Abstract
Escherichia coli O104:H4, an hybrid pathotype of Shiga toxigenic and enteroaggregative E. coli, involved in a major foodborne outbreak in Germany in 2011, has not been detected in cattle feces. Serogroup O104 with H type other than H4 has been reported to cause human illnesses, but their prevalence and characteristics in cattle have not been reported. Our objectives were to determine the prevalence of E. coli O104 in feces of feedlot cattle, by culture and PCR detection methods, and characterize the isolated strains. Rectal fecal samples from a total of 757 cattle originating from 29 feedlots were collected at a Midwest commercial slaughter plant. Fecal samples, enriched in E. coli broth, were subjected to culture and PCR methods of detection. The culture method involved immunomagnetic separation with O104-specific beads and plating on a selective chromogenic medium, followed by serogroup confirmation of pooled colonies by PCR. If pooled colonies were positive for the wzxO104 gene, then colonies were tested individually to identify wzxO104-positive serogroup and associated genes of the hybrid strains. Extracted DNA from feces were also tested by a multiplex PCR to detect wzxO104-positive serogroup and associated major genes of the O104 hybrid pathotype. Because wzxO104 has been shown to be present in E. coli O8/O9/O9a, wzxO104-positive isolates and extracted DNA from fecal samples were also tested by a PCR targeting wbdDO8/O9/O9a, a gene specific for E. coli O8/O9/O9a serogroups. Model-adjusted prevalence estimates of E. coli O104 (positive for wzxO104 and negative for wbdDO8/O9/O9a) at the feedlot level were 5.7% and 21.2%, and at the sample level were 0.5% and 25.9% by culture and PCR, respectively. The McNemar’s test indicated that there was a significant difference (P < 0.01) between the proportions of samples that tested positive for wzxO104 and samples that were positive for wzxO104, but negative for wbdDO8/O9/O9a by PCR and culture methods. A total of 143 isolates, positive for the wzxO104, were obtained in pure culture from 146 positive fecal samples. Ninety-two of the 143 isolates (64.3%) also tested positive for the wbdDO8/O9/O9a, indicating that only 51 (35.7%) isolates truly belonged to the O104 serogroup (positive for wzxO104 and negative for wbdDO8/O9/O9a). All 51 isolates tested negative for eae, and 16 tested positive for stx1 gene of the subtype 1c. Thirteen of the 16 stx1-positive O104 isolates were from one feedlot. The predominant serotype was O104:H7. Pulsed-field gel electrophoresis analysis indicated that stx1-positive O104:H7 isolates had 62.4% homology to the German outbreak strain and 67.9% to 77.5% homology to human diarrheagenic O104:H7 strains. The 13 isolates obtained from the same feedlot were of the same PFGE subtype with 100% Dice similarity. Although cattle do not harbor the O104:H4 pathotype, they do harbor and shed Shiga toxigenic O104 in the feces and the predominant serotype was O104:H7.
Introduction
In the summer of 2011, Germany and other European countries experienced a large outbreak of foodborne illness affecting nearly 4,000 people, with about 900 developing hemolytic uremic syndrome, leading to 54 deaths [1]. The causative agent was identified as Escherichia coli O104:H4, a hybrid serotype possessing characteristics of two pathotypes of E. coli, Shiga toxin-producing E. coli (STEC) and enteroaggregative E. coli (EAEC). The outbreak strain carried Shiga toxin 2 gene (stx2) and genes characteristic of EAEC, such as aatA (pAA virulence plasmid marker gene), aggA (pilin subunit of aggregative adherence fimbriae I), aggR (aggregative adherence fimbriae I transcriptional regulator), but was negative for other enterohemorrhagic E. coli (EHEC) genes, such as stx1 (Shiga toxin 1), eae (intimin) and ehxA (enterohemolysin) [2]. The serogroup O104 has serotypes other than O104:H4 and at least two of them have been implicated in human illnesses. A stx2-carrying O104:H21 serotype that was also negative for eae (similar to the O104:H4 German strain) was implicated in an outbreak of hemorrhagic colitis associated with consumption of raw milk in Helena, Montana in 1994 [3]. Sporadic cases of diarrhea caused by O104:H7, carrying stx1 or stx2, also negative for eae, have been reported [4, 5]. Non-Shiga toxigenic strains of E. coli O104 that were either EAEC or enteropathogenic (EPEC) pathotype have been reported in human patients with diarrhea in South Africa [6].
Because cattle are a primary reservoir of STEC, studies have been conducted to determine whether cattle harbor the O104:H4 serotype. Wieiler et al. [7] tested 100 cattle fecal samples from 34 different farms in the outbreak region of Germany and found that none of the fecal samples were positive for E. coli strains carrying genes characteristic of O104:H4. Auvray et al. [8] analyzed 1,468 cattle fecal samples collected from several slaughter facilities in France by real-time and conventional PCR assays that targeted stx2, wzxO104, fliCH4 (H4 flagellar gene) and aggR and reported that none of the fecal samples were positive for all four genes. We conducted a study to detect E. coli O104:H4 in feedlot cattle fecal samples (n = 248) using a multiplex PCR that targeted O104 (wzxO104), H4 (fliCH4), aggregative adherence fimbriae 1 (aggA), Shiga toxins 1 and 2 (stx1 and stx2), intimin (eae), tellurite resistance (terD), and enterohemolysin (ehxA), characteristic of the outbreak serotype and reported that cattle feces were positive for the O104 serogroup, but negative for the hybrid pathotype [9]. In that study, fecal samples positive for wzxO104 were plated onto several selective and differential media, and only a small number of PCR-positive samples yielded pure cultures of serogroup O104 and none of the isolates carried Shiga toxin genes. The likely reason for the poor recovery of O104 from PCR-positive fecal samples was that the culture method did not have an immunomagnetic separation (IMS) step because O104-specific IMS beads were not available at the time the study was conducted. Therefore, our objectives were to determine the prevalence of E. coli O104 in feedlot cattle feces utilizing a culture method involving an IMS step and a PCR-based method of detection, and to characterize the isolated strains. The culture method utilized in the study involved an enrichment step, followed by IMS with O104-specific IMS beads, plating on a chromogenic selective medium and confirming the O104 serogroup and major virulence genes by a multiplex PCR. The wzxO104 gene that was targeted in the PCR assay to detect O104 has also been reported in O8, O9 and O9a serogroups of E. coli [10]. Therefore, pre- and post-enriched fecal suspensions and putative E. coli O104 isolates were also subjected to a PCR assay with primers that targeted wbdD (which codes for methyl and kinase transferase), specific for E. coli O8, O9 and O9a serogroups [11].
Materials and Methods
Sample collection
The study was approved by the Kansas State University Institutional Animal Care and Use Committee (IACUC # 3172). Rectal content samples were collected from feedlot cattle immediately after slaughter at a Midwest slaughter plant during two visits, one week apart, in July 2013. The permission to collect samples was given under an agreement that the name and location of the abattoir will not be disclosed. Rectums were incised and contents were scooped with a plastic spoon. The spoon with the contents (approximately 10 to 20 g) were placed in a Whirl-Pak bag (Nasco, Ft. Atkinson, WI), transported on ice in a cooler to the Preharvest Food Safety Laboratory at Kansas State University and processed within 24 h. Rectal content samples were collected from a total of 757 cattle (both heifers and steers) originating from 29 feedlots located in six Midwestern States (IA, IL, MN, MO, NE and SD). Sixteen to 38 samples were collected per lot of cattle from a total of 35 lots, with each lot consisting of 36 to 227 animals. Cattle from one feedlot (No. 2) were sampled in both weeks.
Culture method of detection
Approximately two grams of fecal samples were suspended in 18 ml of E. coli broth (EC; Difco™, Becton, Dickinson Co., Sparks, MD) and incubated at 40°C for 6 h [9]. Post-enrichment fecal samples were subjected to a culture-based procedure, which involved immunomagnetic separation with O104 serogroup-specific beads (Abraxis®, Warminster, PA), and plating onto a selective chromogenic Possé medium [12] modified to include novobiocin at 5 mg/l and potassium tellurite at 0.5 mg/l (MP) [13]. After 20–24 h of incubation at 37°C, six chromogenic colonies (mauve, pink, or purple) were picked and streaked onto blood agar and incubated at 37°C for 24 h. The six colonies were pooled, boiled and the lysate was subjected to an eight-plex PCR targeting O-antigen genes of O104 (wzxO104) and the seven major serogroups of STEC (O26, O45, O103, O111, O121, O145 and O157). If pooled colonies were positive for wzxO104, then the six colonies were tested individually by a nine-plex PCR to identify pure culture of putative O104 serogroup (wzxO104) and associated major genes of the STEC and EAEC pathotypes: stx1 (Shiga toxin 1), stx2 (Shiga toxin 2), eae (intimin), ehxA (enterohemolysin), terD (tellurite resistance), aggA (pilin subunit of aggregative adherence fimbriae 1), bfpA (bundle-forming pilus) and flicH4 (H4-specific flagella). The primer pairs used for the eight-plex and nine-plex PCR assays are listed in Table 1. PCR amplification protocol for both assays included an initial denaturation at 94°C for 5 min followed by 25 cycles (pure culture) or 35 cycles (fecal suspension in broth) of 94°C for 30 s, 65°C for 30 s, 68°C for 75 s, and the final extension was 68°C for 7 min [9]. Isolates confirmed as positive for the wzxO104 gene were stored on cryogenic beads (CryoCare™, Key Scientific Products, Round Rock, TX).
Table 1. Target genes, primer sequences used and size of amplicons in PCR assays.
| Target gene | Primer sequences | Amplicon size (bp) | Reference |
|---|---|---|---|
| wzxO26 | F:AGGGTGCGAATGCCATATT | 417 | [33] |
| R:GACATAATGACATACCACGAGCA | |||
| wzxO45 | F: GGGCTGTCCAGACAGTTCAT | 890 | [33] |
| R: TGTACTGCACCAATGCACCT | |||
| wzxO103 | F: GCAGAAAATCAAGGTGATTACG | 740 | [33] |
| R: GGTTAAAGCCATGCTCAACG | |||
| wzxO104 | F: GGTTTTATTGTCGCGCAAAG | 337 | [9] |
| R: TATGCTCTTTTTCCCCATCG | |||
| wzxO111 | F: ACAAGAGTGCTCTGGGCTTC | 230 | [13] |
| R: AAACTAAGTGAGACGCCACCA | |||
| wbqE + wbqFO121 | F: TCAGCAGAGTGGAACTAATTTTGT | 587 | [33] |
| R: TGAGCACTAGATGAAAAGTATGGCT | |||
| wzxO145 | F: TCAAGTGTTGGATTAAGAGGGATT | 523 | [33] |
| R: CACTCGCGGACACAGTACC | |||
| rfbEO157 | F: CAGGTGAAGGTGGAATGGTTGTC | 296 | [34] |
| R: TTAGAATTGAGACCATCCAATAAG | |||
| wbdDO8/O9/O9a | F: GGCATCGGTCGGTATTCC | 1000 | [11] |
| R: TGCGCTAATCGCGTCTAC | |||
| stx1 | F: TGTCGCATAGTGGAACCTCA | 655 | [34] |
| R: TGCGCACTGAGAAGAAGAGA | |||
| stx2 | F: CCATGACAACGGACAGCAGTT | 477 | [34] |
| R: TGTCGCCAGTTATCTGACATTC | |||
| eae | F: CATTATGGAACGGCAGAGGT | 375 | [34] |
| R: ACGGATATCGAAGCCATTTG | |||
| ehxA | F: GCGAGCTAAGCAGCTTGAAT | 199 | [34] |
| R: CTGGAGGCTGCACTAACTCC | |||
| terD | F: AGTAAAGCAGCTCCGTCAAT | 434 | [2] |
| R: CCGAACAGCATGGCAGTCT | |||
| aggA | F: CGTTACAAATGATTGTCCTGTTACTAT | 151 | [9] |
| R: ACCTGTTCCCCATAACCAGAC | |||
| bfpA | F: CAGAAGTAATGAGCGCAACG | 285 | This study |
| R: CGTAGCCTTTCGCTGAAGTA | |||
| flicH4 | F: ACGGCTGCTGATGGTACAG | 244 | [9] |
| R: CGGCATCCAGTGCTTTTAAC | |||
| flicH2 | F: GCAACGGCTGAAACAACCTA | 585 | This study |
| R: TGCAGTTACAACTTCGGTTTTG | |||
| flicH4 | F: ACGGCTGCTGATGGTACAG | 244 | [9] |
| R: CGGCATCCAGTGCTTTTAAC | |||
| flicH7 | F: AGCTGCAACGGTAAGTGATTT | 949 | [34] |
| R: GGCAGCAAGCGGGTTGGTC | |||
| flicH11 | F: TCTGACACAAACATAGCTGGTACA | 228 | This study |
| R: TGTCTCACTCGTAATCAAAGAAGC | |||
| flicH21 | F: TCGATGGCGCGCAGAAAGCA | 419 | [35] |
| R: GGCTGTCGTAGGGGCAACGG |
PCR method of detection
One ml of pre- and post-enriched fecal suspensions were removed, boiled for 10 min and centrifuged at 9,300 x g for 5 min. The DNA in the supernatant was purified using a GeneClean® Turbo Kit (MP Biomedicals LLC., Solon, OH). DNA extracted from pre- and post-enrichment samples were tested by a nine-plex PCR to detect O antigen of O104 serogroup (wzxO104) and associated major genes of the STEC and EAEC (stx1, stx2, eae, ehxA, terD, aggA, bfpA and flicH4) and a single-plex PCR assay to detect wbdDO8/O9/O9a, a gene specific for serogroups O8, O9 and O9a [11].
Characterization of the E. coli O104 isolates
All putative O104 isolates (positive for wzxO104) were tested by a PCR assay targeting O8/O9/O9a (wbdDO8/O9/O9a), and isolates positive for wzxO104 and negative for wbdDO8/O9/O9a were considered as truly O104, and were further characterized. The flagellar types of the O104 isolates were identified by a multiplex PCR targeting five flagellar types (H2, H4, H7, H11 and H21) (Table 1) using the PCR running conditions as described before for the nine- or eight-plex PCR. The subtype of Shiga toxin genes was identified by nucleotide sequencing. Shiga toxin genes of the wzxO104-positive isolates were amplified (F-GCTCAAGGAGTATTGTGTAATATG and R- TCGCTGAATCCCCYTC) by a touchdown PCR method where the annealing temperature of each cycle was lowered gradually to avoid amplification of non-specific sequences [14]. PCR amplification protocol included an initial denaturation at 94°C for 5 min, 10 cycles of touchdown PCR (denature: 94°C for 30 s, annealing: 56–51°C (Δ-0.5°C) for 30 s; extension: 72°C for 1 min 45 s) followed by 30 cycles of regular PCR (denaturation: 94°C for 30 s, annealing: 51°C for 30 s; extension: 72°C for 1 min 45 s). Amplicons (1,233 bp) were purified using QIAquick® PCR Purification Kit. The purity and concentration of purified PCR products were determined using a spectrophotometer (NanoDrop, Thermo Scientific, Wilmington, DE). The products were then shipped to Genewiz Inc., (South Plainfield, NJ) for sequencing. The sequence data were aligned using the CLC Main Workbench 6.8.4 software for further analysis. Shiga toxin subtypes were determined according to the procedure described by Scheutz et al., [15]. Briefly, nucleotide sequences were translated to amino acid sequences after removing intergenic regions. The subtype of Shiga toxins was determined based on the amino acid motifs that define each Shiga toxin subtype [15]. Additionally, the subtyping of Shiga toxin genes was confirmed by PCR-RFLP (restriction fragment length polymorphism) [16]. Briefly, the B subunit of stx1 genes was amplified yielding a 283 bp amplicon. PCR amplification protocol included an initial denaturation at 94.0°C for 5 min, followed by 35 cycles of 94.0°C for 30 s, 51.3°C for 60 s, 72.0°C for 40 s. PCR products were purified using QIAquick® PCR purification Kit, digested separately with BstEII, HaeII and PflmI enzymes. Restriction fragments were visualized by microcapillary electrophoresis using Qiaxcel (Qiagen, Valencia, CA). Additionally, subtyping of stx1was conducted by in silico RFLP. A 283 bp fragment starting with CAGTTGAGGGGGGTAAAATG (forward primer used for amplification of stx1 in PCR-RFLP) and ending with GATTCAGCGAAGTTATTTTCCG (reverse complement of reverse primer used for amplification of stx1 in PCR-RFLP) was digested by BstEII, HaeII and PflmI using CLC Main Workbench 6.8.4 software. Restriction patterns of O104 STEC isolates were compared to reference sequences of three stx1 subtypes from the NCBI database (stx1a: Accession no. M16625.1; stx1c: Accession no. DQ449666.1; stx1d: Accession no. AY170851.1).
Pulsed-field gel electrophoresis (PFGE)
The clonal relationships between O104 STEC strains isolated from cattle feces were assessed by PFGE typing. The human clinical strains, German (O104:H4; BAA-2326) and Montana outbreak (O104:H21; BAA-178) and O104:H7 strains (06–3637, 07–3598, 08–4061, 2011C-3665, 2012C-3400; provided by Nancy A. Strockbine, Centers for Disease Control and Prevention, Atlanta, GA) were also included for comparison. The PFGE was performed according to the Centers for Disease Control and Prevention’s PulseNet protocol. Briefly, agarose embedded DNA of the O104 isolates were digested with XbaI followed by separation of restriction fragments by electrophoresis. Salmonella enterica serotype Braenderup (strain H9812) DNA digested with XbaI was used as DNA marker. Gel images were captured with a Gel Doc 2000 system (Bio-Rad). PFGE patterns were analyzed using the Bionumerics software (Applied Maths, Inc., Austin, TX). Band-based Dice similarity coefficients and unweighted pair-group method for clustering with a position tolerance setting of 1.5% were used for optimization and band comparison. Isolates with 100% homology were grouped as subtypes and those isolates which were > 96% but less than 100% homologous were grouped as types based on the Dice coefficients.
Statistical analysis
Generalized linear mixed models (GLMM) were fitted using a binary or binomial (events/trials) distribution, logit link, Laplace estimation, and Newton-Raphson and Ridging optimization, to estimate fecal prevalence of wzxO104 and wzxO104/wbdDO8/O9/O9a in Proc Glimmix (SAS 9.3, SAS Institute Inc., Cary, NC). Sample-, lot- and feedlot-level prevalence were calculated as the proportion of samples, samples within lots or samples within feedlots testing positive for wzxO104 (serogroup O104 and or O8/O9/O9a) and positive for wzxO104 but negative for wbdDO8/O9/O9a (serogroup O104 only) by PCR or culture methods, and divided by the total number of samples tested per lot or per feedlot, respectively. Random effects were used to account for the hierarchical structure of the data (samples within lots, lots within feedlots and feedlots within states). Prevalence estimates (and 95% confidence intervals) were obtained from model intercepts using the formula p = eβ0 / (1 + eβ0), where β0 is the coefficient of the model intercept. The Cohen’s Kappa statistic was used to compute the agreement beyond that due to chance between PCR- and culture-based methods for detection of serogroup O104 and or O8/O9/O9a (positive for wzxO104) and serogroup O104 only (wzxO104 positive but negative for wbdDO8/O9/O9a) in fecal samples. Interpretation of the Kappa statistic was based on the scale proposed by Landis and Koch [17]. The McNemar’s Chi-square test was used to compare the proportion of positive samples detected by both methods [18]. When the P-value of the McNemar’s test is not significant (P > 0.01), there is little evidence to conclude that the proportion of positives are different, whereas when the P-value is significant (P < 0.01), there is a significant disagreement between tests, indicating little value in assessing agreement. In the latter case, Cohen’s Kappa statistics are provided for reference only.
Results
Culture method of detection
Of the total 757 fecal samples tested, 146 samples (19.3%) tested positive for serogroup O104 (and/or O8/O9/O9a), based on PCR testing (for the wzxO104 gene) of the six-pooled colonies picked from the O104 IMS beads-inoculated MP plates. Fecal samples from 21 of 29 feedlots (72.4%) and 27 of 35 lots (77.1%) tested positive for the serogroup O104 (and/or O8/O9/O9a) (Table 2). At the sample level, the crude prevalence of serogroup O104 (and or O8/O9/O9a) within feedlots ranged from 0 to 93.8%. Because six chromogenic colonies were picked from O104 beads-plated medium, pooled, and tested by a multiplex PCR targeting a total of eight serogroups (O26, O45, O103, O104, O111, O121, O145, and O157), other E. coli serogroups were also detected occasionally. Pooled colonies from fecal samples (n = 757) also tested positive for O26 (3.7%), O45 (0.9%), O103 (5.8%), O145 (0.5%) and O157 (4.9%). Serogroups O111 and O121 were not detected in any of these samples. The testing of individual colonies was designed to identify wzxO104-positive serogroup and associated virulence genes only; therefore, individual isolates of the other seven serogroups were not identified. A total of 143 isolates, positive for the wzxO104, were obtained in pure culture from 146 positive (based on pooled colonies) fecal samples. Ninety-two of the 143 isolates (64.3%) also tested positive for the wbdDO8/O9/O9a (Table 2), indicating that only 51 (35.7%) isolates truly belonged to the O104 serogroup (positive for wzxO104 and negative for wbdDO8/O9/O9a). After excluding the samples that yielded isolates positive for O8/O9/O9a, only 14 of the 29 (48.3%) feedlots, 20 of the 35 lots (57.1%), and 49 of the 757 (6.5%) samples were considered truly positive for the serogroup O104.
Table 2. Number of fecal samples from feedlot cattle positive for wzxO104 possessing E. coli (serogroups O104 and/or O8/O9/O9a) based on the culture method and wzxO104-positive isolates that tested positive for wbdDO8/O9/O9a (serogroups O8/O9/O9a) or negative for wbdDO8/O9/O9a (serogroup O104).
| No. of wzxO104-positive isolates | ||||||
|---|---|---|---|---|---|---|
| Week | Feedlot no. | Lot No. | No. of samples collected | No. of samples positive for wzxO104 (%)a | Positive for wbdDO8/O9/O9a (%)b | Negative for wbdDO8/O9/O9a (%)b |
| 1 | 1 | 1 | 38 | 18 (47.4) | 20 (52.6) | 0 |
| 2 | 2 | 38 | 5 (13.2) | 4 (10.5) | 2 (5.3) | |
| 2 | 3 | 19 | 3 (15.8) | 0 | 1 (5.3) | |
| 3 | 4 | 38 | 0 | 0 | 0 | |
| 4 | 5 | 19 | 6 (31.6) | 4 (21.1) | 2 (10.5) | |
| 4 | 6 | 19 | 9 (47.4) | 4 (21.1) | 5 (26.3) | |
| 5 | 7 | 38 | 1 (2.6) | 0 | 1 (2.6) | |
| 6 | 8 | 19 | 3 (15.8) | 0 | 3 (15.8) | |
| 7 | 9 | 19 | 11 (57.9) | 10 (52.6) | 1 (5.3) | |
| 8 | 10 | 19 | 0 | 0 | 0 | |
| 9 | 11 | 19 | 0 | 0 | 0 | |
| 10 | 12 | 19 | 0 | 0 | 0 | |
| 11 | 13 | 19 | 4 (21.1) | 0 | 3 (15.8) | |
| 12 | 14 | 19 | 1 (5.3) | 0 | 0 | |
| 13 | 15 | 19 | 8 (42.1) | 2 (10.5) | 5 (26.3) | |
| 14 | 16 | 16 | 15 (93.8) | 16 (100.0) | 2 (12.5) | |
| 2 | 2 | 17 | 20 | 2 (10.0) | 0 | 2 (10.0) |
| 2 | 18 | 20 | 5 (25.0) | 3 (15.0) | 1 (5.0) | |
| 15 | 19 | 20 | 1 (5.0) | 0 | 1 (5.0) | |
| 16 | 20 | 20 | 2 (10.0) | 2 (10.0) | 0 | |
| 17 | 21 | 20 | 1 (5.0) | 0 | 1 (5.0) | |
| 18 | 22 | 20 | 2 (10.0) | 0 | 0 | |
| 19 | 23 | 20 | 0 | 0 | 0 | |
| 20 | 24 | 20 | 2 (10.0) | 2 (10.0) | 0 | |
| 21 | 25 | 20 | 10 (50.0) | 9 (45.0) | 0 | |
| 22 | 26 | 20 | 5 (25.0) | 2 (10.0) | 3 (15.0) | |
| 22 | 27 | 20 | 5 (25.0) | 2 (10.0) | 3 (15.0) | |
| 23 | 28 | 20 | 11 (55.0) | 11(55.0) | 0 | |
| 24 | 29 | 20 | 1 (5.0) | 0 | 1 (5.0) | |
| 25 | 30 | 20 | 0 | 0 | 0 | |
| 26 | 31 | 20 | 9 (45.0) | 0 | 9 (45.0) | |
| 26 | 32 | 20 | 5 (25.0) | 1 (5.0) | 4 (20.0) | |
| 27 | 33 | 20 | 1 (5.0) | 0 | 1 (5.0) | |
| 28 | 34 | 20 | 0 | 0 | 0 | |
| 29 | 35 | 20 | 0 | 0 | 0 | |
| Total | 29 | 35 | 757 | 146 (19.3) | 92 (12.2) | 51 (6.7) |
aSamples positive by PCR of pooled colonies
bThe percentages in parentheses are number of wzxO104 isolates that were positive or negative for wbdDO8/O9/O9a from the total number of samples in each lot.
PCR method of detection
Of the nine genes included in the multiplex PCR assay, two genes, aggA and bfpA, which code for aggregative adherence fimbriae 1 and bundle forming pili, respectively, were not detected in any of the fecal samples, either before or after enrichment in EC broth (Table 3). The overall prevalence of wzxO104 gene in samples before and after enrichment was 5% (38/757) and 46.1% (349/757), respectively. Based on the single-plex assay of samples targeting wbdDO8/O9/O9a, 13 (1.7%) and 238 (31.4%) of the 757 samples tested were positive—before and after enrichment, respectively. Thirty-four (4.5%) and 194 (25.6%) fecal samples were positive for wzxO104 and negative for wbdDO8/O9/O9a, suggesting that those fecal samples truly contained E. coli O104 (Table 3). A higher proportion of fecal samples tested positive for the stx2 than stx1 gene (66.8 vs. 27.5%) in enriched samples. Among the genes tested, ehxA that codes for enterohemolysin was the most prevalent in both pre- and post-enriched samples, followed by flicH4, terD, eae, stx2 and stx1 genes (Table 3).
Table 3. Number of fecal samples from feedlot cattle positive for wzxO104 and or wbdDO8/O9/O9a and associated major genes of the O104 hybrid pathotype (O104:H4) in cattle feces based on the PCR method.
| No. of samples (n = 757) positive | |||
|---|---|---|---|
| Genes | Protein or Function | Before enrichment, n (%) | After enrichmenta, n (%) |
| wzxO104 | O104 antigen flippase | 38 (5.0) | 349 (46.1) |
| wbdDO8/O9/O9a | Kinase and methyl transferase | 13 (1.7) | 238 (31.4) |
| Onlyb wzxO104 | 34 (4.5) | 194 (25.6) | |
| stx1 | Shiga toxin 1 | 29 (3.8) | 208 (27.5) |
| stx2 | Shiga toxin 2 | 156 (20.6) | 506 (66.8) |
| eae | Intimin | 112 (14.8) | 549 (72.5) |
| ehxA | Enterohemolysin | 370 (48.9) | 710 (93.8) |
| terD | Tellurite resistance | 339 (44.8) | 624 (82.4) |
| aggA | Aggregative adherence fimbriae 1 | 0 (0) | 0 (0) |
| bfpA | Bundle forming pili | 0 (0) | 0 (0) |
| flicH4 | H4 flagellar antigen | 200 (26.4) | 659 (87.1) |
a Feces were enriched in Escherichia coli broth at 40 C for 6 h.
b Positive for wzxO104 and negative for wbdDO8/O9/O9a.
Based on the PCR assay of post-enriched fecal suspensions, 26 of 29 feedlots (89.7%) and 32 of 35 lots (91.4%) contained one or more fecal samples that tested positive for wzxO104 (serogroup O104 and or O8/O9/O9a). At the sample level, the crude prevalence of wzxO104 within feedlots ranged from 0 to 95%. Of the three feedlots that were negative for wzxO104, two had samples that tested positive for wbdDO8/O9/O9a. Twenty-three of the 29 feedlots (79.3%) and 29 of the 35 lots (82.9%) were positive for wzxO104 and negative for wbdDO8/O9/O9a, indicating that cattle feces from these feedlots can be considered truly positive for serogroup O104. Five of the 29 feedlots were negative for wbdDO8/O9/O9a and of those four were positive for wzxO104 (Table 4).
Table 4. Number of fecal samples from feedlot cattle positive for E. coli O104 and O8/O9/O9a based on PCR assays of wzxO104 and wbdDO8/O9/O9a.
| Positive for: | ||||||
|---|---|---|---|---|---|---|
| Week | Feedlot no. | Lot No. | No. of samples collected | wzxO104 | wbdDO8/O9/O9a | wzxO104 and negative for wbdDO8/O9/O9a |
| 1 | 1 | 1 | 38 | 31 (81.6) | 12 (31.6) | 21 (55.3) |
| 2 | 2 | 38 | 10 (26.3) | 9 (23.7) | 6 (15.8) | |
| 2 | 3 | 19 | 9 (47.4) | 7 (36.8) | 5 (26.3) | |
| 3 | 4 | 38 | 3 (7.9) | 5 (13.2) | 2 (5.3) | |
| 4 | 5 | 19 | 14 (73.7) | 16 (84.2) | 0 | |
| 4 | 6 | 19 | 18 (94.7) | 7 (36.8) | 11 (57.9) | |
| 5 | 7 | 38 | 26 (68.4) | 19 (50.0) | 12 (31.6) | |
| 6 | 8 | 19 | 7 (36.8) | 2 (10.5) | 5 (26.3) | |
| 7 | 9 | 19 | 12 (63.2) | 5 (26.3) | 8 (42.1) | |
| 8 | 10 | 19 | 0 | 1 (5.3) | 0 | |
| 9 | 11 | 19 | 0 | 0 | 0 | |
| 10 | 12 | 19 | 2 (10.5) | 0 | 2 (10.5) | |
| 11 | 13 | 19 | 8 (42.1) | 0 | 8 (42.1) | |
| 12 | 14 | 19 | 5 (26.3) | 1 (5.3) | 4 (21.1) | |
| 13 | 15 | 19 | 10 (52.6) | 1 (5.3) | 10 (52.6) | |
| 14 | 16 | 16 | 10 (62.5) | 3 (18.8) | 8 (50.0) | |
| 2 | 2 | 17 | 20 | 5 (25.0) | 3 (15.0) | 4 (20.0) |
| 2 | 18 | 20 | 11 (55.0) | 3 (15.0) | 10 (50.0) | |
| 15 | 19 | 20 | 5 (25.0) | 0 | 5 (25.0) | |
| 16 | 20 | 20 | 14 (70.0) | 5 (25.0) | 11 (55.0) | |
| 17 | 21 | 20 | 4 (20.0) | 1 (5.0) | 4 (20.0) | |
| 18 | 22 | 20 | 19 (95.0) | 18 (90.0) | 1 (5.0) | |
| 19 | 23 | 20 | 13 (65.0) | 20 (100.0) | 0 | |
| 20 | 24 | 20 | 6 (30.0) | 5 (25.0) | 4 (20.0) | |
| 21 | 25 | 20 | 16 (80.0) | 7 (35.0) | 9 (45.0) | |
| 22 | 26 | 20 | 18 (90.0) | 9 (45.0) | 9 (45.0) | |
| 22 | 27 | 20 | 15 (75.0) | 12 (60.0) | 5 (25.0) | |
| 23 | 28 | 20 | 17 (85.0) | 14 (70.0) | 3 (15.0) | |
| 24 | 29 | 20 | 1 (5.0) | 5 (25.0) | 0 | |
| 25 | 30 | 20 | 2 (10.0) | 15 (75.0) | 1 (5.0) | |
| 26 | 31 | 20 | 9 (45.0) | 9 (45.0) | 4 (20.0) | |
| 26 | 32 | 20 | 9 (45.0) | 10 (50.0) | 5 (25.0) | |
| 27 | 33 | 20 | 4 (20.0) | 13 (65.0) | 1 (5.0) | |
| 28 | 34 | 20 | 0 | 1 (5.0) | 0 | |
| 29 | 35 | 20 | 16 (80.0) | 0 | 16 (80.0) | |
| Total | 29 | 35 | 757 | 349 (46.1) | 238 (31.4) | 194 (25.6) |
Model-adjusted prevalence estimates of wzxO104 -positive fecal samples (serogroups O104 and/or O8/O9/O9a) and wzxO104 -positive fecal samples that were negative for wbdDO8/O9/O9a (serogroup O104 only) at the sample-, lot- and feedlot-levels detected by culture and PCR methods are presented in Table 5. The McNemar’s test indicated that there was a significant difference (P < 0.01) between the proportions of samples that tested positive for wzxO104 and samples that were positive for wzxO104, but negative for wbdDO8/O9/O9a by PCR and culture methods, hence the Kappa statistics (κ = 0.27; κ 95% CI = 0.21–0.32 for samples that tested positive for wzxO104 and κ = 0.10; κ 95% CI = 0.04–0.17 for samples that were positive for wzxO104, but negative for wbdDO8/O9/O9a) are provided for reference only.
Table 5. Model-adjusted prevalence estimates of fecal samples from feedlot cattle positive for wzxO104/wbdDO8/O9/O9a and wzxO104 at the feedlot-, lot- and sample- levels.
| Target genes, serogroups, and detection method | Level of prevalence estimation | Mean Prevalence, % (95% confidence interval) |
|---|---|---|
| Positive for wzxO104 (Positive for O104 and/or O8/O9/O9a) a, b | ||
| Culture method | Feedlot | 17.6 (6.3–40.3) |
| Lot | 13.5 (4.8–32.9) | |
| Sample | 11.8 (6.3–21.0) | |
| PCR method | Feedlot | 49.5 (29.3–69.9) |
| Lot | 42.5 (22.6–65.1) | |
| Sample | 41.7 (27.1–57.8) | |
| Positive for wzxO104 and negative for wbdDO8/O9/O9a (Positive for O104 only) a, c | ||
| Culture method | Feedlot | 5.7 (2.9–10.7) |
| Lot | 2.8 (1.1–7.2) | |
| Sample | 0.50 (0.2–1.2) | |
| PCR method | Feedlot | 21.2 (14.7–29.5) |
| Lot | 20.1 (13.2–29.2) | |
| Sample | 25.9 (17.5–36.6) |
a The proportions of samples that tested positive by culture and PCR methods were significantly different by McNemar’s Chi square test (P < 0.01).
bKappa statistics: κ = 0.27; κ 95% CI = 0.21–0.32
cKappa statistics: κ = 0.10; κ 95% CI = 0.04–0.17
Characteristics of the E. coli O104 isolates
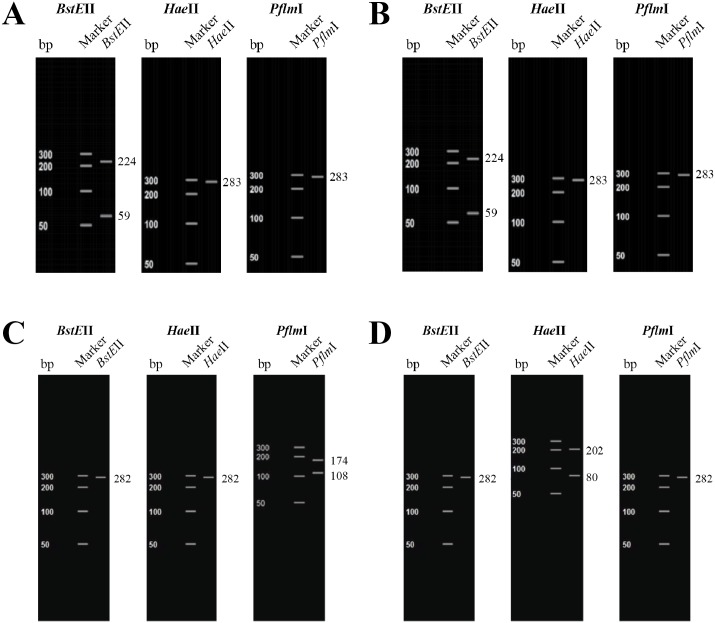
Of the 51 O104 isolates (positive for wzxO104 and negative for wbdDO8/O9/O9a), 16 isolates (31.4%) carried stx1 gene and all 16 also tested positive for ehxA and terD genes (Table 6). None of the O104 isolates carried stx2, flicH4, eae, bfpA and aggA genes (Table 6). Thirteen of the 16 stx1-positive O104 isolates were from one feedlot. Based on PCR assays targeting flagellar genes, 37 isolates tested positive for H7, four for H2, one each for H11 and H21, eight were unidentified, and none of the isolates tested positive for H4. The 16 stx1-positive O104 isolates possessed the H7 flagellar type. Amino acid sequences deduced from nucleotide sequences of stx1 amplicons indicated that the Shiga toxin genes of all O104 isolates (n = 16) were of subtype 1c. Digestion of stx1 amplicon (283 bp) with BstEII yielded two fragments of 224 and 59 bp whereas a single undigested fragment of 283 bp was produced with HaeII and PflMI enzymes. In silico RFLP analysis indicated that restriction patterns of Shiga toxin genes of O104 isolates were similar to that of stx1c of the reference strain (E. coli strain BCN26- Accession no. DQ449666.1; Fig 1) and matched the PCR-RFLP results (data not shown).
Table 6. Virulence gene profiles of strains of Escherichia coli O104 isolated from feedlot cattle feces.
| Genes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Week of sample collection | Feedlot no. | No. of samples collected | Total no. of O104 isolatesa | stx1 | stx2 | eae | ehxA | terD |
| 1 | 2 | 38 | 2 | 1, 0 | 0 | 0 | 1, 0 | 1, 0 |
| 2 | 19 | 1 | 1 | 0 | 0 | 1 | 1 | |
| 4 | 19 | 2 | 1, 0 | 0 | 0 | 1, 0 | 2 | |
| 4 | 19 | 5 | 0 | 0 | 0 | 0 | 0 | |
| 5 | 38 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 6 | 19 | 3 | 0 | 0 | 0 | 0 | 0 | |
| 7 | 19 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 11 | 19 | 3 | 0 | 0 | 0 | 0 | 1, 0, 0 | |
| 13 | 19 | 5 | 0 | 0 | 0 | 0 | 3, 0, 0 | |
| 14 | 16 | 2 | 0 | 0 | 0 | 0 | 1, 0 | |
| 2 | 2 | 20 | 2 | 0 | 0 | 0 | 0 | 0 |
| 2 | 20 | 1 | 0 | 0 | 0 | 0 | 0 | |
| 15 | 20 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 17 | 20 | 1 | 0 | 0 | 0 | 0 | 0 | |
| 22 | 20 | 3 | 0 | 0 | 0 | 0 | 1, 0, 0 | |
| 22 | 20 | 3 | 0 | 0 | 0 | 0 | 0 | |
| 24 | 20 | 1 | 0 | 0 | 0 | 0 | 1 | |
| 26 | 20 | 9 | 9 | 0 | 0 | 9 | 9 | |
| 26 | 20 | 4 | 4 | 0 | 0 | 4 | 4 | |
| 27 | 20 | 1 | 0 | 0 | 0 | 0 | 0 | |
| Total | 425 | 51 | 16 | 0 | 0 | 16 | 27 | |
a Isolates positive for wzxO104 gene and negative for O8/O9/O9a
All isolates were negative for bfpA, aggA and flicH4 genes
Fig 1. In silico restriction fragment length polymorphism (RFLP) subtyping of Shiga toxin genes of O104 isolates.
(A) RFLP pattern of Shiga toxin of an O104 isolate; (B) RFLP pattern of stx1c of a reference sequence (Accession no. DQ449666.1); (C) RFLP pattern of stx1a of a reference sequence (Accession no. M16625.1); (D) RFLP pattern of stx1d of a reference sequence (Accession no. AY170851.1)
Pulsed-field gel electrophoresis (PFGE)
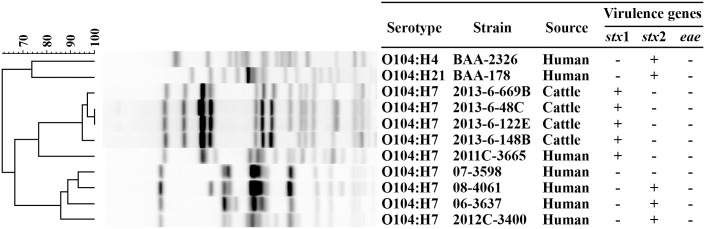
The 16 stx1-positive E. coli O104 isolates obtained in the present study, German (O104:H4) and Montana outbreak (O104:H21) strains and the five human O104:H7 strains formed seven separate PFGE clusters (Fig 2). Bovine O104 strains were 62.4% similar to the German and Montana outbreak strains and 67.9% to 77.5% similar to human O104:H7 strains (Fig 2). The Dice similarity between the German outbreak and the Montana strains was 73%. The stx1-positive O104 strains (n = 13) obtained from the same feedlot were of the same PFGE subtype with 100% similarity, and the remaining three STEC O104 strains from different feedlots were of the same PFGE type (96–100% similarity).
Fig 2. Pulsed-field gel electrophoresis-based clustering of Escherichia coli O104 strains from cattle feces and human clinical strains (O104:H4; O104:H21; and O104:H7).
Discussion
The prevalence of wzxO104-positive E. coli was determined in feedlot cattle feces by culture and PCR methods of detection. Our primary goal was to detect the prevalence of serogroup O104. However, the target gene, wzxO104, used in the culture and PCR methods of detection is also present in E. coli O8, O9, and O9a serogroups [11]. In fact, the O antigen gene cluster of O104 serogroup has the same genes (O antigen polymerase gene, O antigen flippase gene [wzx104], three CMP-sialic acid synthesis genes, and three glycosyl transferase genes) in the same order as that of the gene cluster that codes for the K9 capsular antigen of O8, O9 and O9a serogroups [10, 11]. Therefore, a wzxO104-positive fecal sample could contain E. coli O104 and or O8/O9/O9a. In order to distinguish O104 from O8/O9/O9a, we assayed all fecal samples and pure cultures of wzx104-positive isolates by PCR with primers targeting wbdDO8/O9/O9a, a gene that is specific for O8/O9/O9a [11]. A fecal sample or an O104 isolate that was positive for wzx104 and negative for wbdDO8/O9/O9a was considered as truly positive for serogroup O104.
The fecal samples collected in our study to estimate prevalence of wzxO104-positive E. coli were representative of a large population of cattle originating from 29 feedlots located in six Midwestern states. The prevalence of wzxO104-positive fecal sample, determined by PCR, reported in our study (46.1%) was higher than that reported (20.6%) by Paddock et al.[9], possibly because samples collected were from multiple feedlots (29 vs 8). Based on the culture method, 19.3% of fecal sample were positive for E. coli containing wzxO104 compared to 2.8% reported by Paddock et al [9]. In addition, the use of the O104-specific IMS beads likely increased the sensitivity of detection. None of the previous studies has utilized IMS beads because O104-serogroup specific beads had been commercially available only recently [19].
In the culture method, fecal samples that tested positive (pooled colonies) for E. coli possessing the wzxO104 gene indicated the sample was positive for O104 and/or O8.O9/O9a. However, isolates positive for wzxO104 and negative for wbdDO8/O9/O9a, which could be considered as truly E. coli O104, were obtained from 6.5% fecal samples. Ninety-two of the 143 wzxO104-positive isolates (64.3%) were also positive for wbdDO8/O9/O9a by PCR, which means the isolates were not O104, but could be E. coli O8, O9, or O9a. Isolates positive for wzxO104 and wbdDO8/O9/O9a have been reported in previous studies [9, 20]. Fecal prevalence of E. coli O8/O9/O9a has been previously reported in cattle [21, 22]. Manna et al. (2010) tested cattle feces collected at a slaughter plant in India for E. coli O8 and reported a prevalence of 2% [22]. Some of the chromogenic colonies picked from O104 beads-plated medium tested positive for other E. coli serogroups, such as O26, O45, O103, O145 and O157, which indicates some cross-reactivity of O104 beads with other serogroups. Unfortunately, the multiplex PCR targeting eight serogroup-specific genes (O104, O157 and 6 non-O157) that was used to test pooled colonies did not include the wbdDO8/O9/O9a gene. Therefore, we could not ascertain the prevalence of serogroups of O8/O9/O9a in the pooled colonies. The detection of O157 and six non-O157 serogroups was low suggesting that non-specificity of the O104 beads does not appear to be an issue compared to IMS beads for other serogroups, particularly O103 [13]. We picked colonies with a range of color because chromogenic colonies of pure cultures of O104 serogroup on MP medium were indistinguishable from other serogroups of STEC
Our study showed that feedlot cattle harbor E. coli O104 serogroup (positive for wzxO104 and negative for wbdDO8/O9/O9a) in the gut and shed these organisms in their feces, however only a small proportion of the O104 isolates obtained carried the Shiga toxin gene and none exhibited the enteroaggregative genes of the pathotype of the German outbreak strain. Unlike other predominant STEC serogroups (O157 and non-O157 STEC) causing human illnesses, O104:H4 serotype has never been reported in animals. Previous studies that aimed at detecting the O104:H4 serotype carrying genes characteristic of the German outbreak strain in cattle feces reported that cattle do not harbor the combination of genes (wzxO104, stx1, stx2, flicH4, aggA or aggR) that are unique to this pathotype [7–9]. The present study confirms the absence of the unique pathotype in cattle feces in this population of feedlot cattle, based on both PCR and culture-based detection methods. E. coli O104 strains with H antigen, other than H4, have been isolated from animals [23–25]. Blanco et al. [23] have reported that O104:H7, positive for stx1 and negative for stx2 and eae, was one of the eight non-O157 serotypes more frequently detected among STEC strains in sheep in Spain, and interestingly, in the same study, none of the non-O157 STEC strains isolated from cattle included O104. Serotype O104:H21, positive for stx1 and stx2, but negative for eae, has been isolated from feces of healthy and diarrheic cattle in Spain [24]. None of the O104 isolates in our study tested positive for stx2 and the one isolate of H21 serotype obtained was negative for stx. To our knowledge, this is the first report of Shiga toxin carrying O104 serogroup in feces of cattle in the US.
Of the 51 O104 isolates (positive wzxO104 and negative for wbdDO8/O9/O9a), 16 (31.4%) carried a combination of stx1, terD and ehxA genes. Because the modified MP medium contained potassium tellurite, it is possible that there was a selection pressure exerted for terD-positive isolates. The stx1 of O104 isolates were of subtype stx1c based on nucleotide sequencing. Shiga toxin subtyping based on amino acid sequences were further confirmed by in silico RFLP, which matched results obtained from PCR-RFLP. Our study shows that in silico RFLP, a simple and rapid method, is a reliable alternative to PCR-RFLP for subtyping of stx. None of the O104 isolates obtained in the present study were positive for eae, indicating that serogroup O104 in our study population could be Shiga toxigenic, but not enterohemorrhagic E. coli. The absence of eae appears to be a feature of the serogroup O104 because previously reported serotypes such as O104:H4 (German outbreak strain; [2]), O104:H21 (Montana outbreak strain; [3]), and O104:H7 (CDC strains from sporadic diarrheal cases [5, 20] were all negative for eae. Based on PFGE typing, the O104:H7 strains of cattle origin were only 67.9% to 77.5% similar to human O104:H7 strains. Intimin-negative STEC isolates of serogroups O5, O76, O78, O113, O128, O146, O174, O178, and O181 carrying stx1c have been isolated from stools of asymptomatic carriers and individuals with diarrhea [26]. The O104 strains were also negative for aggA and bfpA, which are responsible for adherence to host cells in enteroaggregative E. coli [27] and enteropathogenic E. coli [28], respectively. All 16 O104 STEC strains isolated in our study carried H7 flagellar type and possessed the same profile of virulence genes tested (ehxA and terD). Miko et al. [5] have reported that STEC strains carrying same flagellar type generally harbor similar virulence genes. Thirteen of the 16 stx1-positive isolates were from the same feedlot and all 13 were of the same PFGE type, suggesting spread of a single clone within a feedlot.
Escherichia coli is a continuously evolving organism with the capacity to acquire virulence genes from other pathogenic organisms and become virulent [29]. Sialic acid which has been reported to be an important component of E. coli O104 antigen and other organisms such as E. coli O24, O56, Campylobacter jejuni, Salmonella enterica, and Citrobacter freundii [30, 31], is also an important component of animal tissues. This trait of bacterial antigens may contribute to evasion of immune system by mimicking the host tissue component [11]. Therefore, STEC O104:H7 serotype has the potential to be a human pathogen. Because the prevalence of O104 is low in cattle and only a small proportion of O104 is STEC, cattle are not likely to be a major reservoir for E. coli O104. Escherichia coli O104:H4 involved in the German outbreak in 2011 is a classic example of the emergence of a highly virulent pathogen by acquisition of prophage encoding Shiga toxin 2 through horizontal gene transfer [32]. Similarly, E. coli O104 with H types other than H4 has the potential to emerge as a virulent pathogen by acquiring Shiga toxins 1 and or 2 via phage-mediated transfer.
Conclusions
Cattle harbor and shed eae-negative serogroup O104 in feces, however, none of the isolated strains in this study carried genes characteristic of the hybrid serotype reported in Germany (stx2, aggA and flicH4) and only a small proportion of O104 strains carried the stx1 gene. The predominant STEC serotype detected in cattle feces was O104:H7, which has been previously isolated from sporadic cases of diarrhea in humans. Based on our results, cattle are not a reservoir of O104:H4 serotype, however, they do harbor other O104 serotypes, such as O104:H2, O104:H7, O104:H11 and O104:21.
Acknowledgments
The authors wish to thank Neil Wallace, Rachael Henderson, Sean Stenseng, Diane Larson and Tiffany Mainini for their assistance with this project. Contribution no. 15-435-J from the Kansas Agricultural Experiment Station, Manhattan.
Data Availability
All relevant data are within the paper.
Funding Statement
This material is based upon work that is supported by the National Institute of Food and Agriculture, U. S. Department of Agriculture, under award number 2012-68003-30155 (to TGN). The funders had no role in study design, data collection and analysis, decision to publish, or presentation of the manuscript.
References
- 1.Karch H, Denamur E, Dobrindt U, Finlay BB, Hengge R, Johannes L, et al. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO molecular medicine. 2012;4(9):841–8. 10.1002/emmm.201201662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bielaszewska M, Mellmann A, Zhang W, Köck R, Fruth A, Bauwens A, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. The Lancet Infectious Diseases. 2011;11(9):671–6. 10.1016/S1473-3099(11)70165-7 [DOI] [PubMed] [Google Scholar]
- 3.Feng P, Weagant SD, Monday SR. Genetic analysis for virulence factors in Escherichia coli O104:H21 that was implicated in an outbreak of hemorrhagic colitis. Journal of clinical microbiology. 2001;39(1):24–8. 10.1128/JCM.39.1.24-28.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussein HS. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. Journal of animal science. 2007;85(13 Suppl):E63–72. [DOI] [PubMed] [Google Scholar]
- 5.Miko A, Delannoy S, Fach P, Strockbine NA, Lindstedt BA, Mariani-Kurkdjian P, et al. Genotypes and virulence characteristics of Shiga toxin-producing Escherichia coli O104 strains from different origins and sources. International journal of medical microbiology: IJMM. 2013;303(8):410–21. 10.1016/j.ijmm.2013.05.006 . [DOI] [PubMed] [Google Scholar]
- 6.Tau NP, Meidany P, Smith AM, Sooka A, Keddy KH, Group for Enteric R, et al. Escherichia coli O104 associated with human diarrhea, South Africa, 2004–2011. Emerging infectious diseases. 2012;18(8):1314–7. 10.3201/eid1808.111616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wieler LH, Semmler T, Eichhorn I, Antao EM, Kinnemann B, Geue L, et al. No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut pathogens. 2011;3(1):17 10.1186/1757-4749-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auvray F, Dilasser F, Bibbal D, Kerouredan M, Oswald E, Brugere H. French cattle is not a reservoir of the highly virulent enteroaggregative Shiga toxin-producing Escherichia coli of serotype O104:H4. Veterinary microbiology. 2012;158(3–4):443–5. 10.1016/j.vetmic.2012.02.029 . [DOI] [PubMed] [Google Scholar]
- 9.Paddock ZD, Bai J, Shi X, Renter DG, Nagaraja TG. Detection of Escherichia coli O104 in the feces of feedlot cattle by a multiplex PCR assay designed to target major genetic traits of the virulent hybrid strain responsible for the 2011 German outbreak. Applied and environmental microbiology. 2013;79(11):3522–5. 10.1128/AEM.00246-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Molecular microbiology. 1999;31(5):1307–19. 10.1046/j.1365-2958.1999.01276.x [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Briggs CE, Rothemund D, Fratamico P, Luchansky JB, Reeves PR. Sequence of the E. coli O104 antigen gene cluster and identification of O104 specific genes. Gene. 2001;270(1–2):231–6. [DOI] [PubMed] [Google Scholar]
- 12.Possé B, De Zutter L, Heyndrickx M, Herman L. Novel differential and confirmation plating media for Shiga toxin-producing Escherichia coli serotypes O26, O103, O111, O145 and sorbitol-positive and -negative O157. FEMS microbiology letters. 2008;282(1):124–31. 10.1111/j.1574-6968.2008.01121.x [DOI] [PubMed] [Google Scholar]
- 13.Noll LW, Belagola Shridhar P, Dewsbury DM, Shi X, Cernicchiaro N, Renter DG, et al. Culture- and PCR-Based Methods to Detect Seven Major Serogroups of Shiga Toxin-Producing Escherichia coli in Cattle Feces. PloS one. 2015;(In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 'Touchdown' PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19(14):4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. Journal of clinical microbiology. 2012;50(9):2951–63. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beutin L, Miko A, Krause G, Pries K, Haby S, Steege K, et al. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Applied and environmental microbiology. 2007;73(15):4769–75. 10.1128/AEM.00873-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33(1):159–74. [PubMed] [Google Scholar]
- 18.McNemar Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 1947;12(2):153–7. [DOI] [PubMed] [Google Scholar]
- 19.Baranzoni GM, Fratamico PM, Rubio F, Glaze T, Bagi LK, Albonetti S. Detection and isolation of Shiga toxin-producing Escherichia coli (STEC) O104 from sprouts. Int J Food Microbiol. 2014;173:99–104. 10.1016/j.ijfoodmicro.2013.12.020 . [DOI] [PubMed] [Google Scholar]
- 20.Delannoy S, Beutin L, Burgos Y, Fach P. Specific detection of enteroaggregative hemorrhagic Escherichia coli O104:H4 strains by use of the CRISPR locus as a target for a diagnostic real-time PCR. Journal of clinical microbiology. 2012;50(11):3485–92. 10.1128/jcm.01656-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amézquita-López BA, Quiñones B, Cooley MB, León-Félix J, Castro-del Campo N, Mandrell RE, et al. Genotypic analyses of Shiga toxin-producing Escherichia coli O157 and non-O157 recovered from feces of domestic animals on rural farms in Mexico. PloS one. 2012;7(12):e51565 10.1371/journal.pone.0051565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manna SK, Manna C, Batabyal K, Das B, Golder D, Chattopadhyay S, et al. Serogroup distribution and virulence characteristics of sorbitol-negative Escherichia coli from food and cattle stool. Journal of applied microbiology. 2010;108(2):658–65. 10.1111/j.1365-2672.2009.04460.x [DOI] [PubMed] [Google Scholar]
- 23.Blanco M, Blanco JE, Mora A, Rey J, Alonso JM, Hermoso M, et al. Serotypes, Virulence Genes, and Intimin Types of Shiga Toxin (Verotoxin)-Producing Escherichia coli Isolates from Healthy Sheep in Spain. Journal of clinical microbiology. 2003;41(4):1351–6. 10.1128/jcm.41.4.1351-1356.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco M, Blanco JE, Mora A, Dahbi G, Alonso MP, Gonzalez EA, et al. Serotypes, Virulence Genes, and Intimin Types of Shiga Toxin (Verotoxin)-Producing Escherichia coli Isolates from Cattle in Spain and Identification of a New Intimin Variant Gene (eae-). Journal of clinical microbiology. 2004;42(2):645–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.European Centre for Disease Prevention and Control EFSA. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2011. EFSA Journal. 2013;11(4):3129. [Google Scholar]
- 26.Friedrich AW, Borell J, Bielaszewska M, Fruth A, Tschape H, Karch H. Shiga Toxin 1c-Producing Escherichia coli Strains: Phenotypic and Genetic Characterization and Association with Human Disease. Journal of clinical microbiology. 2003;41(6):2448–53. 10.1128/jcm.41.6.2448-2453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nataro JP, Yikang D, Giron JA, Savarino SJ, Kothary MH, Hall R. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infection and immunity. 1993;61(3):1126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cleary J, Lai L-C, Shaw RK, Straatman-Iwanowska A, Donnenberg MS, Frankel G, et al. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology. 2004;150(3):527–38. 10.1099/mic.0.26740-0 [DOI] [PubMed] [Google Scholar]
- 29.Moriel DG, Rosini R, Seib KL, Serino L, Pizza M, Rappuoli R. Escherichia coli: great diversity around a common core. mBio. 2012;3(3). 10.1128/mBio.00118-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gamian A, Kenne L. Analysis of 7-substituted sialic acid in some enterobacterial lipopolysaccharides. Journal of bacteriology. 1993;175(5):1508–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kedzierska B. N-Acetylneuraminic acid: a constituent of the lipopolysaccharide of Salmonella toucra. Eur J Biochem. 1978;91(2):545–52. [DOI] [PubMed] [Google Scholar]
- 32.Hao W, Allen VG, Jamieson FB, Low DE, Alexander DC. Phylogenetic incongruence in E. coli O104: understanding the evolutionary relationships of emerging pathogens in the face of homologous recombination. PloS one. 2012;7(4):e33971 10.1371/journal.pone.0033971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai J, Paddock ZD, Shi X, Li S, An B, Nagaraja TG. Applicability of a multiplex PCR to detect the seven major Shiga toxin-producing Escherichia coli based on genes that code for serogroup-specific O-antigens and major virulence factors in cattle feces. Foodborne pathogens and disease. 2012;9(6):541–8. 10.1089/fpd.2011.1082 [DOI] [PubMed] [Google Scholar]
- 34.Bai J, Shi X, Nagaraja TG. A multiplex PCR procedure for the detection of six major virulence genes in Escherichia coli O157:H7. Journal of microbiological methods. 2010;82(1):85–9. 10.1016/j.mimet.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 35.Sekse C, Sunde M, Lindstedt B-A, Hopp P, Bruheim T, Cudjoe KS, et al. Potentially Human-Pathogenic Escherichia coli O26 in Norwegian Sheep Flocks. Applied and environmental microbiology. 2011;77(14):4949–58. 10.1128/aem.00189-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.