Abstract
Amyloid imaging plays an important role in the research and diagnosis of dementing disorders. Substantial variation in quantitative methods to measure brain amyloid burden exists in the field. The aim of this work is to investigate the impact of methodological variations to the quantification of amyloid burden using data from the Dominantly Inherited Alzheimer’s Network (DIAN), an autosomal dominant Alzheimer’s disease population. Cross-sectional and longitudinal [11C]-Pittsburgh Compound B (PiB) PET imaging data from the DIAN study were analyzed. Four candidate reference regions were investigated for estimation of brain amyloid burden. A regional spread function based technique was also investigated for the correction of partial volume effects. Cerebellar cortex, brain-stem, and white matter regions all had stable tracer retention during the course of disease. Partial volume correction consistently improves sensitivity to group differences and longitudinal changes over time. White matter referencing improved statistical power in the detecting longitudinal changes in relative tracer retention; however, the reason for this improvement is unclear and requires further investigation. Full dynamic acquisition and kinetic modeling improved statistical power although it may add cost and time. Several technical variations to amyloid burden quantification were examined in this study. Partial volume correction emerged as the strategy that most consistently improved statistical power for the detection of both longitudinal changes and across-group differences. For the autosomal dominant Alzheimer’s disease population with PiB imaging, utilizing brainstem as a reference region with partial volume correction may be optimal for current interventional trials. Further investigation of technical issues in quantitative amyloid imaging in different study populations using different amyloid imaging tracers is warranted.
Introduction
Alzheimer’s disease (AD) is the most common form of dementia [1] with its prevalence expected to dramatically increase in the next 50 years [2]. AD pathology begins to accumulate at least 10 to 20 years before clinical symptoms appear [1,3,4], and there is a growing consensus that effective treatment of AD will require early intervention [5,6]. The amyloid cascade is the primary target of the largest ongoing clinical trials [7], including DIAN-TU (DIAN Trial Unit) [8], A4 trial (Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease) [9], and API (Alzheimer’s Prevention Initiative) [10] in preclinical population. Well validated biomarkers of amyloid accumulation are needed for these treatment development efforts and trial design [5,6].
The most commonly used quantification technique for amyloid Positron Emission Tomography (PET) imaging involves comparing regional uptake to cerebellar cortex [11,12]. However, the observation of amyloid deposition in the cerebellar cortex of familial AD cases [13,14] had led to the identification and validation of pons as an alternative reference region [15]. A large white matter region of interest (ROI) has been proposed as an reference region to provide improved discrimination between clinically defined groups [16]; and a smaller “core” white matter reference region was reported to improve the detection of changes in tracer retention in longitudinal studies within the Alzheimer’s Disease Neuroimaging Initiative (ADNI) [18F]-florbetapir data [17].
Correction of partial volume effects represents another area of active investigation [18]. PET imaging based measurement is subjected to partial volume effects, because of the low spatial resolution [19]. Although partial volume effects are well-recognized, correction techniques remain controversial. One study comparing two-component (brain and non-brain) vs. three-component (gray matter, white matter, non-brain) reported relative advantages and disadvantages of both methods [20]. Thomas et al. [21] reported improved quantitative accuracy using a region-based voxel-wise partial volume correction (PVC) method. More recently, Brendel et al. [16] and our group [18] found PVC improved the power to detect longitudinal amyloid burden change.
A third methodological choice concerns the quantitative model used to determine amyloid burden. The most commonly used approach is the target-to-reference region standard-uptake-value-ratio (SUVR) [4,16,17]. SUVR evaluation is simple and only requires a short acquisition, which translates to low cost. However, SUVR measurements are sensitive to the choice of temporal window used for evaluation, in part, owing to individual variability in cerebral perfusion [16,22]. Alternatively, kinetic modeling techniques [23,24] can be used to calculate distribution volume ratio (DVR) or binding potential (BPND) as a quantitative measure of amyloid burden [11,22]. These techniques require longer dynamic PET acquisitions initiated in synchrony with tracer administration and are less attractive because of increased participant burden, study cost and complexity of quantification.
It should be pointed out that currently a “gold standard” technique that objectively measures brain amyloid burden is not available for validation of in vivo amyloid PET measurements. Some other criteria are often used to judge the performance of amyloid quantification techniques. These criteria include low inter-subject variability in reference regions [15,22], strong group differences between control subjects and AD patients [15,16,22], and, in longitudinal studies, strong longitudinal changes [16,17,25]. Here, we investigate the impact of 1) reference region selection; 2) correction for partial volume effects; and 3) choice of quantification technique using cross-sectional and longitudinal [11C]-Pittsburgh Compound B (PiB) imaging data from the DIAN study. Regional SUV and volume of distribution (VT) were used to determine the stability of reference regions tracer uptake by comparing mutation carriers against non-carriers. The power of detecting longitudinal changes was also examined to compare different amyloid quantification techniques.
Materials and Methods
Participants
The participants included in this study were recruited as part of the international Dominantly Inherited Alzheimer Network (DIAN) [26]. The DIAN includes individuals from families with known autosomal dominant mutation in amyloid precursor protein [27], presenilin 1 [28], or presenilin 2 [29] genes leading to early onset AD. The current study was based on the eighth semiannual data cutoff with a total of 341 participants who had at least one PiB scan; among them 203 were mutation carriers (APP = 27, PSEN1 = 161, PSEN2 = 15); within these 203 participants, 59 had usable PiB scans for at least two visits on the same scanners and formed the longitudinal mutation carrier cohort (LC). For participants with more than two PiB scans, the first two usable visits with an interval as close to 2 years as possible (range 0.8–3.3 yrs) was included in this study. The same scanner rule was enforced to minimize the impact of scanner difference. Within the LC cohort, a subset of 23 participants (LC_Dyn) had full dynamic scans at both time points. A longitudinal cohort for non-carriers (LNC) was also selected using the same criteria and had a total of 36 participants. A cross-sectional cohort (CC) was selected from the Washington University in St. Louis (WU) site DIAN cohort, including 69 participants (38 mutation carriers) with valid baseline PiB scans on the same PET scanner. Single scanner data minimizes differences in scanner calibration protocols. In addition, 42 participants (21 mutation carriers) from the WU site who had time-of-flight (TOF) MR angiography (MRA) scans and 70-min dynamic PiB PET scans were included for absolute quantification (AQ) of PiB binding using an image-derived arterial input function (IDAIF) technique [30]. Demographic details of these cohorts are provided in Table 1.
Table 1. Demographics Summary.
| Cohort | LC | LC_Dyn | CC_Carrier | CC_Noncarrier | AQ_Carrier | AQ_Noncarrier | LNC |
|---|---|---|---|---|---|---|---|
| N | 59 | 23 | 38 | 31 | 21 | 20 | 36 |
| Age (SD) years | 42.6 (8.7) | 44.8 (10.8) | 38.4 (10.5) | 39.3 (9.5) | 37.4 (11.2) | 39.9 (9.1) | 41.3 (9.6) |
| EYO (SD) years | -2.1 (8.6) | -1.1 (11.3) | -8.8 (9.4) | -5.7 (10.9) | -10.0 (10.3) | -5.5 (9.6) | -4.5 (10.2) |
| Education (SD) years | 14.0 (2.8) | 15.0 (3.2) | 15.5 (3.1) | 15.5 (2.4) | 15.2 (2.6) | 15.5 (2.5) | 14.9 (2.2) |
| Interval (SD) years | 1.7 (0.8) | 1.9 (0.8) | - | - | - | - | 2.4 (0.9) |
| Male (%) | 30 (50.8) | 17 (73.9) | 27 (71.1) | 15 (48.4) | 16 (76.1) | 9 (45.0) | 14 (38.9) |
| APOE4+ (%) | 23 (39.0) | 11 (47.8) | 14 (36.8) | 10 (32.3) | 11 (52.3) | 5 (25.0) | 11 (30.6) |
| CDR>0 (%) | 28 (47.5) | 10 (43.5) | 11 (28.9)* | 0 (0) | 4 (19.0) | 0 (0) | 0 (0) |
| MMSE (SD) | 26.2 (5.2) | 28.2 (2.5)† | 28.2 (3.4)* | 29.6 (0.6) | 28.1 (3.1) | 29.4 (0.7) | 29.1 (1.1) |
LC = longitudinal cohort; CC_Carrier = cross-sectional cohort mutation carrier; CC_Noncarrier = cross-sectional cohort non-carrier; AQ_Carrier = absolute quantification cohort mutation carrier; AQ_Noncarrier = absolute quantification cohort non-carrier; LNC = longitudinal non-carrier cochort; baseline values were reported for LC and LNC; EYO = estimated years to onset; SD = standard deviation; APOE4+ = apolipoprotein E ε4 gene carrier; CDR = clinical dementia rating; MMSE = Mini Mental State Examination
*significantly different from corresponding non-carrier group
†significantly different from the LC group
All assessment and imaging procedures were approved by the WU Human Research Protection Office. Written informed consent was obtained from all individuals or their care-givers. Local institutional review boards (Columbia University Institutional Review Board; University of Pittsburgh Human Research Protection Office; UCLA Institutional Review Boards; Indiana University Institutional Review Boards; Partners Human Research Committee; Butler Hospital's Institutional Review Board for Human Subjects Research; The University of New South Wales Human Research Ethics Committee; University of Melbourne Office for Research Ethics and Integrity; Edith Cowan University Human Research Ethics Committee; and University College London Research Ethics Committee) also approved the collection of scans for archiving and future study at each non-WU study site.
Clinical Assessment
The clinical assessment protocol has been previously described [3,4]. In brief, dementia status was assessed using the clinical dementia rating (CDR) [31]. Estimated year to onset (EYO) was calculated as the difference between the participant’s age at evaluation and the age at which parental cognitive decline began [3]. The ADAD mutation status was determined using polymerase chain reaction based amplification of the appropriate exon followed by Sanger sequencing [3]. Each participant’s apolipoprotein E (APOE) genotype was determined using previously described methods [32]. Clinical evaluators were blind to participant mutation status.
Image Acquisition
Imaging protocol has been described in detail previously [4]. PiB PET acquisition consisted of either a 70-min dynamic scan starting at injection or a 30-min scan with 40 minutes uptake time. Accelerated 3D sagittal T1-weighted images of the head were acquired in each participant. TOF-MRA data was acquired with during the same imaging session as the T1-weighted structural scan for the AQ cohort.
Image Analysis
Our image analysis technique has been previously described in detail [12]. In summary, FreeSurfer v5.1 (Martinos Center for Biomedical Imaging, Charlestown, Massachusetts, USA, https://surfer.nmr.mgh.harvard.edu/fswiki) was used to automatically segment the brain using T1-weighted MR acquired in corresponding visit as the PET scan. A PET Unified Pipeline (PUP) (https://github.com/ysu001/PUP) was used for automated PET data analysis [12,18]. In the default SUVR analysis, regional SUVR was calculated using cerebellar cortex as the reference region and PET data acquired between 40 and 70 minutes post-injection. When a full 70-min dynamic scan was available, regional time-activity curve for each ROI was extracted followed by regional binding potential (BPND) (DVR-1) estimation using Logan graphical analysis with the cerebellar cortex (CER) serving as the reference [23]. Mean cortical binding potentials (MCBP) [11] and mean cortical SUVR (MCSUVR) were calculated based on a selected set of cortical FreeSurfer regions [12]. PVC was also performed using a regional spread function (RSF) technique implemented in PUP [18], which is also known as the geometric transfer matrix (GTM) technique [33]. Regional SUVR and BPND were estimated with and without PVC.
Alternative Reference Region
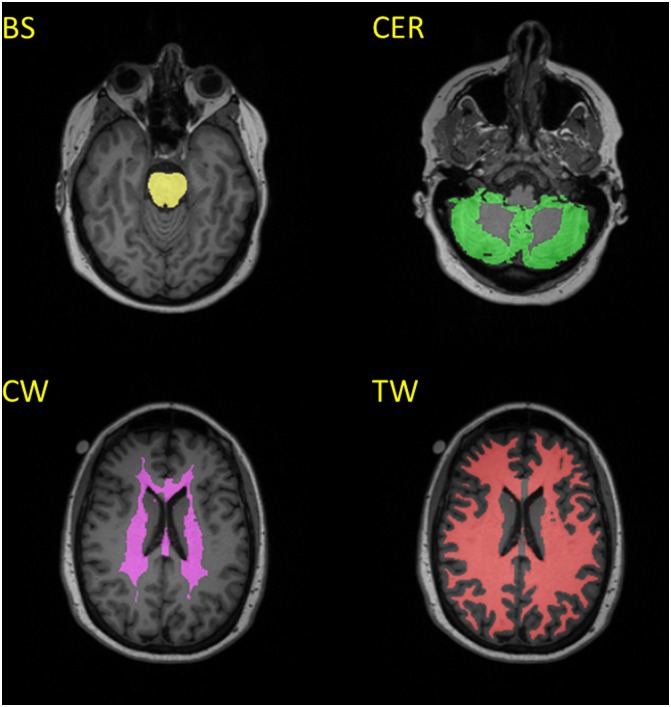
Recent papers [15–17] have proposed three alternative SUVR reference regions. We implemented these alternative reference regions based on FreeSurfer segmentation (Fig 1). The brain stem (BS) region was used to approximate the pontine reference, as proposed by Edison et al. [15]. A core white matter (CW) reference region, as proposed by Chen et al. [17], was approximated by combining corpus callosum and the ROI labelled as “UnsegmentedWhiteMatter” by FreeSurfer. A total white matter (TW) reference region was also constructed using all the cerebral white matter [16].
Fig 1. Example reference regions.
(BS: Brain-Stem; CER: Cerebellar Cortex; CW: Core White Matter; TW: Total White Matter).
Cross-sectional SUV Analysis
For the cross-sectional cohort, regional SUV were estimated by normalizing regional activity concentration with injection dose and body weight. SUV values were calculated for the four candidate reference regions (CER, BS, CW, and TW) and the mean cortical (MC) regions using the same time window as the SUVR analysis.
Absolute Quantification
An IDAIF was obtained using a recently developed technique by combined analysis of TOF-MRA, T1-weighted MR, and dynamic PET imaging data [30,34]. Logan graphical analysis [35] was then applied to obtain regional VT, which is a measurement of absolute tracer binding. The quantification was performed on the same set of regions as the SUV analysis, and results were presented with and without PVC.
Longitudinal Analysis
Longitudinal change in amyloid burden was assessed as the absolute change (delta), percent change (delta%) and annual rate of change (rate) in MCSUVR using all four candidate reference regions with and without RSF PVC. Follow-up MCSUVRs were compared with their corresponding baseline measurements using paired t-test. Cohen’s d effect size measure was calculated based on the mean and standard deviation of the annual rate of change in MCSUVR. Percent change in MCSUVR was also compared between quantification methods using paired t-test. In addition, we also estimated the number of participants per arm needed to detect a 25% and 50% reduction in amyloid accumulation rate due to treatment with 80% power and a two-tailed type-I error of p = 0.05 in a 12-month placebo-controlled randomized clinical trial. The 25% reduction was selected to match a similar study [17]. The 50% reduction was included to approximate hypothesized treatment effects in anti-amyloid treatment trials. The sample size calculation was performed using MATLAB (R2015b) (The Mathworks Inc, Natick, MA) function: sampsizepwr, in the statistics and machine learning toolbox v10.1. The calculation was performed based on the observed annual rate of change in MCSUVR and its standard deviation. LNC cohort data were also analyzed as a test-retest study, since amyloid burden and its change are not expected in this cohort of young participants. A test-retest variability measure was defined as:TRT% = (|M1-M2|/M1)×100%, where M1 was the amyloid burden measurement at baseline and M2 was the follow-up measurement. The mean and standard deviation of TRT% was evaluated for the LNC cohort and compared among different quantification methods.
Results
Cross-sectional SUV Analysis
Significant MCSUV differences were observed between mutation carriers and non-carriers with and without PVC (Table 2). SUVs for the TW region were significantly different between mutation carriers and non-carriers, however, PVC removes this difference. Regional SUVs were not significantly different for any of the other three reference regions regardless of PVC.
Table 2. Regional SUV for the cross-sectional cohort.
| MC | CER | BS | CW | TW | MCRSF | CERRSF | BSRSF | CWRSF | TWRSF | |
|---|---|---|---|---|---|---|---|---|---|---|
| Noncarrier | 0.64±0.11 | 0.60±0.10 | 0.96±0.20 | 0.99±0.21 | 0.87±0.18 | 0.56±0.10 | 0.56±0.09 | 1.06±0.23 | 1.07±0.24 | 1.04±0.24 |
| Carrier | 0.97±0.39 | 0.62±0.12 | 0.97±0.20 | 1.00±0.20 | 1.00±0.27 | 1.26±0.73 | 0.58±0.12 | 1.07±0.24 | 1.02±0.20 | 1.01±0.21 |
| p | 9.10E-06 | 3.55E-01 | 8.53E-01 | 9.29E-01 | 1.43E-02 | 8.76E-07 | 3.37E-01 | 8.92E-01 | 3.27E-01 | 6.60E-01 |
MC = mean cortical regions; CER = cerebellar cortex; BS = brainstem; CW = core white matter; TW = total white matter; MCRSF = mean cortical regions with RSF partial volume correction; CERRSF = cerebellar cortex with RSF partial volume correction; BSRSF = brainstem with RSF partial volume correction; CWRSF = core white matter with RSF partial volume correction; TWRSF = total white matter with RSF partial volume correction; p is the strength of the group difference, i.e. noncarrier vs. carrier, for each region of interest based on student t-test.
Absolute Quantification
VT was significantly different for MC regions between the mutation carriers and non-carriers, and PVC led to smaller p values (Table 3). No group differences in regional VT were observed for any of the reference regions.
Table 3. Regional volume of distribution (VT) estimated using image-derived arterial input function.
| MC | CER | BS | CW | TW | MCRSF | CERRSF | BSRSF | CWRSF | TWRSF | |
|---|---|---|---|---|---|---|---|---|---|---|
| Noncarrier | 2.79±1.00 | 2.75±0.98 | 3.66±1.27 | 3.71±1.49 | 3.35±1.26 | 3.16±1.14 | 2.83±1.02 | 4.20±1.45 | 4.13±1.81 | 3.90±1.56 |
| Carrier | 3.70±1.07 | 2.71±0.62 | 3.55±0.80 | 3.68±0.86 | 3.77±0.87 | 5.20±2.06 | 2.80±0.64 | 4.06±0.93 | 3.79±1.01 | 3.72±0.86 |
| p | 7.78E-03 | 8.83E-01 | 7.28E-01 | 9.22E-01 | 2.27E-01 | 4.17E-04 | 9.07E-01 | 7.14E-01 | 4.69E-01 | 6.54E-01 |
MC = mean cortical regions; CER = cerebellar cortex; BS = brainstem; CW = core white matter; TW = total white matter; MCRSF = mean cortical regions with RSF partial volume correction; CERRSF = cerebellar cortex with RSF partial volume correction; BSRSF = brainstem with RSF partial volume correction; CWRSF = core white matter with RSF partial volume correction; TWRSF = total white matter with RSF partial volume correction; p is the strength of the group difference, i.e. noncarrier vs. carrier, for each region of interest based on student t-test.
Longitudinal Analysis
For the LC cohort, using either cerebellar cortex or brain stem as the reference region, longitudinal MCSUVR change was not significant without partial volume, while PVC revealed the change (Table 4). Using core white matter and total white matter reference regions revealed significant longitudinal change in MCSUVR with or without PVC (Table 4). The MCSUVR change from baseline to follow-up had the smallest p values with (p = 0.0000075) and without (p = 0.000017) PVC when core white matter was used as the reference. PVC and using white matter as reference region also led to larger effect size (Table 4). PVC based analysis generated significantly (p<0.0005) larger longitudinal percent change in MCSUVR regardless of the reference regions used (Table 4). PVC and adoption of white matter as reference led to a considerably smaller estimated sample size needed to achieve an 80% power and two-sided type-I error of p = 0.05 to detect a reduced rate of amyloid accumulation in hypothetical anti-amyloid therapy trials.
Table 4. Longitudinal SUVR analysis for mean cortical regions in mutation carriers.
| MC_CER | MC_BS | MC_CW | MC_TW | MCRSF_CER | MCRSF_BS | MCRSF_CW | MCRSF_TW | |
|---|---|---|---|---|---|---|---|---|
| Baseline | 1.73±0.58 | 1.15±0.35 | 1.08±0.31 | 0.99±0.14 | 2.54±1.28 | 1.43±0.63 | 1.53±0.73 | 1.43±0.56 |
| follow-up | 1.76±0.60 | 1.17±0.35 | 1.12±0.32 | 1.00±0.14 | 2.65±1.36 | 1.48±0.64 | 1.64±0.80 | 1.49±0.57 |
| delta | 0.03±0.11 | 0.01±0.07 | 0.04±0.07 | 0.01±0.03 | 0.11±0.26 | 0.05±0.10 | 0.11±0.17 | 0.06±0.11 |
| delta% | 1.63±7.04 | 1.51±6.30† | 3.34±5.88 | 1.29±3.54 | 4.19±11.07*‡ | 4.08±9.86*‡ | 7.03±11.27* | 5.05±9.82* |
| p (follow-up vs. Baseline) | 7.22E-02 | 1.39E-01 | 1.70E-05 | 8.12E-03 | 8.78E-04 | 6.25E-04 | 7.46E-06 | 1.14E-04 |
| Rate | 0.01±0.08 | 0.00±0.04 | 0.02±0.04 | 0.01±0.02 | 0.07±0.18 | 0.02±0.06 | 0.07±0.13 | 0.04±0.07 |
| Effect Size | 0.15 | 0.07 | 0.55 | 0.38 | 0.38 | 0.38 | 0.51 | 0.50 |
| sample size (25% reduction in Rate) | 5714 | 24165 | 411 | 885 | 858 | 852 | 480 | 502 |
| sample size (50% reduction in Rate) | 1430 | 6043 | 105 | 223 | 216 | 215 | 122 | 127 |
MC_CER = mean cortical region SUVR using cerebellar cortex as reference; MC_BS = mean cortical region SUVR using brainstem as reference; MC_CW = mean cortical region SUVR using core white matter as reference; MC_TW = mean cortical region SUVR using total white matter as reference; MCRSF_CER = mean cortical region SUVR using cerebellar cortex as reference with RSF partial volume correction; MCRSF_BS = mean cortical region SUVR using brainstem as reference with RSF partial volume correction; MCRSF_CW = mean cortical region SUVR using core white matter as reference with RSF partial volume correction; MCRSF_TW = mean cortical region SUVR using total white matter as reference with RSF partial volume correction; delta = change in SUVR from baseline to follow-up; delta% = percent change in SUVR from baseline to follow-up; p is the strength of the difference between follow-up and baseline SUVRs based on a paired t-test; Rate = the annual rate of SUVR change; sample size is the estimated number of participants per arm needed to detect a 25% or a 50% reduction in amyloid accumulation rate due to treatment with 80% power and a two-tailed type-I error of p = 0.05 in a 12-month placebo-controlled randomized clinical trial.
*percent change in MCSUVR significantly greater with PVC than without (p<0.0005)
†percent change in MCSUVR significantly smaller than CW referencing (p<0.01)
‡percent change in MCSUVR with PVC significantly smaller than CW referencing (p<0.05)
For the LNC cohort, no change in MCSUVR was observed, as expected, between baseline and follow-up regardless of the quantification methods used. It is observed that cerebellar cortex referencing led to the smallest test-retest variability (Table 5), while the other reference regions did not differ significantly.
Table 5. Reproducibility of PiB measurements based on the longitudinal non-carriers cohort (LNC).
| MC_CER | MC_BS | MC_CW | MC_TW | MCRSF_CER | MCRSF_BS | MCRSF_CW | MCRSF_TW | |
|---|---|---|---|---|---|---|---|---|
| TRT% (Mean±SD) | 3.0±2.7 | 4.0±3.8 | 4.5±4.0 | 3.0±2.4 | 5.0±3.7 | 7.1±7.3 | 9.2±8.3 | 8.5±7.5 |
MC_CER = mean cortical region SUVR using cerebellar cortex as reference; MC_BS = mean cortical region SUVR using brainstem as reference; MC_CW = mean cortical region SUVR using core white matter as reference; MC_TW = mean cortical region SUVR using total white matter as reference; MCRSF_CER = mean cortical region SUVR using cerebellar cortex as reference with RSF partial volume correction; MCRSF_BS = mean cortical region SUVR using brainstem as reference with RSF partial volume correction; MCRSF_CW = mean cortical region SUVR using core white matter as reference with RSF partial volume correction; MCRSF_TW = mean cortical region SUVR using total white matter as reference with RSF partial volume correction; TRT% = (|M1-M2|/M1)×100% is the test-retest reproducibility of SUVR measurement.
Impact of Dynamic Acquisition
The estimated sample sizes for anti-amyloid therapy trials were smaller based on LC_Dyn data compared to the LC and the observed longitudinal MCSUVR change had smaller p values and larger effect size (Table 6 vs. Table 4). PVC had less of an impact on BPND compared to SUVR analyses.
Table 6. Mean cortical measurement for longitudinal cohort participants with full dynamic PiB.
| MC_CER | MC_BS | MC_CW | MC_TW | MCRSF_CER | MCRSF_BS | MCRSF_CW | MCRSF_TW | MCBP | MCBPRSF | |
|---|---|---|---|---|---|---|---|---|---|---|
| Baseline | 1.83±0.59 | 1.18±0.35 | 1.12±0.31 | 1.02±0.16 | 2.80±1.33 | 1.50±0.64 | 1.60±0.71 | 1.54±0.62 | 0.62±0.45 | 1.33±0.93 |
| follow-up | 1.89±0.58 | 1.22±0.35 | 1.16±0.32 | 1.03±0.15 | 2.93±1.33 | 1.58±0.66 | 1.71±0.75 | 1.60±0.62 | 0.67±0.45 | 1.43±0.95 |
| Delta | 0.05±0.13 | 0.04±0.06 | 0.04±0.05 | 0.01±0.04 | 0.14±0.29 | 0.09±0.09 | 0.11±0.12 | 0.06±0.11 | 0.05±0.10 | 0.10±0.22 |
| p (follow-up vs. Baseline) | 6.34E-02 | 1.98E-03 | 4.47E-04 | 8.39E-02 | 3.33E-02 | 2.02E-04 | 1.68E-04 | 1.19E-02 | 2.91E-02 | 3.84E-02 |
| Rate | 0.02±0.09 | 0.02±0.03 | 0.02±0.03 | 0.00±0.02 | 0.05±0.17 | 0.04±0.05 | 0.05±0.06 | 0.03±0.05 | 0.02±0.05 | 0.04±0.11 |
| Effect Size | 0.23 | 0.62 | 0.71 | 0.25 | 0.33 | 0.85 | 0.82 | 0.49 | 0.33 | 0.34 |
| sample size (25% reduction in Rate) | 2286 | 333 | 251 | 2038 | 1171 | 177 | 188 | 519 | 1177 | 1097 |
| sample size (50% reduction in Rate) | 573 | 85 | 65 | 511 | 295 | 46 | 49 | 132 | 296 | 276 |
MC_CER = mean cortical region SUVR using cerebellar cortex as reference; MC_BS = mean cortical region SUVR using brainstem as reference; MC_CW = mean cortical region SUVR using core white matter as reference; MC_TW = mean cortical region SUVR using total white matter as reference; MCRSF_CER = mean cortical region SUVR using cerebellar cortex as reference with RSF partial volume correction; MCRSF_BS = mean cortical region SUVR using brainstem as reference with RSF partial volume correction; MCRSF_CW = mean cortical region SUVR using core white matter as reference with RSF partial volume correction; MCRSF_TW = mean cortical region SUVR using total white matter as reference with RSF partial volume correction; MCBP = mean cortical binding potential; MCBPRSF = mean cortical binding potential with RSF partial volume correction; delta = change in SUVR from baseline to follow-up; p is the strength of the difference between follow-up and baseline SUVRs based on a paired t-test; Rate = the annual rate of SUVR change; sample size is the estimated number of participants per arm needed to detect a 25% or a 50% reduction in amyloid accumulation rate due to treatment with 80% power and a two-tailed type-I error of p = 0.05 in a 12-month placebo-controlled randomized clinical trial.
Discussion
Reference Region
Both SUV analysis and IDAIF based absolute quantification showed all four reference regions were not different between mutation carriers and non-carriers. White matter referencing, especially using core white matter ROI, improved the sensitivity for detecting longitudinal changes in MCSUVR, however, the cause of this improvement is unclear and requires further investigation. White matter PiB uptake was not more stable in either the non-carrier control population or the mutation carrier participants. In fact, the coefficient of variation was greater for white matter than for cerebellar cortex and brain stem. In the LNC cohort, larger test-retest variability was observed when core white matter was used as reference region in comparison to cerebellar cortex referencing. On the other hand, a (non-significant) trend was observed towards lower white matter uptake (for both core and total white matter regions) for mutation carriers than noncarriers in the IDAIF analysis and partial volume corrected SUV, suggesting potential changes in white matter region properties in the mutation carriers that may alter white matter tracer uptake. We suspect such changes in the white matter region over time may, in part, be the cause of the improved power in detecting longitudinal MCSUVR change using white matter referencing. In addition, the observed longitudinal change in MCSUVR agreed well (Pearson r = 0.66, p = 1.03E-8) when cerebellar cortex and brain stem were used as the reference region. However, the longitudinal MCSUVR changes measured using white matter as reference region did not agree (Pearson r = 0.25, p = 0.052) with the results obtained using cerebellar cortex referencing. These differences suggest that using white matter as the reference region potentially leads to less consistent results.
Partial Volume Correction
We show that PVC enhances sensitivity for the detection of longitudinal changes in PiB binding in the DIAN cohort, in accordance with our previous report in a sporadic AD cohort [18]. In fact, PVC consistently increased the observed longitudinal percent change in measured PiB uptake regardless of the reference region. Two separate characteristics of PVC technique may be the causes of the beneficial effects. Firstly, PVC amplifies the signal we want to detect more than the amplification of noise therefore improve signal to noise ratio. Secondly, without PVC, MCSUVR and its change over time is dependent upon both amyloid deposition and brain atrophy; when PVC is performed we can separate the two processes. Therefore, we strongly recommend PVC especially in longitudinal studies aimed at detecting changes in amyloid burden.
Full Dynamic Acquisition and Modeling
Moderate improvement was observed when full dynamic PET acquisition was available. As previously reported [36], SUVR analysis is more sensitive to the choice of time window used for quantification. Improved statistical power with full dynamic scanning likely is attributable to greater consistency in the selection of the time window used for quantification. In short acquisitions, uptake time may vary from one scan to the next. Regional binding potential estimation provided improved statistical power in comparison to SUVR, as previously reported [36]. Nevertheless, full kinetic modeling requires longer scan time and the quantification requires more PET imaging expertise, both of which could lead to increased cost. However, such increases in cost may be offset by improved accuracy, increased statistical power and reduced sample size. This trade-off should be carefully evaluated at study design.
Observed Longitudinal Change
In this study, moderate annual rate of change in amyloid burden was observed with substantial inter-individual variability. And it appears that the inter-individual variability was larger when cerebellar cortex was used as reference than when white matter were used as reference. Partial volume correction increased the magnitude of the annual rate of change no matter what reference region was used. Similar annual rate of change was observed in this cohort (0.02±0.05 in MCBP) in comparison to our previous observations in a sporadic AD cohort (0.016±0.03 in MCBP)[18]. Using ADNI [18F]-florbetapir data, Chen et al. [17] reported an annual rate of SUVR change of 0.013±0.011 in the asymptomatic amyloid positive group, and an annual rate of 0.012±0.014 in the MCI amyloid positive group using cerebral white matter as the reference. A similar rate of change was also reported by Brendel et al. [16] using white matter as reference region based on the same dataset. In comparison, in this current study in the DIAN cohort using PiB as the tracer, we observed a substantially larger annual rate of 0.02±0.03 in SUVR in a cohort that is a mixture of MCI (CDR = 0.5) and asymptomatic participants using similar reference region. The estimated sample size needed for anti-amyloid trials were similar between the two studies. The difference in annual rate of change and similarity of estimated sample size is likely a combined effects of cohorts, tracer, and quantification methods, and further investigation is necessary.
Limitations
The analysis performed in this study is based on the DIAN cohort using PiB as the imaging tracer. Whether our observations can be translated to other cohorts and/or using other amyloid tracers remains to be determined. We focused our comparison of quantification methodology on those utilizing structural MR obtained on the same participant. While MR data is generally available for most of the ongoing neuroimaging studies, quantification methods that does not require MR [37–39] have their advantages and warrant further investigation. In this study, we focused on a single PVC technique in based on our previous experience [18]. This technique depends on segmentation of high resolution structural MR data. Other techniques that do not rely on structural MR data are available, such as deconvolution based approaches [40] and resolution recovery techniques incorporated into image reconstruction [41], although they generally cannot achieve full recovery of the resolution. Nevertheless, these techniques is worth investigating further as well because they can be applied in cases where only PET data are available. One approach we took to assess the stability and noise properties of reference region tracer uptake was based on an IDAIF technique using population based parent compound ratio for metabolites correction [30]. It should be pointed out that there is potentially group difference in tracer metabolism as well as inter individual variability which may introduce noise into the estimated tracer uptake. This can be investigated by performing actual metabolites measurement in a similar cohort. Nevertheless, the observation we obtained using this approach is consistent with SUV analysis. Additional investigation is also warranted on the feasibility of using an IDAIF approach for absolute quantification of amyloid burden in general.
Conclusion
We examined several technical variations in the approach to obtain quantitative and semi-quantitative measurement of amyloid burden using PET imaging. PVC emerged as the strategy that most consistently improved statistical power for the detection of both longitudinal changes and across-group differences. This result accords with several previously reported studies [16,18,21]. Among several reference regions, core white matter provided the greatest sensitivity for the detection of longitudinal changes. However, the observed longitudinal change using white matter reference may be confounded by processes unrelated to amyloid burden and requires further investigation. We observed some advantages of acquiring full dynamic PET rather than shorter scans. However, given the countervailing costs, it remains unclear whether the advantages of full dynamic scanning outweigh the disadvantages. For the ADAD population with PiB imaging, utilizing brainstem as a reference region with PVC may be optimal for current interventional trials. Further investigation of technical issues in quantitative amyloid imaging in different study populations using different amyloid imaging tracers is warranted.
Acknowledgments
We acknowledge the altruism of the DIAN participants and their families and contributions of the DIAN research. We acknowledge the DIAN genetics core for APOE genotype and mutation status; the DIAN clinical core for providing CDR status; and the DIAN biostatistics, informatics, neuropathology, and biomarker cores for their assistance in the DIAN project. We acknowledge each of the participating sites (list follows in alphabetical order) and their staff for their contributions to this study (see DIAN website (www.DIAN-info.org) for a list of DIAN sites and other DIAN related information). We also acknowledge Alzheimer’s Disease Cooperative Study (ADCS) for coordination of clinical and neuropsychological evaluations and monitoring of the acquisition of imaging studies and collection of biological fluids; Mayo Clinic Rochester for MRI site training and standardization; University of Michigan for PET site training and standardization; and San Francisco Veteran’s Administration Medical Center and Alzheimer’s Disease Neuroimaging Initiative (ADNI) for the assistance in the conduct of project research. Support was also provided by the NIHR Queen Square Dementia Biomedical Research Unit.
Data Availability
In accordance with policies put in place by the DIAN Steering Committee, patient and imaging data are available upon request. The full policy regarding data access as well as instructions for submitting requests for data access are available here: http://www.dian-info.org/resourcedb/default.htm.
Funding Statement
YS received funds from the Knight Alzheimer's Disease Research Center (http://knightadrc.wustl.edu/) pilot award, Washington University Institute of Clinical and Translational Sciences (http://icts.wustl.edu/) Pilot Grant supported by the Clinical and Translational Science Award (CTSA) program (https://www.ctsacentral.org/) of National Institute of Health (http://www.nih.gov/): UL1TR000448. AZS and DSM received funds from National Institute of Neurological Disorders and Stroke (http://www.ninds.nih.gov/): P30NS048056; JCM and TLSB received funds from National Institute of Ageing (http://www.nia.nih.gov/): P01AG026276,U19AG032438, P50AG005681, P01AG003991. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Holtzman DM, Morris JC, Goate AM Alzheimer's Disease: The Challenge of the Second Century. Science Translational Medicine 2011;3: 77sr71–77sr71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer's A 2014 Alzheimer's disease facts and figures. Alzheimers Dement 2014;10: e47–92. [DOI] [PubMed] [Google Scholar]
- 3.Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer's disease. N Engl J Med 2012;367: 795–804. 10.1056/NEJMoa1202753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benzinger TL, Blazey T, Jack CR Jr., Koeppe RA, Su Y, Xiong C, et al. Regional variability of imaging biomarkers in autosomal dominant Alzheimer's disease. Proc Natl Acad Sci U S A 2013;110: E4502–4509. 10.1073/pnas.1317918110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aisen PS, Andrieu S, Sampaio C, Carrillo M, Khachaturian ZS, Dubois B, et al. Report of the task force on designing clinical trials in early (predementia) AD. Neurology 2011;76: 280–286. 10.1212/WNL.0b013e318207b1b9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aisen PS Alzheimer's disease therapeutic research: the path forward. Alzheimers Res Ther 2009;1: 2 10.1186/alzrt2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider LS, Mangialasche F, Andreasen N, Feldman H, Giacobini E, Jones R, et al. Clinical trials and late-stage drug development for Alzheimer's disease: an appraisal from 1984 to 2014. J Intern Med 2014;275: 251–283. 10.1111/joim.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills SM, Mallmann J, Santacruz AM, Fuqua A, Carril M, Aisen PS, et al. Preclinical trials in autosomal dominant AD: implementation of the DIAN-TU trial. Rev Neurol (Paris) 2013;169: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, et al. The A4 study: stopping AD before symptoms begin? Sci Transl Med 2014;6: 228fs213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiman EM, Langbaum JB, Fleisher AS, Caselli RJ, Chen K, Ayutyanont N, et al. Alzheimer's Prevention Initiative: a plan to accelerate the evaluation of presymptomatic treatments. J Alzheimers Dis 2011;26 Suppl 3: 321–329. 10.3233/JAD-2011-0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mintun MA, Larossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, et al. [11C]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology 2006;67: 446–452. [DOI] [PubMed] [Google Scholar]
- 12.Su Y, D'Angelo GM, Vlassenko AG, Zhou G, Snyder AZ, Marcus DS, et al. Quantitative analysis of PiB-PET with FreeSurfer ROIs. PLoS One 2013;8: e73377 10.1371/journal.pone.0073377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lippa CF, Saunders AM, Smith TW, Swearer JM, Drachman DA, Ghetti B, et al. Familial and sporadic Alzheimer's disease: neuropathology cannot exclude a final common pathway. Neurology 1996;46: 406–412. [DOI] [PubMed] [Google Scholar]
- 14.Mann DM, Pickering-Brown SM, Takeuchi A, Iwatsubo T Amyloid angiopathy and variability in amyloid beta deposition is determined by mutation position in presenilin-1-linked Alzheimer's disease. Am J Pathol 2001;158: 2165–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edison P, Hinz R, Ramlackhansingh A, Thomas J, Gelosa G, Archer HA, et al. Can target-to-pons ratio be used as a reliable method for the analysis of [(11)C]PIB brain scans? Neuroimage 2012;60: 1716–1723. 10.1016/j.neuroimage.2012.01.099 [DOI] [PubMed] [Google Scholar]
- 16.Brendel M, Hogenauer M, Delker A, Sauerbeck J, Bartenstein P, Seibyl J, et al. Improved longitudinal [(18)F]-AV45 amyloid PET by white matter reference and VOI-based partial volume effect correction. Neuroimage 2015;108: 450–459. 10.1016/j.neuroimage.2014.11.055 [DOI] [PubMed] [Google Scholar]
- 17.Chen K, Roontiva A, Thiyyagura P, Lee W, Liu X, Ayutyanont N, et al. Improved power for characterizing longitudinal amyloid-beta PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J Nucl Med 2015;56: 560–566. 10.2967/jnumed.114.149732 [DOI] [PubMed] [Google Scholar]
- 18.Su Y, Blazey TM, Snyder AZ, Raichle ME, Marcus DS, Ances BM, et al. Partial volume correction in quantitative amyloid imaging. Neuroimage 2015;107: 55–64. 10.1016/j.neuroimage.2014.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soret M, Bacharach SL, Buvat I Partial-volume effect in PET tumor imaging. J Nucl Med 2007;48: 932–945. [DOI] [PubMed] [Google Scholar]
- 20.Meltzer CC, Kinahan PE, Greer PJ, Nichols TE, Comtat C, Cantwell MN, et al. Comparative evaluation of MR-based partial-volume correction schemes for PET. J Nucl Med 1999;40: 2053–2065. [PubMed] [Google Scholar]
- 21.Thomas BA, Erlandsson K, Modat M, Thurfjell L, Vandenberghe R, Ourselin S, et al. The importance of appropriate partial volume correction for PET quantification in Alzheimer's disease. Eur J Nucl Med Mol Imaging 2011;38: 1104–1119. 10.1007/s00259-011-1745-9 [DOI] [PubMed] [Google Scholar]
- 22.Lopresti BJ, Klunk WE, Mathis CA, Hoge JA, Ziolko SK, Lu X, et al. Simplified quantification of Pittsburgh Compound B amyloid imaging PET studies: a comparative analysis. J Nucl Med 2005;46: 1959–1972. [PubMed] [Google Scholar]
- 23.Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, Alexoff DL Distribution volume ratios without blood sampling from graphical analysis of PET data. Journal of Cerebral Blood Flow and Metabolism 1996;16: 834–840. [DOI] [PubMed] [Google Scholar]
- 24.Lammertsma AA, Hume SP Simplified reference tissue model for PET receptor studies. Neuroimage 1996;4: 153–158. [DOI] [PubMed] [Google Scholar]
- 25.Landau SM, Fero A, Baker SL, Koeppe R, Mintun M, Chen K, et al. Measurement of Longitudinal beta-Amyloid Change with 18F-Florbetapir PET and Standardized Uptake Value Ratios. J Nucl Med 2015;56: 567–574. 10.2967/jnumed.114.148981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris JC, Aisen PS, Bateman RJ, Benzinger TL, Cairns NJ, Fagan AM, et al. Developing an international network for Alzheimer research: The Dominantly Inherited Alzheimer Network. Clin Investig (Lond) 2012;2: 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 1991;349: 704–706. [DOI] [PubMed] [Google Scholar]
- 28.Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer's disease. Nature 1995;375: 754–760. [DOI] [PubMed] [Google Scholar]
- 29.Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, et al. Candidate gene for the chromosome 1 familial Alzheimer's disease locus. Science 1995;269: 973–977. [DOI] [PubMed] [Google Scholar]
- 30.Su Y, Blazey TM, Snyder AZ, Raichle ME, Hornbeck RC, Aldea P, et al. Quantitative amyloid imaging using image-derived arterial input function. PLoS One 2015;10: e0122920 10.1371/journal.pone.0122920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris JC The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 1993;43: 2412–2414. [DOI] [PubMed] [Google Scholar]
- 32.Pastor P, Roe CM, Villegas A, Bedoya G, Chakraverty S, Garcia G, et al. Apolipoprotein Eepsilon4 modifies Alzheimer's disease onset in an E280A PS1 kindred. Ann Neurol 2003;54: 163–169. [DOI] [PubMed] [Google Scholar]
- 33.Rousset OG, Collins DL, Rahmim A, Wong DF Design and implementation of an automated partial volume correction in PET: application to dopamine receptor quantification in the normal human striatum. J Nucl Med 2008;49: 1097–1106. 10.2967/jnumed.107.048330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Y, Arbelaez AM, Benzinger TL, Snyder AZ, Vlassenko AG, Mintun MA, et al. Noninvasive estimation of the arterial input function in positron emission tomography imaging of cerebral blood flow. J Cereb Blood Flow Metab 2013;33: 115–121. 10.1038/jcbfm.2012.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Logan J, Fowler JS, Volkow ND, Wolf AP, Dewey SL, Schlyer DJ, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab 1990;10: 740–747. [DOI] [PubMed] [Google Scholar]
- 36.van Berckel BN, Ossenkoppele R, Tolboom N, Yaqub M, Foster-Dingley JC, Windhorst AD, et al. Longitudinal amyloid imaging using 11C-PiB: methodologic considerations. J Nucl Med 2013;54: 1570–1576. 10.2967/jnumed.112.113654 [DOI] [PubMed] [Google Scholar]
- 37.Joshi AD, Pontecorvo MJ, Clark CM, Carpenter AP, Jennings DL, Sadowsky CH, et al. Performance characteristics of amyloid PET with florbetapir F 18 in patients with alzheimer's disease and cognitively normal subjects. J Nucl Med 2012;53: 378–384. 10.2967/jnumed.111.090340 [DOI] [PubMed] [Google Scholar]
- 38.Bilgel M, Carass A, Resnick SM, Wong DF, Prince JL Deformation field correction for spatial normalization of PET images. Neuroimage 2015;119: 152–163. 10.1016/j.neuroimage.2015.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raniga P, Bourgeat P, Fripp J, Acosta O, Villemagne VL, Rowe C, et al. Automated (11)C-PiB standardized uptake value ratio. Acad Radiol 2008;15: 1376–1389. 10.1016/j.acra.2008.07.006 [DOI] [PubMed] [Google Scholar]
- 40.Tohka J, Reilhac A Deconvolution-based partial volume correction in Raclopride-PET and Monte Carlo comparison to MR-based method. Neuroimage 2008;39: 1570–1584. [DOI] [PubMed] [Google Scholar]
- 41.Panin VY, Kehren F, Michel C, Casey M Fully 3-D PET reconstruction with system matrix derived from point source measurements. IEEE Trans Med Imaging 2006;25: 907–921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
In accordance with policies put in place by the DIAN Steering Committee, patient and imaging data are available upon request. The full policy regarding data access as well as instructions for submitting requests for data access are available here: http://www.dian-info.org/resourcedb/default.htm.