Abstract
Investigating crop origins is a priority to understand the evolution of plants under domestication, develop strategies for conservation and valorization of agrobiodiversity and acquire fundamental knowledge for cultivar improvement. The date palm (Phoenix dactylifera L.) belongs to the genus Phoenix, which comprises 14 species morphologically very close, sometimes hardly distinguishable. It has been cultivated for millennia in the Middle East and in North Africa and constitutes the keystone of oasis agriculture. Yet, its origins remain poorly understood as no wild populations are identified. Uncultivated populations have been described but they might represent feral, i.e. formerly cultivated, abandoned forms rather than truly wild populations. In this context, this study based on morphometrics applied to 1625 Phoenix seeds aims to (1) differentiate Phoenix species and (2) depict the domestication syndrome observed in cultivated date palm seeds using other Phoenix species as a “wild” reference. This will help discriminate truly wild from feral forms, thus providing new insights into the evolutionary history of this species. Seed size was evaluated using four parameters: length, width, thickness and dorsal view surface. Seed shape was quantified using outline analyses based on the Elliptic Fourier Transform method. The size and shape of seeds allowed an accurate differentiation of Phoenix species. The cultivated date palm shows distinctive size and shape features, compared to other Phoenix species: seeds are longer and elongated. This morphological shift may be interpreted as a domestication syndrome, resulting from the long-term history of cultivation, selection and human-mediated dispersion. Based on seed attributes, some uncultivated date palms from Oman may be identified as wild. This opens new prospects regarding the possible existence and characterization of relict wild populations and consequently for the understanding of the date palm origins. Finally, we here describe a pipeline for the identification of the domestication syndrome in seeds that could be used in other crops.
Introduction
Global food security is facing challenges posed by the sharp reduction in the diversity of cultivated plants associated with planetary changes and the increasing food demand [1–4]. The implementation of crop improvement programs is expected to boost food production and food security and thus to help rising to the current and future challenges of crop cultivation. The identification of wild populations is one of the important prerequisites for breeding programs, as it is long known that they represent a genetic resource for cultivar improvement [5]. Moreover, it opens the possibility of comparing wild and domesticates to identify selected traits and understand evolution patterns of phenotypic traits, some of them defining the domestication syndrome [6–12]. For most crops, especially annuals, the wild ancestor is well known and populations are characterized, so that their domestication histories have been intensively studied [7–9]. In perennials, in contrast, we have a much less comprehensive knowledge due to their long life, ongoing crop-wild gene flow and clonal propagation that contribute to mild domestication bottlenecks and thus weak domestication syndrome [13,14]. Escaped individuals (called feral) from more or less distant cultivation areas may survive and reproduce without human intervention (e.g. olive tree [15] and grapevine [16]). It is therefore difficult to identify truly wild populations, as demonstrated in olive trees [17]. More strikingly, in date palms (Phoenix dactylifera L., Arecaceae) no wild population has been characterized to date [18–20].
The date palm belongs to the Old World genus Phoenix L. (Arecaceae) composed of 14 inter-fertile species distributed from the Atlantic islands, through Southern Europe, Africa and Southern Asia to the Philippines [21,22]. The whole genus is economically very important as most species are cultivated or exploited for many purposes such as ornamentation, food or construction. Recent barcoding studies based on nuclear and chloroplastic sequences allowed to identify unambiguously nine of the 13 Phoenix species included and identified the date palm sister species as Phoenix sylvestris and Phoenix atlantica [18,23,24]. However, Phoenix species are morphologically close and sometimes hardly distinguishable as there are only few systematically useful morphological and anatomical characters [18,21]. Additional features are therefore required to easily distinguish Phoenix species.
The most important species of the genus, the date palm, constitutes the main element in oasis agro-ecosystems and has assumed a nutritional, economic and symbolic role for millennia [25]. It not only provides dates, a highly nutritious fruit [26], but it also allows the cultivation of other crops by protecting them from sun, heat and wind: this is the oasis polyculture system [27].
Traditional areas of cultivation are North Africa and the Middle East stretching as far as Pakistan and North-Western India [21]. In recent centuries, it was introduced in America, sub-Saharan Africa, Southern Europe and Oceania as a fruit crop or for ornamental and religious purposes [21]. Despite the importance of its cultivation, we possess little data about the date palm origins of domestication, historical biogeography and evolutionary history. According to archaeological data, date palm cultivation, also known as phoeniciculture, seems to emerge between the 5th and the 3rd millennium BC in the Middle East, more precisely around the Persian Gulf [25]. The cultivated date palm derives from wild populations of the same species, but in the current state of research none is securely identified [18,19]. Indeed, spontaneously growing or uncultivated populations are found within its whole distribution area [28,29] but no tangible element to differentiate wild from feral date palms has been evidenced [28]. Therefore, the status of the mentioned uncultivated date palm populations remains to be clarified.
Traditional (study of size) and geometric morphometrics (outline analysis) applied to seeds appear as two attractive and complementary tools to differentiate distinct species [30–34], distinguish wild from domesticated crops (e.g. in the olive tree [17,35], grapevine [36] or caimito [37]) and detect or suspect feral individuals [17,38]. Focusing on seeds rather than other plant organs is interesting because seeds are easily sampled and stored, and keep well. Very interestingly, they are intimately related to the fruit, i.e. the main object of selection in a fruit crop like the date palm, since the increase in seed size is likely linked allometrically to increase in fruit size [39–41]. In addition, seeds are the most abundant archaeological remains reflecting, in Egypt and the Persian Gulf area, the traditional use of date palm for over 6,000 years [25] and can thus be used to study past agrobiodiversity and the emergence of cultivation [38,42]. Phoenix seeds display a hard endosperm and are characterized by a deeply grooved raphe [21]. Seeds of Phoenix have been previously described [19,21,43,44]. They are of varying size and shape [19,21,43,44]. Length ranges from 7 mm in Phoenix roebelenii to 30 mm in cultivated date palm [21]. They are elongated in date palm cultivars while they are rounded in other Phoenix species [19]. Nevertheless, a comprehensive study combining size and shape analysis of Phoenix seeds is still required. Indeed, the aforementioned studies use qualitative descriptors for size and shape or focus only on size or shape rather than combining both information types. In addition, a recent study suggests the possible wild status of some date palm individuals spontaneously growing in Oman, based on the wild morphotype of their seeds [19]. Thus, the capability of distinguishing feral, wild and cultivated date palms based on seed morphology needs to be carefully assessed as it represents a major challenge in the understanding of date palm domestication history.
The objective of this study is to improve our knowledge of the origins of the cultivated date palm and of the morphological changes that occurred under domestication, i.e. infer the domestication syndrome affecting the seeds. A morphometric study of seeds belonging to different Phoenix species was carried out. Firstly, it aimed at evaluating the potentiality of seed size and shape to distinguish Phoenix species. Secondly, seed comparison between cultivated date palms and other Phoenix species was expected to help predicting seed size and shape in wild date palms. Because morphology expresses an essential part of the phenotype, it is an important indicator of the nature of selection pressures, including environmental constraints and anthropogenic factors. In the case of wild Phoenix species, as in that of wild date palm, the environmental context including both abiotic and biotic factors represents a set of natural selection pressures, while size and shape of cultivated date palm seeds, subjected to artificial selection, were differentiated under domestication. As a consequence, we used Phoenix non dactylifera species as a wild reference to anticipate seed size and shape in wild date palms. Our results first demonstrate that it is possible to differentiate most Phoenix species based on their seed size and shape and that seed morphometrics is a reliable tool to corroborate the species delimitation of the Phoenix species previously derived from nuclear and chloroplastic data [18,23,24]. We showed that the cultivated date palms have distinct seed features compared to wild Phoenix species and that we expect feral and wild date palms to have different phenotypes. Based on this, the uncultivated samples from Oman included in this analysis could be truly wild date palms.
Materials and Methods
Sampling
Collection of Phoenix seeds
Seeds of 13 Phoenix species (all species of the genus except P. atlantica) were analysed in this work (Table 1; Table 2).
Table 1. Phoenix dactylifera seed samples for morphometric analyses.
When different from the country of sampling, the country of origin of the cultivar is given in parenthesis. Acc. Nb.: Accession number; Nb. seed: Number of seeds.
| Cultivar name | Acc. Nb. | Country of sampling (Origin) | Sampling authorized by | Nb. seed |
|---|---|---|---|---|
| La Confitera | 0081_CON1 | Spain | Estación Phoenix | 20 |
| Iberica | 0072_IBE4 | Spain | Estación Phoenix | 20 |
| Bou Feggous | 0076_BFE1 | Spain (Morocco) | Estación Phoenix | 20 |
| Medjoul | 0083_MED1 | Spain (Morocco) | Estación Phoenix | 20 |
| Thorry | 0080_THO1 | Spain (Algeria) | Estación Phoenix | 20 |
| Ghars Mettig | 0212_GME2 | Tunisia (Algeria) | Centre de Recherches sur l’Elevage et le Pâturage, Kébili | 20 |
| Ahmar | 1249_AHM4 | Mauritania | Market | 20 |
| Tijib | 1254_TIJ2 | Mauritania | Market | 20 |
| Deglet Nour | 0186_DEG2 | Tunisia | Centre Régional de Recherches sur l’Agriculture d’Oasis (Ministry of Agriculture), Degache | 20 |
| Lagou | 0216_LAG2 | Tunisia | Private land | 20 |
| Tiswin | 1550_TIS1 | Libya | Market | 20 |
| Digla | 1552_DIG1 | Libya | Market | 20 |
| Shelabi | 0097_SHE1 | Syria (Egypt) | Market | 20 |
| Siwi | 0007_SIW3 | Egypt | Private land | 20 |
| Ibrahimi | 0093_IBR1 | Syria | Market | 20 |
| Om Asal | 0094_OMA1 | Syria | Market | 20 |
| Halaoui | 0198_HAL2 | Tunisia (Iraq) | Centre Régional de Recherches sur l’Agriculture d’Oasis (Ministry of Agriculture), Degache | 20 |
| Zaydi | 0079_ZAY1 | Spain (Iraq) | Estación Phoenix | 20 |
| Khalass | 0077_KHA1 | Spain (Saudi Arabia) | Estación Phoenix | 11 |
| Qadi | 0095_QAD1 | Syria (Saudi Arabia) | Market | 20 |
| Nashu Al Khasba | 0122_NBA1 | Oman | Wadi Qurayat Collection, Research Department of the Ministry of Agriculture | 20 |
| Khasab | 0139_KAB1 | Oman | Wadi Qurayat Collection, Research Department of the Ministry of Agriculture | 20 |
| Mozafati | 1549_MAZ1 | France (Iran) | Market | 20 |
| Iswid | 0107_ISW1 | Syria (Iran) | Market | 20 |
| Seedling | 1601_DAC492 | India | Private land | 20 |
| Seedling | 1625_DAC514 | India | Collected in the wild. No permission required. | 20 |
| Feral | 2431-DAC832 | Egypt | Collected in the wild. No permission required. | 20 |
| Feral | 2433-DAC834 | Egypt | Collected in the wild. No permission required. | 20 |
| Uncultivated | 344-WILD63 | Oman | Research Department of the Ministry of Agriculture | 20 |
| Uncultivated | 403-WILD82 | Oman | Research Department of the Ministry of Agriculture | 20 |
Table 2. Phoenix non dactylifera seed samples for morphometric analyses.
Acc. Nb.: Accession number; Nb. seed: Number of seeds.
| Species | Acc. Nb. | Location of sampling (Origin) | Nb. seed. |
|---|---|---|---|
| Phoenix acaulis | 1267_ACA4 | Ordered via internet (India) | 15 |
| Phoenix acaulis | 1720_ACA6 | Millenium Seed Bank, Kew, UK (India) | 20 |
| Phoenix acaulis | 1867_ACA7 | Herbarium Palmarum, Florence, Italy (India) | 20 |
| Phoenix acaulis | 1871-ACA8 | Herbarium Palmarum, Florence, Italy (India) | 7 |
| Phoenix andamanensis | 2139_AND2 | Royal Botanic Garden, Kew, UK (North Andaman, India) | 13 |
| Phoenix caespitosa | 1322_CAE3 | Royal Botanic Garden, Kew, UK (Somalia) | 20 |
| Phoenix caespitosa | 1878_CAE4 | Centro Studi Erbario Tropicale, Florence, Italy (Somalia) | 20 |
| Phoenix caespitosa | 1879_CAE5 | Centro Studi Erbario Tropicale, Florence, Italy (Somalia) | 20 |
| Phoenix canariensis | 0721_CAN8 | Bordighera, Italy | 20 |
| Phoenix canariensis | 0880_CAN37 | San Remo, Italy | 20 |
| Phoenix canariensis | 0092_CAN1 | Palavas, France | 20 |
| Phoenix canariensis | 1870_CAN62 | Herbarium Palmarum, Florence, Italy (Canary Islands) | 20 |
| Phoenix canariensis | 1875_CAN63 | Seed reference collection, Florence, Italy (Canary Islands) | 20 |
| Phoenix loureiroi var. pedunculata | 1722_LOR12 | Millenium Seed Bank, Kew, UK (India) | 20 |
| Phoenix loureiroi var. pedunculata | 1863_LOR14 | Herbarium Palmarum, Florence, Italy (India) | 20 |
| Phoenix loureiroi var. pedunculata | 1864_LOR15 | Herbarium Palmarum, Florence, Italy (India) | 20 |
| Phoenix loureiroi var. pedunculata | 2140_LOR17 | Millenium Seed Bank, Kew, UK (Bhutan) | 20 |
| Phoenix loureiroi var. loureiroi | 1865_LOR16 | Herbarium Palmarum, Florence, Italy (Batanes Islands, Philippines) | 20 |
| Phoenix loureiroi var. loureiroi | 2143_LOR18 | Millenium Seed Bank, Kew, UK (Thailand) | 20 |
| Phoenix paludosa | 1868_PAL4 | Herbarium Palmarum, Florence, Italy | 20 |
| Phoenix paludosa | 1869_PAL5 | Herbarium Palmarum, Florence, Italy | 20 |
| Phoenix paludosa | 1872_PAL6 | Herbarium Palmarum, Florence, Italy (Vietnam) | 10 |
| Phoenix paludosa | 2144_PAL7 | Millenium Seed Bank, Kew, UK (Thailand) | 20 |
| Phoenix pusilla | 1873_PUS5 | Herbarium Palmarum, Florence, Italy | 20 |
| Phoenix pusilla | 1874_PUS6 | Herbarium Palmarum, Florence, Italy (Sri Lanka) | 20 |
| Phoenix pusilla | 2141_PUS7 | Millenium Seed Bank, Kew, UK (Sri Lanka) | 20 |
| Phoenix pusilla | 2142_PUS8 | Millenium Seed Bank, Kew, UK (India) | 20 |
| Phoenix pusilla | 2145_PUS9 | Millenium Seed Bank, Kew, UK (Sri Lanka) | 20 |
| Phoenix reclinata | 0441_REC1 | Madagascar | 20 |
| Phoenix reclinata | 0443_REC2 | Montpellier herbarium (Benin) | 20 |
| Phoenix reclinata | 0766_REC14 | San Remo, Italy | 20 |
| Phoenix reclinata | 0771_REC15 | San Remo, Italy | 20 |
| Phoenix reclinata | 1321_REC40 | Millenium Seed Bank, Kew, UK (Tanzania) | 20 |
| Phoenix reclinata | 1719_REC42 | Millenium Seed Bank, Kew, UK (Gabon) | 20 |
| Phoenix roebelenii | 0906_ROE4 | San Remo, Italy | 20 |
| Phoenix rupicola | 1721_RUP10 | Millenium Seed Bank, Kew, UK (India) | 9 |
| Phoenix rupicola | 1866_RUP11 | Herbarium Palmarum, Florence, Italy (India) | 20 |
| Phoenix rupicola | 1876_RUP12 | Seed reference collection, Florence, Italy (India) | 20 |
| Phoenix rupicola | 1877_RUP13 | Seed reference collection, Florence, Italy (India) | 20 |
| Phoenix sylvestris | 1653_SYL20 | Kathiawar peninsula, Gujarat, India | 20 |
| Phoenix sylvestris | 1680_SYL47 | Aravalli range, Rajasthan, India | 20 |
| Phoenix sylvestris | 1688_SYL55 | Aravalli range, Rajasthan, India | 20 |
| Phoenix sylvestris | 1689_SYL56 | Aravalli range, Rajasthan, India | 20 |
| Phoenix sylvestris | 1696_SYL63 | Marwar region, Rajasthan, India | 20 |
| Phoenix sylvestris | 1702_SYL69 | Marwar region, Rajasthan, India | 20 |
| Phoenix sylvestris | 1709_SYL76 | Udaipur district, Rajasthan, India | 20 |
| Phoenix sylvestris | 1712_SYL79 | Udaipur district, Rajasthan, India | 20 |
| Phoenix sylvestris | 1714_SYL81 | Udaipur district, Rajasthan, India | 20 |
| Phoenix sylvestris | 1718_SYL85 | Udaipur district, Rajasthan, India | 20 |
| Phoenix theophrasti | 1461_THE19 | Vai, Crete, Greece | 20 |
| Phoenix theophrasti | 1462_THE20 | Vai, Crete, Greece | 20 |
| Phoenix theophrasti | 1478_THE36 | Martsalos, Crete, Greece | 20 |
| Phoenix theophrasti | 1479_THE37 | Martsalos, Crete, Greece | 20 |
| Phoenix theophrasti | 2146_THE82 | Millenium Seed Bank, Kew, UK (Crete, Greece) | 20 |
Samples belonging to P. dactylifera consist in 26 cultivated date palms among which 24 are cultivars (clones) and two are seedlings. They originate from 30 different countries spanning the whole date palm distribution. The origin of these samples is stated as the country of sampling for seedlings and the country where it supposedly originates for cultivars (i.e. although the Deglet Noor cultivar is grown in Arabia, it is well known that it originates from Tunisia). It is important to note that the seed shape of a cultivar is only slightly affected by environmental conditions as it was previously evidenced [19]. Indeed, the same cultivar grown in two different countries display more closely related seeds than other cultivars studied [19]. The utilization of samples grown in a different region than the country of natural origin is therefore not of concern for this study. Seeds from four uncultivated date palms were also included: two feral date palms from an ancient and abandoned Egyptian palm grove as well as two potentially wild date palms growing in Oman [19] (Table 1). Most samples were collected on private lands or in collections and for any location, the landowner or the authority responsible for the collection gave us permission. For samples bought on markets or collected in the wild, no permission was required as the date palm is not an endangered or protected species and the collection was not carried out in national parks or other protected areas.
Material from other Phoenix species mostly comes from approved herbaria and collections (Royal Botanic Gardens, Millennium Seed Bank, and Carpological Collection, Kew, UK; Herbarium Palmarum, seed reference collection, Centro Studi Erbario Tropicale, Firenze, Italy; Montpellier herbarium of the Institute of Botany, France) and each location issued a specific permission to sample for this study. The material from collections was photographed on site and the photographs were subsequently used in the analyses. Some seeds were however directly collected in the field and identified without any doubt (Table 2). In those cases, specific permission was not required as the collected species are not protected or endangered and the collection was not carried out in national parks or other protected areas. For samples collected in herbaria, the region/country of origin attributed was defined as the original country of origin as stated on the herbarium voucher and when the information was missing or ambiguous, the origin was set as missing. Phoenix atlantica is the only missing species in this analysis, despite our tremendous effort in sampling. Previously considered as feral date palms or the product of hybridization between several Phoenix species [21,45], it has been only recently recognized as a distinct species [45]. Given its close morphology to the date palm [45,46] and its only recent status as a distinct species, sampling this species would necessitate a careful examination of specimens on its endemic Islands, Cape Verde, rather than sampling in herbarium where Phoenix atlantica samples might be date palms or hybrids.
As previously reported, non dactylifera Phoenix species (Table 2) are used as a “wild” reference compared to cultivated date palms in order to infer morphometrical features of wild date palm seeds. Although these species may be used by Human, they are not subject to artificial selection (undomesticated) compared to cultivated date palms and thus represents a reliable “wild” reference. They are referred as the “wild” Phoenix. Seeds were photographed in both dorsal and lateral sides in order to appreciate the real three-dimensional shapes.Number of seeds analysed per individual for a reliable characterization of morphological features
The number of Phoenix seeds to be analysed for an optimal evaluation of intra-individual shape variation when using Fourier coefficients method has already been tested and set at 20 [19]. The sample size necessary to correctly assess the seed dimension for one individual has not been tested and was therefore assessed in this study with five individuals of different species. For each of these five accessions, we randomly sampled one to 30 seeds. For each of these 30 subsets of different sample size, the average of the four size parameters was calculated. The number of seeds required for studying the size parameters was evaluated as the minimum number of seeds required to stabilize the mean of the dimension parameters that is the minimum number of seeds from which the averages are stable.
Describing seed size and shape using traditional and geometric morphometrics
Size analysis of seeds
Four parameters representing the seed dimensions were measured using ImageJ version 1.42 [47] (Fig 1). Length and width of seeds were measured on the dorsal view. Thickness was measured as the maximum width of the seed lateral side. The surface of the dorsal side of the seed was also measured. The correlations between each pair of size parameters were plotted and assessed using Pearson product-moment correlation tests.
Fig 1. Overview of the seed size parameters measured in this study.
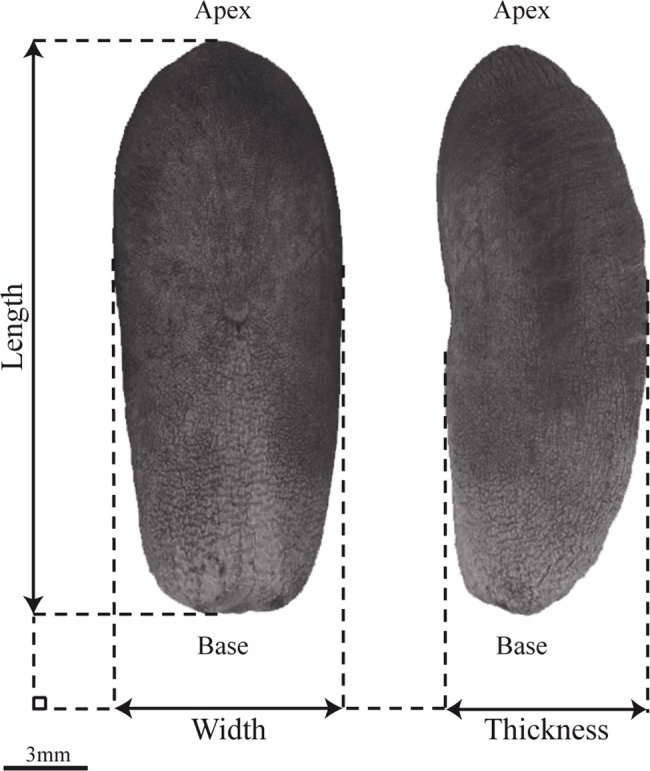
Left: dorsal view; right: lateral view.
Fourier analysis of seed outlines
Seed shape was quantified using outline analyses based on Fourier method following the protocol developed on Phoenix seeds [19,48] and implemented in R software (Momocs package [49,50]). The x and y coordinates of 64 points equally spaced along the outline of each seed were extracted semi-automatically using an image analysis system, the starting-point being the seed base. Coordinates present a high quantity of redundant information and they therefore need to be standardised for size and orientation in order to retain shape information only [51]. For this purpose, they are transformed using the Elliptic Fourier Transform (EFT) method. It is based on the separate Fourier decomposition of the incremental changes of the x and y coordinates as a function of the cumulative length along the outline [48,52]. To each harmonic n correspond four coefficients: An and Bn for x, Cn and Dn for y, defining an xy-plane. In order to retain shape information mainly, the size is standardized and seeds are oriented using the coefficients of the best-fitting ellipse of any outline, that is the first harmonic (H1) [48].
The outline is described by a maximum of 32 harmonics (in case of an outline defined by 64 points) but the information added by each harmonic decreases with the rank of the harmonic while the measurement noise increases [53–55]. The number of harmonics for an optimal description of Phoenix seed outlines was evaluated as eight (H2 to H9 after the exclusion of H1 whose coefficients correspond to residuals after standardization) [19]. As a consequence, a set of 64 Fourier coefficients (i.e. four Fourier coefficients for each of the eight harmonics for both lateral and dorsal sides) was retained and exploited in the following statistical analysis.
Discrimination of Phoenix species based on seed size and shape
The following statistical analyses were performed using the R software v2.15.3 [50].
Intra-specific variability of seed size and shape
For each species and each parameter of size, the range of values was computed as it represents the most obvious measure of variability. In order to visually inspect the variation of shape within species, a reconstruction of the mean seed outline of each sample was obtained using the inverse Fourier Transform method, following processes inverse to those used in calculating the Fourier coefficients [56].
Furthermore, the variability of size and shape within species was quantified by the dispersion of seed points around species centroid in two PCA (Principal Component Analysis, dudi.pca function) spaces; the first PCA was performed according to the four size parameters and the second according to the 64 Fourier coefficients related to both dorsal and lateral sides. For each species, the mean of the distances of seed points from the species centroid was computed. The distances were calculated as the sum of the squared distances in each PCA component, weighted by the variance explained by that component. Measure of intra-specific variability may be correlated with the number of seeds and individuals included, especially for small sample size. To standardize this measurement, we used the rarefaction method: a fixed number of seeds were randomly sampled one hundred times and the mean distance calculated over the one hundred replicates. This method allows quantification of the intra-specific variability among equal-sized samples drawn from the different species. The number of seeds to sample was evaluated at 20 (S2 Appendix). The intra-specific variability was thus calculated as the average of 100 mean distances between 20 randomly sampled seeds and the species centroid. Species represented by a single sample (Phoenix andamanensis and P. roebelenii) were excluded, as the calculation of intra-specific variability has no meaning in this case. The difference of variability of size and shape among species was tested with post-hoc Tukey’s test (HSD.test function).
Size and shape specificity of each Phoenix species
The homoscedasticity and the normality of each seed measurements were tested using Bartlett’s test and Shapiro-Wilk’s test respectively (bartlett.test and shapiro.test functions). The difference in seed dimensions between Phoenix species was tested using first nested ANOVAs (Analysis of variance) on each dimension parameter (aov function) with individual accession nested in species in order to take into account the non-independence of seeds and secondly post-hoc Tukey’s test. To evaluate the among-species differentiation of seed shape variation, a PCA was carried out on the 64 Fourier coefficients and the homoscedasticity and the normality of the coordinates were tested using Bartlett’s test and Shapiro-Wilk’s test respectively. A nested MANOVA (Multivariate analysis of variance, manova function) was performed on the first five coordinates, the explanatory variable being the species. To test the discrimination between the different Phoenix species in relation with the seed size and shape, three Linear Discriminant Analyses (LDA, lda function) were performed according to (1) size parameters, (2) 64 Fourier coefficients associated with both dorsal and lateral sides, (3) the combination of dimension parameters and 64 Fourier coefficients associated with both dorsal and lateral sides. To estimate the discriminant power of the LDAs, leave-one-out cross-validations were performed: posterior species assignations were executed for each seed (lda function with option CV = T). The discriminating rate of each species was calculated as the percentage of positive allocation.
Seed comparison between cultivated date palms and “wild” Phoenix
The seed dimension was compared among 4 groups: cultivated date palms, feral date palms, Oman uncultivated date palms of unknown status and Phoenix non dactylifera species, referred as the “wild” group, using boxplots, nested ANOVAs as well as Tukey’s test. Additionally, Student tests were undertaken for each of the four size parameters in order to compare cultivars and cultivated seedlings. The differentiation of shape among the four groups was appraised by a nested MANOVA carried out on the five first components of a PCA performed on the 64 Fourier coefficients. An LDA was undertaken on both size and shape variables on all samples except the four uncultivated date palms, in order to distinguish two groups: cultivated date palms and “wild” Phoenix. The distinction between them was assessed using the discriminant power computed with leave-one-out cross-validations as previously explained. The seeds from the four uncultivated date palms (two feral and two of unknown status) were included in the study as supplementary individuals. These individuals did not participate in the construction of the discriminant model but were projected onto the discriminant functions that were previously computed in order to predict which of the two groups they more probably belong to.
Results
Estimation of intra-individual seed sample size
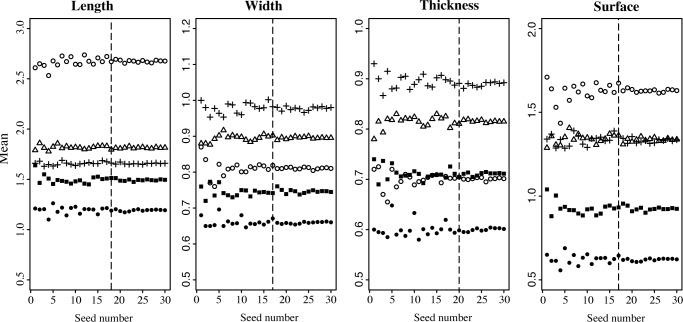
The seed number to analyse in order to stabilize the mean of the dimension parameters was quantified by randomly sampling 1 to 30 seeds in 5 individuals (Fig 2). The length mean appears stabilised, that is it stops fluctuating, with a minimum of 18 seeds. The width and surface means are stabilised with a minimum of 17 seeds. The thickness is stabilised with a minimum of 20 seeds. These results indicate that using 20 seeds is enough to describe the variability of size in Phoenix seeds, the same number as previously calculated to describe their shape [19]; the subsequent statistical analyses will thus be performed on 20 seeds per individual when available, that is a total of 1625 seeds (Table 1; Table 2).
Fig 2. Mean of seed dimensions (in mm) calculated with an increasing number of randomly sampled seeds.
White dot: 186_DEG2; Triangle: 1653_SYL20; Black square: 1876_RUP12; Black dot: 441_REC1; Cross: 1870_CAN62.
Description and comparison of seed size and shape in Phoenix species
Discrete measurements and Fourier coefficients for each seed can be found in S1 Appendix.
Difference in seed size between Phoenix species
Statistical properties on size of seeds (Fig 1) are given in Table 3 for each Phoenix species. Assumptions of data normality and homoscedasticity for the subsequent nested ANOVA were met (Shapiro-Wilk’s test: p-values < 0.05 and Bartlett's test: p-values < 0.05). The different Phoenix species display differences in size parameters (nested ANOVA, p-values < 2.10−16). The date palm displays the greatest seed length (Tukey’s test: p-value <0.05). It ranges from 1.12 to 3.16 cm with a mean of 2.08 cm while other Phoenix seeds have a maximum length of 2.02 cm, observed in P. sylvestris. It also displays the widest range of values for each size parameter, especially the length: the difference between the shortest and the longest seeds is greater than 2 cm while for other species this difference is lower than 1, except for Phoenix reclinata (Table 3). The variability in size, computed as the average dispersion of one hundred replicates of 20 randomly sampled seed points around the species centroids in a PCA space, is higher in P. dactylifera than in all other species (Table 4, Tukey’s test: p-value < 0.05) except P. reclinata.
Table 3. Mean, standard deviation, minimum (min) and maximum (max) of size parameters measured on each Phoenix species and groups derived from Tukey’s test.
| Number | Sample | Length (cm) | Width (cm) | Thickness (cm) | Surface (cm²) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| of seeds | number | mean ± sd | min | max | Tukey's group(s) | mean ± sd | min | max | Tukey's group(s) | mean ± sd | min | max | Tukey's group(s) | mean ± sd | min | max | Tukey's group(s) | |
| P. acaulis | 62 | 4 | 1.01 ± 0.12 | 0.79 | 1.33 | gh | 0.58 ± 0.06 | 0.45 | 0.76 | g | 0.51 ± 0.07 | 0.35 | 0.68 | g | 0.47 ± 0.11 | 0.32 | 0.75 | gh |
| P. andamanensis | 13 | 1 | 1.41 ± 0.06 | 1.29 | 1.5 | de | 0.75 ± 0.03 | 0.69 | 0.81 | d | 0.66 ± 0.03 | 0.63 | 0.75 | e | 0.81 ± 0.06 | 0.68 | 0.91 | e |
| P. caespitosa | 60 | 3 | 1.18 ± 0.11 | 0.93 | 1.36 | f | 0.87 ± 0.07 | 0.71 | 1.01 | c | 0.77 ± 0.06 | 0.64 | 0.88 | c | 0.84 ± 0.17 | 0.52 | 1.18 | de |
| P. canariensis | 100 | 5 | 1.51 ± 0.16 | 1.25 | 1.78 | c | 0.95 ± 0.10 | 0.74 | 1.15 | a | 0.89 ± 0.09 | 0.69 | 1.09 | a | 1.17 ± 0.23 | 0.72 | 1.64 | c |
| P. dactylifera | 591 | 30 | 2.08 ± 0.43 | 1.12 | 3.16 | a | 0.85 ± 0.10 | 0.61 | 1.14 | c | 0.77 ± 0.10 | 0.49 | 1.06 | c | 1.34 ± 0.33 | 0.627 | 2.51 | a |
| P. loureiroi | 120 | 6 | 1.05 ± 0.13 | 0.8 | 1.32 | g | 0.61 ± 0.05 | 0.52 | 0.7 | f | 0.55 ± 0.04 | 0.46 | 0.64 | f | 0.51 ± 0.09 | 0.37 | 0.71 | g |
| P. paludosa | 70 | 4 | 0.99 ± 0.08 | 0.78 | 1.23 | hi | 0.69 ± 0.06 | 0.58 | 0.78 | e | 0.54 ± 0.05 | 0.45 | 0.64 | f | 0.52 ± 0.09 | 0.35 | 0.75 | g |
| P. pusilla | 100 | 5 | 0.95 ± 0.12 | 0.7 | 1.23 | i | 0.58 ± 0.08 | 0.44 | 0.71 | g | 0.52 ± 0.08 | 0.39 | 0.64 | g | 0.45 ± 0.11 | 0.25 | 0.68 | h |
| P. reclinata | 120 | 6 | 1.16 ± 0.31 | 0.73 | 1.9 | f | 0.70 ± 0.12 | 0.52 | 1.02 | e | 0.63 ± 0.12 | 0.44 | 0.94 | e | 0.68 ± 0.31 | 0.27 | 1.57 | f |
| P. roebelenii | 20 | 1 | 0.80 ± 0.03 | 0.75 | 0.86 | j | 0.43 ± 0.02 | 0.39 | 0.47 | h | 0.35 ± 0.02 | 0.32 | 0.37 | h | 0.27 ± 0.02 | 0.23 | 0.31 | i |
| P. rupicola | 69 | 4 | 1.44 ± 0.15 | 1.2 | 1.68 | d | 0.74 ± 0.05 | 0.67 | 0.88 | d | 0.71 ± 0.06 | 0.62 | 0.88 | d | 0.88 ± 0.15 | 0.66 | 1.16 | d |
| P. sylvestris | 200 | 10 | 1.65 ± 0.20 | 1.11 | 2.02 | b | 0.92 ± 0.10 | 0.73 | 1.13 | b | 0.83 ± 0.08 | 0.65 | 1.03 | b | 1.24 ± 0.22 | 0.67 | 1.71 | b |
| P. theophrasti | 100 | 5 | 1.33 ± 0.18 | 0.97 | 1.62 | e | 0.76 ± 0.08 | 0.59 | 0.93 | d | 0.72 ± 0.07 | 0.57 | 0.85 | d | 0.81 ± 0.18 | 0.5 | 1.17 | e |
Table 4. Variability of seed dimensions and shape within Phoenix species.
It is calculated as the dispersion of seeds around the related species’ centroid in two PCA spaces obtained from size parameters (Size Var.) and 64 Fourier coefficients related to dorsal and lateral seed shapes (Shape Var.) using the rarefaction method. The values are the average over the mean distance between 20 randomly sampled seeds in one hundred replicates and the standard deviation over the one hundred replicates. The groups derived from Tukey’s test are given into parentheses.
| Species | Number of seeds | Size Var. (Tukey’s group) | Shape Var. (Tukey’s group) |
|---|---|---|---|
| Phoenix acaulis | 62 | 31.22 ± 6.59 (g) | 284.91 ± 37.20 (c) |
| Phoenix caespitosa | 60 | 39.15 ± 8.94 (fg) | 238.57 ± 18.52 (d) |
| Phoenix canariensis | 100 | 91.09 ± 20.14 (c) | 224.32 ± 33.58 (e) |
| Phoenix dactylifera | 591 | 128.02 ± 33.05 (b) | 504.32 ± 76.44 (a) |
| Phoenix loureiroi | 120 | 17.57 ± 3.25 (e) | 205.71 ± 34.83 (e) |
| Phoenix paludosa | 70 | 20.49 ± 5.21 (h) | 178.87 ± 23.21 (f) |
| Phoenix pusilla | 100 | 46.35 ± 6.12 (f) | 151.48 ± 19.24 (g) |
| Phoenix reclinata | 20 | 177.85 ± 43.14 (a) | 269.30 ± 40.33 (c) |
| Phoenix rupicola | 69 | 34.73 ± 6.90 (g) | 171.17 ± 19.78 (fg) |
| Phoenix sylvestris | 200 | 72.40 ± 22.30 (d) | 432.62 ± 58.67 (b) |
| Phoenix theophrasti | 100 | 57.43 ± 12.11 (e) | 217.14 ± 34.99 (e) |
Difference in seed shape between Phoenix species
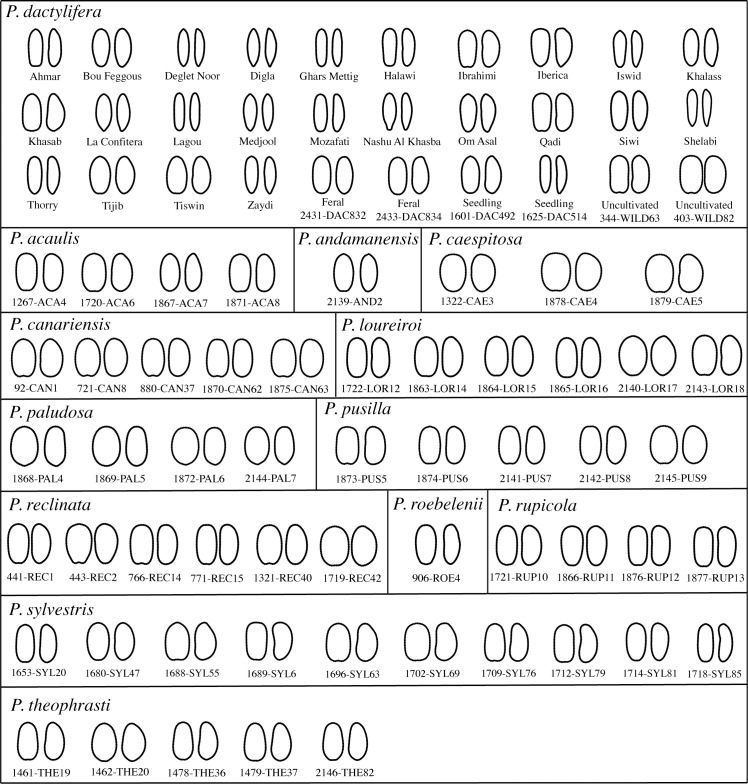
The mean outline of the 20 seeds for each individual was reconstructed using the inverse Fourier Transform method [56] (Fig 3). The existence of a seed shape difference among the different Phoenix species was tested with a nested MANOVA applied to the first five components over the components of a PCA analysis (explaining 51.38% of the variability) carried out on 64 Fourier coefficients after the homoscedasticity and the normality of the data were checked (Shapiro-Wilk’s test: p-values < 0.05 and Bartlett's test: p-values < 0.05). It indicates that a seed shape differentiation exists among the 13 Phoenix species included in this study (p-value < 0.01).
Fig 3. Reconstructed mean outlines of each individual included in this study using the inverse Fourier Transform method.
Left: dorsal side; right: lateral side.
Like in the case of size parameters, we visually observe that the seed shape of date palms is greatly diversified compared to other Phoenix species’ (Fig 3) and this variability is reflected in the value of dispersion computed for each species (Table 4). This species appears more variable in shape than any other in the genus Phoenix (Tukey’s test: p-values<0.05). Within Phoenix loureiroi, Phoenix rupicola, P. reclinata, P. sylvestris and P. dactylifera, several morphotypes can be visually evidenced (Fig 3). Two sub-species of P. loureiroi are included here but the different morphotypes are not allocated to either of them so that the distinction between subspecies based on seed shape is not possible from these samples.
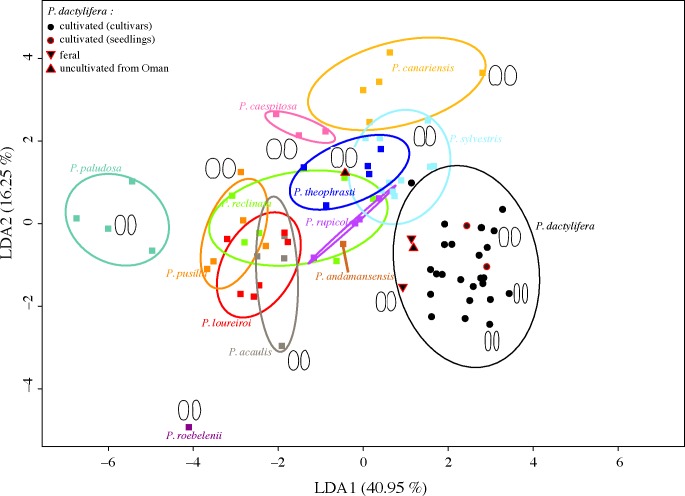
Characterizing Phoenix species based on seed size and shape
The LDA performed on both size and shape variables with the species being the discriminant parameter is plotted in Fig 4. The first axis represents the shape of seeds: on the left, species with rounded seeds like Phoenix paludosa are found, while at the right end is the date palm, displaying elongated seeds (Fig 4). The second axis (16.25%) is related to both size and shape. Indeed, Phoenix non dactylifera species are distributed upwards from the species with the smallest seeds (P. roebelenii) to the one with the largest (Phoenix canariensis), while the date palm, the species with the largest but elongated seeds, is found in the middle. Some species like P. paludosa and P. canariensis constitute a distinct group/cloud while the cloud of some species are overlapping like those of Phoenix acaulis and P. loureiroi. The addition of a third axis does not solve the overlapping problem. The cultivated date palm seeds constitute a distinguishable group although close to P. sylvestris. Seeds from uncultivated date palms are not found within the cultivated group but rather between the “wild” group and the cultivated group or in one case within the “wild” group. However, when the LDA is performed on shape only, the feral individuals are found within the points cloud of P. dactylifera while one of the uncultivated individuals form Oman is found within the “wild” Phoenix cloud (S1 Fig).
Fig 4. Linear Discriminant Analysis biplot of axis 1 (40.95% of variance explained) and axis 2 (16.25% of variance explained) performed on discrete measurements and 64 Fourier coefficients representing the dorsal and lateral seed shapes of 13 Phoenix species.
Dorsal (left) and lateral (right) mean outlines are plotted for several individuals.
The mean percentage of correct allocation for each species computed with three different LDAs is given in Table 5. The size parameters alone do not allow a good distinction of species (36.4% on average). When using shape (dorsal and lateral sides combined), the average discriminant power increases substantially, reaching 68.8%. The addition of the dimension parameters to shape further increases the positive allocation for each species, with a mean of 79.5%. P. paludosa, P. caespitosa, P. canariensis and P. roebelenii appear as the easiest species to differentiate with a percentage of positive allocation above 90%. On the contrary, P. acaulis, P. loureiroi and P. reclinata are hardly differentiated from other Phoenix (percentage of positive allocation under 65%). Phoenix dactylifera is distinguished from other species at 87.8%. More specifically, when taking into account only cultivated date palms by discarding the uncultivated samples (feral from Egypt and uncultivated from Oman) this number reaches 92.9%.
Table 5. Discriminant power (in percentage) of the dimensions and/or shape of seeds in the genus Phoenix.
| Size | Outline | Size and outline | |
|---|---|---|---|
| P. acaulis | 17.7 | 36.2 | 48.1 |
| P. andamanensis | 6 | 64.4 | 82.9 |
| P. caespitosa | 29.9 | 89.9 | 94.5 |
| P. canariensis | 41.8 | 81.7 | 93 |
| P. dactylifera | 68.8 | 84.3 | 87.8 |
| P. loureiroi | 30.7 | 45.9 | 64.3 |
| P. paludosa | 74.7 | 97.2 | 97.4 |
| P. pusilla | 29.3 | 56.6 | 73.5 |
| P. reclinata | 16.1 | 44.3 | 50.2 |
| P. roebelenii | 59 | 84.6 | 95 |
| P. rupicola | 21.9 | 69.1 | 79.8 |
| P. sylvestris | 46.9 | 57.1 | 77.9 |
| P. theophrasti | 30.2 | 82.8 | 88.8 |
| Mean | 36.4 | 68.8 | 79.5 |
Morphological features of cultivated date palm seeds compared to other Phoenix species
Correlation between size parameters
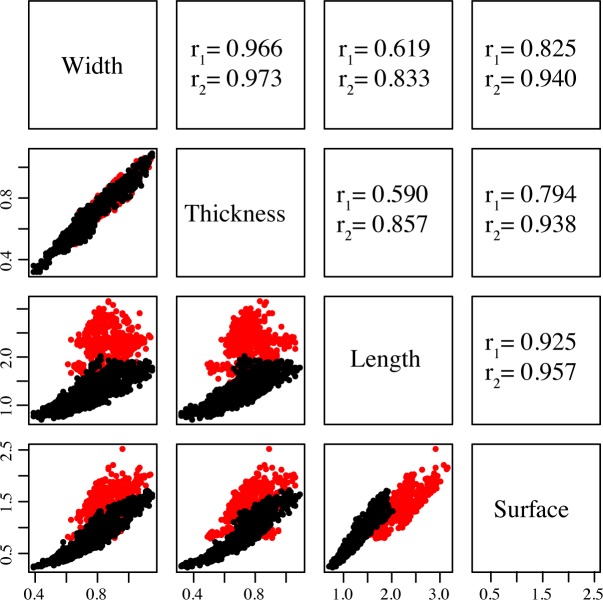
Each pair of correlations between size parameters is significant (Fig 5; p-values << 0.01). When all the Phoenix species are included, the width and the thickness are highly correlated (r = 0.966) as well as the length and the surface of the dorsal side (r = 0.925). When discarding P. dactylifera from the correlation tests, both the thickness and the width were highly correlated with the length (r = 0.857 and r = 0.833 respectively).
Fig 5. Correlation of size parameters.
Red: Phoenix dactylifera, black: all Phoenix except P. dactylifera, in mm. Each pair of correlations is plotted on the lower left side while the coefficients of correlation r are given on the upper right-hand side: r1 corresponds to the correlation coefficients when all Phoenix species are included while r2 were calculated after discarding the date palm P. dactylifera.
Difference in seed size and shape in cultivars versus “wild” species
Seed length of cultivated date palms, feral date palms from Egypt, uncultivated date palms of unknown status from Oman and other Phoenix species (“wild”) is plotted in Fig 6. The boxplots related to the other three parameters measured may be found in S2 Fig. Within the cultivated date palm group, the seed size of seedlings (1601_DAC492 and 1625_DAC514) is comparable to the seed size of cultivars (Student tests: p-values >> 0.05). The nested ANOVAs performed on each parameter indicate that size is different according to the group they belong to (p-values < 0.01). Post-hoc Tukey’s tests between the 4 different statuses for the four size parameters are all significant (p-values < 0.05) except the width between uncultivated date palms from Oman and wild Phoenix. The cultivated date palm seeds display the greatest length, width, thickness and surface (Tukey’s test: p-value < 0.05). The feral date palm seeds from Egypt are smaller than the cultivated date palms seeds and larger than seeds of uncultivated individuals from Oman and of wild Phoenix species (Tukey’s test: p-value < 0.05). Uncultivated date palms from Oman have smaller seeds than cultivated and feral ones (Tukey’s test: p-value < 0.05). Additionally, the shape is also influenced by the status (nested MANOVA, p-value < 0.01). The LDA combining both size and shape features and performed to differentiate cultivated date palms from “wild” Phoenix provides a mean discriminant power of 94.7%: 90.5% of the cultivated seeds and 99.0% of the “wild” seeds are a posteriori positively allocated to their group. Within the two feral samples (individuals) from Egypt, 12 out of 40 seeds (30.0%) were allocated to the cultivated group while others were allocated to the “wild” group. The seeds from the supposed spontaneous date palm originating from Oman were all but one (97.5%) allocated to the “wild” group.
Fig 6. Comparison of seed length (in mm) between date palms from cultivated, feral and uncultivated populations of unknown status with wild Phoenix.
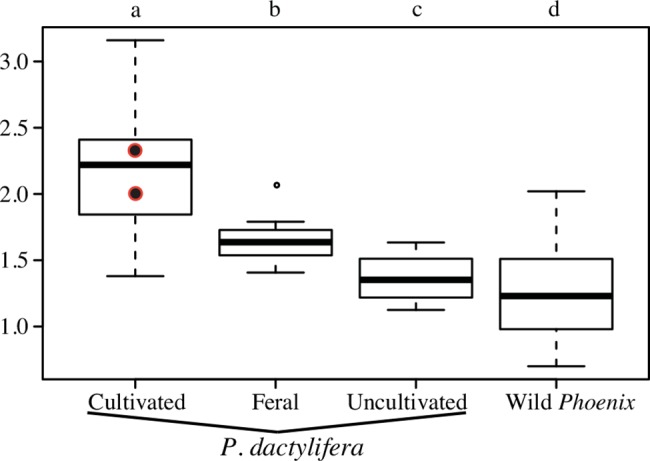
The two black dots with red contour represent the average of seed length for the two cultivated date palm seedlings from India (1601_DAC492 and 1625_DAC514).
Discussion
Distinction of Phoenix species based on seed morphological features in the light of molecular data
While size seems weakly discriminant, the shape of seeds is highly distinctive for some species such as P. paludosa, almost a hundred percent discriminated from the other Phoenix species (Table 5). The combination of both seed size and shape provides a good rate of discrimination among Phoenix species (79.5%). However, some of them remain poorly discriminated. This is probably because of a high intraspecific variability (P. reclinata, Table 4) and/or a strong morphological similarity between species (P. acaulis and P. loureiroi). The high variability in size observed in the seeds of P. reclinata (Table 4) questions the existence of different ecotypes as previously proposed [21] and remains to be investigated with extensive sampling in relation to environmental parameters. Morphometrics of seeds thus appears as an efficient tool to differentiate most Phoenix species and should be considered for the identification of hybrids as previously stated [57].
Based on the analysis of chloroplastic sequences, P. sylvestris and P. atlantica are the closest relatives of the date palm P. dactylifera [18]. On the basis of seed size and shape results, P. dactylifera appears close to P. sylvestris (Figs 3 and 4), thus in agreement with genetic data. Phoenix atlantica is absent from the morphometric analysis thus its morphological proximity with the date palm remains to be assessed. This morphometric study thus corroborate genetic data [18,23,24] since these two methods allow to distinguish most Phoenix species and identify the date palm’s closest relatives.
Seed peculiarities of date palms in the genus Phoenix and emphasis on the domestication syndrome
The variability in seed size and shape was assessed among equal-sized samples drawn from the different Phoenix species with a rarefaction method. The great variability of seed size and shape within the cultivated date palm was evidenced (Tables 3 and 4; Fig 3). The pattern of great phenotypic variability in cultivated species is well documented as a consequence of varietal diversification through space and time [58]. For the date palm, it may reflect its long-term history of cultivation associated with selection of traits (including fruit size and correlatively seed size), breeding and human-mediated diffusion [19].
The seeds from cultivated date palms are easily discriminated from those of wild other Phoenix species. On the one hand, seeds of “wild” species are smaller (Table 3; Fig 6) and rounded (Fig 3) and a strong correlation between their width/thickness and length was shown (Fig 5). On the other hand, seeds of cultivated date palms, whether they are seedlings or cultivars vegetatively propagated with offshoots, are longer (Table 3; Fig 6) and elongated (Fig 3), and they show no correlation pattern between thickness/width and length (Fig 5). These differences may be explained by divergent selection pressures leading to different patterns of morphological changes through time. Indeed, wild Phoenix are subject to a set of selection pressures including environmental constraints conditioning morphological evolution through time. Cultivated date palms are rather the subject of repeated strong human constraints related to cultivation practices that explain these particular phenotypes [19]. Moreover, seeds of wild Phoenix species and date palms growing without human influence (referred here as uncultivated date palms) seem to be submitted to constraints tending to minimize seed size and to standardize their phenotype. The canalization process, i.e. the ability of the organism to produce a constant phenotype despite genetic and/or environmental effects [59,60] leading in such palms to produce seeds with a similar phenotype (small and rather rounded), may be involved. As a corollary, the increase in seed length between wild progenitor and domesticated plant is a pattern observed for cereals [10,12], beans (e.g. soybean [61]) and fruit trees (e.g. Caimito [37], olive tree [35] and grapevine [36]). It has been shown to be correlated with an increase in fruit size [39–41]. Therefore, morphological changes or phenotypic trajectories from "wild" Phoenix species to cultivated date palm morphotype may be interpreted as a drastic shift related to selection pressures, and may be considered as a syndrome of domestication. As a result, we expect wild date palms to display small and rounded seeds; in addition, their length should be correlated to their width, thickness and surface.
The seeds of the four uncultivated individuals from Oman and Egypt appear smaller than that of the cultivated date palms and longer than that of “wild” Phoenix (Fig 6). Although the shift in seed size between wild and domesticated plants is related to artificial selection as stated before, the size of the seeds is also influenced by environmental and developmental factors as demonstrated for other models such as the olive tree [35]. Therefore, in the case of a search for distinctive criteria at the intra-specific level, seed size seems to be uninformative to distinguish feral from wild date palms as both may display small seeds as a result of constraining environmental conditions, while seeds from domesticated individuals develop large seeds as a consequence of selection and cultivation practices (irrigation and fertilization). In contrast, shape descriptors such as those used in this work were shown to be only slightly affected by environmental parameters and more powerful in a biosystematic point of view [19,34,38]. Feral date palms from Egypt display seeds presenting genuine affinity with cultivated date palms (Figs 3, 4 and 6). On the other hand, the uncultivated date palms from Oman show a morphology converging toward a wild Phoenix morphotype both in seed size and shape (Figs 3, 4 and 6). Thus, on the basis of morphometric data, we suggest that the individuals from Oman studied in this work may be true wild individuals even if some may have been introgressed by varieties cultivated in the region. Genetic analysis of these Oman populations are required to validate their wild status.
Conclusion & Prospects
Through a morphometric approach combining traditional and geometric morphometrics, this study provides new and accurate insights into morphological changes of seed that occurred under domestication (i.e. syndrome of domestication). It allows us to discuss the possible existence of wild Phoenix dactylifera populations in the Middle East and thus the origins of the date palm. This study opens up exciting prospects for research and exploration of wild date palm populations that will represent a great challenge in preservation and conservation of biological resources.
In the future, predictive morphometric models applied to seed and previously validated by genetics will be applied to archaeological seeds such as those found in Miri Qalat and Shahi Tump, Pakistan [25]. A collaborative morphometric, genetic and archaeological approach will allow us to unravel the origins, the history, the historical biogeography and the evolution of the date palm through space and time.
This study includes the description of a pipeline of statistical analyses for (1) selecting the accurate number of seeds per sample, (2) quantifying and comparing seed size and shape and (3) studying the variability using a rarefaction method to equalize sample size. It could be applied to other crops and it thus constitutes a comprehensive methodology for the study of the domestication syndrome in seeds.
Supporting Information
First sheet “Discrete measures” contains the measure of length, width, thickness (cm) and surface (cm2). Second sheet “Fourier coefficients” contains the Fourier coefficients obtained from the outline analyses based on Fourier method. Column C to AH contains the measures for dorsal view (name of the column suffixed with VD) and columns AI to BN contain the measures of the lateral side (suffixed with VL). In the name of those columns, A, B, C and D refers to the four coefficients of each harmonic and the following number to the number of the harmonic. For both sheets, first column (“Sample”) is the name of the sample; second column (“Seed”) is the number of the seed (D1 to D20 given that 20 seeds, when available, were analyzed for each sample) prefixed by the name of the sample.
(XLSX)
(DOCX)
Based on (A) four seed size parameters and (B) seed shape (64 normalized elliptic Fourier coefficients).
(PDF)
(PDF)
Acknowledgments
Dr. Jean-Christophe Pintaud passed away before the submission of the final version of this manuscript. This article is dedicated to our colleague and friend who died prematurely. We are immensely grateful to Jean-Christophe Pintaud. Dr. Muriel Gros-Balthazard accepts responsibility for the integrity and validity of the data collected and analyzed. We thank the Royal Botanic Gardens, the Millennium Seed Bank, and the Carpological Collection, Kew (UK); the Herbarium Palmarum, the seed reference collection and the Centro Studi Erbario Tropicale, Firenze (Italy); the Montpellier herbarium of the Institute of Botany, France; the Centre de Recherches sur l’Elevage et le Pâturage, Kébili, Tunisia; the Centre Régional de Recherches sur l’Agriculture d’Oasis (Ministry of Agriculture), Degache, Tunisia; the Research Department of the Ministry of Agriculture of Oman who issued the permission of seed collection for this study. We acknowledge Susi Gomez and Michel Ferry, Phoenix station, Elche, Spain and Vincent Battesti, CNRS-MNHN, France. We are grateful to the many collaborators who gave us permission to collect seeds on their private lands. This work was supported by the Agence Nationale de la Recherche (http://www.agence-nationale-recherche.fr/): programme blanc, ANR-06-BLAN-0212-02-PHOENIX project (dir. M. Tengberg; WP coordinated by J.-F. Terral) and ANR-07-BLAN-003301 –FRUCTIMEDHIS project (dir. A. Durand; WP coordinated by J.-F. Terral). This work was also supported by PhD fellowships from the French Ministry of Research granted to M.G.-B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This article is the ISEM contribution n° ISEM 2016–032.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Agence Nationale de la Recherche (http://www.agence-nationale-recherche.fr/): programme blanc, ANR-06-BLAN-0212-02-PHOENIX project (dir. M. Tengberg; WP coordinated by J.-F. Terral) and ANR-07-BLAN-003301 – FRUCTIMEDHIS project (dir. A. Durand; WP coordinated by J.-F. Terral). This work was also supported by PhD fellowships from the French Ministry of Research granted to M.G.-B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haussmann BIG, Parzies HK, Presterl T, Susic Z, Miedaner T. Plant genetic resources in crop improvement. Plant Genet Resour Charact Util. 2004; 2: 3–21. 10.1079/PGR200430 [DOI] [Google Scholar]
- 2.IPCC. Emissions Scenarios—A Special Report of Working Group III of the Intergovernmental Panel on Climate Change. Nakicenovi N, Swart R, editors. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- 3.Lutz W, Sanderson W, Scherbov S. The end of world population growth. Nature. 2001; 412: 543–545. 10.1038/35087589 [DOI] [PubMed] [Google Scholar]
- 4.Rosegrant MW, Cline SA. Global food security: challenges and policies. Science. 2003; 302: 1917–9. 10.1126/science.1092958 [DOI] [PubMed] [Google Scholar]
- 5.Harlan JR. Genetic Resources in Wild Relatives of Crops. Crop Sc; 1976; 16: 329 10.2135/cropsci1976.0011183X001600030004x [DOI] [Google Scholar]
- 6.Breton C, Terral J-F, Pinatel C, Medail F, Bonhomme F, Berville A. The origins of the domestication of the olive tree. C R Biol. 2009; 332: 1059–1064. 10.1016/j.crvi.2009.08.001 [DOI] [PubMed] [Google Scholar]
- 7.Gao L-Z, Innan H. Nonindependent domestication of the two rice subspecies, Oryza sativa ssp. indica and ssp. japonica, demonstrated by multilocus microsatellites. Genetics. 2008; 179: 965–976. 10.1534/genetics.106.068072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mamidi S, Rossi M, Annam D, Moghaddam S, Lee R, Papa R, et al. Investigation of the domestication of common bean (Phaseolus vulgaris) using multilocus sequence data. Funct Plant Biol. 2011; 38: 953–967. 10.1071/fp11124 [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka Y, Vigouroux Y, Goodman MM, Sanchez GJ, Buckler E, Doebley J. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci U S A. 2002; 99: 6080–6084. 10.1073/pnas.052125199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature. 2009; 457: 843–848. 10.1038/nature07895 [DOI] [PubMed] [Google Scholar]
- 11.Fuller DQ. Contrasting patterns in crop domestication and domestication rates: Recent archaeobotanical insights from the old world. Ann Bot. 2007; 100: 903–924. 10.1093/aob/mcm048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willcox G. Measuring grain size and identifying Near Eastern cereal domestication: evidence from the Euphrates valley. J Archaeol Sci. 2004; 31: 145–150. 10.1016/j.jas.2003.07.003 [DOI] [Google Scholar]
- 13.Miller AJ, Gross BL. From forest to field: perennial fruit crop domestication. Am J Bot. 2011; 98: 1389–1414. 10.3732/ajb.1000522 [DOI] [PubMed] [Google Scholar]
- 14.Meyer RS, DuVal AE, Jensen HR. Patterns and processes in crop domestication: an historical review and quantitative analysis of 203 global food crops. New Phytol. 2012; 196: 29–48. 10.1111/j.1469-8137.2012.04253.x [DOI] [PubMed] [Google Scholar]
- 15.Bronzini de Caraffa V, Maury J, Gambotti C, Breton C, Bervillé A, Giannettini J. Mitochondrial DNA variation and RAPD mark oleasters, olive and feral olive from Western and Eastern Mediterranean. Theor Appl Genet. 2002; 104: 1209–1216. [DOI] [PubMed] [Google Scholar]
- 16.Levadoux L. Les populations sauvages et cultivées de Vitis vivifera L. Ann l’amélioration des plantes. 1956; 1: 59–118. [Google Scholar]
- 17.Newton C, Christine L, Caroline S, Sarah I, Terral J-F. On the origins and spread of Olea europaea L. (olive) domestication: evidence for shape variation of olive stones at Ugarit, Late Bronze Age, Syria—a window on the Mediterranean Basin and on the westward diffusion of olive varieties. Veg Hist Archaeobot. 2014; 23: 567–575. [Google Scholar]
- 18.Pintaud J-C, Zehdi S, Couvreur T, Barrow S, Henderson S, Aberlenc-Bertossi F, et al. Species delimitation in the genus Phoenix (Arecaceae) based on SSR markers, with emphasis on the identity of the Date Palm (Phoenix dactylifera L.) Seberg O, Petersen G, Barford A, Davis J, editors. Divers phylogeny, Evol monocotyledons. Aarhus Univ Press, Denmark: 2010; 267–286. [Google Scholar]
- 19.Terral J-F, Newton C, Ivorra S, Gros-Balthazard M, Tito de Morais C, Picq S, et al. First insights into the complex structure of date palm agrobiodiversity (Phoenix dactylifera L.) and history of ancient Egyptian cultivated forms assessed by geometric morphometrical analysis of modern and archaeological seeds. J Biogeogr. 2012; 39: 929–941. [Google Scholar]
- 20.Gros-Balthazard M, Newton C, Ivorra S, Tengberg M, Pintaud J-C, Terral J-F. Origines et domestication du palmier-dattier (Phoenix dactylifera L.) : Etat de l’art et perpectives d'étude. Rev d’ethnoécologie. 2013; 4. [Google Scholar]
- 21.Barrow S. A revision of Phoenix L. (Palmae: Coryphoideae). Kew Bull. 1998;53: 513–575. [Google Scholar]
- 22.Govaerts R, Dransfield J. World checklist of palms. Kew, U.K.: Royal Botanic Gardens. Royal Botanic Gardens, Kew; 2005. [Google Scholar]
- 23.Ballardini M, Mercuri A, Littardi C, Abbas S, Couderc M, Ludeña B, et al. The chloroplast DNA locus psbZ-trnfM as a potential barcode marker in Phoenix L. (Arecaceae). Zookeys. Pensoft Publishers; 2013; 365: 71–82. 10.3897/zookeys.365.5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pintaud J-C, Ludena B, Zehdi S, Gros-Balthazard M, Ivorra S, Terral J-F, et al. Biogeography of the date palm (Phoenix dactylifera L., Arecaceae): insights on the origin and on the structure of modern diversity. ISHS Acta Hortic. 2013; 994: 19–36. [Google Scholar]
- 25.Tengberg M. Beginnings and early history of date palm garden cultivation in the Middle East. J Arid Environ. Elsevier Ltd. 2012; 86: 139–147. 10.1016/j.jaridenv.2011.11.022 [DOI] [Google Scholar]
- 26.Habib HM, Ibrahim WH. Nutritional quality of 18 date fruit varieties. Int J Food Sci Nutr. Informa HealthcareLondon. 2011; 62: 544–51. 10.3109/09637486.2011.558073 [DOI] [PubMed] [Google Scholar]
- 27.Ferry M, Toutain G. Concurrence et complémentarité des espèces végétales dans les oasis Dollé V, Toutain G, editors. Options Méditerranéennes. Montpellier: : CIHEAM; 1990; 11: 261–270. [Google Scholar]
- 28.Tengberg M. Research into the origins of date palm domestication The date palm: from traditional resource to green wealth. Abu Dhabi: Emirates Center for Strategic Studies and Research; 2003; 51–64. [Google Scholar]
- 29.Zohary D, Hopf M, E. W. Domestication of plants in the Old World (3rd Ed.). Oxford, UK Oxford Univ Press; 2012. [Google Scholar]
- 30.Wilkin P. A morphometric study of Dioscorea quartiniana (Dioscoreaceae). Kew Bull. 1999; 54: 1–18. [Google Scholar]
- 31.Lanta V, Havranek P, Ondrej V. Morphometry analysis and seed germination of Amaranthus cruentus, A. retroflexus and their hybrid (A. x turicensis). Plant Soil Environ. 2003; 49: 364–369. [Google Scholar]
- 32.Aybeke M. Pollen and seed morphology of some Ophrys L. (Orchidaceae) taxa. J Plant Biol. 2007; 50: 387–395. [Google Scholar]
- 33.Chemisquy MA, Prevosti FJ, Morrone O. Seed morphology in the tribe Chloraeeae (Orchidaceae): combining traditional and geometric morphometrics. Bot J Linn Soc. 2009; 160: 171–183. 10.1111/j.1095-8339.2009.00968.x [DOI] [Google Scholar]
- 34.Cope JS, Corney D, Clark JY, Remagnino P, Wilkin P. Plant species identification using digital morphometrics: A review. Expert Syst Appl. Elsevier Ltd. 2012; 39: 7562–7573. 10.1016/j.eswa.2012.01.073 [DOI] [Google Scholar]
- 35.Terral JF, Alonso N, Capdevila RBI, Chatti N, Fabre L, Fiorentino G, et al. Historical biogeography of olive domestication (Olea europaea L.) as revealed by geometrical morphometry applied to biological and archaeological material. J Biogeogr. 2004; 31: 63–77. [Google Scholar]
- 36.Bouby L, Figueiral I, Bouchette A, Rovira N, Ivorra S, Lacombe T, et al. Bioarchaeological insights into the process of domestication of grapevine (Vitis vinifera L.) during Roman times in Southern France. PLoS One; 2013; 8: e63195 10.1371/journal.pone.0063195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker IM, Lopez I, Petersen JJ, Anaya N, Cubilla-Rios L, Potter D. Domestication Syndrome in Caimito (Chrysophyllum cainito L.): Fruit and Seed Characteristics. Econ Bot. 2010; 64: 161–175. 10.1007/s12231-010-9121-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terral J-F, Tabard E, Bouby L, Ivorra S, Pastor T, Figueiral I, et al. Evolution and history of grapevine (Vitis vinifera) under domestication: new morphometric perspectives to understand seed domestication syndrome and reveal origins of ancient European cultivars. Ann Bot. 2010; 105: 443–455. 10.1093/aob/mcp298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuller D. New archaeobotanical information on plant domestication from macro-remains: Tracking the evolution of domestication syndrome traits In: Gepts P, Famula TR, Bettinger RL, Brush SB, Damania AB, McGuire PE, et al. , editors. Biodiversity in Agriculture: Domestication, Evolution, and Sustainability. Cambridge University Press; 2012; 110–135. [Google Scholar]
- 40.Bolmgren K, Eriksson O. Seed mass and the evolution of fleshy fruits in angiosperms. Oikos. 2010; 119: 707–718. 10.1111/j.1600-0706.2009.17944.x [DOI] [Google Scholar]
- 41.Alcantara JM, Rey PJ. Conflicting selection pressures on seed size: evolutionary ecology of fruit size in a bird-dispersed tree, Olea europaea. J Evol Biol. 2003; 16: 1168–1176. 10.1046/j.1420-9101.2003.00618.x [DOI] [PubMed] [Google Scholar]
- 42.Burger P, Terral J-FF, Ruas M-PP, Ivorra S, Picq S. Assessing past agrobiodiversity of Prunus avium L. (Rosaceae): a morphometric approach focussed on the stones from the archaeological site Htel-Dieu (16th century, Tours, France). Veg Hist Archaeobot. 2011; 20: 447–458. 10.1007/s00334-011-0310-6 [DOI] [Google Scholar]
- 43.Beccari O. Rivista monografica delle specie del genero Phoenix. Malesia. 1890; 3: 345–416. [Google Scholar]
- 44.Rivera D, Obón C, García-Arteaga J, Egea T, Alcaraz F, Laguna E, et al. Carpological analysis of Phoenix (Arecaceae): contributions to the taxonomy and evolutionary history of the genus. Bot J Linn Soc. 2014; 175: 74–122. 10.1111/boj.12164 [DOI] [Google Scholar]
- 45.Henderson SA, Billotte N, Pintaud JC. Genetic isolation of Cape Verde Island Phoenix atlantica (Arecaceae) revealed by microsatellite markers. Conserv Genet. 2006; 7: 213–223. 10.1007/s10592-006-9128-7 [DOI] [Google Scholar]
- 46.Henderson S, Gomes I, Gomes S, Baker WJ. Phoenix in the Cape Verde islands. Palms. 2003; 47. [Google Scholar]
- 47.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004; 11: 36–42. [Google Scholar]
- 48.Kuhl FP, Giardina CR. ELLIPTIC FOURIER FEATURES OF A CLOSED CONTOUR. Comput Graph Image Process. 1982; 18: 236–258. [Google Scholar]
- 49.Bonhomme V, Picq S, Gaucherel C, Claude J. Momocs: Outline Analysis Using R. J Stat Softw. 2014; 56. [Google Scholar]
- 50.R Development Core Team. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria; 2015. [Google Scholar]
- 51.Rohlf FJ. Fitting curves to outlines. Pp 167–177 F J Rohlf, F L Bookstein, eds Proc Michigan morphometrics Work Spec Publ number 2 Univ Michigan Museum Zool Ann Arbor. 1990.
- 52.Renaud S, Michaux JR. Adaptive latitudinal trends in the mandible shape of Apodemus wood mice. J Biogeogr. 2003; 30: 1617–1628. [Google Scholar]
- 53.Crampton JS. Elliptic fourier shape-analysis of fossil bivalves—some practical considerations. Lethaia. 1995; 28: 179–186. [Google Scholar]
- 54.Renaud S, Michaux J, Jaeger J, Auffray J. Paleontological Society Fourier Analysis Applied to Stephanomys (Rodentia, Muridae) Molars : Nonprogressive Evolutionary Pattern in a Gradual Lineage Fourier analysis applied to Stephanomys (Rodentia, Muridae) molars : nonprogressive evolutionary pa. 2012.
- 55.Renaud S, Millien V. Intra- and interspecific morphological variation in the field mouse species. 2001; 557–569.
- 56.Rohlf FJ, Archie JW. A comparison of Fourier methods for the description of wing shape in mosquitos (Diptera, Culicidae). Syst Zool. 1984; 33: 302–317. [Google Scholar]
- 57.Gros-Balthazard M. Hybridization in the genus Phoenix: A review. Emirates J Food Agric. 2013; 25: 831–842. [Google Scholar]
- 58.Glemin S, Bataillon T. A comparative view of the evolution of grasses under domestication. New Phytol. 2009; 183: 273–290. 10.1111/j.1469-8137.2009.02884.x [DOI] [PubMed] [Google Scholar]
- 59.Siegal ML, Bergman A. Waddington’s canalization revisited: Developmental stability and evolution. Proc Natl Acad Sci U S A. 2002; 99: 10528–10532. 10.1073/pnas.102303999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waddington CH. Experiments on canalyzing selection. Genet Res. 1960; 1: 140–150. [Google Scholar]
- 61.Chen Y, Nelson RL. Genetic Variation and Relationships among Cultivated, Wild, and Semiwild Soybean. Crop Sci. Crop Science Society of America; 2004; 44: 316–325. 10.2135/cropsci2004.3160 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
First sheet “Discrete measures” contains the measure of length, width, thickness (cm) and surface (cm2). Second sheet “Fourier coefficients” contains the Fourier coefficients obtained from the outline analyses based on Fourier method. Column C to AH contains the measures for dorsal view (name of the column suffixed with VD) and columns AI to BN contain the measures of the lateral side (suffixed with VL). In the name of those columns, A, B, C and D refers to the four coefficients of each harmonic and the following number to the number of the harmonic. For both sheets, first column (“Sample”) is the name of the sample; second column (“Seed”) is the number of the seed (D1 to D20 given that 20 seeds, when available, were analyzed for each sample) prefixed by the name of the sample.
(XLSX)
(DOCX)
Based on (A) four seed size parameters and (B) seed shape (64 normalized elliptic Fourier coefficients).
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.