Abstract
The emerging field of behavioral epigenetics is producing a growing body of evidence that early life experience and social exposure can alter the way by which genes are marked with DNA methylation. We hypothesize that changes in DNA methylation as well as other epigenetic markers could generate stable phenotypes. Early life adversity appears to result in altered DNA methylation of genes in the brain and peripheral tissues, and these changes are associated with adverse phenotypic changes. Although the data are still sparse, early epigenetic studies have provided a proof of principle that experiences and the environment leave marks on genes, and thus suggest molecular and physical mechanisms for the epidemiological concept of gene-environment interaction. The main attraction of DNA methylation for type I (TI) translational prevention science is the fact that, different from genetic changes that are inherited from our ancestors, DNA methylation is potentially preventable and reversible and, therefore, there is a prospect of epigenetically targeted interventions. In addition, DNA methylation markers might provide an objective tool for assessing effects of early adverse experience on individual risks as well as providing objective measures of progress of an intervention. In spite of this great potential promise of the emerging field of social and translational epigenetics, many practical challenges remain that must be addressed before behavioral epigenetics could become translational epigenetics.
Keywords: DNA methylation, Epigenetics, Early life stress, Translational science, Interventions, Gene environment, Transgenerational, Prevention science
INTRODUCTION
The understanding that behavior and human mental activity are tied to gene function has important implications for understanding not only neuropsychiatric disease but also the natural variation in social and intellectual achievements of humans. Animal studies have identified genes that influence animal behavior, and human genome-wide association studies (GWASs) have linked certain neuropsychiatric diseases to interindividual differences in sequences of genes [1–3]. Moreover, certain common differences in genetic sequences were associated with human behavioral phenotypes such as anxiety [4], stress [5], and aggression [6]. However, although GWASs were able to associate several genetic differences in people with behavioral disorders and neuropsychiatric disease, the effects in most cases are small and explain only a small fraction of the interindividual variation in the population. It is possible that the missing explanation resides in the fact that variations in these phenotypes are determined by sequence differences in a combination of a large number of genes. An additional possible mechanism is that other factors are critical in stable programming of gene function involved in human behavior. The fundamental questions are how much of our behavior is predetermined exclusively by genetic sequences and thus resilient to interventions and whether there are other factors that influence interindividual differences in stable gene programs which could be altered by interventions. The answer to these long-standing questions has wide-ranging implications not only to the quest for understanding fundamental biological and pathological mechanisms but also to almost all areas of human activity from medicine to education, politics, and social policy.
The idea that the relationship between genotype and phenotype is not always directly immediate has been noted in the previous century by Waddington who was trying to understand how one genome could encode the multiple phenotypes of a multicellular organisms [7]. Waddington coined the term epigenetics to describe the mechanisms that are involved in programming identical genes differently in different organs during embryogenesis. The biochemical mechanisms underlying epigenetic programming have been elucidated in the last few decades. However, the dominant idea in the field has been that these epigenetic programs are highly predictable and evolutionarily programmed during embryogenesis and are driven by predetermined (innate) driving forces that are fixed after birth. Are similar mechanisms utilized to program the genome to respond to external environmental factors? Could epigenetics confer experiential identity on DNA in addition to cell type identity? Are the epigenetic changes in response to the environment limited to a period after birth or could these be operative throughout life? Obviously, the answer for this question has immediate implications for either prevention or intervention.
The question of whether epigenetic mechanisms could be programmed by external environments has been a topic of extensive research in the last decade. The first set of experiments demonstrating postnatal epigenetic programming by postnatal social environments was by Weaver et al., who showed that differences in maternal care in rodents after birth determine differences in their adult offspring behavior by epigenetic programming [8]. Importantly, these studies also showed that programming by maternal care could be reversed in adulthood by cross-fostering as well as epigenetic therapeutics [8]. These studies and others established that epigenetic programs are reset in response to external cues including social environments and, as a result, they define stable phenotypes without changing the gene sequence. Epigenetic processes provide experiential identity onto DNA in addition to the cell type identity. These processes are proposed to be active during the early life period. However, it is possible to reverse epigenetic programming later in life with the appropriate interventions. These studies imply that it should be possible to prevent the emergence of certain phenotypes by targeting the social environments that trigger adverse epigenetic programming. Implications for the role of epigenetics in prevention research and practice fall into the rubric of type I translation (see Table 1), which pertains to the development of basic science-informed preventive interventions. In this paper, we will discuss epigenetic mechanisms with particular focus on DNA methylation and address the questions of how epigenetic information might instruct translational science by providing mechanistic concepts, diagnostic tools, and guides for prevention.
Table 1.
Translational research stages
| Type | Definition |
|---|---|
| Type 0 translation (T0) | The fundamental process of translating findings and discoveries from social and biomedical sciences into research with human subjects |
| Type 1 translation (T1) | Moving from bench to bedside. Translation of applied theory to methods and program development |
| Type 2 translation (T2) | Moving from bedside to practice and involves translation of program development to implementation |
| Type 3 translation (T3) | Determining whether efficacy and effectiveness trial outcomes can be replicated under real-world settings |
| Type 4 translation (T4) | Wide-scale implementation, adoption, and institutionalization of new guidelines, practices, and policies |
| Type 5 translation (T5) | Translation to global communities. Involves fundamental and universal change in attitudes, policies, and social systems |
EPIGENETIC MECHANISMS
RNA is transcribed from DNA by the cellular transcription machinery which is composed of several proteins that can read the DNA sequence template and write or transcribe a chain of RNA bases, a transcript. Some transcripts called messenger RNA (mRNA) could be further translated into proteins. An mRNA transcript is then transported into a different machinery in the cell that translates the RNA sequence and assembles, based on the information in the transcript, a sequence of amino acids which are the building blocks of proteins. Proteins are responsible for both the structure as well as the “workings” of our body. A genetic change in the sequence can change the identity of the protein produced or change its activity, and this could result in a change in phenotype. Epigenetic modifications consist of changes on top of (epi) the genome but do not change the inherited genomic structure itself. During the last three decades, biochemistry has been characterizing several levels of epigenetic mechanisms. Two ways this modification commonly occurs include (1) the attachment of a chemical structure called a methyl group to the backbone of a DNA molecule effectively turning off a transcription of an associated gene [9, 10] or (2) the modification of the histones around which the DNA is wrapped, which then affects the accessibility of the DNA for transcription [11, 12]. DNA is packaged in the nucleus of a cell into a structure termed chromatin. This structure includes, in addition to DNA, a scaffold of proteins called histones. Histones are chemically modified, and the modification of histones determines the accessibility of genes to the machinery that transcribes genes into RNA. DNA methylation is thought to be more on/off, while chromatin modification is more graduated, like a knob ratcheting up or down transcription [13]. Epigenetic modification of DNA functioning is a natural process. Epigenetic modification through exposure to the cellular environment plays a large role in tissue differentiation during fetal development [14]. Evidence has also been growing about the role of epigenetic modification in response to the social environment. The lesson that we have learned from studying DNA methylation patterns in different tissues is that identical DNAs could have different DNA methylation patterns and divergent gene expression programs. Vast phenotypic (including behavioral) differences could therefore be expressed by identical genetic sequences. Phenotypic differences could be generated by epigenetic rather than inherited genetic differences. If a similar mechanism operates in people that were exposed to different environments, then we have a mechanism explaining how environment could generate different phenotypes even when the genetic sequence is identical. This area of research addresses age-old questions about phenotypic variability given similar genomic structures and potentially furthers our understanding of the mechanisms by which environments alter phenotypic expressions.
DNA methylation and regulation of gene expression
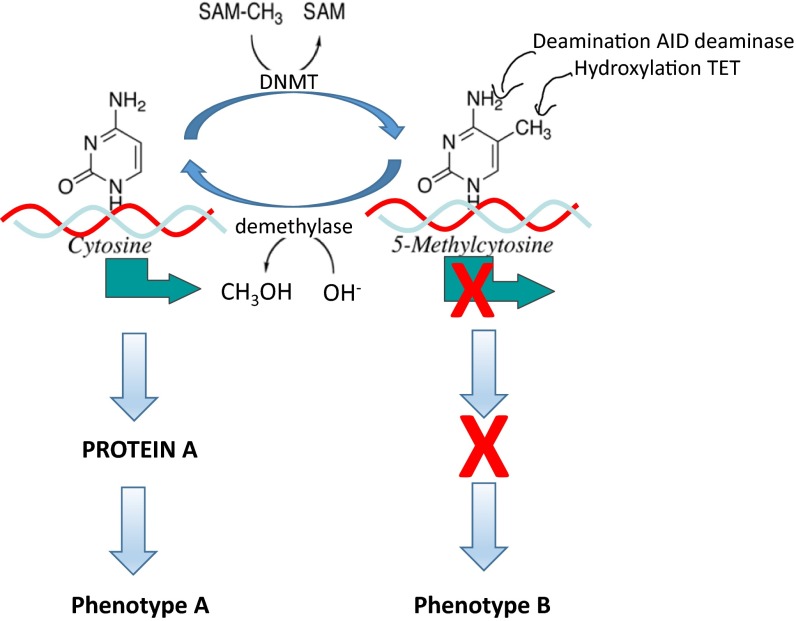
DNA methylation alters the way in which genes express themselves without inducing change in the actual sequence of the genes but having functional consequences. The same sequence of DNA could either be expressed or silenced based on its state of methylation (Fig. 1). Methylation of critical regulatory regions of genes can silence gene expression by blocking access to factors that recruit the transcription machinery that transcribes the genes [15, 16] or through recruitment of proteins that modify the chromatin and close the chromatin around the gene [17]. Modification of histones determines the accessibility of genes to the machinery that transcribes genes into RNA. A gene found in a closed chromatin is inaccessible and silent.
Fig 1.
DNA methylation and demethylation equilibrium; impact on gene expression and the phenotype. The state of DNA methylation is a balance of methylation and demethylation. DNA methylation reactions are catalyzed by DNA methyltransferases (DNMT) which transfer the methyl group from S-adenosylmethionine (SAM) releasing S-adenosylhomocysteine (SAH). Demethylation is catalyzed by a bona fide demethylase which removes a methyl group from DNA which reacts with OH– from water and is released as methanol (CH3OH). Methyl cytosine in critical positions in DNA silences gene transcription (X over green horizontal arrow), while the unmethylated gene is transcribed (green horizontal arrow). The transcript is translated to protein A which is responsible for phenotype A. When the gene is methylated, no transcript is produced and no protein is translated, leading to phenotype B. Alternative mechanisms for demethylation involve either hydroxylation of the methyl group by TET enzymes or deamination by deaminase enzymes followed with repair
Reversibility of DNA methylation
If DNA methylation plays a role in responding to experience in fully differentiated tissue, it must be biochemically reversible. That is, DNA should be either methylated or demethylated in response to environmental signals. Reversibility of DNA methylation is also critical for an intervention aimed at resetting epigenetic programming.
Although the mechanisms responsible for demethylation are yet unclear, there is evidence that DNA methylation is potentially reversible even in mature fully differentiated neurons [8, 18]. This knowledge has two important implications for our discussion. First, DNA methylation could change in adult tissues and, therefore, even adult tissue should be potentially responsive to environmental cues and readjustment of phenotypes could theoretically occur even in adults. Second, it should be possible to intervene to reverse DNA methylation and alleviate adverse phenotypes.
Epigenetic programming by early life adversity
It was originally believed that a loss of methyl marks could occur only when cells divide. During cell division, new unmethylated DNA is synthesized; if DNA methylation is blocked at the time of synthesis, the new DNA will be unmethylated. Most neurons in our brain do not divide. It was therefore believed that DNA methylation in neurons remains fixed after neurons mature and that changes in DNA methylation were unlikely to occur in the brain. The reversibility of DNA methylation suggests that DNA methylation changes may be feasible in cells that do not divide, such as neurons. The fact that the biochemistry is consistent with changes in DNA methylation even after embryonal development is completed opens up the opportunity for additional changes in DNA methylation in mature cells that would affect the phenotype. Cues triggering such changes could come from the environment. It is conceivable that chemical exposures could alter DNA methylation by interacting with the enzymes that either methylate or demethylate DNA. A more challenging hypothesis, however, is that changes in DNA methylation could be brought about in response to social exposures.
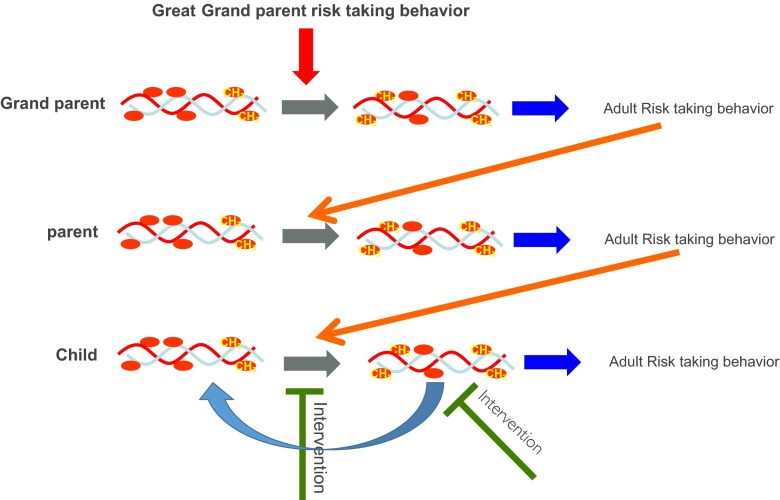
The first evidence that social exposure could alter DNA methylation in the brain came from studies on maternal care in rats [8]. The postnatal mother-neonate interaction is the earliest social environment encountered by the developing mammal, and it plays an important role in determining its future health. In rodents, maternal care involves pup licking and grooming (LG) by the mother. Pups that experience high intensity of maternal LG develop into adults that are less stressed and less anxious than pups that experience low maternal LG. Interestingly, the phenotypic differences between offspring of high and low LG are not genetic and are not inherited from the biological mother through germline transmission (the maternal egg that formed the offspring); cross-fostering experiments demonstrate that the stress responsivity and anxiety phenotype of the low-LG offspring are transmitted by the fostering mother [8, 19]. In addition to the stress and anxiety phenotype, the mother transmits her nurturing behavior to her female offspring [19]. The low/high LG stress and anxiety phenotypes are behaviorally transmitted across multiple generations since female offspring of low-LG mothers develops into low-LG mothers and offspring of high-LG mothers develops into high-LG mothers and sire high-LG offspring. This serves as a perfect example of the impact that a postnatal social environment has on the establishment of stable phenotypes that are socially inherited. This study also illustrates that early life interventions such as cross-fostering [19] could prevent the development of adverse phenotypes and block their transgenerational transmission. It should be noted here that that there are two phenotypic outcomes potentially affected. First is the impact on the offspring’s overall predisposition or behavior (stress/anxiety, aggression, etc.) that could possibly be epigenetically transmitted. And second, that the actual behavior of the parent could be transmitted to the offspring (e.g., nurturance). It stands to reason that this response is not limited to cross-fostering and to rodents only and that other early life interventions in humans such as parent training [20, 21] could have a similar impact (see Fig. 2).
Fig 2.
Multigenerational gamete-independent transmission of risk behavior phenotype. Exposure of the grandparent during early childhood to a risk-taking behavior leads to methylation of critical genes (CH 3), resulting in a risk-taking behavior phenotype in the grand parent. The grandparent risk-taking behavior alters the parent DNA methylation, leading to a risk-taking behavior in the parent. The parent risk-taking behavior triggers change in methylation in his offspring that results in a risk-taking behavior in the child. Intervention could happen either by protecting the child from parental risk-taking behavior, thus preventing the development of risk-taking behavior in the child or using epigenetic therapy or behavioral therapy later in life to reverse the methylation state responsible for a risk-taking behavior
What is the mechanism that mediates the long-term effects of parental care and other early life experiences? Understanding these mechanisms is critical for developing more targeted preventive and intervention strategies as well as for identifying people at risk of developing adverse phenotypes later in life. Weaver et al. focused on the glucocorticoid receptor (nr3c1) gene in the hippocampus, a nodal point of negative feedback regulation of the hypothalamic-pituitary-adrenal (HPA) axis and stress. Weaver et al. demonstrated a higher level of DNA methylation in adult offspring of the low LG in the CG dinucleotide sequence site in the nr3c1 gene contained in the sequence that binds the transcription factor NGFIA, a transcription factor that activates the nr3c1 gene [8]. The difference in DNA methylation emerged after birth concurrently with the exposure of the pups to maternal licking and grooming, remained into adulthood, and was transmitted by the fostering mother and not the biological mother in cross-fostering experiments [8]. These studies provided the first evidence that social exposure results in a change in DNA methylation in a critical gene that acts in the brain. Moreover, this change alters gene expression of a gene with a key role in regulating the body’s stress response, establishing a stable phenotype of heightened stress responsivity. Multigenerational transmission of phenotypes triggered by parental behavior and mediated by DNA methylation in the offspring could explain gamete-independent (unrelated to inherited genetic material) multigenerational transmission of other behaviors such as risk taking or impulsivity (see Fig. 2).
In summary, these results serve as a paradigm for other adverse as well as positive social exposures early in life, providing a proof of principle that (1) adverse environments in the early life period change the chemical properties of gene modifications and their mode of action resulting in a long-term phenotypic impact and (2) altering an adverse environment in very early life could prevent emergence of adverse phenotypes. This has obviously important implications for translational science in humans. An important caveat should be mentioned here, however, which is that the responses of DNA methylation to adverse experience early in life might also be adaptive by preparing the offspring to an anticipated harsher environment; the context in many respects determines whether the response is pathological or functional. Such adaptive processes could turn maladaptive when the experienced environment later in life is inconsistent with the anticipated environment early in life.
The breadth and scope of DNA methylation changes driven by early life experience
The experiments described above focused on candidate genes, candidate physiological pathways, and candidate brain regions. The question, nevertheless, remains whether the response to early life adversity is limited to just classical stress pathways. It is well known from human studies that early life adversity impacts several physiological systems in adults and has broad and varied effects on human health [22, 23]. Differences in maternal care also affect the expression of hundreds of genes in the hippocampus, suggesting a broad response of the DNA to early life experience [18]. DNA methylation and chromatin changes affect broad genomic regions [24]. For example, McGowan et al. showed that a large gene cluster containing the entire family of the protocadherin genes (associated with many neurodevelopmental processes like axon guidance and dendrite branching) is epigenetically programmed by maternal care; the entire cluster exhibited reduced expression, increased DNA methylation, and altered chromatin modification in low-LG adult offspring [24]. It is tempting to speculate that these responses evolved to adapt animals to anticipated dynamic challenges in the environment [25].
SYSTEM-WIDE RESPONSES TO EARLY LIFE ADVERSITY IN NON-HUMAN PRIMATES: IMPLICATIONS FOR TRANSLATIONAL SCIENCE
It is almost impossible to define a causal relationship between early life adversity and DNA methylation changes in humans since the DNA methylation changes detected might reflect underlying genetic differences or past histories of environmental exposures rather than exposure to early life adversity. Addressing this question requires a randomized design, but it is ethically impossible to randomize child adversity in humans. Nonhuman primates offer the closest model to humans to address this question. Another critical question for translation of epigenetic data to human prevention and intervention studies is the question of whether informative methylation differences associated with early life adversity are limited to the brain or whether they exist in other peripheral tissues as well. Studies comparing methylation in brain and peripheral tissues in rhesus macaque monkeys raised maternally or with a surrogate/peer-rearing group support the hypothesis that the response to maternal care is not limited to one tissue or one brain region but that it is system wide as well as genome wide [26]. The data are consistent with the idea that the phenotypic impact of early life adversity is not limited to the brain and involves other tissues such as the immune system [27]. These data also provide a proof of principle for the feasibility of studying the epigenetic consequences of social exposures in peripheral T cells which should have important implications for translational science.
Reversibility of epigenetic programming by early life adversity
Phenotypes fixed by epigenetic changes are potentially biochemically reversible [28]. Is it possible to reverse early life adversity-programmed epigenetic alterations later in life? This question is critical for translational science and for translating epigenetic concepts to interventions in humans. It has been proposed that DNA methylation is a reversible biological signal [28]. Several biochemical mechanisms were proposed to be involved in active DNA demethylation that does not require DNA synthesis [29–31]. Weaver et al. [8, 18] showed that differences in rat licking and grooming of pups resulted in differences in methylation of a gene related to HPA responses to stress in the pups and that this epigenetic modification was reversible by cross-fostering and epigenetic therapeutics in adult animals [8, 18]. It was shown that a DNA demethylation inhibitor reprogrammed cocaine addiction in a rat model of cocaine craving [32]. These data provide a proof of principle for the reversibility of phenotypic changes triggered by experience-driven epigenetic programming. It is consistent with the idea that the changes that define the phenotype are not caused only by inherited genetic polymorphisms but also by reversible epigenetic marks such as DNA methylation. The remaining challenge is to determine whether these conclusions could be translated to humans and whether behavioral interventions could substitute for pharmacological interventions (for a recent review of epigenetic approaches that target brain functions, see [33]).
TRANSLATION TO HUMANS; EVIDENCE FOR EPIGENETIC CONSEQUENCES OF EARLY LIFE ADVERSITY IN HUMANS
Establishing causal relationship between early life adversity and offspring phenotypes in humans in the same way that it is done in animals is much more challenging. The first line of evidence in humans for DNA methylation differences associated with adversity came from examining postmortem hippocampi of humans who experienced adverse experiences in childhood and controls. McGowan et al. showed that the adult suicide victims who experienced childhood abuse had higher overall methylation in their ribosomal RNA (rRNA) genes and NR3C1 gene and expressed less rRNA and NR3C1 mRNA [34]. These data were the first demonstration of evolutionary conservation of the epigenetic responses to early life experiences.
As discussed above, the changes in DNA methylation in rodents and monkeys are broad and clustered and are not limited to one gene. Remarkably, Suderman et al. showed that the response in human brains to early life experience is broad as it is in rats and monkeys and is evolutionary conserved with a striking overall similarity between rat and human brains [35]. The PROTOCADHERIN gene family cluster which showed cluster-wide DNA methylation differences between high- and low-LG offspring adult rats [24] is differentially methylated between adult humans who were abused as children and control adults as well [35]. This evolutionary conservation of the epigenetic response of the same cluster of a gene family presents a strong argument that early life experience alters epigenetic programming in the brain in humans.
SUMMARY AND PROSPECTIVE
Epigenetic studies in animals have provided evidence that early life experiences and adversity could stably affect the phenotype in the absence of genetic difference. They also provided evidence that prevention of early life adversities would prevent the development of adverse phenotypes later in life. Moreover, there is evidence from animal experiments that phenotypes that were epigenetically programmed by early life adversity could be reversed by epigenetic therapeutics [8, 18]. This suggests that it might be possible not only to prevent but also to reverse the consequences of early life experiences. Obviously, the most critical problem with pharmacological approaches is lack of specificity. An important question is therefore whether behavioral interventions would be as effective as pharmacological interventions in reversing epigenetic programming by early life experience. In addition, studies in animals and preliminary studies in humans suggest that DNA methylation markers of early life adversity might serve as objective diagnostic tools. An important goal would be to develop early DNA methylation markers of vulnerability and resilience for behavioral disorders later in life.
Many epigenetic challenges remain that need to be addressed: How can we capture DNA methylation markers of behavioral disorders before the phenotype is detected using either prospective or cross-sectional studies? We need to map the normal longitudinal trajectories of DNA methylation in human peripheral cells that could serve as bookmarks for more dedicated studies. Also needed is to define the temporal order of early experience, early DNA methylation changes, late experience, late DNA methylation changes, and the phenotype. We must further address genetic, ethnic, and gender heterogeneity in these responses. A huge challenge is how do we define causality between experience, DNA methylation alterations, the role of these changes in gene programming, and the resulting behavioral disorders in humans in the absence of a randomized experimental design? How do we design epigenetically informed interventions that tap into epigenetic mechanisms? What do we do about transgenerational effects? Is pharmacology an option when there are no behavioral strategies? How do we translate new intervention strategies from animal experiments to humans?
In addition, many critical questions in translational science remain that must be addressed in properly designed studies. Could we map DNA methylation changes early in life that predict risk and resilience and provide mechanistic insight as well? Does consistent exposure to positive, supportive environments produce a methylation pattern that places children on a positive developmental trajectory, more resilient to subsequent adversities [36]? Are DNA methylation patterns defined by early life experience altered with childhood, adolescence, and adult experience and how do these changes relate to environmental exposures? When is the best time for intervention? How narrow is the window of opportunity for prevention? How do we use epigenetic concepts to design interventions? Prevention research has shown that early life preventive interventions can have long-lasting effects on the children into adulthood [37–40]. Could epigenetic markers be used to follow up these interventions? Are the critical periods absolutely critical or could we intervene later in life as well?
The data in humans are still sparse, and none of the differentially DNA methylation markers described to date could possibly serve as biomarkers in human studies. The data to date provide, nevertheless, sufficient justification for integrating DNA methylation mapping and epigenetic intervention strategies into our experimental designs. Future studies should focus on discovery of a panel of DNA methylation-based biomarkers that could guide epigenetically informed interventions.
Acknowledgments
The work on this transdisciplinary manuscript resulted from a National Institute of Nursing Research-funded R13 conference “Advancing Transdisciplinary Translation for Prevention of High-Risk Behaviors: Critical Thinking to Overcome Individual and Institutional Barriers” (R13NR013623-01-02). Additional support for this paper was provided in part by a grant from the National Institute on Drug Abuse (R01DA024411 01-06). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NINR or NIDA. MS is supported by CIHR-MOP-42411
Compliance with ethical standards
The paper is a review article and discusses published studies. No primary studies with ethics requirements are discussed.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Implications
Researchers: provides a review and insight into the science of epigenetics and its implications in neuroscience.
Practitioners: introduce the practitioners to implications of the science of epigenetics on understanding, preventing, and treating behavioral disorders
Policy makers: the implications that epigenetics has on social policy
References
- 1.Cohen-Woods S, Craig IW, McGuffin P. The current state of play on the molecular genetics of depression. Psychol Med. 2013;43(4):673–87. doi: 10.1017/S0033291712001286. [DOI] [PubMed] [Google Scholar]
- 2.Schwab SG, Wildenauer DB. Genetics of psychiatric disorders in the GWAS era: an update on schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2013;263(Suppl 2):147–54. doi: 10.1007/s00406-013-0450-z. [DOI] [PubMed] [Google Scholar]
- 3.Stergiakouli E, Hamshere M, Holmans P, et al. Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am J Psychiatry. 2012;169(2):186–94. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murakami F, Shimomura T, Kotani K, et al. Anxiety traits associated with a polymorphism in the serotonin transporter gene regulatory region in the Japanese. J Hum Genet. 1999;44(1):15–7. doi: 10.1007/s100380050098. [DOI] [PubMed] [Google Scholar]
- 5.Binder EB, Salyakina D, Lichtner P, et al. Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet. 2004;36(12):1319–25. doi: 10.1038/ng1479. [DOI] [PubMed] [Google Scholar]
- 6.Eisenberger NI, Way BM, Taylor SE, et al. Understanding genetic risk for aggression: clues from the brain’s response to social exclusion. Biol Psychiatry. 2007;61(9):1100–8. doi: 10.1016/j.biopsych.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183(4676):1654–5. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- 8.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 9.Jin SG, Wu X, Li AX, et al. Genomic mapping of 5-hydroxymethylcytosine in the human brain. Nucleic Acids Res. 2011;39(12):5015–24. doi: 10.1093/nar/gkr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science. 2009;324(5929):929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11(6):266–73. doi: 10.1016/S0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- 12.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 13.Szyf M, McGowan PO, Turecki J, et al. The social environment and the epigenome. In: Worthman CM, Plotsky PM, Schechter DS, Cummings CA, et al., editors. Formative experiences the interaction of caregiving, culture, and developmental psychobiology. Cambridge: Cambridge University Press; 2013. pp. 53–82. [Google Scholar]
- 14.Razin A, Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984;782(4):331–42. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 15.Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18(13):3975–82. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inamdar NM, Ehrlich KC, Ehrlich M. CpG methylation inhibits binding of several sequence-specific DNA- binding proteins from pea, wheat, soybean and cauliflower. Plant Mol Biol. 1991;17(1):111–23. doi: 10.1007/BF00036811. [DOI] [PubMed] [Google Scholar]
- 17.Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88(4):471–81. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 18.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006;103(9):3480–5. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis D, Diorio J, Liu D, et al. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286(5442):1155–8. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- 20.Goyal NK, Teeters A, Ammerman RT. Home visiting and outcomes of preterm infants: a systematic review. Pediatrics. 2013;132(3):502–16. doi: 10.1542/peds.2013-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaminski JW, Valle LA, Filene JH, et al. A meta-analytic review of components associated with parent training program effectiveness. J Abnorm Child Psychol. 2008;36(4):567–89. doi: 10.1007/s10802-007-9201-9. [DOI] [PubMed] [Google Scholar]
- 22.Power C, Hertzman C. Social and biological pathways linking early life and adult disease. Br Med Bull. 1997;53(1):210–21. doi: 10.1093/oxfordjournals.bmb.a011601. [DOI] [PubMed] [Google Scholar]
- 23.Power C, Jefferis BJ, Manor O, et al. The influence of birth weight and socioeconomic position on cognitive development: does the early home and learning environment modify their effects? J Pediatr. 2006;148(1):54–61. doi: 10.1016/j.jpeds.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 24.McGowan PO, Suderman M, Sasaki A, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6(2):e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szyf M. The early-life social environment and DNA methylation. Clin Genet. 2012;81(4):341–9. doi: 10.1111/j.1399-0004.2012.01843.x. [DOI] [PubMed] [Google Scholar]
- 26.Provencal N, Suderman MJ, Guillemin C, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci: Off J Soc Neurosci. 2012;32(44):15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coe CL, Lubach GR. Prenatal influences on neuroimmune set points in infancy. Ann N Y Acad Sci. 2000;917:468–77. doi: 10.1111/j.1749-6632.2000.tb05411.x. [DOI] [PubMed] [Google Scholar]
- 28.Ramchandani S, Bhattacharya SK, Cervoni N, et al. DNA methylation is a reversible biological signal. Proc Natl Acad Sci U S A. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya SK, Ramchandani S, Cervoni N, et al. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- 30.Gavin DP, Chase KA, Sharma RP. Active DNA demethylation in post-mitotic neurons: a reason for optimism. Neuropharmacology. 2013;75:233–245. doi: 10.1016/j.neuropharm.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rai K, Huggins IJ, James SR, et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massart R, Barnea R, Dikshtein Y, et al. Role of DNA methylation in the nucleus accumbens in incubation of cocaine craving. J Neurosci. 2015;35:8042–8058. doi: 10.1523/JNEUROSCI.3053-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szyf M. Prospects for the development of epigenetic drugs for CNS conditions. Nature Rev. 2015;14:461–474. doi: 10.1038/nrd4580. [DOI] [PubMed] [Google Scholar]
- 34.McGowan PO, Sasaki A, Huang TC, et al. Promoter-wide hypermethylation of the ribosomal RNA gene promoter in the suicide brain. PLoS One. 2008;3(5):e2085. doi: 10.1371/journal.pone.0002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suderman M, McGowan PO, Sasaki A, et al. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc Natl Acad Sci U S A. 2012;109(Suppl 2):17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masten AS, Burt KB, Coatsworth JD. Competence and psychopathology. Hoboken: Wiley; 2006. pp. 696–738. [Google Scholar]
- 37.Eckenrode J, Campa M, Luckey DW, et al. Long-term effects of prenatal and infancy nurse home visitation on the life course of youths: 19-year follow-up of a randomized trial. Arch Pediatr Adolesc Med. 2010;164(1):9–15. doi: 10.1001/archpediatrics.2009.240. [DOI] [PubMed] [Google Scholar]
- 38.Zielinski DS, Eckenrode J, Olds DL. Nurse home visitation and the prevention of child maltreatment: impact on the timing of official reports. Dev Psychopathol. 2009;21(2):441–53. doi: 10.1017/S0954579409000248. [DOI] [PubMed] [Google Scholar]
- 39.Hawkins JD, Kosterman R, Catalano RF, Hill KG, Abbott RD. Effects of social development intervention in childhood 15 years later. Arch Pediatr Adolesc Med. 2008; 162(12): 1133-1141. [DOI] [PMC free article] [PubMed]
- 40.Hill KG, Bailey JA, Hawkins JD, Catalano RF, Kosterman R, Oesterle S, Abbott RD. The onset of STI diagnosis through age 30: results from the Seattle Social Development Project intervention. Prev Sci. 2014; 15(Suppl 1): S19-S32. [DOI] [PMC free article] [PubMed]