Abstract
Identifying homology between sex chromosomes of different species is essential to understanding the evolution of sex determination. Here, we show that the identity of a homomorphic sex chromosome pair can be established using a linkage map, without information on offspring sex. By comparing sex-specific maps of the European tree frog Hyla arborea, we find that the sex chromosome (linkage group 1) shows a threefold difference in marker number between the male and female maps. In contrast, the number of markers on each autosome is similar between the two maps. We also find strongly conserved synteny between H. arborea and Xenopus tropicalis across 200 million years of evolution, suggesting that the rate of chromosomal rearrangement in anurans is low. Finally, we show that recombination in males is greatly reduced at the centers of large chromosomes, consistent with previous cytogenetic findings. Our research shows the importance of high-density linkage maps for studies of recombination, chromosomal rearrangement and the genetic architecture of ecologically or economically important traits.
Introduction
One of the central questions in sex chromosome research is whether the sex chromosomes found in different species are homologous (Ohno, 1967; Miura, 2007; Graves and Peichel, 2010; Pease and Hahn, 2012; O'Meally et al., 2012). Despite recent progress (see, for example, Gamble et al., 2014; Gamble and Zarkower, 2014; Chen et al., 2014; Nanda et al., 2014; Srikulnath et al., 2014), identifying the homomorphic sex chromosomes found in many cold-blooded vertebrates, invertebrates and some plants remains a major challenge. Researchers are currently interested in the factors that maintain homomorphy in these sex chromosome systems. Two key hypotheses have been put forward: either sex chromosome turnovers occur too frequently to allow substantial degeneration of the nonrecombining Y (or W) chromosome, or a low level of recombination is maintained between the X and Y (or Z and W) chromosomes in these systems (Perrin, 2009; Beukeboom and Perrin, 2014). Distinguishing between these two hypotheses requires us to quantify rates of sex chromosome turnover across the phylogeny, and this can only be done by identifying the sex chromosomes in a large number of species. Here, we propose a novel approach to identify sex chromosomes through high-density linkage mapping. Importantly, the siblings used to prepare this linkage map do not need to be sexed.
Shortly after the onset of reduced recombination between a proto-X and proto-Y chromosome (Bachtrog, 2013), we expect divergence in allele frequencies between the two gametologs. In this case, an X and Y chromosome sampled from a population are expected to have more differences between them than two X chromosomes. In a full-sib mapping cross, more markers will be heterozygous and therefore map informative in the XY male parent than in the XX female. In contrast, the number of heterozygous markers on each autosome is expected to be similar between the two parents. In a comparison of the two sex-specific maps, if one linkage group has a strong difference in marker number between the male and female maps, that linkage group is likely to correspond to the sex chromosome.
We demonstrate this method in the European tree frog Hyla arborea. Previous work on this species has shown that its sex determination system is at least 5 million years old, but that occasional X–Y recombination prevents degeneration of the Y chromosome (Stöck et al., 2011). The sex chromosome is largely homologous to chromosome 1 in Xenopus tropicalis, and contains the candidate sex determination gene Dmrt1 (Brelsford et al., 2013). Males and females differ strongly in recombination rate (Berset-Brändli et al., 2008; Dufresnes et al., 2014a, b), but X–Y recombination is higher and X–Y differentiation is lower in Balkan glacial refugia than in recently colonized northwestern Europe (Dufresnes et al., 2014b). In this study, we validate our method for identifying sex chromosomes by showing that only linkage group 1, homologous to X. tropicalis chromosome 1, has a strongly different number of markers between paternal and maternal linkage maps. This approach also enables us to localize sex differences in recombination to specific parts of the genome, and extends to the entire genome our previous work on conserved synteny in anurans that until now was largely limited to a single chromosome.
Materials and methods
Samples
We constructed a linkage map based on 90 offspring from a single H. arborea cross collected in Krk, Croatia (family K10 in Dufresnes et al., 2014b). DNA was collected from both parents using buccal swabs. Fertilized eggs were kept until shortly after hatching, when tadpoles were preserved in ethanol. DNA from parents and offspring was extracted using a BioSprint robotic workstation (Qiagen, Venlo, Netherlands).
Genotyping by sequencing
We collected genotyping-by-sequencing data using a novel molecular protocol adapted from Parchman et al. (2012), Peterson et al. (2012) and Purcell et al. (2014). The complete protocol is available in Supplementary Materials. Briefly, we digested DNA with restriction enzymes SbfI and MseI, ligated barcoded adapters, amplified each individual sample in four separate PCR reactions, pooled all PCR products and selected fragments between 400 and 500 bp using agarose gel. To increase sequence depth for the two parental DNA samples, PCR reactions for these samples were run at triple the volume of offspring samples. The library of 92 samples was sequenced on a single Illumina (San Diego, CA, USA) HiSeq 2000 lane by the Lausanne Genetic Technology Facility (Lausanne, Switzerland), with single-end 100 bp reads.
Genotype calling and filtering
We demultiplexed the raw reads using the process_radtags module of Stacks (Catchen et al., 2013), removing low-quality reads, reads with uncalled bases and reads that failed the Illumina ‘chastity' filter. Adapters were trimmed from demultiplexed reads using a custom shell script. Reads were then aligned to the H. arborea low-coverage draft genome (Dryad, doi: 10.5061/dryad.n856c) using Bowtie2 (Langmead and Salzberg, 2012). Variants were called using the mpileup command of Samtools (Li et al., 2009), using only reads with mapping quality of at least 20. We used VCFtools (Danecek et al., 2011) to filter the resulting raw variants. First, genotypes with a quality score <20 were converted to missing data. Loci with >20% missing data, offspring heterozygosity >0.8 or a minor allele frequency <0.15 were removed.
We then generated separate lists of paternal-informative and maternal-informative markers, excluding loci that were heterozygous in both parents or in neither parent. For example, a male-informative marker with two alleles, A and G, could have genotypes AG in the male and GG in the female, with offspring expected to show the AG and GG genotypes in equal numbers. A female-informative marker could have genotypes GG in the male and AG in the female, offspring showing the same pattern as in the previous example. A marker with AG genotypes in both parents would be excluded, as in heterozygous offspring we would be unable to determine which parent provided the A allele and which provided the G allele. Because the H. arborea reference genome is highly fragmented (N50, 1.2 kbp), we retained only the marker with the lowest proportion of missing data from each genome scaffold for each of the two sex-specific data sets, assuming no recombination within the short genome fragments. Finally, we searched for and corrected Mendelian segregation errors. For example, if 45 offspring were homozygous for the C allele at a locus, 44 heterozygous for C and T and one homozygous for T, the rare homozygous genotype was corrected to a heterozygous call. Markers with >5% of offspring having the homozygous genotype unexpected based on parent genotypes were removed. For markers with unexpected homozygous genotypes at a frequency <5%, these were corrected to heterozygous genotypes. Genotypes were then exported in 012 format using VCFtools.
Linkage mapping
We constructed sex-specific genetic maps using MSTmap (Wu et al., 2008), using cross type ‘DH.' Following Gadau et al. (2001), to account for the unknown phase of markers in each parent we added each marker to the data set twice, once with each possible phase. Each linkage group then appeared in the output twice with opposite marker phases; duplicate linkage groups were removed manually.
Assessment of conserved synteny with Xenopus tropicalis
We searched the X. tropicalis genome sequence (available at ftp://ftp.xenbase.org/pub/Genomics/JGI/Xentr7.1/) for each H. arborea scaffold that contained a marker on the linkage map using blastn. Blast hits were retained only if the e-value of the best hit was five orders of magnitude better than that of the second hit (Purcell et al., 2014). Homology between the X. tropicalis genome and H. arborea linkage map was visualized using an Oxford grid plot.
Results
Genotyping by sequencing
We obtained 153 million demultiplexed, quality-filtered Illumina reads, with number of reads per individual ranging from 1.1 to 1.9 M in offspring, and 5.8 and 6.5 M in the parents. Approximately 21% of these reads mapped uniquely to the H. arborea genome draft; this low rate is likely because of the incompleteness of the draft genome assembly (1.2 Gbp of an expected 5 Gbp). After genotype calling and filtering, we retained 1608 male-informative and 1461 female-informative single-nucleotide polymorphisms or small indel markers, with an average sequence depth of 16.5.
Linkage mapping
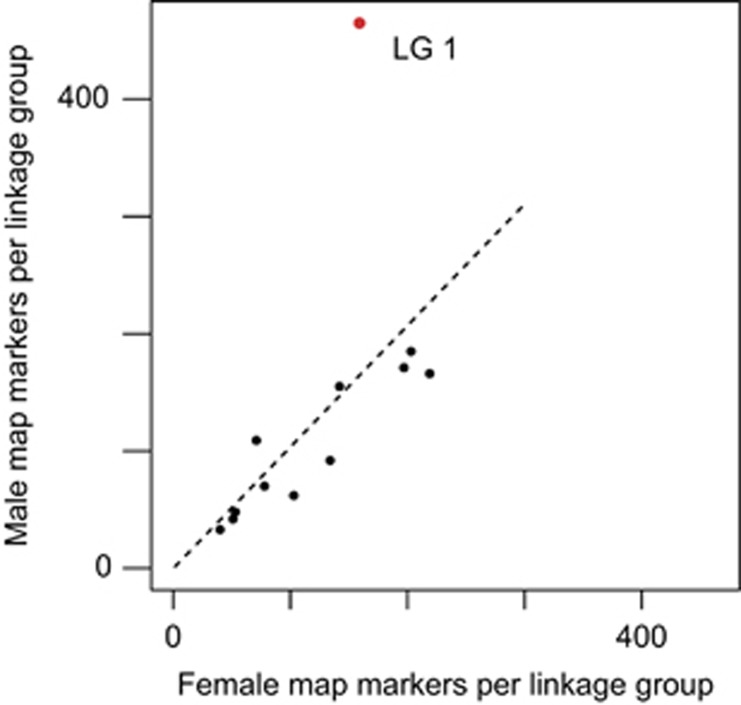
We recovered 12 linkage groups for both sex-specific maps, consistent with the species karyotype (both maps available in Supplementary Materials). The number of informative markers per linkage group was similar between male and female maps for 11 of the 12 linkage groups. In contrast, linkage group 1, the sex chromosome pair, contained nearly three times as many male-informative as female-informative markers (Figure 1).
Figure 1.
Linkage group 1, the sex chromosome in H. arborea, contains nearly three times as many informative markers on the male-specific map compared with the female-specific map. The 11 autosomal linkage groups fall close to the dashed line, illustrating equal marker numbers between the sexes.
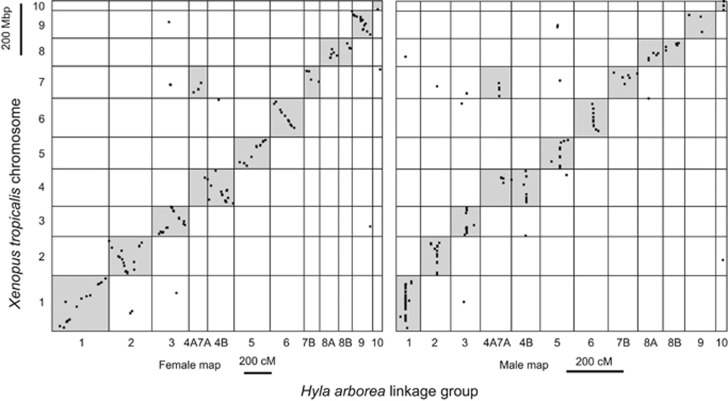
A subset (10.7%) of mapped markers could be assigned positions in the X. tropicalis genome sequence (Table 1). These markers revealed strongly conserved synteny between Hyla and Xenopus, with most Hyla linkage groups matching a single Xenopus chromosome (Figure 2). Because no data are available to associate linkage groups with the Hyla karyotype, we named the Hyla linkage groups based on their homologous chromosomes in X. tropicalis. In several cases, even the ordering of markers along the chromosome is largely conserved across the entire chromosome (for example, Hyla linkage groups 5, 6 and 9) or large chromosomal regions (for example, Hyla linkage groups 2 and 3). Marker order is conserved on the linkage group 1, with the exception of a small region. Brelsford et al. (2013) noted that a region of X. tropicalis chromosome 1 corresponding to avian chromosome 26 was not found on the same chromosome in H. arborea; in this study, we map that rearranged region to linkage group 2.
Table 1. Number of mapped markers, and number available for comparisons between sex-specific Hyla arborea maps and between H. arborea and Xenopus tropicalis.
| Male map | Female map | |
|---|---|---|
| Total markers genotyped | 1608 | 1461 |
| Markers successfully mapped | 1598 | 1450 |
| Markers present on both maps | 96 | 96 |
| Markers aligning to X. tropicalis | 173 | 152 |
Figure 2.
Oxford grid comparison of H. arborea linkage map and X. tropicalis genome shows blocks of conserved synteny, often extending across entire chromosomes. Marker order is largely conserved on linkage group 1 that is sex linked in several frog species but autosomal in X. tropicalis.
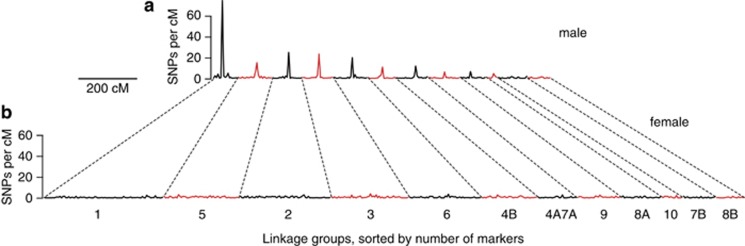
Most male linkage groups contained a cluster of markers near the center of the linkage group with an identical segregation pattern, suggesting a large region of suppressed recombination (Figure 3a). This pattern was most pronounced in the larger linkage groups. In contrast, the number of markers per cM was close to uniform across the female map (Figure 3b).
Figure 3.
Marker density is very high at the centers of male linkage groups (a), and approximately uniform across female linkage groups (b). Linkage groups containing more markers also show a larger difference in map length between the male and female maps. Taken together, this suggests that recombination in males occurs only near the ends of large chromosomes.
Discussion
Linkage maps provide an opportunity to investigate recombination and serve as a resource for research on genome structure, dynamics and rearrangements. Our female- and male-specific maps respectively contain 1450 and 1598 markers, with an average intermarker distance of 1.65 and 0.72 cM. This marker density compares favorably with other published amphibian maps; to our knowledge, only the model species X. tropicalis has a higher marker density (2886 markers, intermarker distance 0.38 cM; Wells et al., 2011). Availability of a high-density linkage map will enable future research on how population genetic patterns vary across different regions of the genome, essential for understanding the genomics of sex chromosome evolution and speciation. Immediately, it also demonstrates the potential of linkage mapping for identification of sex chromosomes without knowledge of offspring sexes. The sex-specific linkage maps we present here show that the previously documented absence of recombination in males is restricted to the central parts of the chromosome. In contrast, chromosome ends have similar rates of recombination between the two sexes. In addition, we identify a high level of synteny between the whole genome of Hyla and the distantly related model frog species X. tropicalis.
A new method for identifying sex chromosomes
Using high-density linkage maps, we demonstrate that we can identify the sex chromosome from families with sex-unknown offspring. When paternal and maternal chromosomes are drawn from the same population, the number of heterozygous single-nucleotide polymorphisms per chromosome pair is expected to be equal for the father and mother. If the X and Y chromosomes show significant genetic differentiation, heterozygosity will be higher in the male than the female for that chromosome only. By plotting male-informative markers against female-informative markers for each of 12 linkage groups, we show that linkage group 1 is an outlier with nearly three times as many male-information as female-informative markers, consistent with previous studies (Berset-Brändli et al., 2008; Brelsford et al., 2013) identifying this as the sex chromosome.
Our approach will serve as a powerful tool to identify the sex chromosome of species with large numbers of offspring available, but for which juvenile sex identification is difficult or impossible. It does, however, have several limitations. First, it requires significant differentiation in allele frequencies across a large region of the sex chromosome. The important parameter for detecting the sex chromosome is the ratio of differentiation between the X and Y chromosomes (DXY) to polymorphism on the X (πX). In the parents of this cross, our estimated DXY/πX is 2.9. Among the 11 autosomal linkage groups, LG 9 has the largest deviation from equal marker number between the sexes, with 1.7 × more female-informative than male-informative markers. The method's power will be highly dependent on polymorphism and linkage disequilibrium in the parental source population as well as the level of differentiation between the X and Y. However, to the extent that other species are similar to this H. arborea population, we suggest that a DXY/πX ratio of >2 will be necessary to detect the sex chromosome. Second, it requires identifying X-linked and Y-linked markers as different alleles of the same locus, and not different loci. If a Y chromosome is missing large regions that are present on the X, or if sequence divergence is too high to recognize orthologous X and Y sequence reads, the number of male-informative markers on the sex chromosome will decline, reducing the method's power. Finally, a large autosomal supergene with two differentiated haplogroups (see, for example, Purcell et al., 2014) could produce a false positive signal, if one parent of the mapping cross is heterozygous for the inversion and the other is homozygous. Further testing of our method will be necessary to determine how often its requirements are met in other species.
Synteny between Hyla and Xenopus
Chromosomes are largely conserved between H. arborea and X. tropicalis, despite divergence time estimates ranging from 165 to 305 million years (Aris-Brosou and Yang, 2002; Wiens, 2007). A previous study demonstrated conserved synteny across H. arborea, Rana temporaria, Bufo siculus and X. tropicalis at chromosome 1 (Brelsford et al., 2013), with one small segment of the X. tropicalis chromosome found on a different chromosome in H. arborea. In H. arborea, Dufresnes et al. (2014a) inferred two other linkage groups that each contained two markers mapped to the same X. tropicalis chromosome (chromosomes 2 and 8), hinting at conserved synteny in at least two other chromosomes. More recently, Sun et al. (2015) reported conservation of large synteny blocks between X. tropicalis and the Tibetan frog Nanorana parkeri, although the Nanorana genome assembly did not allow comparison of entire chromosomes. We extend these previously published results by assigning each Hyla linkage group to one Xenopus chromosome (or in the case of LG 4A7A, two chromosomes), and by identifying large blocks of conserved marker order on several chromosomes (Figure 2a). Marker order is similar in four of the 12 Hyla chromosomes (1, 5, 6 and 9), and a single large-scale inversion is apparent on two others (2 and 3). Of the 10 X. tropicalis chromosomes, three (4, 7 and 8) are split in two, with 4A and 7A reunited into a new Hyla chromosome.
A similar level of synteny has been observed in birds based on comparison of chicken and zebra finch (Stapley et al., 2008), representing close to 100 Myr of divergence. In contrast, some systems have substantial levels of interchromosomal rearrangement even with much shorter divergence times, as in ants (Purcell et al., 2014). Interestingly, the Z chromosome in birds has undergone more intrachromosomal rearrangement than autosomes (Griffin et al., 2006; Stapley et al., 2008). The X. tropicalis chromosome 1 is partially homologous to the avian Z and is recurrently used as the sex chromosome in multiple frog species (Brelsford et al., 2013). Thus, it is surprising to find little structural variation in chromosome 1 between Xenopus and Hyla.
Differences in male and female recombination rate
Heterochiasmy, or difference between sexes in recombination rate, is a well-documented phenomenon in many taxa. Although Haldane (1922) and Huxley (1928) noted that recombination tends to be lower in the heterogametic sex, many exceptions have been observed and comprehensive explanations for this phenomenon are still lacking (Mank, 2009). In frogs, previous low-density linkage maps documented a strong asymmetry in recombination between males and females, with males having greatly reduced recombination (see, for example, Nishioka and Sumida, 1994; Sumida and Nishioka, 1994; Berset-Brändli et al., 2008; Cano et al., 2011; Rodrigues et al., 2013). Our high-density linkage map allows us to localize the recombination events along each linkage group. The differences in map length between the male and female are particularly striking. In females, the map length is roughly proportional to the marker number, suggesting that recombination rate scales linearly with chromosome length. On the other hand, the male map lengths of all linkage groups are close to 100 cM, implying approximately one crossover per chromosome regardless of the physical size of the chromosome. It should be noted that these calculations are based on single-nucleotide polymorphism density per cM. This measure is expected to be only a rough proxy for the physical length per cM, as genotyping-by-sequencing markers and heterozygosity may not be uniformly distributed across the genome. Our finding that male recombination is restricted to chromosome ends, whereas female recombination is relatively constant across the full length of each linkage group, suggests that chromosome ends experience more recombination and genes in these regions may evolve differently (Cutter and Payseur, 2013). Our localization of heterochiasmy is fully consistent with previous cytogenetic findings. In female frogs, chiasmata have been shown to occur in proportion to chromosome size (being more frequent in large bivalents than in small ones) and uniformly along the chromosomes. Male frogs, in contrast, typically show two chiasmata per meiosis (independent of bivalent size), and always localized in terminal regions, conferring a characteristic ring shape to chromosomes during diakinesis (see, for example, Morescalchi and Galgano, 1973).
Most frogs with known systems of sex determination are male heterogametic. In Hyla, Bufo and Rana, recombination is reduced in males (Berset-Brändli et al., 2008; Stöck et al., 2013; Rodrigues et al., 2013), consistent with the Haldane–Huxley rule. However, we still do not know whether one of these patterns is the cause of the other. Insights into the basis of these patterns will likely emerge from exceptional systems. For instance, investigating recombination rates in frogs with ZW sex determination will help us to disentangle the causal relationships between sex determination and heterochiasmy.
Conclusions
Overall, we have used our high-density linkage maps to achieve three goals. First, we have evaluated a novel method to infer the sex chromosome from the comparison of sex-specific maps. Second, we have demonstrated conserved synteny between the whole genomes of Hyla and Xenopus. Finally, we have identified the relative positions and frequency of recombination events in male and female H. arborea. These maps will also serve as vital tools for future population genomic work.
Data archiving
Demultiplexed genotyping-by-sequencing reads and whole-genome reads are available on NCBI Sequence Read Archive (BioProjects PRJNA292823 and PRJNA292815). The draft low-coverage genome assembly is available on Dryad (DOI 10.5061/dryad.n856c).
Acknowledgments
We thank R Sermier for laboratory assistance. J Purcell suggested improvements to the manuscript. The computations were performed at the Vital-IT (http://www.vital-it.ch) Center for high-performance computing of the SIB Swiss Institute of Bioinformatics. Funding was provided by the Swiss National Science Foundation (Grant 31003A_129894 to NP) and by the University of Lausanne (PhD fellowship from the Faculty of Biology and Medicine, grants from the Fondation Agassiz and the Fondation du 450e to CD).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Aris-Brosou S, Yang Z. (2002). Effects of models of rate evolution on estimation of divergence dates with special reference to the metazoan 18S ribosomal RNA phylogeny. Syst Biol 51: 703–714. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. (2013). Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berset-Brändli L, Jaquiéry J, Broquet T, Ulrich Y, Perrin N. (2008). Extreme heterochiasmy and nascent sex chromosomes in European tree frogs. Proc R Soc B 275: 1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beukeboom LW, Perrin N. (2014) The Evolution of Sex Determination. Oxford University Press: Oxford. [Google Scholar]
- Brelsford A, Stöck M, Betto-Colliard C, Dubey S, Dufresnes C, Jourdan-Pineau H et al. (2013). Homologous sex chromosomes in three deeply divergent anuran species. Evolution 67: 2434–2440. [DOI] [PubMed] [Google Scholar]
- Cano JM, Li M-H, Laurila A, Vilkki J, Merilä J. (2011). First-generation linkage map for the common frog Rana temporaria reveals sex-linkage group. Heredity 107: 530–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. (2013). Stacks: an analysis tool set for population genomics. Mol Ecol 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang G, Shao C, Huang Q, Liu G, Zhang P et al. (2014). Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat Genet 46: 253–260. [DOI] [PubMed] [Google Scholar]
- Cutter AD, Payseur B. (2013). Genomic signatures of selection at linked sites: unifying the disparity among species. Nat Rev Genet 14: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA et al. (2011). The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresnes C, Brelsford A, Perrin N. (2014. a). First-generation linkage map for the European tree frog (Hyla arborea with utility in congeneric species. BMC Res Notes 7: 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresnes C, Bertholet Y, Wassef J, Ghali K, Savary R, Pasteur B et al. (2014. b). Sex-chromosome differentiation parallels postglacial range expansion in European tree frogs (Hyla arborea. Evolution 68: 3445–3456. [DOI] [PubMed] [Google Scholar]
- Gadau J, Gerloff CU, Krüger N, Chan H, Schmid-Hempel P, Wille A et al. (2001). A linkage analysis of sex determination in Bombus terrestris (L.) (Hymenoptera: Apidae). Heredity 87: 234–242. [DOI] [PubMed] [Google Scholar]
- Gamble T, Geneva AJ, Glor RE, Zarkower D. (2014). Anolis sex chromosomes are derived from a single ancestral pair. Evolution 68: 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, Zarkower D. (2014). Identification of sex-specific molecular markers using restriction site associated DNA sequencing. Mol Ecol Resour 14: 902–913. [DOI] [PubMed] [Google Scholar]
- Graves JAM, Peichel CL. (2010). Are homologies in vertebrate sex determination due to shared ancestry or limited options? Genome Biol 11: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM. (2006). The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res 117: 64–77. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. (1922). Sex ratio and unisexual sterility in hybrid animals. J Genet 12: 101–109. [Google Scholar]
- Huxley JS. (1928). Sexual differences of linkage in Gammarus chevreuxi. J Genet 20: 145–156. [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. (2009). The evolution of heterochiasmy: the role of sexual selection and sperm competition in determining sex-specific recombination rates in eutherian mammals. Genet Res 91: 355–363. [DOI] [PubMed] [Google Scholar]
- Miura I. (2007). An evolutionary witness: the frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex Dev 1: 323–331. [DOI] [PubMed] [Google Scholar]
- Morescalchi A, Galgano M. (1973). Meiotic chromosomes and their taxonomic value in Amphibia Anura. Caldasia 11: 41–50. [Google Scholar]
- Nanda I, Schories S, Tripathi N, Dreyer C, Haaf T, Schmid M et al. (2014). Sex chromosome polymorphism in guppies. Chromosoma 123: 373–383. [DOI] [PubMed] [Google Scholar]
- Nishioka M, Sumida M. (1994). The differences in recombination rate between the male and female in Rana nigromaculata and Rana brevipoda. Sci Rep Lab Amphibian Biol Hiroshima Univ 13: 99–136. [Google Scholar]
- Ohno S. (1967) Sex Chromosomes and Sex Linked Genes. Springer-Verlag: New York. [Google Scholar]
- O'Meally D, Ezaz T, Georges A, Sarre SD, Graves JAM. (2012). Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res 20: 7–19. [DOI] [PubMed] [Google Scholar]
- Parchman TL, Gompert Z, Mudge J, Schilkey FD, Benkman CW, Buerkle C. (2012). Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol Ecol 21: 2991–3005. [DOI] [PubMed] [Google Scholar]
- Pease JB, Hahn MW. (2012). Sex chromosomes evolved from independent ancestral linkage groups in winged insects. Mol Biol Evol 29: 1645–1653. [DOI] [PubMed] [Google Scholar]
- Perrin N. (2009). Sex reversal: a fountain of youth for sex chromosomes? Evolution 63: 3043–3049. [DOI] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. (2012). Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7: e37135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell J, Brelsford A, Wurm Y, Perrin N, Chapuisat M. (2014). Convergent genetic architecture underlies social organization in ants. Curr Biol 24: 2728–2732. [DOI] [PubMed] [Google Scholar]
- Rodrigues N, Betto-Colliard C, Jourdan-Pineau H, Perrin N. (2013). Within-population polymorphism of sex-determination systems in the common frog (Rana temporaria. J Evol Biol 26: 1569–1577. [DOI] [PubMed] [Google Scholar]
- Srikulnath K, Matsubara K, Uno Y, Nishida C, Olsson M, Matsuda Y. (2014). Identification of the linkage group of the Z sex chromosomes of the sand lizard (Lacerta agilis, Lacertidae) and elucidation of karyotype evolution in lacertid lizards. Chromosoma 123: 563–575. [DOI] [PubMed] [Google Scholar]
- Stapley J, Birkhead TR, Burke T, Slate J. (2008). A linkage map of the zebra finch Taeniopygia guttata provides new insights into avian genome evolution. Genetics 179: 651–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck M, Horn A, Grossen C, Lindtke D, Sermier R, Betto-Colliard C et al. (2011). Ever-young sex chromosomes in European tree frogs. PLoS Biol 9: e1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöck M, Savary R, Betto-Colliard C, Biollay S, Jourdan-Pineau H, Perrin N. (2013). Low rates of X-Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup). J Evol Biol 26: 674–682. [DOI] [PubMed] [Google Scholar]
- Sumida M, Nishioka M. (1994). A pronounced sex difference when two linked loci of the Japanese brown frog Rana japonica are recombined. Biochem Genet 32: 361–369. [DOI] [PubMed] [Google Scholar]
- Sun YB, Xiong ZJ, Xiang XY, Liu SP, Zhou WW, Tu XL et al. (2015). Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci USA 112: E1257–E1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells DE, Gutierrez L, Xu Z, Krylov V, Macha J, Blankenburg KP et al. (2011). A genetic map of Xenopus tropicalis. Dev Biol 354: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiens JJ. (2007). Global patterns of diversification and species richness in amphibians. Am Nat 170: S86–S106. [DOI] [PubMed] [Google Scholar]
- Wu Y, Bhat PR, Close TJ, Lonardi S. (2008). Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet 4: e1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.