Abstract
Background and Aims: The hepatitis C virus (HCV) genotype 1b is known to exhibit treatment resistance with respect to interferon (IFN) therapy. Substitution of amino acids 70 and 91 in the core region of the 1b genotype is a significant predictor of liver carcinogenesis and poor response to pegylated-IFN-α and ribavirin therapy. However, the molecular mechanism has not yet been clearly elucidated because of limitations of the HCV genotype 1b infectious model. Recently, the TPF1-M170T HCV genotype 1b cell culture system was established, in which the clone successfully replicates and infects Huh-7-derived Huh7-ALS32.50 cells. Therefore, the purpose of this study was to compare IFN resistance in various HCV clones using this system. Methods: HCV core amino acid substitutions R70Q and L91M were introduced to the TPF1-M170T clone and then transfected into Huh7-ALS32.50 cells. To evaluate the production of each virus, intracellular HCV core antigens were measured. Results were confirmed with Western blot analysis using anti-NS5A antibodies, and IFN sensitivity was subsequently measured. Results: Each clone was transfected successfully compared with JFH-1, with a significant difference in intracellular HCV core antigen (p < 0.05), an indicator of continuous HCV replication. Among all clones, L91M showed the highest increase in the HCV core antigen and HCV protein. There was no significant resistance against IFN treatment in core substitutions; however, IFN sensitivity was significantly different between the wildtype core and JFH-1 (p < 0.05). Conclusions: A novel genotype 1b HCV cell culture was constructed with core amino acid substitutions, which demonstrated IFN resistance of genotype 1b. This system will be useful for future analyses into the mechanisms of HCV genotype 1b treatment.
Keywords: Cell culture system, Core Amino acid substitutions 70 and 91, Genotype 1b, Hepatitis C virus
Introduction
Hepatitis C virus (HCV) infection is the major cause of chronic liver diseases that lead to cirrhosis and hepatocellular carcinoma. An estimated 130–170 million people worldwide are chronically infected with HCV.1 Furthermore, HCV infection has been implicated in 28% of cirrhosis cases and 26% of liver cancer cases each year; thus, accounting for almost 500,000 deaths per year globally.2,3
Previous HCV therapies comprised interferon (IFN)-based regimens, such as pegylated-IFN-α and ribavirin, for up to 72 weeks, which lead to sustained virologic response (SVR) rates in approximately 50% of compliant patients infected with HCV genotype 1.4 Among the direct-acting antiviral agents, the first generation NS3/4A protease inhibitor telaprevir, in combination with pegylated-IFN-α and ribavirin, has been approved since 2011 for the treatment of patients infected with HCV genotype 1. Increasing cure rates of approximately 70% have been achieved; however, adverse events of telaprevir were frequent, sometimes severe, and in some cases, treatment limiting.5 In 2014, the first IFN-free and all-oral regimen combined with NS3/4A protease and an NS5A inhibitor was initiated in Japan.6 This phase 3 trial reported that all-oral regimens were well tolerated with a low incidence of serious adverse effects and high SVR rates within the range of 80–90%. This early evidence suggests that combination therapy with direct-acting antiviral agents and without IFN may offer fewer side effects and higher SVR rates, but also may introduce issues on affordability and accessibility, including the emergence of drug-resistant HCV mutants and hepatocarcinogenesis after achieving SVR.7 Therefore, treatment with IFN in combination with direct-acting antiviral agents is still of great value because of its cost effectiveness, lower drug–drug interactions, lower emergence of resistant mutants, and suppressive effect of carcinogenesis.
Molecular studies on HCV have been hampered by the lack of efficient in vitro and in vivo models of infection, which have been partly overcome by the development of HCV subgenomic replicons8,9 and an HCV–JFH1 genotype 2a cell culture system.10 Recent developments have led to the establishment of efficient culture-adapted full-length systems for isolates of genotypes 1a (strain TN),11 2a, and 2b, but have never led to the generation of a robust genotype 1b system.12
Our previous study described viral infection, replication kinetics, and drug resistance of HCV core mutants using the HCV–JFH1 cell culture system.13 With this system, IFN resistance of HCV core amino acid 70/91 substitutions could be determined by cellular expression levels of interleukin-6 and upregulation of SOCS3 that lead to the suppression of the JAK/STAT pathway. However, this system was only established for genotype 2a, which is different from the clinically reported IFN-resistant genotype 1b. In the clinical setting, pathology and treatment responses in chronic hepatitis C vary among genotypes.14
Recently, a genotype 1b HCV cell culture system, TPF1-M170T, has been established (data in submission). The strain was cloned from a patient with HCV-related cirrhosis who developed fibrosing cholestatic hepatitis after liver transplantation. In this system, replication-enhancing mutations were introduced into the NS2 and NS4B regions to enable abundant replication. Simultaneously, Huh7-ALS32.50 cells were cloned as one of the most adapted Huh7 cell lines for the TPF1-M170T strain. The cell line was established by treating replicon-transfected Huh7 cells with IFN-αA/D.
The aim of the present study was to compare IFN sensitivity in various clones, including genotype 1 and 2 with or without core amino acid substitutions that are clinically important for treatment resistance and liver carcinogenesis.15,16 A novel HCV cell culture system was established by introducing core amino acid 70/91 substitutions in genotype 1b. Virus replication and production, and IFN sensitivity were subsequently evaluated.
Materials and methods
Reagents and antibodies
Recombinant human IFN-α-2b was obtained from Schering–Plough (Kenilworth, NJ, USA). Antibodies used were: HCV core (Abcam, Cambridge, UK), NS5A (BioDesign, Saco, ME, USA), and β-actin (Sigma-Aldrich, St. Louis, MO, USA). Secondary antibodies were peroxidase-labeled anti-mouse (GE Healthcare; Little Chalfont, UK) and Alexa Fluor 488-labeled goat anti-mouse (Invitrogen of Thermo Fisher Scientific, Waltham, MA, USA) IgG antibodies.
Cell lines and culture conditions
Huh7-ALS32.50 cells were maintained in Dulbecco’s modified medium (Sigma–Aldrich) supplemented with 10% fetal bovine serum, 100 IU/mL penicillin, 100 µg/mL streptomycin (Nacalai Tesque, Kyoto, Japan), and non-essential amino acids (Gibco of Thermo Fisher Scientific) at 37°C under 5% CO2. Cells were maintained at a confluence range of 60–70%. For culture passages, cells were incubated in trypsin with 0.05% EDTA at 37°C for 4 min for cell detachment; the cells were then rinsed 4–5 times with the culture medium to prevent further enzymatic degradation due to trypsin exposure. The cells were collected in a conical tube and then subjected to low-speed centrifugation at 700 rpm for 5 min at 4°C to isolate the pellet. The supernatant was discarded, and the cells were resuspended in fresh culture media. HCV RNA transcribed from pTPF1-M170T and pJFH-1 was transfected into Huh7-ALS32.50 cells.
Introduction of core amino acid substitutions
Based on the pTPF1-M170T (GenBank Accession Number: LC011929), which is a 12,526 bp plasmid, full-length 1b clones were constructed with core amino acid mutations that are clinically known for their treatment resistance. A full-length pTPF1-M170T was digested with restriction enzyme (NotI; New England Biolabs, Ipswich, MA, USA), and the DNA fragments encompassing the full sequence of the core region were subcloned into pcDNA3.1. Mutations were introduced into the DNA fragment of the subcloning vector using a Quickchange II XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). Finally, these mutants were sequenced (Hokkaido System Science, Hokkaido, Japan).
The pTPF1-M170T plasmids, which encode the full-length genotype1b sequence, were linearized at their 3′ ends and used as templates for HCV RNA synthesis using the RiboMax large-scale RNA production system (Promega, Madison, WI, USA). After undergoing DNaseI treatment (RQ-1 RNase-free DNase; Promega), the transcribed HCV RNA was purified using ISOGEN reagent (Nippon Gene, Tokyo, Japan).
Electroporation
For RNA transfection, Huh7-ALS32.50 cells were cultured for 7–14 days after thawing to 90% confluence. The cells were washed twice in phosphate-buffered saline (PBS); 4 × 106 cells were then suspended in cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4 [pH 7.6], 25 mM HEPES [pH 7.6], 2 mM EGTA [pH 7.6], 5 mM MgCl2, filtered) containing 10 µg of HCV RNA. These cells were transferred into a 4 mm electroporation cuvette (Gene Pulser Cuvette; Bio-Rad Laboratories, Inc., Hercules, CA, USA) and subjected to an electric pulse (250 V and 950 F) using a Gene Pulser Xcell (Bio-Rad Laboratories, Inc.). These transfected cells were slowly added to 1 mL of culture medium placed in a cuvette and were left to stand for 15 min at room temperature, followed by incubation at normal culture conditions. Approximately 24 h post-transfection, the cells were washed twice in PBS and then incubated with the new culture medium, which was replaced every other day.
Quantification of intracellular HCV core antigen (Ag)
Transfected cells in each culture dish were lysed in 100 µL lysis buffer (10 mM Tris-HCl [pH7.4], 0.15M NaCl, 1 mM EDTA, 0.5% NP40, 0.1% SDS, 0.04% 1 × cOmplete Mini: Rishe Diagnostics, Tokyo, Japan). Next, cells were sonicated for 10 min (Biorupter UCW-201; BM Equipment Co., Tokyo, Japan) and centrifuged; the resulting supernatant was aspirated from these lysates and 270 μL fetal calf serum was added to 30 µL of each sample liquid. Samples were stored at −80°C and measured using a chemical reaction immunoassay (CLIA) following the manufacturer’s protocol (SRL, Tokyo, Japan).
Identification of IL28B single nucleotide polymorphisms (SNPs) in Huh7-ALS32.50 cells
Identification of IL28B SNPs (rs8099917, rs11881222, and rs8103142) in Huh7-ALS32.50 cells was performed using an invader assay, following the manufacturer’s protocol (SRL, Tokyo, Japan).
RNA extraction, cDNA synthesis, and real-time PCR
HCV-transfected Huh7-ALS32.50 cells were cultured with various concentrations of IFN-α-2b, such that the final DMSO concentration was < 1%. For HCV detection, RNAs were isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany); the concentration of RNA products was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). Total cellular RNA was used to generate cDNA from each sample using SuperScript II reverse transcriptase (Invitrogen). mRNA expression levels were quantified using the Taq Man Universal Master Mix II, no UNG (Applied Biosystems of Thermo Fisher Scientific) and the StepOnePlus Real-Time PCR System (Applied Biosystems).
The primers used for genotype 1b core were: sense, 5′-GGT ACT GCC TGA TAG GGT GCT T-3′, and antisense, 5′-TGG TTT TTC TTT GAG GTT TAG GAT TC-3′. The amount of HCV RNA was normalized to GAPDH using huGAPDH probe dye (Applied Biosystems). Assays were performed in triplicate, and the results are expressed as mean percentage of the control ± SD. The 50% effective concentration (EC50) for each compound was calculated three times using the probit method and expressed as mean ± SD.
Western blot analysis
Western blot was performed as previously described.17,18 Briefly, 10 µg of total cell lysate was separated using NuPAGE4-12% Bis-Tris Midi Gel (Life Technologies of Thermo Fisher Scientific) and then blotted onto a polyvinylidene fluoride membrane (Roche Holding AG Basel, Switzerland). The membrane was incubated with the primary antibodies for HCV core and NS5A followed by a peroxidase-labeled anti-IgG antibody; the results were visualized by chemiluminescence using the ChemiDoc MP Imaging System (Bio-Rad Laboratories, Inc.).
Immunohistochemistry
HCV-transfected Huh7-ALS32.50 cells were cultured on 22-mm-round micro cover glasses (Matsunami, Tokyo, Japan), which were coated with 0.1% gelatin. For the detection of endoplasmic HCV core protein, cells were fixed with 4% paraformaldehyde for 10 min and permeabilized with 1% PBS-Tween for 10 min. The cells were incubated overnight at 4°C with the primary antibodies. The fluorescent secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG. The cells were mounted with a Vectashield mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA) and visualized using a confocal laser-scanning microscope (FV10i; Olympus Corp., Tokyo, Japan).
Statistical analysis
Statistical analyses were performed by Welch’s t test, with p < 0.05 considered as statistically significant.
Results
Introduction of core amino acid 70/91 substitutions to TPF1-M170T
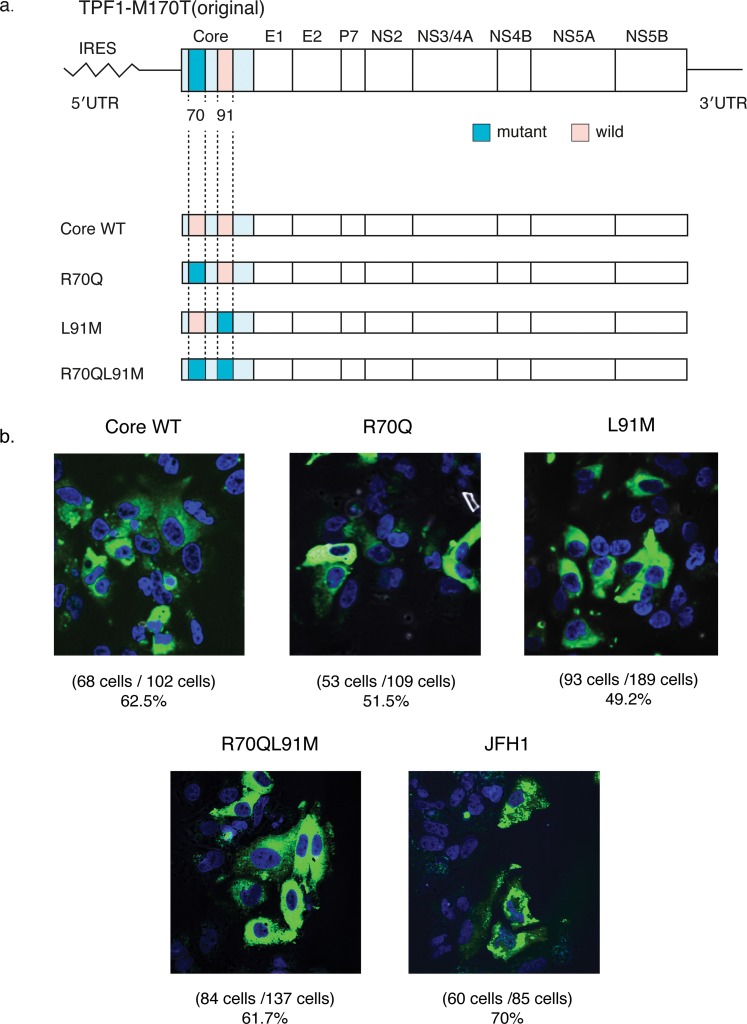
The IL28B SNPs (rs8099917, rs11881222, and rs8103142) of Huh7-ALS32.50 cells were all treatment-resistant heterozygous genotypes (TG, AG, and TC, respectively). The following new clones were constructed with core amino acid substitutions: Core wildtype, R70Q, L91M, and R70QL91M. R70Q and L91M (Fig. 1a), which were reported as clinically resistant to pegylated-IFN-α and ribavirin therapy.17 The R70Q strain is the same as the pTPF1-M170T, which harbors an R70Q mutation.
Fig. 1. Construction of TPF1-M170T clones and evaluation of transfection efficiency.
(a) Genomic structures of the original pTPF1-M170T and their developed clones: Core WT, R70Q, L91M, and R70QL91M. As the pTPF1-M170T already harbors R70Q at the core region, the R70Q clone is the same as the original one. (b) Immunofluorescence assay of the developed clones. Hepatitis C virus-positive cells at 3 d post-transfection are visualized with anti-core antibody (green); nuclei are visualized with DAPI (blue) (magnification, ×60).
Immunohistochemistry was performed to clarify the transfection efficiency of these mutants. The transfected cells were fixed 72 h after electroporation, and the infected foci were visualized by immunostaining against core antibodies. All cells in high-power fields (~100 cells) were counted. The proportion of core-positive cells were 62.5%, 51.5%, 49.2%, and 61.7% for WT, R70Q, L91M, and R70QL91M, respectively, which was similar to that of the JFH-1 strain, which was 70.0% (Fig. 1b).10
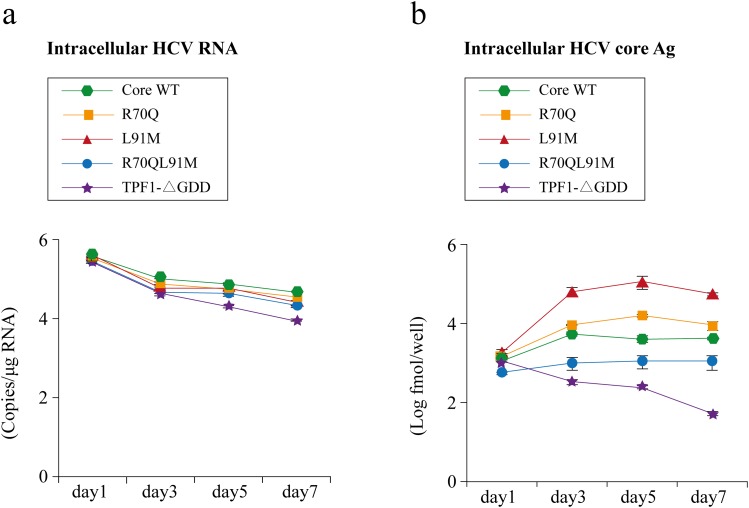
Fig. 2. Comparing replications of TPF1-M170T clones.
(a) Time course of intracellular hepatitis C virus (HCV)-RNA replications after in vitro-transcribed mutant and wild-type RNAs were transfected into Huh7-ALS32.50 cells. (b) Expression of intracellular HCV core antigen (Ag) as quantified by chemical reaction immunoassay. Data are presented as mean ± SD.
Comparison of virus replication between core amino acid 70/91 substitutions
After transfecting TPF1-M170T clones, intracellular HCV RNA was quantified by real-time PCR. The RNA replication-defective mutant TPF1-ΔGDD kindly provided by Mori et al. (data in submission) was used as the negative control. The construct was created by in-frame deletion of the active-site motif GDD in NS5B polymerase. There were no significant differences in HCV RNA between TPF1-M170T clones and TPF1-ΔGDD (Fig. 2a).
Intracellular HCV core Ag was measured from extracted protein lysates to evaluate virus replication (Fig. 2b). The levels of intracellular HCV core Ag in each clone increased from day 1 to 5, in contrast to the levels of TPF1-ΔGDD, which gradually declined. These results indicate that these clones replicated and were efficiently synthesized inside cells. L91M demonstrated the highest HCV core Ag levels among the clones, but there were no significant differences in the core amino acid 70/91 substitutions.
Comparison of protein synthesis between core amino acid 70/91 substitutions
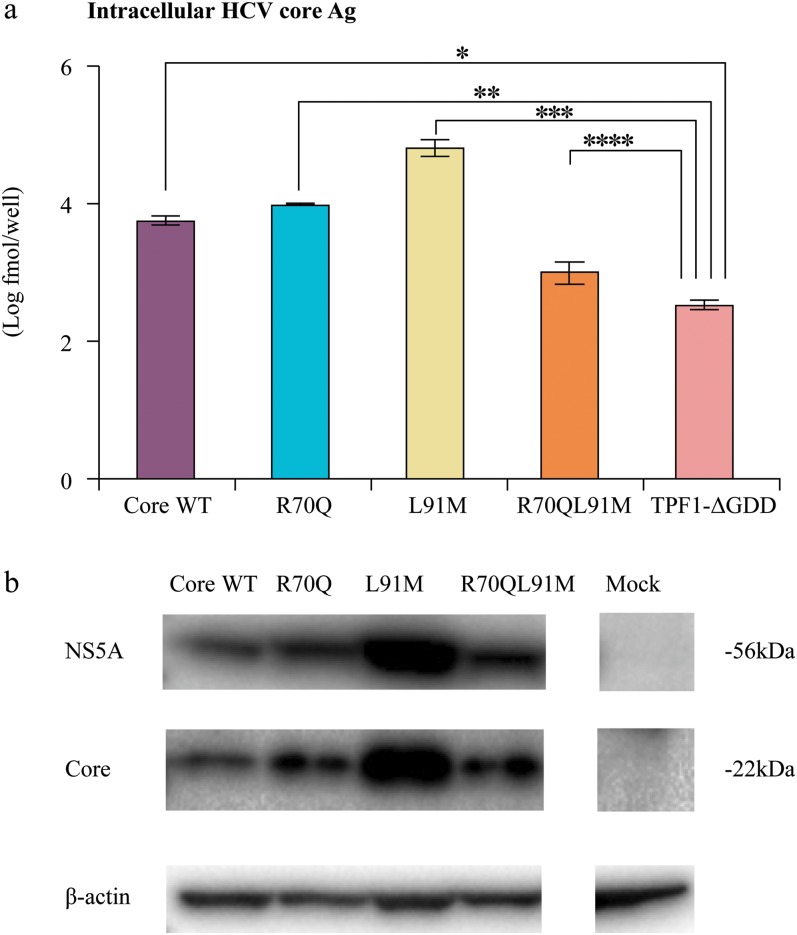
Virus replication was further confirmed by measuring HCV protein by Western blot analysis with anti-NS5A antibodies. The intensity of all clones was higher than that of the negative control (Fig. 3). The intensity of NS5A protein of the L91M clone was the highest among these clones, which corresponds to the results obtained from the HCV core Ag.
Fig. 3. Comparison of protein synthesis among the TPF1-M170T clones.
(a) Comparison of intracellular hepatitis C virus (HCV) core antigen (Ag) at day 3. (b) Representative Western blot for NS5A and core proteins;*p < 0.05.
Difference in IFN resistance between genotypes 1b and 2a
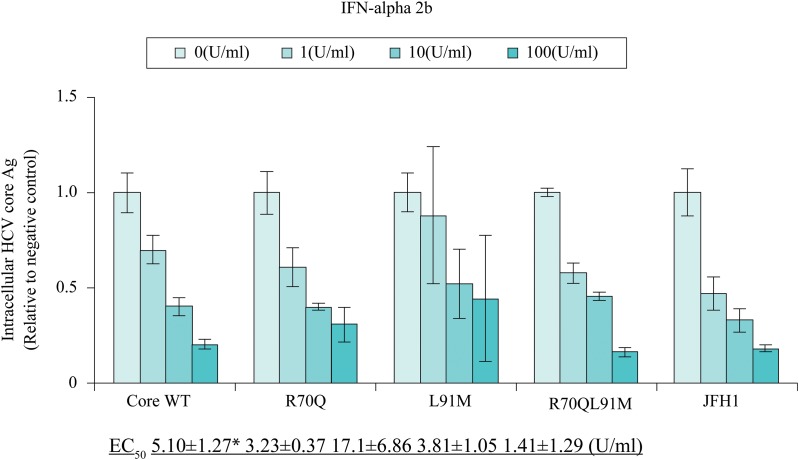
Sensitivity of the TPF1-M170T clones with core substitutions to IFN-α-2b treatment was assessed (Fig. 4). IFN-α-2b is clinically effective for HCV and was reported to have inhibitory effects on HCV replication in subgenomic replicons.19 Each clone was transfected into Huh7-ALS32.50 cells by electroporation, which were then seeded into 12-well dishes. Four days post-transfection, cells were cultured for 48 h in various concentrations of IFN-α-2b. Protein was extracted from the cells, and the level of HCV core Ag was quantified by CLIA.
Fig. 4. Comparison of interferon (IFN) sensitivity between TPF1-M170T and JFH-1.
Expression of intracellular HCV core Ag was measured by chemical reaction immunoassay at 72 h post-transfection of Core wild type (WT), R70Q, L91M, R70QL91M, and JFH-1. Cells were treated for 48 h with 0–100 IU/mL. The values are displayed as percentages of the IFN-untreated control. *p < 0.05 compared with JFH1.
JFH-1 was the most sensitive to IFN-α-2b, with an EC50 of 1.41 ± 1.29 IU/mL. The values of EC50 for Core WT, R70Q, and R70QL91M were 5.10 ± 1.27 IU/mL, 3.23 ± 0.37 IU/mL, and 3.81 ± 1.05 IU/mL, respectively. L91M demonstrated resistance at an EC50 of 17.1 ± 6.86 IU/mL. Compared with the Core WT, L91M did not significantly differ in the level of EC50, but JFH-1 did (p < 0.05), confirming that the 1b clone was more resistant than the 2a clone.
Discussion
This study describes a novel cell culture system for HCV genotype 1b with core amino acid 70/91 substitutions. This system could successfully demonstrate, for the first time, the IFN resistance of HCV genotype 1b as clinically observed, after confirming HCV replication and translation. In this analysis, the TPF1-ΔGDD clone, which is the deletion mutant of the active site (GDD motif) of the NS5B polymerase, was used as a negative control. Although the quantity of intracellular HCV RNA in each of the clones tended to be higher than that of the control TPF1-ΔGDD clone, the differences were not statistically significant. One of the possible reasons is the low replication efficiency of the control clone. Even in the JFH-1, an evident difference with the GDD mutant does not emerge until approximately ten days after transfection.8 Moreover, an apparent difference in the genotype 1b replicon was previously observed at only 72 hours post-transfection.20 Another possibility is that the amount of input HCV RNA exceeded the replicated RNA. To solve this problem, we measured HCV core Ag, which is not influenced by input RNA, to verify higher production of HCV core Ag in each clone compared with TPF1-ΔGDD clone. Furthermore, this result was validated by Western blotting using anti-NS5A antibodies. We therefore concluded that our clone could be successfully transfected, replicated, and translated. Next, these clones were assessed for IFN resistance. The results demonstrate the IFN resistance of HCV genotype 1b. To the best of our knowledge, this is the first report that compared IFN sensitivity of HCV genotypes 1b and 2a using the HCV cell culture system.
Compared with our previous results, the order of the replication efficiency of each clone was different. Funaoka et al.13 reported that R70Q and L91M replicated significantly better than the wild type, whereas Tasaka-Fujita et al.21 found that R70Q replicated less and L91M replicated more. The former report used genotype 2a, whereas the latter used 1b/2a chimeric virus. As both reports used genotype 2a-derived sequences for NS proteins, which comprise a replication complex that is indispensable for virus replication, the differences in NS proteins may account for the contrasting results.
In this study, no significant difference in IFN response was observed between the core amino acid 70/91 substitutions, which is also in contrast to our previous report.13 One of the reasons may be the different replication efficiency of the JFH-1 HCV cell culture, which enabled robust replication in the former study. The other possibility may be an extracellular IFN response that is different from intracellular mechanisms. However, in the study by Tasaka-Fujita et al.,21 there was also no difference in IFN response between core 70/91 substitutions using 1b/2a chimeric clones. Nevertheless, a difference in the cell surface expression of MHC Class I in HCV-transfected cells was observed, which was considered a possible cause for IFN resistance. MHC class I is the molecule that is recognized by cytotoxic T cells to induce the elimination of infected cells. Therefore, when HCV hinders expression of MHC class I, infected cells may be able to continue replicating after the cessation of treatment. An alternative possibility concerns the type of cells used in the present study. IL28B SNPs in Huh7-ALS32.50 cells are all treatment-resistant heterotypes. Therefore, it might be difficult to find the difference between core amino acid 70/91 mutants and the wild type because of the pre-existing IFN-resistant environment. A different outcome may result if a treatment-sensitive cell line expressing the IL28B major homozygote is used, which would show a favorable response to exogenous IFN, whereas IFN-sensitive clones would be eliminated. In such a case, a significant difference may then be detectable. As it was clinically reported that a significant difference in treatment response among core substitutions was only observed in the patients with unfavorable IL28B SNPs, further experiments will be required to validate our results.
Conclusions
The results of this study from newly developed HCV cell culture system demonstrated, for the first time, that HCV genotype 1b was significantly resistant to IFN treatment compared with genotype 2a. This result indicates that this system reproduce clinical characteristics of HCV genotype 1b in vitro and will be useful for developing novel antiviral chemotherapy against HCV.
Acknowledgments
The authors would like to thank Takaji Wakita for providing the pJFH1 full. This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology-Japan, Japan Agency for Medical Research and Development, the Japan Society for the Promotion of Science, Ministry of Health, Labor Welfare-Japan, the Japan Health Science Foundation, and National Institute of Biomedical Innovation.
Abbreviations
- Ag
antigen
- CLIA
chemical reaction immunoassay
- HCV
hepatitis C virus
- IFN
interferon
- PBS
phosphate-buffered saline
- SNP
single nucleotide polymorphism
- SVR
sustained virologic response
- WT
wildtype
References
- 1.Global Burden Of Hepatitis C Working Group. Global burden of disease (GBD) for hepatitis C. J Clin Pharmacol. 2004;44:20–29. doi: 10.1177/0091270003258669. doi: 10.1177/0091270003258669. [DOI] [PubMed] [Google Scholar]
- 2.Jayasekera CR, Barry M, Roberts LR, Nguyen MH. Treating hepatitis C in lower-income countries. N Engl J Med. 2014;370:1869–1871. doi: 10.1056/NEJMp1400160. doi: 10.1056/NEJMp1400160. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manns MP1, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, et al. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. doi: 10.1016/S0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 5.Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, et al. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405–2416. doi: 10.1056/NEJMoa1012912. doi: 10.1056/NEJMoa1012912. [DOI] [PubMed] [Google Scholar]
- 6.Kumada H, Suzuki Y, Ikeda K, Toyota J, Karino Y, Chayama K, et al. Daclatasvir plus asunaprevir for chronic HCV genotype 1b infection. Hepatology. 2014;59:2083–2091. doi: 10.1002/hep.27113. doi: 10.1002/hep.27113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manns M, Pol S, Jacobson IM, Marcellin P, Gordon SC, Peng CY, et al. All-oral daclatasvir plus asunaprevir for hepatitis C virus genotype 1b: a multinational, phase 3, multicohort study. Lancet. 2014;384:1576–1605. doi: 10.1016/S0140-6736(14)61059-X. doi: 10.1016/S0140-6736(14)61059-X. [DOI] [PubMed] [Google Scholar]
- 8.Blight KJ1, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 9.Lohmann V1, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 10.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Medicine. 2005;11:791–796. doi: 10.1038/nm1268. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YP1, Ramirez S, Jensen SB, Purcell RH, Gottwein JM, Bukh J. Highly efficient full-length hepatitis C virus genotype 1 (strain TN) infectious culture system. Proc Natl Acad Sci U S A. 2012;109:19757–19762. doi: 10.1073/pnas.1218260109. doi: 10.1073/pnas.1218260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheel TK1, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837–849. doi: 10.1038/nm.3248. doi: 10.1038/nm.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funaoka Y, Sakamoto N, Suda G, Itsui Y, Nakagawa M, Kakinuma S, et al. Analysis of Interferon Signaling by Infectious Hepatitis C Virus Clones with Substitutions of Core Amino Acids 70 and 91. J Virol. 2011;85:5986–5994. doi: 10.1128/JVI.02583-10. doi: 10.1128/JVI.02583-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshioka K, Kakumu S, Wakita T, Ishikawa T, Itoh Y, Takayanagi M, et al. Detection of hepatitis C virus by polymerase chain reaction and response to interferon-alpha therapy:relationship to genotypes of hepatitis C virus. Hepatology. 1992;16:293–299. doi: 10.1002/hep.1840160203. doi: 10.1002/hep.1840160203. [DOI] [PubMed] [Google Scholar]
- 15.Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, et al. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology. 2007;46:1357–1364. doi: 10.1002/hep.21836. doi: 10.1002/hep.21836. [DOI] [PubMed] [Google Scholar]
- 16.Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, et al. Substitution of amino acid 70 in the hepatitis C virus core region of genotype 1b is an important predictor of elevated alpha-fetoprotein in patients without hepatocellular carcinoma. J Med Virol. 2008;80:1354–1362. doi: 10.1002/jmv.21202. doi: 10.1002/jmv.21202. [DOI] [PubMed] [Google Scholar]
- 17.Yokota T, Sakamoto N, Enomoto N, Tanabe Y, Miyagishi M, Maekawa S, et al. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO Reports. 2003;4:602–608. doi: 10.1038/sj.embor.embor840. doi: 10.1038/sj.embor.embor840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanabe Y, Sakamoto N, Enomoto N, Kurosaki M, Ueda E, Maekawa S, et al. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. Journal of Infectious Diseases. 2004;189:1129–1139. doi: 10.1086/382595. doi: 10.1086/382595. [DOI] [PubMed] [Google Scholar]
- 19.Fridell RA, Wang C, Sun JH, O’Boyle DR, Nower P, Valera L, et al. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology. 2011;54:1924–1935. doi: 10.1002/hep.24594. doi: 10.1002/hep.24594. [DOI] [PubMed] [Google Scholar]
- 20.Kanazawa N, Kurosaki M, Sakamoto N, Enomoto N, Itsui Y, Yamashiro T, et al. Journal of Virology. 2004;78:9713–9720. doi: 10.1128/JVI.78.18.9713-9720.2004. doi: 10.1128/JVI.78.18.9713-9720.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tasaka-Fujita M, Sugiyama N, Kang W, Masaski T, Murayama A, Yamada N, et al. Amino Acid Polymorphisms in Hepatitis C Virus Core Affect Infectious Virus Production and Major Histocompatibility Complex Class I Molecule Expression. Sci Rep. 2015;5:139942015. doi: 10.1038/srep13994. doi: 10.1038/srep13994. [DOI] [PMC free article] [PubMed] [Google Scholar]