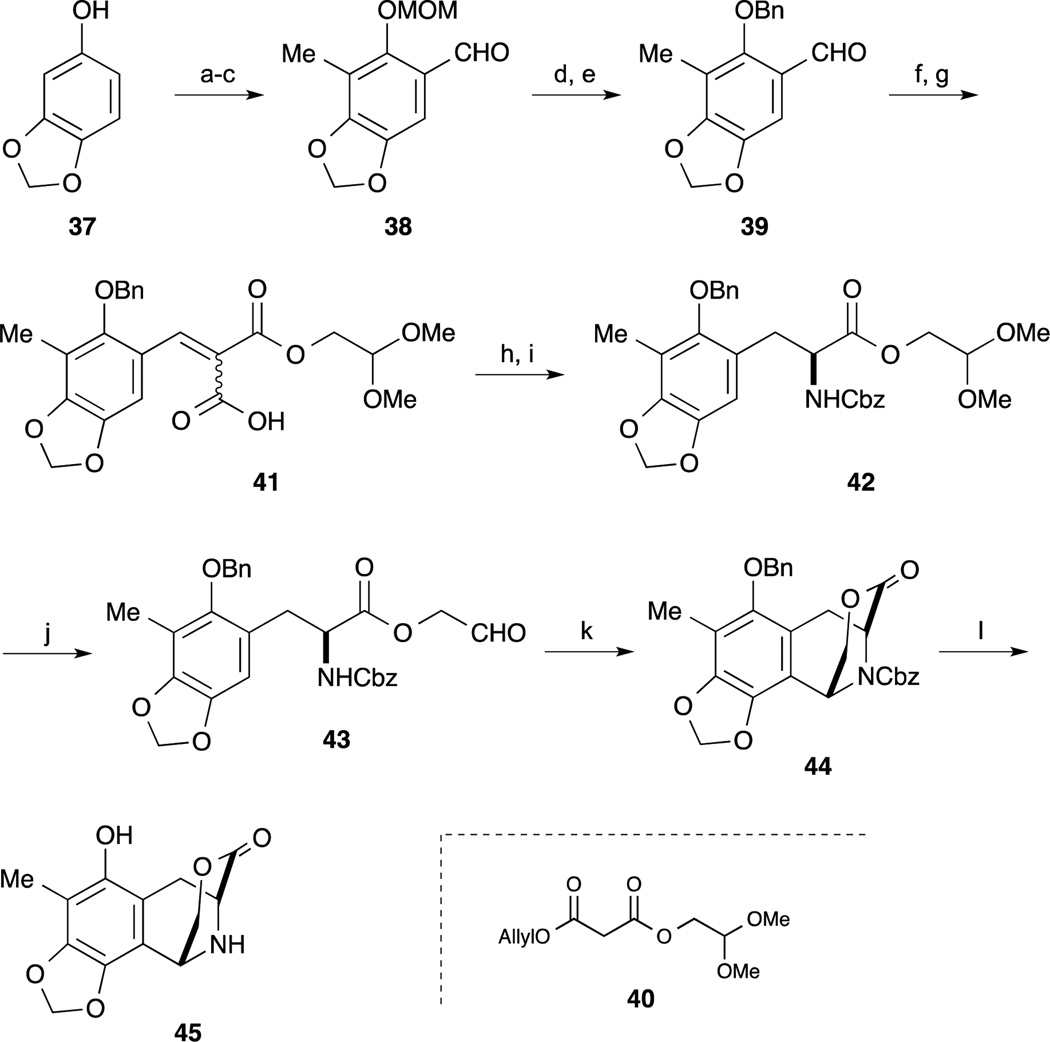
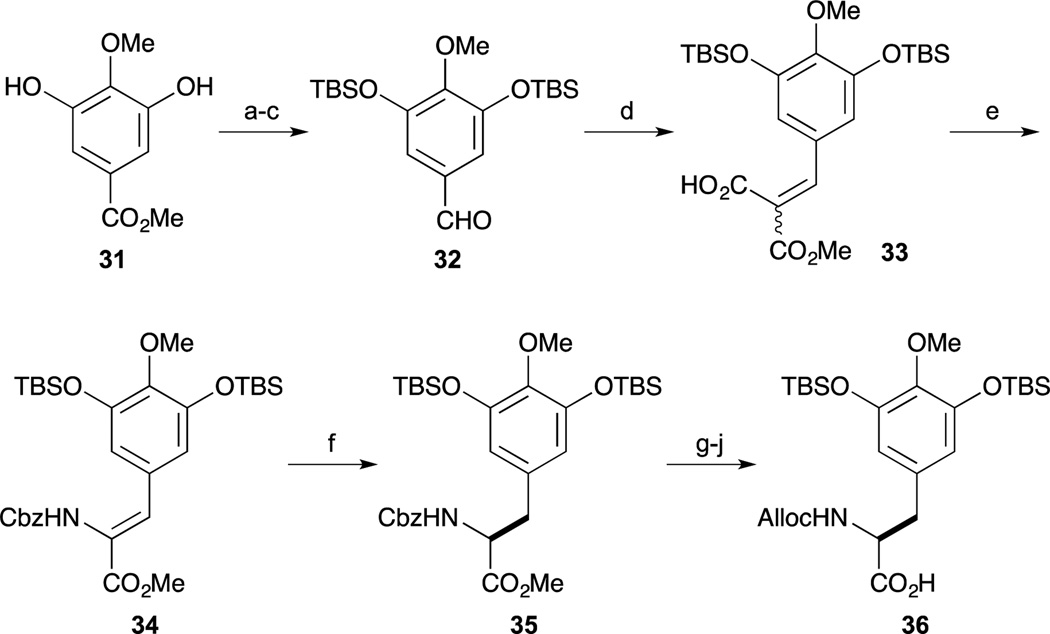
The first, and landmark total synthesis of Et-743 was reported by Corey and Gin et al. in 1996.62,63 Their synthesis featured a biomimetic condensation of two tyrosine derivatives 36 and 45. Synthesis of the right hand segment 36 commenced with gallic ester 31. After protection of 31 as the TBS ether, the methyl ester was converted to the aldehyde 32 by reduction and oxidation. Condensation of aldehyde 32 and monomethylmalonate proceeded smoothly to give 33 as a 1 : 1 mixture. Upon heating 33 and DPPA, isocyanate formation by Curtius rearrangement and subsequent trapping with benzyl alcohol proceeded to give the (Z)-enamide ester as a single isomer. With the desired compound 34 in hand, an asymmetric hydrogenolysis reaction was accomplished by treatment with H2 in the presence of Rh-DiPAMP catalyst. After changing the nitrogen-protecting group of 35 from Cbz to the Alloc group, acidic hydrolysis of the methyl ester provided the carboxylic acid 36.
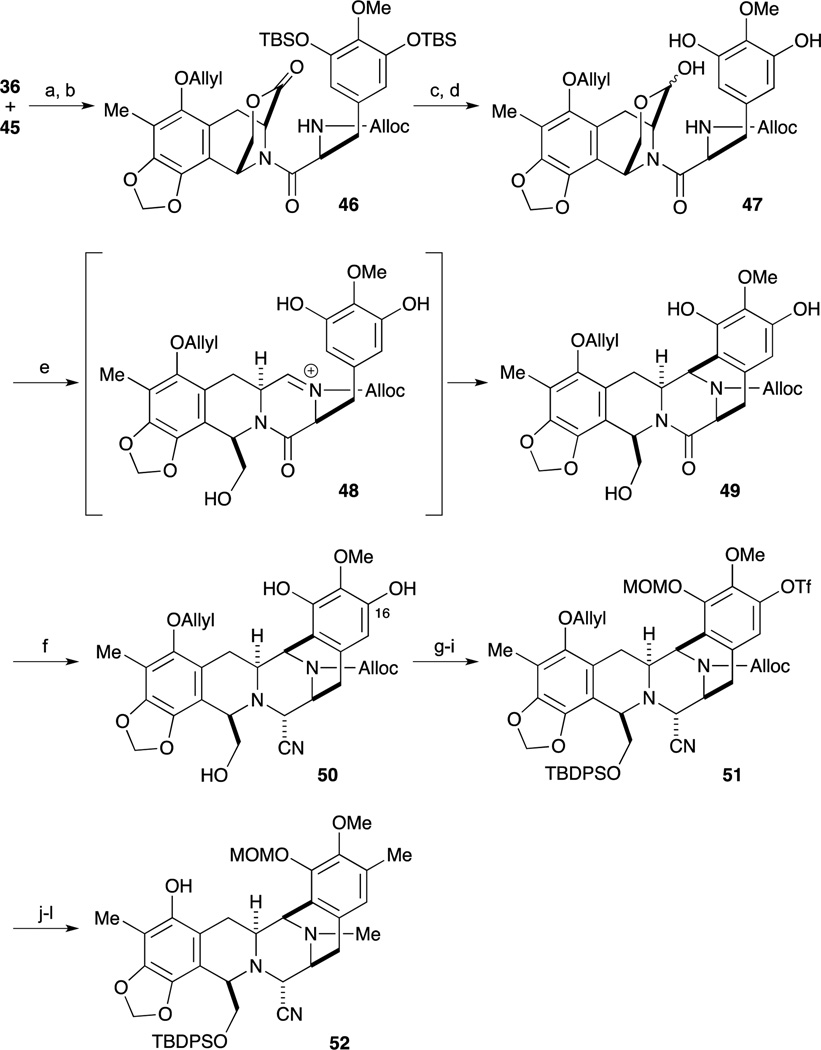
As shown in Scheme 3, the left-hand segment 45 was also synthesized by a Rh-catalyzed asymmetric hydrogenolysis reaction of a dehydroamino ester derived from 41. After protection of sesamol 37 as the MOM ether, regioselective lithiation with n-BuLi in the presence of TMEDA and following treatment with MeI the desired alkylation reaction proceeded smoothly. Subsequent ortho-lithiation of the MOM ether and addition of DMF gave the benzaldehyde derivative 38. After changing the protecting group of 38 from the MOM ether to the corresponding benzyl ether, Knoevenagel reaction of 39 with malonate 40 and cleavage of the protecting group provided the carboxylic acid 41. After incorporation of the nitrogen atom by Curtius rearrangement of 41, Rh-DiPAMP catalyst-mediated asymmetric hydrogenation of the Cbz-protected Z-enamide ester proceeded smoothly to provide the desired substance 42 in 95% yield and 96% ee. After deprotection of the dimethyl acetal by treatment with BF3·OEt2 in the presence of H2O, treatment with BF3·OEt2 and MS 4A, the desired intramolecular Pictet-Spengler reaction proceeded to give the tricyclic lactone 44 as a single isomer. Subsequent cleavage of the Cbz and Bn groups under hydrogenolysis conditions gave the left-hand segment 45.
Scheme 3.
Reagents and conditions (a) NaH, DMF/Et2O, 0 °C; MOMBr, 0 °C (90%); (b) n-BuLi, TMEAD, hexane 0 °C; MeI, –78 to 23 °C (87%); (c) n-BuLi, THF –30 °C; DMF, 0 °C (64%); (d) MeSO3H, CH2Cl2, 0 °C; (e) NaH, 23 °C; BnBr, 23 °C (86% 2steps); (f) 40, piperidine, AcOH, MS4A, benzene, 23 °C (99%); (g) Et3N/HCO2H, Pd(PPh3)4, THF, 23 °C (93%); (h) (PhO)2P(O)N3, Et3N, MS4A, toluene, 70 °C; BnOH, 23 °C; (i) H2 (20 psi), Rh[(COD)]-(R,R)-DiPAMP]+BF4, MeOH, CH2Cl2 23 °C (97%, 96% ee); (j) BF3·OEt2, H2O, CH2Cl2, 0 °C; (k) BF3·OEt2, MS4A, CH2Cl2, 23 °C; (73% 2 steps); (l) H2, 10% Pd/C, EtOAc, 23 °C.
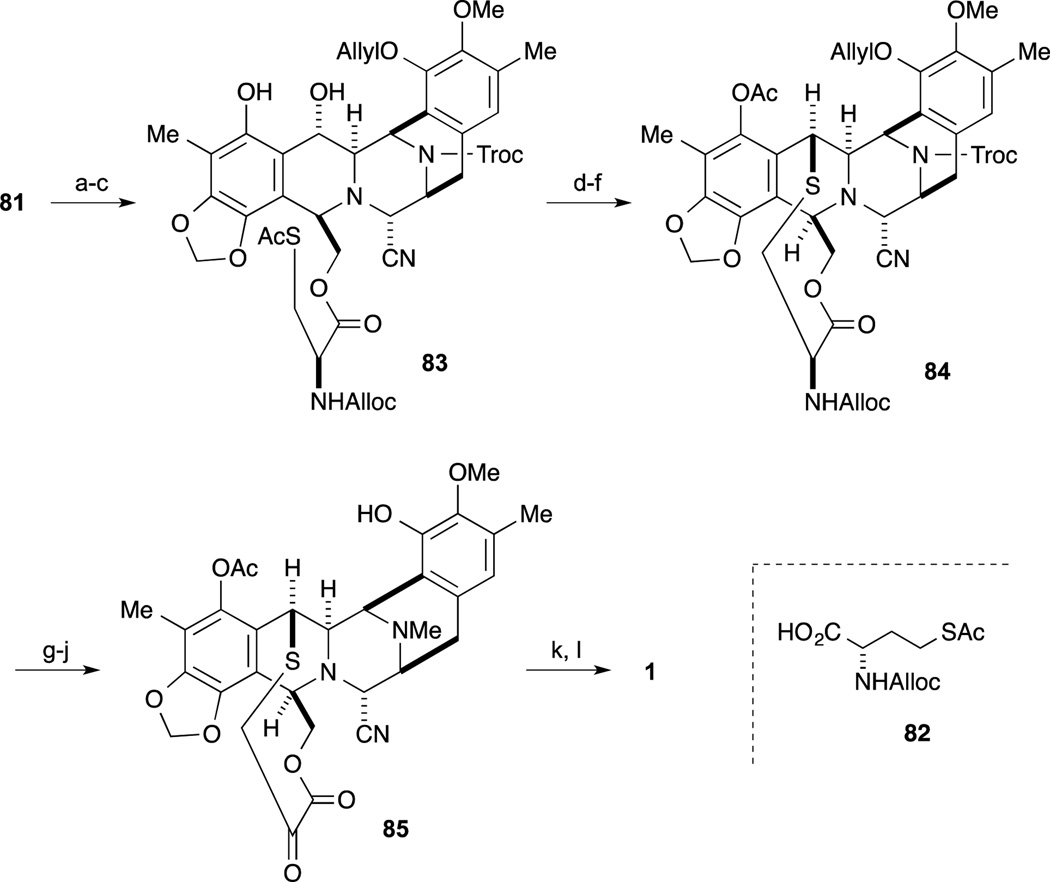
With both desired segments in hand, condensation of 36 and 45 was accomplished by treatment with CIP and HOAt, followed by protection of the phenolic residue as the O- ether furnishing 46 in good yield. After partial reduction of lactone 46 with LiAlH2(OEt)2, deprotection of both TBS ethers gave the cyclization precursor 47. Upon treatment of 47 with CF3SO3H in a 3:2 mixture of H2O : CF3CH2OH, the desired intramolecular Mannich bis-annulation reaction proceeded smoothly through the N-acyl-imine to provide the desired pentacyclic intermediate 49. After partial reduction of lactone 49 with LiAlH2(OEt)2 and subsequent treatment with KCN and AcOH, aminonitrile formation of the incipient aminal derivative proceed to afford 50. After regioselective incorporation of the trifluoromethyl sulfonyl group on the less-hindered phenol of 50, the resultant primary alcohol and phenol were protected as their TBDPS and MOM ethers respectively. After spontaneous deprotection of the Alloc group and ether by treatment with a Pd-catalyst and Bu3SnH, reductive amination of the secondary amine with formaldehyde was carried out by treatment with NaBH3CN and AcOH. The incorporation of the aryl methyl group via the triflate was accomplished by a Stille cross-coupling to give 52.
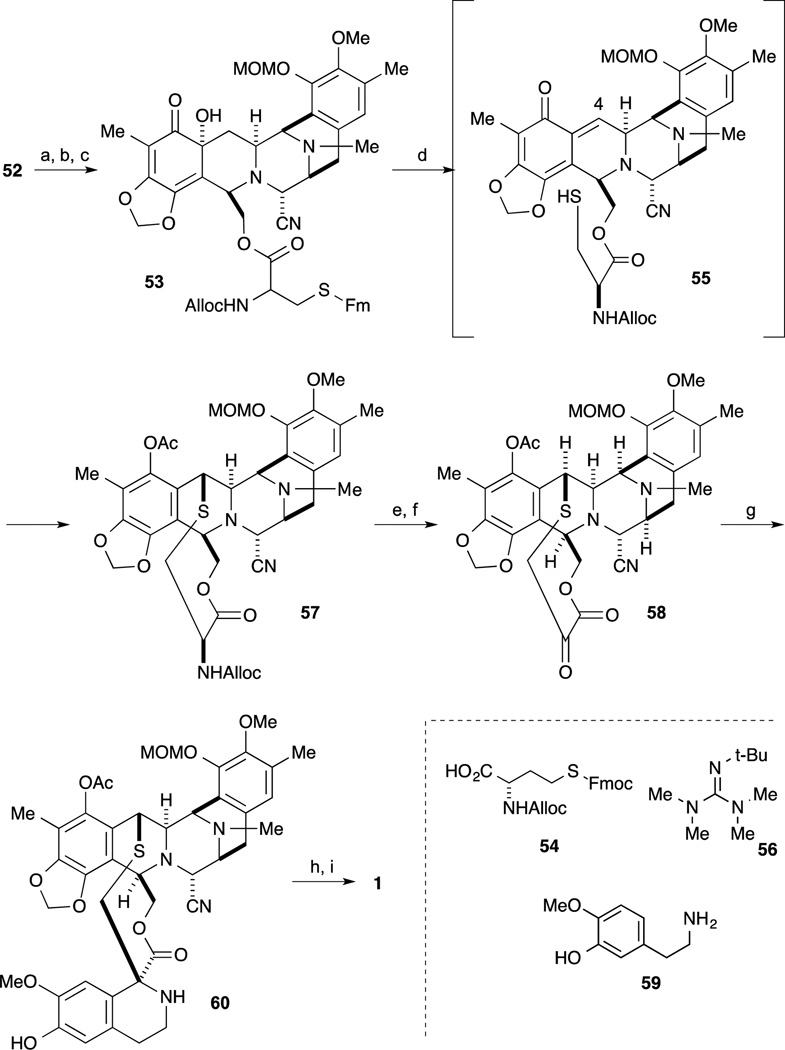
For the final transformation, oxidation of the phenol 52 to the quinone and incorporation of the side chain was accomplished, as shown in Scheme 5. Oxidation of phenol 52 with benzeneseleninic anhydride proceeded to give the tertiary α-hydroxyl ketone derivative. After deprotection of the TBS ether, incorporation of protected cysteine derivative 54 afforded 53. The crucial step for construction of 57 into the ten-membered lactone was accomplished in the following sequence. First, Swern-type oxidation proceeded by treatment of 53 with Tf2O and DMSO followed by the addition of the non-nucleophilic base i-Pr2NEt, resulted in the interesting o-quinone methide intermediate 55. Subsequent addition of Barton’s base 56,64 deprotection of the Fmoc group and in situ cyclization to construct the ten-membered ring proceeded with final trapping of the incipient phenolate by acetate furnishing 57. After deprotection of the alloc group, oxidative deamination was accomplished employing Rappaport’s conditions to give the keto-lactone 58. Condensation of amine 59 with 58 in the presence of silica gel, the desired Pictet-Spengler reaction proceeded to give 60 as a single isomer. Finally, deprotection of the MOM ether and conversion of the α-amino-nitrile to the carbinolamine by exposure to AgNO3 afforded Et-743 (1).
Scheme 5.
Reagents and conditions (a) (PhSeO)2O, CH2Cl2, 23 °C, 15 min (82%); (b) TBAF, THF, 23 °C (91%); (c) 54, EDCI, DMAP, CH2Cl2, 23 °C (91%); (d) Tf2O, DMSO, DMSO, –40 °C; DIPEA, 0 °C; t-BuOH, 0 °C; 56, 23 °C; Ac2O, 23 °C; (e) n-Bu3SnH, PdCl2(PPh3)2, AcOH, CH2Cl2, 23 °C (84%); (f) [N-methylpyridinium-4-carboxaldehyde]+I, DBU, DMF, CH2Cl2, 23 °C (70%); (g) 59, silica gel, EtOH (58%); (h) CF3CO2H, THF/H2O; (i) AgNO3, H2O, (77%, 2 step).
4-2. Fukuyama and Kan’s total synthesis of Et-743
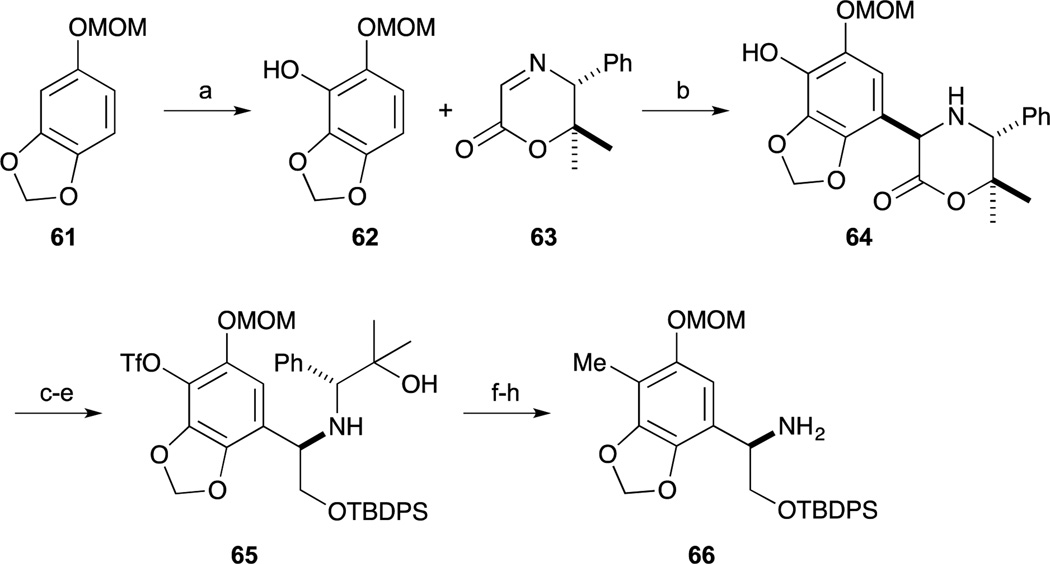
In 2002, Fukuyama and Kan reported the total synthesis of Et-743 (1).65 Their synthesis featured the powerful Ugi four-component condensation and intramolecular Heck reactions that rapidly assembled the tetrahydroisoquinoline ring. As shown in Scheme 6, phenyl glycinol derivative 66 was synthesized by Mannich-type reaction with chiral template 63, which was developed by the author’s group.66 The phenol 62, which was readily obtained from MOM-protected sesamol 61, was reacted with 63 to provide 64. After conversion to the triflate, reduction of the lactone ring and protection of the resulting primary alcohol as the TBDPS ether provided 65. After incorporation of the methyl group,67 removal of the chiral auxiliary was performed by Pb(OAc)4-mediated oxidation of the amino-alcohol and aminolysis of the imine intermediate by treatment with hydroxylamine yielding 66.
Scheme 6.
Reagents and conditions (a) n-BuLi, THF, 0°C to rt; B(OMe)3, H2O2, AcOH, rt (92%); (b) TFA, CH2Cl2, –10 °C (89%); (c) Tf2O, pyridine, CH2Cl2, 0 °C (90%); (d) NaBH4, MeOH, 0 °C (85%); (e) TBDPSCl, imidazole, DMF, rt (91%); (f) MeZnCl, PdCl2(dppf) (3 mol%), THF, reflux (97%); (g) Pb(OAc)4, MeCN, 0 °C; (h) NH2OH·HCl, NaOAc, EtOH, rt (89% in 2 steps).
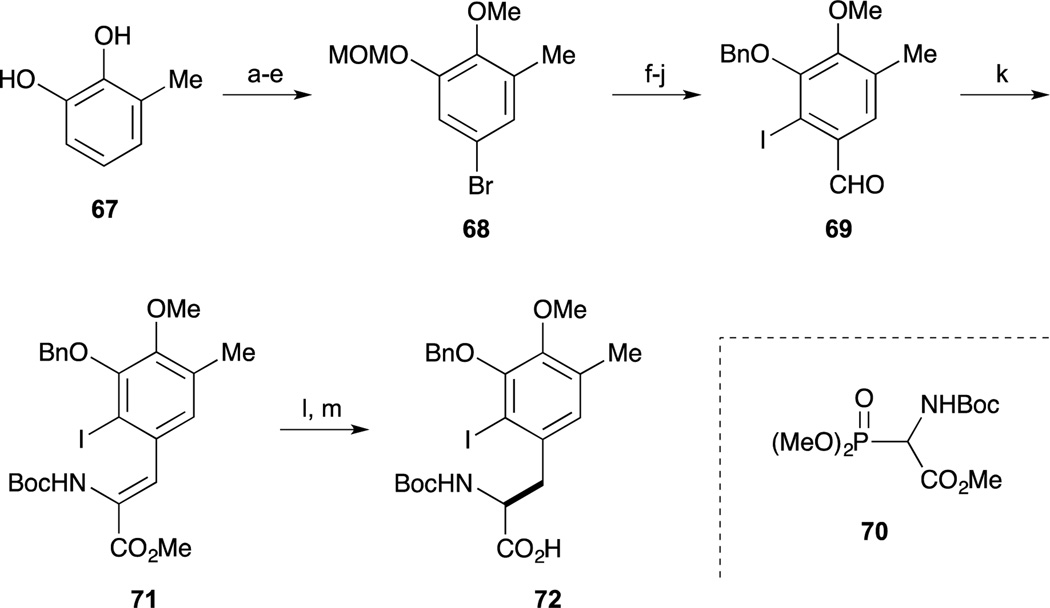
As shown in Scheme 7, the right-hand segment was synthesized from inexpensive 3-methylcatechol by employing the DuPHOS-mediated asymmetric hydrogenation. Regioselective protection of 67 was performed by treatment with TsCl. Incorporation of a bromine atom was carried out at the p-position of the phenol and adjustment of phenolic protecting groups furnished 68. The bromide 68 was converted to the benzaldehyde by halogen-lithium exchange and subsequent treatment with DMF. Regioselective introduction of the iodo substituent was next achieved by directed ortho-lithiation of the corresponding dimethylacetal followed by quenching with I2. Simultaneous cleavage of the MOM ether and the dimethylacetal and subsequent benzylation of the phenol afforded the iodobenzaldehyde 69, which was then subjected to Horner-Emmons reaction with the phosphonate 70 to give the (Z)-dehydrophenylalanine derivative 71.68 Catalytic asymmetric hydrogenation of 71 proceeded smoothly in the presence of DuPHOS catalyst to afford the aminoester and subsequent basic hydrolysis gave the desired carboxylic acid 72.69
Scheme 7.
Reagents and conditions (a) TsCl, TEA, CH2Cl2, 0 °C (83%); (b) Br2, AcOH, CH2Cl2, rt (97%); (c) MeI, K2CO3, acetone, reflux (96%); (d) NaOH, EtOH/H2O, reflux (97%); (e) MOMCl, DIPEA, CH2Cl2, 0 °C (97%); (f) n-BuLi, THF, –60 °C; DMF (79%); (g) HC(OMe)3, CSA, MeOH, rt (94%); (h) n-BuLi, Et2O, 0 °C to rt; I2; (i) conc. HCl, THF, rt (72%, 2 steps); (j) BnBr, K2CO3, CH3CN, reflux (98%); (k) 70, TMG, CH2Cl2, rt (93%); (l) Rh[(COD)-(S,S)-Et-DuPHOS]+OTf−, H2 (500 psi), EtOAc, 50 °C (99%, 94% ee); (m) LiOH, MeOH/H2O/THF, 0 °C to rt (quant).
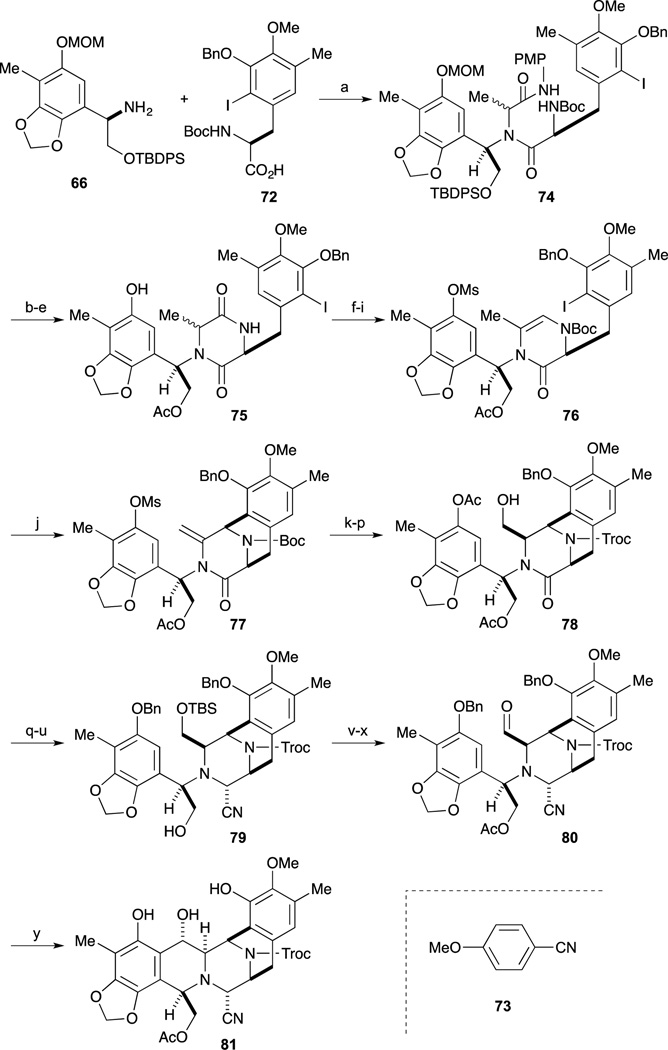
With both desired segments 66 and 72 in hand, condensation was next examined a mixture of amine 66, carboxylic acid 72, p-methoxyphenyl isocyanide (73) and acetaldehyde was heated in MeOH, the desired Ugi’s four-component condensation reaction proceeded to afford the dipeptide 74.70 After switching from the TBDPS ether to the acetate, simultaneous cleavage of the Boc group and the MOM ether gave the aminophenol, which was readily cyclized to afford 75. The dioxopiperazine 75 was converted to the cyclization precursor 76 by a four-step sequence involving mesylation, introduction of a Boc group onto the lactam nitrogen, partial reduction of the ring carbonyl group, and dehydration. The crucial Heck reaction of 76 was performed by treatment with a catalytic amount of Pd2(dba)3 and P(o-tol)3 to afford the desired tricycle 77 in good yield.71–73 After switching the protecting groups of the amine and the phenol of 77 to the corresponding N-Troc-O-Ac compound, the enamide was oxidized with DMDO and immediately treated with CSA to afford the corresponding methoxy alcohol. The subsequent acyliminium ion-mediated reduction under acidic conditions afforded alcohol 78 as a single product. After silylation of the alcohol 78, cleavages of the two acetyl groups and selective benzylation of the phenolic hydroxyl group, the partial reduction of the lactam carbonyl was performed by treatment with Red-Al to afford the corresponding oxazolidine. Cleavage of the oxazolidine with TMSCN and BF3·OEt2 afforded the aminonitrile 79 as a single stereoisomer. Conversion from 79 to aldehyde 80 was accomplished by an acetylation of the regenerated hydroxyl group, cleavage of the TBS ether, and oxidation of the resultant alcohol with Dess-Martin periodinane. During the hydrogenolysis reaction of the benzyl ethers 80 a spontaneous cyclization reaction proceeded to give the desired pentacycle 81.
As shown in Scheme 9, selective allylation of the phenol, cleavage of the acetyl group, and condensation of the resultant alcohol with L-cysteine derivative 82 furnished ester 83. Chemoselective hydrazinolysis of the thioacetate gave the thiol, which, upon exposure to TFA, smoothly underwent cyclization to give the ten-membered sulfide. Subsequent acetylation of the resultant phenol gave 84. Cleavage of the Troc group followed by reductive alkylation afforded the N-methyl amine, whose Alloc group and ether were simultaneously cleaved with a palladium catalyst to give the aminophenol. According to the protocol reported by Corey, conversion to keto-lactone 85 and Pictet-Spengler reaction furnished Ecteinascidin 770. Finally, conversion from the aminonitrile to the labile hemiaminal was effected by treatment with AgNO3 to give Ecteinascidin 743 (1).
Scheme 9.
Reagents and conditions (a) bromide, DIPEA, CH2Cl2, reflux (89%); (b) K2CO3, MeOH, rt (99%); (c) 82, EDCI, DMAP, CH2Cl2, rt (94%); (d) H2NNH2, CH3CN, rt (98%); (e) TFA, CF3CH2OH, rt; (f) Ac2O, pyridine, DMAP, rt (71% in 2 steps); (g) Zn, AcOH, Et2O, rt (92%); (h) HCHO, AcOH, NaBH3CN, MeOH, rt (96%); (i) PdCl2(PPh3)2, AcOH, n-Bu3SnH, CH2Cl2, rt (89%); (j) [N-methylpyridinium-4-carboxaldehyde]+I, DMF/CH2Cl2, rt; DBU; citric acid (54%); (k) 59, NaOAc, EtOH, rt (96%); (l) AgNO3, CH3CN/H2O, rt (93%).
4-3. Zhu’s total synthesis of Et-743
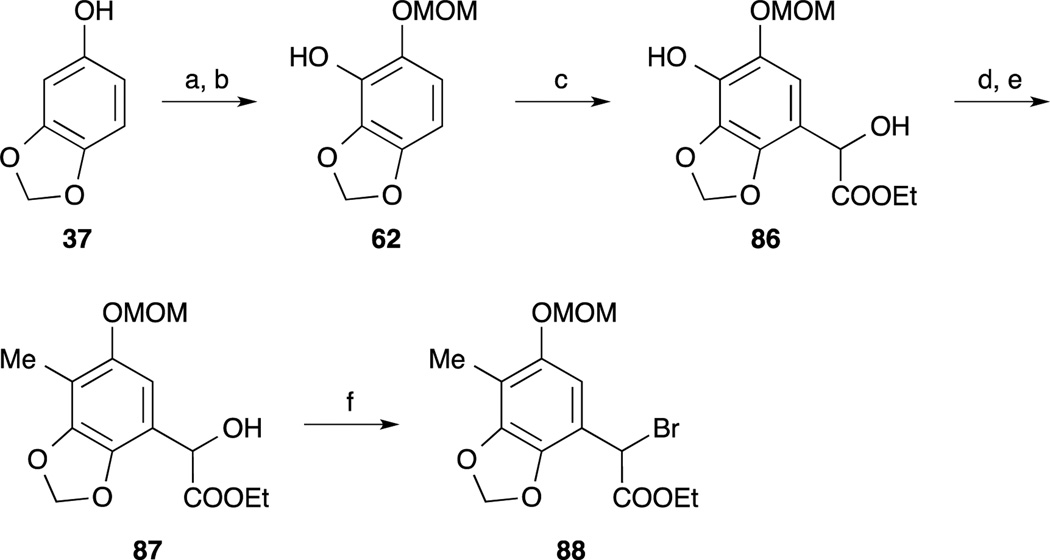
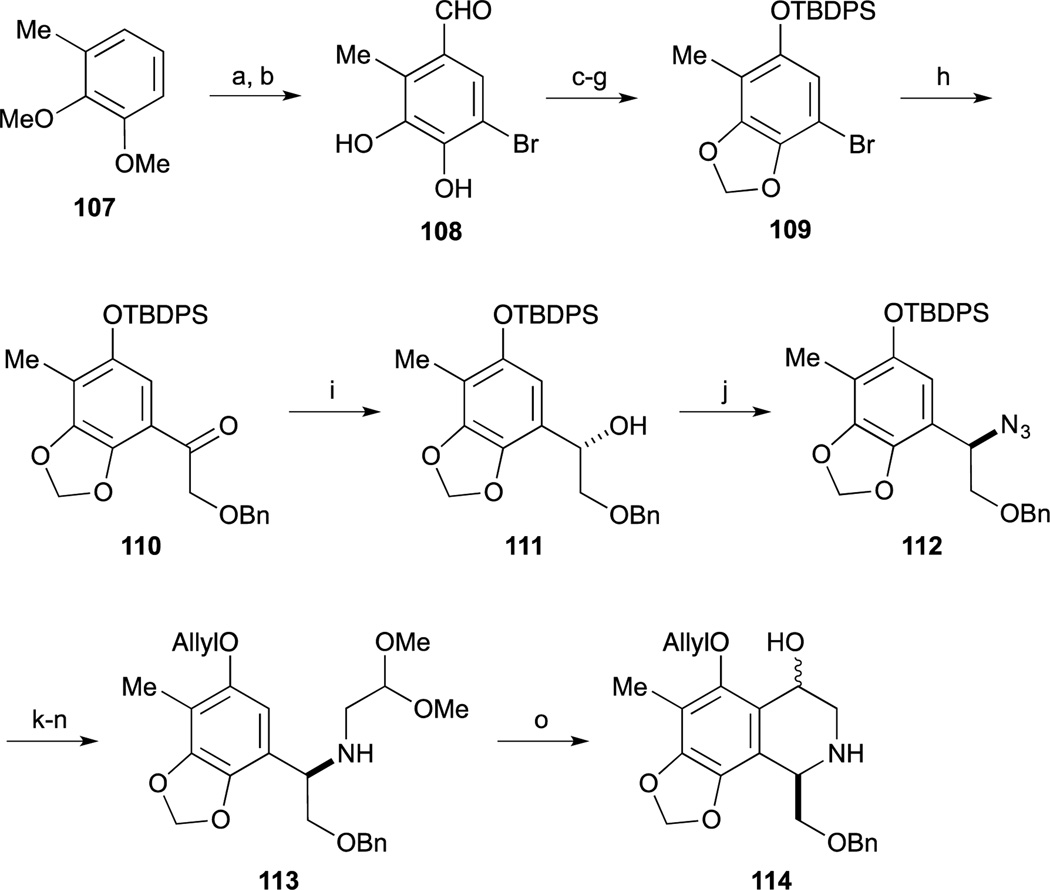
In 2005, Zhu and co-workers reported the third total synthesis of Et-743.74 The left-hand benzyl bromide 88 was synthesized from sesamol 61, which was converted to the phenol 62 according to Fukuyama’s protocol.65 Friedel-Crafts type reaction of 62 and ethyl glyoxalate provided α-hydroxy ester 86. Incorporation of the methyl group on to the phenol 86 was performed by a two-step sequence including chemoselective conversion to the triflate by using the author’s nitrophenyl triflate74 and Suzuki-Miyaura cross coupling with boroxine.75 Treatment of benzyl alcohol 87 with thionyl bromide and benzotriazole provided the benzyl bromide 88.
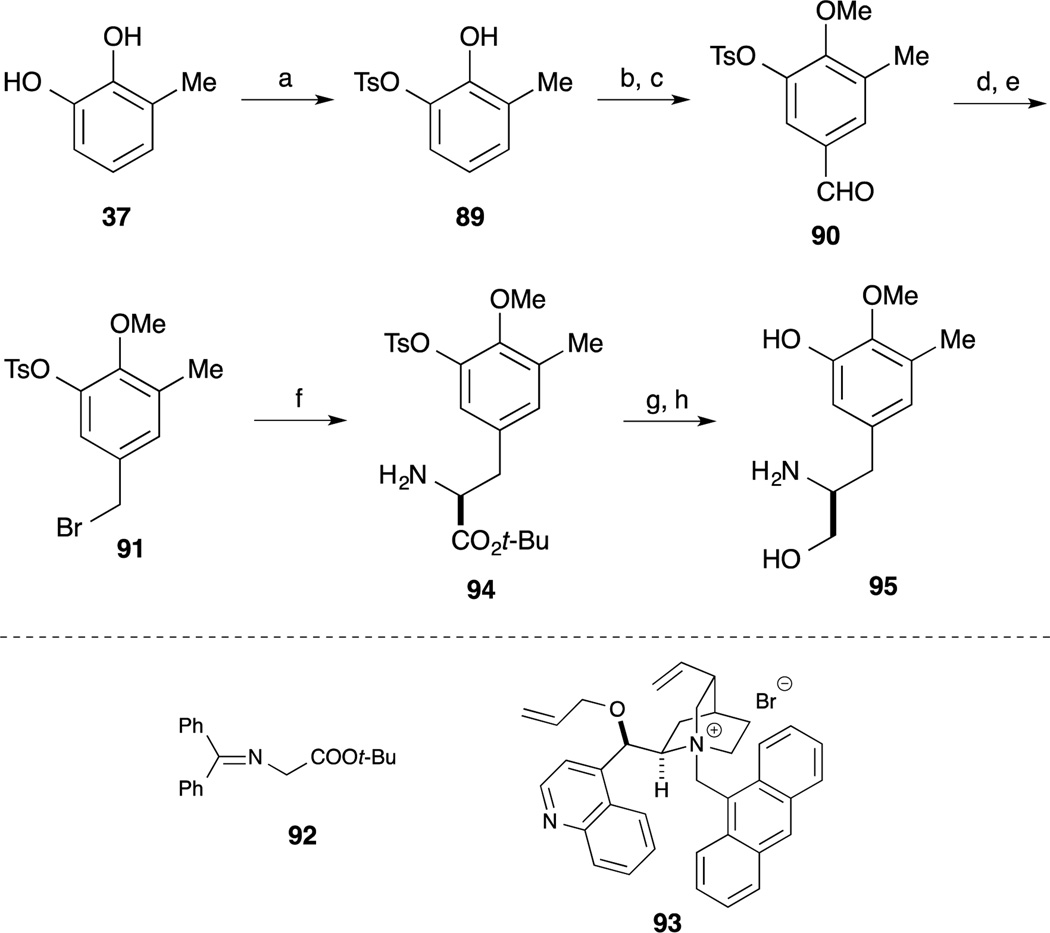
The right-hand segment phenyl alaninol derivative 95 was synthesized from 3-methylcatechol 67 by utilizing an asymmetric alkylation with glycine derivative 92.76 Regioselective incorporation of the Ts group, methyl group and formyl group provided aldehyde 90. After reduction of 90, benzyl alcohol was converted to the corresponding bromide 91. Upon treatment of 91 and glycine derivative 92 in the presence of the Corey’s catalyst,77–79 the desired alkylation reaction proceeded smoothly and subsequent acidic hydrolysis of the imine to provide the amino ester 94. After reduction of the ester moiety of 94 and basic hydrolysis of the Ts group provided the phenyl alaninol derivative 95.
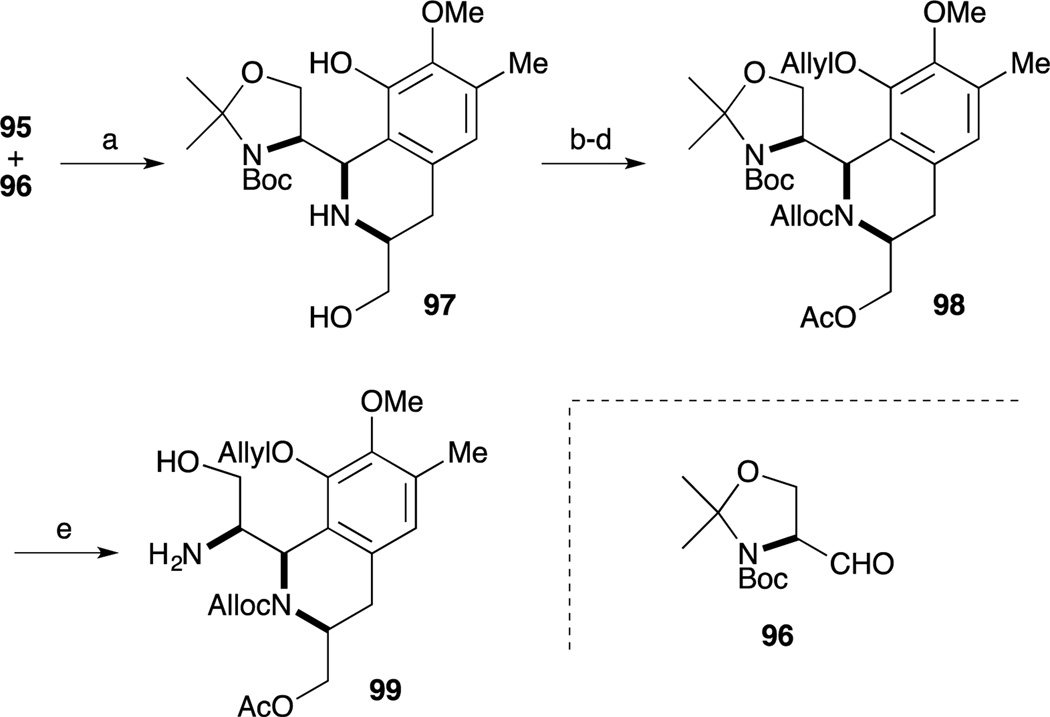
Construction of the DE-ring was accomplished by Pictet-Spengler reaction of amino-phenol 95 and Garner’s aldehyde (96).80–82 Upon treatment of 95 and 96 with AcOH in CH2Cl2 and CF3CH2OH, the desired tetrahydroisoquinoline formation reaction proceeded to give 97 as a single isomer.83–85 Employment of these optimized conditions, the cyclization reaction was accomplished with high regio- and diastereoselectivity. After protection of secondary amine with the Alloc group, chemoselective allylation of the phenol and acetylation of resultant primary alcohol provided 98. Acidic hydrolysis of 98 by TFA provided amino-alcohol 99.
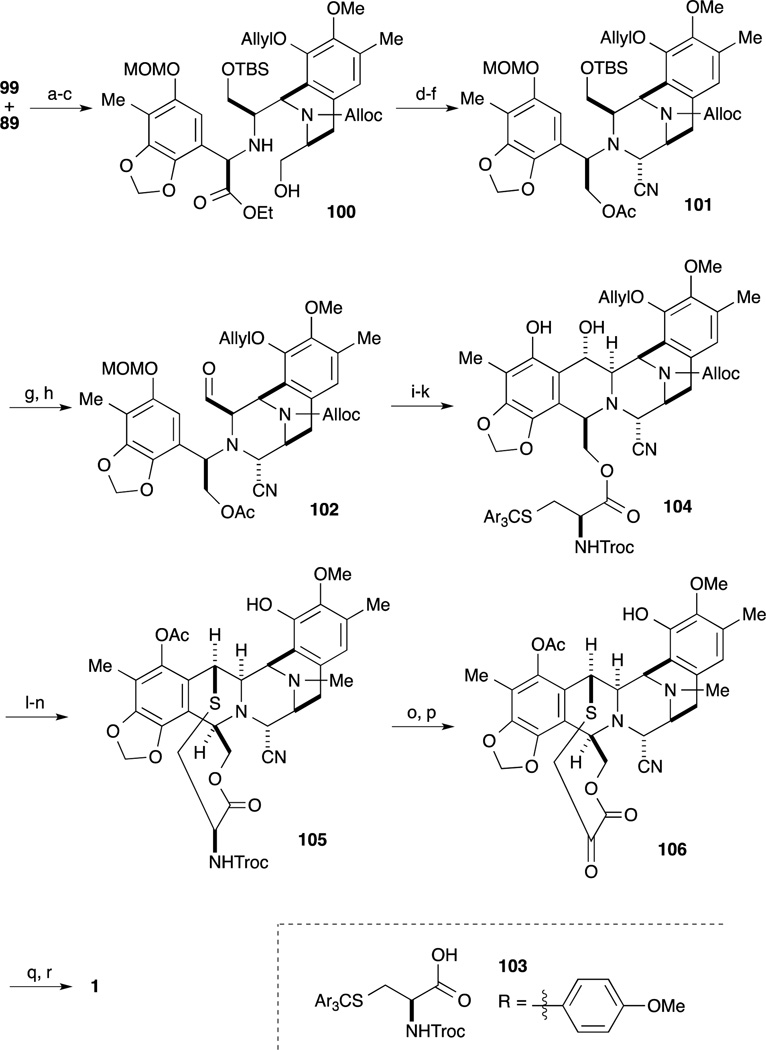
With the desired A-ring bromide 89 and DE ring amine 99 in hand, the next challenge in their synthesis was condensation of these two segments. Upon treatment of 89 and 99 with base, secondary amine formation proceeded to provide desired product 100 and its β-isomer in 68% and 23% yield, respectively. The C-ring precursor 100 was prepared by silylation of the alcohol and cleavage of the acetyl group. After oxidation of the primary alcohol with Dess-Martin periodinane, treatment with TMSCN and a catalytic amount of ZnCl2 afforded the aminonitrile as a single stereoisomer. For construction of the B-ring, the precursor aldehyde was converted by a four-step sequence involving reduction of the ester moiety, introduction of the acetyl group onto the primary alcohol, cleavage of the TBS ether, and Dess-Martin oxidation of the resultant alcohol. Upon treatment of 102 with TFA, cleavage of the MOM ether and subsequent spontaneous cyclization ensued to give the desired pentacyclic intermediate. After cleavage of the acetyl group, regioselective condensation of the primary alcohol with L-cysteine derivative 103 furnished ester 104. Upon exposure 104 to 1% TFA in CF3CH2OH, cleavage of Troc group and cyclization of the ten-membered sulfide ring proceeded; subsequent acetylation of the resultant phenol gave the desired acetate. Simultaneous deprotection of the Alloc group and ether, followed by reductive N-methylation gave 105. Finally, according Corey’s protocol,62 key intermediate 105 was readily converted to 1.
4-4. Danishefsky’s formal synthesis of Et-743
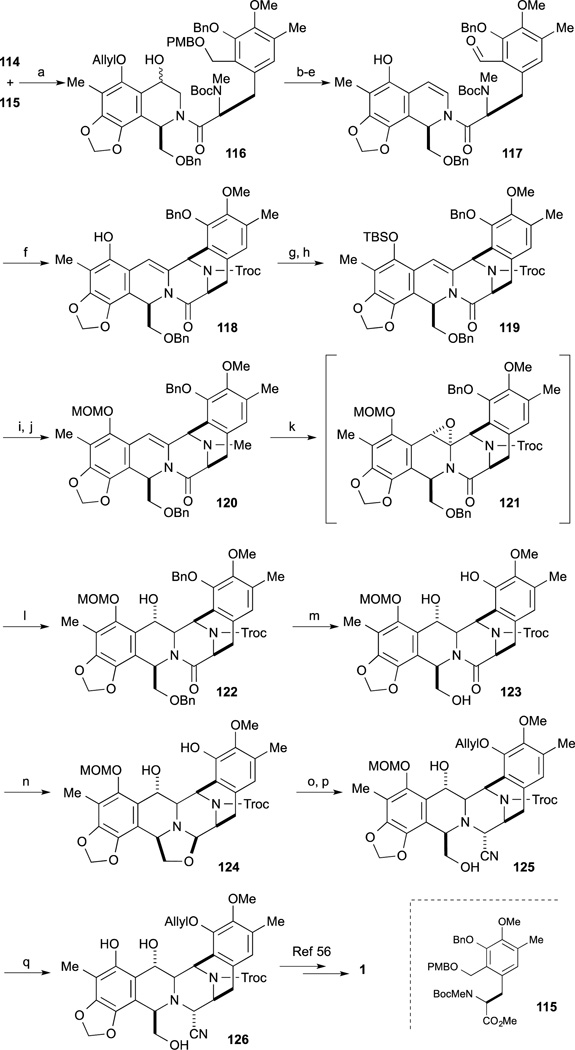
In 2006, Danishefsky and co-workers reported a formal total synthesis of Et-743.86 The synthesis started from Borchardt catechol, readily obtained by formylation of 107.87 After cleavage of two methyl ethers, mono-bromination was carried out regioselectively. Subsequent methylenation, Bayer-Villiger oxidation, acidic hydrolysis, and incorporation of the TBDPS group onto phenol provided 109. After lithiation of 109, treatment with the Weinreb amide derivative gave the ketone 110.88 Asymmetric reduction of 110 was accomplished by the combination of Noyori transfer-hydrogenation reaction89 and treatment of 111 with DPPA to give azide 112. After hydrogenation of azide of 112, reductive amination provided the two-carbon homologated product. Subsequent protecting group swap of the TBDPS group to the O-ether gave 113. Upon treatment of 113 with HCl, hydrolysis of the dimethyl acetal and cyclization reaction proceeded to afford tetrahydroisoquinoline 114.
Condensation reaction of 114 and tyrosine derivative 11590 was performed by treatment with BOPCl in the presence of Et3N to provide 116. Preparation of the intramolecular Pictet-Spengler cyclization precursor 117 was carried out by a four-step sequence, involving the deprotection of PMB group, dehydration to the enamide, oxidation to the aldehyde, and Pd-mediated cleavage of ether. Upon treatment of 117 with thirty equivalents of difluoroacetic acid, the desired cyclization proceeded to give the pentacycle 118. Conversion to Fukuyama’s synthetic intermediate 126 from phenol 118 was accomplished by the following sequence. After incorporation of the TBS residue to the phenol 118, conversion of the N-Me amine to the Troc-protected species was carried out by treatment with TrocCl in the presence of TBAI. After changing the protecting group of 119 from TBS to the MOM group, hydration of enamide 120 was employed. DMDO oxidation of 120 provided unstable epoxide 121 and without isolation treatment with NaBH3CN provided benzyl alcohol 122 as a predominant product. After removal of the two benzyl ethers of 122, treatment with a 1:1 complex of n-BuLi and DIBAL, partial reduction of amide 123 proceeded to provide the oxazolidine 124. After selective protection of phenol 124 with the group, exposure to KCN and acetic acid provided amino-nitrile 125. Finally cleavage of the MOM group of 125 with TFA gave Fukuyama’s synthetic intermediate 126, constituting a formal total synthesis.
4-5. Williams’ formal synthesis of Et-743
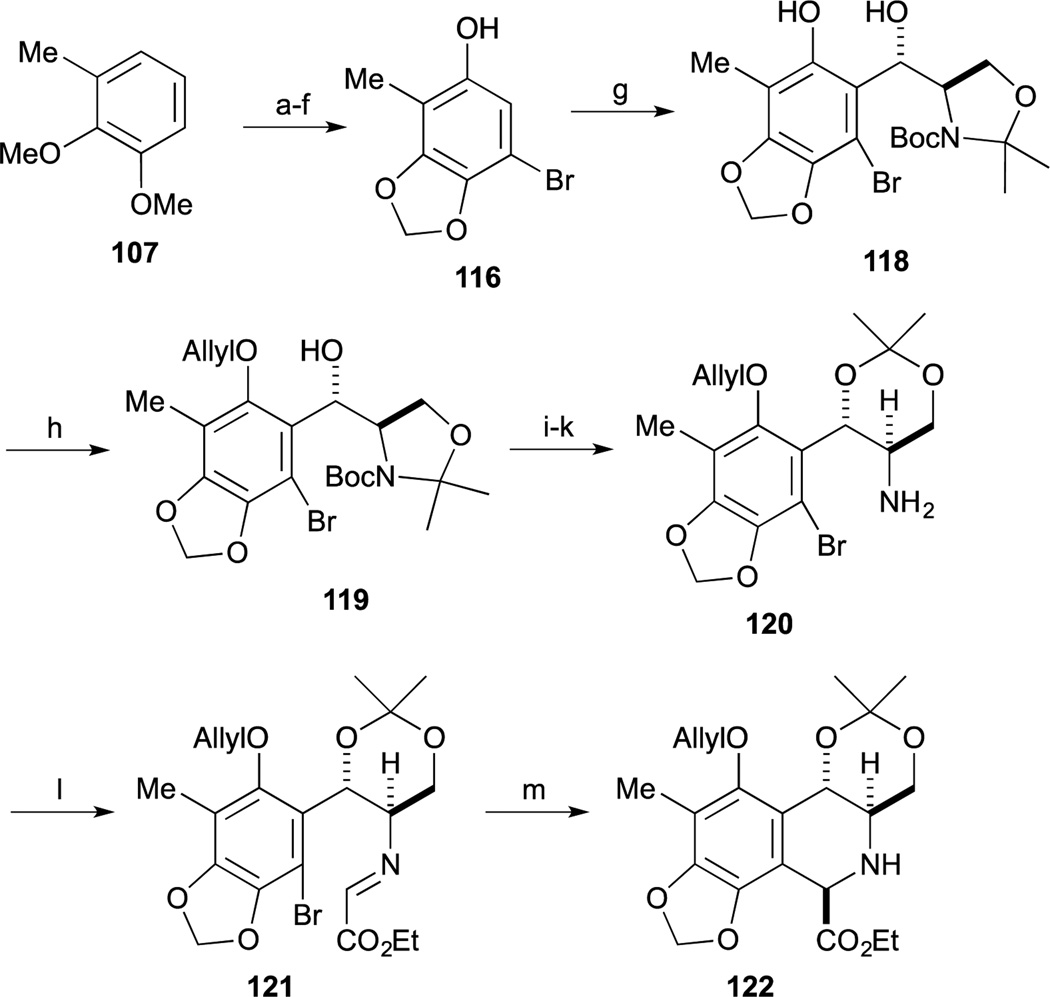
Williams and Fishlock reported the formal total synthesis of 1 in 2008, that relayed into Fukuyama’s synthesis.91 For this effort, they developed a new, radical-based cyclization reaction to form the left-hand tetrahydroisoquinoline. The synthesis commenced with phenol derivative 116, which was readily obtained from 107 by using a similar procedure employed by Danishefsky.86 Upon treatment of phenol 116 with Garner’s aldehyde (117)80–82 with titanium phenolate, the Friedel-Crafts reaction proceeded smoothly to give the desired anti-product 118. After protection of the phenolic group as the corresponding O- ether, hydrolysis of the oxazolidine ring of 119, formation of the trans-acetonide and deprotection of the Boc group via Ohfune’s protocol provided amine 120.92 Construction of the B-ring of 121 was accomplished by a novel glyoxal-imine radical cyclization. Condensation between amine 120 and ethyl glyoxalate proceeded smoothly to provide the imine intermediate 122. Upon treatment of the glyoxal-imine intermediate 121 with slow addition of n-Bu3SnH and AIBN, the desired radical cyclization reaction proceeded smoothly to provide tetrahydroisoquinoline 122 as a single isomer.
After reduction of the ester group of 122, subsequent protection of the resulting primary alcohol as a benzyl ether afforded 123. Acylation of the tetrahydroisoquinoline amine 123 was achieved by conversion to the acid chloride from 12493 to give the desired amide 125 without epimerization. After swapping out the amine protecting group from Fmoc to Boc group, treatment of 126 with Dowex in methanol, the desired removal of acetonide group proceeded smoothly with incorporation of methanol at the benzylic position (as an inconsequential 1:1 diastereomer mixture).
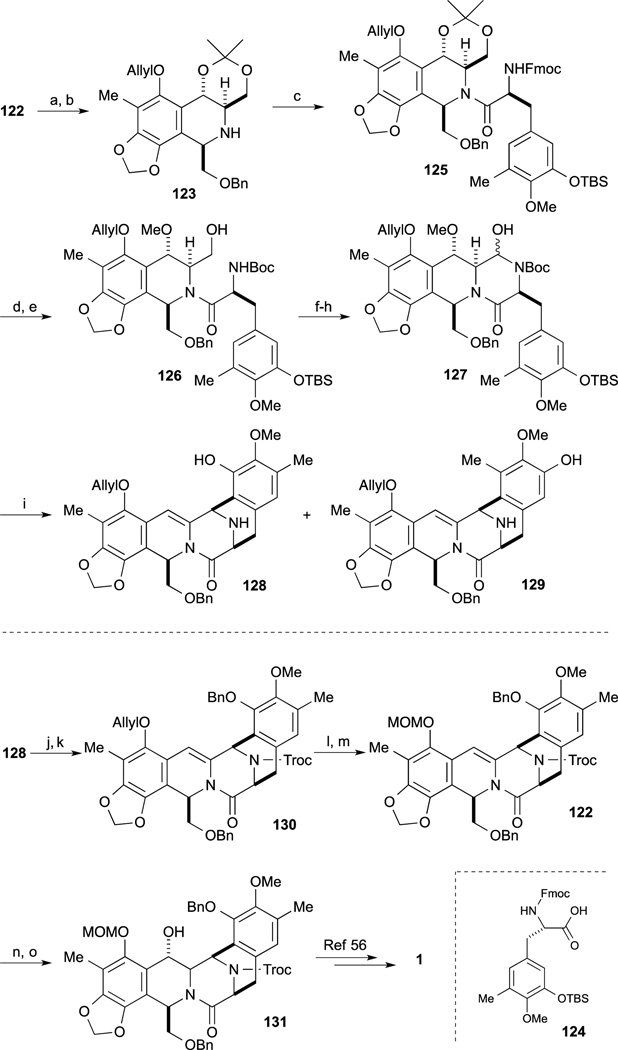
Deprotection of the phenolic O-TBS group and oxidation of the primary alcohol under Swern conditions provided the aminal 127. Upon treatment of 127 with TFA, deprotection of the Boc group and Pictet-Spengler reaction proceeded with elimination concomitant elimination of the methoxy group at the benzylic position to afford 128 and 129 as a 1:1 regioisomeric mixture. After separation, the desired pentacycle 128 was converted to the Fukuyama synthetic intermediate 122 also intercepted by Danishefsky.65 Protection of the amine with the Troc group and the phenolic group as the benzyl ether provided 130. Changing the protecting group from the allyl ether of 130 to the corresponding MOM ether afforded 120 which constituted a formal total synthesis. 4–6. Fukuyama’s formal synthesis of Et-743
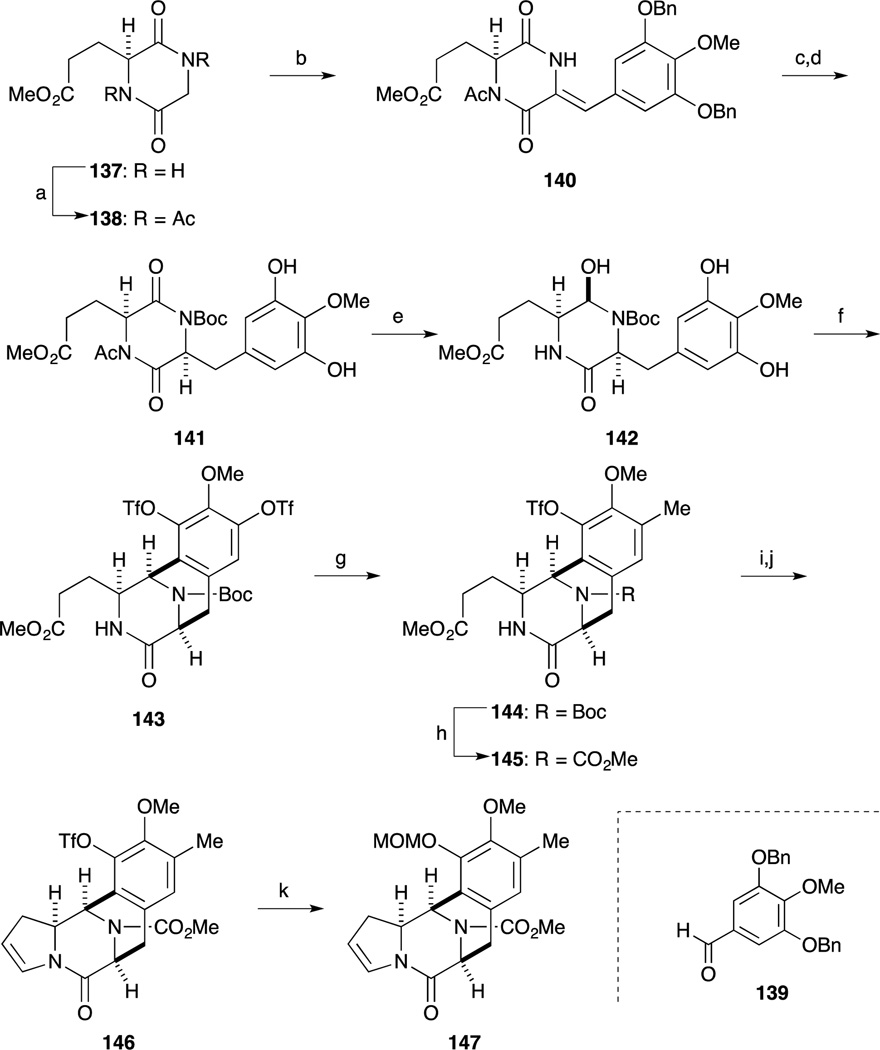
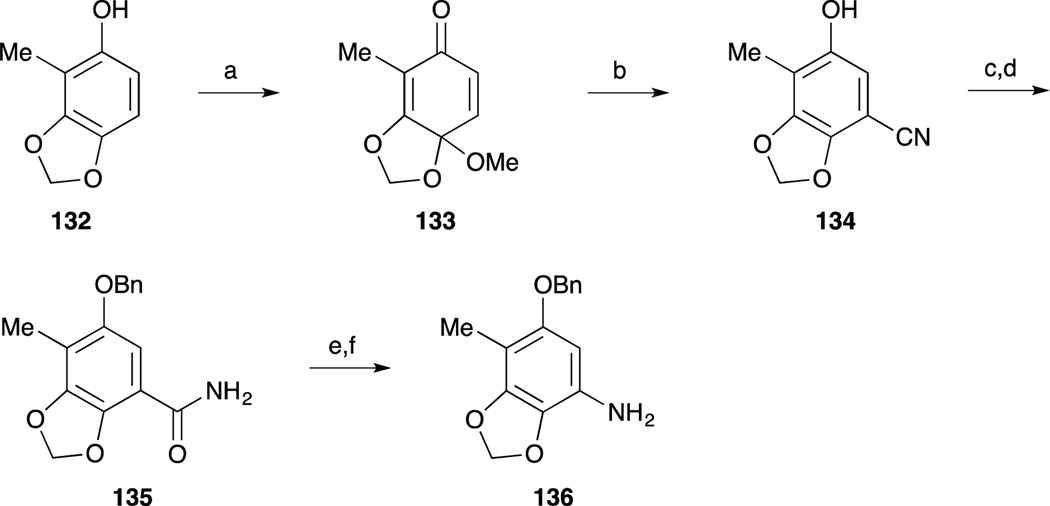
In 2013, Fukuyama and co-workers reported a second-generation total synthesis of Et-743. The A-ring moiety 136 as the precursor for diazonium salt was prepared from known phenol 132.94,95 Oxidation of 132 with PhI(OAc)2, and treatment with sodium cyanide gave nitrile 134. After benzylation of the phenolic hydroxy group, the resulting nitrile was hydrolyzed to furnish carboxamide 135. Hofmann rearrangement followed by hydrolysis afforded amine 136.
The construction of the CDE ring by N-acyliminium ion-mediated cyclization reaction is detailed in Scheme 19. Perkin condensation of known dioxopiperazine 13896 with aldehyde 139 proceeded smoothly to give 140. After introduction of a Boc group at the lactam, cleavage of the benzyl group, stereoselective reduction of the double bond, hydrazinolysis of the acetyl group in 141 followed by selective reduction of the imide carbonyl group with sodium borohydride afforded 142. Upon treatment of 142 with TFA, the N-acyliminium ion-mediated cyclization reaction proceeded smoothly, and subjection of the product to PhNTf2 under basic conditions afforded bis-triflate 143 in 88% yield. Suzuki-Miyaura coupling of 143 with trimethylboroxine took place selectively at the less-hindered triflate to produce 144 in 92% yield. After the Boc group was switched to a methoxycarbonyl group, partial reduction of the ester moiety in 145 with L-Selectride and subsequent dehydration of the resulting hemi-aminal under acidic conditions afforded enamide 146. The Tf group was replaced with a MOM group in a one-pot process to afford 147.
Scheme 19.
Reagents and conditions (a) Ac2O, 130 °C (80%); (b) 139 t-BuOK, THF, –78 to 0 °C; DBU, 0 °C; (c) Boc2O, DMAP, THF, rt (quant, 2 steps); (d) H2 (750 psi), Pd/C, EtOAc, rt; (e) H2NNH2·H2O, THF, rt; evaporation; NaBH4, MeOH, 0 °C (57%, 2 steps); (f) TFA, CF3CH2OH, rt; evaporation; PhNTf2, DMAP, Cs2CO3, MeCN, rt (88%); (g) trimethylboroxine, Pd(PPh3)4, K3PO4, 1,4-dioxane, 100 °C (92%); (h) HCl, EtOAc, rt; ClCO2Me, NaHCO3, H2O, 0 °C (91%); (i) L-Selectride, THF, –42 °C; (j) CSA, toluene, reflux (55%, 2 steps); (k) aq KOH, 1,4-dioxane, rt; MOMCl, 0 °C (95%).
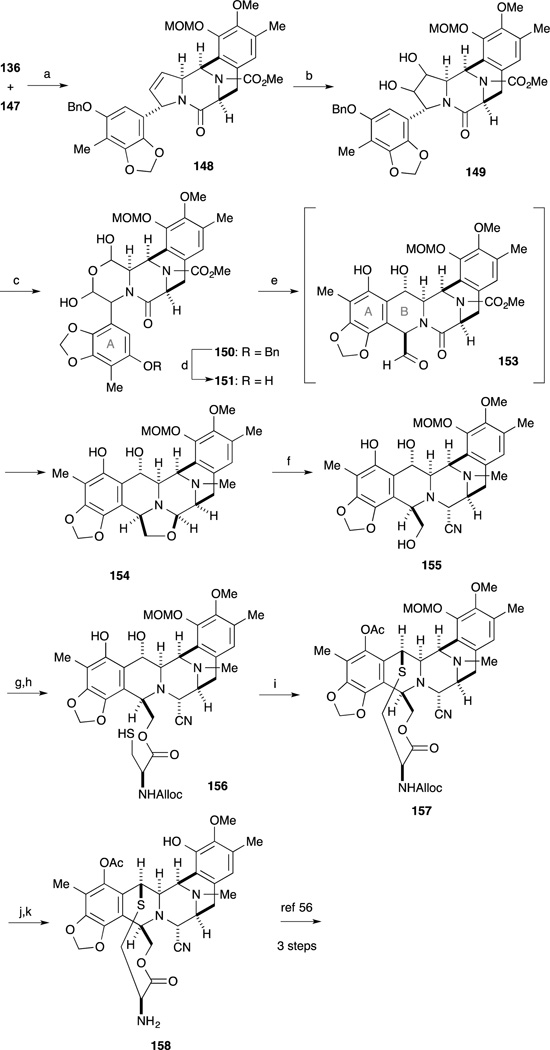
After treatment of amine 136 with t-butyl nitrite and BF3·OEt2,97 Heck reaction of the resulting diazonium salt and enamide 147 from the less-hindered face of the enamide produced coupling product 148 with the desired stereo- and regiochemistry. The dihydroxylation of the highly hindered double bond in 148 and subsequent oxidative cleavage of the resulting 1,2-diol with H5IO6 formed a dialdehyde, which underwent facile hydration to afford 150. Hydrogenolysis of the benzyl ether in 150 gave phenol 151. Heating 151 in m-xylene promoted liberation of the dialdehyde, which was trapped intramolecularly by the electron-rich A-ring moiety to furnish 153. Subsequent reduction of 153 with Red-Al afforded 154 in 76% yield over the two steps. Treatment of 154 with KCN in acetic acid induced cleavage of the oxazolidine ring, forming aminonitrile 155. Condensation of the primary hydroxyl group in 155 with cysteine derivative 82 followed by selective cleavage of the S-acetyl group with hydrazine furnished thiol 156 in good yield. Upon treatment of 156 with TFA, the cyclic sulfide formation occurred presumably via the generation of an ortho-quinone methide to give, after acetylation of the phenolic hydroxyl group, compound 157 in 55% yield. Sequential cleavage of the MOM and Alloc protecting groups furnished 158, which was identical to the intermediate of their previous synthesis, and was converted into Ecteinascidin 743 (1) via the published three-step sequence.
5. Development of Et-743 (Yondelis®) as a Clinical Drug: Therapeutic Applications
The pharmacological development and commercialization of Et-743 are performed by PharmaMar (Zeltia group) and Ortho Biotech Products (Johnson & Johnson) under the commercial name Trabectedin or Yondelis®. Phase I clinical trials of Trabectedin were performed in the United States, France, and by the European Organisation for the Research and Treatment of Cancer (EORTF).98 The most common toxicity observed in these studies was hematological, including grade 3–4 neutropenia, febrile neutropenia and thrombocytopenia. Grade 3–4 transaminitis was observed, and this adverse event is reversible. Nausea/vomiting, fatigue and hyperbilirubinemia were also associated with Trabectedin treatments. In the American study, the maximum tolerated dose (MTD) was recommended to be 1.2 mg/m2 as a continuous infusion over 72 hours every three weeks for phase II studies.99 The proposed MTD was 1.5 mg/m2 as a continuous infusion over 24 hours every three weeks in the French study, and 1 mg/m2 over 1-hour infusion or 1.65 mg/m2 over 3-hour infusion every three weeks in the EORTF study.100, 101
The safety and toxicity profile of Trabectedin was obtained from phase II studies on a total of 1,132 patients (all had received prior treatments, including surgery, radiotherapy and/or chemotherapy) with a wide variety of malignancies.102 These patients were administered with Tracbectedin according to one of the following schedules: twenty-four-hour infusion every three weeks at 1.5 mg/m2, three-hour infusion every three weeks at 1.3 mg/m2, or three-hour infusion every week at 0.58 mg/m2. Nineteen deaths were reported only during the first two cycles of the treatments. 27.7% of the cycles were delayed over five days, 10.2% of the patients were discontinued, and dose reduction was necessary to 9% of the patients due to severe adverse events. Hematological toxicity was common, with predictable and reversible neutropenia and thrombocytopenia occurring the most frequently. Elevation of liver enzymes (AST/ALT) was the most common non-hematological event; however, liver damage by Trabectedin can be managed by administration of Dexamethasone thirty minutes prior to Trabectedin infusion. Nine patients experienced rhabdomyolysis, leading to neutropenia, sepsis, renal failure and elevated liver enzymes, and this adverse event was the most fatal among all of the drug-related toxicities. Liver damage associated with Trabectedin treatment can be reduced by Dexamethasone pretreatment. Other Trabectedin-related toxicities, including fatigue, nausea, vomiting, alopecia, mucositis/stomatitis, periphery sensory neuropathy, cardiac disorder, and renal toxicity, were also reported in these studies.
In a randomized phase II study of Trabectedin in patients with advanced or metastatic liposarcoma and leiomyosarcoma after failure of standard treatments, a reduction in tumor progression was recognized, and the overall survival was favorable.103 Following these trials, Trabectedin was approved for the treatment of advanced or metastatic soft tissue sarcoma after failure of anthracyclines and ifosfamide, or in patients that are not suited for these treatments in 2007 in Europe.102
The combination of Trabectedin with Cisplatin or Doxorubicin is well-tolerated among patients. Also, synergistic activity was observed with little or no cross-resistance of Trabectedin to Cisplatin- or Doxorubicin-resistant models,104–110 supporting the use of Trabectedin in combination with other antitumor agents. A randomized phase III study of Trabectedin/pegylated liposomal Doxorubicin (PLD) combination on recurrent ovarian cancer showed a significant improvement in progression-free survival and overall response rates compared to PLD alone.111 Moreover, this combination was well-tolerated among patients with manageable and non-cumulative adverse events, and a reduction in PLD-related toxicity was observed.112 Therefore, in 2009, Trabectedin was granted approval for treatment of relapsed platinum-sensitive ovarian cancer in combination with PLD in Europe.113
In 2012, the Children’s Oncology Group reported a phase II clinical trial of Trabectedin in children with recurrent rhabdomyosarcoma, Ewing sarcoma and non-rhabdomyosarcoma soft tissue sarcoma.114 Fifty patients ranging from four to twenty-four years old were administered Trabectedin as 24-hour infusion every three weeks at 1.5 mg/m2 or three-hour infusion every three weeks at 1.3 mg/m2.115 Reported adverse events were less serious compared to the adults’ studies with neutropenia being the most common toxicity, where grade 3–4 neutropenia and febrile neutropenia were rare. Other adverse events were also observed, including ALT/AST and GGT elevation and hematological toxicity. Despite the less severe toxicity in this group of patients, the development of Trabectedin to treat these malignancies was not continued due to the lack of meaningful response rates.
Trabectedin was not approved for cancer treatment by the FDA due to its hepatotoxicity and not statistically improving survival rate.96 However, it is certified as an orphan drug for soft tissue sarcoma and ovarian cancer in the United States.116 Currently, it is in phase II and III clinical trial for treatment of breast cancer, other pediatric tumors, and first-line treatment of soft tissue sarcoma with translocation. In Japan, trabectedin is developing as a novel treatment of malignant soft tissue sarcoma with translocation and is gaining supportive results from a phase II clinical trials.
It is significant that, this drug, which will likely become more widely adopted as on-going clinical studies advance, was made possible through the pioneering synthetic chemistry efforts of the Corey laboratory, enabling the semi-synthetic manufacturing route from Cyanosafracin B. Emerging studies on the biosynthesis of the Ecteinascidins, along with many of the additional synthetic studies reviewed here, will likely play a role in the evolution of this potent and fascinating marine alkaloid as a life-saving medicine for various forms of refractory cancers. The story of the Ecteinascidins, serves to underscore the tremendous and continuing importance of natural products discovery, total synthesis efforts and engineered biosynthesis as enabling technologies for the discovery, development and advance of powerful new therapeutic agents. Future chapters in the history of this complex family of alkaloids, will no doubt be exciting.
Scheme 2.
Reagents and conditions (a) TBSCl, Et3N, DMAP, CH2Cl2, 23 °C (98%); (b) DIBAL-H, CH2Cl2, –78 °C (quant); (c) PDC, MS4A, CH2Cl2, 23 °C (99%); (d) monomethylmalonate, piperidine, AcOH, MS3A, toluene, 23 °C (92%); (e) (PhO)2P(O)N3, Et3N, MS4A, toluene, 70 °C; BnOH, 70 °C (89%); (f) H2 (45 psi), Rh[(COD)]-(R,R)-DiPAMP]+BF4, MeOH, 23 °C (100%, 96% ee); (g) H2, 10% Pd/C, EtOAc, 23 °C (quant); (h) AllocCl, pyridine, 23 °C (93%); (i) 0.2 M HCl, AcOH, 110 °C; (j) TBSCl, imidazole, DMF, 23 °C (95%).
Scheme 4.
Reagents and conditions (a) 2-chloro-1,3-dimethyl-imidazolidium hexafluorophosphate (CIP), HOAt, Et3N, THF/CH2Cl2, 23 °C; (b) bromide, Cs2CO3, DMF, 23 °C (81%, 2 steps); (c) LiAlH2(OEt)2, Et2O, –78 °C (95%); (d) KF, MeOH, 23 °C; (e) 0.6 M TfOH, H2O/CF3CH2OH, BHT, 45 °C (89%, 2 steps); (f) LiAlH2(OEt)2, THF, 0 °C; AcOH, 4.8 M aq KCN, 23 °C (87%); (g) PhNTf2, Et3N, DMAP, CH2Cl2, –30 °C (74%); (h) TBDPSCl, DMAP, CH2Cl2, 23 °C (89%); (i) MOMBr, DIPEA, CH2Cl2, 23 °C (92%); (j) n-Bu3SnH, PdCl2(PPh3)2, AcOH, CH2Cl2, 23 °C (quant); (k) CH2O, NaBH3CN, AcOH, MeCN, 23 °C (95%); (l) SnMe4, PdCl2(PPh3)2, LiCl, DMF, 80 °C (83%).
Scheme 8.
Reagents and conditions (a) 73, MeCHO, MeOH, reflux (90%); (b) TBAF, THF, rt (89%); (c) Ac2O, pyridine, DMAP, rt (93%); (d) TFA, anisole, CH2Cl2, rt; (e) EtOAc, reflux (87%, 2 steps); (f) MeSO2Cl, pyridine, CH2Cl2, 0 °C (91%); (g) (Boc)2O, DMAP, CH3CN, rt (97%); (h) NaBH4, H2SO4, EtOH/CH2Cl2, 0 °C; (i) CSA, quinoline, toluene, reflux (88% in 2 steps); (j) Pd2(dba)3 (5 mol%), P(o-tol)3 (20 mol%), TEA, CH3CN, reflux (83%); (k) NaOH, MeOH/H2O, reflux; (l) Ac2O, pyridine, DMAP, rt (93%, 2 steps); (m) TFA, CH2Cl2, rt; (n) TrocCl, aq. NaHCO3/CH2Cl2, rt (74%, 2 steps); (o) DMDO, MeOH/acetone, 0 °C; cat. CSA (90%); (p) NaBH3CN, TFA/THF, 0 °C (94%); q) TBSCl, imidazole, DMF, rt (92%); (r) guanidinium nitrate, NaOMe, MeOH/CH2Cl2, 40 °C (85%); (s) BnBr, K2CO3, CH3CN, reflux (91%); (t) Red-Al, THF, 0 °C (82%); (u) TMSCN, BF3·OEt2, CH2Cl2, 0 °C (73%); (v) Ac2O, pyridine, DMAP, rt (92%); (w) HF, CH3CN, rt (quant); (x) Dess-Martin periodinane, CH2Cl2, rt (92%); (y) Pd/C, H2, THF, rt (84%).
Scheme 10.
Reagents and conditions (a) MOMCl, NaH, Et2O/DMF, 0 °C to rt (96%); (b) n-BuLi, B(OMe)3, THF then AcOH, H2O2, 0 °C to rt (95%); (c) LiCl, MS 3A, HFIP/toluene, ethyl glyoxalate, rt (97%); (d) 4-nitrophenyltriflate, K2CO3, DMF, rt (94%); (e) trimethyl boroxine, K3PO4, Pd(PPh3)4, 1,4-dioxane, reflux (93%); (f) SOBr2, benzotriazole, CH2Cl2, rt (91%).
Scheme 11.
Reagents and conditions (a) TsCl, Et3N, CH2Cl2, –70 °C; (b) MeI, K2CO3, acetone, 55 °C (84%, 2 steps) (c) TiCl4 in CH2Cl2, Cl2CHOCH3, 0 °C to rt (85%); (d) NaBH4, MeOH/THF/H2O, 0 °C (quant); (e) PBr3, toluene/CH2Cl2, 0 °C to rt (96%); (f) 92, cat. 93 (10 mol%), CsOH·H2O, CH2Cl2, –78 °C, then after work up THF/H2O/AcOH (85%); (g) LiBH4, MeOH/Et2O, rt; (b) 2M NaOH, EtOH, reflux.
Scheme 12.
Reagents and conditions (a) 96 AcOH, CH2Cl2/CF3CH2OH (7:1), MS3A, 20 h (84%); (b) AllocCl, NaHCO3, CH2Cl2, rt, 2 h (88%); (c) bromide, Cs2CO3, DMF, rt, 3 h (86%); (d) Ac2O, pyridine, DMAP, CH2Cl2, rt, 1 h (92%); (e) TFA, CH2Cl2, rt (72%).
Scheme 13.
Reagents and conditions (a) TEA, MeCN, 0 °C (α-isomer: 68%, β-isomer: 23%); (b) TBSCl, imidazole, DMF, rt (97%); (c) K2CO3, MeOH, rt (94%); (d) Dess-Martin periodinane, rt; TMSCN, ZnCl2, rt (78%); (e) LiBH4, MeOH, THF, 0 °C (80%); (f) Ac2O, pyridine, DMAP, CH2Cl2 (92%); (g) HF, H2O, MeCN, rt (91%); (h) Dess-Martin periodinane, rt (93%); (i) TFA, CH2Cl2, rt (95%); (j) K2CO3, MeOH, rt (96%); (k) 103, EDCI, DMAP, CH2Cl2, rt (95%); (l) TFA, TFE, rt; Ac2O, pyridine, DMAP, CH2Cl2 (77%); (m) n-Bu3SnH, PdCl2(PPh3)2, AcOH, CH2Cl2, rt (87%); (n) NaBH3CN, AcOH, HCHO, rt (96%); (o) Zn, AcOH, rt (92%); (p) 4-formyl-1-methylpyridinium benzenesulfonate, DBU, sat. oxalic acid, DMF/CH2Cl2, rt (53%); (q) 59, NaOAc, EtOH, rt (97%); (r) AgNO3, MeCN/H2O, rt (92%).
Scheme 14.
Reagents and conditions (a) Cl2CHOMe, TiCl4, CH2Cl2; (b) BBr3, CH2Cl2 (c) Br2, NaOAc, AcOH, 0 to 25 °C; (d) BrCH2Cl, Cs2CO3, DMF, 105 °C (66%, 2 steps); (e) mCPBA, 25 to 64 °C; (f) HCl, 0 to 25 °C (78%, 2 steps); (g) TBDPSCl, TEA, DMAP, 25 °C (89%); (h) n-BuLi, toluene/THF, –78 °C; neat Me-(MeO)NC(O)CH2OBn, –78 °C (80 %); (i) RuCl[(R,R)-Ts-DPEN](p-cymene), HCO2H, TEA, DMF, 0 to 40 °C (78%, 95% ee); (j) DPPA, DBU, toluene/DMF, 50 °C (89%, 95% ee); (k) H2 (1 atm), Pd/C, EtOAc, 25 °C (80%); (l) (MeO)2CHCHO, AcOH, NaBH3CN, MgSO4, MeOH, 0 to 65 °C; (m) TBAF, THF, 0 °C (94%, 2 steps); (n) allyl bromide, NaH, DMF, 0 to 25 °C (84%); (o) 7 M HCl, 1,4-dioxane, 0 to 25 °C (90%).
Scheme 15.
Reagents and conditions (a) BOPCl, TEA, CH2Cl2, 0 to 25 °C (85%); (b) DDQ, CH2Cl2/pH 7.0 buffer, 25 °C (90 %); (c) Cu(OTf)2, benzene, 85 °C (61%); (d) Dess-Martin periodinane, CH2Cl2, 25 °C (94%); (e) PdCl2(PPh3)2n-Bu3SnH, AcOH, 0 to 25 °C (93%); (f) CHF2CO2H, MgSO4, benzene, 100 °C, (42–58%); (g) TBSOTf, TEA, CH2Cl2, 0 °C (quant); (h) TrocCl, TBAI, toluene, 110 °C (92%); (i) TBAF, CH2Cl2, 0 °C, 2 min; (j) MOMCl, DIPEA, (79%); (k) DMDO, CH2Cl2, 0 to 25 °C; (l) NaCNBH3 (78%) (m) H2 (1 atm), Pd/C, EtOAc, 25 °C (77%); (n) DIBAL-H/n-BuLi, THF, 0 °C (78%); o) bromide, DIPEA, CH2Cl2, 50 °C (66%); (p) KCN, AcOH, 25 °C (79%); (q) TFA, CH2Cl2, 20 °C (54%).
Scheme 16.
Reagents and conditions (a) Cl2CHOMe, TiCl4, CH2Cl2; (b) BBr3, CH2Cl2; (c) Br2, NaOAc, AcOH/CH2Cl2 (92%); (d) BrCH2Cl, Cs2CO3, MeCN, 110 °C (69%); (e) mCPBA, CHCl3, reflux; (f) 4 M HCl, MeOH/CH2Cl2, rt (73%, 2 steps); (g) R-Garner’s aldehyde (117) Ti(Oi-Pr)4, toluene, rt; (h) bromide, Cs2CO3, DMF, rt (65%, 2 steps); (i) TsOH·H2O, MeOH, rt; (j) 2,2-dimethoxypropane, TsOH·H2O, DMF, rt (84% 2 steps); (k) TBSOTf, 2,6-lutidine; MeOH; KF·2H2O, MeOH (76%); (l) ethyl glyoxalate, MS4A, toluene; (m) n-Bu3SnH, AIBN, benzene, reflux.
Scheme 17.
Reagents and conditions (a) LiAlH4, THF, 0 °C; (b) NaH, BnBr, DMF/THF, 0 °C (77%, 2 steps); (c) 121, (COCl)2, CH2Cl2, rt; 120, DMF, rt (70%); (d) Et2NH, CH2Cl2, rt; (e) Boc2O, EtOH/CH2Cl2, rt; (f) Dowex 50W–X8, MeOH (90%); (g) TBAF, THF, rt (95%); (h) Swern Oxidation; (i) TFA, anisole, CH2Cl2, rt (72%, 128 : 129 = 0.72 : 1); (j) TrocCl, pyridine, CH2Cl2, 0 °C; (k) BnBr, K2CO3, TBAI, acetone, rt (85%, 2 steps); (l) Pd(PPh3)4, pyrrolidine, CH2Cl2, rt; (m) MOMBr, DIPEA, CH2Cl2, rt (56%, 2 steps); (n) DMDO; (o) NaBH3CN.
Scheme 18.
Reagents and conditions (a) PhI(OAc)2, MeOH, 0 °C; (b) NaCN, DMF/H2O, 0 °C to rt (37%, 2 steps); (c) BnBr, K2CO3, DMF, rt; (d) aq H2O2, K2CO3, DMSO, rt; (e) PhI(OAc)2, KOH, MeOH, 0 °C; (f) LiOH, EtOH/H2O, reflux (83%, 4 steps).
Scheme 20.
Reagents and condition: (a) BF3·OEt2t-BuONO, THF, –15 to 0 °C; 136, Pd2(dba)3, NaOAc, MeCN/THF, 0 °C to rt; (b) OsO4, K3[Fe(CN)6], K2CO3, quinuclidine·HCl, MeSO2NH2t-BuOH, H2O, rt (93%, 2 steps); (c) H5IO6, THF, 0 °C (87%); (d) H2, Pd/C, MeOH, rt; (e) m-xylene, 120 °C; Red-Al, –42 to 60 °C (76%, 2 steps); (f) KCN, AcOH, rt (98%); (g) 82, EDCI, DMAP, CH2Cl2, rt (92%); (h) H2NNH2·H2O, MeCN, rt (85%); (i) TFA, CF3CH2OH, 25 °C; toluene, evaporation; Ac2O, pyridine, rt (55%); (j) TFA, CH2Cl2, rt (64%); (k) PdCl2(Ph3P)2, AcOH, n-Bu3SnH, CH2Cl2, rt (95%).
Notes and References
- 1.Rinehart KL. Med. Res. Rev. 2000;20:1–27. doi: 10.1002/(sici)1098-1128(200001)20:1<1::aid-med1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. J. Org. Chem. 1990;55:4512–4515. [Google Scholar]
- 3.Rath CM, Janto B, Earl J, Ahmed A, Hu FZ, Hiller L, Dahlgren M, Kreft R, Yu FA, Wolff JJ, Kweon HK, Christiansen MA, Hakansson K, Williams RM, Ehrlich GD, Sherman DH. ACS Chem. Biol. 2011;6:1244–1256. doi: 10.1021/cb200244t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss C, Green DH, Perez B, Velasco A, Henriquez R, McKenzie JD. Marine Biology. 2003;143:99–110. [Google Scholar]
- 5.Perez-Matos AE, Rosado W, Govind NS. Antonie Van Leeuwenhoek International Journal of General and Molecular Microbiology. 2007;92:155–164. doi: 10.1007/s10482-007-9143-9. [DOI] [PubMed] [Google Scholar]
- 6.Sakai R, Rinehart KL, Guan Y, Wang AHJ. Proc. Natl. Acad. Sci. U. S. A. 1992;89:11456–11460. doi: 10.1073/pnas.89.23.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore BM, Seaman FC, Hurley LH. J. Am. Chem. Soc. 1997;119:5475–5476. [Google Scholar]
- 8.David-Cordonnier MH, Gajate C, Olmea O, Laine W, de la Iglesia-Vicente J, Perez C, Cuevas C, Otero G, Manzanares I, Bailly C, Mollinedo F. Chem. Biol. 2005;12:1201–1210. doi: 10.1016/j.chembiol.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Application: US US Pat., 2003-406997 20040059112. 2004 [Google Scholar]
- 10.Izbicka E, Lawrence R, Raymond E, Eckhardt G, Faircloth G, Jimeno J, Clark G, Von Hoff DD. Ann Oncol. 1998;9:981–987. doi: 10.1023/A:1008224322396. [DOI] [PubMed] [Google Scholar]
- 11.Hendriks HR, Fiebig HH, Giavazzi R, Langdon SP, Jimeno JM, Faircloth GT. Ann Oncol. 1999;10:1233–1240. doi: 10.1023/a:1008364727071. [DOI] [PubMed] [Google Scholar]
- 12.Bonfanti M, La Valle E, Fernandez Sousa Faro JM, Faircloth G, Caretti G, Mantovani R, D’Incalci M. Anticancer Drug Des. 1999;14:179–186. [PubMed] [Google Scholar]
- 13.Martinez EJ, Owa T, Schreiber SL, Corey EJ. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3496–3501. doi: 10.1073/pnas.96.7.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebayashi Y, Pourquier P, Yoshida A, Kohlhagen G, Pommier Y. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7196–7201. doi: 10.1073/pnas.96.13.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Rocha M, Garcia-Gravalos MD, Avila J. Br. J. Cancer. 1996;73:875–883. doi: 10.1038/bjc.1996.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zwicker J, Muller R. Trends Genet. 1997;13:3–6. doi: 10.1016/s0168-9525(96)30112-1. [DOI] [PubMed] [Google Scholar]
- 17.Simoens C, Korst AE, De Pooter CM, Lambrechts HA, Pattyn GG, Faircloth GT, Lardon F, Vermorken JB. Br. J. Cancer. 2003;89:2305–2311. doi: 10.1038/sj.bjc.6601431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin S, Gorfajn B, Faircloth G, Scotto KW. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6775–6779. doi: 10.1073/pnas.97.12.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW. Biochemistry. 1996;35:13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 20.Moore BM, Seaman FC, Wheelhouse RT, Hurley LH. J. Am. Chem. Soc. 1998;120:2490–2491. [Google Scholar]
- 21.Garcia-Nieto R, Manzanares I, Cuevas C, Gago F. J. Am. Chem. Soc. 2000;122:7172–7182. [Google Scholar]
- 22.Zewail-Foote M, Hurley LH. J. Med. Chem. 1999;42:2493–2497. doi: 10.1021/jm990241l. [DOI] [PubMed] [Google Scholar]
- 23.Zewail-Foote M, Hurley LH. J. Am. Chem. Soc. 2001;123:6485–6495. doi: 10.1021/ja004023p. [DOI] [PubMed] [Google Scholar]
- 24.Seaman FC, Hurley LH. J. Am. Chem. Soc. 1998;120:13028–13041. [Google Scholar]
- 25.Garcia-Nieto R, Manzanares I, Cuevas C, Gago F. J. Med. Chem. 2000;43:4367–4369. doi: 10.1021/jm000322d. [DOI] [PubMed] [Google Scholar]
- 26.Kanzaki A, Takebayashi Y, Ren XQ, Miyashita H, Mori S, Akiyama S, Pommier Y. Mol. Cancer Ther. 2002;1:1327–1334. [PubMed] [Google Scholar]
- 27.Beumer JH, Buckle T, Ouwehand M, Franke NE, Lopez-Lazaro L, Schellens JH, Beijnen JH, van Tellingen O. Invest. New Drugs. 2006;25:1–7. doi: 10.1007/s10637-006-7773-9. [DOI] [PubMed] [Google Scholar]
- 28.Feuerhahn S, Giraudon C, Martinez-Diez M, Bueren-Calabuig JA, Galmarini CM, Gago F, Egly JM. Chem. Biol. 2011;18:988–999. doi: 10.1016/j.chembiol.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 29.Di Giandomenico S, Frapolli R, Bello E, Uboldi S, Licandro SA, Marchini S, Beltrame L, Brich S, Mauro V, Tamborini E, Pilotti S, Casali PG, Grosso F, Sanfilippo R, Gronchi A, Mantovani R, Gatta R, Galmarini CM, Sousa-Faro JM, D’Incalci M. Oncogene. 2013 doi: 10.1038/onc.2013.462. [DOI] [PubMed] [Google Scholar]
- 30.Grohar PJ, Griffin LB, Yeung C, Chen QR, Pommier Y, Khanna C, Khan J, Helman LJ. Neoplasia. 2011;13:145–153. doi: 10.1593/neo.101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez-Garcia I, Rabbitts TH. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7869–7873. doi: 10.1073/pnas.91.17.7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Losada J, Pintado B, Gutierrez-Adan A, Flores T, Banares-Gonzalez B, del Campo JC, Martin-Martin JF, Battaner E, Sanchez-Garcia I. Oncogene. 2000;19:2413–2422. doi: 10.1038/sj.onc.1203572. [DOI] [PubMed] [Google Scholar]
- 33.Owen LA, Kowalewski AA, Lessnick SL. PLoS One. 2008;3:e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takebayashi Y, Pourquier P, Zimonjic DB, Nakayama K, Emmert S, Ueda T, Urasaki Y, Kanzaki A, Akiyama SI, Popescu N, Kraemer KH, Pommier Y. Nat Med. 2001;7:961–966. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- 35.Herrero AB, Martin-Castellanos C, Marco E, Gago F, Moreno S. Cancer Res. 2006;66:8155–8162. doi: 10.1158/0008-5472.CAN-06-0179. [DOI] [PubMed] [Google Scholar]
- 36.Damia G, Silvestri S, Carrassa L, Filiberti L, Faircloth GT, Liberi G, Foiani M, D’Incalci M. Int. J. Cancer. 2001;92:583–588. doi: 10.1002/ijc.1221. [DOI] [PubMed] [Google Scholar]
- 37.Takebayashi Y, Goldwasser F, Urasaki Y, Kohlhagen G, Pommier Y. Clin Cancer Res. 2001;7:185–191. [PubMed] [Google Scholar]
- 38.Soares DG, Escargueil AE, Poindessous V, Sarasin A, de Gramont A, Bonatto D, Henriques JA, Larsen AK. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13062–13067. doi: 10.1073/pnas.0609877104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavecchio M, Simone M, Erba E, Chiolo I, Liberi G, Foiani M, D’Incalci M, Damia G. Eur J Cancer. 2008;44:609–618. doi: 10.1016/j.ejca.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Zewail-Foote M, Li VS, Kohn H, Bearss D, Guzman M, Hurley LH. Chem. Biol. 2001;8:1033–1049. doi: 10.1016/s1074-5521(01)00071-0. [DOI] [PubMed] [Google Scholar]
- 41.Guirouilh-Barbat J, Redon C, Pommier Y. Mol. Biol. Cell. 2008;19:3969–3981. doi: 10.1091/mbc.E08-02-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiloh Y. Trends Biochem. Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Germano G, Frapolli R, Belgiovine C, Anselmo A, Pesce S, Liguori M, Erba E, Uboldi S, Zucchetti M, Pasqualini F, Nebuloni M, van Rooijen N, Mortarini R, Beltrame L, Marchini S, Fuso Nerini I, Sanfilippo R, Casali PG, Pilotti S, Galmarini CM, Anichini A, Mantovani A, D’Incalci M, Allavena P. Cancer Cell. 2013;23:249–262. doi: 10.1016/j.ccr.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 44.Arai T, Takahashi K, Nakahara S, Kubo A. Experientia. 1980;36:1025–1027. doi: 10.1007/BF01965946. [DOI] [PubMed] [Google Scholar]
- 45.Irschik H, Trowitzsch-Kienast W, Gerth K, Hofle G, Reichenbach H. J Antibiot (Tokyo) 1988;41:993–998. doi: 10.7164/antibiotics.41.993. [DOI] [PubMed] [Google Scholar]
- 46.Ikeda Y, Shimada Y, Honjo K, Okumoto T, Munakata T. J Antibiot (Tokyo) 1983;36:1290–1294. doi: 10.7164/antibiotics.36.1290. [DOI] [PubMed] [Google Scholar]
- 47.Kluepfel D, Baker HA, Piattoni G, Sehgal SN, Sidorowicz A, Singh K, Vezina C. J. Antibiot. 1975;28:497–502. doi: 10.7164/antibiotics.28.497. [DOI] [PubMed] [Google Scholar]
- 48.Tomita F, Takahashi K, Shimizu K. J. Antibiot. 1983;36:463–467. doi: 10.7164/antibiotics.36.463. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi K, Tomita F. J. Antibiot. 1983;36:468–470. doi: 10.7164/antibiotics.36.468. [DOI] [PubMed] [Google Scholar]
- 50.Kerr RG, Miranda NF. J. Nat. Prod. 1995;58:1618–1621. [Google Scholar]
- 51.Peng C, Pu JY, Song LQ, Jian XH, Tang MC, Tang GL. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8540–8545. doi: 10.1073/pnas.1204232109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jeedigunta S, Krenisky JM, Kerr RG. Tetrahedron. 2000;56:3303–3307. [Google Scholar]
- 53.Li L, Deng W, Song J, Ding W, Zhao QF, Peng C, Song WW, Tang GL, Liu W. J. Bacteriol. 2008;190:251–263. doi: 10.1128/JB.00826-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pospiech A, Cluzel B, Bietenhader J, Schupp T. Microbiology. 1995;141(Pt 8):1793–1803. doi: 10.1099/13500872-141-8-1793. [DOI] [PubMed] [Google Scholar]
- 55.Velasco A, Acebo P, Gomez A, Schleissner C, Rodriguez P, Aparicio T, Conde S, Munoz R, de la Calle F, Garcia JL, Sanchez-Puelles JM. Mol. Microbiol. 2005;56:144–154. doi: 10.1111/j.1365-2958.2004.04433.x. [DOI] [PubMed] [Google Scholar]
- 56.Fu CY, Tang MC, Peng C, Li L, He YL, Liu W, Tang GL. J Microbiol Biotechnol. 2009;19:439–446. doi: 10.4014/jmb.0808.484. [DOI] [PubMed] [Google Scholar]
- 57.Finking R, Marahiel MA. Annu. Rev. Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 58.Fischbach MA, Walsh CT. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 59.Koketsu K, Minami A, Watanabe K, Oguri H, Oikawa H. Methods Enzymol. 2012;516:79–98. doi: 10.1016/B978-0-12-394291-3.00026-5. [DOI] [PubMed] [Google Scholar]
- 60.Arroyo M, de la Mata I, Acebal C, Castillon MP. Appl. Microbiol. Biotechnol. 2003;60:507–514. doi: 10.1007/s00253-002-1113-6. [DOI] [PubMed] [Google Scholar]
- 61.Cuevas C, Pérez M, Martín MJ, Chicharro JL, Fernández-Rivas C, Flores M, Francesch A, Gallego P, Zarzuelo M, de la Calle F, Garcıía J, Polanco C, Rodríguez I, Manzanares I. Org. Lett. 2000;2:2545–2548. doi: 10.1021/ol0062502. [DOI] [PubMed] [Google Scholar]
- 62.Corey EJ, Gin DY, Kania RS. J. Am. Chem. Soc. 1996;118:9202–9203. [Google Scholar]
- 63.Martinez EJ, Corey EJ. Org. Lett. 2000;2:993–996. doi: 10.1021/ol0056729. [DOI] [PubMed] [Google Scholar]
- 64.Barton DHR, Elliott JD, Gero SD. Journal of the Chemical Society-Perkin Transactions 1. 1982:2085–2090. [Google Scholar]
- 65.Endo A, Yanagisawa A, Abe M, Tohma S, Kan T, Fukuyama T. J. Am. Chem. Soc. 2002;124:6552–6554. doi: 10.1021/ja026216d. [DOI] [PubMed] [Google Scholar]
- 66.Tohma S, Endo A, Kan T, Fukuyama T. Synlett. 2001:1179–1181. [Google Scholar]
- 67.Hayashi T, Konishi M, Kobori Y, Kumada M, Higuchi T, Hirotsu K. J. Am. Chem. Soc. 1984;106:158–163. [Google Scholar]
- 68.Gokel GW, Widera RP, Weber WP. Org. Synth. 1988;50–9:232–235. [Google Scholar]
- 69.Burk MJ, Feaster JE, Nugent WA, Harlow RL. J. Am. Chem. Soc. 1993;115:10125–10138. [Google Scholar]
- 70.Gokel G, Ludke G, Ugi I. Isonitrile Chemistry. New York: Acadiemic Press; 1971. [Google Scholar]
- 71.Link JT, Overman LE. Metal-Catalyzed Cross-Coupling Reactions. Weinheim, Germany: Wiley-VCH Verlag GmbH; 1998. [Google Scholar]
- 72.Shibasaki M, Boden CDJ, Kojima A. Tetrahedron. 1997;53:7371–7395. [Google Scholar]
- 73.Brase S, De Meijere A. Metal-Catalyzed Cross-Coupling Reactions. Weinheim, Germany: Wiley-VCH Verlag GmbH; 1998. [Google Scholar]
- 74.Neuville L, Bigot A, Dau METH, Zhu JP. J. Org. Chem. 1999;64:7638–7642. [Google Scholar]
- 75.Miyaura N, Suzuki A. Chemical Reviews. 1995;95:2457–2483. [Google Scholar]
- 76.De Paolis M, Chen XC, Zhu JP. Synlett. 2004:729–731. [Google Scholar]
- 77.Corey EJ, Xu F, Noe MC. J. Am. Chem. Soc. 1997;119:12414–12415. [Google Scholar]
- 78.Lygo B, Wainwright PG. Tetrahedron Lett. 1997;38:8595–8598. [Google Scholar]
- 79.Odonnell MJ, Bennett WD, Wu SD. J. Am. Chem. Soc. 1989;111:2353–2355. [Google Scholar]
- 80.Garner P, Park JM. J. Org. Chem. 1987;52:2361–2364. [Google Scholar]
- 81.Garner P, Park JM. J. Org. Chem. 1988;53:2979–2984. [Google Scholar]
- 82.Garner P, Park JM, Malecki E. J. Org. Chem. 1988;53:4395–4398. [Google Scholar]
- 83.Cox ED, Cook JM. Chemical Reviews. 1995;95:1797–1842. [Google Scholar]
- 84.Massiot G, Mulamba T. Journal of the Chemical Society-Chemical Communications. 1983:1147–1149. [Google Scholar]
- 85.Bailey PD, Hollinshead SP, Mclay NR, Morgan K, Palmer SJ, Prince SN, Reynolds CD, Wood SD. Journal of the Chemical Society-Perkin Transactions 1. 1993:431–439. [Google Scholar]
- 86.Zheng S, Chan C, Furuuchi T, Wright BJ, Zhou B, Guo J, Danishefsky SJ. Angew Chem Int Ed Engl. 2006;45:1754–1759. doi: 10.1002/anie.200503983. [DOI] [PubMed] [Google Scholar]
- 87.Sinhababu AK, Ghosh AK, Borchardt RT. J. Med. Chem. 1985;28:1273–1279. doi: 10.1021/jm00147a027. [DOI] [PubMed] [Google Scholar]
- 88.Williams RM, Ehrlich PP, Zhai WX, Hendrix J. J. Org. Chem. 1987;52:2615–2617. [Google Scholar]
- 89.Fujii A, Hashiguchi S, Uematsu N, Ikariya T, Noyori R. J. Am. Chem. Soc. 1996;118:2521–2522. [Google Scholar]
- 90.Chan C, Heid R, Zheng SP, Guo JS, Zhou BS, Furuuchi T, Danishefsky SJ. J. Am. Chem. Soc. 2005;127:4596–4598. doi: 10.1021/ja050203t. [DOI] [PubMed] [Google Scholar]
- 91.Fishlock D, Willlams RM. J. Org. Chem. 2008;73:9594–9600. doi: 10.1021/jo801159k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sakaitani M, Ohfune Y. J. Org. Chem. 1990;55:870–876. [Google Scholar]
- 93.Jin W, Williams RM. Tetrahedron Lett. 2003;44:4635–4639. [Google Scholar]
- 94.Hussain HH, Babic G, Durst T, Wright JS, Flueraru M, Chichirau A, Chepelev LL. J. Org. Chem. 2003;68:7023–7032. doi: 10.1021/jo0301090. [DOI] [PubMed] [Google Scholar]
- 95.Chen XC, Chen JC, De Paolis M, Zhu JP. J. Org. Chem. 2005;70:4397–4408. doi: 10.1021/jo050408k. [DOI] [PubMed] [Google Scholar]
- 96.Weigl M, Wunsch B. Tetrahedron. 2002;58:1173–1183. [Google Scholar]
- 97.Doyle MP, Bryker WJ. J. Org. Chem. 1979;44:1572–1574. [Google Scholar]
- 98.Cioffi A, Italiano A. Expert Opin Drug Metab Toxicol. 2012;8:113–122. doi: 10.1517/17425255.2012.636353. [DOI] [PubMed] [Google Scholar]
- 99.Ryan DP, Supko JG, Eder JP, Seiden MV, Demetri G, Lynch TJ, Fischman AJ, Davis J, Jimeno J, Clark JW. Clin Cancer Res. 2001;7:231–242. [PubMed] [Google Scholar]
- 100.Taamma A, Misset JL, Riofrio M, Guzman C, Brain E, Lopez Lazaro L, Rosing H, Jimeno JM, Cvitkovic E. J Clin Oncol. 2001;19:1256–1265. doi: 10.1200/JCO.2001.19.5.1256. [DOI] [PubMed] [Google Scholar]
- 101.Twelves C, Hoekman K, Bowman A, Vermorken JB, Anthoney A, Smyth J, van Kesteren C, Beijnen JH, Uiters J, Wanders J, Gomez J, Guzman C, Jimeno J, Hanauske A. Eur J Cancer. 2003;39:1842–1851. doi: 10.1016/s0959-8049(03)00458-1. [DOI] [PubMed] [Google Scholar]
- 102.Le Cesne A, Yovine A, Blay JY, Delaloge S, Maki RG, Misset JL, Frontelo P, Nieto A, Jiao JJ, Demetri GD. Invest. New Drugs. 2012;30:1193–1202. doi: 10.1007/s10637-011-9662-0. [DOI] [PubMed] [Google Scholar]
- 103.Demetri GD, Chawla SP, von Mehren M, Ritch P, Baker LH, Blay JY, Hande KR, Keohan ML, Samuels BL, Schuetze S, Lebedinsky C, Elsayed YA, Izquierdo MA, Gomez J, Park YC, Le Cesne A. J Clin Oncol. 2009;27:4188–4196. doi: 10.1200/JCO.2008.21.0088. [DOI] [PubMed] [Google Scholar]
- 104.Takahashi N, Li WW, Banerjee D, Scotto KW, Bertino JR. Clin Cancer Res. 2001;7:3251–3257. [PubMed] [Google Scholar]
- 105.Erba E, Muratore I, Tiozzo G, Ubezio P, Colombo T, Nicoletti I, Giavazzi R, Zucchetti M, Damia G, Meco D, Riccardi R, Faircloth GT, Jimeno J, D’Incalci M. Clinical Cancer Research. 2001;7:3735s–3735s. [Google Scholar]
- 106.Scotlandi K, Perdichizzi S, Manara MC, Serra M, Benini S, Cerisano V, Strammiello R, Mercuri M, Reverter-Branchat G, Faircloth G, D’Incalci M, Picci P. Clinical Cancer Research. 2002;8:3893–3903. [PubMed] [Google Scholar]
- 107.Takahashi N, Li WW, Banerjee D, Guan YB, Wada-Takahashi Y, Brennan MF, Chou TC, Scotto KW, Bertino JR. Cancer Res. 2002;62:6909–6915. [PubMed] [Google Scholar]
- 108.Meco D, Colombo T, Ubezio P, Zucchetti M, Zaffaroni M, Riccardi A, Faircloth G, Jose J, D’Incalci M, Riccardi R. Cancer Chemotherapy and Pharmacology. 2003;52:131–138. doi: 10.1007/s00280-003-0636-6. [DOI] [PubMed] [Google Scholar]
- 109.Erba E, Colombo T, Ubezio P, Ferrarese L, Matera G, Falcioni C, Faircloth G, Jimeno J, D’Incalci M. Clinical Cancer Research. 2003;9:6264s–6264s. [Google Scholar]
- 110.D’Incalci M, Colombo T, Ubezio P, Nicoletti I, Giavazzi R, Erba E, Ferrarese L, Meco D, Riccardi R, Sessa C, Cavallini E, Jimeno J, Faircloth GT. European Journal of Cancer. 2003;39:1920–1926. doi: 10.1016/s0959-8049(03)00490-8. [DOI] [PubMed] [Google Scholar]
- 111.Monk BJ, Herzog TJ, Kaye SB, Krasner CN, Vermorken JB, Muggia FM, Pujade-Lauraine E, Lisyanskaya AS, Makhson AN, Rolski J, Gorbounova VA, Ghatage P, Bidzinski M, Shen K, Ngan HY, Vergote IB, Nam JH, Park YC, Lebedinsky CA, Poveda AM. J Clin Oncol. 2010;28:3107–3114. doi: 10.1200/JCO.2009.25.4037. [DOI] [PubMed] [Google Scholar]
- 112.Krasner CN, Poveda A, Herzog TJ, Vermorken JB, Kaye SB, Nieto A, Claret PL, Park YC, Parekh T, Monk BJ. Gynecol Oncol. 2012;127:161–167. doi: 10.1016/j.ygyno.2012.06.034. [DOI] [PubMed] [Google Scholar]
- 113.Lebedinsky C, Gomez J, Park YC, Nieto A, Soto-Matos A, Parekh T, Alfaro V, Roy E, Lardelli P, Kahatt C. Cancer Chemother Pharmacol. 2011;68:1223–1231. doi: 10.1007/s00280-011-1614-z. [DOI] [PubMed] [Google Scholar]
- 114.Baruchel S, Pappo A, Krailo M, Baker KS, Wu B, Villaluna D, Lee-Scott M, Adamson PC, Blaney SM. Eur J Cancer. 2012;48:579–585. doi: 10.1016/j.ejca.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 115.Lau L, Supko JG, Blaney S, Hershon L, Seibel N, Krailo M, Qu W, Malkin D, Jimeno J, Bernstein M, Baruchel S. Clin Cancer Res. 2005;11:672–677. [PubMed] [Google Scholar]
- 116.Drugs R D. 2006;7:317–328. [Google Scholar]