Abstract
Lysine methylation of non-histone proteins has emerged as a key regulator of many cellular functions. Although less studied than other post-translational modifications such as phosphorylation and acetylation, the number of known methylated non-histone proteins is rapidly expanding. We have identified the p21-activated kinase 4 (PAK4) as a new substrate for methylation by the protein lysine methyltransferase SETD6. Our data demonstrate that SETD6 methylates PAK4 both in vitro and at chromatin in cells. Interestingly, depletion of SETD6 in various cellular systems significantly hinders the activation of the Wnt/β-catenin target genes. PAK4 was recently shown to regulate β-catenin signaling, and we show that SETD6 is a key mediator of this pathway. In the presence of SETD6, the physical interaction between PAK4 and β-catenin is dramatically increased, leading to a significant increase in the transcription of β-catenin target genes. Taken together, our results uncover a new regulatory layer of the Wnt/β-catenin signaling cascade and provide new insight into SETD6 biology.
Keywords: β-catenin (β-catenin), chromatin, gene transcription, post-translational modification (PTM), S-adenosylmethionine (SAM), Wnt pathway, Wnt signaling, PAK4, SETD6, methylation
Introduction
Post-translational modifications (PTMs)2 contribute to extended protein network complexity and serve as a major mechanism for the regulation of many signaling pathways and biological processes (1). In recent years, it has become evident that like other PTMs, lysine methylation may be important not only for chromatin regulation but also for regulating key cellular signal transduction pathways (2, 3) via methylation of non-histone proteins (4, 5).
Lysine methylation is carried out by protein lysine methyltransferases (PKMTs) (6, 7). There are ∼50 members of this family, of which all but one contains a conserved SET domain that carries out the enzymatic activity (8). In recent years, we have characterized the enzymatic activity of the PKMT SETD6 (9, 10). SETD6 is a mono-methyltransferase that directly methylates RelA at Lys-310 to suppress the activation of NF-κB target genes. It was recently shown that mono-methylation of H2AZ by SETD6 at lysine 7 is required for the maintenance of embryonic stem cell self-renewal (11) and that SETD6 associates with several factors that control the expression of estrogen-responsive genes in the nuclear hormone receptor signaling pathway (12).
Using a proteomic approach (13), we identified a number of novel SETD6 substrates, one of which was PAK4 (p21-activated kinase 4). PAK4 is a member of a family of serine/threonine protein kinases and an effector of activated Cdc42. Cdc42 directly regulates PAK4 activity in mammalian cells through an auto-inhibitory domain (AID), which inhibits PAK4 catalytic activity (14, 15). PAK4 is involved in many cellular processes, playing a role in cytoskeletal organization, cell motility, and cell cycle regulation. Moreover, constitutive knock-out of PAK4 in mice is lethal during embryonic development (16–20). Recent work has shown that PAK4 regulates the Wnt signaling pathway by directly phosphorylating β-catenin at Ser-675, leading to its stabilization and to the enhancement of β-catenin-dependent transcriptional activity by interaction with the TCF/LEF transcriptional complex leading, to activation of Wnt target genes (21). The Wnt/β-catenin signaling pathway is critical for normal embryonic developmental patterning, cellular proliferation, and homeostasis, and its aberrant activity is linked to the progression of different human malignancies (22).
In this study we characterized SETD6 activity on PAK4 and linked this cross-talk to the regulation of the Wnt/β-catenin pathway. We show that SETD6 binds and methylates PAK4 both in vitro and in cells at chromatin. Depletion of SETD6 using the CRISPR/CAS9 system in various cell lines leads to a dramatic reduction in the expression of Wnt/β-catenin target genes. The presence of SETD6 on chromatin enhances the physical interaction between PAK4 and β-catenin. Simultaneously, PAK4 serves as the mediator of SETD6-β-catenin association. Together, our data suggest that the functional interplay between SETD6, PAK4, and β-catenin is critical for the activation of Wnt/β-catenin target genes.
Experimental Procedures
Cell Lines, Transfection, and Treatment
Human embryonic kidney cells (HEK-293T), human lung carcinoma cells (A549) and human breast adenocarcinoma cells (MDA-MB-231) were maintained in Dulbecco's modified Eagle's medium (Sigma, D5671) with 10% fetal bovine serum (Gibco), 2 mg/ml l-glutamine (Sigma, G7513), penicillin-streptomycin (Sigma, P0781), and non-essential amino acids (Sigma, M7145). Cells were cultured at 37 °C in humidified incubator with 5% CO2. For cell transfection, cells were plated in 5- or 10-cm plates, and transfected using Mirus transfection reagents (TransIT®-LT1 for HEK-293T cells, or TransIT®-X2 for A549 and MDA-MB-231 cells), according to the manufacturer's instructions. LRP6 ΔN plasmid was used to activate the Wnt signaling pathway, and was transfected for 24–48 h. For CRISPR/CAS9-mediated gene disruption, four different sgRNAs for SETD6 were cloned to the lentiCRISPR plasmid (Addgene, #49535). Following transduction and puromycin selection, single clones were isolated and expanded.
Plasmids
The β-catenin coding sequence was excised from pFLAG-CMV-2 β-catenin (kindly provided by Inon Ben-Neriah) and cloned into pcDNA3.1 3×FLAG and pET-Duet plasmids for overexpression and protein purification, respectively. TOP-GFP and LRP6 ΔN plasmids were kindly provided by Amir Orian. For MBT pull-down assays, GST-3xMBT (from pGEX-3xMBT, provided by Or Gozani) was subcloned into pET-Duet. PAK4 and SETD6 sequences were amplified by PCR and were subcloned into pcDNA3.1 3×FLAG and pcDNA3.1 3×HA plasmids. For recombinant protein purification, PAK4 was cloned into pET-Duet and pET-Sumo plasmids.
Recombinant Protein Expression and Purification
Escherichia coli BL21 transformed with a plasmid expressing a protein of interest were grown in LB medium. Bacteria were harvested by centrifugation after IPTG induction and lysed by sonication on ice (18% amplitude, 1 min total, 10 s on/off). The GST fusion proteins were purified on a glutathione-Sepharose 4B (GE) and eluted with 10 mg/ml reduced glutathione (Sigma). His-tagged purified proteins were purified on Ni-NTA beads (Pierce), eluted with 500 mm imidazole buffer followed by an overnight dialysis. Recombinant SETD6 was overexpressed and purified from insect cells as previously described at Levy et al. (10).
Antibodies, Immunoprecipitation, MBT Pull-down, and Western Blot Analysis
Rabbit polyclonal antibodies used are: anti-PAK4 (Cell Signaling, C-3242S), anti-β-catenin (Abcam, ab32572), anti-pan-methyl (Abcam, ab23366), and anti-GFP (MBL, 598). Mouse monoclonal antibodies used are: anti-SETD6 (Genetex, GTX629891), anti-FLAG M2 (Sigma, F1804), anti-HA (Millipore, 05–904), anti-actin (Abcam, ab3280), anti-tubulin (Abcam, ab44928), and anti-histone3 (H3) (Abcam, ab10799). HRP-conjugated secondary antibodies (goat anti-rabbit and goat anti-mouse) were purchased from Jackson ImmunoResearch (111-035-144 and 115-035-062, respectively). Antibodies were diluted and prepared in TBST with 5% BSA or in PBST with 10% skim milk, according to manufacturer's recommendations. For Western blot analysis, cells were homogenized and lysed in RIPA buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm DTT, and 1:100 protease inhibitor mixture (Sigma)). Samples were heated at 95 °C for 5 min in Laemmli sample buffer and resolved by SDS-PAGE, followed by Western blot analysis.
Immunoprecipitation was performed using anti-FLAG M2 Affinity gel (Sigma, A2220) or A/G-agarose beads (Santa Cruz Biotechnology, SC-2003) conjugated to the antibody of interest. Briefly, ∼200 μg of proteins extracted from cells using RIPA buffer were incubated overnight at 4 °C with 15 μl of pre-washed FLAG M2 beads or A/G beads. The beads were then washed three times with RIPA buffer, heated for 5 min in Laemmli sample buffer at 95 °C and resolved on 7–10% SDS-PAGE gel followed by Western blot analysis. MBT pull-downs were performed as previously described (23, 24). To increase the solubility of the chromatin, cells were treated with benzonase nuclease enzyme (Sigma) for 30 min at 4 °C prior to the pull-down procedure.
RNA Extraction, Reverse Transcription, and Real-time qPCR
RNA extraction was performed using the NucleoSpin RNA Kit (Macherey-Nagel). Extracted RNA was reverse-transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturer's instructions. Real-time qPCR was carried out using the UPL probe library system (Roche). The real-time qPCR primers were the following; GAPDH: forward, 5′-AGCCACATCGCTCAGACAC-3′, reverse, 5′-GCCCAATACGACCAAATCC-3′; CCND1:forward, 5′-GCTGTGCATCTACACCGACA-3′, reverse, 5′-TTGAGCTTGTTCACCAGGAG-3′; MET: forward, 5′-TCGATCAGGACCATCAACC-3′, reverse, 5′-TCAATGGGATCTTCGTGATCT-3′; MYC: forward, 5′-CGGTTTTCGGGGCTTTAT-3′, reverse, 5′-GGCTCTTCCACCCTAGCC-3′; SOX9: forward, 5′-GTACCCGCACTTGCACAAC-3′, reverse, 5′-TCTCGCTCTCGTTCAGAAGTC-3′; YAP: forward, 5′-AATCCCACTCCCGACAGG-3′, reverse, 5′-GACTACTCCAGTGGGGGTCA-3′; CTBP1: forward, 5′-CGAGTCGGAACCCTTCAG-3′, reverse, 5′-CAGATGAGGTTGGGTGCAT-3′; JUN: forward, 5′-CCAAAGGATAGTGCGATGTTT-3′, reverse, 5′-CTGTCCCTCTCCACTGCAAC-3′; SENP2:forward, 5′-TCACTGGCTCAATGATGAAGTC-3′, reverse, 5′-AGTGCTGGATAGCCTTGCTT-3′.
All samples were amplified in triplicates in a 384-well plate using the following cycling conditions: 10 min at 95 °C, followed by 45 cycles of amplification at 95 °C for 10 s, 30 s at 60 °Cand 1 s at 72 °C. The reaction was performed using 0.2 μl of cDNA, 0.45 μl of each primer, and LightCycler® 480 Probes Master Mix for total volume reaction of 12 μl. Gene expression levels were analyzed relative to the housekeeping gene GAPDH and controls of the experiment.
In Vitro Methylation Assay
For assays with recombinant proteins, the methylation assay reaction (total volume of 25 μl) contained 4 μg of a substrate protein and 4 μg of GST-SETD6, 2mCi of 3H-labeled S-adenosyl-methionine (AdoMet, Perkin-Elmer) and PKMT buffer (20 mm Tris-HCl, pH 8, 10% glycerol, 20 mm KCl, 5 mm MgCl2). The reaction tube was incubated overnight at 30 °C. For assays using proteins immunoprecipitated from mammalian cells, FLAG-PAK4 or empty plasmid transfections were immunoprecipitated using FLAG M2 beads, and then incubated overnight at 30 °C in a 25-μl reaction, containing 4 μg of recombinant GST-SETD6, 2 mCi 3H-labeled S-adenosyl-methionine and PKMT buffer. These reactions were resolved by SDS-PAGE for Coomassie staining (Expedeon, InstantBlue) and exposed to autoradiogram.
Enzyme-linked Immunosorbent Assay (ELISA)
2 μg of recombinant proteins (BSA, His-PAK4 or His-β-catenin) diluted in phosphate-buffered saline (PBS) were added to a “sticky surface” (Greiner Microlon) 96-well plate and incubated for 1 h at room temperature followed by overnight incubation at 4 °C with 3% BSA in PBST. After three washes with PBST, the plate was covered with 0.5 μg of GST-SETD6 or GST protein (negative control) diluted in 1% BSA in PBST for 1 h at room temperature. Plates were then washed and incubated with primary antibody (anti-GST, 1:4000 dilution) followed by incubation with HRP-conjugated secondary antibody (goat anti-rabbit, 1:2000 dilution) for 1 h. Finally, TMB reagent and then 1N H2SO4 were added, and the absorbance at 450 nm was detected using a Tecan Infinite M200 plate reader. Results are represented as relative absorbance fold compared with GST or BSA.
Biochemical Fractionation
Biochemical fractionation was performed as previously described (25), with a final step added. Briefly, the chromatin pellet was resuspended for 30 min on ice in buffer C (RIPA buffer, 1 mm MgCl2 and benzonase nuclease enzyme (Sigma)), and supernatant was collected by low-speed centrifugation (5 min, 1,700 × g, 4 °C).
Protein-Protein Chromatin Immunoprecipitation (ChIP)
Protein-protein ChIP was modified from a published protocol (26). After cross-linking, cells were harvested and washed twice with PBS and then lysed in 1 ml of lysis buffer (20 mm Tris-HCl, pH 8, 85 mm KCl, 0.5% Nonidet P-40, and 1:100 protease inhibitor mixture) for 10 min on ice. Nuclear pellets were resuspended in 200 μl of nuclei lysis buffer (50 mm Tris-HCl pH 8, 10 mm EDTA, 1% SDS, 1:100 protease inhibitor mixture) for 10 min on ice, and then sonicated (Bioruptor, Diagenode) with high power settings for 3 cycles, 6 min each cycle (30 s on/off). Samples were centrifuged (17 min, 13,000 rpm, 4 °C), and the soluble chromatin fraction was collected. The soluble chromatin was immunoprecipitated as described above, washed according to the published protocol, resolved by SDS-PAGE gel, and analyzed by Western blot.
Results
SETD6 Methylates and Binds PAK4 in Vitro
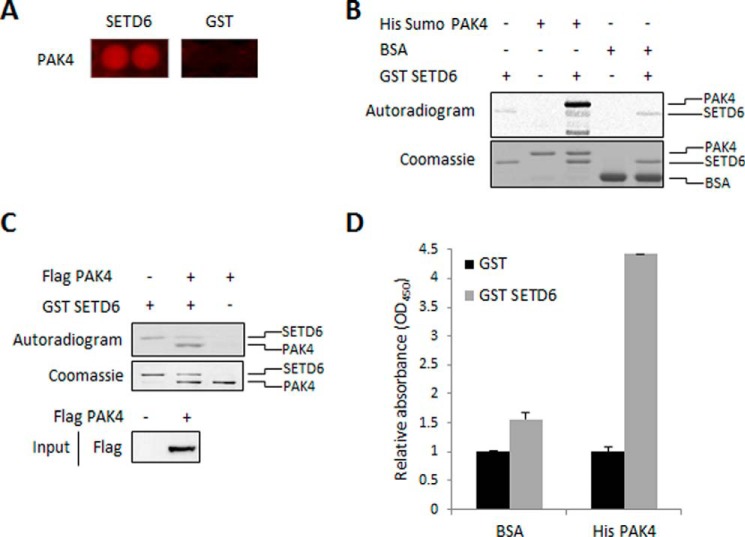
In a proteomic screen for novel SETD6 substrates, we previously identified 118 proteins that were methylated by SETD6 (13). One of these newly identified SETD6 substrates was PAK4 (Fig. 1A). To validate this finding we performed an in vitro methylation assay in the presence of purified recombinant His-Sumo-PAK4 and GST-SETD6. As shown in Fig. 1B, recombinant PAK4 is methylated by GST-SETD6. BSA served as a negative control for the experiment. Consistent with these results, we found that immunoprecipitated FLAG-PAK4 from HEK-293T cells is methylated in vitro by recombinant SETD6 (Fig. 1C). Furthermore, a direct interaction between the two proteins was demonstrated by ELISA experiment (Fig. 1D). From these data we concluded that SETD6 interacts with and methylates PAK4 in vitro.
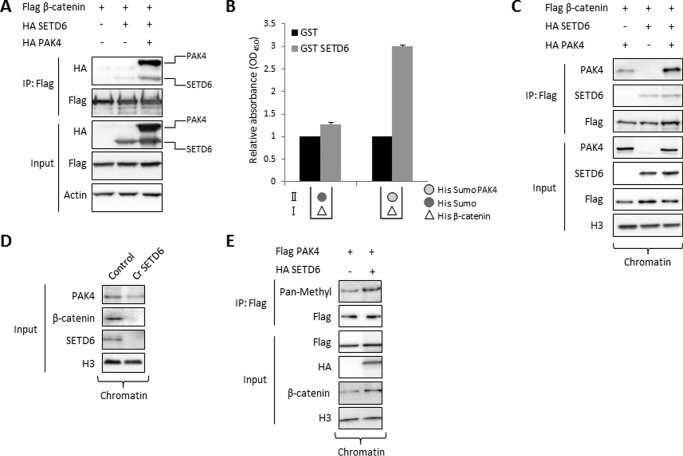
FIGURE 1.
SETD6 interacts and methylates PAK4 in vitro. A, protoarray experiment showed a positive signal for methylation of recombinant PAK4 protein by GST-SETD6, contrary to GST protein as a negative control (13). Protoarrays were incubated overnight under PKMT reaction conditions with GST-SETD6 and GST as a negative control. Arrays were then probed with a pan-methyl antibody followed by a fluorophore-conjugated secondary antibody. B, in vitro methylation assay in the presence of 3H-labeled SAM with recombinant His-Sumo-PAK4, BSA (negative control), and GST-SETD6. Coomassie stain of the recombinant proteins used in the reactions is shown on the bottom. C, HEK-293T cells were transfected with FLAG-PAK4 or empty plasmid. Cell extracts were immunoprecipitated (IP) with FLAG M2 beads, followed by a radioactive in vitro methylation assay in the presence of GST-SETD6. Coomassie Blue stain shows the immunoprecipitated FLAG-PAK4 and the added GST-SETD6. Anti-FLAG Western blot represents input of FLAG-PAK4 transfected to HEK-293T cells (bottom panel). D, ELISA-based analysis of the interaction between recombinant GST-SETD6 and His-PAK4. The 96-well plate was coated with 2 μg of His-PAK4 or BSA as control, and then covered with 0.5 μg of GST-SETD6 or GST. Signal detection was achieved using primary anti-GST antibody and secondary HRP-conjugated anti-rabbit antibody and normalized to GST signal. Data are from at least three experiments (error bars, S.E.).
SETD6 Methylates and Binds PAK4 in Cells
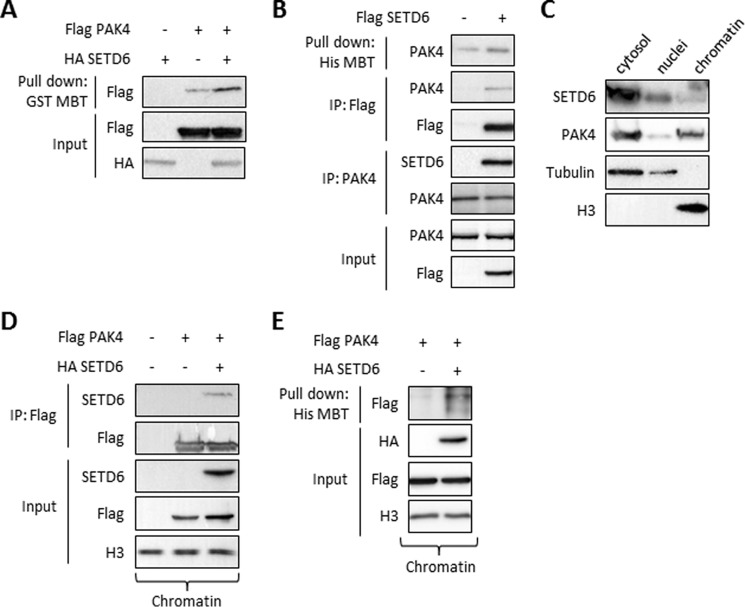
To determine whether SETD6 methylates PAK4 in cells, we have utilized the MBT pull-down assay (23, 24). MBT serves as an affinity reagent to perform direct pull-downs of methylated lysine residues in cells followed by Western blot analysis (23, 24). Overexpression of FLAG-PAK4 and HA-SETD6 in HEK-293T cells, followed by protein extraction and MBT pull-down, revealed that PAK4 is methylated in cells and that methylation is significantly increased in the presence of SETD6 (Fig. 2A). This was also the case for endogenous PAK4 (Fig. 2B, upper panel). In a co-immunoprecipitation we observed that overexpressed SETD6 interacts with endogenous PAK4 in cells (Fig. 2B). The interaction between SETD6 and PAK4 was also confirmed in a reciprocal experiment where we immunoprecipitated endogenous PAK4 (Fig. 2B). Based on this set of experiments, we have concluded that SETD6 methylates and binds PAK4 in cells.
FIGURE 2.
SETD6 binds and methylates PAK4 in cells. A, HEK-293T cells were transfected with HA-SETD6 or FLAG-PAK4 or both. Methylated proteins from cellular extracts were pulled-down with GST-MBT to detect methylation differences on FLAG-PAK4 with or without HA-SETD6. B, HEK-293T cells were transfected with FLAG-SETD6 or empty plasmid. Cell extracts were then taken for His-MBT pull-down assay (His MBT panel), IP with FLAG M2 beads (IP: FLAG panel), and a reciprocal IP using anti-PAK4 antibody (IP: PAK4 panel). Input of endogenous PAK4 and FLAG-SETD6 is shown in the bottom panel. C, immunoblot analysis of HEK-293T cells biochemically separated into cytoplasmic (cytosol), nucleoplasmic (nuclei), or chromatin-enriched (chromatin) fractions; tubulin and H3 signals serve as controls for fractionation integrity. D, HEK-293T cells were transfected with empty plasmid or FLAG-PAK4 alone or with HA-SETD6. Cells were submitted to chromatin isolation using a protein-protein ChIP protocol. The chromatin fraction was immunoprecipitated with FLAG M2 beads, followed by Western blot analysis with the indicated antibodies. E, HEK-293T cells were transfected with FLAG-PAK4 alone or with HA-SETD6 plasmid. Chromatin fraction was taken for His-MBT pull-down assay as described in B.
SETD6 and PAK4 Interact at Chromatin
In biochemical fractionation experiments, that separate the cytosol from the nucleus we observed that endogenous SETD6 and PAK4 localized both to the cytosol and the chromatin fractions (Fig. 2C). Based on our previous findings that SETD6 plays a key role in regulating NF-κB transcriptional activity (10) and along with our new finding that SETD6 is localized to chromatin, we postulated that the interaction and the methylation of PAK4 by SETD6 takes place at chromatin. Indeed, chromatin isolation, followed by co-immunoprecipitation of overexpressed FLAG-PAK4 and HA-SETD6 from HEK-293T cells, revealed a strong interaction between the two proteins (Fig. 2D). Furthermore, MBT pull-down assay on chromatin-isolated fraction revealed that PAK4 is methylated by SETD6 at chromatin (Fig. 2E). Collectively these data demonstrate that chromatin associated PAK4 is bound to and methylated by SETD6.
SETD6 Is a Positive Regulator of the Wnt/β-Catenin Pathway
PAK4 constitutively cycles between the nucleus and cytoplasm and modulates the dynamic nucleo-cytoplasmic trafficking and signaling activity of β-catenin in the Wnt pathway (21). We therefore hypothesized that SETD6 regulates Wnt/β-catenin signaling. To investigate the relationship between SETD6 and the Wnt signaling pathway we took advantage of the TOP-GFP system which consists of multimerized TCF/LEF binding sites fused to GFP and allows the monitoring of β-catenin transcriptional activity (27, 28).
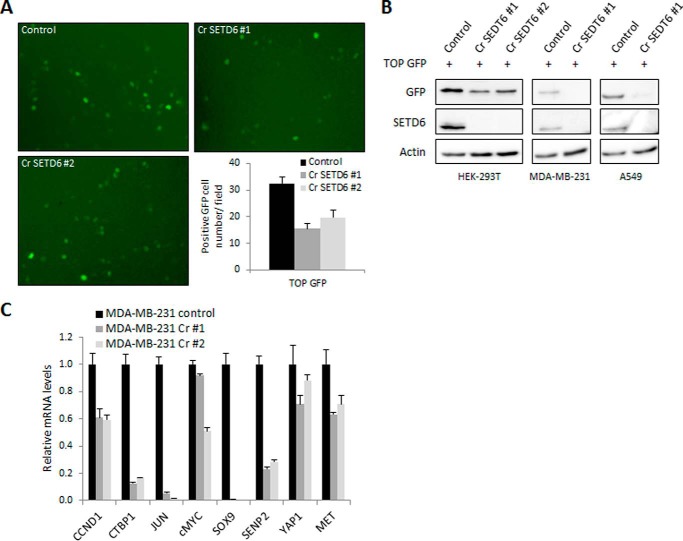
We used CRISPR/CAS9 in HEK-293T cells to generate two SETD6 knock-out cell lines (Fig. 3B). When we transfected wild-type cells and the two knock-out cell lines with the TOP-GFP plasmid, we observed a 50% reduction in the expression of GFP-positive cells in CRISPR-SETD6 cells compared with wild-type cells (Fig. 3A, microscopic images and quantification, respectively). Western blot analysis of the TOP-GFP expression in wild-type compared with CRISPR-SETD6 knock-out HEK-293T, MDA-MB-231, and A549 cells revealed the same trend (Fig. 3B). Next, RNA was extracted from MDA-MB-231 cells, and the expression of various β-catenin target genes was measured by qPCR. As shown in Fig. 3C, the basal expression levels of various β-catenin target genes were dramatically down-regulated in the CRISPR-SETD6 cells compared with the wild-type MDA-MB-231 cells. Taken together, these data strongly suggest that SETD6 plays a role in the regulation of the Wnt/β-catenin transcriptional program under basal conditions, and that SETD6 and β-catenin participate at the same pathway.
FIGURE 3.
SETD6 is a positive regulator of the Wnt signaling pathway. A, HEK-293T wild-type and CRISPR-SETD6 knock-out cells (Cr SETD6 #1 and #2) were transfected with TOP-GFP. 48 h post-transfection cells were visualized by fluorescence microscopy. The graph (bottom right) represents the quantification of GFP-positive cell numbers per field. Data are from at least three experiments (error bars, S.E.). B, HEK-293T, MDA-MB-231, and A549 wild-type and CRISPR-SETD6 knock-out cells were transfected with TOP-GFP plasmid and subjected to Western blot analysis with the indicated antibodies. C, qPCR of the indicated Wnt signaling target genes, from mRNA extracted from MDA-MB-231 CRISPR-SETD6 wild-type or knock-out cells. mRNA levels were normalized to GAPDH and to control cells. Data are from at least three experiments (error bars, S.E.).
SETD6 Does Not Methylate but Weakly Binds β-Catenin
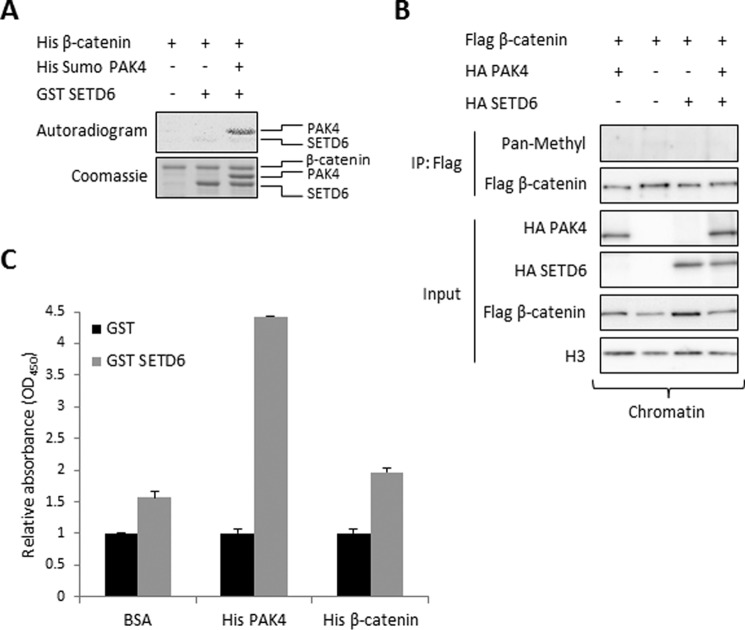
These results led us to hypothesize that SETD6 methylates β-catenin, and therefore we were encouraged to look for a direct interaction between them. In an in vitro methylation assay (Fig. 4A), we observed that β-catenin is not methylated by SETD6. To test whether PAK4 mediates β-catenin methylation, we added PAK4 to the reaction (Fig. 4A, lane 3). We found that while PAK4 is specifically methylated by SETD6, β-catenin is not.
FIGURE 4.
SETD6 does not methylate but weakly binds β-catenin. A, in vitro methylation assay with 3H-labeled SAM and the indicated recombinant proteins. Coomassie Blue stain represents all the proteins used in each reaction. B, immunoblot analysis of immunoprecipitated FLAG-β-catenin isolated from the chromatin fraction, which was co-expressed with the indicated plasmids. Methylation was detected with a pan-methyl antibody. C, ELISA-based analysis of the interaction between recombinant GST-SETD6 and His-PAK4 or His-β-catenin. The signal detection was achieved using primary anti-GST antibody and secondary HRP-conjugated anti-rabbit antibody and normalized to GST signal. Data are from at least three experiments (error bars, S.E.).
To determine whether β-catenin is methylated by SETD6 in cells we immunoprecipitated chromatin associated FLAG-β-catenin with or without overexpression of HA-SETD6 followed by a Western blot with a pan-methyl antibody. As shown in Fig. 4B, SETD6 did not methylate β-catenin also in the presence of PAK4. To test if there is a direct physical interaction between SETD6 and β-catenin, we performed an ELISA experiment using recombinant proteins (Fig. 4C). In this experiment, we confirmed the strong association between SETD6 and PAK4. Surprisingly, we could also observe a weak interaction between SETD6 and β-catenin. Taken together, these data indicate that SETD6 does not methylate β-catenin; however a weak interaction is observed between these two proteins.
SETD6 Promotes the Interaction of PAK4 and β-Catenin at Chromatin
It was previously shown that PAK4 binds and phosphorylates β-catenin at Ser-675, contributing to its stability and promoting its transcriptional activity (29). Having demonstrated that (i) SETD6 methylates and interacts with PAK4 at chromatin (Figs. 1 and 2), (ii) SETD6 positively regulates Wnt/β-catenin signaling (Fig. 3) and (iii) β-catenin is not methylated by SETD6 in vitro and in cells (Fig. 4), we hypothesized that a three-protein SETD6-PAK4-β-catenin complex is formed at chromatin and that PAK4 serves as a linker. To address this hypothesis we tested the interaction between SETD6 and β-catenin in the presence or absence of PAK4 (Fig. 5A). Consistent with our ELISA experiment (Fig. 4C) we could identify a very weak interaction between SETD6 and β-catenin in cells. However, a significantly stronger physical interaction between FLAG-β-catenin and HA-SETD6 was observed in the presence of HA-PAK4. We confirmed this result by ELISA (Fig. 5B). In this experiment, the plate was covered with 3 layers of different recombinant proteins as follows: (i) His-β-catenin; (ii) His-Sumo-PAK4 or His-Sumo; and (iii) GST-SETD6 or GST. We found that the interaction between His-β-catenin and GST-SETD6 was significantly elevated only upon His-Sumo-PAK4 addition. Furthermore, the interaction between PAK4 and β-catenin at chromatin in CRISPR-SETD6 knock-out HEK-293T cells was significantly elevated after overexpressing SETD6 (Fig. 5C).
FIGURE 5.
SETD6 stabilizes the interaction of PAK4 and β-catenin at chromatin. A, HEK-293T cells were transfected with FLAG-β-catenin alone, with HA-SETD6, or with HA-SETD6 and HA-PAK4. Cell extracts were immunoprecipitated with FLAG M2 beads, and proteins in IP and input samples were detected by Western blot. B, 96-well plate was coated with 3 layers: first with 2 μg of His-β-catenin (white triangle), blocked with 3% BSA in PBST; then 3 μg of His-Sumo-PAK4 or His-Sumo (light and dark circle, respectively); and finally with 0.5 μg of GST-SETD6 or GST. Data are from at least three experiments (error bars, S.E.). C, HEK-293T CRISPR-SETD6 knock-out cells were transfected with the indicated plasmids. Isolated chromatin was immunoprecipitated with FLAG M2 beads, followed by Western blot analysis. Input samples of the chromatin fraction are also shown. D, chromatin fraction derived from wild-type and the CRISPR-SETD6 knock-out MDA-MB-231 cells were subjected to a Western blot analysis with the indicated antibodies. E, HEK-293T cells were transfected with the indicated plasmids and the methylation of immunoprecipitated FLAG-PAK4 at chromatin was detected with a pan-methyl antibody. The level of the chromatin-associated proteins (input) were detected with the indicated antibodies.
We then compared the expression level of endogenous PAK4 and β-catenin in the wild-type and the CRISPR-SETD6 knock-out MDA-MB-231 cells and found that the expression of both proteins at chromatin was dramatically reduced in the absence of SETD6 (Fig. 5D). In a reciprocal experiment we found that an increased methylation of PAK4 by overexpression of SETD6 correlates with higher levels of β-catenin at chromatin (Fig. 5E). Taken together, these data support a suggested model in which PAK4 methylation physically links SETD6 and β-catenin. Based on these results we hypothesized that SETD6 may be required for β-catenin transcriptional activity at chromatin.
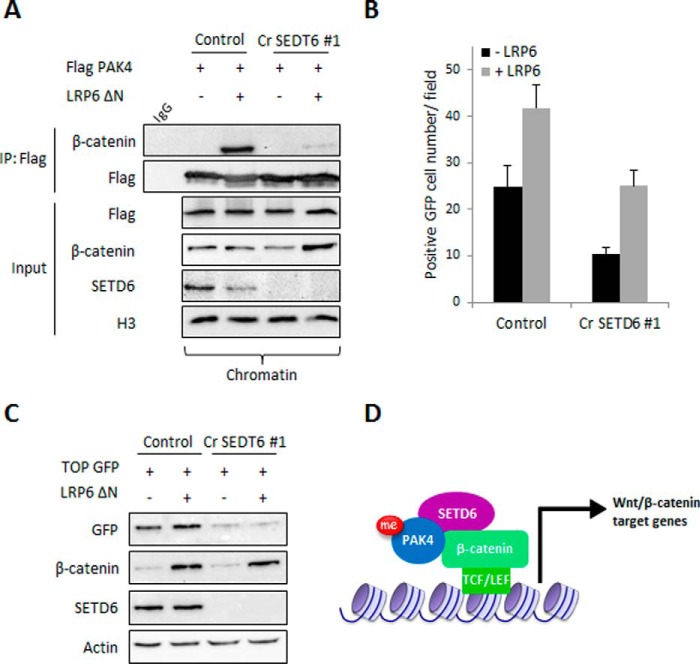
To determine whether SETD6 is an essential component of the transcription activation of the Wnt/β-catenin/PAK4 signaling pathway, control and CRISPR-SETD6 knock-out HEK-293T cells were transfected with FLAG-PAK4 and stimulated with LRP6 ΔN, a constitutively active receptor that induces β-catenin target genes activation (30). Chromatin-associated proteins were then immunoprecipitated by FLAG M2 beads, and the interaction between PAK4 and β-catenin was detected by Western blot (Fig. 6A). The interaction between PAK4 and endogenous β-catenin at chromatin was significantly elevated in response to activation by LPR6. Strikingly, in the SETD6 knock-out cells this interaction was almost completely abolished. These data suggest that the presence of SETD6 at chromatin is required for PAK4-mediated β-catenin target gene activation. To address this hypothesis, wild-type or CRISPR-SETD6 knock-out HEK-293T cells were transfected with TOP-GFP plasmid and activated with LRP6 ΔN. As seen in Fig. 6, B and C, stimulation of control cells led to a significant elevation of TOP-GFP expression. In contrast, a significant reduction in GFP signal was observed in the SETD6 knock-out cells. Interestingly, the number of GFP-positive cells in the SETD6 knock-out cells was higher after stimulation; however, this elevation is not beyond the basal activity level of the control cells. These results suggest that SETD6 is required for the activation of β-catenin target genes also in the absence of stimulation. Collectively, these data support a model in which SETD6 methylates PAK4 at chromatin. PAK4 and SETD6 cross-talk at chromatin promotes the formation of active β-catenin complex, which in turn leads to the efficient transcription activation of Wnt/β-catenin target genes (Fig. 6D).
FIGURE 6.
SETD6 is required for β-catenin transcriptional activity at chromatin. A, HEK-293T control and SETD6 knock-out cells were transfected with FLAG-PAK4 and with a control plasmid or LRP6 ΔN plasmid to activate the Wnt pathway. After 48 h, chromatin extracts were incubated with FLAG M2 beads and A/G beads conjugated to IgG as a control. Input shows the levels of FLAG-PAK4, β-catenin, SETD6, and H3 in the chromatin fraction. B, quantification of GFP-positive cell numbers in HEK-293T control and SETD6 knock-out cells transfected with TOP-GFP that were activated with LRP6 ΔN or transfected with empty plasmid. Data are from at least three experiments (error bars, S.E.). C, same as in B, except that after transfection, cells were harvested and subjected to Western blot analysis with the indicated antibodies. D, suggested model for how SETD6 regulates Wnt/β-catenin signaling.
Discussion
The Wnt signaling pathway is fundamental in controlling cell proliferation, embryonic development, and cell homeostasis (31, 32). One of the main mechanisms through which Wnt signaling triggers downstream physiological effects is by the modulation of β-catenin/TCF target genes activity (33). Thus, the regulation of such complex biological processes requires a fine-tuned mode of activity to ensure proper cellular homeostasis. In the present study we have added another layer of regulation that modulates the Wnt signaling transcription activity by β-catenin/TCF.
We show that the SETD6 methyltransferase plays a key role in regulating this pathway, as depletion of SETD6 hampers β-catenin transcriptional activity. We suggest that this process is mediated by SETD6-dependent interaction and methylation of PAK4, a kinase that phosphorylates β-catenin to regulate its stability and cellular localization (21). Our data indicate that the interaction between SETD6 and β-catenin is mediated by PAK4. Strikingly, PAK4 association with β-catenin is significantly stronger in the presence of SETD6. Therefore, we postulate that formation of a SETD6-PAK4-β-catenin complex at chromatin enhances the transcription of various Wnt/β-catenin target genes.
Like many other post-translational modifications, methylation-based signal transduction cellular mechanisms are fundamental in the regulation of many cellular signaling pathways (10, 34–39). In a recent report, Shen et al. (40) have shown that SETD7 methylates β-catenin on Lys-180 to regulate its stability and transcriptional activity. Based on our findings, SETD6 also serves as a positive regulator of β-catenin. Unlike SETD7, SETD6 does not methylate β-catenin (Fig. 4) but promotes β-catenin activity through PAK4. It is still not clear if these two methylation-based signaling regulations mediated by SETD7 and SETD6 work in concert or are mutually exclusive. Nevertheless, both modes of regulation demonstrate that the enzymatic activity of methyltransferase family members is required for the fine tuning of the Wnt signaling pathway.
Our results indicate that PAK4 and SETD6 are localized to the chromatin under un-stimulated conditions. In the absence of Wnt activation, cytosolic β-catenin is phosphorylated at multiple sites, triggering ubiquitination and proteasomal degradation (29). In these conditions, β-catenin localization to the nucleus is thus inhibited, leading to repression of target genes. Interestingly, under lack of activation we observed basal transcription activity of Wnt/β-catenin target genes in a SETD6-dependent manner. Basal transcription activity is required to maintain cellular homeostasis and here we show for the first time that SETD6 plays a key role in this physiological state. Nevertheless, we cannot exclude the possibility that other transcription factors that physically localize to β-catenin-specific target genes promoters at chromatin exert the same effect under nonstimulated conditions.
We have previously shown that SETD6 negatively regulates RelA activity at chromatin (10). This methylation signaling cascade is initiated by site-specific methylation of the nuclear factor NF-κB subunit RelA at Lys-310 by SETD6. Methylated RelA serves as a scaffold for subsequent recruitment and stabilization of the H3K9 PKMT GLP at chromatin which results in de novo histone methylation and leads to a constitutive repression of cell proliferation and inflammatory gene expression programs. In contrast, in this study we show that SETD6 promotes the transcription of Wnt/β-catenin target genes. These findings suggest that SETD6 can regulate transcription in two opposite manners. It is still not fully understood what determines SETD6's preference toward repression or activation of transcription; however, this can be explained by alterations of SETD6 expression and SETD6-mediated methylation of non-histone proteins, which can lead to activation or repression of transcription, depending on the cellular or physiological context.
In summary, we have identified a new lysine methylation regulatory mechanism that modulates the activity of the Wnt/β-catenin signaling pathway. SETD6-PAK4 cellular cross-talk enhances the association between PAK4 and β-catenin atchromatin which modulates the transcriptional activity of β-catenin. These findings reveal a new and important level of regulation of the Wnt/β-catenin pathway, providing fundamental insight into Wnt signaling and raising new possibilities for therapeutic intervention in disease states in which this pathway is dysregulated.
Author Contributions
Z. V., M. F., and D. L. conceived and designed the experiments. Z. V. performed the majority of the experiments. A. C. and D. L. generated the CRISPR cell lines. Z. V. and D. L. wrote the paper. All authors read and approved the final manuscript.
Acknowledgments
We thank Inon Ben-Neriah for the pFLAG-CMV-2 β-catenin plasmid; Amir Orian for providing the TOP-GFP and LRP6 ΔN plasmids; and Or Gozani for the pGEX-3xMBT plasmid. We also thank the Levy laboratory for technical assistance and Ruth Tennen for critical reading of the manuscript.
This work was supported by grants (to D. L.) from The Israel Science Foundation (285/14), The Research Career Development Award from the Israel Cancer Research Fund, and a Marie Curie Career Integration Grant (PCIG12-GA-2012–333242). The authors declare that they have no conflicts of interest with the contents of this article.
- PTM
- post-translational modifications
- PKMT
- protein lysine methyltransferase
- PAK4
- p21-activated kinase 4
- ChIP
- protein-protein chromatin immunoprecipitation
- SAM
- S-adenosylmethionine.
References
- 1. Pawson T., and Warner N. (2007) Oncogenic re-wiring of cellular signaling pathways. Oncogene 26, 1268–1275 [DOI] [PubMed] [Google Scholar]
- 2. Hamamoto R., Saloura V., and Nakamura Y. (2015) Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat. Rev. Cancer 15, 110–124 [DOI] [PubMed] [Google Scholar]
- 3. Alam H., Gu B., and Lee M. G. (2015) Histone methylation modifiers in cellular signaling pathways. Cell Mol. Life Sci. 72, 4577–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bremang M., Cuomo A., Agresta A. M., Stugiewicz M., Spadotto V., and Bonaldi T. (2013) Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. Biosyst. 9, 2231–2247 [DOI] [PubMed] [Google Scholar]
- 5. Guo A., Gu H., Zhou J., Mulhern D., Wang Y., Lee K. A., Yang V., Aguiar M., Kornhauser J., Jia X., Ren J., Beausoleil S. A., Silva J. C., Vemulapalli V., Bedford M. T., and Comb M. J. (2014) Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteomics 13, 372–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greer E. L., and Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 8. Arrowsmith C. H., Bountra C., Fish P. V., Lee K., and Schapira M. (2012) Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 [DOI] [PubMed] [Google Scholar]
- 9. Chang Y., Levy D., Horton J. R., Peng J., Zhang X., Gozani O., and Cheng X. (2011) Structural basis of SETD6-mediated regulation of the NF-κB network via methyl-lysine signaling. Nucleic Acids Res. 39, 6380–6389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Levy D., Kuo A. J., Chang Y., Schaefer U., Kitson C., Cheung P., Espejo A., Zee B. M., Liu C. L., Tangsombatvisit S., Tennen R. I., Kuo A. Y., Tanjing S., Cheung R., Chua K. F., Utz P. J., Shi X., Prinjha R. K., Lee K., Garcia B. A., Bedford M. T., Tarakhovsky A., Cheng X., and Gozani O. (2011) Lysine methylation of the NF-κB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat. Immunol. 12, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Binda O., Sevilla A., LeRoy G., Lemischka I. R., Garcia B. A., and Richard S. (2013) SETD6 monomethylates H2AZ on lysine 7 and is required for the maintenance of embryonic stem cell self-renewal. Epigenetics 8, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. O'Neill D. J., Williamson S. C., Alkharaif D., Monteiro I. C., Goudreault M., Gaughan L., Robson C. N., Gingras A. C., and Binda O. (2014) SETD6 controls the expression of estrogen-responsive genes and proliferation of breast carcinoma cells. Epigenetics 9, 942–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Levy D., Liu C. L., Yang Z., Newman A. M., Alizadeh A. A., Utz P. J., and Gozani O. (2011) A proteomic approach for the identification of novel lysine methyltransferase substrates. Epigenetics Chromatin 4, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Selamat W., Tay P. L., Baskaran Y., and Manser E. (2015) The Cdc42 Effector Kinase PAK4 Localizes to Cell-Cell Junctions and Contributes to Establishing Cell Polarity. PLoS ONE 10, e0129634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ha B. H., Davis M. J., Chen C., Lou H. J., Gao J., Zhang R., Krauthammer M., Halaban R., Schlessinger J., Turk B. E., and Boggon T. J. (2012) Type II p21-activated kinases (PAKs) are regulated by an autoinhibitory pseudosubstrate. Proc. Natl. Acad. Sci. U.S.A. 109, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Abo A., Qu J., Cammarano M. S., Dan C., Fritsch A., Baud V., Belisle B., and Minden A. (1998) PAK4, a novel effector for Cdc42Hs, is implicated in the reorganization of the actin cytoskeleton and in the formation of filopodia. EMBO J. 17, 6527–6540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Baskaran Y., Ng Y. W., Selamat W., Ling F. T., and Manser E. (2012) Group I and II mammalian PAKs have different modes of activation by Cdc42. EMBO Rep. 13, 653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bompard G., Rabeharivelo G., Cau J., Abrieu A., Delsert C., and Morin N. (2012) P21-activated kinase 4 (PAK4) is required for metaphase spindle positioning and anchoring. Oncogene [DOI] [PubMed] [Google Scholar]
- 19. Park M. H., Lee H. S., Lee C. S., You S. T., Kim D. J., Park B. H., Kang M. J., Heo W. D., Shin E. Y., Schwartz M. A., and Kim E. G. (2012) p21-Activated kinase 4 promotes prostate cancer progression through CREB. Oncogene 32, 2475–2482 [DOI] [PubMed] [Google Scholar]
- 20. Whale A., Hashim F. N., Fram S., Jones G. E., and Wells C. M. (2011) Signalling to cancer cell invasion through PAK family kinases. Front. Biosci. 16, 849–864 [DOI] [PubMed] [Google Scholar]
- 21. Li Y., Shao Y., Tong Y., Shen T., Zhang J., Gu H., and Li F. (2012) Nucleo-cytoplasmic shuttling of PAK4 modulates β-catenin intracellular translocation and signaling. Biochim. Biophys. Acta 1823, 465–475 [DOI] [PubMed] [Google Scholar]
- 22. Bikkavilli R. K., Feigin M. E., and Malbon C. C. (2008) p38 mitogen-activated protein kinase regulates canonical Wnt-β-catenin signaling by inactivation of GSK3β. J. Cell Sci. 121, 3598–3607 [DOI] [PubMed] [Google Scholar]
- 23. Carlson S. M., Moore K. E., Green E. M., Martin G. M., and Gozani O. (2014) Proteome-wide enrichment of proteins modified by lysine methylation. Nat. Protoc. 9, 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore K. E., Carlson S. M., Camp N. D., Cheung P., James R. G., Chua K. F., Wolf-Yadlin A., and Gozani O. (2013) A general molecular affinity strategy for global detection and proteomic analysis of lysine methylation. Mol. Cell 50, 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Méndez J., and Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nelson J. D., Denisenko O., and Bomsztyk K. (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 27. Vermeulen L., De Sousa E. M. F., van der Heijden M., Cameron K., de Jong J. H., Borovski T., Tuynman J. B., Todaro M., Merz C., Rodermond H., Sprick M. R., Kemper K., Richel D. J., Stassi G., and Medema J. P. (2010) Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 12, 468–476 [DOI] [PubMed] [Google Scholar]
- 28. Reya T., Duncan A. W., Ailles L., Domen J., Scherer D. C., Willert K., Hintz L., Nusse R., and Weissman I. L. (2003) A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 [DOI] [PubMed] [Google Scholar]
- 29. MacDonald B. T., Tamai K., and He X. (2009) Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu G., Bafico A., Harris V. K., and Aaronson S. A. (2003) A novel mechanism for Wnt activation of canonical signaling through the LRP6 receptor. Mol. Cell. Biol. 23, 5825–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herbst A., Jurinovic V., Krebs S., Thieme S. E., Blum H., Göke B., and Kolligs F. T. (2014) Comprehensive analysis of β-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/β-catenin signaling. BMC Genomics 15, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lien W. H., and Fuchs E. (2014) Wnt some lose some: transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 28, 1517–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin T., George Fantus I., and Sun J. (2008) Wnt and beyond Wnt: multiple mechanisms control the transcriptional property of β-catenin. Cell Signal. 20, 1697–1704 [DOI] [PubMed] [Google Scholar]
- 34. Mazur P. K., Reynoird N., Khatri P., Jansen P. W., Wilkinson A. W., Liu S., Barbash O., Van Aller G. S., Huddleston M., Dhanak D., Tummino P. J., Kruger R. G., Garcia B. A., Butte A. J., Vermeulen M., Sage J., and Gozani O. (2014) SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shi X., Kachirskaia I., Yamaguchi H., West L. E., Wen H., Wang E. W., Dutta S., Appella E., and Gozani O. (2007) Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol. Cell 27, 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang J., Huang J., Dasgupta M., Sears N., Miyagi M., Wang B., Chance M. R., Chen X., Du Y., Wang Y., An L., Wang Q., Lu T., Zhang X., Wang Z., and Stark G. R. (2010) Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. U.S.A. 107, 21499–21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X., Tanaka K., Yan J., Li J., Peng D., Jiang Y., Yang Z., Barton M. C., Wen H., and Shi X. (2013) Regulation of estrogen receptor α by histone methyltransferase SMYD2-mediated protein methylation. Proc. Natl. Acad. Sci. U.S.A. 110, 17284–17289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chuikov S., Kurash J. K., Wilson J. R., Xiao B., Justin N., Ivanov G. S., McKinney K., Tempst P., Prives C., Gamblin S. J., Barlev N. A., and Reinberg D. (2004) Regulation of p53 activity through lysine methylation. Nature 432, 353–360 [DOI] [PubMed] [Google Scholar]
- 39. Kontaki H., and Talianidis I. (2010) Lysine methylation regulates E2F1-induced cell death. Mol. Cell 39, 152–160 [DOI] [PubMed] [Google Scholar]
- 40. Shen C., Wang D., Liu X., Gu B., Du Y., Wei F. Z., Cao L. L., Song B., Lu X., Yang Q., Zhu Q., Hou T., Li M., Wang L., Wang H., Zhao Y., Yang Y., and Zhu W. G. (2015) SET7/9 regulates cancer cell proliferation by influencing β-catenin stability. FASEB J. 29, 4313–4323 [DOI] [PubMed] [Google Scholar]