Abstract
The genome of the extremely thermophilic bacterium Caldicellulosiruptor kronotskyensis encodes 19 surface layer (S-layer) homology (SLH) domain-containing proteins, the most in any Caldicellulosiruptor species genome sequenced to date. These SLH proteins include five glycoside hydrolases (GHs) and one polysaccharide lyase, the genes for which were transcribed at high levels during growth on plant biomass. The largest GH identified so far in this genus, Calkro_0111 (2,435 amino acids), is completely unique to C. kronotskyensis and contains SLH domains. Calkro_0111 was produced recombinantly in Escherichia coli as two pieces, containing the GH16 and GH55 domains, respectively, as well as putative binding and spacer domains. These displayed endo- and exoglucanase activity on the β-1,3-1,6-glucan laminarin. A series of additional truncation mutants of Calkro_0111 revealed the essential architectural features required for catalytic function. Calkro_0402, another of the SLH domain GHs in C. kronotskyensis, when produced in E. coli, was active on a variety of xylans and β-glucans. Unlike Calkro_0111, Calkro_0402 is highly conserved in the genus Caldicellulosiruptor and among other biomass-degrading Firmicutes but missing from Caldicellulosiruptor bescii. As such, the gene encoding Calkro_0402 was inserted into the C. bescii genome, creating a mutant strain with its S-layer extensively decorated with Calkro_0402. This strain consequently degraded xylans more extensively than wild-type C. bescii. The results here provide new insights into the architecture and role of SLH domain GHs and demonstrate that hemicellulose degradation can be enhanced through non-native SLH domain GHs engineered into the genomes of Caldicellulosiruptor species.
Keywords: biofuel, cell surface, cell surface enzyme, enzyme, glycoside hydrolase, plant cell wall, Caldicellulosiruptor, S-layer, lignocellulose
Introduction
Many bacteria (1, 2) and archaea (3) produce a two-dimensional, para-crystalline array of protein that covers the outside of the cell, referred to as a surface layer (S-layer).5 In bacteria, the majority of S-layer proteins are non-covalently associated with the bacterial cell surface via specialized domains at their N or C terminus, typically from one of three distinct but analogous domain categories: surface layer homology (SLH) (4, 5), CWB2 (pfam04122) (6), or NCAD (previously SLAP, pfam03217) (7, 8). In archaea, S-layer proteins are most often attached to the cell membrane. S-layer proteins can be anchored to the membrane via a C-terminal transmembrane helix domain (3, 9) or covalently attached to cell membrane lipid via an archaeosortase, as shown in Haloferax volcanii (10, 11). In most microorganisms, the S-layer is composed entirely of one or two proteins (SLPs), which self-assemble. In addition to these SLPs, some Firmicutes produce a family of proteins that also contain SLH, CWB2, or NCAD domains and, as such, are predicted to be associated with the S-layer (12). SLPs and S-layer associated proteins can also contain domains with specialized functions allowing the S-layer to play a role in adherence and biofilm formation (13–16), biogenesis of the cell envelope (17), cell division (18), motility (19, 20), and, of interest here, plant cell wall degradation (21, 22).
Many lignocellulose-degrading bacteria utilize SLH domain-containing proteins to display plant biomass-degrading carbohydrate active enzymes (CAZymes) (23) on the cell surface. In cellulosomal bacteria, SLH domain proteins attached to the cell surface interact with the scaffoldin protein of the multienzyme cellulosome complex to anchor it to the cell surface (24–26). In many non-cellulosomal, biomass-degrading bacteria, multidomain CAZymes containing SLH proteins are implicated in lignocellulose deconstruction. About 5% of the 20,710 predicted SLH domain proteins, represented in 1889 genomes from the Integrated Microbial Genomes database (27), contain at least one identifiable CAZyme-related component: glycoside hydrolase (GH), polysaccharide lyase (PL), carbohydrate esterase, or carbohydrate binding module (CBM).
Multidomain CAZymes containing SLH domains have been characterized from Caldanaerobius polysaccharolyticus (28, 29), Bacillus sp. (30), Paenibacillus sp. (31–36), Clostridium sp. (37–44), Thermoanaerobacterium sp. (45–51), and Caldicellulosiruptor sp. (21, 52). Across these species, the number of identifiable CAZyme domain-containing SLH proteins and total predicted SLH proteins vary significantly. Examples include Paenibacillus sp. strain JDR-2 (23 CAZyme domain containing of 78 total SLH domain proteins), Clostridium thermocellum ATCC 27405 (4 of 25), Thermoanaerobacterium saccharolyticum strain JW/SL-YS485 DSM 8691 (7 of 12), Caldicellulosiruptor bescii (1 of 12), and Caldicellulosiruptor kronotskyensis (6 of 19).
The genus Caldicellulosiruptor is composed of extremely thermophilic, Gram-positive, anaerobic bacteria that are able to attach to and degrade the variety of polysaccharides found in lignocellulosic biomass. Currently, the genomes of 12 members of the genus have been sequenced, representing species isolated from globally distributed terrestrial hot springs (53–57). Caldicellulosiruptor species produce many extracellular CAZymes, including some that have SLH domains (58). The enzyme inventory produced by individual species, and thus the capacity to degrade plant biomass polysaccharides, varies significantly across the genus (59, 60). C. kronotskyensis has the largest inventory of CAZymes of any Caldicellulosiruptor species, with 31 CAZymes compared with 20 CAZymes in C. bescii, which is to date the most studied member of the genus (61).
During growth on complex carbohydrates, Caldicellulosiruptor species physically associate with the substrate. This attachment is mediated in part by non-catalytic cellulose-binding proteins, called tāpirins, that were recently identified and characterized (62). In addition, proteins anchored to the cell surface within the S-layer via SLH domains are also believed to play a role in the attachment of Caldicellulosiruptor species to insoluble plant biomass substrates (21, 63). Two such SLH domain proteins from C. saccharolyticus, Csac_0678 and Csac_2722, were shown to bind to insoluble substrates and could be identified in the S-layer protein cell fraction, implying a role in cell-substrate attachment (21). However, more information is needed to understand the role of SLH domain proteins in plant biomass deconstruction, especially how their function relates to other CAZymes produced for this purpose. In this study, the localization, biochemical characteristics, and physiological role of two SLH domain enzymes from C.kronotskyensis, xylanase Calkro_0402 and laminarinase Calkro_0111, were examined from this perspective.
Experimental Procedures
Bacterial Strains, Plasmids, and Reagents
Cloning and expression of recombinant proteins used various E. coli strains: NovaBlue GigaSinglesTM (EMD Millipore), RosettaTM 2(DE3) SinglesTM (EMD Millipore), NEB 10-beta electrocompetent E. coli (New England Biolabs), and Arctic Express (DE3)RIL E. coli (Agilent Technologies). Axenic strains of C. bescii and C. kronotskyensis were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. C. bescii strain JWCB018 (64) and non-replicating vector pDCW121 (65) were obtained from J. Westpheling (University of Georgia, Athens, Georgia). Genes of interest were amplified by polymerase chain reaction (PCR) and cloned using the pET46 Ek/LIC vector kit (EMD Millipore) for protein expression. All other vectors were constructed using one-step isothermal assembly of overlapping dsDNA, as described previously (66). Plasmids were isolated using Qiaprep miniprep kits (Qiagen), and plasmid sequences were confirmed by sequencing (Genewiz). Oligonucleotide primer sequences used are listed in Table 1. Carbohydrates and biomasses used included the following: laminarin from Laminaria digitata (Sigma L9634), xylan from birchwood (Sigma X0502), xylan from oat spelts (Sigma X0627), and dilute acid-pretreated Populus trichocarpa × Populus deltoides (National Renewable Energy Laboratory, Golden, CO).
TABLE 1.
Primers used in this study
Underlined portions are vector-specific or overlap regions for isothermal assembly.
| Primer | Sequence | Use |
|---|---|---|
| Calkro_0111 TM1 F | GACGACGACAAGATAAATAAAGCAGGCACA | pET46 cloning |
| Calkro_0111 TM1/TM3 R | GAGGAGAAGCCCGGTTAATCTGACATATCTGATTC | pET46 cloning |
| Calkro_0111 TM2 F | GACGACGACAAGATGGTTGAAAAAGGATATTTGATTGG | pET46 cloning |
| Calkro_0111 TM2 R | GAGGAGAAGCCCGGTTAACTTGGAGGGTTAGGATAAA | pET46 cloning |
| Calkro_0111 TM3/TM4 F | GACGACGACAAGATGGGTGAATGGAAACTTGT | pET46 cloning |
| Calkro_0111 TM4 R | GAGGAGAAGCCCGGTTACGGTGTAGAATAGATGATGAA | pET46 cloning |
| Calkro_0111 TM5 F | GACGACGACAAGATTCCTTCCGTGGGTGAATGGAAACTTGTATGG | pET46 cloning |
| Calkro_0111 TM5 R | GAGGAGAAGCCCGGTTAACCTTCTCTTTGGTATAC | pET46 cloning |
| Calkro_0111 TM6 F | GACGACGACAAGATGATTTTAGGATTAGGT | pET46 cloning |
| Calkro_0111 TM6 R | GAGGAGAAGCCCGGTTACAATTTCTTTTCTAAAGAT | pET46 cloning |
| Calkro_0111 TM7 F | GACGACGACAAGATCTCTGGAGGCAGGTCTTA | pET46 cloning |
| Calkro_0111 TM7 R | GAGGAGAAGCCCGGTTAGGTTTCTCCATTTTCATTG | pET46 cloning |
| Calkro_0111 TM8 F | GACGACGACAAGATGACAGATACAGGTTTGAG | pET46 cloning |
| Calkro_0111 TM8 R | GAGGAGAAGCCCGGTTACTTGCTCCCATACAC | pET46 cloning |
| Calkro_0111 TM9 F | GACGACGACAAGATGTCAGAGCCTATATCT | pET46 cloning |
| Calkro_0111 TM9 R | GAGGAGAAGCCCGGTTACTTGCTCCCATACACTTCA | pET46 cloning |
| Calkro_0111 TM10 F | GACGACGACAAGATGTTTGGTCCAAATGTCTA | pET46 cloning |
| Calkro_0111 TM10 R | GAGGAGAAGCCCGGTTAGTTGCAATATTCTGTTAC | pET46 cloning |
| Calkro_0402 F | GACGACGACAAGATTCAAAGCAGCCAAACTAA | pET46 cloning |
| Calkro_0402 R | GAGGAGAAGCCCGGTTACTTTTTGAGATTGTTAAGTGCTCTG | pET46 cloning |
| Calkro_0402 TM1 F | GACGACGACAAGATGTCAGATAGCATAAAG | pET46 cloning |
| Calkro_0402 TM1 R | GAGGAGAAGCCCGGTTATTGAGCATTTCTTGTAACTG | pET46 cloning |
| ΔAthe_2438 5′ F | GAAGTTAGGCTGGTGGTACCCATACACCAAAATGATATAATCTCC | pJMC009 |
| ΔAthe_2438 5′ R | TAAATCCTGTTTATCATGTTGGAAGGCAAATTG | pJMC009 |
| Cbescii Pslp (Athe_2303 promoter) F | AACATGATAAACAGGATTTAAAAGAGGCTATG | pJMC009 |
| Cbescii Pslp (Athe_2303 promoter) R | GCTTTTTCATAACTACTCACCAAACCTC | pJMC009 |
| Calkro_0402 + Calkro Terminator F | GTGAGTAGTTATGAAAAAGCGACTTATAGC | pJMC009 |
| Calkro_0402 + Calkro Terminator R | CTACCTAATCGGAAGACTAAAGATTTTCTCC | pJMC009 |
| ΔAthe_2438 3′ F | TTAGTCTTCCGATTAGGTAGGTGGTTTTAAC | pJMC009 |
| ΔAthe_2438 3′ R | CACTGAGCGTCAGAGAATTCGGATTTATCGCCTGAGTTG | pJMC009 |
| pDCW121 backbone F | GAATTCTCTGACGCTCAG | pJMC009 |
| pDCW121 backbone R | GGTACCACCAGCCTAACTTC | pJMC009 |
| ΔAthe_2438 locus F | GGCAACCTTCAGTAGGGCTGC | PCR |
| ΔAthe_2438 locus R | CATTTGATATCAGAATGTTCTCGTTGATT | PCR |
Growth Media and Culture Conditions
All E. coli cultures were maintained in Luria-Bertani (LB) medium (5 g/liter yeast extract, 10 g/liter tryptone, 10 g/liter NaCl) or LB medium with 1.5% (w/v) agar plates supplemented with 50 μg/ml carbenicillin (Corning Cellgro), 34 μg/ml chloramphenicol (Sigma), 20 μg/ml gentamycin (Chem-Impex International Inc.), 50 μg/ml streptomycin (Sigma), and/or 50 μg/ml apramycin (Fisher), as appropriate. Protein expression was performed in either Terrific Broth medium (containing 12 g/liter tryptone, 24 g/liter yeast extract, 4 ml/liter glycerol, 2.31 g/liter KH2PO4, and 12.54 g/liter K2HPO4) or ZYM-5052 medium (67) modified to contain 0.625% glycerol, 0.0625% glucose, and 0.25% lactose.
Caldicellulosiruptor strains were grown anaerobically at 70 °C in modified DSM640 medium (DSMZ), containing 0.9 g/liter NH4Cl, 0.9 g/liter NaCl, 0.4 g/liter MgCl2·6H2O, 0.75 g/liter KH2PO4, 1.5 g/liter K2HPO4, 1 g/liter yeast extract, 1 ml/liter trace element solution SL-10, 5 g/liter cellobiose, 0.5 mg/liter resazurin, and 0.75 g/liter l-cysteine-HCl×H2O). Caldicellulosiruptor genomic DNA for cloning and vector construction was extracted, as described previously (68). For genetic manipulations of C. bescii, strains were grown anaerobically at 70 °C in low osmolality defined (LOD) or low osmolality complex (LOC) medium (69). C. bescii was plated embedded in LOD medium with 1.5% agar and grown at 70 °C in anaerobic tanks with N2 head space. For solubilization experiments and immunofluorescence microscopy, Caldicellulosiruptor strains were grown on a modified defined version of DSMZ671 medium (671d medium), described previously (70), with 5 g/liter of the specific biomass substrate as the carbon source. For growth of all uracil auxotroph mutants, growth medium was supplemented with 40 mm uracil.
Expression of Recombinant Proteins in E. coli
All Calkro_0111 truncation mutants (TMs) and Calkro_0402 TM1 were cloned into the pET46 Ek/LIC vector maintained in NovaBlue E. coli. Sequence confirmed plasmids were transformed into Rosetta 2(DE3) E. coli for protein expression. Proteins were expressed in modified ZYM-5052 autoinduction medium at 37 °C 250 rpm. Cells were harvested 18–24 h after induction by centrifugation at 6,000 × g for 10 min. For the expression of Calkro_0402, the gene was cloned into the pET46 Ek/LIC vector without the predicted signal peptide, as determined by the SignalP 4.1 server (71), and maintained in NEB 10-beta E. coli. The sequence-confirmed plasmid was transformed into Arctic Express (DE3)RIL E. coli for protein expression. Expression was performed in Terrific Broth medium by growing cells at 30 °C and 250 rpm until A600 reached 1.0 followed by induction at 13 °C with 1 mm isopropyl-β-d-thiogalactopyranoside. Cells were harvested 16–20 h later by centrifugation at 6,000 × g for 10 min. All cell pellets were frozen at −20 °C prior to purification.
Purification of Recombinant Protein and Truncation Mutants
Cell pellets were resuspended in 5 ml of 20 mm sodium phosphate, pH 7.4, 0.5 m NaCl, 20 mm imidazole, 0.1% (v/v) Nonidet P-40, and 1 mg/ml lysozyme per g of wet cell pellet. Cells were lysed in a French press at 16,000 p.s.i. Cell lysates were heat-treated at 65 °C for 20 min (except for the full-length Calkro_0402 protein product which was not heat-treated) to remove heat-labile proteins from E. coli. The cell lysate was then centrifuged at 25,000 × g for 20–60 min, and the supernatant was passed through a 0.22-μm filter to prepare cell-free cell extract. Proteins were purified using 1- or 5-ml HisTrap HP nickel-Sepharose (GE Healthcare) immobilized metal affinity chromatography columns, operated according to the manufacturer's instructions. For full-length Calkro_0402 and Calkro_0111 TM8, size exclusion chromatography was used after immobilized metal affinity chromatography on a HiLoad 26/600 Superdex 200 pg column (GE Healthcare) in 50 mm sodium phosphate, 150 mm NaCl, pH 7.2, buffer. All chromatography steps were performed on a Biologic DuoFlow FPLC (Bio-Rad). Purity of the recombinant proteins was evaluated by SDS-PAGE using NuPAGE Novex 4–12% BisTris protein gels (Life Technologies, Inc.) or 4–15% Mini-PROTEAN TGX gels (Bio-Rad) with a Benchmark protein ladder (Life Technologies) and staining by GelCode Blue Safe Protein Stain (Thermo Scientific). The concentration of purified proteins was determined using the bicinchoninic acid assay (Pierce).
Optimal pH and Temperature Determination
The pH optimum for each recombinant enzyme was determined at 70 °C in buffers at pH 2.5–10 (50 mm citrate buffer, pH 2.5 and 3.5; 50 mm sodium acetate buffer, pH 4.5, 5, 5.5, and 6; 50 mm sodium phosphate, pH 6.5, 7, and 8; 50 mm sodium bicarbonate, pH 9.2 and 10). The temperature optimum was determined at the optimal pH between 30 and 100 °C. Enzyme reactions and no enzyme/substrate controls were prepared in triplicate with 100 μl of 1% substrate and incubated in PCR multiwell plates in a thermocycler (Eppendorf). Activity was assessed by measuring reducing sugar released using a 3,5-dinitrosalicylic acid (DNS) assay (modified from Refs. 72 and 73 and adapted to a 96-well microplate format (74, 75)). The DNS reagent used contained 1.6% (w/v) sodium hydroxide, 30% (w/v) sodium potassium tartrate, and 0.1% (w/v) 3,5-dinitrosalicylic acid. Briefly, 30 μl of sample was mixed with 60 μl of DNS reagent and incubated in a 96-well PCR plate in a thermocycler (Eppendorf) using the following program: 95 °C for 5 min, 48 °C for 1 min, hold at 20 °C. Then the DNS reaction was diluted with distilled water, and absorbance was measured at 540 nm. Glucose or xylose was used as a standard for glucooligosaccharide or xylooligosaccharide release.
Analysis of Oligosaccharide Products from Enzymatic Hydrolysis
To analyze oligosaccharides released by the various enzymes, enzyme was added to 500 μl of 1% substrate in buffer at the enzyme's optimum pH and incubated between 0 and 240 min at 70 °C at 800 rpm in a mixing thermocycler (Eppendorf). Reactions without substrate and/or enzyme were also assessed. After incubation, tubes were spun at 16,000 × g for 5 min at 4 °C, and the supernatant was applied to 0.5 ml of 10,000 molecular weight cut-off protein concentrators (Pierce). The protein concentrator eluent was diluted 10-fold with distilled water and analyzed by high performance liquid chromatography (HPLC) (Alliance e2695 separations module, Waters) with refractive index (model 2414, Waters) and photodiode array (model 2998, Waters) detectors. Columns were either the KS-801 (xylooligosaccharides) or KS-802 (glucooligosaccharides) (Shodex) operated with water as the mobile phase (0.6 ml/min and 80 °C). Laminarioligosaccharides (L2–L6) (Seikagaku Corp.), individual xylooligosaccharides (X2–X6) (Megazyme), glucose (Sigma), and xylose (Acros Organics) were used as standards.
Confocal and Epifluorescence Microscopy
All centrifugation steps for the preparation of labeled cells were carried out at 6,000 × g for 10 min. Cells harvested from a 50-ml culture in 671d medium were washed one time with sterile 1× phosphate-buffered saline (PBS), and cells were fixed in 1× PBS containing 4% formaldehyde (methanol-free; Fisher) for 30 min at room temperature with gentle shaking, followed by washing three times with 50 ml of PBS. Cells were resuspended in 3 ml of antibody-blocking solution containing 5% (v/v) normal goat serum (Immunoreagents), 1% (w/v) bovine serum albumin (protease- and nuclease-free; Fisher), and 0.05% (v/v) Tween 20 (Fisher) in PBS and incubated on a Boeckle rocker for 60 min at room temperature. After this blocking step, cells were harvested by centrifugation and resuspended in antibody blocking solution containing 100 μg/ml primary antibody. Polyclonal antibodies were raised against Calkro_0111 TM7 or Calkro_0402 TM1 in chickens and supplied as total IgY with preimmune total IgY controls by GeneTel Laboratories (Madison, WI). Cells were incubated with the primary antibody for 18 h at 4 °C on a Boeckle rocker. After this incubation, cells were washed three times with 1 ml of PBS containing 1% (w/v) bovine serum albumin and 0.05% (v/v) Tween 20 and resuspended in antibody blocking solution containing a 1:400 dilution of goat anti-chicken DyLight488-conjugated secondary antibody (Immunoreagents) and incubated on a Boeckle rocker for 60 min at room temperature. Cells were harvested and washed three times with PBS, resuspended in 100 μl of 3.6 μm DAPI in PBS, and incubated at 4 °C for 18 h. Following this incubation, cells were washed three times, resuspended with PBS, and vacuum-filtered onto 0.22-μm polycarbonate hydrophilic isopore membrane filters (EDM Millipore) and mounted in 15 μl of SlowFade Diamond anti-fade mountant (Life Technologies). For Calkro_0111 and Calkro_0402 localization in C. kronotskyensis, slides were imaged on a Zeiss LSM 710 confocal work station using a ×63 oil immersion plan-apochromat (numerical aperture 1.4) objective (North Carolina State University Cellular and Molecular Imaging Facility, Raleigh, NC). Image processing was performed using Zeiss Zen Blue software.
Epifluorescence imaging and cell counting were performed using a Nikon eclipse 50i microscope with a Plan Fluor ×100 (numerical aperture 1.3) oil emersion objective and Nikon DS-Fi1 camera. For routine cell counting, cells were stained with acridine orange, as described previously (76). For immunofluorescence images of C. bescii genetic mutant strains, cells were labeled and slides were prepared as described above for confocal imaging. Images were captured as a z-stack of images through the sample with 0.33-μm steps controlled by the ES10ZE Focus controller (Prior Scientific) and processed into a single focused image using the Nikon Elements software extended depth of field processing methods. Transmission electron microscopy was performed at the Laboratory for Advanced Electron and Light Optical Methods (College of Veterinary Science, North Carolina State University).
Genetic Manipulation of C. bescii
To prepare competent cells, a 500-ml culture of C. bescii strain JWCB018 was grown to an optical density at 680 nm of 0.06–0.07 in LOD medium supplemented with 1× 19-amino acid solution (77). The culture was cooled to room temperature, and cells were harvested by centrifugation. All centrifugation steps were carried out at 6,000 × g for 10 min. The cell pellet was washed three times with 10% sucrose and resuspended with 10% sucrose to a total volume of 100–120 μl. Fifty μl of competent cells were mixed with 1 μg of pJMC009 plasmid DNA at room temperature and electroporated using 1-mm gap cuvettes (USA Scientific) and a Gene Pulser II system with a Pulse Controller PLUS module (Bio-Rad) operated at 2.25 kV, 600 ohms, and 25 microfarads. Immediately following electroporation, cells were transferred to 10 ml of prewarmed LOC medium and incubated at 70 °C for 1 h. After this incubation, the culture was cooled to room temperature, and cells were harvested by centrifugation, transferred to 100 ml of prewarmed LOD medium lacking uracil, and incubated at 70 °C for 2–4 days to select for first crossover transformants. This culture was passaged and then plated on LOD medium lacking uracil to select individual colonies. Integration of the pJMC009 vector was confirmed by PCR, and a successful first crossover mutant strain was plated on LOD medium supplemented with 40 mm uracil and 4 mm 5-fluoroorotic acid to select for second crossover mutants. Isolated colonies were screened by PCR, and successful second crossovers were identified. A successful second crossover was plated on LOD medium supplemented with uracil, and individual colonies were isolated. This plating and isolation of colonies was repeated two times to obtain the final strain and ensure its purity. All PCR screening steps were performed using genomic DNA isolated using the ZymoBeadTM genomic DNA kit (Zymo Research).
Hydrolysis of Hemicellulose Substrates
Washed biomass substrates (oat spelt xylan, birchwood xylan) were prepared by washing 1 g of substrate/100 ml of distilled water overnight at 70 °C to remove soluble sugars, centrifuging at 6,000 × g for 10 min, and drying at 70 °C. Caldicellulosiruptor strains were passaged on modified 671d medium with respective biomass substrates 3–4 times. Solubilization cultures were prepared in triplicate as 50-ml 671d medium cultures with 5 g/liter substrate, inoculated to 1 × 106 cells/ml, and grown at 70 °C for 45.5 h. Residual substrate was harvested by centrifugation at 6,000 × g for 10 min and drying at 70 °C until constant mass. The extent of solubilization was determined from the difference in mass between the biomass used to prepare each culture and the residual remaining after harvest. For time course analysis of oat spelt xylan solubilization, cultures were prepared as above except that cultures were sampled by removing 2 ml of culture and centrifuging the samples at 6,000 × g for 10 min. The supernatant was stored at −20 °C prior to HPLC analysis.
Analysis of Solubilization in Culture Supernatants
Supernatant samples were analyzed for xylose content, as described previously (70). Briefly, the samples were brought to 4% (w/w) sulfuric acid and autoclaved for 1 h on the liquid cycle. Samples were cooled to room temperature and spun at 18,000 × g for 5 min, and supernatant was analyzed by HPLC as described above except using an Aminex-87H (300 × 7.8 mm; Bio-Rad) column with a mobile phase of 5 mm sulfuric acid at 0.6 ml/min and 60 °C.
C. kronotskyensis and C. bescii RNA Microarray
Transcriptomic data, which were acquired previously (70) and deposited in the NCBI Gene Expression Omnibus database (accession number GSE68810), were reanalyzed with respect to the SLH domain-containing proteins from C. bescii and C. kronotskyensis.
Results
SLH Domain Proteins in Caldicellulosiruptor Species
The genomes of the 12 sequenced Caldicellulosiruptor species collectively encode 34 different groups of SLH domain-containing proteins (supplemental Table S1). About half of the SLH domain-containing protein groups in these genomes have no additional identifiable domains other than SLH domains, highlighting the limited understanding of this group of proteins. Caldicellulosiruptor species produce between 10 (C. saccharolyticus and Caldicellulosiruptor acetigenus) and 19 (C. kronotskyensis) SLH domain proteins.
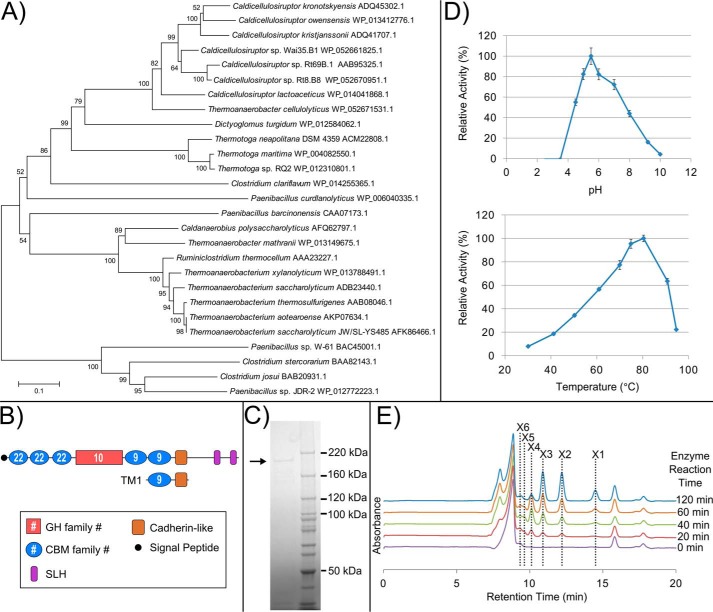
Eight of the 34 SLH domain-containing protein groups contain identifiable catalytic GH or PL CAZyme domains (Fig. 1). Csac_0678 (GH5), which has been characterized previously (21), has homologs in all 12 sequenced Caldicellulosiruptor species. Three of the 12 Caldicellulosiruptor species (Caldicellulosiruptor owensensis, Caldicellulosiruptor obsidiansis, and Caldicellulosiruptor sp. strain Rt8.B8) only produce a truncated homolog of Csac_0678 (see supplemental Table S1). Homologs of Calkro_0402 (GH10) are found in seven sequenced Caldicellulosiruptor species, with 65–83% amino acid sequence identity. One ortholog of Calkro_0402 (76% identity) has been characterized from Caldicellulosiruptor sp. Rt69.B1 (52), which does not have a fully sequenced genome. Calkro_0402 and its Caldicellulosiruptor sp. homologs belong to a larger group of GH10 enzymes with similar domain architecture, both with and without C-terminal SLH domains, that have been characterized from several xylan-degrading microbes (28, 31, 33, 34, 36, 37, 39–46, 48–51, 78).
FIGURE 1.
Catalytic CAZyme SLH domain proteins from the genus Caldicellulosiruptor. Domain arrangement of these multidomain proteins is drawn to scale based on a prediction from the NCBI conserved domain database (93) and Pfam protein families database (94). Representative genes for each group containing more than one member are shown on the left. The number of Caldicellulosiruptor species genomes from the 12 completely sequenced to date that contain a member from each group is shown to the right. BIg, bacterial immunoglobulin-like (CL0159); Cadherin-like (pfam12733).
The other six groups of catalytic SLH domain proteins (Fig. 1) have not been characterized from Caldicellulosiruptor species. Calkro_0111 (GH16/GH55) is the largest of all of these at 2,435 amino acids and is also the largest CAZyme of any kind identified so far in the genus Caldicellulosiruptor. Calkro_0121 is a second truncated paralog of Calkro_0111, which has 60% amino acid identity to Calkro_0111 and identical domain architecture but is lacking the final CBM32 domain. The arrangement of the domains in Calkro_0111 and Calkro_0121 is entirely unique to C. kronotskyensis, and no homologs of the full protein (Calkro_0111) can be identified in any other sequenced microorganism. Calkro_0072 (GH16) has homologs in seven Caldicellulosiruptor species and is very similar to the β-1,3-glucanase Lic16A characterized from C. thermocellum (38). C. hydrothermalis produces two catalytic SLH domain proteins that are unique to this species, putative xylanase Calhy_1629 (GH43) and putative α-glucanase Calhy_2383 (GH87). Calkro_0550 (PL11) from C. kronotskyensis has one additional homolog in C. owensensis and is a putative rhamnogalacturonan lyase. C. owensensis also produces one other putative pectate lyase SLH domain protein, Calow_2109 (PL9), which is unique to this species.
These catalytic SLH domain proteins encompass a range of enzymatic activities localized in the S-layer, presumably used to break down plant polysaccharide components that are in proximity of the cell surface. C. kronotskyensis produces six CAZyme SLH domain proteins, the most of any species; C. bescii only produces one, a homolog of Csac_0678, which is conserved in all Caldicellulosiruptor species. These SLH-localized CAZymes accompany a host of other CAZyme domains found in extracellular free enzymes produced by Caldicellulosiruptor sp. for plant biomass degradation (58).
C. kronotskyensis Produces an S-layer and SLH Enzymes Calkro_0111 and Calkro_0402 Are Localized on the C. kronotskyensis Cell Surface
Transmission electron microscopy of C. kronotskyensis growing on dilute acid-treated poplar (P. trichocarpa × P. deltoides) shows the cell membrane, peptidoglycan layer, and S-layer of C. kronotskyensis cells (Fig. 2A), with the cells appearing to attach to granules of the substrate mediated by appendages at the cell surface. To determine the localization of the SLH CAZymes Calkro_0402 and Calkro_0111, polyclonal antibodies were raised against portions of these proteins and used for immunofluorescence microscopy in C. kronotskyensis. Fig. 2, panels B and C and panels D and E, shows confocal microscopy of C. kronotskyensis labeled with anti-Calkro_0111 and anti-Calkro_0402 antibodies, respectively. Controls labeled with preimmune total IgY antibodies show minimal labeling (data not shown). In each case, these enzymes are localized on the cell surface within the S-layer, as would be predicted by the presence of SLH domains. Because of the high similarity between Calkro_0111 and Calkro_0121, the antibodies are probably labeling both of these proteins in Fig. 2 (B and C).
FIGURE 2.

Transmission electron microscopy and immunofluorescence microscopy of C. kronotskyensis. A, transmission electron microscopy of C. kronotskyensis grown on dilute acid-pretreated P. trichocarpa × P. deltoides showing the cytoplasmic membrane (cm), peptidoglycan (pg), and S-layer (s). B and C, confocal microscopy of C. kronotskyensis grown on laminarin labeled with total IgY antibodies raised against Calkro_0111 TM7. C and D, confocal microscopy of C. kronotskyensis grown on xylooligosaccharides labeled with total IgY antibodies raised against Cakro_0402 TM1. The cell counterstain (DAPI) is shown in magenta, and secondary antibody labeling (DyLight488 goat anti-chicken) is shown in green. Scale bars, 100 nm (A), 4 μm (B), 1 μm (C), 4 μm (D), and 1 μm (E).
Laminarinase Calkro_0111 Displays Endo- and Exoglucanase Activity in Separate Catalytic Domains
Calkro_0111 contains 15 predicted domains, including the two catalytic units: GH16 and GH55 (Fig. 3A). Because the full enzyme could not be produced recombinantly in E. coli, the enzyme was split into several TMs for characterization. Two of these TMs were used to characterize the enzymatic activity: TM1, containing the GH16 domain and covering the first half of Calkro_0111, and TM8, containing the GH55 domain and covering the second half of Calkro_0111. For both TM1 and TM8, the optimal pH was 5, whereas the optimal temperature was 75 °C (Fig. 3B). Analysis of the oligosaccharides released from laminarin by these two halves of Calkro_0111 showed that the GH55 domain of TM8 generates primarily glucose and a small amount of laminaribiose from laminarin, whereas the GH16 domain of TM1 released laminooligosaccharides of size L3 (laminaritriose) and greater, which slowly accumulated with time (Fig. 3C). Taken together, these findings suggest that the GH16 domain is an endoglucanase, whereas the GH55 domain is an exoglucanase. Considering the full-length enzyme, coordination between the GH16 and GH55 domains is expected, such that the GH16 cuts new chain ends for the GH55 to digest, thereby liberating glucose. This synergism is not uncommon in freely secreted Caldicellulosiruptor enzymes with two catalytic domains (76, 79, 80), but synergism had yet to be demonstrated in any CAZyme SLH domain protein. This is primarily due to the fact that very few SLH domain proteins have two catalytic CAZyme domains, further highlighting the uniqueness of Calkro_0111.
FIGURE 3.
Biochemical characterization of Calkro_0111. A, domain organization for full-length Calkro_0111 and TMs produced in E. coli used for characterization. B, optimum pH at 70 °C and optimum temperature at the optimum pH were determined for Calkro_0111 TM1 (0.9 mg/ml, incubated for 5 h) and TM8 (0.005 mg/ml, incubated for 30 min) on the substrate laminarin. Error bars, S.D. (n = 3). C, oligosaccharides released from laminarin by TM1 and TM8. HPLC chromatograms for each enzyme reaction time are aligned with respect to retention time and graphed together. The location of the peaks for standards glucose (G), laminaribiose (L2), laminaritriose (L3), laminaritetraose (L4), laminaripentaose (L5), and laminarihexaose (L6) are shown. D, truncation mutant activity as measured by the DNS reducing sugar assay. TM1 to TM5 and TM7 (4.2 μm) were incubated for 5 h, TM6 (12.3 μm) was incubated for 10 h, and TM8 to TM10 (0.2 μm) were incubated for 30 min with 1% (w/v) laminarin at 70 °C and pH 5 or 7. Activity is calculated in μmol of reducing sugar released/h/nmol of protein to compare activity for the proteins of different mass. Error bars, S.D. (n = 3).
To further explore the domain arrangement and the contribution of the non-catalytic domains to activity, a range of TMs (Fig. 3A) were produced in E. coli and analyzed. Activity of these TMs at 70 °C on laminarin was evaluated at pH 5, the enzyme optimum pH, and pH 7, the growth optimum pH for Caldicellulosiruptor (Fig. 3D). TM3 and TM4 had the highest activity at both pH levels tested, whereas TM1 was slightly less active. TM2, which is truncated between the GH16 and the first actin cross-linking-like (ACL) domain, was significantly less active. TM5, which contains the GH16 domain alone, and TM6 and TM7, which contain the ACL and fibronectin type-III (FN3) domains and not the GH16, all displayed little to no activity (Fig. 3D). TM4, which contains the GH16 and first ACL domain, represents the smallest portion of the GH16 side of the enzyme that remains fully active under these conditions. This suggests that the ACL domain plays an important role in GH16 function. The ACL domain could stabilize the GH16 domain, a role that has been shown before for accessory domains in other GH multidomain enzymes. For example, two FN3 domains (also previously called X1 modules) of CbhA (C. thermocellum cellobiohydrolase A) increased the thermostability of the adjacent GH9 domain (81). For the Calkro_0111 GH55 domain, removal of the ACL and FN3 domains from TM8 to TM9 and then removal of the two CBM32 domains from TM9 to TM10 resulted in successive reductions in activity (Fig. 3D). This suggests that both the ACL-FN3-FN3 grouping and two CBM32 domains play an important role in the activity of the Calkro_0111 GH55.
Biochemical Characterization of Calkro_0402 and Relatedness to Other CBM22/GH10/CBM9 Enzymes
Calkro_0402 belongs to a large group of homologous CBM22-, GH10-, and CBM9-containing proteins, from a variety of bacteria, primarily Firmicutes. An unrooted neighbor-joining phylogenetic tree constructed from the full amino acid sequences of top blastp hits to Calkro_0402 and similar xylanases that have been previously characterized shows the relatedness of xylanase enzymes from this group (Fig. 4A).
FIGURE 4.
Biochemical characterization of Calkro_0402. A, MEGA version 6 (95) was used to align, using MUSCLE (96), full protein amino acid sequences and construct an unrooted neighbor-joining phylogenetic tree of close blastp homologs to Calkro_0402 and previously characterized CBM22/GH10/CBM9-containing enzymes. Species and protein accession number are shown for each enzyme branch. B, domain organization for Calkro_0402 and TM1, which was used as the antigen for antibody production. Cadherin-like (pfam12733). C, purified full-length Calkro_0402 (∼180 kDa) produced in E. coli and purified by immobilized metal affinity and size exclusion chromatography. D, optimum pH at 70 °C and the optimum temperature at this optimum pH were determined for recombinant Calkro_0402 (0.02 mg/ml incubated 30 min). Error bars, S.D. (n = 3). E, oligosaccharides released from birchwood xylan by Calkro_0402. HPLC chromatograms for each enzyme reaction time are aligned with respect to retention time and graphed together. The location of the peaks for the standards xylose (X1), xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6) are shown.
Calkro_0402 contains three CBM22, one catalytic GH10, two CBM9 domains, and one cadherin-like domain, in addition to C-terminal SLH domain repeats (Fig. 4B). Full-length Calkro_0402 was produced recombinantly in E. coli (Fig. 4C), and the optimal pH and temperature for activity on birchwood xylan were found to be pH 5.5 and 80 °C, respectively (Fig. 4D). Calkro_0402 releases primarily xylotriose and xylobiose from birchwood xylan but also a small amount of xylose (Fig. 4E). This type of activity is consistent with other xylanase homologs that have been characterized previously (28, 31). Activity of Calkro_0402 was also detected on oat spelt xylan, wheat arabinoxylan, pachyman 1,3-β-glucan, barley β-glucan, and lichenan in addition to birchwood xylan, all at pH 5.5 and 70 °C (data not shown).
Transcriptomic Analysis of SLH Domain Proteins in C. bescii and C. kronotskyensis
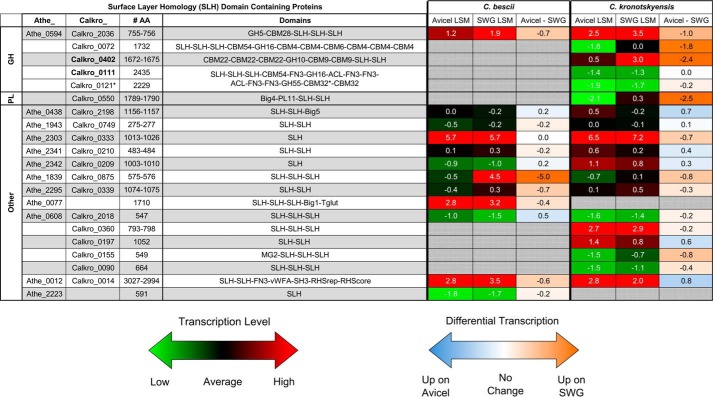
Transcriptomic analysis was performed on C. kronotskyensis and C. bescii, the Caldicellulosiruptor species with the most and least catalytic CAZyme SLH domain proteins, respectively, when these species were grown on Avicel (crystalline cellulose) and, the more heterogeneous substrate, switchgrass. All CAZyme SLH domain proteins from both species, except Calkro_0111 and Calkro_0121, were up-regulated on switchgrass. This transcriptional response to switchgrass suggests that these genes respond to components of switchgrass, like hemicellulose polysaccharides, not found in crystalline cellulose Avicel. The transcriptional level of these genes on switchgrass is similar to the free enzymes in the glucan degradation locus, known to be a genomic feature of cellulolytic Caldicellulosiruptor sp. (59, 61, 70). Calkro_0333 and Athe_2303 are very highly transcribed under both conditions. These proteins are homologs of Csac_2451, which was previously identified as the main SLP (59, 60). A number of other non-catalytic SLH domain proteins are transcribed at moderate to high levels, but because domains other than SLH domains are not predicted, the role of these proteins is unknown.
Manipulation of the C. bescii S-layer by the Insertion (Knock-in) of the Gene Encoding Calkro_0402
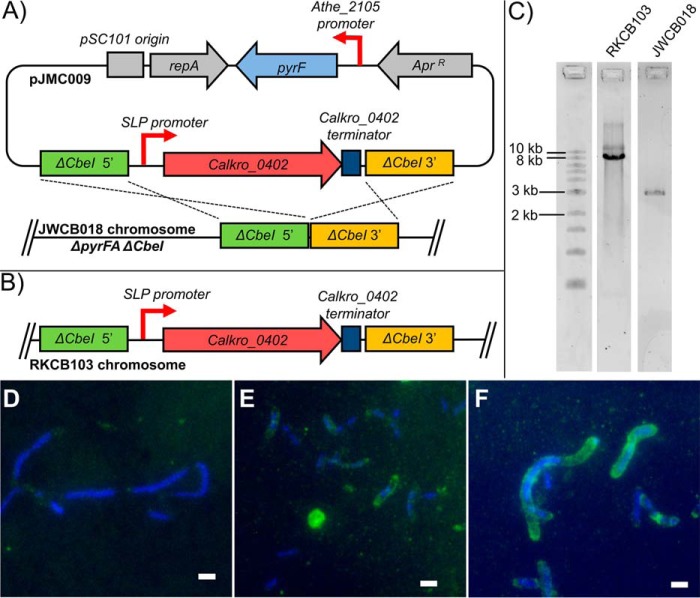
Because Calkro_0402 is very highly transcribed in C. kronotskyensis (Fig. 5) and utilized by a variety of xylan-degrading bacteria (Fig. 4A), Calkro_0402 was “knocked in” to genetically tractable C. bescii to examine its potential contribution to plant biomass degradation in vivo. C. bescii is a good genetic background for testing SLH proteins in vivo, because it produces only 12 SLH domain proteins in total and does not produce a homolog of Calkro_0402. Calkro_0402 was inserted into C. bescii uracil auxotroph strain JWCB018 using pyrF complementation and 5-fluoroorotic acid counterselection. The knock-in construct contained Calkro_0402, including the native signal peptide and the native predicted terminator sequence from C. kronotskyensis, under the control of the slp promoter from the main SLP (Athe_2303) in C. bescii (Fig. 6, A and B). As shown in Fig. 5, the main SLP (Athe_2303) is very highly and constitutively transcribed. Thus, its promoter should direct very high levels of transcription of Calkro_0402 in the knock-in construct. The knock-in was targeted at the ΔCbeI (Athe_2438) deletion locus in C. bescii strain JWCB018, because genetic manipulation in this area of the genome was successful previously (64). The PCR amplicon for the knock-in C. bescii strain RKCB103 compared with strain JWCB018 is shown in Fig. 6C.
FIGURE 5.
Transcriptional response of genes encoding SLH domain proteins from C. bescii and C. kronotskyensis. Shown is the log squared mean (LSM) transcriptomic level of the 19 SLH domain proteins from C. kronotskyensis and 12 SLH domain proteins from C. bescii when each species is grown on crystalline cellulose (Avicel) and switchgrass (SWG). A log squared mean value of 0 represents average transcript abundance (black). Genes transcribed at levels higher than average have positive log squared mean values (red), whereas genes transcribed at levels lower than average have negative log squared mean values (green). Differential transcription is shown as Avicel minus switchgrass with negative values (orange) up-regulated on switchgrass relative to Avicel. Positive values (blue) are up-regulated on Avicel relative to switchgrass. Analysis is based on whole-genome oligonucleotide microarray experiments deposited in the NCBI Gene Expression Omnibus database with accession number GSE68810 (70). Big, bacterial immunoglobulin-like (CL0159); Tglut, transglutaminase-like superfamily (pfam01841); MG2, macroglobulin 2 (pfam01835); vWFA, von Willebrand factor type A (pfam00092); SH3, bacterial Src homology 3 (pfam08239); RHSrep, RHS repeat (pfam05593); RHScore, RHS-associated core domain (TIGR03696). *, Calkro_0121 is truncated after the first CBM32.
FIGURE 6.
Construction of Calkro_0402 knock-in C. bescii strain RKCB103. A, Calkro_0402 knock-in vector pJMC009 homologous recombination with the chromosome of parent C. bescii strain JWCB018. pJMC009 was constructed using the pDCW121 (65) vector backbone, which contains the pSC101 origin, repA, and apramycin resistance marker for maintenance in E. coli as well as C. bescii wild type pyrF under the control of the Athe_2105 promoter for selection of uracil prototrophy and 5-fluoroorotic acid counterselection. Cross-over regions 1 kb in length 5′ and 3′ to the ΔCbeI locus (ΔAthe_2438) in strain JWCB018 were used to target Calkro_0402 for insertion via homologous recombination to that locus. The Calkro_0402 insertion construct included Calkro_0402 and an additional 81 bp downstream of the gene to include its predicted transcriptional terminator element amplified from C. kronotskyensis genomic DNA. High, constitutive expression of Calkro_0402 was driven by the 200-bp promoter element upstream of the C. bescii main SLP gene (Athe_2303). B, the Calkro_0402 construct is inserted in the chromosome of RKCB103 after first crossover vector integration and second crossover 5-fluoroorotic acid counterselection to remove the vector backbone containing pyrF. C, using primers outside of the 5′ and 3′ crossover regions (Table 1, ΔAthe_2438 locus F and R), the ΔCbeI locus was amplified and sequenced to verify the Calkro_0402 knock-in. Strain JWCB018 has the expected 2.9-kb product, whereas strain RKCB103 has the expected 8.2-kb product with the 5.3-kb insertion of the SLP promoter and Calkro_0402 construct. A New England Biolabs 1-kb ladder was run alongside these PCR products. D–F, anti-Calkro_0402 immunofluorescence microscopy of C. bescii strain JWCB018, C. kronotskyensis, and C. bescii strain RKCB103 grown on birchwood xylan. D, C. bescii strain JWCB018, the genetic parent strain, which does not contain Calkro_0402, shows minimal labeling with the anti-Calkro_0402 antibodies. C. kronotskyensis (E) and Calkro_0402 knock-in C. bescii strain RKCB103 (F) are both extensively labeled with antibody. E and F, Calkro_0402 is shown localized on the cell surface in both C. kronotskyensis and C. bescii strain RKCB103 and appears primarily at the poles of the cells. All cells are labeled with chicken anti-Calkro_0402 total IgY primary antibody and DyLight 488-conjugated goat anti-chicken secondary antibody (green) with DAPI as a counterstain (blue). Scale bars, 2 μm.
Using this Calkro_0402 knock-in C. bescii strain RKCB103, immunofluorescence microscopy was performed to investigate the expression of Calkro_0402 within the S-layer of C. bescii. For comparison, immunofluorescence microscopy was also performed on parent C. bescii strain JWCB018 and C. kronotskyensis, in addition to C. bescii strain RKCB103 (Fig. 6, D–F). C. bescii strain JWCB018, which does not have Calkro_0402, had minimal labeling with the anti-Calkro_0402 antibodies (Fig. 6D), whereas C. bescii strain RKCB103 was extensively labeled on the cell surface (Fig. 6F). Labeling of the native expression of Calkro_0402 in C. kronotskyensis is also shown (Fig. 6E). Based on transcription levels of Calkro_0402 and Athe_2303, the main SLP, (Fig. 5), the Calkro_0402 transcript in C. bescii strain RKCB103 should be >10-fold higher, using the slp promoter, compared with levels observed in wild-type C. kronotskyensis. This clearly relates to an increase in Calkro_0402 protein expression in C. bescii strain RKCB103 compared with native expression in C. kronotskyensis, as seen via the immunofluorescent labeling. Also of particular note is that the native signal peptide from C. kronotskyensis on Calkro_0402 appeared to function in C. bescii to route the protein for secretion. The C. bescii-produced Calkro_0402 also localized to the surface of the C. bescii strain RKCB103 cells and in Fig. 6F appears to localize primarily at the poles of the cell. The organization of SLH proteins on the cell surface is generally thought to be coordinated in bacteria that produce multiple SLH proteins, but the mechanisms by which this occurs have not been determined (82). This is the first example of manipulation of the Caldicellulosiruptor S-layer through genetic modification.
C. bescii Strain RKCB103 Xylan Solubilization
To understand the role of Calkro_0402 in vivo, the ability of C. bescii strains RKCB103 and JWCB018 to solubilize plant biomass in culture was examined. Fig. 7A shows the solubilization of oat spelt and birchwood xylans. Strain RKCB103 is able to solubilize significantly more washed oat spelt xylan than strain JWCB018; the xylose equivalents from the soluble xylooligosaccharides measured in the supernatant are more than double for strain RKCB103 compared with strain JWCB018 (Fig. 7B). For both the washed and unwashed birch xylan substrates, strain RKCB103 performed slightly better and released more xylose equivalents to the supernatant than strain JWCB018. Other biomass substrates tested (diluted acid-pretreated P. trichocarpa × P. deltoides, dilute acid-pretreated switchgrass, and unpretreated Panicum virgatum switchgrass) showed no significant difference in solubilization and also showed <30 μg/ml xylose equivalents in the culture supernatant (data not shown). This suggests that on these lignocellulosic substrates, C. bescii strain JWCB018 produced enough xylanase activity to remove xylan at the rate at which it was exposed from the complex polysaccharide matrix in the substrate, and cellulose solubilization or occlusion of the polysaccharides by lignins was probably limiting the overall solubilization. However, in the substrates with high xylan content, strain RKCB103 with the addition of Calkro_0402 outperformed strain JWCB018.
FIGURE 7.
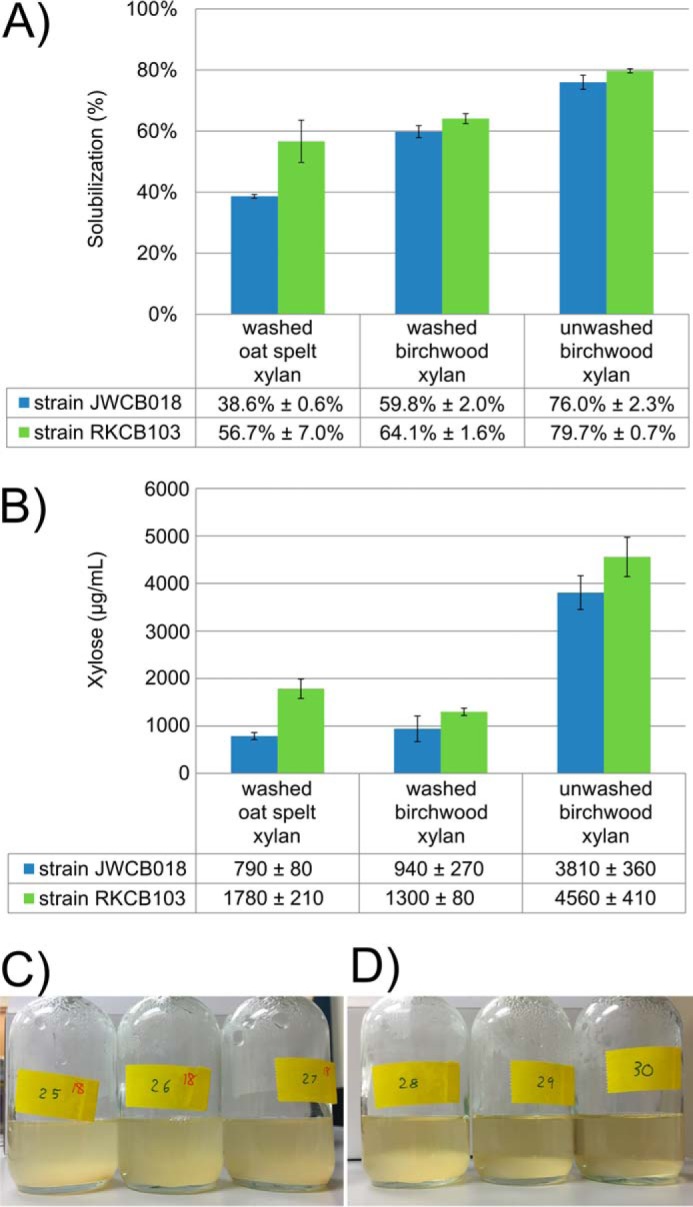
Increased xylan solubilization and xylan attachment by Calkro_0402 knock-in C. bescii strain RKCB103. A, total biomass solubilization of washed oat spelt xylan, washed birchwood xylan, and unwashed birchwood xylan after 45.5 h of growth of C. bescii strain JWCB018 or RKCB103. B, xylose measured in the acid-hydrolyzed culture supernatant harvested at 45.5 h. Error bars, S.D. (n = 3). C. bescii strain JWCB018 cultures on washed birchwood xylan appear very turbid (C), and C. bescii strain RKCB103 cultures appear much less turbid (D) while having the same cell density/ml of culture. C. bescii strain RKCB103 appears to be more readily attached to the insoluble washed birchwood xylan substrate, suggesting that the expression of Calkro_0402 in this strain is playing a role in tethering the cells to the xylan substrate.
Immunofluorescence microscopy (Fig. 6F) shows that Calkro_0402 is localized on the surface of strain RKCB103 cells. While the enzyme is tethered to the cell surface, the GH and CBM domains of Calkro_0402 interact with the xylan substrate. This suggests that Calkro_0402 plays a role in mediating cell attachment to xylan substrates. Cultures of strain JWCB018 and strain RKCB103 (Fig. 7, C and D, respectively) grown on washed birchwood xylan support this role for Calkro_0402. These cultures contained approximately the same number of cells as determined by cell counts (data not shown), but the supernatant in Fig. 7D for strain RKCB103 with Calkro_0402 is relatively clear, because the cells appear to be attached to the insoluble xylan at the bottom of the culture bottle. The strain JWCB018 cultures (Fig. 7C) were more turbid, suggesting that the cells did not as readily attach to the biomass substrate.
To understand the role of Calkro_0402 in improved xylan solubilization, a time course experiment was performed on washed oat spelt xylan. Fig. 8A shows the planktonic cell density for strains JWCB018 and RKCB103 over the duration of the culture. The cell densities of the two strains track each other closely. When monitoring xylose release, roughly 9–12 h postinoculation, the xylose hydrolyzed from xylooligosaccharides by strain RKCB103 was measurable above the baseline abiotic control, whereas strain JWCB018 took between 16 and 22 h postinoculation (Fig. 8B). It is important to note that the strains were consuming xylose as they degraded the xylan substrate, and these values represent the xylose from excess xylooligosaccharides released by the enzymatic action of the cells that has not been consumed for growth. As with the closed bottle total solubilization experiment, strain RKCB103 has about double the xylose equivalents in the supernatant samples as strain JWCB018 at 45.5 h, the point at which the cultures were harvested.
FIGURE 8.
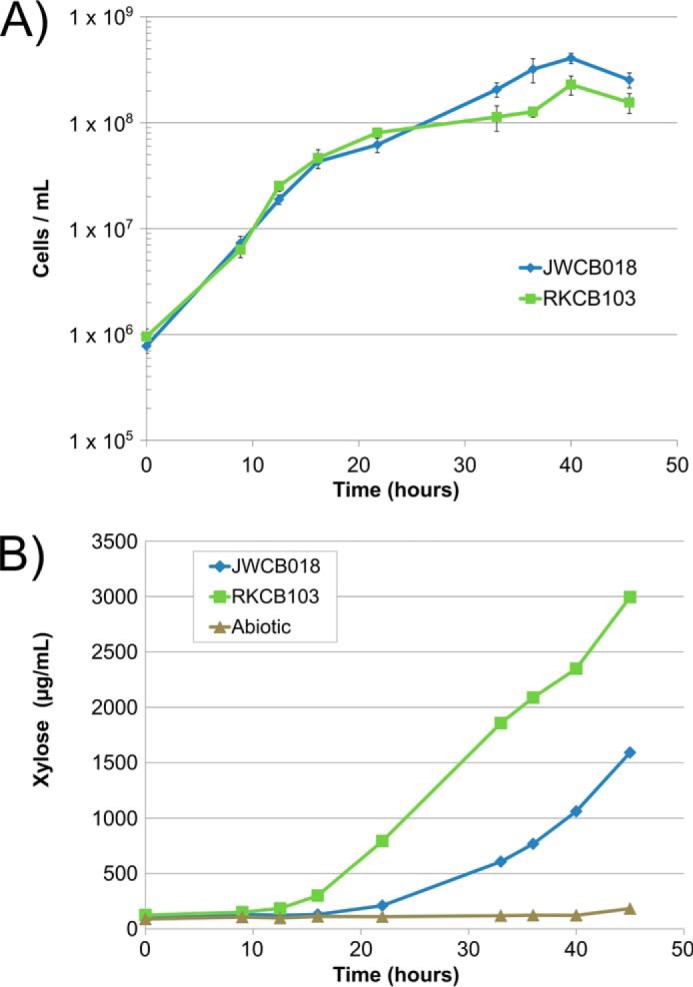
Time course of washed oat spelt xylan solubilization. A, cell densities of C. bescii strains JWCB018 (blue) and RKCB103 (green) over time while growing on washed oat spelt xylan. B, xylose measured in acid-hydrolyzed supernatant samples for C. bescii strains JWCB018 (blue) and RKCB103 (green) and an abiotic control (brown) (B). Error bars, S.D.
Discussion
Catalytic CAZyme-containing SLH domain proteins are produced by a variety of lignocellulose-degrading bacteria (21, 22, 45). The genomes of 12 Caldicellulosiruptor species encode eight different groups of these (Fig. 1), a subset of the 34 groups of Caldicellulosiruptor SLH domain proteins (supplemental Table S1). These proteins are one of several mechanisms by which Caldicellulosiruptor species effect plant biomass degradation (21, 59, 61–63).
Calkro_0111 from C. kronotskyensis is the largest glycoside hydrolase from all of the biomass degradation enzymes in the Caldicellulosiruptor genus, free or cell-associated, and is entirely unique to C. kronotskyensis. The GH16 domain of Calkro_0111 is an endoglucanase, whereas the GH55 is an exoglucanase (Fig. 3). Thus, the two GH domains probably work synergistically, as has been shown for other Caldicellulosiruptor enzymes without SLH domains but with two catalytic domains (76, 79, 80). Various truncation mutants of Calkro_0111 (Fig. 3D) were examined, revealing that the non-catalytic ACL domain adjacent to the GH16 domain is necessary for full activity. Furthermore, for the GH55 domain of Calkro_0111, both the ACL-FN3-FN3 domain grouping and two CBM32 domains improved the activity of the GH55 domain.
The very large multidomain structure of Calkro_0111 is characteristic of many Caldicellulosiruptor polysaccharide-degrading enzymes, where this architecture is thought to have arisen from domain shuffling (83, 84). Specifically, for Calkro_0111, the N terminus GH16 portion is similar to the N terminus of Calkro_0072 (45% amino acid identity), and the C terminus GH55 portion is similar to Calkro_0113 (58% amino acid identity). These protein segments may have combined by domain shuffling to form Calkro_0111. However, the ACL-FN3-FN3-ACL-FN3-FN3 motif in the middle of Calkro_0111 and in homolog Calkro_0121 is completely unique to these two genes. These ACL domains belong to the same protein superfamily (CL22458 RICIN superfamily) as fascins, which play a role in bundling actin filaments for cell adhesion and migration in Drosophila and vertebrate species (85). Plants and brown algae, such as Laminaria sp., produce actin and actin bundling proteins that are distinct from fascins for a variety of cellular roles (86, 87). The ACL domains of Calkro_0111 may play a role in attachment of this enzyme to actin found in algal or plant cells, similar to the role of a CBM for carbohydrate binding, to help localize the enzyme to laminarin-containing substrates.
Many GH16- and GH55-containing enzymes are active on β-1,3-glucans. Calkro_0111 activity was detected on a model substrate, laminarin (β-1,3-1,6-glucan), albeit at modest levels such that this is not likely to be its natural substrate. In fact, β-1,3-glucans, similar to laminarin, are produced in plant reproductive and wound tissue (callose) (88, 89), exopolysaccharide from bacteria (curdlan) (90), algae such as L. digitata (laminarin), and lichen such as Certraria islandica (lichenan). In addition to this unique substrate preference of Calkro_0111, its genomic neighborhood is also unique, containing other extracellular biomass-degrading enzymes that have no homologs within other species in the genus. These include GH16/GH55 Calkro_0121, GH55 Calkro_0113, and GH81 Calkro_0114. This locus also encodes two ABC transporters, one of which is homologous to a putative glucooligosaccharide transporter in C. saccharolyticus (91). This entire genomic region is transcribed at relatively low levels when C. kronotskyensis is grown on Avicel or switchgrass. Taken together, this genomic locus, including Calkro_0111, probably degrades unique β-1,3-glucans from lichens, algae, or other microorganisms that may be more abundant in the natural environment of Kamchatka, Russia, from which C. kronotskyensis was isolated.
The large size of the gene encoding Calkro_0111 demonstrates that this unique multidomain arrangement is beneficial to C. kronotskyensis for it to be maintained and even duplicated as Calkro_0121. More broadly in the genus Caldicellulosiruptor, the unique multidomain architecture of enzymes, both with and without SLH domains, has probably evolved to appropriately space the CAZyme domains for increased synergy and activity. Specifically, for SLH domain proteins, domain spacing must also accommodate the tethering of the SLH domains at one end of the protein to the cell surface while it is attaching to and degrading the plant biomass substrate with the other. The large size of these catalytic SLH proteins may reflect the length needed to swing out away from the cell to reach biomass substrates to which they can attach. Although domain spacing and arrangement appear to be critical factors in the activity of Calkro_0111, no pattern for domain arrangement through all of the catalytic Caldicellulosiruptor SLH proteins is readily apparent, based on amino acid sequence. Lacking structural data, which are difficult to obtain for these large multidomain proteins, the orientation of each domain relative to another cannot be determined precisely. It does seem likely, however, that each protein has evolved its domain spacing to enable its unique combination of binding and catalytic domains to function properly while being tethered to the cell.
Whereas Calkro_0111 and Calkro_0121 are transcribed at low levels when grown on plant biomass substrates, all of the other SLH GH and PL enzymes from C. kronotskyensis and C. bescii are up-regulated when these species are grown on switchgrass compared with Avicel (Fig. 5), including the xylanase Calkro_0402. This suggests an important role in plant biomass degradation. Calkro_0402 belongs to a large group of homologous CBM22/GH10/CBM9 xylanases (Fig. 4A). While Calkro_0402 releases primarily xylobiose and xylotriose from birchwood xylan (Fig. 4E), similar to previously characterized homologs (28, 31), the optimal temperature of 80 °C for Calkro_0402 (Fig. 4D) makes it one of the most thermophilic versions of this family of xylanase enzymes characterized to date. As is predicted by the presence of SLH domains, Calkro_0402 is localized to the cell surface of C. kronotskyensis (Fig. 2, D and E), which is a common feature of many homologs of Calkro_0402, including some shown in Fig. 4A associated with the cell by means other than SLH domains. The Calkro_0402 homolog from Thermotoga maritima (and presumably homologs in other Thermotoga sp.) is cell-associated by an N-terminal hydrophobic peptide anchor and not SLH domains (92). Furthermore, the Clostridium clariflavum xylanase lacks SLH domains but has a dockerin domain on its C terminus, which would allow this enzyme to associate via protein-protein interactions into a cellulosome multienzyme complex that is also typically anchored to the cell surface via SLH domains (61). The fact that these homologs without SLH domains can remain associated with the cell implies that there is particular utility for having this type of CBM22/GH10/CBM9 xylanase associated with the cell.
Using genetic manipulation of C. bescii to knock in Calkro_0402, the role of this SLH protein in vivo for substrate attachment (Fig. 7, C and D) and degradation (Figs. 7 (A and B) and 8 (A and B)) could be examined. Wild-type C. bescii has previously been shown to attach to xylan substrates (63). Our observations suggest that modification of the C. bescii S-layer to contain SLH domain xylanase Calkro_0402 probably improves this attachment (Fig. 7, C and D). Truncation mutants of Xyn10B from Clostridium stercorarium and Xyn10A from Clostridium josui showed that the CBM9 and CBM22 domains mediate the attachment of these enzymes to xylan and other substrates (42, 43). While these two xylanases are phylogenetically distant from Calkro_0402 (Fig. 4A), the role of these CBM22 and CBM9 domains is probably very similar in Calkro_0402 in the functional role of degrading xylan substrates. The attachment of the Calkro_0402 knock-in strain RKCB103 is also probably a significant factor in its improved solubilization of xylans (Fig. 7A). Both C. bescii and C. kronotskyensis produce at least five other extracellular xylanases targeted for the secretome with GH10, GH11, or GH43 domains. If the cell is attached to xylan by Calkro_0402, these enzymes are secreted from the cell proximate to their substrate, probably making them more effective in the degradation process as well.
Very few of the diverse catalytic CAZyme-containing SLH domain proteins have been characterized, but their role in biomass deconstruction clearly merits closer examination. There are more than 20,000 SLH domain-containing proteins predicted in over 1,800 microbial genomes. These unusual proteins on the surface of bacterial cells contain a variety of functional protein domains, localized at the interface between the cell and its environment. Although a full understanding of bacterial S-layer proteins and their varied and important roles at the cell surface is not yet available, the results reported here provide new insights into the role of SLH GHs as agents of plant polysaccharide degradation.
Author Contributions
J. M. C., W. S. P., G. W. H., A. L. T., and S. E. B.-S. contributed to protein cloning, expression, and enzymatic characterization. J. M. C., W. S. P., J. H. L., J. H. W., L. L. L., and S. E. B.-S. contributed to construction of strain RKCB103. J. M. C., G. W. H., and S. E. B.-S. contributed to microscopy. J. M. C., J. H. L., G. W. H., and J. V. Z. contributed to biomass solubilization assays and HPLC analysis. J. M. C. and R. M. K. designed and directed the research and drafted the manuscript. All authors provided comments on and approved the final manuscript.
Supplementary Material
Acknowledgments
We acknowledge the North Carolina State University Cellular and Molecular Imaging Facility and Dr. Eva Johannes for assistance in operating the Zeiss LSM 710 confocal microscope.
This work was supported by the BioEnergy Science Center, a United States Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
- S-layer
- surface layer
- SLH
- surface layer homology (pfam00395)
- SLP
- S-layer protein
- CAZyme
- carbohydrate active enzyme
- GH
- glycoside hydrolase
- PL
- polysaccharide lyase
- CBM
- carbohydrate binding module
- FN3
- fibronectin type III (pfam00041)
- ACL
- actin cross-linking-like RICIN superfamily (CL22458)
- LOD
- low osmolality defined
- LOC
- low osmolality complex
- TM
- truncation mutant
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- DNS
- 3,5-dinitrosalicylic acid.
References
- 1. Sára M., and Sleytr U. B. (2000) S-Layer proteins. J. Bacteriol. 182, 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sleytr U. B., and Beveridge T. J. (1999) Bacterial S-layers. Trends Microbiol. 7, 253–260 [DOI] [PubMed] [Google Scholar]
- 3. Albers S.-V., and Meyer B. H. (2011) The archaeal cell envelope. Nat. Rev. Microbiol. 9, 414–426 [DOI] [PubMed] [Google Scholar]
- 4. Kern J., Wilton R., Zhang R., Binkowski T. A., Joachimiak A., and Schneewind O. (2011) Structure of surface layer homology (SLH) domains from Bacillus anthracis surface array protein. J. Biol. Chem. 286, 26042–26049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mesnage S., Fontaine T., Mignot T., Delepierre M., Mock M., and Fouet A. (2000) Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19, 4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fagan R. P., Janoir C., Collignon A., Mastrantonio P., Poxton I. R., and Fairweather N. F. (2011) A proposed nomenclature for cell wall proteins of Clostridium difficile. J. Med. Microbiol. 60, 1225–1228 [DOI] [PubMed] [Google Scholar]
- 7. Boot H. J., Kolen C. P., and Pouwels P. H. (1995) Identification, cloning, and nucleotide sequence of a silent S-layer protein gene of Lactobacillus acidophilus ATCC 4356 which has extensive similarity with the S-layer protein gene of this species. J. Bacteriol. 177, 7222–7230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson B. R., Hymes J., Sanozky-Dawes R., Henriksen E. D., Barrangou R., and Klaenhammer T. R. (2015) Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl. Environ. Microbiol. 82, 134–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veith A., Klingl A., Zolghadr B., Lauber K., Mentele R., Lottspeich F., Rachel R., Albers S. V., and Kletzin A. (2009) Acidianus, Sulfolobus and Metallosphaera surface layers: structure, composition and gene expression. Mol. Microbiol. 73, 58–72 [DOI] [PubMed] [Google Scholar]
- 10. Abdul Halim M. F., Pfeiffer F., Zou J., Frisch A., Haft D., Wu S., Tolić N., Brewer H., Payne S. H., Paša-Tolić L., and Pohlschroder M. (2013) Haloferax volcanii archaeosortase is required for motility, mating, and C-terminal processing of the S-layer glycoprotein. Mol. Microbiol. 88, 1164–1175 [DOI] [PubMed] [Google Scholar]
- 11. Abdul Halim M. F., Karch K. R., Zhou Y., Haft D. H., Garcia B. A., and Pohlschroder M. (2015) Permuting the PGF-CTERM signature motif blocks both archaeosortase-dependent C-terminal cleavage and prenyl lipid attachment for the Haloferax volcanii S-layer glycoprotein. J. Bacteriol. 10.1128/JB.00849-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fagan R. P., and Fairweather N. F. (2014) Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 12, 211–222 [DOI] [PubMed] [Google Scholar]
- 13. Pham T. K., Roy S., Noirel J., Douglas I., Wright P. C., and Stafford G. P. (2010) A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia. Proteomics 10, 3130–3141 [DOI] [PubMed] [Google Scholar]
- 14. Honma K., Inagaki S., Okuda K., Kuramitsu H. K., and Sharma A. (2007) Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb. Pathog. 42, 156–166 [DOI] [PubMed] [Google Scholar]
- 15. Reynolds C. B., Emerson J. E., de la Riva L., Fagan R. P., and Fairweather N. F. (2011) The Clostridium difficile cell wall protein CwpV is antigenically variable between strains, but exhibits conserved aggregation-promoting function. PLoS Pathog. 7, e1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Z., Kong J., Hu S., Kong W., Lu W., and Liu W. (2013) Characterization of a S-layer protein from Lactobacillus crispatus K313 and the domains responsible for binding to cell wall and adherence to collagen. Appl. Microbiol. Biotechnol. 97, 1941–1952 [DOI] [PubMed] [Google Scholar]
- 17. de la Riva L., Willing S. E., Tate E. W., and Fairweather N. F. (2011) Roles of cysteine proteases Cwp84 and Cwp13 in biogenesis of the cell wall of Clostridium difficile. J. Bacteriol. 193, 3276–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderson V. J., Kern J. W., McCool J. W., Schneewind O., and Missiakas D. (2011) The SLH-domain protein BslO is a determinant of Bacillus anthracis chain length. Mol. Microbiol. 81, 192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brahamsha B. (1996) An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 93, 6504–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarren J., and Brahamsha B. (2007) SwmB, a 1.12-megadalton protein that is required for nonflagellar swimming motility in Synechococcus. J. Bacteriol. 189, 1158–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozdemir I., Blumer-Schuette S. E., and Kelly R. M. (2012) S-layer homology domain proteins Csac_0678 and Csac_2722 are implicated in plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 78, 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Egelseer E., Schocher I., Sára M., and Sleytr U. B. (1995) The S-layer from Bacillus stearothermophilus DSM 2358 functions as an adhesion site for a high-molecular-weight amylase. J. Bacteriol. 177, 1444–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lemaire M., Ohayon H., Gounon P., Fujino T., and Béguin P. (1995) OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177, 2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fujino T., Béguin P., and Aubert J. P. (1993) Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175, 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fontes C. M., and Gilbert H. J. (2010) Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 27. Markowitz V. M., Chen I. M., Palaniappan K., Chu K., Szeto E., Pillay M., Ratner A., Huang J., Woyke T., Huntemann M., Anderson I., Billis K., Varghese N., Mavromatis K., Pati A., Ivanova N. N., and Kyrpides N. C. (2014) IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han Y., Agarwal V., Dodd D., Kim J., Bae B., Mackie R. I., Nair S. K., and Cann I. K. (2012) Biochemical and structural insights into xylan utilization by the thermophilic bacterium Caldanaerobius polysaccharolyticus. J. Biol. Chem. 287, 34946–34960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han Y., Dodd D., Hespen C. W., Ohene-Adjei S., Schroeder C. M., Mackie R. I., and Cann I. K. (2010) Comparative analyses of two thermophilic enzymes exhibiting both β-1,4 mannosidic and β-1,4 glucosidic cleavage activities from Caldanaerobius polysaccharolyticus. J. Bacteriol. 192, 4111–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S. P., Morikawa M., Takagi M., and Imanaka T. (1994) Cloning of the aapT gene and characterization of its product, α-amylase-pullulanase (AapT), from thermophilic and alkaliphilic Bacillus sp. strain XAL601. Appl. Environ. Microbiol. 60, 3764–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waeonukul R., Pason P., Kyu K. L., Sakka K., Kosugi A., Mori Y., and Ratanakhanokchai K. (2009) Cloning, sequencing, and expression of the gene encoding a multidomain endo-β-1,4-xylanase from Paenibacillus curdlanolyticus B-6, and characterization of the recombinant enzyme. J. Microbiol. Biotechnol. 19, 277–285 [PubMed] [Google Scholar]
- 32. Cheng Y. M., Hong T. Y., Liu C. C., and Meng M. (2009) Cloning and functional characterization of a complex endo-β-1,3-glucanase from Paenibacillus sp. Appl. Microbiol. Biotechnol. 81, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 33. Stjohn F. J., Rice J. D., and Preston J. F. (2006) Paenibacillus sp. strain JDR-2 and XynA1: a novel system for methylglucuronoxylan utilization. Appl. Environ. Microbiol. 72, 1496–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ito Y., Tomita T., Roy N., Nakano A., Sugawara-Tomita N., Watanabe S., Okai N., Abe N., and Kamio Y. (2003) Cloning, expression, and cell surface localization of Paenibacillus sp. strain W-61 xylanase 5, a multidomain xylanase. Appl. Environ. Microbiol. 69, 6969–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Itoh T., Sugimoto I., Hibi T., Suzuki F., Matsuo K., Fujii Y., Taketo A., and Kimoto H. (2014) Overexpression, purification, and characterization of Paenibacillus cell surface-expressed chitinase ChiW with two catalytic domains. Biosci. Biotechnol. Biochem. 78, 624–634 [DOI] [PubMed] [Google Scholar]
- 36. St John F. J., Preston J. F., and Pozharski E. (2012) Novel structural features of xylanase A1 from Paenibacillus sp. JDR-2. J. Struct. Biol. 180, 303–311 [DOI] [PubMed] [Google Scholar]
- 37. Kim H., Jung K. H., and Pack M. Y. (2000) Molecular characterization of xynX, a gene encoding a multidomain xylanase with a thermostabilizing domain from Clostridium thermocellum. Appl. Microbiol. Biotechnol. 54, 521–527 [DOI] [PubMed] [Google Scholar]
- 38. Fuchs K.-P., Zverlov V. V., Velikodvorskaya G. A., Lottspeich F., and Schwarz W. H. (2003) Lic16A of Clostridium thermocellum, a non-cellulosomal, highly complex endo-β-1,3-glucanase bound to the outer cell surface. Microbiology 149, 1021–1031 [DOI] [PubMed] [Google Scholar]
- 39. Adelsberger H., Hertel C., Glawischnig E., Zverlov V. V., and Schwarz W. H. (2004) Enzyme system of Clostridium stercorarium for hydrolysis of arabinoxylan: reconstitution of the in vivo system from recombinant enzymes. Microbiology 150, 2257–2266 [DOI] [PubMed] [Google Scholar]
- 40. Feng J. X., Karita S., Fujino E., Fujino T., Kimura T., Sakka K., and Ohmiya K. (2000) Cloning, sequencing, and expression of the gene encoding a cell-bound multi-domain xylanase from Clostridium josui, and characterization of the translated product. Biosci. Biotechnol. Biochem. 64, 2614–2624 [DOI] [PubMed] [Google Scholar]
- 41. Ali M. K., Fukumura M., Sakano K., Karita S., Kimura T., Sakka K., and Ohmiya K. (1999) Cloning, sequencing, and expression of the gene encoding the Clostridium stercorarium xylanase C in Escherichia coli. Biosci. Biotechnol. Biochem. 63, 1596–1604 [DOI] [PubMed] [Google Scholar]
- 42. Ali E., Araki R., Zhao G., Sakka M., Karita S., Kimura T., and Sakka K. (2005) Functions of family-22 carbohydrate-binding modules in Clostridium josui Xyn10A. Biosci. Biotechnol. Biochem. 69, 2389–2394 [DOI] [PubMed] [Google Scholar]
- 43. Ali M. K., Hayashi H., Karita S., Goto M., Kimura T., Sakka K., and Ohmiya K. (2001) Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci. Biotechnol. Biochem. 65, 41–47 [DOI] [PubMed] [Google Scholar]
- 44. Ali M. K., Kimura T., Sakka K., and Ohmiya K. (2001) The multidomain xylanase Xyn10B as a cellulose-binding protein in Clostridium stercorarium. FEMS Microbiol. Lett. 198, 79–83 [DOI] [PubMed] [Google Scholar]
- 45. Brechtel E., Matuschek M., Hellberg A., Egelseer E. M., Schmid R., and Bahl H. (1999) Cell wall of Thermoanaerobacterium thermosulfurigenes EM1: isolation of its components and attachment of the xylanase XynA. Arch. Microbiol. 171, 159–165 [DOI] [PubMed] [Google Scholar]
- 46. Liu S. Y., Gherardini F. C., Matuschek M., Bahl H., and Wiegel J. (1996) Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J. Bacteriol. 178, 1539–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matuschek M., Burchhardt G., Sahm K., and Bahl H. (1994) Pullulanase of Thermoanaerobacterium thermosulfurigenes EM1 (Clostridium thermosulfurogenes): molecular analysis of the gene, composite structure of the enzyme, and a common model for its attachment to the cell surface. J. Bacteriol. 176, 3295–3302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Matuschek M., Sahm K., Zibat A., and Bahl H. (1996) Characterization of genes from Thermoanaerobacterium thermosulfurigenes EM1 that encode two glycosyl hydrolases with conserved S-layer-like domains. Mol. Gen. Genet. 252, 493–496 [DOI] [PubMed] [Google Scholar]
- 49. Shao W., Deblois S., and Wiegel J. (1995) A high-molecular-weight, cell-associated xylanase isolated from exponentially growing Thermoanaerobacterium sp. strain JW/SL-YS485. Appl. Environ. Microbiol. 61, 937–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang X., Li Z., Du C., Wang J., and Li S. (2015) Improved expression and characterization of a multidomain xylanase from Thermoanaerobacterium aotearoense SCUT27 in Bacillus subtilis. J. Agric. Food Chem. 63, 6430–6439 [DOI] [PubMed] [Google Scholar]
- 51. Hung K. S., Liu S. M., Fang T. Y., Tzou W. S., Lin F. P., Sun K. H., and Tang S. J. (2011) Characterization of a salt-tolerant xylanase from Thermoanaerobacterium saccharolyticum NTOU1. Biotechnol. Lett. 33, 1441–1447 [DOI] [PubMed] [Google Scholar]
- 52. Morris D. D., Gibbs M. D., Ford M., Thomas J., and Bergquist P. L. (1999) Family 10 and 11 xylanase genes from Caldicellulosiruptor sp. strain Rt69B. 1. Extremophiles 3, 103–111 [DOI] [PubMed] [Google Scholar]
- 53. Blumer-Schuette S. E., Ozdemir I., Mistry D., Lucas S., Lapidus A., Cheng J.-F., Goodwin L. A., Pitluck S., Land M. L., Hauser L. J., Woyke T., Mikhailova N., Pati A., Kyrpides N. C., Ivanova N., Detter J. C., Walston-Davenport K., Han S., Adams M. W. W., and Kelly R. M. (2011) Complete genome sequences for the anaerobic, extremely thermophilic plant biomass-degrading bacteria Caldicellulosiruptor hydrothermalis, Caldicellulosiruptor kristjanssonii, Caldicellulosiruptor kronotskyensis, Caldicellulosiruptor owensensis, and Caldicellulosiruptor lactoaceticus. J. Bacteriol. 193, 1483–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee L. L., Izquierdo J. A., Blumer-Schuette S. E., Zurawski J. V., Conway J. M., Cottingham R. W., Huntemann M., Copeland A., Chen I. M., Kyrpides N., Markowitz V., Palaniappan K., Ivanova N., Mikhailova N., Ovchinnikova G., Andersen E., Pati A., Stamatis D., Reddy T. B., Shapiro N., Nordberg H. P., Cantor M. N., Hua S. X., Woyke T., and Kelly R. M. (2015) Complete genome sequences of Caldicellulosiruptor sp. Strain Rt8.B8, Caldicellulosiruptor sp. Strain Wai35.B1, and “Thermoanaerobacter cellulolyticus”. Genome Announc. 10.1128/genomeA.00440-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Elkins J. G., Lochner A., Hamilton-Brehm S. D., Davenport K. W., Podar M., Brown S. D., Land M. L., Hauser L. J., Klingeman D. M., Raman B., Goodwin L. A., Tapia R., Meincke L. J., Detter J. C., Bruce D. C., Han C. S., Palumbo A. V., Cottingham R. W., Keller M., and Graham D. E. (2010) Complete genome sequence of the cellulolytic thermophile Caldicellulosiruptor obsidiansis OB47T. J. Bacteriol. 192, 6099–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van de Werken H. J., Verhaart M. R., VanFossen A. L., Willquist K., Lewis D. L., Nichols J. D., Goorissen H. P., Mongodin E. F., Nelson K. E., van Niel E. W., Stams A. J., Ward D. E., de Vos W. M., van der Oost J., Kelly R. M., and Kengen S. W. (2008) Hydrogenomics of the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Appl. Environ. Microbiol. 74, 6720–6729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kataeva I. A., Yang S. J., Dam P., Poole F. L. 2nd, Yin Y., Zhou F., Chou W. C., Xu Y., Goodwin L., Sims D. R., Detter J. C., Hauser L. J., Westpheling J., and Adams M. W. (2009) Genome sequence of the anaerobic, thermophilic, and cellulolytic bacterium “Anaerocellum thermophilum” DSM 6725. J. Bacteriol. 191, 3760–3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Conway J. M., Zurawski J. V., Lee L. L., Blumer-Schuette S. E., and Kelly R. M. (2015) in Thermophilic Microorganisms (Li F., ed) pp. 91–120, Caister Academic Press, Norfolk, UK [Google Scholar]
- 59. Blumer-Schuette S. E., Giannone R. J., Zurawski J. V., Ozdemir I., Ma Q., Yin Y., Xu Y., Kataeva I., Poole F. L. 2nd, Adams M. W., Hamilton-Brehm S. D., Elkins J. G., Larimer F. W., Land M. L., Hauser L. J., Cottingham R. W., Hettich R. L., and Kelly R. M. (2012) Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J. Bacteriol. 194, 4015–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Blumer-Schuette S. E., Lewis D. L., and Kelly R. M. (2010) Phylogenetic, microbiological, and glycoside hydrolase diversities within the extremely thermophilic, plant biomass-degrading genus Caldicellulosiruptor. Appl. Environ. Microbiol. 76, 8084–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blumer-Schuette S. E., Brown S. D., Sander K. B., Bayer E. A., Kataeva I., Zurawski J. V., Conway J. M., Adams M. W. W., and Kelly R. M. (2014) Thermophilic lignocellulose deconstruction. FEMS Microbiol. Rev. 38, 393–448 [DOI] [PubMed] [Google Scholar]
- 62. Blumer-Schuette S. E., Alahuhta M., Conway J. M., Lee L. L., Zurawski J. V., Giannone R. J., Hettich R. L., Lunin V. V., Himmel M. E., and Kelly R. M. (2015) Discrete and structurally unique proteins (tāpirins) mediate attachment of extremely thermophilic Caldicellulosiruptor species to cellulose. J. Biol. Chem. 290, 10645–10656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dam P., Kataeva I., Yang S. J., Zhou F., Yin Y., Chou W., Poole F. L. 2nd, Westpheling J., Hettich R., Giannone R., Lewis D. L., Kelly R., Gilbert H. J., Henrissat B., Xu Y., and Adams M. W. (2011) Insights into plant biomass conversion from the genome of the anaerobic thermophilic bacterium Caldicellulosiruptor bescii DSM 6725. Nucleic Acids Res. 39, 3240–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chung D., Farkas J., and Westpheling J. (2013) Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement. Biotechnol. Biofuels 6, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cha M., Chung D., Elkins J. G., Guss A. M., and Westpheling J. (2013) Metabolic engineering of Caldicellulosiruptor bescii yields increased hydrogen production from lignocellulosic biomass. Biotechnol. Biofuels 6, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gibson D. G. (2011) Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 498, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 68. Geslin C., Le Romancer M., Erauso G., Gaillard M., Perrot G., and Prieur D. (2003) PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote, “Pyrococcus abyssi”. J. Bacteriol. 185, 3888–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Farkas J., Chung D., Cha M., Copeland J., Grayeski P., and Westpheling J. (2013) Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J. Ind. Microbiol. Biotechnol. 40, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zurawski J. V., Conway J. M., Lee L. L., Simpson H. J., Izquierdo J. A., Blumer-Schuette S., Nookaew I., Adams M. W. W., and Kelly R. M. (2015) Comparative analysis of extremely thermophilic Caldicellulosiruptor species reveals common and differentiating cellular strategies for plant biomass utilization. Appl. Environ. Microbiol. 81, 7159–7170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Petersen T. N., Brunak S., von Heijne G., and Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 72. Ghose T. K. (1987) Measurement of cellulase activities. Pure Appl. Chem. 59, 257–268 [Google Scholar]
- 73. Miller G. L. (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 31, 426–428 [Google Scholar]
- 74. Xiao Z., Storms R., and Tsang A. (2005) Microplate-based carboxymethylcellulose assay for endoglucanase activity. Anal. Biochem. 342, 176–178 [DOI] [PubMed] [Google Scholar]
- 75. Xiao Z., Storms R., and Tsang A. (2004) Microplate-based filter paper assay to measure total cellulase activity. Biotechnol. Bioeng. 88, 832–837 [DOI] [PubMed] [Google Scholar]
- 76. VanFossen A. L., Ozdemir I., Zelin S. L., and Kelly R. M. (2011) Glycoside hydrolase inventory drives plant polysaccharide deconstruction by the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus. Biotechnol. Bioeng. 108, 1559–1569 [DOI] [PubMed] [Google Scholar]
- 77. Lipscomb G. L., Stirrett K., Schut G. J., Yang F., Jenney F. E. Jr., Scott R. A., Adams M. W., and Westpheling J. (2011) Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zhao Y., Meng K., Luo H., Huang H., Yuan T., Yang P., and Yao B. (2013) Molecular and biochemical characterization of a new alkaline active multidomain xylanase from alkaline wastewater sludge. World J. Microbiol. Biotechnol. 29, 327–334 [DOI] [PubMed] [Google Scholar]
- 79. Yi Z., Su X., Revindran V., Mackie R. I., and Cann I. (2013) Molecular and biochemical analyses of CbCel9A/Cel48A, a highly secreted multi-modular cellulase by Caldicellulosiruptor bescii during growth on crystalline cellulose. PLoS One 8, e84172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ye L., Su X., Schmitz G. E., Moon Y. H., Zhang J., Mackie R. I., and Cann I. K. O. (2012) Molecular and biochemical analyses of the GH44 Module of CbMan5B/Cel44A, a bifunctional enzyme from the hyperthermophilic bacterium Caldicellulosiruptor bescii. Appl. Environ. Microbiol. 78, 7048–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Brunecky R., Alahuhta M., Bomble Y. J., Xu Q., Baker J. O., Ding S. Y., Himmel M. E., and Lunin V. V. (2012) Structure and function of the Clostridium thermocellum cellobiohydrolase A X1-module repeat: enhancement through stabilization of the CbhA complex. Acta Crystallogr. D Biol. Crystallogr. 68, 292–299 [DOI] [PubMed] [Google Scholar]
- 82. Schneewind O., and Missiakas D. M. (2012) Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1123–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Bergquist P. L., Gibbs M. D., Morris D. D., Te'o V. S. J., Saul D. J., and Morgan H. W. (1999) Molecular diversity of thermophilic cellulolytic and hemicellulolytic bacteria. FEMS Microbiol. Ecol. 28, 99–110 [Google Scholar]
- 84. Gibbs M. D., Reeves R. A., Farrington G. K., Anderson P., Williams D. P., and Bergquist P. L. (2000) Multidomain and multifunctional glycosyl hydrolases from the extreme thermophile Caldicellulosiruptor isolate Tok7B.1. Curr. Microbiol. 40, 333–340 [DOI] [PubMed] [Google Scholar]
- 85. Kureishy N., Sapountzi V., Prag S., Anilkumar N., and Adams J. C. (2002) Fascins, and their roles in cell structure and function. Bioessays 24, 350–361 [DOI] [PubMed] [Google Scholar]
- 86. Thomas C., Tholl S., Moes D., Dieterle M., Papuga J., Moreau F., and Steinmetz A. (2009) Actin bundling in plants. Cell Motil. Cytoskeleton 66, 940–957 [DOI] [PubMed] [Google Scholar]
- 87. Charrier B., Le Bail A., and de Reviers B. (2012) Plant proteus: brown algal morphological plasticity and underlying developmental mechanisms. Trends Plant Sci. 17, 468–477 [DOI] [PubMed] [Google Scholar]
- 88. Verma D. P., and Hong Z. (2001) Plant callose synthase complexes. Plant Mol. Biol. 47, 693–701 [DOI] [PubMed] [Google Scholar]
- 89. Dumas C., and Knox R. B. (1983) Callose and determination of pistil viability and incompatibility. Theor. Appl. Genet. 67, 1–10 [DOI] [PubMed] [Google Scholar]
- 90. Harada T. (1992) The story of research into curdlan and the bacteria producing it. Trends Glycosci. Glycotechnol. 4, 309–317 [Google Scholar]
- 91. Vanfossen A. L., Verhaart M. R., Kengen S. M., and Kelly R. M. (2009) Carbohydrate utilization patterns for the extremely thermophilic bacterium Caldicellulosiruptor saccharolyticus reveal broad growth substrate preferences. Appl. Environ. Microbiol. 75, 7718–7724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Liebl W., Winterhalter C., Baumeister W., Armbrecht M., and Valdez M. (2008) Xylanase attachment to the cell wall of the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 190, 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., Wang Z., Yamashita R. A., Zhang D., Zheng C., and Bryant S. H. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., and Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S. (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.