Abstract
Polynucleotide phosphorylase (PNPase), a 3′-to-5′ phosphorolytic exoribonuclease, is thought to be the primary enzyme responsible for turnover of Bacillus subtilis mRNA. The role of PNPase in B. subtilis mRNA decay has been analyzed previously by comparison of mRNA profiles in a wild-type strain versus a strain that is deleted for pnpA, the gene encoding PNPase. Recent studies have provided evidence for a degradosome-like complex in B. subtilis that is built around the major decay-initiating endonuclease, RNase Y, and there is ample evidence for a strong interaction between PNPase and RNase Y. The role of the PNPase-RNase Y interaction in the exonucleolytic function of PNPase needs to be clarified. We sought to construct a B. subtilis strain containing a catalytically active PNPase that could not interact with RNase Y. Mapping studies of the PNPase-RNase Y interaction were guided by a homology model of B. subtilis PNPase based on the known structure of the Escherichia coli PNPase in complex with an RNase E peptide. Mutations in B. subtilis residues predicted to be involved in RNase Y binding showed a loss of PNPase-RNase Y interaction. Two mRNAs whose decay is dependent on RNase Y and PNPase were examined in strains containing full-length PNPase that was either catalytically active but unable to interact with RNase Y, or catalytically inactive but able to interact with RNase Y. At least for these two mRNAs, disruption of the PNPase-RNase Y interaction did not appear to affect mRNA turnover.
Keywords: bacterial genetics, endonuclease, mRNA decay, phosphorylase, RNA turnover, Bacillus subtilis, PNPase, RNase Y
Introduction
Bacterial mRNA turnover is an essential process that replenishes the ribonucleotide pool and plays a role in regulation of gene expression. We have been studying 3′-exonuclease activity in Bacillus subtilis, the Gram-positive model organism. Although at least four 3′-exonucleases are known to exist in B. subtilis (1), the phosphorolytic enzyme, polynucleotide phosphorylase (PNPase,3 encoded by the pnpA gene), has long been thought to play the major role in mRNA turnover in this organism (2, 3). The B. subtilis pnpA gene was originally identified in a transposon insertion screen of competence-deficient mutants, and was also shown to be required for cold adaptation (4). A deletion strain lacking most of the pnpA coding sequence (the ΔpnpA strain) was constructed, and additional phenotypes were observed: growth as non-motile chains, tetracycline sensitivity (5), and accumulation of mRNA decay intermediates (1, 6). More recently, reliance of global mRNA turnover on PNPase was examined in an RNA sequencing analysis of the B. subtilis transcriptome in wild-type and ΔpnpA strains, and this demonstrated the broad impact of PNPase on mRNA turnover (7).
Many of the above mentioned studies were performed before the discovery of RNase Y, a B. subtilis endoribonuclease that appears to function in initiation of decay similarly to the more well known RNase E of Escherichia coli (8, 9). Endonucleolytic cleavage in the body of a message by RNase E in E. coli or RNase Y in B. subtilis generates mRNA fragments that are susceptible to further steps in mRNA turnover. In addition, just as RNase E of E. coli coordinates a complex of several proteins known as the “degradosome,” so, too, there is evidence that RNase Y of B. subtilis is the organizing protein for a complex that includes, at least, PNPase, CshA (an RNA helicase), and the glycolytic enzymes, enolase and phosphofructokinase (8, 10, 11). A PNPase-RNase Y interaction was first indicated by bacterial-2-hybrid (B2H) assay and in vivo protein crosslinking experiments (8). This interaction has more recently been confirmed by surface plasmon resonance analysis, which showed a strong PNPase-RNase Y interaction with a Kd of 5 nm (12). Structural mapping of regions of PNPase that interact with an RNase E peptide has been done for the E. coli enzyme (13). Structural studies have not been reported for the B. subtilis PNPase-RNase Y interaction.
The existence of a PNPase-RNase Y complex raises the question of whether this interaction is relevant to the ability of PNPase to rapidly degrade upstream decay intermediates that are created by RNase Y cleavage. In addition, prior results of experiments in the strain that contained no PNPase may be confounded by effects on RNase Y; the complete absence of PNPase protein in the ΔpnpA strain may not only affect 3′-exonucleolytic turnover directly, but may also affect RNase Y-mediated initiation of decay indirectly. As such, we sought to map the regions of PNPase that interact with RNase Y, to construct a B. subtilis mutant strain containing a catalytically active PNPase that is unable to interact with RNase Y.
Experimental Procedures
Bacterial Strains
E. coli DH5α (14) and BTH101 (15) were used for cloning experiments and B2H analysis. The wild-type B. subtilis strain was BG1, a trpC2 thr-5 derivative of B. subtilis strain 168. In BG546, a large portion of the pnpA coding sequence (CDS) was replaced with a neomycin resistance cassette, as described previously (5). In this study, the wild-type pnpA CDS was replaced with a CDS containing the RNase Y interaction domain E331A change, to give BG1039. Similarly, the wild-type pnpA CDS was replaced with a CDS containing the active site D493A change, to give BG1030.
Homology Modeling
A B. subtilis PNPase model was constructed based on the crystal structure of the E. coli PNPase trimer in complex with an RNase E peptide (Protein Data Bank identifier 3GCM (13)). The alignment between the B. subtilis and E. coli PNPase sequences was generated with the Promals3D server (16) using the default parameters. We used MODELLER-9v14 (17) to construct 100 models in the presence of the RNase E peptide. The models were assessed and ranked with Z-DOPE, a normalized atomic distance-dependent statistical potential based on known protein structures (18). For each PNPase chain, we modeled the residues that are included in the E. coli PNPase structure (13). The models were visualized using PyMOL (Schrödinger, LLC). The electrostatic potential of the homology model and template structure surfaces were calculated using the Adaptive Poisson-Boltzmann Solver (APBS) plug-in in PyMOL.
Hotspot Analysis
To prioritize residues for mutagenesis, various “hotspot” predictors on the E. coli PNPase/RNase E template structure were run, and the predicted residues were evaluated according to their conservation between E. coli and B. subtilis PNPase sequences, as well as their location in the B. subtilis PNPase model. In particular, hotspots were predicted using the KFC2 server (19), HotPOINT (20), and the Computational Interface Alanine Scanning Server (21). ConSurf (22) was used for estimating the evolutionary conservation of amino acid positions in a protein, based on the phylogenetic relation between homologous sequences.
Site-directed Mutagenesis and Allele Replacement
Plasmids for the B2H assay carrying the pnpA and rny coding sequences (pGP984 and pGP978, respectively) were constructed previously (8). Point mutations in the pnpA CDS were constructed using the QuikChange II site-directed mutagenesis protocol (Agilent Technologies). Primer sequences are available on request. All gene mutations were verified by DNA sequencing of the complete pnpA CDS. For replacement of the wild-type pnpA allele with mutant versions, we used the pMiniMAD vector (23). A 2.1-kb fragment containing the pnpA gene was amplified by PCR with an upstream primer that contained an EcoRI site and a downstream primer that contained a BamHI site. The amplified fragment was cloned into the EcoRI and BamHI sites of the pMiniMAD plasmid. Replacement of the wild-type pnpA allele with a mutated allele carried on the pMiniMAD plasmid derivative was performed as described (23).
Bacterial Two-hybrid Screening
Plasmids containing wild-type (pGP984) or mutant PNPase CDSs were co-transformed with the RNase Y-containing plasmid (pGP798) into E. coli strain BTH101. Strains carrying the PNPase and RNase Y plasmids were grown overnight in Luria-Bertani broth (LB, 10 g of Tryptone, 5 g of yeast extract, 5 g of NaCl/liter) and diluted for plating on MacConkey agar plates (Difco, BD Biosciences) containing maltose (1%), ampicillin (100 μg/ml), kanamycin (50 μg/ml), and isopropyl β-d-1-thiogalactopyranoside (0.5 mm). The plates were incubated for 30 h at 30 °C and then at room temperature overnight to enhance the colony color. As a negative control, cells were co-transformed with pGP984 and pKT25::zip (24), which contains the CDS for the yeast GCN4 transcription factor.
Protein Isolation and Western Blotting Analysis
Pellets from 40 ml of E. coli overnight cultures (∼0.3 g of dry weight) were frozen/thawed and treated with 1.2 ml of B-PER reagent (Pierce), according to the manufacturer's protocol. 3 μl of soluble protein extract were loaded on a 10% Bis-Tris NuPAGE gel (Invitrogen) and electrophoresed under reducing conditions. The gel was electroblotted onto a PVDF membrane (Thermo Scientific) for 1 h at 30 V. Membranes were probed with either mouse anti-cyaA (adenylate cyclase) antibody at a 1:1000 dilution (Santa Cruz Biotechnology, catalog number sc-13582, lot number C2715) or anti-GAPDH antibody at a 1:5000 dilution (Antibodies-online, catalog number L001, lot number 01016), followed by goat anti-mouse antibody conjugated to horseradish peroxidase (Bio-Rad). The signal was detected by ECL Western blotting substrate (Pierce) and quantitated on a Typhoon TRIO variable-mode imager (GE Healthcare).
RNA Extractions and Northern Blotting Analysis
For RNA isolation, a 1:40 dilution of an overnight culture was inoculated into LB broth. Cells were harvested at mid-logarithmic stage (A600 0.4–0.5), and total RNA was extracted by the hot phenol method (25). RNA concentrations were determined using a Qubit fluorimeter (Invitrogen), and RNA integrity was checked on a 1% denaturing agarose MOPS gel. To determine mRNA half-life, total RNA was isolated at times after the addition of 150 μg/ml rifampin. RNA was either fractionated on a 6% denaturing polyacrylamide gel and electroblotted, or fractionated on a 1.2% denaturing agarose MOPS gel and blotted by wicking. T4 polynucleotide kinase and [γ-32P] ATP were used to prepare 5′-end-labeled oligonucleotide probes. The rpsO probe was complementary to the sequence from 13 nt upstream to 10 nt downstream of the rpsO start codon. The cggR probe was complementary to nt 37–66 of the cggR CDS. To control for RNA loading, membranes were stripped and probed either for 5S rRNA, as described (26), or for 16S rRNA, using an oligonucleotide complementary to nt 1405–1424 (27). Results were the average of two or more biological replicates. Quantitation of the radioactivity in bands on Northern blots was performed with a Typhoon TRIO variable-mode imager (GE Healthcare). To determine the half-life of full-length RNA, exponential regression analysis (R2 > 0.9) was performed on the percentage of RNA remaining versus time.
Results and Discussion
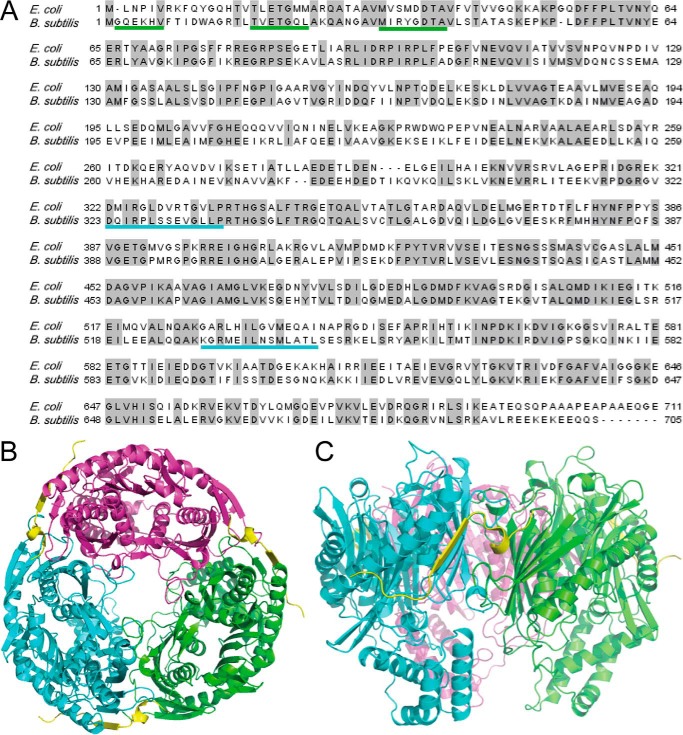
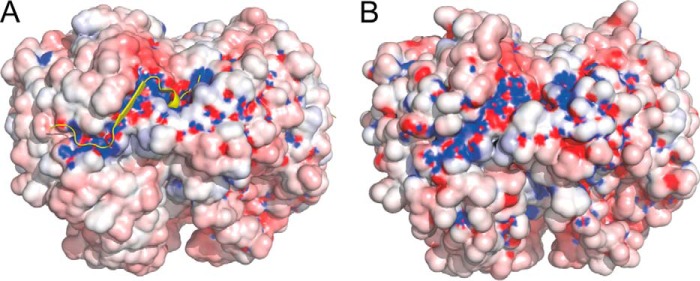
Bacterial PNPase exists primarily as a trimer (28). B. subtilis and E. coli PNPase share 52% identity and 69% similarity (Fig. 1A). A homology model of the B. subtilis PNPase trimer was generated based on the trimeric structure of the E. coli PNPase (Fig. 1, B and C) (see “Experimental Procedures”). The Z-DOPE score of the top model was −0.3, indicating that this model would likely be sufficiently accurate to guide further structural and functional studies. The E. coli crystal structure also contained a 21-amino acid peptide of the exonuclease RNase E (Fig. 1, B and C, yellow) that is in contact with E. coli PNPase (13). The interacting peptide binds at a site that is formed by residues from adjacent monomers. Although there is no sequence similarity between E. coli RNase E and B. subtilis RNase Y, these two proteins are thought to play functionally equivalent roles in the initiation of mRNA decay (29). We therefore hypothesized that RNase Y binds to B. subtilis PNPase in a similar region as RNase E binds to E. coli PNPase. In support of this hypothesis, the overall shape and electrostatic potential of both the E. coli structure and the B. subtilis model were analyzed (Fig. 2, A and B). The surface of E. coli PNPase in the region that interacts with the RNase E peptide showed a pronounced electrostatic potential (called the “electrostatic channel” below), as well as a deep binding pocket for the helical domain of the RNase E peptide (Fig. 2A). The overall shape, electrostatic potential, and binding pocket were conserved in the corresponding location of the B. subtilis PNPase (Fig. 2B), suggesting that RNase Y binds in a similar manner to PNPase.
FIGURE 1.
Homology modeling of B. subtilis PNPase. A, alignment of E. coli and B. subtilis PNPase amino acid sequences. Identical residues are indicated by the gray highlighting. Underlining indicates regions of B. subtilis PNPase that interact with RNase Y, as suggested by the modeling and mutagenesis experiments; colors are those used for PNPase monomers in subsequent figures. B, top view of B. subtilis PNPase trimer, with three subunits (green, cyan, and magenta) surrounding the central channel in which RNA enters to reach the PNPase active sites (28). For modeling purposes, the interacting peptide is that of RNase E (in yellow), and its coordinates were derived from the crystal structure via superposition to the B. subtilis PNPase model. C, side view of two PNPase monomers and the RNase E interacting peptide.
FIGURE 2.
Electrostatic channel for binding of interacting peptide. A, surface representation of the E. coli PNPase/RNase E trimer structure showing the electrostatic channel, with positive charge in blue and negative charge in red. Bound RNase E peptide is represented by yellow graphic. B, surface representation of the B. subtilis PNPase model, showing a similar predicted electrostatic channel.
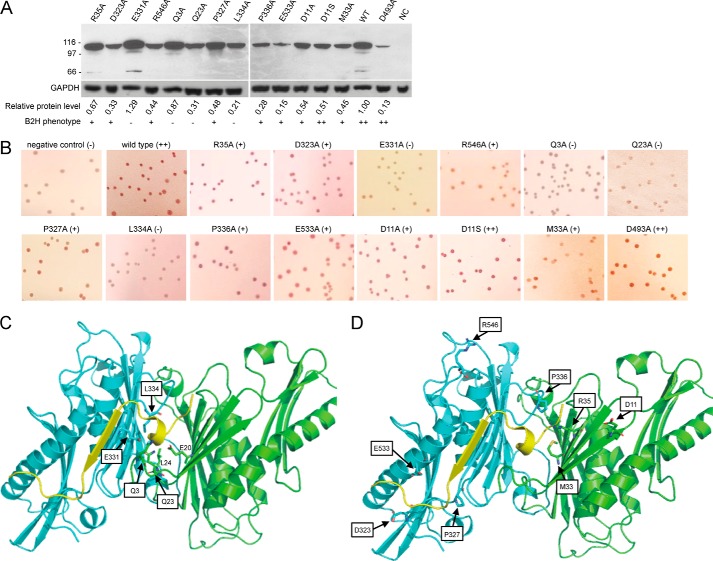
Several B. subtilis PNPase residues, Glu-20, Arg-35, Asp-323, Glu-331, and Arg-546, were hypothesized to be involved in the RNase Y interaction. These are charged residues located on the protein surface that are predicted to be far from the catalytic site and that correspond with residues of E. coli PNPase that appear to interact with the RNase E peptide (13). The B2H assay system (24) was used to screen for the interaction of RNase Y with PNPase mutant proteins. For this, we used plasmids pGP798, which contains the RNase Y CDS fused to the N-terminal fragment of the adenylate cyclase catalytic domain, and pGP984, which contains the PNPase CDS fused to the C-terminal fragment of the adenylate cyclase catalytic domain (8). Derivatives of pGP984 were constructed that had the above mentioned PNPase residues mutated to alanine. The B2H assay of PNPase-RNase Y interaction was performed on MacConkey agar plates (see “Experimental Procedures”). To ensure that the introduction of mutations did not cause drastic changes in protein stability, Western blotting analysis was performed using antibody to the C-terminal fragment of the adenylate cyclase catalytic domain. The data in Fig. 3A show the presence of full-length PNPase/adenylate cyclase fusion protein for all mutants examined. Although there was as much as an 8-fold difference in protein amounts relative to wild type, the absolute level of protein did not affect the results of the B2H assay. Thus, several mutants showed a high level of fusion protein on the Western blot but were negative for interaction in the B2H assay, and several mutants showed a low level of fusion protein on the Western blot but were positive for interaction in the B2H assay. Apparently, the readout is sensitive to protein function, but is not strongly dependent on the amount of protein.
FIGURE 3.
B2H assay and location of mutated residues. A, Western blotting analysis to demonstrate presence of PNPase/adenylate cyclase fusion protein. The antibody used to detect the fusion protein was directed against the C-terminal portion of the Bordetella pertussis adenylate cyclase catalytic domain (35). To control for loading and blotting, antibody to E. coli GAPDH was used (36). The amount of mutant fusion protein, relative to wild type, is shown below each lane (average of two experiments), as well as the result of the B2H assay (from Table 1). NC, negative control. B, results of B2H assays for PNPase-RNase Y interaction. The first two images in the top row are results from negative (no PNPase, no pink color) and positive (wild-type PNPase, dark pink color) controls. The last image in the bottom row is the result from the catalytically inactive D493A mutant that interacts with RNase Y as well as wild type. C, close-up of PNPase residues that resulted in loss of interaction with RNase Y when mutated to alanine. Boxed residues were mutated. Two residues that are also likely to contribute to the binding site for RNase Y are indicated without boxes. D, close-up of PNPase residues that resulted in a weak effect on the interaction with RNase Y when mutated to alanine.
We mutated one of the two proposed glutamate residues, Glu-331, to alanine. The B2H assay indicated a clear loss of interaction of the E331A mutant with RNase Y (Table 1, first round); although wild-type PNPase gave an intense pink color on the B2H assay, the E331A mutant showed no pink color (Fig. 3B). In a close-up of B. subtilis PNPase regions that correspond to E. coli PNPase regions that interact with the RNase E peptide (Fig. 3C), the Glu-331 side chain is seen to be facing, and a short distance (∼3 Å) from, the helical domain of the peptide. As shown in Fig. 3C, the position of the Glu-20 side chain suggests that this residue is also an important contributor to the binding site.
TABLE 1.
B2H screening results
| Mutated residue | Residue characteristics | Colony colora |
|---|---|---|
| Controls and wild type | ||
| Negative control | − | |
| Wild type | ++ | |
| D493A | ++ | |
| First round | ||
| R35A | Surface residue, positive electrostatic potential | ++ |
| D323A | Conserved surface residue, negative electrostatic potential | + |
| E331A | Surface residue, negative electrostatic potential | − |
| R546A | Conserved surface residue, positive electrostatic potential | + |
| Second round | ||
| Q3A | Predicted to affect RNase Y interaction; located in N-terminal loop region | − |
| Q23A | Located in predicted RNase Y binding pocket | − |
| P327A | Test bottom boundary of electrostatic channel | + |
| L334A | Faces RNase Y binding pocket | − |
| P336A | Close proximity to RNase E peptide loop; P may be structurally significant | + |
| E533A | Predicted not to affect RNase Y interaction; located close to boundary of electrostatic channel | + |
| Third round | ||
| D11A | Predicted not to affect RNase Y interaction | + |
| D11S | Predicted to affect RNase Y interaction less than D11A | ++ |
| M33A | Possible hydrophobic effect | + |
a −, no pink color (no interaction); +, light pink color (weak interaction); ++, dark pink color (strong interaction).
The other three mutants, R35A, D323A, and R546A, showed a weaker effect in the B2H assay (Table 1), and the close-up in Fig. 3D provides the rationale for this observation. The Arg-35 side chain faces the non-helical domain of the binding peptide, and its mutation may therefore have less effect. Arg-546 is located farther away from the binding peptide, perhaps in an unstructured region. The third residue, Asp-323, sits near the C-terminal end of the peptide sequence, which likely explains the weaker effect of mutating this residue.
To prioritize additional residues for mutagenesis studies, we used three different hotspot predictor servers on the E. coli PNPase/RNase E template structure (see “Experimental Procedures”). Residues predicted to be hotspots were evaluated further based on their location in the B. subtilis PNPase model, and their evolutionary conservation as calculated by ConSurf. Six additional PNPase residues were selected to be mutated to alanine (Table 1, second round) and were tested by B2H analysis for the RNase Y interaction. Mutation of three residues, Gln-3, Gln-23, and Leu-334, resulted in a loss of RNase Y interaction. As can be seen in Fig. 3C, these three residues sit in close proximity (∼3 Å) to the helical domain of the binding peptide. Although we have no data yet, it is likely that Leu-24 (Fig. 3C) is also important for formation of the binding site. From these results, and according to the models in Fig. 3, C and D, the binding site for the interacting peptide is formed by N-proximal residues of one PNPase monomer and two central regions of an adjacent monomer (Fig. 1A, underlined regions).
Pro-327 and Pro-336 were chosen to test the boundaries of the electrostatic channel that interacts with the binding peptide (Fig. 2B). Mutation of Pro-327 and Pro-336 had a weak effect on the RNase Y interaction, with a light pink color on the B2H assay (Fig. 3B). This could be due to alteration of the binding site shape, or due to loss of potential hydrophobic interactions with the binding peptide. Glu-533 is located in a helical domain that is near the C-terminal end of the binding peptide. From the structure, it seems reasonable that mutation of Glu-533 and D323A (Table 1, first round) would have similar effects.
In a final round of mutagenesis, two additional residues were selected for mutagenesis (Table 1, third round). Mutagenesis of Met-33, which is conserved from E. coli PNPase and is located close to the helical domain of the binding peptide, had only a weak effect on binding. Perhaps the change from a medium-sized hydrophobic residue (Met) to a small hydrophobic residue (Ala) at this location was not substantial enough to cause a large effect. Asp-11 is located close to the N-terminal end of the binding peptide. We predicted that a D11A mutation may not affect the RNase Y interaction; however, the change to a hydrophobic, uncharged reside could affect the overall structure. Indeed, although the D11A mutation affected RNase Y binding weakly (Fig. 3B), mutation of Asp-11 to a polar neutral residue, D11S, did not affect the interaction with RNase Y (Table 1, Fig. 3B).
Overall, the modeling and mutagenesis results suggest that adjacent PNPase monomers form a binding site for interaction with RNase Y. The site includes a channel with extended electrostatic potential, as well as a binding pocket for a helical domain of the interacting peptide. Modeling of the B. subtilis PNPase-RNase Y interaction after the E. coli PNPase-RNase E interaction appeared to be successful. As has been pointed out, it is remarkable that, despite the absence of similarity at the amino acid sequence level, RNase E and RNase Y have corresponding functionality (29).
The PNPase E331A mutant, which could not interact with RNase Y and which we assumed was catalytically active, was chosen for further studies. A pnpA CDS with the E331A change was cloned into the pMiniMAD vector (23) and was used to replace the endogenous pnpA CDS to give the pnpAE331A strain. As a control, we wished to construct a B. subtilis strain that expressed a full-length PNPase protein that could interact with RNase Y but was catalytically inactive. An earlier study of E. coli PNPase mutants indicated that the conserved Asp-492 residue was required for catalytic activity (30). The corresponding B. subtilis PNPase Asp-493 residue was mutated to alanine, and this change did not affect the interaction with RNase Y (Table 1, Fig. 3B). A pnpA CDS with the D493A change was used to replace the endogenous pnpA allele to give the pnpAD493A strain.
Known phenotypes of the ΔpnpA strain, cold sensitivity, tetracycline resistance, competence, and growth in chains, were tested with the newly constructed strains. Reassuringly, all four mutant phenotypes were recapitulated with the pnpAD493A strain (data not shown). On the other hand, the pnpAE331A strain was wild-type for these same phenotypes. These results indicated that loss of PNPase exonucleolytic activity in the ΔpnpA strain is responsible for the observed phenotypes, rather than disruption of RNase Y activity by the loss of its binding partner. These results also suggested that the PNPase-RNase Y interaction was not required, at least at a gross phenotypic level, for the required level of PNPase exonuclease activity.
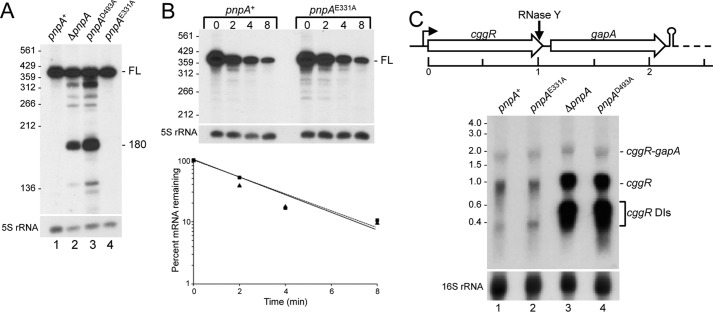
To examine the effect of these mutations on mRNA turnover in more detail, we analyzed two mRNAs, rpsO and cggR, whose decay has been studied previously (7, 31). rpsO is a monocistronic gene encoding a small-subunit ribosomal protein, and this mRNA has been used as a model to study the mechanism of B. subtilis mRNA turnover. Earlier work demonstrated that efficient turnover of rpsO RNA decay intermediates was dependent on PNPase (1). Decay of full-length rpsO mRNA initiates with an endonuclease cleavage mediated by RNase Y, followed by turnover of upstream RNA by PNPase, as well as turnover of downstream RNA by RNase J1, a 5′-exonuclease (31). Northern blotting analysis, using an oligonucleotide probe complementary to a sequence that overlaps the rpsO start codon, was performed on total RNA isolated from wild type and from three PNPase mutant strains (Fig. 4A). In the pnpA+ and pnpAE331A strains (active PNPase), only the full-length rpsO mRNA (388 nt) was detected (Fig. 4A, lanes 1 and 4). In the ΔpnpA and pnpAD493A strains (none or inactive PNPase), decay intermediates were detected (Fig. 4A, lanes 2 and 3), including the prominent 180-nt fragment, as observed previously (1). Accumulation of decay intermediates in the ΔpnpA strain is thought to be due to the 3′-terminal sequences of such fragments, which are predicted to form secondary structures that can be degraded by PNPase but are resistant to decay by other 3′-exonucleases present in the ΔpnpA strain (1).
FIGURE 4.
Northern blotting analysis of mRNA processing in PNPase mutants. A, steady-state pattern of rpsO mRNA in wild-type and mutant PNPase strains, in a 6% denaturing polyacrylamide gel. Migration of full-length mRNA (FL), as well as a prominent 180-nt decay intermediate detected in the strains without PNPase activity (lanes 2 and 3), is indicated at the right. Migration of 5′-end-labeled fragments of TaqI-digested pSE420 (37) is indicated at the left. B, Northern blotting analysis of rpsO mRNA half-life in the pnpA+ and pnpAE331A strains. Above each lane is time (in min) after rifampin addition. Half-life from blot is shown below. C, cggR RNA levels. At the top is a schematic of the first two genes of the gapA operon. The dashed line indicates the rest of the operon (four genes). Transcription termination downstream of the gapA gene is indicated by stem-loop structure. Below the schematic is a Northern blotting analysis of the steady-state pattern of cggR mRNA in wild-type and mutant PNPase strains. RNAs were separated on a 1.2% agarose, MOPS-formaldehyde gel. Indicated at the right is migration of dicistronic cggR-gapA mRNA (∼2 kb), its RNase Y cleavage product that produces cggR mRNA (∼1 kb), and decay intermediates (DIs) of cggR mRNA. Migration of unlabeled marker RNAs (sizes in kb) is indicated at the left. On a higher resolution polyacrylamide gel (not shown), the cggR decay intermediates resolve into several distinct bands ranging from ∼230 to ∼460 nt.
We found earlier that the half-life of the full-length rpsO mRNA was dependent on RNase Y activity and independent of subsequent exonucleolytic decay (31). If the presence or absence of an RNase Y-PNPase interaction was not affecting RNase Y activity, we predicted that rpsO full-length mRNA half-life would be similar in the pnpA+ and pnpAE331A strains. This was indeed the case. Northern blotting analysis of rpsO mRNA decay after inhibition of new transcription by rifampin showed a half-life close to 2 min in either strain (average of two experiments) (Fig. 4B).
Although the pattern of decay intermediates in the pnpAD493A strain was similar to that of the ΔpnpA strain, there was a somewhat higher level of several decay intermediates (Fig. 4A, lanes 3 and 4). For example, for the 180-nt fragment, we measured a 2.2-fold increase in the level of this fragment in the pnpAD493A strain, relative to the ΔpnpA strain (average of five experiments). This may be because the PNPase D493A protein is able to bind the 3′-end of the upstream fragment generated by RNase Y but cannot proceed in the 3′-to-5′ direction. Thus, the 3′-end of this fragment is protected against decay by other 3′-exonucleases, more so than in the strain that is lacking PNPase altogether.
A second RNA that was analyzed was that encoded by cggR, the first gene in the 6-kb hexacistronic gapA operon, which encodes a transcriptional repressor that regulates expression of the gapA operon; see Ref. 11 for references. Transcription termination also occurs downstream of the first structural gene, gapA, giving an ∼2.2-kb dicistronic cggR-gapA mRNA (see gene schematic in Fig. 4C). Cleavage at a site several nucleotides upstream of the cggR stop codon by RNase Y results in an ∼1-kb cggR RNA fragment, and we showed previously that decay of the cggR RNA fragment was PNPase-dependent (7). Timely turnover of cggR RNA appears to be required for appropriate regulation of gapA operon expression (32). Northern blotting analysis using a cggR probe was performed on total RNA isolated from wild type and from three PNPase mutant strains, and the results are shown in Fig. 4C. The predicted cggR-gapA 2.2-kb mRNA was detected, with no significant difference in amount between any of the strains (Table 2). This is consistent with decay of cggR-gapA mRNA being initiated by RNase Y (8), and with RNase Y activity not being affected by the lack of a PNPase binding partner. When PNPase activity was lacking (the ΔpnpA and pnpAD493A strains), a 6-fold accumulation of the 1-kb cggR RNA was observed, as well as a >20-fold accumulation of cggR decay intermediates (Fig. 4C, Table 2). We attempted to determine half-lives of the cggR RNAs in the wild-type and pnpAE331A strains, but the low level of expression and rapid degradation did not make this possible.
TABLE 2.
cggR RNA levels in pnpA mutant strains
RNA levels are relative to level in the pnpA+ strain. Data are average of three experiments; p values in parentheses.
| Strain | cggR-gapA | cggR | cggR decay intermediates |
|---|---|---|---|
| pnpAE331A | 1.24 ± 0.57 (0.54) | 0.83 ± 0.36 (0.50) | 1.40 ± 0.62 (0.38) |
| ΔpnpA | 1.25 ± 0.23 (0.20) | 6.36 ± 0.98 (0.01) | 23.28 ± 4.52 (0.01) |
| pnpAD493A | 1.04 ± 0.13 (0.68) | 5.62 ± 1.73 (0.04) | 25.75 ± 3.48 (0.01) |
The fact that we did not detect the rpsO 180-nt fragment in the pnpAE331A strain, even at steady state (Fig. 4A, lane 4), suggested that it is degraded efficiently by PNPase E331A, and the loss of interaction with RNase Y does not affect this activity. Similarly, the fact that we saw no difference in the cggR RNA pattern or amount between the pnpA+ and pnpAE331A strains (Fig. 4C, lanes 1 and 2; Table 2) suggested that PNPase E331A was as efficient as wild-type PNPase in cggR RNA turnover. To explain why decay of many mRNAs is strongly dependent on PNPase, despite the presence of several other 3′-exonucleases in B. subtilis, one could hypothesize that the presence of PNPase in a degradosome with RNase Y localizes PNPase to the site of RNase Y cleavage. This localization allows for efficient transfer of the 3′-hydroxyl end generated by RNase Y into the central channel formed by the PNPase trimer (28), where it then enters the active sites for phosphorolysis. The results presented here argue against this hypothesis, as disruption of the PNPase-RNase Y interaction had no effect on decay of upstream fragments of RNase Y cleavage. It is possible that only a fraction of the PNPase pool is actually in the degradosome complex, and free PNPase is dominant in the degradative function. Indeed, in E. coli, it has been estimated that PNPase is present in 5–10-fold excess over RNase E, its degradosome interaction partner (33). Co-immunoprecipitation experiments are planned to probe the interaction of RNase Y with wild-type and E331A mutant proteins and to determine how much of the total PNPase protein is in a complex with RNase Y in B. subtilis. The antibody used in the Western blot shown in Fig. 3A was directed against the adenylate cyclase domain of the fusion protein, and so cannot be used for co-immunoprecipitation experiments involving native PNPase protein. As there is no antibody available for B. subtilis PNPase, we are planning to do the co-immunoprecipitation experiments with newly constructed strains that have the native PNPase and RNase Y CDSs replaced with FLAG-tagged and HA-tagged versions, respectively.
Our results suggest, in addition, that RNase Y cleavage of full-length rpsO and cggR mRNAs is not affected by the loss of the interaction of RNase Y with PNPase. It may therefore be the case that the B. subtilis degradosome interaction plays a regulatory role for specific growth conditions, as has been suggested for Streptococcus pyogenes (34). Clearly, analyzing a larger sampling of mRNAs will be needed to assess the role of the PNPase-RNase Y interaction. RNA sequencing experiments on the strains constructed in this study are planned.
Author Contributions
D. H. B. designed the study. E. S. carried out the mutagenesis and B2H assays, constructed mutant strains, and performed Northern blotting analyses. S. A. did the structure modeling, and A. S. interpreted the modeling data. B. L. performed some of the Northern blotting experiments, analyzed the Northern blotting data, and performed the Western blotting experiments.
Acknowledgments
We thank Dr. Jörg Stülke for generously sharing the plasmids carrying pnpA and rny constructs used in the B2H screening. We thank Dr. R. J. Lewis for the first suggestions on PNPase residues that might be involved in the interaction with RNase Y.
This work was supported by National Institutes of Health Grants R01 GM100137 (to D. H. B.) and R01 GM108911 (to A. S.) and United States Department of Defense Grant W81XWH-15-1-0539 (to A. S.) This work was also supported by Post-baccalaureate Research Education Program Grant R25 GM06118 (to S .A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
- PNPase
- polynucleotide phosphorylase
- B2H
- bacterial-2-hybrid
- CDS
- coding sequence
- nt
- nucleotide(s)
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Oussenko I. A., Abe T., Ujiie H., Muto A., and Bechhofer D. H. (2005) Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 187, 2758–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duffy J. J., Chaney S. G., and Boyer P. D. (1972) Incorporation of water oxygens into intracellular nucleotides and RNA. I. Predominantly non-hydrolytic RNA turnover in Bacillus subtilis. J. Mol. Biol. 64, 565–579 [DOI] [PubMed] [Google Scholar]
- 3. Deutscher M. P., and Reuven N. B. (1991) Enzymatic basis for hydrolytic versus phosphorolytic mRNA degradation in Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 88, 3277–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luttinger A., Hahn J., and Dubnau D. (1996) Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19, 343–356 [DOI] [PubMed] [Google Scholar]
- 5. Wang W., and Bechhofer D. H. (1996) Properties of a Bacillus subtilis polynucleotide phosphorylase deletion strain. J. Bacteriol. 178, 2375–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bechhofer D. H., and Wang W. (1998) Decay of ermC mRNA in a polynucleotide phosphorylase mutant of Bacillus subtilis. J. Bacteriol. 180, 5968–5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu B., Deikus G., Bree A., Durand S., Kearns D. B., and Bechhofer D. H. (2014) Global analysis of mRNA decay intermediates in Bacillus subtilis wild-type and polynucleotide phosphorylase-deletion strains. Mol. Microbiol. 94, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Commichau F. M., Rothe F. M., Herzberg C., Wagner E., Hellwig D., Lehnik-Habrink M., Hammer E., Völker U., and Stülke J. (2009) Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol. Cell. Proteomics 8, 1350–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shahbabian K., Jamalli A., Zig L., and Putzer H. (2009) RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 28, 3523–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lehnik-Habrink M., Pförtner H., Rempeters L., Pietack N., Herzberg C., and Stülke J. (2010) The RNA degradosome in Bacillus subtilis: identification of CshA as the major RNA helicase in the multiprotein complex. Mol. Microbiol. 77, 958–971 [DOI] [PubMed] [Google Scholar]
- 11. Lehnik-Habrink M., Lewis R. J., Mäder U., and Stülke J. (2012) RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol. Microbiol. 84, 1005–1017 [DOI] [PubMed] [Google Scholar]
- 12. Newman J. A., Hewitt L., Rodrigues C., Solovyova A. S., Harwood C. R., and Lewis R. J. (2012) Dissection of the network of interactions that links RNA processing with glycolysis in the Bacillus subtilis degradosome. J. Mol. Biol. 416, 121–136 [DOI] [PubMed] [Google Scholar]
- 13. Nurmohamed S., Vaidialingam B., Callaghan A. J., and Luisi B. F. (2009) Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J. Mol. Biol. 389, 17–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grant S. G., Jessee J., Bloom F. R., and Hanahan D. (1990) Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karimova G., Pidoux J., Ullmann A., and Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pei J., Tang M., and Grishin N. V. (2008) PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 36, W30–W34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sali A., and Blundell T. L. (1993) Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 18. Shen M. Y., and Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Darnell S. J., Page D., and Mitchell J. C. (2007) An automated decision-tree approach to predicting protein interaction hot spots. Proteins 68, 813–823 [DOI] [PubMed] [Google Scholar]
- 20. Tuncbag N., Gursoy A., and Keskin O. (2009) Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics 25, 1513–1520 [DOI] [PubMed] [Google Scholar]
- 21. Kortemme T., Kim D. E., and Baker D. (2004) Computational alanine scanning of protein-protein interfaces. Sci. STKE 2004, pl2. [DOI] [PubMed] [Google Scholar]
- 22. Ashkenazy H., Erez E., Martz E., Pupko T., and Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patrick J. E., and Kearns D. B. (2008) MinJ (YvjD) is a topological determinant of cell division in Bacillus subtilis. Mol. Microbiol. 70, 1166–1179 [DOI] [PubMed] [Google Scholar]
- 24. Karimova G., Ullmann A., and Ladant D. (2000) A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 328, 59–73 [DOI] [PubMed] [Google Scholar]
- 25. Bechhofer D. H., Oussenko I. A., Deikus G., Yao S., Mathy N., and Condon C. (2008) Analysis of mRNA decay in Bacillus subtilis. Methods Enzymol. 447, 259–276 [DOI] [PubMed] [Google Scholar]
- 26. Sharp J. S., and Bechhofer D. H. (2003) Effect of translational signals on mRNA decay in Bacillus subtilis. J. Bacteriol. 185, 5372–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sulthana S., and Deutscher M. P. (2013) Multiple exoribonucleases catalyze maturation of the 3′ terminus of 16S ribosomal RNA (rRNA). J. Biol. Chem. 288, 12574–12579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Symmons M. F., Jones G. H., and Luisi B. F. (2000) A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8, 1215–1226 [DOI] [PubMed] [Google Scholar]
- 29. Laalami S., Zig L., and Putzer H. (2014) Initiation of mRNA decay in bacteria. Cell. Mol. Life Sci. 71, 1799–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jarrige A., Bréchemier-Baey D., Mathy N., Duché O., and Portier C. (2002) Mutational analysis of polynucleotide phosphorylase from Escherichia coli. J. Mol. Biol. 321, 397–409 [DOI] [PubMed] [Google Scholar]
- 31. Yao S., and Bechhofer D. H. (2010) Initiation of decay of Bacillus subtilis rpsO mRNA by endoribonuclease RNase Y. J. Bacteriol. 192, 3279–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerwig J., and Stülke J. (2014) Caught in the act: RNA-Seq provides novel insights into mRNA degradation. Mol. Microbiol. 94, 5–8 [DOI] [PubMed] [Google Scholar]
- 33. Liou G. G., Jane W. N., Cohen S. N., Lin N. S., and Lin-Chao S. (2001) RNA degradosomes exist in vivo in Escherichia coli as multicomponent complexes associated with the cytoplasmic membrane via the N-terminal region of ribonuclease E. Proc. Natl. Acad. Sci. U.S.A. 98, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kang S. O., Caparon M. G., and Cho K. H. (2010) Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect. Immun. 78, 2754–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schechter L. M., Roberts K. A., Jamir Y., Alfano J. R., and Collmer A. (2004) Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186, 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu Y., Wu M., He G., Zhang X., Li W., Gao Y., Li Z., Wang Z., and Zhang C. (2012) Glyceraldehyde-3-phosphate dehydrogenase: a universal internal control for Western blots in prokaryotic and eukaryotic cells. Anal. Biochem. 423, 15–22 [DOI] [PubMed] [Google Scholar]
- 37. Brosius J. (1992) Compilation of superlinker vectors. Methods Enzymol. 216, 469–483 [DOI] [PubMed] [Google Scholar]