Abstract
Recognition of double-stranded DNA with a mixed nucleotide sequence by oligonucleotide is a long-term challenge. This aim can be achieved via formation of the recombination R-triplex, accommodating two identical DNA strands in parallel orientation, and antiparallel complementary strand. In the absence of proteins the R-triplex stability is low, however, so that intermolecular R-triplex is not formed by three DNA strands in a ligand-free system. Recently, recognition of DNA with mixed base sequence by single-stranded oligonucleotide in the presence of bis-intercalator YOYO was reported. Here, we describe thermodynamic characteristics of YOYO complexes with the model oligonucleotides 5'-GT-2AP-GACTGAG TTTT CTCAGTCTACGC GAA GCGTAGACTGAG-3' (R2APCW) bearing a single reporting 2-aminopurine (2AP) in place of adenine and 5'- CTCAGTCTACGC GAA GCGTAGACTGAG-3' (CW). We found that each oligonucleotide is able to bind two YOYO molecules via intercalation mode in 0.5 M LiCl. Fluorescence intensity of YOYO intercalated in triplex R2APCW and in CW hairpin increased 40-fold compared to the free YOYO. Remarkably, the melting temperature of the triplex (determined using temperature dependence of the 2AP fluorescence) increased from 19° C to 33° C upon binding two YOYO molecules. Further increase in the YOYO concentration resulted in binding of up to five YOYO molecules to R2APCW triplex and up to six YOYO molecules to CW hairpin.
Keywords: R-triplex DNA, stabilization, YOYO, melting temperature
Introduction
Recognition of a mixed-sequence DNA by a single-stranded (ss) oligonucleotide is a longstanding goal in molecular biotechnology. Employing the formation of a DNA triplex with Hoogsteen-type hydrogen bonding scheme met a major limitation. Namely, the requirement of oligopurine/oligopyrimidine nucleotide sequence is a structural basis for the classical H- triplex formation (1, 2). Recent efforts of many laboratories partially reduced the strictness of this requirement (3-6).
Alternatively, recognition of DNA duplex with a mixed sequence may potentially be achieved via formation of the recombination-type R-triplex (7). Earlier, specially designed oligonucleotides able to fold as R-triplex were used for studying of the R-triplex characteristics and its complexes with intercalators (8-13). The R-triplex-assisted recognition of a 14-bp mixed DNA sequence with a homologous single strand bearing intercalator acridine covalently attached to its sugar-phosphate backbone was observed (14). Nevertheless, in the absence of proteins and other ligands, the R-triplex stability is low, so that separate DNA strand cannot bind to homologous duplex to form such a triplex (8). Therefore, a search for ligands which could stabilize R-triplex to such an extent that the structural analysis becomes possible, seems a commendable task.
Recognition of a mixed DNA sequence with homologous ss oligonucleotides in the presence of bis-intercalator YOYO was reported recently, though no structural implications were made in that study (15). Here, we analyze the YOYO binding to the intramolecular R-triplex in comparison with binding to double-stranded and ss oligonucleotides. We deduce thermodynamic characteristics of YOYO binding to R-triplex and discuss its potential to serve as a structure-selective stabilizer.
Materals and Methods
Oligonucleotides
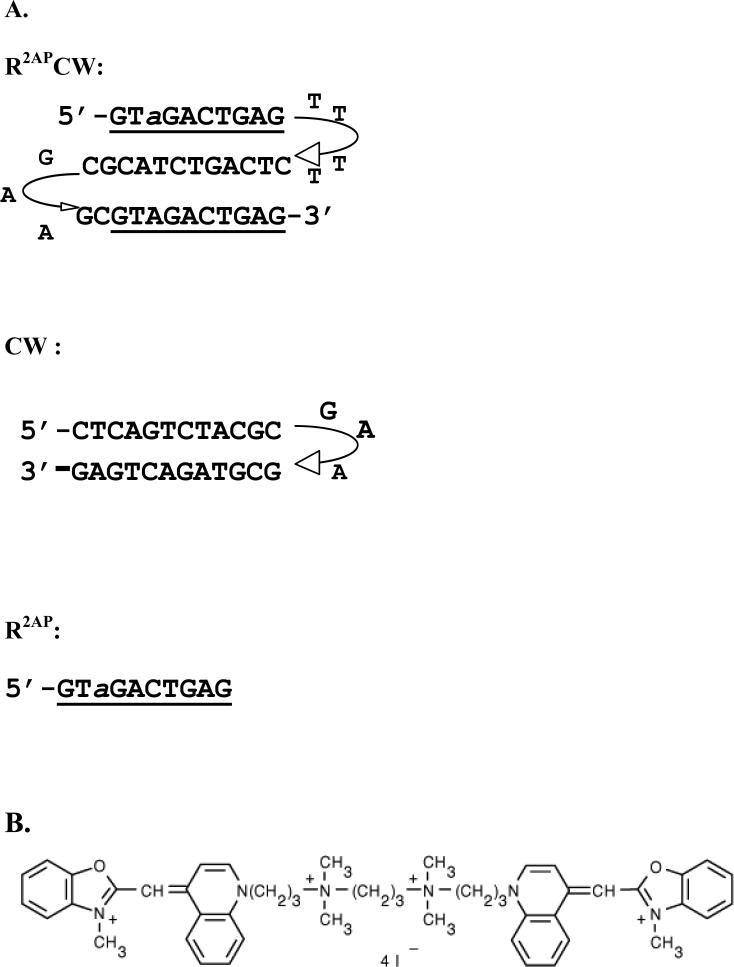
Oligonucleotides R2APCW, CW and R2AP were synthesized and purified with HPLC by Syntol (Moscow). The oligonucleotide sequences and folding schemes are presented in Figure 1A. Earlier, R2APCW was shown to form intramolecular sequence-specific triple-stranded structure, while CW formed a hairpin under the experimental conditions (9, 10). The scheme of YOYO molecule is given in Figure 1B. Samples contained 1 or 0.5 μM of oligonucleotides, 0.5 M LiCl, 10 mM Tris HCl buffer, pH 8, temperature was 0°C, if not stated otherwise.
FIGURE 1.
(A) Oligonuclotide sequences and folding schemes. R2APCW, three- stranded fold; CW, double-stranded hairpin; R2AP, single strand. 2-aminopurine is designated as a. At 0° C, in 0.5 M LiCl, 10 mM Na phosphate of Tris HCl buffer, pH8, the oligonucleotides has been proved to form the intramolecular folds shown here (9). (B) scheme of YOYO dye.
Analysis of the ligand binding curves
Parameters of YOYO binding to the olgonucelotide folds were derived by fitting the McGhee and Von Hippel equation to experimental binding curves (16):
| [1] |
Here K is the binding constant for the isolated bound dye and n the number of base pairs (or base triplets) per one bound dye; C1 and C2 are the free and bound dye concentrations; r is the ratio of the bound dye concentration to the DNA concentration. (The DNA concentration is presented in base triplets, base pairs or nucleotides in case of a triplex, duplex and a single DNA strand, respectively.)
Fluorescence measurements
Measurement of the temperature profiles of R2APCW steady-state fluorescence emission was carried out using a Cary Eclipse (Varian) spectrofluorimeter equipped with a thermostatted cuvette holder. The excitation wavelength was 310 nm, the emission wavelength was 370 nm.
Triplex melting temperatures
We denote the triplex formation as the intramolecular binding of the dangling third strand of the triplex-forming oligonucleotide to its double helical CW part. The temperature dependence of the R2APCW steady-state fluorescence emission depicts formation/disruption of a single triplet 2AP*(T·A). As the R-triplex formation is a cooperative process, the triplex formation as a whole may be monitored with R2APCW “fluorescence” melting curve (9). Thermodynamic analysis of the transition curves was performed as described earlier and the melting temperatures was derived by fitting theoretical “fluorescence” melting curves to experimental temperature profiles (9).
Results and Discussion
Binding of YOYO to the model R-triplex, duplex hairpin and single strand
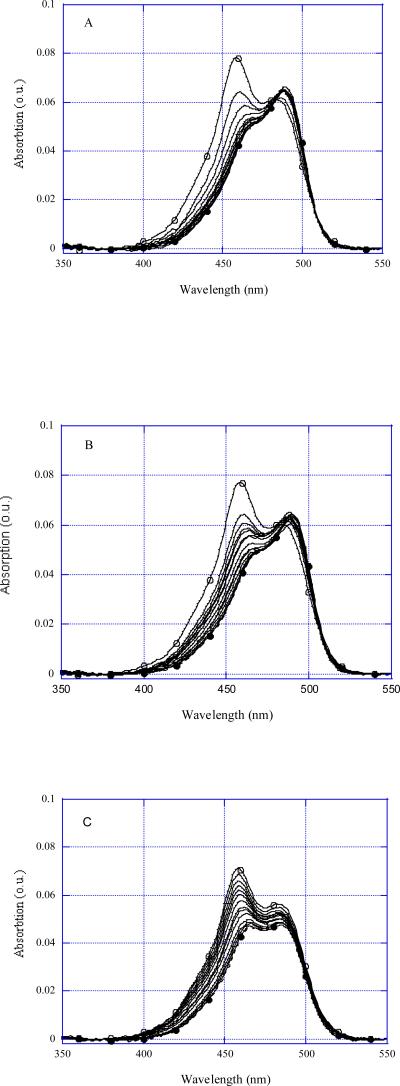
Although YOYO is brightly fluorescing, we found its absorption spectra to be the most sensitive characteristics for studying the ligand binding to DNA structures. The intensities of absorption bands at 458 nm and at 490 nm vary depending on the ligand state, the band at 458 nm reaching its maximum value for free YOYO (Figure 2, open circles) and the band at 490 nm reaching its maximum value for YOYO bound to DNA (Figure 2, filled circles). The isosbestic point is close to 481 nm when YOYO is bound to R-triplex and CW hairpin (Figures 2A, 2B). In the case of ss oligonucleotide R2AP there was no isosbestic point (Figure 2C), as several imperfect hairpins or duplexes were involved in complexes with the dye rather than a single distinct DNA structure.
FIGURE 2.
YOYO absorption spectra at different additions of olighonucleotides. (A) R2APCW, (B) CW, (C) R2AP. Free YOYO concentrations were 10−6 M, R2APCW concentrations were from 1 to 20 μM; CW concentrations were from 1.5 to 41 μM, R2AP concentrations were from 0.5 to 82 μM. Solution dilutions were taken into account.
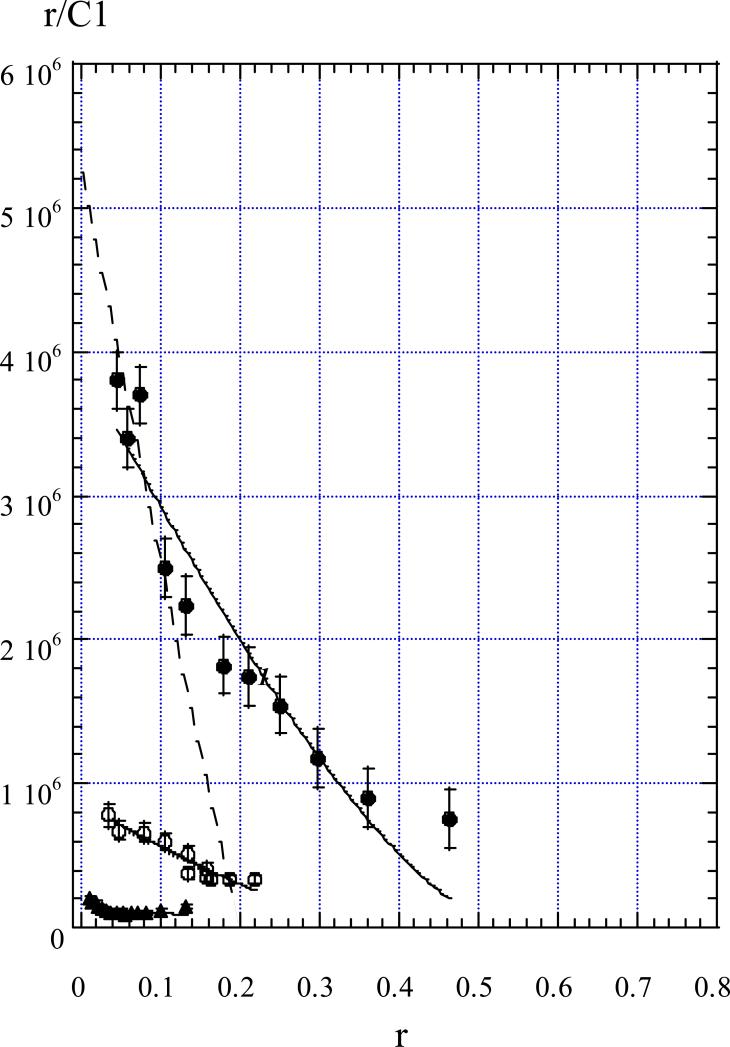
By use of the absorption values at a given wavelength, concentrations of free and bound ligand were calculated at each added oligonucleotide concentration, and the binding curves were plotted (Figure 3). A considerable difference in YOYO affinity to different DNA conformations is evident, the affinity for R-triplex being the greatest. Straightforward fitting of McGhee and Von Hippel equation (see Methods) to the experimental points yielded the Kass values given in Table I. The YOYO binding constant Kass to R-triplex is ~4 times greater than that to the double-stranded hairpin of the same sequence and length. The maximal number of base pairs (or base triplets) per bound ligand appeared to be about two base pairs for CW and two triplets for R2APCW (Figure 3). Unlike this, in NMR experiments (17), single YOYO molecule was found to cover 3-4 base pairs in a bis-intercalating complex, with two DNA base pairs positioned between the oxazole chromophores.
FIGURE 3.
Scatchard plot depicting YOYO binding to R2APCW (filled circles), CW (open circles) and R2AP (triangles). r values were calculated as the ratio of the bound YOYO concentration to oligonucleotide concentrations taken in base triplets (filled circles), base pairs (open circles) and nucleotides (triangles). Solid curves are theoretical fits (see eq. 1). For the dashed line see the text.
TABLE I.
Association constants of YOYO complexes with the triplex, duplex hairpin and single-stranded oligonucleotides
| Oligonucleotide | Kass (M−1) | Max. number of bound YOYO per oligonucleotide |
|---|---|---|
| triplex R2APCW | 3.9±0.2·106 | 5 |
| hairpin CW | 8.9±0.5 ·105 | 6 |
| single-stranded R2AP | ~2· 105 | - |
In fact, experimental dependence of YOYO binding to R2APCW (Figure 3, filled circles) has a more complex shape than the theoretical curve defined by [1] (Figure 3, solid curve 1). At the low occupancies, below r= 0.2, the first and the second YOYO molecules bind to R2APCW with a higher apparent binding constant than the association constant Kass determined with equation [1]. The stoichiometry of such a binding suggests a bis-intercalation mode. Therefore, the first and the second YOYO molecules bind to R2APCW triplex by bis-intercalation in accord with the NMR data (17) (see the dashed line in Figure 3). The dashed line intersections with the X and Y axis allow estimating the apparent association constant Kapp for the first and second YOYO molecules binding to R-triplex, Kapp ≈3·107 M−1. Importantly, these YOYO molecules bind to R-triplex with ~7 times higher affinity than to double helical or to ss DNA (Table I). Fluorescence experiments (see the next section) support the intercalation character of YOYO binding. At the higher YOYO occupancies, r > 0.2, triplex binds YOYO molecules with a lower affinity. This binding may occur either in the triplex grooves or by a mono-intercalation of one of the two YOYO oxasoles in the DNA structure. In the latter case, another YOYO chromophore may intercalate into another oligonucleotide fold. Thereby, YOYO may presumably form a “bridge” between oligonucleotides.
Note that the experimental binding dependencies do not have a tendency to reach the X axis (Figure 3). On the contrary, at the higher occupancies all the dependencies tend to ascend. Such a cooperative-type behaviour may reflect a complex mode of the YOYO-DNA binding, consistent with formation of the possible “bridges” bringing different oligonucleotides together.
Intercalation type of YOYO binding to R-triplex and duplex DNA
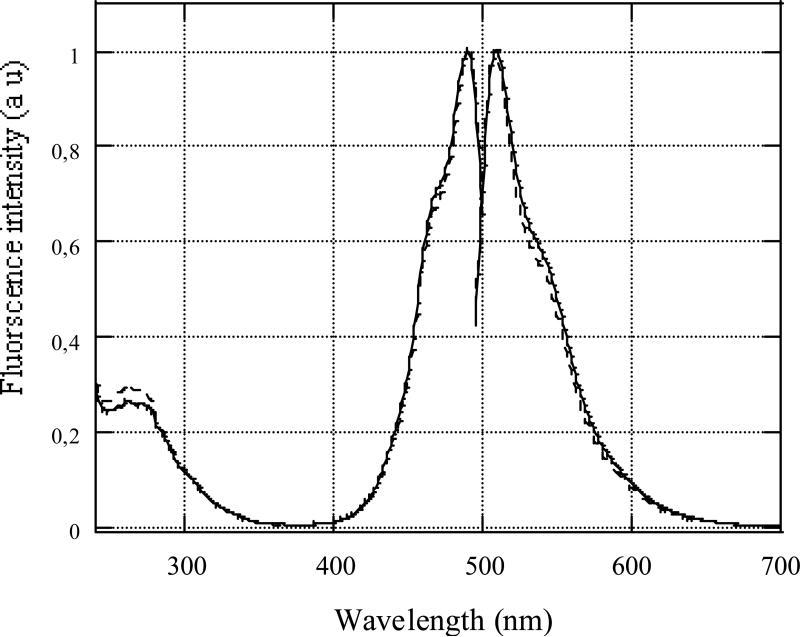
In the fluorescence experiments, such YOYO concentrations were chosen that no more than one YOYO molecule per triplex or duplex fold was intercalated (Figure 3). Fluorescence intensity of YOYO intercalated in R2APCW triplex and in CW hairpin increased ~40-fold compared to free YOYO. Normalized excitation and emission spectra of fully bound YOYO to the two DNA structures are compared in Figure 4. Excitation maximum was centered at 490 nm, and emission maximum at 510 nm; the shapes of the curves were very close. The great enhancement of YOYO fluorescence upon binding to DNA indicates the highly hydrophobic dye environment in the complexes. This is consistent with the more than tenfold increase in the quantum yield of fluorescence of many ligands upon their intercalation (bis-intercalation) into DNA double- and multi-stranded helices (18-20). Relative quantum yield of YOYO fluorescence bound to R-triplex was estimated and found to be similar to that bound to DNA duplex within experimental error. Thus, we believe that at low occupancies (r < 0.1), YOYO binds to R-triplex, as well as to DNA duplex, via intercalation (bis-intercalation) mode.
FIGURE 4.
Normalized excitation and emission fluorescence spectra of YOYO bound to R2APCW triplex (bold curves) and duplex hairpin CW (dashed curves). Temperature was 0° C, other conditions are given in Materials.
Stabilization of R-triplex with YOYO
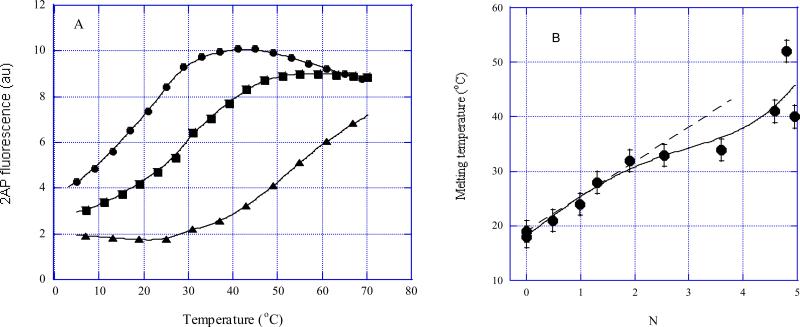
Complexes of R2APCW fold with various YOYO concentrations were formed at 0° C and the bound YOYO concentration was calculated for each sample using the binding curve (Figure 3, filled circles). The melting curves for N = 0, 2 and 4.9 are shown in Figure 5A. In this figure N is the ratio of the bound YOYO concentration to concentration of oligonucleotides, or a number of YOYO molecules bound to R-triplex. The solid curves in Figure 5A are the theoretical fits (see Methods). The triplex melting temperatures (derived by fitting theoretical curves to the experimental points) are plotted versus N values in Figure 5B. The first two bound YOYO molecules increase the triplex Tm by 14° C. These two YOYO molecules are likely to bind via bis-intercalation mode (Figure 3, circles). Thus, each bis-intercalating YOYO molecule stabilizes R-triplex by 7° C under the experimental conditions. For comparison, binding of a single mono-intercalating molecule, such as EtBr or acridine orange, has been shown to increase thermostability of R-triplex 5’-d(CATGCTAACT)-L-(AGTTAGCATG)-L-(CATGCTAACT)-3’ by ~5° C (13). Further addition of YOYO, up to four bound molecules, does not cause a detectable stabilization (Figure 5B). Surprisingly, a sharp Tm rise occurred at N > 4. Such a steep increase in thermostability may be due to formation of non-specific “bridges” between oligonucleotides at saturating YOYO concentrations, as discussed above.
FIGURE 5.
Thermal denaturation of R2APCW in the presence of different amounts of bound YOYO as monitored with 2-aminopurine steady-state fluorescence intensity. The N values were calculated per oligonucleotide (using the data in Figure 3) and thus represent the number of YOYO molecules bound to one R2APCW fold. (A) Only three R2APCW:YOYO melting curves are shown for clarity, for N = 0, 2 and 4.9 (circles, squares and triangles, respectively). (B) Tm values were derived by fitting of equation [1] (see Methods) to the experimental melting curves (three of the melting curves are shown in (A)).
Conclusions
We detected a preferential binding of YOYO to R-triplex DNA in comparison with the double- and single-stranded DNA. YOYO may interact with the triple- and double-helical oligonucleotide folds as a bis-intercalator at the low ligand concentrations, and in a more complex manner at the higher concentrations. Thermostability of R-triplex bound to the bis-intercalator at 0.1 M LiCl inreases by 7° C per one ligand. The ratio Kasstriplex / Kassds ≤ 10 may be small to ensure the selective YOYO binding to the R-triplex versus ds DNA. Taking this into account as well as the low stability of R-triplex (2, 4, 12), formation of an intermolecular R-triplex by targeting DNA with a homologous single strand in the presence of YOYO (15) seems questionable and requires further investigation. Furthermore, formation of the second type complex at the higher YOYO concentrations may facilitate inter-molecular DNA-DNA aggregates which would hamper specific recognition of ds DNA by a homologous single strand. Nonetheless, the bis-intercalation of YOYO into the intramolecular R-triplex markedly increases its thermostability and may assist in structural NMR studies of this DNA conformation.
Acknowledgements
The authors appreciate helpful discussions with T.M. Jovin, D.J. Arndt-Jovin and V.A. Livshits. The study was supported by RFBR grants 07-04-00645-a, 07-04-00691-a.
References
- 1.M Mirkin S. Front Biosci. 2008;13:1064–1071. doi: 10.2741/2744. [DOI] [PubMed] [Google Scholar]
- 2.Shafer RH. Prog. Nucleic Acid Res. Mol. Biol. 1998;59:55–94. doi: 10.1016/s0079-6603(08)61029-6. [DOI] [PubMed] [Google Scholar]
- 3.Fox KR, Brown T. Q. Rev. Biophys. 2005;38:311–320. doi: 10.1017/S0033583506004197. [DOI] [PubMed] [Google Scholar]
- 4.Kalish JM, Glazer PM. Ann. N. Y. Acad. Sci. 2005;1058:151–161. doi: 10.1196/annals.1359.023. [DOI] [PubMed] [Google Scholar]
- 5.Grunweller A, Hartmann RK. BioDrugs. 2007;21:235–243. doi: 10.2165/00063030-200721040-00004. [DOI] [PubMed] [Google Scholar]
- 6.Kuan JY, Glazer PM. Methods Mol. Biol. 2004;262:173–194. doi: 10.1385/1-59259-761-0:173. [DOI] [PubMed] [Google Scholar]
- 7.Zhurkin VB, Raghunathan G, Ulyanov NB, Camerini-Otero RD, Jernigan RL. J. Mol. Biol. 1994;239:181–200. doi: 10.1006/jmbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- 8.Shchyolkina AK, Timofeev EN, Borisova OF, Il'icheva IA, Minyat EE, Khomyakova EB, Florentiev VL. FEBS Letters. 1994;339:113–118. doi: 10.1016/0014-5793(94)80396-x. [DOI] [PubMed] [Google Scholar]
- 9.Shchyolkina AK, Kaluzhny DN, Borisova OF, Hawkins ME, Jernigan RL, Jovin TM, Arndt-Jovin DJ, Zhurkin VB. Nucleic Acids Res. 2004;32:432–440. doi: 10.1093/nar/gkh158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shchyolkina AK, Kaluzhny DN, Arndt-Jovin DJ, Jovin TM, Zhurkin VB. Nucleic Acids Res. 2006;34:3239–3245. doi: 10.1093/nar/gkl431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dagneaux C, Gousset H, Shchyolkina AK, Ouali M, Letellier R, Liquier J, Florentiev VL, Taillandier E. Nucleic Acids Res. 1996;24:4506–4512. doi: 10.1093/nar/24.22.4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borisova OF, Shchyolkina AK, Timofeev EN, Tsybenko S, Mirzabekov AD, Florentiev VL. J. Biomol. Struct. Dyn. 1995;13:15–27. doi: 10.1080/07391102.1995.10508818. [DOI] [PubMed] [Google Scholar]
- 13.Shchyolkina AK, Borisova OF. FEBS Letters. 1997;419:27–31. doi: 10.1016/s0014-5793(97)01417-8. [DOI] [PubMed] [Google Scholar]
- 14.Shchyolkina AK, Timofeev EN, Lysov YP, Florentiev VL, Jovin TM, Arndt-Jovin DJ. Nucleic Acids Res. 2001;29:986–995. doi: 10.1093/nar/29.4.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daksis JI, Erikson GH. Genet. Test. 2005;9:111–120. doi: 10.1089/gte.2005.9.111. [DOI] [PubMed] [Google Scholar]
- 16.McGhee JD, von Hippel PH. J. Mol. Biol. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 17.Johansen F, Jakobsen JP. J. Biomol. Struct. Dyn. 1998;16:205–222. doi: 10.1080/07391102.1998.10508240. [DOI] [PubMed] [Google Scholar]
- 18.LePecq JB, Paoletti C. J. Mol. Biol. 1967;27:87–106. doi: 10.1016/0022-2836(67)90353-1. [DOI] [PubMed] [Google Scholar]
- 19.Shchyolkina AK, Borisova OF, Livshits MA, Pozmogova GE, Chernov BK, Klement R, Jovin TM. Biochemistry. 2000;39:10034–10044. doi: 10.1021/bi9913909. [DOI] [PubMed] [Google Scholar]
- 20.Mergny JL, Collier D, Rougee M, Montenay-Garestier T, Helene C. Nucleic Acids Res. 1991;19:1521–1526. doi: 10.1093/nar/19.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]