Abstract
HOX transcript antisense intergenic RNA (HOTAIR) is a long non-coding RNA (lncRNA) that functions as an oncogenic molecule in different cancer cells. Genetic variants of HOTAIR may affect the activity of certain regulatory factors and further regulate the aberrant expression of HOTAIR, which might be underlying mechanisms that affect tumour susceptibility and prognosis. Recently, several studies have been performed to examine the possible link between polymorphisms in HOTAIR and cancer risk; however, the results have been inconclusive. Therefore, we performed a meta-analysis to estimate the associations between HOTAIR polymorphisms (rs920778, rs4759314 and rs1899663) and cancer risk. Eight studies comprising 7,151 cases and 8,740 controls were included in our study. Overall, no significant associations between the HOTAIR polymorphisms (rs920778, rs4759314 and rs1899663) and cancer risk were observed. However, in further stratified analyses, the variant T allele of rs920778 exhibited a significant increased risk of developing digestive cancers (dominant model: OR = 1.44; 95% CI = 1.31–1.59). These findings provided evidence that HOTAIR rs920778 may modify the susceptibility to certain cancer types. Further studies incorporating subjects with different ethnic backgrounds combined with re-sequencing of the marked region and functional evaluations are warranted.
Introduction
Cancers are the leading cause of deaths worldwide. According to WHO estimates, there were 14.1 million new cancer cases in 2012, and over 20 million new cancer cases will be expected annually as early as 2025, indicating an ever-increasing global cancer burden [1]. Generally, cancers are considered to be multifactorial diseases, and the occurrence of cancers are related to environmental, genetic and lifestyle factors. Among these factors, the genetic factors are of particular interest, especially because recent genome-wide association studies (GWAS) and next-generation sequencing (NGS) have greatly broadened our understanding of the genetic variations that confer risks for cancers.
Long non-coding RNAs (lncRNAs) are a type of non-coding RNA (ncRNA) that contain more than 200 nucleotides and do not encode proteins but have pivotal roles in numerous biological functions. Recent reports suggest that aberrant expression of lncRNAs might play important roles in the development and progression of tumours [2,3,4] that are likely mediated through changes at the chromatin, transcriptional or post-transcriptional levels that influence target gene expression [5]. Additionally, GWASs have successfully identified several lncRNA polymorphisms that are associated with the risks of developing different types of cancer. However, similar to the other reported GWAS, the majority of the lncRNA SNPs that have been identified by GWAS have been mapped to intergenic regions or introns that do not encode proteins, and the potential functions of these SNPs in the pathogenesis of cancer remain undefined. Furthermore, due to the stringent screening criteria of GWASs, some putative causal lncRNAs and variants that are associated with carcinogenesis may have been be largely ignored. Thus, additional efforts directed toward candidate lncRNAs that function in the development of cancer may be expedient to uncover part of the missing heritability. One such lncRNA is the HOX transcript antisense intergenic RNA (HOTAIR), the function of which has been demonstrated to be closely related to the development and progression of some cancers [6,7,8].
To date, the relationship between the aberrant expression of HOTAIR and cancer prognosis has been explored by many researchers. Moreover, meta-analyses have demonstrated that the aberrant expression of HOTAIR may serve as an indicator that predicts poor prognoses both in cancer overall [9] and in particular types of cancers (e.g., digestive system cancers and oestrogen-dependent malignant tumours) [10,11,12]. Additionally, a meta-analysis performed by Cai et al. revealed that the overexpression of HOTAIR is significantly associated with lymph node metastasis in cancer patients, which might further affect cancer prognoses [13].
In 2015, Bayram S et al. demonstrated that rs920778 SNP of HOTAIR is significantly associated with advanced TNM stage, distant metastasis and poor histological grade in breast cancer patients, which indicates that this polymorphism may be associated with breast cancer prognosis [14]. Similarly, it has been reported that another SNP in HOTAIR (rs12826786) is associated with the clinicopathological features involved in gastric cardia adenocarcinoma progression [15]. Additionally, relationships of HOTAIR polymorphisms (including rs4759314 and rs920778) with the expression of HOTAIR and various cancer risks have been observed [15,16,17,18]. Moreover, Zhang et al. reported that SNP rs920778 in HOTAIR may alter the activity of a novel intronic HOTAIR enhancer [17]. Thus, it is biologically conceivable that the genetic variants of HOTAIR may affect the activities of certain regulatory factors and further regulate the aberrant expression of HOTAIR, which might be one of the underlying mechanisms that affect tumour susceptibility and prognosis. As expected, the relationships HOTAIR of polymorphisms with sensitivities to cancers have attracted much interest [14,15,16,17,18,19,20,21]. However, the results of the studies that have explored this association are inconclusive. For example, a previous study report that the rs920778 variant genotype significantly increases the risk of gastric cancer in Chinese people [16]; however, among Turkish people, the same variant exhibited no significant association, and the effect values were even in the opposite direction relative to the previous studies of Chinese populations [19]. Therefore, based on all of the currently published data, we performed a meta-analysis to more precisely characterize the associations of HOTAIR polymorphisms (rs920778, rs4759314 and rs1899663) with cancer risk.
Materials and Methods
Identification and eligibility of the relevant studies
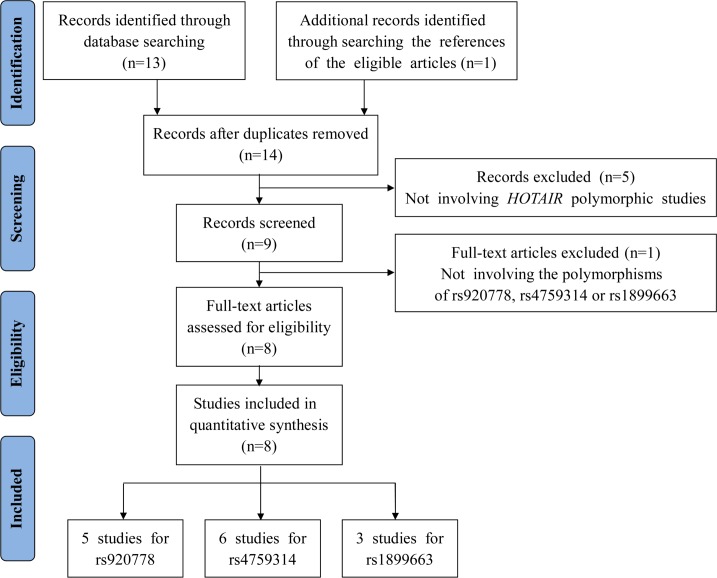
A comprehensive literature search of PubMed and Embase up to November 30, 2015 was performed using the following keywords: ("HOTAIR" or "HOX transcript antisense intergenic RNA") and ("cancer", "carcinoma", "tumor", "tumour", or "neoplasm") and ("polymorphism", "variation", "variant", or "mutation"). The references in retrieved articles were also reviewed for possible inclusion. Only publications written in English with available full-text articles were included in this meta-analysis. Studies were included if they met the following eligibility criteria: (1) case-control studies focused on the relationship between HOTAIR polymorphisms and any type of cancer, (2) more than two articles for each studied HOTAIR polymorphism, (3) available information about the genotype frequency of each included HOTAIR SNP (i.e., rs920778, rs4759314 or rs1899663), and (4) published as a full paper in English. The main reasons for the exclusion of studies were the following: (1) not focused on cancer risk, (2) did not study the HOTAIR SNPs (rs920778, rs4759314 or rs1899663), (3) did not report the relevant genotype frequency data, (4) not published in English, and (5) non-human research. Ultimately, a total of 8 articles including 7,151 cases and 8,740 controls were included in this meta-analysis (Fig 1).
Fig 1. Flow diagram of the study selection process.
Data extraction
Two investigators (T.T. and L.J.) independently extracted the data and reached consensus regarding all of the items. The following information was sought from each article: the first author’s name, year of publication, country of origin, ethnicity, type of cancer, numbers of cases and controls, genotyping platform and genotyped SNPs. We categorized the different ethnicities as Caucasians and Asians.
Statistical analysis
The risk of cancer associated with each HOTAIR polymorphism was estimated for each study using the odds ratio (OR) and its 95% confidence interval (95%CI). The between-study heterogeneity was examined with a chi-square-based Q statistic test, and P ≤ 0.05 was considered as statistically significant. We pooled the results using fixed-effect models when the heterogeneity between studies was absent. Otherwise, a random-effects model was selected. Subsequently, we evaluated the risks of the heterozygous and variant homozygous genotypes relative to the wild-type homozygous genotype and then evaluated the risks of the combined heterozygous and variant homozygous genotypes relative to the wild-type homozygous genotype while assuming the dominant effects of the variant allele. For rs920778, we also performed a stratification analyses based on ethnicity (divided into Caucasians and Asians) and cancer types. Funnel plots and Begg’s test were utilized to evaluate the publication bias. All analyses were performed using the Stata version 12.0 software (Stata Corporation, College Station, TX, USA).
Functional annotation based on publically available databases
The SNPs in high LD (r2 ≥ 0.80) with the marker SNP rs920778 were identified according with SNAP 2.2 (http://www.broadinstitute.org/mpg/snap/ldsearch.php/). Expression quantitative trait loci (eQTL) analysis was performed based on the Genotype-Tissue Expression project (GTEx) database (http://www.gtexportal.org/home/). The influences of the SNPs on miRNA binding were predicted with an online service (http://www.bioguo.org/miRNASNP2/). Additionally, other functional annotation results were derived from the ENCODE database (http://genome.ucsc.edu/ENCODE/).
Results
Characteristics of the published studies
After the application of the strict screening criteria, 8 articles that included a total of 7,151 cases and 8,740 controls harbouring gastric cancer, breast cancer, colorectal cancer and oesophageal squamous cell carcinomas (ESCCs) were ultimately included in the current quantitative analysis. The general characteristics of the included studies are listed in Table 1, and the excluded studies are listed in S1 Table. Among the included studies, six studies were conducted in Asians (Chinese) populations, and two studies were conducted in Caucasian (Turkish) populations. The publication years of the included studies were between 2014 and 2015, and the range of the sample sizes was 245 to 4,248. A total of four articles reported the effects of HOTAIR polymorphisms in gastric cancer, two in breast cancer, one in oesophageal squamous cell carcinoma (ESCC), and one in colorectal cancer. Among the studies that explored the relationships of HOTAIR SNPs with cancer risk, five focused on the rs920778 SNP, six on the rs4759314 SNP and three on the rs1899663 SNP. Genotyping was performed using TaqMan in 4 studies, PCR-RFLP in 3 studies and CRS–RFLP in 1 study. The distributions of the genotypes and alleles of the HOTAIR polymorphisms (rs920778, rs4759314 and rs1899663) in the individual studies are listed in S2–S4 Tables.
Table 1. Characteristics of the studies included in the meta-analysis.
| First Author | Year | Country | Ethnicity | Type of cancer | Case/Control | Platform | Genotyped SNPs |
|---|---|---|---|---|---|---|---|
| Zhang | 2014 | China | Asian | ESCC a | 2098/2150 | PCR-RFLP | rs920778, rs4759314, rs1899663 |
| Bayram | 2015 | Turkey | Caucasian | gastric cancer | 104/209 | TaqMan | rs920778 |
| Pan | 2015 | China | Asian | gastric cancer | 800/1600 | PCR-RFLP | rs920778, rs4759314, rs1899663 |
| Xue | 2015 | China | Asian | colorectal cancer | 1734/1855 | TaqMan | rs4759314 |
| Du | 2015 | China | Asian | gastric cancer | 1275/1646 | TaqMan | rs4759314 |
| Guo | 2015 | China | Asian | GCA b | 515/654 | PCR-RFLP | rs4759314 |
| Bayram | 2015 | Turkey | Caucasian | breast cancer | 123/122 | TaqMan | rs920778 |
| Yan | 2015 | China | Asian | breast cancer | 502/504 | CRS–RFLP/PCR-RFLP | rs920778, rs4759314, rs1899663 |
a oesophageal squamous cell carcinoma (ESCC)
b gastric cardia adenocarcinoma (GCA)
Quality assessments of the included studies
The methodological quality of each included study was evaluated using the Newcastle-Ottawa quality assessment scale (NOS). Using this method, each study was judged on standard criteria and subsequently categorized based on three factors: selection, comparability, and exposure. Summary scores ranging from 0 to 9 points were calculated, and higher scores indicate lower risks of bias (S5 Table).
Quantitative synthesis
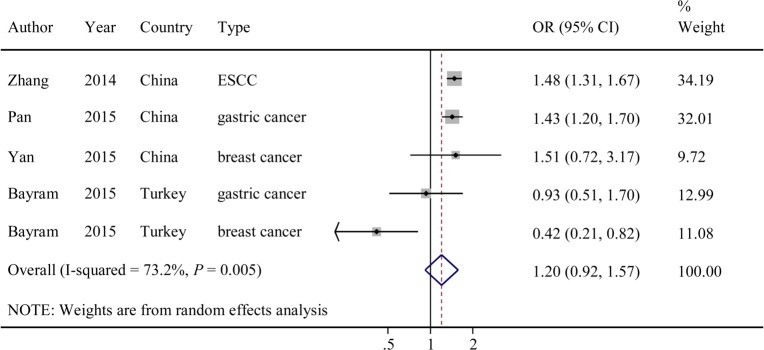
The evaluations of the associations of rs920778 with cancer risks are presented in Table 2. Overall, the T allele variant exhibited no significant association with cancer risk in any of the tested models (CT versus CC: OR = 1.11; 95% CI = 0.86–1.44; P = 0.014 for the heterogeneity test, I2 = 67.8%; TT versus CC: OR = 1.55; 95% CI = 0.84–2.85; P = 0.000 for the heterogeneity test, I2 = 85.8%; dominant model: OR = 1.20; 95% CI = 0.92–1.57, P = 0.005 for the heterogeneity test, I2 = 73.2%; Fig 2).
Table 2. Summary ORs of the HOTAIR rs920778 polymorphism and cancer risk.
| Variables | Studies | Sample size | CT versus CC | TT versus CC | Dominant model | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR(95%CI) | P a | I 2 | OR (95%CI) | P a | I 2 | OR(95%CI) | P a | I 2 | |||
| Total | 5 | 8,212 | 1.11(0.86–1.44) | 0.014 | 67.8% | 1.55(0.84–2.85) | 0.000 | 85.8% | 1.20(0.92–1.57) | 0.005 | 73.2% |
| Ethnicity | |||||||||||
| Asians | 3 | 7,654 | 1.32(1.19–1.47) | 0.938 | 0.0% | 2.76(2.22–3.43) | 0.403 | 0.0% | 1.46(1.32–1.61) | 0.953 | 0.0% |
| Caucasians | 2 | 558 | 0.63(0.39–1.00) | 0.064 | 70.8% | 0.68(0.41–1.12) | 0.199 | 39.4% | 0.65(0.42–1.00) | 0.080 | 67.4% |
| Cancer type | |||||||||||
| digestive cancer b | 3 | 6,961 | 1.31(1.19–1.46) | 0.564 | 0.0% | 2.17(1.26–3.75) | 0.008 | 79.4% | 1.44(1.31–1.59) | 0.339 | 7.6% |
| breast cancer | 2 | 1,251 | 0.67(0.22–2.04) | 0.032 | 78.1% | 0.90(0.25–3.20) | 0.017 | 82.4% | 0.79(0.22–2.78) | 0.012 | 84.3% |
a P for heterogeneity (a random-effects model was used when the P value for heterogeneity test was < 0.05; otherwise, a fixed-effect model was used.)
b including gastric cancer and ESCC
Fig 2. Forest plot of OR with 95%CI for the HOTAIR rs920778 with cancer risk under dominant model.
We next evaluated the effect of the rs920778 polymorphism on cancer risk among the subgroups (Table 2). In a stratified analyses, a significantly increased cancer risk was observed among Asians (Chinese; dominant model: OR = 1.46; 95% CI = 1.32–1.61; P = 0.953 for the heterogeneity test, I2 = 0.0%). In contrast, a significantly decreased cancer risk was observed among Caucasians (Turkish; dominant model: OR = 0.65; 95%CI = 0.42–1.00; P = 0.080 for the heterogeneity test, I2 = 67.4%). Additionally, because the majority (60%, 3/5) of the studies included in our meta-analysis involved digestive cancers (gastric cancer and ESCC), we then performed a separate analysis for digestive cancers. Interestingly, the rs920778 variant exhibited a significant association with an increased risk of digestive cancers (dominant model: OR = 1.44; 95% CI = 1.31–1.59; P = 0.339 for the heterogeneity test, I2 = 7.6%; Table 2).
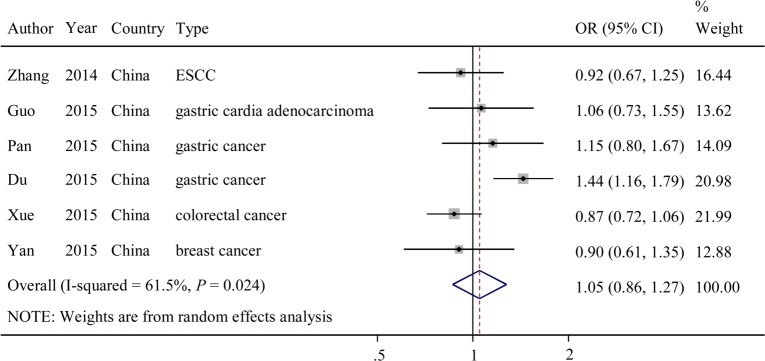
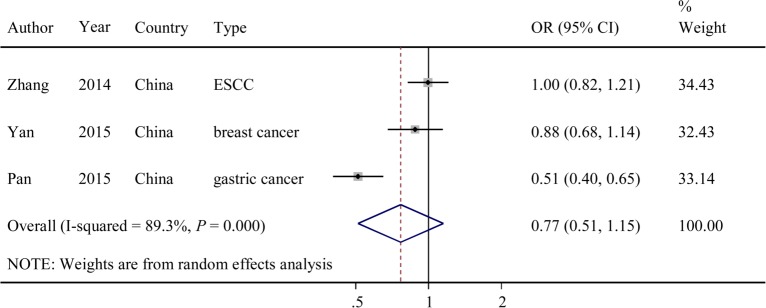
The evaluations of the associations between the other 2 SNPs (rs4759314 and rs1899663) and cancer risk are presented in Figs 3 and 4. Overall, the G variant allele of rs4759314 exhibited no significant association with cancer risk (dominant model: OR = 1.05; 95% CI = 0.86–1.27, P = 0.024 for the heterogeneity test, I2 = 61.5%). The T variant allele of rs1899663 also exhibited no significant association with cancer risk (dominant model: OR = 0.77; 95% CI = 0.51–1.15, P = 0.000 for the heterogeneity test, I2 = 89.3%).
Fig 3. Forest plot of OR with 95%CI for the HOTAIR rs4759314 with cancer risk under dominant model.
Fig 4. Forest plot of OR with 95%CI for the HOTAIR rs1899663 with cancer risk under dominant model.
Test of heterogeneity
For rs920778, significant heterogeneity was observed after the data were pooled (dominant model: P for heterogeneity = 0.005, I2 = 73.2%). (Table 2) When the subjects were stratified based on ethnicity, the heterogeneity obviously disappeared in the Asians (dominant model: P for heterogeneity = 0.953, I2 = 0.0%); however, heterogeneity was still present among the Caucasians (dominant model: P for heterogeneity = 0.080, I2 = 67.4%), which might have been due to the genetic heterogeneity between the different included ethnicities. Additionally, in stratified analyses based on cancer types, the heterogeneity was significantly reduced for the digestive cancers (dominant model: P for heterogeneity = 0.339, I2 = 7.6%).
Sensitivity analysis
To test the stability of the rs920778 results, we performed sensitivity analyses by sequentially removing each eligible study (S6 Table). The study by Bayram et al. that focused on breast cancer was the major contributor of heterogeneity in the dominant model (I2 = 73.2%, P for heterogeneity = 0.005). After the removal of this study, the heterogeneity was significantly reduced (I2 = 0.0%, P for heterogeneity = 0.536). As expected, similar results were observed in the other genetic models (i.e., CT versus CC and TT versus CC) and indicated that the study by Bayram et al. that focused on breast cancer markedly changed the pooled OR.
Publication bias
We used funnel plots and Begg’s test to evaluate potential publication biases of the studied literature. As illustrated in S1–S3 Figs, the shapes of the funnel plots were symmetrical, and a Begg’s test provided further statistical evidence for the absence of publication bias (P = 0.14 for rs920778, P = 0.85 for rs4759314, and P = 0.60 for rs1899663).
Discussion
Recently, an increasing number of studies have investigated the associations of different HOTAIR expression levels with cancer survival, but fewer studies have focused specific attention on the relationships of HOTAIR polymorphisms with cancer susceptibility and survival. Nevertheless, the relationships of HOTAIR polymorphisms with cancer sensitivities have attracted much interest; however, the results are controversial. In the present study, we performed a meta-analysis by pooling 8 studies with totals of 7,151 cases and 8,740 controls and demonstrated that the T allele of rs920778 was associated with a significant increased risk of digestive cancers. However, the rs4759314 and rs1899663 variant alleles exhibited no significant associations with cancer risk.
The rs920778 SNP at 12q13.13 is located in intron 2 of HOTAIR. Based on public datasets and tools (see Methods; S7 Table), we performed further functional annotations of the marker SNP rs920778 and the SNPs that are in strong LD with rs920778 (r2> 0.8). Among the 11 SNPs that were found to be in strong LD with rs920778, significant genotype-specific effects of 5 SNPs (i.e., rs10783618, rs11170775, rs4759059, rs4237809 and rs2366150) on the mRNA expression of HOTAIR were observed (eQTL analysis). Subsequently, we explored the possible mechanisms by which these SNPs modulate the expression of HOTAIR. According to the ChIP-Seq data from the ENCODE database (http://genome.ucsc.edu/ENCODE/), a total of 10 SNPs were located in motifs that may affect the binding activities of numerous transcription factors, including EZH2, CTBP2, CHD1, ZNF143, SUZ12, TCF7L2, CTCF, RAD21, and YY1. The transcription factors mentioned above may be intimately connected with the occurrence and progression of many types of tumours in human. For example, enhancer of zeste homologue 2 (EZH2) is over-expressed in several human tumours and accounts for the aggressiveness and unfavourable prognoses of various tumours [22,23]. Furthermore, the suppressor of zeste-12 protein (SUZ12) is of great importance in the tumourigenesis of several human cancers [24,25] and is involved in the progression of non-small cell lung cancer via its role in promoting cell proliferation and metastasis [26]. Additionally, as predicted by a miRNA-binding analysis website (http://www.bioguo.org/miRNASNP2/), the rs7958904 SNP may affect the binding activity of hsa-miR-615. This miRNA is a newly identified tumour suppressor that can regulate the proliferation, migration, invasion, and apoptosis of various types of cancer cells [27,28]. Thus, these functional annotations suggest that the above-mentioned SNPs (including the marker SNP rs920778 and the tagged SNPs) might separately or jointly influence the aberrant activities of certain transcription factors or miRNAs and further affect the occurrence and progression of certain tumours through different mechanisms. However, experimental evidence validating this hypothesis is limited, and future functional studies are needed to clarify the possible mechanisms.
In this meta-analysis, we did not identify a significant relationship of the rs920778 SNP with cancer risk. However, according to the analyses stratified by population ethnicity, the rs920778 SNP was significantly associated with an increased risk of cancer in Asians (Chinese). In contrast, we found that the rs920778 polymorphism exhibited the opposite association with the risk of cancer in Caucasians (Turkish). The possible reasons for the different results between Asians and Caucasians are as follows. First, the difference may have resulted from differences in the genetic backgrounds of the studied populations. For example, based on the HapMap data (International HapMap Project), the allele frequencies of the rs920778 SNP are different between Asian and Caucasian populations. Second, the difference may owe to the utilization of different genotyping methods, which included PCR-RFLP, CRS-RFLP, TaqMan Real-Time PCR, etc. Third, compared with Asian populations, the sample sizes of the Caucasian populations might not have been sufficiently large to reach a convincing conclusion regarding the association of the rs920778 SNP with cancer risk. Additionally, the different types of cancers involved and random errors may also be potential reasons for the differences in the findings between Asians and Caucasians.
In 2014, Deng et al. conducted a meta-analysis that revealed that HOTAIR abundance might serve as a novel predictive factor for poor cancer prognoses [9]. The results of this study and our results both demonstrated significant relationships between HOTAIR and cancer. The findings of our study provided evidence that HOTAIR polymorphisms might modify cancer susceptibility. Unfortunately, in their meta-analysis of the prognostic value of HOTAIR in cancer, Deng et al. did not explore the associations of HOTAIR polymorphisms with cancer prognoses, and the relevant research on HOTAIR polymorphisms and cancer prognoses is limited. However, based on both findings and functional annotation results, we speculate that HOTAIR polymorphisms might regulate the expression of HOTAIR and further affect cancer development and progression via certain underlining mechanisms, which may open new avenues for the prevention and treatment of cancer.
The strength of this meta-analysis is that we systemically reviewed the relationships between HOTAIR polymorphisms and tumour susceptibility for the first time, and we identified different associations of the rs920778 SNP with cancer risk in different ethnic populations. Additionally, the well-designed functional annotations that further verified our findings are another strength of this study. However, there are also some limitations that need to be addressed. First, significant heterogeneity between studies was observed. Among the 8 published studies included in our meta-analysis, some of the studies were population-based, while others were hospital-based. Second, we did not search Chinese databases, which will result in publication bias. Third, in some of the studies, detailed information (e.g., age, gender, smoking status, and alcohol consumption) was not provided, which limited further stratification analyses. Additionally, if we had been able to acquire more detailed information, we would have achieved more precise estimations by adjusting for other potential covariates. Finally, for this study, we collected only 8 reports. The majority of the subjects were Asians, and the number of Caucasian subjects included in this study was relatively small. Thus, well-conducted large-sample studies are needed to further explore the cancer risks associated with HOTAIR SNPs, especially in Caucasians.
Conclusions
This meta-analysis provided evidence that HOTAIR rs920778 may modify the susceptibility to certain cancer types. Further studies incorporating subjects with different ethnic backgrounds combined with re-sequencing of the marked region and functional evaluations are warranted.
Supporting Information
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the National Natural Science Foundation of China (81502876), the Natural Science Research of Jiangsu Higher Education Institutions (15KJB330006), the Science and Technology Program of Nantong City (MS22015088) and the Doctoral Scientific Research Foundation of Nantong University (14R16). The funding sources had no role to play in the study design, the collection and interpretation of the data, writing of the report, or decision to submit this paper for publication.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136: E359–386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Guttman M, Rinn JL (2012) Modular regulatory principles of large non-coding RNAs. Nature 482: 339–346. 10.1038/nature10887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sang H, Liu H, Xiong P, Zhu M (2015) Long non-coding RNA functions in lung cancer. Tumour Biol 36: 4027–4037. 10.1007/s13277-015-3449-4 [DOI] [PubMed] [Google Scholar]
- 4.Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, et al. (2015) Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut. [DOI] [PubMed] [Google Scholar]
- 5.Angrand PO, Vennin C, Le Bourhis X, Adriaenssens E (2015) The role of long non-coding RNAs in genome formatting and expression. Front Genet 6: 165 10.3389/fgene.2015.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajjari M, Salavaty A (2015) HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med 12: 1–9. 10.7497/j.issn.2095-3941.2015.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, et al. (2015) Long non-coding RNA HOTAIR promotes tumor cell invasion and metastasis by recruiting EZH2 and repressing E-cadherin in oral squamous cell carcinoma. Int J Oncol 46: 2586–2594. 10.3892/ijo.2015.2976 [DOI] [PubMed] [Google Scholar]
- 8.Wu Y, Zhang L, Zhang L, Wang Y, Li H, Ren X, et al. (2014) Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol 35: 9531–9538. 10.1007/s13277-014-2523-7 [DOI] [PubMed] [Google Scholar]
- 9.Deng Q, Sun H, He B, Pan Y, Gao T, Chen J, et al. (2014) Prognostic value of long non-coding RNA HOTAIR in various cancers. PLoS One 9: e110059 10.1371/journal.pone.0110059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Wang Z (2015) Prognostic value of long noncoding RNA HOTAIR in digestive system malignancies. J Gastroenterol Hepatol 30: 1123–1133. 10.1111/jgh.12940 [DOI] [PubMed] [Google Scholar]
- 11.Li J, Wen W, Zhao S, Wang J, Chen J, Wang Y, et al. (2015) Prognostic role of HOTAIR in four estrogen-dependent malignant tumors: a meta-analysis. Onco Targets Ther 8: 1471–1482. 10.2147/OTT.S84687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma G, Wang Q, Lv C, Qiang F, Hua Q, Chu H, et al. (2015) The prognostic significance of HOTAIR for predicting clinical outcome in patients with digestive system tumors. J Cancer Res Clin Oncol 141: 2139–2145. 10.1007/s00432-015-1980-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai B, Wu Z, Liao K, Zhang S (2014) Long noncoding RNA HOTAIR can serve as a common molecular marker for lymph node metastasis: a meta-analysis. Tumour Biol 35: 8445–8450. 10.1007/s13277-014-2311-4 [DOI] [PubMed] [Google Scholar]
- 14.Bayram S, Sumbul AT, Batmaci CY, Genc A (2015) Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol 36: 3863–3870. 10.1007/s13277-014-3028-0 [DOI] [PubMed] [Google Scholar]
- 15.Guo W, Dong Z, Bai Y, Guo Y, Shen S, Kuang G, et al. (2015) Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour Biol 36: 2845–2854. 10.1007/s13277-014-2912-y [DOI] [PubMed] [Google Scholar]
- 16.Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, et al. (2015) A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog 55: 90–96. 10.1002/mc.22261 [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wen J, et al. (2014) The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis 35: 2062–2067. 10.1093/carcin/bgu103 [DOI] [PubMed] [Google Scholar]
- 18.Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, et al. (2015) The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget 6: 31255–31262. 10.18632/oncotarget.5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayram S, Ulger Y, Sumbul AT, Kaya BY, Rencuzogullari A, Genç A, et al. (2015) A functional HOTAIR rs920778 polymorphism does not contributes to gastric cancer in a Turkish population: a case-control study. Fam Cancer 14: 561–567. 10.1007/s10689-015-9813-0 [DOI] [PubMed] [Google Scholar]
- 20.Xue Y, Gu D, Ma G, Zhu L, Hua Q, Chu H, et al. (2015) Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis 30: 303–310. 10.1093/mutage/geu076 [DOI] [PubMed] [Google Scholar]
- 21.Yan R, Cao J, Song C, Chen Y, Wu Z, Wang K, et al. (2015) Polymorphisms in lncRNA HOTAIR and susceptibility to breast cancer in a Chinese population. Cancer Epidemiol 39: 978–985. 10.1016/j.canep.2015.10.025 [DOI] [PubMed] [Google Scholar]
- 22.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, et al. (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629. [DOI] [PubMed] [Google Scholar]
- 23.Arisan S, Buyuktuncer ED, Palavan-Unsal N, Caskurlu T, Cakir OO, Ergenekon E (2005) Increased expression of EZH2, a polycomb group protein, in bladder carcinoma. Urol Int 75: 252–257. [DOI] [PubMed] [Google Scholar]
- 24.Xia R, Jin FY, Lu K, Wan L, Xie M, Xu TP, et al. (2015) SUZ12 promotes gastric cancer cell proliferation and metastasis by regulating KLF2 and E-cadherin. Tumour Biol 36: 5341–5351. 10.1007/s13277-015-3195-7 [DOI] [PubMed] [Google Scholar]
- 25.Li H, Cai Q, Wu H, Vathipadiekal V, Dobbin ZC, Li T, et al. (2012) SUZ12 promotes human epithelial ovarian cancer by suppressing apoptosis via silencing HRK. Mol Cancer Res 10: 1462–1472. 10.1158/1541-7786.MCR-12-0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Shi X, Wang L, Wu Y, Jin F, Bai C, et al. (2014) SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol 35: 6073–6082. 10.1007/s13277-014-1804-5 [DOI] [PubMed] [Google Scholar]
- 27.Bai Y, Li J, Liu Y, Zhang B (2015) MiR-615 inhibited cell proliferation and cell cycle of human breast cancer cells by suppressing of AKT2 expression. Int J Clin Exp Med 8: 3801–3808. [PMC free article] [PubMed] [Google Scholar]
- 28.Gao W, Gu Y, Li Z, Cai H, Peng Q, Tu M, et al. (2015) miR-615-5p is epigenetically inactivated and functions as a tumor suppressor in pancreatic ductal adenocarcinoma. Oncogene 34: 1629–1640. 10.1038/onc.2014.101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.