Abstract
Across a diversity of animals, male seminal fluid coagulates upon ejaculation to form a hardened structure known as a copulatory plug. Previous studies suggest that copulatory plugs evolved as a mechanism for males to impede remating by females, but detailed investigations into the time course over which plugs survive in the female's reproductive tract are lacking. Here, we cross males from eight inbred strains to females from two inbred strains of house mice (Mus musculus domesticus). Plug survival was significantly affected by male genotype. Against intuition, plug survival time was negatively correlated with plug size: long-lasting plugs were small and relatively more susceptible to proteolysis. Plug size was associated with divergence in major protein composition of seminal vesicle fluid, suggesting that changes in gene expression may play an important role in plug dynamics. In contrast, we found no correlation to genetic variation in the protein-coding regions of five genes thought to be important in copulatory plug formation (Tgm4, Svs1, Svs2, Svs4 and Svs5). Our study demonstrates a complex relationship between copulatory plug characteristics and survival. We discuss several models to explain unexpected variation in plug phenotypes.
Introduction
Sexual selection is thought to play a central role in driving the rapid evolution of animal reproductive traits (Andersson, 1994; Eberhard, 2009). Diverse aspects of ejaculate composition (volume, sperm count and abundance of accessory proteins) and biochemical function (coagulation and induction of female immune response) can evolve rapidly, especially in species where females mate with multiple males (Pilch and Mann, 2006; Poiani, 2006; Cameron et al, 2007; Robertson, 2007). These patterns suggest that characteristics of the ejaculate mediate outcomes of female choice, sperm competition among males and/or antagonistic conflict between males and females (Chapman, 2001; Birkhead and Pizzari, 2002; Arnqvist and Rowe, 2005).
In many mammals, a large portion of the male's seminal fluid coagulates to form a hardened plug that fills the vaginal–cervical region (Devine, 1975; Martan and Shepherd, 1976; Voss, 1979; Williams-Ashman, 1984; Dixson and Anderson, 2002). A large body of data suggest that plugs evolved to impede remating by females (Mosig and Dewsbury, 1970; Martan and Shepherd, 1976; Hartung and Dewsbury, 1978; Voss, 1979), although additional plug functions may include ejaculate transport through the female's reproductive tract (Blandau, 1945a; Matthews and Adler, 1978; Toner et al, 1987; Carballada and Esponda, 1992; Rogers et al, 2009), stimulation required for proper implantation and pregnancy (Ball, 1934; Dean, 2013) and slow release of sperm (Asdell, 1946).
As remating likely benefits females (Jennions and Petrie, 2000; Zeh and Zeh, 2001; Fedorka and Mousseau, 2002; Slatyer et al, 2012; but see Bilde et al, 2009), the copulatory plug may exist as a source of sexual conflict. Consistent with a hypothesis of sexual conflict (Stockley, 1997), recently mated females upregulate proteases thought to assist in plug degradation (Kelleher and Pennington, 2009; Dean et al, 2011), whereas male seminal fluid is enriched for protease inhibitors (Dean et al, 2009), although proteases and their inhibitors have additional roles in reproduction (Wolfner, 2002; Kawano et al, 2010). Also, plug-forming proteins, proteases and protease inhibitors all tend to evolve rapidly (Dorus et al, 2004; Clark and Swanson, 2005; Kelleher et al, 2007; Lawniczak and Begun, 2007; Ramm et al, 2008; Wong et al, 2008; Dean and Nachman, 2009; Dean et al, 2011) as predicted for genes involved in sexual conflict (Swanson and Vacquier, 2002; Clark et al, 2006). In primates, the evolutionary rate of a key copulatory plug gene, SEMG2, is positively correlated with the inferred intensity of sexual selection (Dorus et al, 2004; Ramm et al, 2009).
In both rodents (Ramm et al, 2005) and primates (Dixson, 1998b), males from species inferred to experience relatively intense sperm competition develop relatively large seminal vesicles compared with their body mass, which has been associated with large copulatory plugs (Ramm et al, 2005). Plugs are more prominent, and molecular studies suggest are more durable, in species that experience relatively intense sperm competition (Dixson and Anderson, 2002; Ramm et al, 2009). Males with relatively larger seminal were more successful under sperm competition (Stockley et al, 2013). In contrast, some primarily monogamous species have lost the ability to make plugs (Dixson, 1998a; Jensen-Seaman and Li, 2003; Kingan et al, 2003; Carnahan and Jensen-Seaman, 2008). Traditionally, these data have suggested that under intense sperm competition, males are selected to make larger, more durable plugs, but such hypotheses remain speculative because we do not know the genetic basis or functional consequences of standing variation in plug phenotypes.
House mice provide a powerful system to investigate the formation, function and evolutionary dynamics of copulatory plugs. Female house mice regularly mate with multiple males while in estrus, creating ample opportunity for sperm competition and sexual conflict in nature (Dean et al, 2006; Firman and Simmons, 2008). Anecdotal accounts suggest that copulatory plugs last ∼24 h after mating (Stockard and Papanicolaou, 1919; Parkes, 1926; Silver, 1995), but it remains unknown if and how this time scale varies. Here we use crosses between eight inbred strains of mice to better understand the genetic contributions to and phenotypic correlates of copulatory plug survival in mice. These experiments represent the first systematic examination of copulatory plug dynamics in mice.
Materials and methods
Study organisms
All husbandry and experimental methods, as well as all personnel involved, were approved by the University of Southern California's Institute for Animal Care and Use Committee, protocols 11394 and 11777. Males were derived from eight genetically distinct strains of mice. Six of these—BIK, DCA, DGA, DIK, DJO and DOT—were originally founded from natural populations—Kefar Galim Israel, Akrotiri Cyprus, Ajdarie Georgia, Keshet Israel, Orcetto Italy and Tahiti, respectively—and maintained under brother–sister mating for more than 20 generations by F Bonhomme and colleagues (University of Montpellier, Montpellier, France). The probability of an initially heterozygous site remaining heterozygous after 20 generations of inbreeding is <10−6, and hence individuals from within a strain are considered genetically identical. As any two of these collection localities are >100 km away from each other, the strains are unrelated and can be viewed as independent snapshots of genetic diversity from those particular places and times. We also included two classical inbred strains, FVB/NJ (hereafter FVB) and C57BL/6N (hereafter C57) available from Jackson Labs (Bar Harbor, ME, USA). Males from all eight strains were crossed to female FVB and C57. The latter two strains were chosen as females because they respond well to hormonal induction of estrus (Byers et al, 2006). Furthermore, they carry divergent serotypes at the major histocompatibility complex locus, which has been shown to affect female choice dynamics (Potts et al, 1991; Roberts and Gosling, 2003; Leinders-Zufall et al, 2004) probably through chemical signals in the urine (Yamaguchi et al, 1981). Specifically, FVB carries the q serotype and C57 carries the b serotype. Though females were not given a choice between males in our crosses, cryptic female choice acting after copulation could in principle affect copulatory plug characteristics, for example, through adjustment of proteolytic responses.
To breed experimental mice, sire and dam were paired for 1–2 weeks, and then separated so that the dam could give birth in isolation. Males and females were weaned at 3–4 weeks postpartum. Females were weaned with up to three individuals per cage and were used in experiments at 4–6 weeks of age. Males were weaned with one individual per cage to avoid dominance interactions and reduced fertility (Snyder, 1967), and were used in experiments at 8–12 weeks of age. The colony was kept at 14:10 h of dark/light and provided food ad libitum.
Experimental matings and plug survival
At 4–6 weeks of age, virgin female mice (FVB or C57) were induced into estrus using established protocols (Nagy et al, 2003). Briefly, an intraperitoneal injection of 5 U pregnant mare's serum gonadotropin, followed ∼48 h later with an intraperitoneal injection of 5 U human chorionic gonadotropin, ensured ovulation ∼12 h later. Approximately 14 h after human chorionic gonadotropin injection, each female was placed into the cage of a randomly assigned experimental male for 4 h. Mating was confirmed by the presence of a copulatory plug in the vaginal–cervical region, observed visually or after very slight probing. Once mating was confirmed, females were randomly assigned to an early (24 h) or late (48 h) time point and housed alone. After the assigned time period, females were killed via carbon dioxide overexposure and copulatory plugs dissected and weighed. Experimental males mated no more than once per week to allow rejuvenation of seminal fluid and sperm stores. We scored 418 successful matings across the entire experiment, roughly 40 crosses per male genotype (10 crosses per male genotype per time point (24 vs 48 h) per female genotype (FVB vs C57)) (Supplementary Table 1), plus additional follow-up experiments.
Some studies regressed plug mass onto female body mass to control for differently sized female reproductive tracts (Ramm et al, 2005). However, plug mass was not significantly correlated to female body mass in our study, and hence we instead analyzed absolute plug mass. Conclusions remain unaltered if we instead analyze residual plug mass, but we present analyses based on absolute plug mass for simplicity. The proportion of successful matings that still had a plug was analyzed with a binomial model with logit link, using the Generalized Linear Model implemented in the GLM function in R (Dalgaard, 2008). Factors included time (24 vs 48 h), male genotype (8 genotypes), female genotype (2 genotypes) and the male × female interaction term.
We compared plug mass from the 24 h time point using several linear mixed models implemented in the LMER function of the R package LME4, followed by likelihood ratio tests (LRTs). The most complex model included male and female genotypes and their interaction as fixed effects, and individual male as a random effect.
Male morphology
To test whether plug survival correlated with male morphological features, we raised approximately five 8-week-old virgin males from each strain, and then took full body measurements including the mass of the testes and one lobe of the paired seminal vesicles (excluding the anterior lobe of the prostate, also known as the coagulating gland) (Supplementary Table 2). In a linear model that incorporated male genotype, both seminal vesicle mass and testes mass correlated to male body mass (F1, 31=78.37 and F1, 31=221.94, P<10−9 and P<10−15, respectively). To control for differences in male body size, seminal vesicle and testes mass were separately regressed onto male body mass using the LM function in R, with genotypes weighted by the inverse of their sample size. Residual seminal vesicle and testes mass were employed in downstream analyses. Although these males were not the same individuals as used in the experimental matings, they were genetically identical as they derived from the same respective strains.
Proteomic analyses
The relative abundance of copulatory plug proteins might play an important role in the length of time the plug lasts. From the same males used for anatomical measurements, we dissected the other lobe of the paired seminal vesicles (excluding the anterior lobe of the prostate) into 100 μl 8 M guanidine and carefully pushed out the luminal fluids to minimize any cellular damage. Proteins were then quantified using a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Then, 5 μg was mixed with sample buffer (urea 8 M, Tris-HCl (pH 6.8) 200 mM, EDTA (pH 8.0) 0.1 mM, DTT 100 mM and Tris Base 100 mM), heated at 37 °C for 15 min and then run in 12% polyacrylamide gel containing 3.5 mM SDS at 110 V for 90 min in SDS–Tris–Glycin buffer pH 8.8 and stained with Coomassie blue. Increasing amounts of bovine serum albumin were run the same way to produce a standard curve for quantification. Gels were scanned and protein bands were quantified with IMAGEJ (Schneider et al, 2012) by two different observers (CM and ZF); the average of these two measurements was taken. To adjust for slight differences in absolute protein amount, we calculated the proportion of protein bands rather than their absolute amount. Using the PRCOMP function in R, with the scale option set to True, principal component (PC) analysis was employed to remove the correlation of abundances among protein bands.
Thrombin assays
In mice, serine endopeptidases are upregulated by females in response to mating, and serine endopeptidase inhibitors are enriched in male seminal fluid (Dean et al, 2009, 2011). Thrombin is a serine endopeptidase that mimics such proteolytic activity. We modified a previously published thrombin fluorescence assay to quantify the amount of serine endopeptidase activity in copulatory plugs (Hengst et al, 2001; Murer et al, 2001). Individual copulatory plugs were homogenized into a fine powder using a mortar and pestle with liquid nitrogen. Plug homogenate was collected into a preweighed 1.7 ml Eppendorf tube (Hauppauge, NY, USA), and then 0.01 g plug powder was added to 400 μl thrombin assay buffer (50 mM Trizma Base, 130 mM sodium chloride, pH 8.3), combined and vortexed for 1 min. Homogenates were then incubated at room temperature for 30 min, with tubes inverted 10 times at 10-min intervals. After incubation, samples were centrifuged at 29 700 g for 3 min and supernatant collected.
We added 15 μl human α-thrombin (0.431 μg ml−1; Sigma-Aldrich, St Louis, MO,USA) to 80 μl of each plug homogenate and incubated for 30 min at 37 °C. After incubation, 5 μl of chromogenic substrate for thrombin, S-2238 (1.25 mg ml−1; Chromogenix, Bedford, MA, USA) was added. Amidolytic activity was quantified by recording fluorescence at 405 nm at 37 °C every min over 2.5 h with a BioTek ELx808 plate reader (BioTek, Winooski, VT, USA). Higher fluorescence indicates higher hydrolysis of the chromogenic substrate that in turn indicates higher activity of serine endopeptidases and/or reduced serine endopeptidase inhibition. Each plate assay was accompanied by two replicate standards, where 80 μl of thrombin assay buffer (no plug homogenate) was added. Fluorescence plotted against time (1–150 min) asymptotes at varying rates (Supplementary Figure 1). Using customized R scripts, we estimated the slope of the line before the asymptote and then subtracted the average slope of the two standard curves from each plug's estimated slope. These methods are presented visually in Supplementary Figure 1.
Exome sequencing
For the six wild-derived strains, we characterized DNA sequence variation at five genes thought to be important in copulatory plug formation—Tgm4, Svs1, Svs2, Svs4 and Svs5—using an exome enrichment and resequencing strategy. The full exomes will be published as part of a larger study elsewhere, but we focus here on these five genes as they are either necessary for copulatory plug formation or present in seminal vesicles and plugs at high abundance (Lundwall et al, 1997; Dean et al, 2009, 2011; Dean, 2013; Kawano et al, 2014). DNA was sheared using a Bioruptor UCD-200 (Diagenode, Denville, NJ, USA) with 7 rounds of sonication (7 min per round on high, 30 s on and 30 s off) and genomic DNA libraries were constructed using a previously described protocol designed to facilitate multiplexed exome capture (Rohland and Reich, 2012). To reduce molecular interference during enrichment, we used truncated adaptors containing unique P5 ‘internal' barcodes (Rohland and Reich, 2012). PCR primers were designed according to Rohland and Reich (2012).
In-solution sequence capture was performed using Nimblegen SeqCap EZ Mouse Exome probes (Roche NimbleGen, Madison, WI, USA) as described in Nimblegen's SeqCap EZ Lbrary User's Guide. Libraries were pooled equally to obtain 1 μg total DNA for each hybridization experiment. Libraries were then enriched using two separate capture reactions with eight libraries each, including blocking oligonucleotides specific to our custom adapters (Rohland and Reich, 2012) and mouse COT-1 DNA (Invitrogen, Carlsbad, CA, USA) to reduce nonspecific hybridization. The capture reactions were hybridized for 68 h at 47 °C in an Eppendorf Mastercycler Pro (Eppendorf) then washed, eluted and PCR enriched. Capture enrichment success was verified using real-time quantitative PCR analysis of three targeted regions on pre- and post-capture library pools. Sequencing was performed using the Illumina Hi-seq 2000 platform (San Diego, CA, USA) at the Epigenome Center at the University of Southern California.
Sequences were mapped to the mm10 reference genome with BWA (Li and Durbin, 2009), allowing for seven mismatches. Samtools (Li et al, 2009) was used to remove PCR duplicates and filter reads mapping with a quality score of at least 20, and then varscan (Koboldt et al, 2009) was used to call variants from the pileup files. To exclude sequencing error, bases different from reference were only accepted if they had a depth of at least two reads, each with a Phred score of at least 20. We did not observe any heterozygous sites, as expected given we were using inbred strains. Gene and transcript annotations were downloaded from Ensembl version 78 (www.ensembl.org). Sequences from FVB (Wong et al, 2012) and C57 (Keane et al, 2011) were downloaded and added to our data set. We applied Mantel's tests to assess whether pairwise DNA distance matrices were correlated to pairwise phenotype distance matrices, using the MANTEL.TEST function in the APE package in R (Paradis and Claude, 2004).
Results
Plug survival
Across the whole data set, 48.8% (102/209) of copulatory plugs were present after 24 h of incubation in the female, and 15.4% (29/188) after 48 h. Time and male genotype had significant effects on plug survival (χ2=62.8, 45.7, d.f.=1, 7, P=10−7, 10−14, respectively), but neither female genotype nor the male × female interaction term did (χ2=0.09, 8.81, d.f.=1, 7, P=0.76, 0.27, respectively; Table 1). A Hosmer–Lemeshow test (Hosmer and Lemeshow, 2000) showed that a binomial model including only male and time as factors fit the data well, with no significant overdispersion (χ2=20.07, d.f.=14, P=0.13). To further test for a male × female interaction term, we repeated the analyses using only FVB and C57 males and females. We did not detect an interaction term in this subset of data.
Table 1. Results of generalized linear model on plug survival.
| Model | d.f. | Deviance (χ2) | Residual d.f. | Residual deviance | P-value |
|---|---|---|---|---|---|
| Null | 31 | 131.49 | |||
| Male genotype | 7 | 45.74 | 24 | 85.75 | 9.83E−08 |
| Female genotype | 1 | 0.09 | 23 | 85.66 | 0.76 |
| Time | 1 | 62.82 | 22 | 22.84 | 2.23E−15 |
| Male × female | 7 | 8.81 | 15 | 14.03 | 0.27 |
Plug mass after 24 h of incubation differed between male and female genotypes. Male genotype had a significant effect on plug mass (LRT, χ2=33.2, d.f.=7, P=10−4) (Supplementary Figure 2). Including female in addition to male genotype fit the data significantly better than male genotype alone (LRT, χ2=6.56, d.f.=1, P=0.01), but including a male × female interaction term did not (LRT, χ2=8.28, d.f.=7, P=0.31).
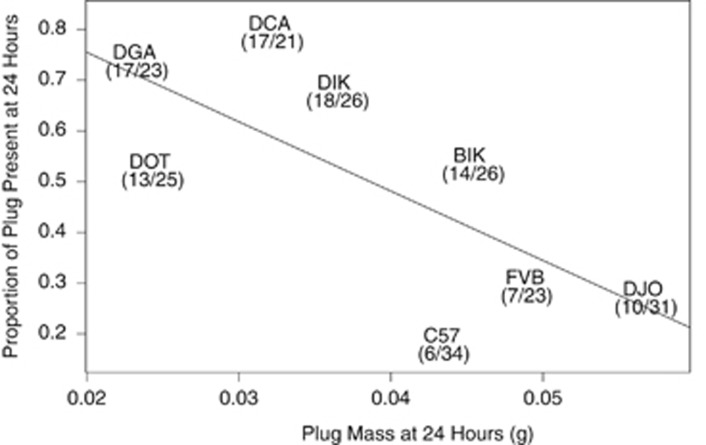
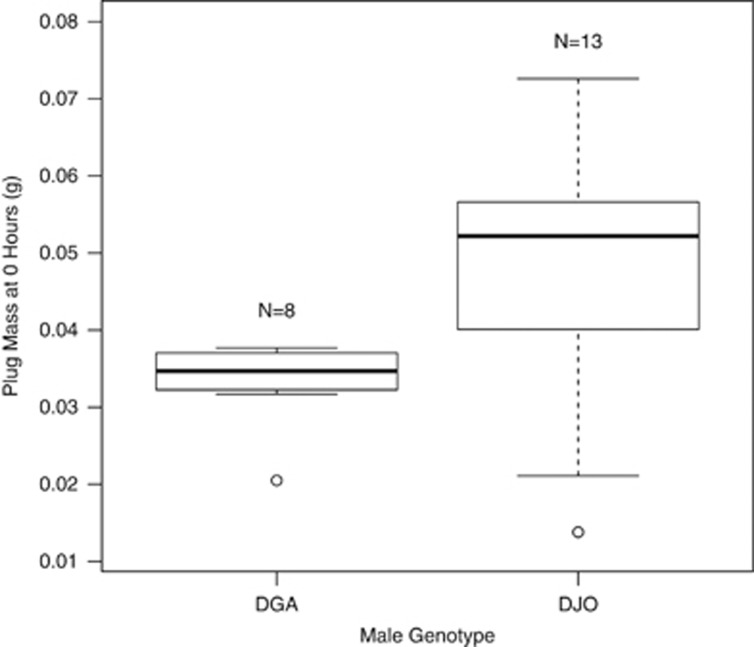
The proportion of plugs present at 24 h was negatively correlated with plug size (Figure 1, F1, 6=5.78, P=0.05). In other words, male genotypes that made smaller plugs also produced plugs that tended to last longer. To further clarify this result, we repeated this portion of the experiment with two extreme genotypes: DGA (N=13 crosses), a genotype that makes relatively long-lasting and small plugs, and DJO (N=8 crosses), a genotype that makes relatively short-lasting and large plugs. For this experiment we dissected plugs immediately following mating rather than 24 or 48 h later, thus minimizing the potential influence of female degradation on plug size. As above, DGA males made significantly smaller plugs than DJO males (Figure 2, Welch's t-test=2.74, d.f.=16.95, P=0.01, accounting for unequal variance as an F-test for equal variances (F=0.12, d.f.=7, P=0.01) was rejected). Plug survival could be influenced by plug density; for example, small plugs may be denser and more difficult for females to degrade. We lack the data to evaluate this hypothesis.
Figure 1.
The proportion of plugs present at 24 h post mating was negatively correlated with plug mass. Mouse genotype (text) and the number of plugs present (numerator) out of the total number of successful matings (denominator) are indicated. The exact location of each strain on the plot is the center of all text.
Figure 2.
In a focused experiment of two extreme genotypes, DGA and DJO, plug size differed significantly when plugs were collected immediately following mating (0 h).
Male morphology
Although male genotypes differed in the size of their seminal vesicles, they did not vary in a way related to plug survival. Residual seminal vesicle mass differed significantly among male genotypes (F7, 32=3.76, P=0.004), but residual seminal vesicle mass did not covary with the proportion of plugs present at 24 h (F1, 6=0.049, P=0.83). Similarly, although residual testes mass differed significantly among male genotypes (F7, 32=39.78, P <10−14), it did not covary with the proportion of plugs present at 24 h (F1, 6=0.125, P=0.735). Plug mass did not correlate with residual seminal vesicle mass or residual testes mass (F1, 6=0.43, 0.08, P=0.54, 0.79, respectively).
Proteomic analyses
The major protein composition of plugs was correlated to plug survival. Coomassie-stained polyacrylamide gels of seminal vesicle fluids revealed four very abundant proteins (Supplementary Figure 3 and Table 2). We did not attempt to identify these four bands using mass spectrometry, but based on previous studies (Lundwall et al, 1997; Lin et al, 2002; Dean et al, 2011; Tseng et al, 2011) and the match to known molecular mass, these four bands most likely consisted of the seminal vesicle-secreted proteins Svs1, Svs2, Svs4 and Svs5 (Supplementary Figure 3). The exact identity of the proteins is not critical as we are only using them as a biomarker of plug composition.
The proportions of these four protein bands (abundance of each protein band divided by the sum of the abundances of the four main protein bands) were reduced to two PCs that explained 84% and 13% of the variance, respectively (Table 2). The loadings of the Svs1-like, Svs4-like and Svs5-like proteins (−0.801, 0.362 and 0.476, respectively) indicated these three protein bands contributed highly to PC1, with the proportion of Svs1-like protein negatively correlated with the proportion of Svs4-like and Svs5-like proteins. PC2 primarily consisted of remaining variation in Svs1-like, Svs2-like and Svs5-like proteins (loadings=0.329, −0.827 and 0.454, respectively). Plug mass was negatively correlated to PC1 (F1, 6=10.3, P=0.02), but not PC2 (F1, 6=1.87, P=0.22). Plug survival was not correlated with either PC1 or PC2 (F1, 6=4.25, 1.55; P=0.09, 0.26, respectively). Variation in Svs2 did not seem to explain variation in plug mass, and this is surprising given its importance in plug formation (Kawano et al, 2014).
Table 2. Principal components (PCs) of protein concentrations of the four major bands detected.
| PC1 (0.84) | PC2 (0.13) | |
|---|---|---|
| SVS1-like | −0.801 | 0.329 |
| SVS2-like | −0.037 | −0.827 |
| SVS4-like | 0.362 | 0.045 |
| SVS5-like | 0.476 | 0.454 |
Thrombin assays
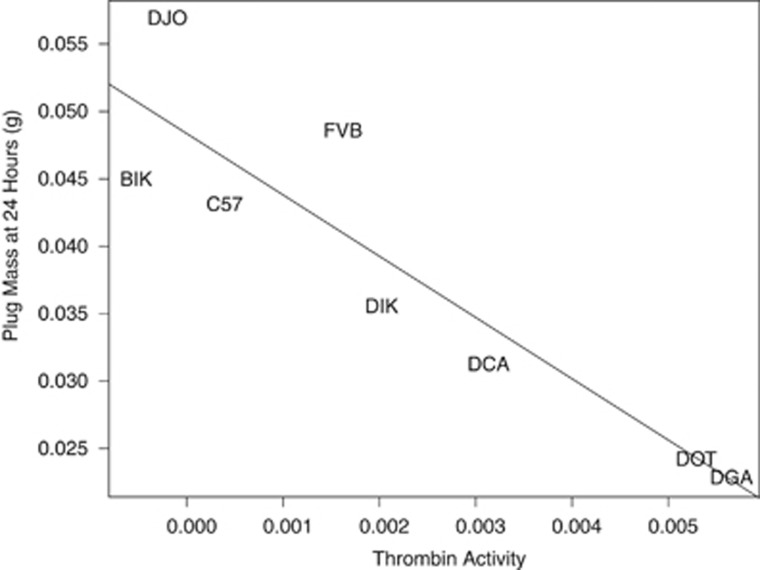
Small plugs showed more protease activity. There was a significant effect of male genotype on protease activity (F7, 53=4.06, P=0.001) but neither female genotype (F1, 53=0.005, P=0.94) nor the male × female interaction term (F6, 53=0.95, P=0.47) had an effect (Table 3). Protease activity was significantly negatively correlated with plug size (Figure 3, F1, 6=29.26, P=0.002). As plug size was negatively correlated to plug survival (Figure 1), we predicted that protease activity correlated to plug survival, but this was not the case (F1, 6=3.37, P=0.11).
Table 3. Results of linear model on protease activity.
| Factor | d.f. | SumSq | F-value | P-value |
|---|---|---|---|---|
| Male genotype | 7 | 3.89E−04 | 4.06 | 0.001 |
| Female genotype | 1 | 7.00E−08 | 0.005 | 0.94 |
| Male × female | 6 | 7.80E−05 | 0.95 | 0.47 |
| Residuals | 53 | 7.26E−04 |
Figure 3.
Protease activity was significantly negatively correlated with plug size. Mouse genotype indicated by text; the exact location of each strain on the plot is the center of all text.
Across 19 technical replicates (9 replicated standard curves and 10 replicated plugs), the median coefficient of variation of the estimated slopes of fluorescence (unbiased s.d. divided by the mean) was 0.058, indicating a high level of repeatability.
Exome variation
There was no variation at five copulatory plug genes that related to plug survival. The proportion of bases in the coding part of the transcript that were covered by at least two (one) reads averaged 0.80 (0.91), 0.73 (0.88), 0.76 (0.81), 0.33 (0.53) and 0.76 (0.84) for Tgm4, Svs1, Svs2, Svs4 and Svs5, respectively. The number of nonsynonymous (synonymous) variants observed in 7 of our 8 genotypes was 0 (2), 4 (8), 1 (2), 1 (0) and 0 (3) for the five genes, respectively. Because of technical difficulties, we were unable to generate sequence from DJO. Plugs form when Tgm4 crosslinks glutamine and lysine residues in Svs2 (Williams-Ashman, 1984). The single nonsynonymous site in Svs2 led to a tyrosine/phenylalanine polymorphism, suggesting it did not contribute to variation in plug survival. There was no association between pairwise genetic distance and pairwise phenotypic difference in plug size or survival (all Mantel's tests, P>0.05).
Two proteases, Ltf and Klk14, were previously shown to be produced by females in response to mating (Dean et al, 2011) and could be important in plug degradation. The two female strains used here, FVB and C57, are identical at both genes (http://www.informatics.jax.org). Therefore, genetic variation at plug genes or genes that potentially degrade plugs cannot explain variation in plug survival.
Discussion
Copulatory plugs are a prominent feature in many internally fertilizing organisms, including nematodes (Barker, 1994; Palopoli et al, 2008), insects (Rogers et al, 2009), reptiles (Devine, 1975, 1977; Moreira and Birkhead, 2004), rodents (Voss, 1979; Dewsbury, 1984) and primates (Hartung and Dewsbury, 1978; Dixson and Anderson, 2002). Comparative studies suggest plugs evolved in the context of sperm competition, as a means for males to inhibit remating by females. The goal of the present study was to better understand survival dynamics of copulatory plugs. Our primary finding was that variation at several copulatory plug phenotypes (size, major protein composition, protease activity and survival) covaried with male genotype, revealing standing genetic variation for diverse male traits that are likely to play important roles in mouse reproductive ecology.
Plug survival
If large plugs are indeed adaptive responses to sperm competition (see Introduction), then we might expect that larger plugs survive longer in the female's reproductive tract. In contrast to this prediction, our study revealed that male genotypes that make long-lasting plugs tended to make smaller plugs. Furthermore, these small but long-lasting plugs were less able to inhibit thrombin proteolysis. In other words, smaller plugs that seemed more susceptible to proteolytic degradation actually survived longer in the female's reproductive tract. There are at least two hypotheses to explain why small plugs survived longer in the female's reproductive tract.
First, it is possible that smaller plugs trigger a less intense female proteolytic response. To address this possibility, we analyzed an additional thrombin assay where no thrombin was added before fluorescence detection (instead of 15 μl). Any fluorescence in this ‘no thrombin' assay must arise from endogenous thrombin-like proteases already present in the plug extract that could be either male or female derived. If smaller plugs triggered a less intense female response, we would predict smaller plugs have less fluorescence in these ‘no thrombin' assays. However, this was not the case as small plugs actually had more protease activity immediately after copulation than large plugs (Supplementary Figure 4), just as they did after 24 h of incubation (Figure 3). Thus, small plugs do not induce a less intense proteolytic response from the female.
Second, small plugs may last longer in the female because they are more difficult to remove. In some rodents, females bite the plug and actively remove it (Koprowski, 1992). In house mice, the plug tightly adheres to the female's epithelium in the vaginal–cervical region. Over time, the epithelium begins to slough off and the fact that plugs can often be found in the bottom of cages suggests that it is not fully degraded in situ but perhaps degraded to a point where it can be expelled (R Mangels and MD Dean, personal observation) and sometimes eaten (Dewsbury, 1984). It is possible that small plugs are more difficult for females to remove through contractions of her reproductive tract, if they provide less traction for female contractions.
Why would males make large plugs?
Male mate choice (Dewsbury, 1982; Drickamer et al, 2003; Edward and Chapman, 2011; Ramm and Stockley, 2014) and the dynamic adjustment of ejaculate allocation (Wedell et al, 2002; Delbarco-Trillo and Ferkin, 2004) suggest that ejaculates are costly to produce and conserved when possible. Plug-forming proteins account for nearly one third of the total protein abundance of the ejaculate in mice, suggesting that this structure is a major reproductive investment for males (Lundwall et al, 1997; Lin et al, 2002; Dean et al, 2011). As large plugs also seem to survive shorter periods of time, and ejaculates are likely to be costly, our study begs the question of why males would ever invest in large plugs. Answering this question requires further experimentation as our study did not specifically link copulatory plug characteristics to fitness traits like number of offspring sired, but potential explanations include tradeoffs between plug size and other aspects of reproductive fitness. For example, small plugs may be more difficult for females to remove, as suggested here, but easier for competitor males to remove. Spines on the penis as well as repeated intromissions without ejaculation may be male adaptations to remove other males' plugs (Wallach and Hart, 1983; Dewsbury, 1984; O'Hanlon and Sachs, 1986). The intensity of sperm competition varies across populations of house mice (Firman and Simmons, 2008), and probably across time as a function of fluctuations in density (Dean et al, 2006), which could potentially sway the balance of selection toward plugs with different benefits. Spatial or temporal variation in the intensity or form of sperm competition could lead to standing variation (Felsenstein, 1976; Siepielski et al, 2009; Bell, 2010).
Sexual conflict could also preserve genetic variation in copulatory plug characteristics. For example, different alleles of copulatory plug genes could be better at avoiding degradation in some but not all females in the population. Statistically, this type of dynamic predicts a male × female interaction term. Although we did not detect such an interaction term in any of our assays, the female genotypes used here did not differ at two candidate protease genes, and our study was probably underpowered to detect it.
Conclusions
The genetic basis of male reproductive phenotypes that are targets of sexual selection remain poorly characterized. We found that male genotype explained a significant amount of variation in plug size and plug survival, demonstrating there is standing genetic variation in this ecologically important trait. Interestingly, small plugs tended to last longer in the female reproductive tract, opposite to the predictions derived from previous comparative studies. Our study reveals that the dynamics of copulatory plugs are more complex than previously appreciated, and suggests that there could be tradeoffs between plug size and specific aspects of house mouse reproductive ecology.
DATA ARCHIVING
Data available from the Dryad Digital Repository: http://doi.org/10.5061/dryad.n48q0.
Acknowledgments
We thank Annie Orth and François Bonhomme (University of Montpellier) for supplying the wild-derived inbred strains. Norm Arnheim provided founder stock for FVB mice, and the KOMP provided C57. Ian Ehrenreich, Kim Siegmund and Peter Ralph provided statistical advice. Ian Ehrenreich provided access to the BioTek plate reader. Dominic Edward and four anonymous reviewers provided many valuable comments on the manuscript. This study was funded by NSF CAREER award 1150259 (to MDD) and NIH Grants R01GM098536 (to MDD) and R01HD73439 (to JMG).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Andersson M. (1994) Sexual Selection. Princeton University Press: Princeton, NJ. [Google Scholar]
- Arnqvist G, Rowe L. (2005) Sexual Conflict. Princeton University Press: Princeton, NJ. [Google Scholar]
- Asdell SA. (1946) Patterns of Mammalian Reproduction. Comstock Publishing Company: Ithaca, New York.
- Ball J. (1934). Demonstration of a quantitative relation between stimulus and response in pseudopregnancy in the rat. Am J Physiol 107: 698–703. [Google Scholar]
- Barker DM. (1994). Copulatory plugs and paternity assurance in the nematode Caenorhabditis elegans. Anim Behav 48: 147–156. [Google Scholar]
- Bell G. (2010). Fluctuating selection: the perpetual renewal of adaptation in variable environments. Phil Trans R Soc London 365: 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilde T, Foged A, Schilling N, Arnqvist G. (2009). Postmating sexual selection favors males that sire offspring with low fitness. Science 324: 1705–1706. [DOI] [PubMed] [Google Scholar]
- Birkhead TR, Pizzari T. (2002). Postcopulatory sexual selection. Nat Rev Genet 3: 262–273. [DOI] [PubMed] [Google Scholar]
- Blandau RJ. (1945. a). Is the copulation plug necessary for the en masse transport of spermatozoa into the uterine cornua of the albino rat? Anat Rec 91: 266–267. [Google Scholar]
- Blandau RJ. (1945. b). On the factors involved in sperm transport through the cervix uteri of the albino rat. Am J Anat 77: 253–272. [Google Scholar]
- Byers SL, Payson SJ, Taft RA. (2006). Performance of ten inbred mouse strains following assisted reproductive technologies (ARTs). Theriogenology 65: 1716–1726. [DOI] [PubMed] [Google Scholar]
- Cameron E, Day T, Rowe L. (2007). Sperm competition and the evolution of ejaculate composition. Am Nat 169: E158–E172. [DOI] [PubMed] [Google Scholar]
- Carballada R, Esponda P. (1992). Role of fluid from seminal vesicles and coagulating glands in sperm transport into the uterus and fertility in rats. J Reprod Fertil 95: 639–648. [DOI] [PubMed] [Google Scholar]
- Carnahan SJ, Jensen-Seaman MI. (2008). Hominoid seminal protein evolution and ancestral mating behavior. Am J Primatol 70: 939–948. [DOI] [PubMed] [Google Scholar]
- Chapman T. (2001). Seminal fluid-mediated fitness traits in Drosophila. Heredity (Edinb) 87: 511–521. [DOI] [PubMed] [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. (2006). Evolution of reproductive proteins from animals and plants. Reproduction 131: 11–22. [DOI] [PubMed] [Google Scholar]
- Clark NL, Swanson WJ. (2005). Pervasive adaptive evolution in primate seminal proteins. PLoS Genet 1: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard P. (2008) Introductory Statistics with R. Springer: New York, NY. [Google Scholar]
- Dean MD. (2013). Genetic disruption of the copulatory plug in mice leads to severely reduced fertility. PLoS Genet 9: e1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Ardlie KG, Nachman MW. (2006). The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus. Mol Ecol 15: 4141–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Clark NL, Findlay GD, Karn RC, Yi X, Swanson WJ et al. (2009). Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus. Mol Biol Evol 26: 1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Findlay GD, Hoopmann MR, Wu CC, MacCoss MJ, Swanson WJ et al. (2011). Identification of ejaculated proteins in the house mouse (Mus domesticus via isotopic labeling. BMC Genomics 12: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Nachman MW. (2009). Faster fertilization rate in conspecific versus heterospecific matings in house mice. Evolution 63: 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbarco-Trillo J, Ferkin MH. (2004). Male mammals respond to a risk of sperm competition conveyed by odours of conspecific males. Nature 431: 446–449. [DOI] [PubMed] [Google Scholar]
- Devine MC. (1975). Copulatory plugs in snakes: enforced chastity. Science 187: 844–845. [DOI] [PubMed] [Google Scholar]
- Devine MC. (1977). Copulatory plugs, restricted mating opportunities and reproductive competition among male garter snakes. Nature 267: 345–346. [DOI] [PubMed] [Google Scholar]
- Dewsbury DA. (1982). Ejaculate cost and male choice. Am Nat 119: 601–610. [Google Scholar]
- Dewsbury DA. (1984) Sperm competition in muroid rodents. In: Smith RL (ed). Sperm Competition and the Evolution of Animal Mating Systems. Academic Press: New York. pp 547–571. [Google Scholar]
- Dixson AF. (1998. a) Primate Sexuality. Oxford University Press: New York. [Google Scholar]
- Dixson AF. (1998. b). Sexual selection and evolution of the seminal vesicles in primates. Folia Primatol (Basel) 69: 300–306. [DOI] [PubMed] [Google Scholar]
- Dixson AF, Anderson MJ. (2002). Sexual selection, seminal coagulation and copulatory plug formation in primates. Folia Primatol (Basel) 73: 63–69. [DOI] [PubMed] [Google Scholar]
- Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. (2004). Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet 36: 1326–1329. [DOI] [PubMed] [Google Scholar]
- Drickamer LC, Gowaty PA, Wagner DM. (2003). Free mutual mate preferences in house mice affect reproductive success and offspring performance. Anim Behav 65: 105–114. [DOI] [PubMed] [Google Scholar]
- Eberhard W. (2009). Evolution of genitalia: theories, evidence, and new directions. Genetica 138: 5–18. [DOI] [PubMed] [Google Scholar]
- Edward DA, Chapman T. (2011). The evolution and significance of male mate choice. Trends Ecol Evol 26: 647–654. [DOI] [PubMed] [Google Scholar]
- Fedorka KM, Mousseau TA. (2002). Material and genetic benefits of female multiple mating and polyandry. Anim Behav 64: 361–367. [Google Scholar]
- Felsenstein J. (1976). The theoretical population genetics of variable selection and migration. Annu Rev Genet 10: 253–280. [DOI] [PubMed] [Google Scholar]
- Firman RC, Simmons LW. (2008). The frequency of multiple paternity predicts variation in testes size among island populations of house mice. J Evol Biol 21: 1524–1533. [DOI] [PubMed] [Google Scholar]
- Hartung TG, Dewsbury DA. (1978). A comparative analysis of copulatory plugs in muroid rodents and their relationship to copulatory behavior. J Mammal 59: 717–723. [Google Scholar]
- Hengst U, Albrecht H, Hess D, Monard D. (2001). The phosphatidylethanolamine-binding protein is the prototype of a novel family of serine protease inhibitors. J Biol Chem 276: 535–540. [DOI] [PubMed] [Google Scholar]
- Hosmer DW, Lemeshow S. (2000) Applied Logistic Regression. John Wiley and Sons: New York. [Google Scholar]
- Jennions MD, Petrie M. (2000). Why do females mate multiply? A review of the genetic benefits. Biol Rev Camb Philos Soc 75: 21–64. [DOI] [PubMed] [Google Scholar]
- Jensen-Seaman MI, Li WH. (2003). Evolution of the hominoid semenogelin genes, the major proteins of ejaculated semen. J Mol Evol 57: 261–270. [DOI] [PubMed] [Google Scholar]
- Kawano N, Araki N, Yoshida K, Hibino T, Ohnami N, Makino M et al. (2014). Seminal vesicle protein SVS2 is required for sperm survival in the uterus. Proc Natl Acad Sci USA 111: 4145–4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano N, Kang W, Yamashita M, Koga Y, Yamazaki T, Hata T et al. (2010). Mice lacking two sperm serine proteases, ACR and PRSS21, are subfertile, but the mutant sperm are infertile in vitro. Biol Reprod 83: 359–369. [DOI] [PubMed] [Google Scholar]
- Keane TM, Goodstadt L, Danecek P, White MA, Wong K, Yalcin B et al. (2011). Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 477: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Pennington JE. (2009). Protease gene duplication and proteolytic activity in Drosophila female reproductive tracts. Mol Biol Evol 26: 2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Swanson WJ, Markow TA. (2007). Gene duplication and adaptive evolution of digestive proteases in Drosophila arizonae female reproductive tracts. PLoS Genet 3: e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingan SB, Tatar M, Rand DM. (2003). Reduced polymorphism in the chimpanzee semen coagulating protein, semenogelin I. J Mol Evol 57: 159–169. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER et al. (2009). VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25: 2283–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski JL. (1992). Removal of copulatory plugs by female tree squirrels. J Mammal 73: 572–576. [Google Scholar]
- Lawniczak MKN, Begun DJ. (2007). Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol Biol Evol 24: 1944–1951. [DOI] [PubMed] [Google Scholar]
- Leinders-Zufall T, Brennan P, Widmayer P, S PC, Maul-Pavicic A, Jager M et al. (2004). MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 306: 1033–1037. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-J, Luo C-W, Chen Y-H. (2002). Localization of the transglutaminase cross-linking site in SVS III, a novel glycoprotein secreted from mouse seminal vesicle. J Biol Chem 277: 3632–3639. [DOI] [PubMed] [Google Scholar]
- Lundwall Å, Peter A, Lovgren J, Lilja H, Malm J. (1997). Chemical characterization of the predominant proteins secreted by mouse seminal vesicles. Eur J Biochem 249: 39–44. [DOI] [PubMed] [Google Scholar]
- Martan J, Shepherd BA. (1976). The role of the copulatory plug in reproduction of the guinea pig. J Exp Zool 196: 79–83. [DOI] [PubMed] [Google Scholar]
- Matthews MK Jr, Adler NT. (1978). Systematic interrelationship of mating, vaginal plug position, and sperm transport in the rat. Physiol Behav 20: 303–309. [DOI] [PubMed] [Google Scholar]
- Moreira PL, Birkhead TR. (2004). Copulatory plug displacement and prolonged copulation in the Iberian rock lizard (Lacerta monticola. Behav Ecol Sociobiol 56: 290–297. [Google Scholar]
- Mosig DW, Dewsbury DA. (1970). Plug fate in the copulatory behavior of rats. Psychon Sci 20: 315–316. [Google Scholar]
- Murer V, Spetz JF, Hengst U, Altrogge LM, de Agostini A, Monard D. (2001). Male fertility defects in mice lacking the serine protease inhibitor protease nexin-1. Proc Natl Acad Sci USA 98: 3029–3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. (2003) Manipulating The Mouse Embryo, 3rd edn. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, New York. [Google Scholar]
- O'Hanlon JK, Sachs BD. (1986). Fertility of mating in rats (Rattus norvegicus: contributions of androgen-dependent morphology and actions of the penis. J Comp Psychol 100: 178–187. [PubMed] [Google Scholar]
- Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A et al. (2008). Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature 454: 1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. (2004). APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Parkes A. (1926). Observations on the oestrous cycle of the albino mouse. Proc R Soc B 100: 151–170. [Google Scholar]
- Pilch B, Mann M. (2006). Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol 7: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiani A. (2006). Complexity of seminal fluid: a review. Behav Ecol Sociobiol 60: 289–310. [Google Scholar]
- Potts WK, Manning CJ, Wakeland EK. (1991). Mating patterns in seminatural populations of mice influenced by MHC genotype. Nature 352: 619–621. [DOI] [PubMed] [Google Scholar]
- Ramm SA, McDonald L, Hurst JL, Beynon RJ, Stockley P. (2009). Comparative proteomics reveals evidence for evolutionary diversification of rodent seminal fluid and its functional significance in sperm competition. Mol Biol Evol 26: 189–198. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. (2008). Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol 25: 207–219. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Parker GA, Stockley P. (2005). Sperm competition and the evolution of male reproductive anatomy in rodents. Proc R Soc B 272: 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm SA, Stockley P. (2014). Sequential male mate choice under sperm competition risk. Behav Ecol 25: 660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SC, Gosling LM. (2003). Genetic similarity and quality interact in mate choice decisions by female mice. Nat Genet 35: 103–106. [DOI] [PubMed] [Google Scholar]
- Robertson SA. (2007). Seminal fluid signaling in the female reproductive tract: lessons from rodents and pigs. J Anim Sci 85: E36–E44. [DOI] [PubMed] [Google Scholar]
- Rogers DW, Baldini F, Battaglia F, Panico M, Dell A, Morris HR et al. (2009). Transglutaminase-mediated semen coagulation controls sperm storage in the malaria mosquito. PLoS Biol 7: e1000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohland N, Reich D. (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepielski AM, DiBattista JD, Carlson SM. (2009). It's about time: the temporal dynamics of phenotypic selection in the wild. Ecol Lett 12: 1261–1276. [DOI] [PubMed] [Google Scholar]
- Silver L. (1995) Mouse Genetics: Concepts and Applications. Oxford University Press: New York, NY, USA. [Google Scholar]
- Slatyer RA, Mautz BS, Backwell PRY, Jennions MD. (2012). Estimating genetic benefits of polyandry from experimental studies: a meta-analysis. Biol Rev 87: 1–33. [DOI] [PubMed] [Google Scholar]
- Snyder RL. (1967) Fertility and reproductive performance of grouped male mice. In: Benirschke K (ed). Comparative Aspects of Reproductive Failure. Springer-Verlag: New York, pp 458–472. [Google Scholar]
- Stockard CR, Papanicolaou GN. (1919). The vaginal closure membrane, copulation, and the vaginal plug in the guinea-pig, with further considerations of the œstrous rhythm. Biol Bull 37: 222–245. [Google Scholar]
- Stockley P. (1997). Sexual conflict resulting from adaptations to sperm competition. Trends Ecol Evol 12: 154–159. [DOI] [PubMed] [Google Scholar]
- Stockley P, Ramm SA, Sherborne AL, Thom MD, Paterson S, Hurst JL. (2013). Baculum morphology predicts reproductive success of male house mice under sexual selection. BMC Biol 11: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. (2002). The rapid evolution of reproductive proteins. Nat Rev Genet 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Toner JP, Attas AI, Adler NT. (1987). Transcervical sperm transport in the rat: the roles of pre-ejaculatory behavior and copulatory plug fit. Physiol Behav 39: 371–375. [DOI] [PubMed] [Google Scholar]
- Tseng H-C, Tang J-B, Sudhakar Gandhi P, Luo C-W, Ou C-M, Tseng C-J et al. (2011). Mutual adaptation between mouse transglutaminase 4 and its native substrates in the formation of copulatory plug. Amino Acids 42: 951–960. [DOI] [PubMed] [Google Scholar]
- Voss R. (1979). Male accessory glands and the evolution of copulatory plugs in rodents. Occas Pap Mus Zool Univ Mich 689: 1–27. [Google Scholar]
- Wallach SJR, Hart BL. (1983). The role of the striated penile muscles of the male rat in seminal plug dislodgement and deposition. Physiol Behav 31: 815–821. [DOI] [PubMed] [Google Scholar]
- Wedell N, Gage MJG, Parker GA. (2002). Sperm competition, male prudence and sperm-limited females. Trends Ecol Evol 17: 313–320. [Google Scholar]
- Williams-Ashman HG. (1984). Transglutaminases and the clotting of mammalian seminal fluids. Mol Cell Biochem 58: 51–61. [DOI] [PubMed] [Google Scholar]
- Wolfner MF. (2002). The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88: 85–93. [DOI] [PubMed] [Google Scholar]
- Wong A, Turchin MC, Wolfner MF, Aquadro CF. (2008). Evidence for positive selection on Drosophila melanogaster seminal fluid protease homologs. Mol Biol Evol 25: 497–506. [DOI] [PubMed] [Google Scholar]
- Wong K, Bumpstead S, Van Der Weyden L, Reinholdt LG, Wilming LG, Adams DJ et al. (2012). Sequencing and characterization of the FVB/NJ mouse genome. Genome Biol 13: R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Yamazaki K, Beauchamp G, Bard J, Thomas L, Boyse E. (1981). Distinctive urinary odors governed by the major histocompatibility locus of the mouse. Proc Natl Acad Sci USA 78: 5817–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh JA, Zeh DW. (2001). Reproductive mode and the genetic benefits of polyandry. Anim Behav 61: 1051–1063. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.