Abstract
Cofactor F420 is an electron carrier with a major role in the oxidoreductive reactions of Mycobacterium tuberculosis, the causative agent of tuberculosis. A γ-glutamyl ligase catalyzes the final steps of the F420 biosynthesis pathway by successive additions of l-glutamate residues to F420-0, producing a poly-γ-glutamate tail. The enzyme responsible for this reaction in archaea (CofE) comprises a single domain and produces F420-2 as the major species. The homologous M. tuberculosis enzyme, FbiB, is a two-domain protein and produces F420 with predominantly 5–7 l-glutamate residues in the poly-γ-glutamate tail. The N-terminal domain of FbiB is homologous to CofE with an annotated γ-glutamyl ligase activity, whereas the C-terminal domain has sequence similarity to an FMN-dependent family of nitroreductase enzymes. Here we demonstrate that full-length FbiB adds multiple l-glutamate residues to F420-0 in vitro to produce F420-5 after 24 h; communication between the two domains is critical for full γ-glutamyl ligase activity. We also present crystal structures of the C-terminal domain of FbiB in apo-, F420-0-, and FMN-bound states, displaying distinct sites for F420-0 and FMN ligands that partially overlap. Finally, we discuss the features of a full-length structural model produced by small angle x-ray scattering and its implications for the role of N- and C-terminal domains in catalysis.
Keywords: crystal structure, high-performance liquid chromatography (HPLC), mass spectrometry (MS), Mycobacterium tuberculosis, x-ray crystallography, FbiB, cofactor F420, gamma-glutamyl ligase, poly-gamma-glutamate tail
Introduction
The cofactor F420 is a flavin derivative that is sporadically distributed among microorganisms, mainly archaea and actinobacteria (including mycobacteria). F420 has been emerging as a new player in the biology of mycobacteria (1), with increasing numbers of F420-utilizing proteins characterized from different mycobacterial species (2–8). This cofactor has been suggested to protect Mycobacterium tuberculosis, the causative agent of tuberculosis, against oxidative and nitrosative stress during pathogenesis (9–11). At the biochemical level, cofactor F420 functions as a hydride transfer agent in oxidoreductive reactions with a lower redox potential than that of NAD(P)+ (12).
The biosynthesis pathway of cofactor F420 has been investigated in both archaeal and mycobacterial species. In the current view of the proposed pathway, the first intermediate with the complete chromophore (7,8-didemethyl-8-hydroxy-5-deazariboflavin (FO)4) is produced by FO synthase (FbiC in mycobacteria (13) and CofGH in archaea (14)). A transferase enzyme (FbiA in mycobacteria (15) and CofD in archaea (16)) subsequently catalyzes the addition of a 2-phospho-l-lactate moiety to FO to produce F420-0 (F420 with no poly-γ-glutamate tail). The final step of the pathway is performed by a γ-glutamyl ligase (FbiB in mycobacteria (15) and CofE in archaea (17)) that catalyzes successive additions of l-glutamate residues to F420-0 (Fig. 1A).
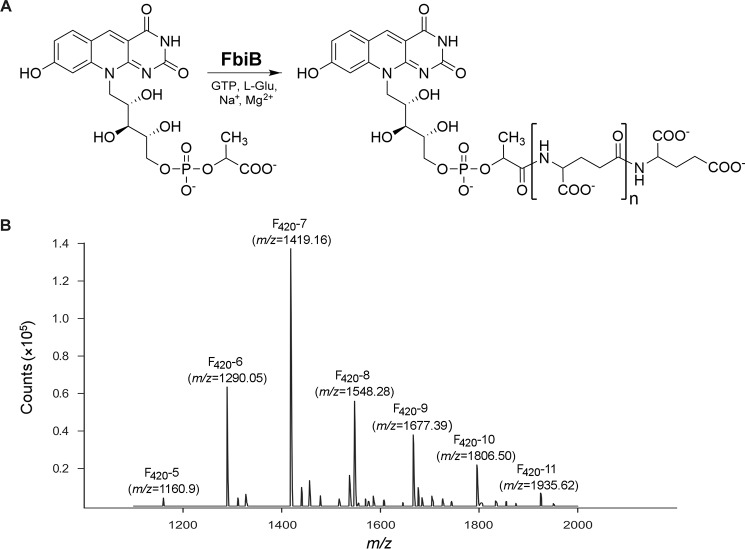
FIGURE 1.
The reaction catalyzed by FbiB. A, molecular structures of F420-0 and the reaction products, produced by the action of FbiB, with different numbers of l-glutamate residues in the poly-γ-glutamate tail. B, mass spectrometry analysis of F420 species co-purified with full-length M. tuberculosis-FbiB produced in M. smegmatis host cells. Selected peaks are labeled by species (F420-5 through F420-11) with the corresponding m/z values indicated in parentheses.
The length of the poly-γ-glutamate tail varies between archaeal and mycobacterial species; in archaea, two l-glutamate residues are seen (18), whereas in mycobacteria, up to nine residues are present (3, 19). There exists an intriguing difference between the enzymes responsible for this reaction in these microorganisms, with CofE having only one domain, whereas FbiB is a two-domain protein. The N-terminal domain of FbiB is annotated as a γ-glutamyl ligase with sequence similarities to CofE, whereas the C-terminal domain has sequence similarity to an FMN-dependent family of nitroreductase enzymes. Functional homology to nitroreductases, however, seems unlikely, and it is reasonable to hypothesize that the C-terminal domain of FbiB facilitates elongation of the poly-γ-glutamate tail of cofactor F420 in mycobacterial species.
Here we describe the structural and functional characterization of the FbiB protein from M. tuberculosis, demonstrating that the full-length enzyme is capable of adding multiple l-glutamate residues to F420-0 in vitro. Our results also indicate that communication between the two domains of FbiB is critical for full γ-glutamyl ligase activity, because the N-terminal domain is capable of producing only F420-1. We have also determined the crystal structure of the C-terminal domain of FbiB in apo-, F420-0-, and FMN-bound states (1.9, 2.05, and 2.1 Å resolution, respectively). These structures indicate specific and distinct binding sites for F420-0 and FMN ligands that are adjacent and partially overlapped. Despite the presence of a conserved FMN binding site in this domain, FMN does not appear to have a role in the reaction catalyzed by FbiB and is probably a remnant of the ancestral FMN-dependent nitroreductase enzyme. A full-length structural model of FbiB produced by small angle x-ray scattering (SAXS), using our x-ray crystal structure of the C-terminal domain and an N-terminal homology model, shows that the two domains are separated in space and are linked by a flexible α-helical segment. We discuss the implications of this full-length model for our functional understanding of the C-terminal domain.
Experimental Procedures
PCR Amplification and Cloning
The open reading frame encoding the full-length FbiB protein (Rv3262) (20) was amplified from M. tuberculosis H37Rv genomic DNA using the primers outlined in Table 1. The cloning was conducted using the Gateway® cloning system (21). The amplified PCR product was used to produce entry clones by performing a BP reaction. Positive entry clones were selected on LB agar medium supplemented with 50 μg/ml kanamycin and were then verified using BsrGI digestion and sequencing. The resulting entry clones were used to clone the full-length construct into pDEST17 (21) and pDESTsmg (22) vectors using an LR reaction. The expression construct for pDEST17 was selected on LB agar plates containing 100 μg/ml ampicillin. Selection of pDESTsmg construct was performed on low salt LB agar plates, pH 8.0, containing 50 μg/ml hygromycin B. All expression constructs were subsequently verified using BsrGI digestion. Two additional constructs were also prepared, using the same protocol, to express N-terminal (FbiB(11–249)) and C-terminal (FbiB(249–448)) domains of FbiB in pDESTsmg vector (Table 1).
TABLE 1.
Primers used in the amplification of FbiB (Rv3262) constructs
| Construct | Primer Sequences (5′-3′) |
|---|---|
| Full-length FbiB | |
| Forward | GGCAGCGGCGCGTTGACCGGCCCCGAACATGGC |
| Reverse | GAAAGCTGGGTGTCACTTCAGGATCAGCAAATC |
| FbiB N-terminal domain (FbiB(11–249)) | |
| Forward | GGCAGCGGCGCGATGACCATCGAGATCCTG |
| Reverse | GAAAGCTGGGTGTTAGAGCGCTTCGGCGGT |
| FbiB C-terminal domain (FbiB(249–448)) | |
| Forward | GGCAGCGGCGCGATGACCGCCGAAGCGCTC |
| Reverse | GAAAGCTGGGTGTTATCACTTCAGGATCAGCAA |
Expression and Purification
FbiB constructs were expressed in Escherichia coli BL21(DE3)pRP and Mycobacterium smegmatis mc2 4517 (23) cells. In both cases, protein expression was performed in autoinduction medium as described previously (24). Protein expression in E. coli was started at 37 °C for 4 h, followed by overnight incubation at 18 °C. M. smegmatis cultures were supplemented with 0.05% (v/v) Tween 80, and protein expression was carried out for 3–4 days at 37 °C (24).
All three FbiB constructs were cloned with an N-terminal His6 tag to facilitate the subsequent purification steps. The His6 tag on both pDEST vectors is cleavable using tobacco etch virus (TEV) protease. All constructs were purified from E. coli and M. smegmatis cells using the same procedure, as described below. The cells were harvested and resuspended in 20 mm HEPES, pH 7.0, 150 mm NaCl, 20 mm imidazole, 1 mm β-mercaptoethanol. The cells were then lysed using a cell disruptor (Microfluidizer M-110P) in the presence of Complete protease inhibitor mixture mini EDTA-free tablets (Roche Applied Science). The lysate was centrifuged at 20,000 × g to separate the insoluble material. The recombinant proteins were first purified using an immobilized metal affinity chromatography step by loading the supernatant onto a HisTrap FF 5-ml nickel-affinity column (GE Healthcare) that had been pre-equilibrated in the lysis buffer. The column was washed with the lysis buffer, and the protein was subsequently eluted using a gradient of imidazole in the buffer. Appropriate protein fractions were pooled and dialyzed at 4 °C in the presence of rTEV protease (25) to remove the N-terminal His6 tag. The His6-tagged rTEV protease, encoded in a pProEX HTa expression vector, was produced earlier from E. coli RosettaTM(DE3)pLysS cells (Novagen), as described previously (25). After overnight incubation of the purified FbiB proteins with rTEV protease, a subtractive immobilized metal affinity chromatography step was performed to remove the cleaved protein from uncut protein and rTEV protease. The resulting protein fraction was concentrated and then injected onto a size exclusion Superdex 200 10/300 column (GE Healthcare) pre-equilibrated in 20 mm HEPES, pH 7.0, 150 mm NaCl, 1 mm β-mercaptoethanol.
Binding Assays
Fluorescence spectroscopy was performed using an EnSpire® multimode plate reader (PerkinElmer Life Sciences). The fluorescence intensity was measured using a black 96-well plate with total reaction volumes of 100 μl in triplicates. Excitation and emission wavelengths of 420 and 480 nm, respectively, were used to monitor the intrinsic F420 fluorescence. For FMN binding assays, the excitation wavelength was set at 445 nm, and emission wavelength was set at 525 nm. To determine the dissociation constant, the protein samples (0.1 μm) were incubated with either F420 (0.001–20 μm) or FMN (0.01–100 μm) and left for 30 min at ambient temperature before fluorescence measurements. The binding reactions contained 20 mm HEPES, pH 7.0, 150 mm NaCl, 1 mm β-mercaptoethanol and were corrected against a control lacking the FbiB protein. Ligand binding data were fitted using one- or two-site binding models (SigmaPlot version 12.5).
Activity Assays
The γ-glutamyl ligase activity (17) was measured in 50-μl reactions containing different FbiB constructs (1 μg). The optimized reaction mixture included 50 mm HEPES, pH 8.5, 100 mm NaCl, 5 mm MnCl2, 10 mm l-glutamate, 5 mm GTP, and 2 μm F420-0. The F420-0 substrate was prepared by enzymatic hydrolysis of the poly-γ-glutamate tail of F420 using carboxypeptidase G (in 50 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 0.2 mm ZnSO4) (17). The possible effect of FMN on the polyglutamylation reaction was also tested by including a range of FMN concentrations (0–1 mm) in duplicate activity assays. The reactions were incubated at 37 °C and stopped using 20 mm EDTA after various time points.
HPLC Analysis
Separation of F420 species was performed on an Agilent HP 1100 HPLC system equipped with photodiode array and fluorescence detectors (Agilent Technologies). Samples were kept at 4 °C, and the injection volume was 20 μl. Samples were separated on a Phenomenex Luna C18 column (150 × 3 mm, 5 μm) with a 0.2-μm in-line filter that was maintained at 30 °C. The mobile phase consisted of 100% methanol (A) and 25 mm sodium acetate buffer, pH 6.0 (B), with a gradient elution at a flow rate of 0.5 ml/min and a run time of 30 min. The gradient profile was performed as follows: 0–25 min 95–80% B, 25–26 min 80% B, 26–27 min 95% B, 27–30 min 95% B, and a post-run of 2 min. The wavelengths used for photodiode array were 280 and 420 nm (bandwidth 20 nm) using a reference of 550 nm (bandwidth 50 nm). The wavelengths used for the fluorescence detector were 420 nm (excitation) and 480 nm (emission).
LC-MS Analysis
After HPLC separation, F420 samples were analyzed using an Agilent series 1200 liquid chromatography instrument coupled with an Agilent 6460 jet stream triple quadrupole mass spectrometer (Agilent Technologies, Santa Clara, CA). The injection volume and sample separation were performed as described above, with the mobile phase consisting of 100% methanol with 0.02% ammonia (A) and 25 mm ammonium acetate buffer, pH 6.0 (B). Ionization was achieved using electrospray ionization in either negative or positive mode.
Crystallization and Data Collection
The C-terminal domain of FbiB (FbiB(249–448)) was crystallized as described previously (26). The best diffracting crystals were obtained in 25% (w/v) PEG 3350, 0.35 m Li2SO4. The crystals were cryo-protected in 70% N-paratone and 30% mineral oil (v/v) before flash-freezing in liquid nitrogen.
For experimental phasing, preformed apo-crystals were soaked for 10 min in 0.5 m KBr solution that was prepared in the cryo-solution (25% PEG 3350 (w/v), 0.35 m LiSO4, 25% glycerol (v/v)) before being flash-cooled in liquid nitrogen. Bromide-multiwavelength anomalous diffraction data sets were collected at three wavelengths at the Australian Synchrotron. Data collection statistics are summarized in Table 2.
TABLE 2.
Data collection and processing statistics
| Br-FbiB |
Apo-FbiB | FbiB-F420 | FbiB-FMN | |||
|---|---|---|---|---|---|---|
| Peak | Inflection | Remote | ||||
| Wavelength (Å) | 0.91906 | 0.91935 | 0.89841 | 1.54179 | 0.95370 | 0.95370 |
| Space group | P41212 | P41212 | P41212 | P41212 | P41212 | P41212 |
| Cell dimensions | ||||||
| a (Å) | 137.03 | 136.95 | 137.08 | 136.94 | 136.61 | 137.14 |
| b (Å) | 137.03 | 136.95 | 137.08 | 136.94 | 136.61 | 137.14 |
| c (Å) | 101.96 | 102.13 | 102.13 | 102.09 | 101.74 | 101.40 |
| α, β, γ (degrees) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution rangea (Å) | 96.9–2.60 (2.74–2.60) | 96.8–2.60 (2.74–2.60) | 96.9–2.60 (2.74–2.60) | 96.8–1.90 (1.94–1.90) | 48.3–2.05 (2.10–2.05) | 97.0–2.10 (2.16–2.10) |
| Rmergea | 0.175 (0.752) | 0.179 (0.879) | 0.194 (0.911) | 0.140 (1.44) | 0.181 (1.90) | 0.166 (2.80) |
| Rpima | 0.027 (0.115) | 0.057 (0.280) | 0.030 (0.140) | 0.018 (0.270) | 0.050 (0.542) | 0.031 (0.522) |
| Observed reflectionsa | 2,462,836 (362,156) | 615,997 (90,387) | 2,469,097 (363,254) | 5,046,328 (124,358) | 884,894 (62,351) | 1,679,900 (137,324) |
| Unique reflectionsa | 30,387 (4372) | 30,409 (4378) | 30,464 (4384) | 76,162 (4378) | 60,778 (4431) | 56,408 (4534) |
| Multiplicitya | 81.0 (82.8) | 20.3 (20.6) | 81.0 (82.9) | 66.3 (28.4) | 14.6 (14.1) | 29.8 (30.3) |
| Mean I/σIa | 40.0 (10.7) | 19.1 (4.2) | 36.7 (8.6) | 36.2 (2.7) | 18.5 (2.0) | 25.8 (1.7) |
| Completeness (%)a | 99.8 (100.0) | 99.8 (100.0) | 99.8 (100.0) | 99.5 (97.8) | 100.0 (99.8) | 99.4 (98.7) |
| CC(1/2)b | 1.0 (0.74) | 0.99 (0.66) | 1.0 (0.53) | |||
| Phasing power iso/ano | 0/0.83 | 0.34/0.46 | 0.64/0.60 | |||
| MapCCc original/inverted (%) | 0.54/0.13 | |||||
a Values in parentheses are for the outermost resolution shell.
b Pearson correlation coefficient.
c Map correlation coefficient.
Crystals of the C-terminal domain of FbiB in complex with ligands were obtained by soaking preformed apo crystals in precipitant solutions containing the ligands. The FMN complex was obtained by overnight soaking of apo-crystals in the cryo-solution (25% PEG 3350 (w/v), 0.35 m Li2SO4, 25% glycerol (v/v)) containing 5 mm FMN. For preparation of crystals in complex with F420-0, apo-crystals were soaked with F420-0 that was prepared for activity assays as mentioned earlier. F420-0 solution in water was concentrated to near dryness and redissolved in the cryo-solution (25% PEG 3350 (w/v), 0.35 m Li2SO4, 25% glycerol (v/v)) to a final concentration of 4.5 mm, after which the crystals were soaked for 2 h.
Structure Determination and Refinement
All data sets were indexed and processed using XDS (27), re-indexed using POINTLESS, and scaled with SCALA from the CCP4 program suite (28, 29). For structure determination using bromide-multiwavelength anomalous diffraction, the bromide sites and their occupancies were found using SHELXD (30), as implemented in the autoSHARP program suite (31). SHARP was then used for substructure refinement and phasing (32), followed by phase improvements by the SOLOMON density modification program (33). Cycles of automatic model building by ARP/wARP (34) resulted in a partial protein model that was then used for further automated model building with the application of non-crystallographic symmetry restraints on the four molecules of the asymmetric unit. The final model was completed manually using COOT (35). Water molecules were identified by their spherical electron density and appropriate hydrogen bond geometry with the surrounding structure. Following each round of manual model building, the model was refined using REFMAC5 (36), against the data to 1.9 Å resolution. The autoBUSTER refinement program (37) was used in the final stages of refinement, both with and without TLS refinement parameters. Full refinement statistics are shown in Table 3.
TABLE 3.
Crystal structure refinement statistics
| Apo-FbiB | FbiB-F420 | FbiB-FMN | |
|---|---|---|---|
| Protein Data Bank code | 4XOM | 4XOQ | 4XOO |
| Resolution range (Å) | 96.8–1.90 | 48.3–2.05 | 97.0–2.10 |
| Rwork/Rfree (%) | 23.3/26.7 | 21.2/24.5 | 23.2/26.9 |
| No. of atoms (non-hydrogen) | |||
| Protein | 6059 | 6445 | 5996 |
| Ligand | 10 | 127 | 124 |
| Solvent | 240 | 398 | 50 |
| Root mean square deviation from ideality | |||
| Bonds (Å) | 0.010 | 0.011 | 0.015 |
| Angles (degrees) | 1.35 | 1.46 | 1.73 |
| Average B factors (Å2) | |||
| Protein | 32.5 | 33.5 | 44.5 |
| F420-0 (n = 4) | 40.1 | ||
| FMN (n = 4) | 43.0 | ||
| Sulfates (n = 2) | 53.7 | 59.2 | |
| Waters | 31.3 | 34.7 | 37.3 |
| Ramachandran statistics | |||
| Favored (%) | 97.9 | 97.6 | 96.8 |
| Allowed (%) | 100.0 | 99.5 | 100.0 |
| Outliers (%) | 0.0 | 0.50 | 0.0 |
| Molprobity score; percentile | 1.30; 100th | 1.51; 100th | 3.31; 99th |
Structures of the C-terminal domain of FbiB in complex with FMN and F420-0 were solved by molecular replacement using PHASER (38) with the apo-FbiB structure as a search model. All structures were refined by cycles of manual building using COOT (35) and refinement using REFMAC5 (36) and BUSTER (37). Full refinement statistics are shown in Table 3.
Full-length Model of FbiB from SAXS and Homology Modeling
Protein samples were extensively dialyzed against 20 mm HEPES, pH 7.5, 150 mm NaCl, 5% glycerol (v/v), and 1 mm tris(2-carboxyethyl)phosphine. This buffer was also used as the buffer control and to dilute protein samples. SAXS data were collected on the Australian Synchrotron SAXS/WAXS beamline and processed using the scatterBrain software package (39). In brief, protein samples and buffer controls were diluted and aliquoted into 96-well plates, and rubber lids placed on the wells to prevent evaporation. The 96-well plates were mounted on the SAXS/WAXS beamline on a temperature-controlled mount at 283 K for autosampling and capillary flow data acquisition. Solutions were flowed through thin walled quartz capillaries, and data acquisition comprised consecutive 1-s x-ray exposures of the flowing samples to minimize radiation damage. Appropriate images were combined, and buffer scattering was carefully subtracted using the scatterBrain software (39). Data quality and derived parameters were calculated using the ATSAS software package (40). Details of SAXS data collection parameters and statistics are shown in Table 4.
TABLE 4.
Small angle x-ray scattering parameters and statistics
| Data collection parameters | |
| Beamlinea | AS SAXS/WAX |
| Wavelength (Å) | 1.0322 |
| Detector | Dectris-Pilatus 1M |
| Camera length (mm) | 1600 |
| q range (Å−1) | 0.006 – 0.6 |
| Sample capillary flow rate (ml/min) | 0.5 |
| Total sample volume (μl) | 100 |
| Exposure time/image (s) | 1 |
| No. of images/sample | 20 |
| Concentration range (mg/ml) | 0.25–5.0 |
| Temperature (K) | 283 |
| Structural parametersb | |
| I(0) (cm−1) (from P(r)) | 0.557 ± 0.002 |
| Rg (Å) (from P(r)) | 39.82 ± 0.01 |
| I(0) (cm−1) (from Guinier) | 0.561 ± 0.001 |
| Rg (Å) (from Guinier) | 39.8 ± 0.2 |
| Dmax (Å) | 130.0 |
| Porod volume estimate (Å3) | 56,501 |
| Dry volume calculated from sequence (Å3) | 57,567 |
| Software | |
| Primary data collection | scatterBrain |
| Data processing | scatterBrain |
| Rigid body modelling | SASREF |
| Computation of model intensities | CRYSOL |
a Full details of the beamline specifications are available at the Australian Synchrotron website.
b For a 5 mg/ml sample.
A structure of the N-terminal domain of FbiB was produced by homology modeling using the Phyre2 software (41) and the Protein Data Bank coordinate set 2PHN. The N-terminal homology model and the C-terminal crystal structure were combined to produce a full-length model using the SAXS data and a rigid body minimization protocol using SASREF (40) with a distance restraint to keep the termini of the two domains in appropriate proximity, effectively linked. Numerous possible models were assessed against the SAXS scattering profile using the CRYSOL software (40) before, eventually, the best fit was discovered as a 50:50 combination of two complete full-length models, as described fully below.
Results
FbiB Expression and Purification
The full-length, N-terminal, and C-terminal constructs of FbiB were expressed as soluble proteins in both E. coli and M. smegmatis cells using an autoinduction protocol. All three constructs were purified using immobilized metal affinity chromatography and size exclusion chromatography steps.
Functional Characterization
The FbiB constructs expressed in E. coli were used for functional studies to alleviate the complications arising from the presence of co-purified F420 in the proteins expressed in M. smegmatis (Fig. 1B). The γ-glutamyl ligase activity of FbiB constructs was investigated by monitoring the addition of l-glutamate residues to F420-0 using HPLC, as described previously for the CofE protein (16). The F420-0 substrate was prepared by enzymatic hydrolysis of the poly-γ-glutamate tail of F420 using carboxypeptidase G (17), and the resulting F420-0 was used as a substrate for FbiB activity experiments and also for crystallographic binding studies.
The intrinsic fluorescence of F420 was used to monitor the addition of l-glutamate residues to F420-0 during enzymatic reactions. A range of different conditions was optimized for the γ-glutamyl ligase activity of the full-length FbiB protein, including pH (6.0–9.0), monovalent (Na+ and K+) and divalent cation (Mg2+ and Mn2+) composition, nucleotides (GTP, dGTP, ATP, and dATP), and also various time points and temperatures. FbiB showed the highest activity at pH 8.5, the same pH dependence displayed by the CofE protein (17). A combination of Na+ and Mn2+ produced the highest activity in FbiB, in contrast to the previously reported dependence on K+ and Mg2+ for CofE (17). The enzyme was only active in the presence of GTP, with no observed activity for dGTP, ATP, and dATP nucleotides. The enzymatic assays for all FbiB constructs were subsequently conducted using the optimized condition (50 mm HEPES, pH 8.5, 100 mm NaCl, 5 mm MnCl2, 10 mm l-glutamate, 5 mm GTP, and 2 μm F420-0) and were incubated for periods of up to 1 week at 37 °C.
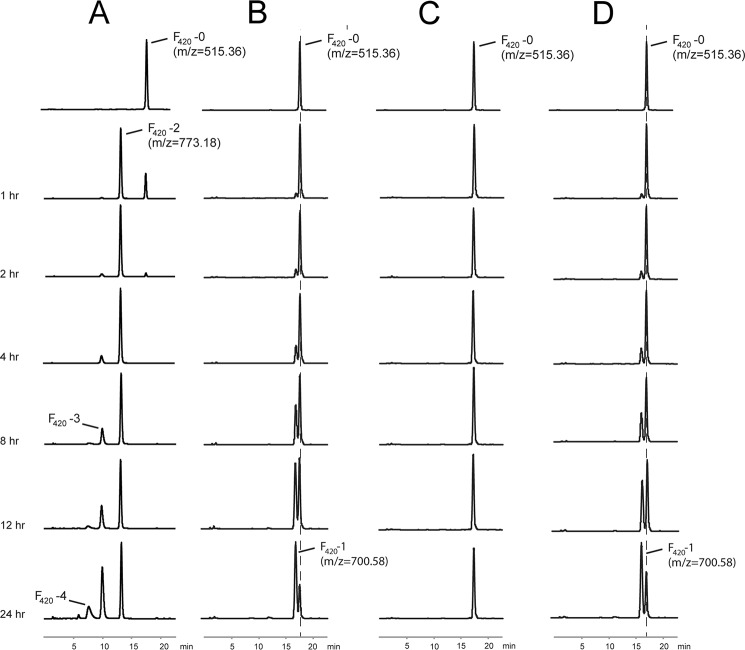
The functional assays show that full-length FbiB can convert F420-0 to F420 molecules with varying numbers of residues in the poly-γ-glutamate tail (Fig. 2A). F420-2 production could be detected within 1 h, with no higher order F420 molecules appearing within 2 h. Incubation for 24 h showed species as large as F420-5, and by 72 h (data not shown), the products resolved into two species with much earlier retention time than that of F420-5; although attempted, mass spectrometry analysis of these two large species was unsuccessful. This result, however, is not achieved by either the N- or C-terminal domains alone (Fig. 2, B and C, respectively), nor by a mixture of these two separate domains (Fig. 2D). Our results show that the N-terminal domain of FbiB adds one l-glutamate residue (to produce F420-1), albeit in a very slow manner compared with that of the full-length protein.
FIGURE 2.
Analysis of FbiB reaction products in vitro. The HPLC traces of the reactions performed using different FbiB constructs (fluorescence versus elution time). In the presence of the full-length construct (A), F420 molecules with various lengths of poly-γ-glutamate tail appear over time, whereas the N-terminal domain alone (B) shows only the production of F420-1. The C-terminal domain alone does not catalyze the reaction (C). The reactions using a combination of separate N- and C-terminal domains produce only F420-1 (D). The HPLC traces are shown for a time course up to 24 h as indicated. Selected peaks are labeled with the corresponding m/z values and F420 species.
Ligand Binding
Expression of FbiB constructs in M. smegmatis host cells resulted in co-purification of F420 with a distinct coloration of the full-length and N-terminal domain constructs. Mass spectrometry analysis confirmed that F420 species containing up to 11 l-glutamate residues in the poly-γ-glutamate tail co-purify with the full-length protein (Fig. 1B). Given that E. coli does not produce F420, the proteins expressed in E. coli cells were not expected to have any ligands co-purified. The C-terminal domain, however, showed a faint yellow color upon purification suggesting co-purification of flavin-like ligands from E. coli.
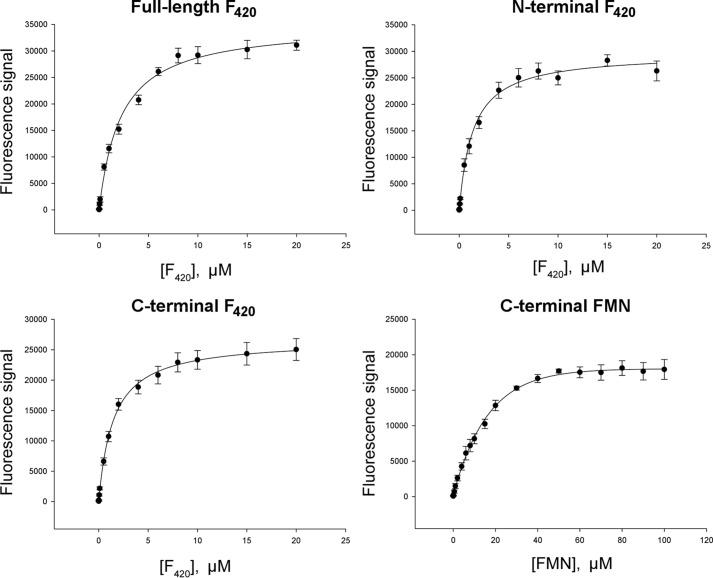
Fluorescence spectroscopy was used to investigate binding of F420 and other flavins (e.g. FMN) to FbiB constructs and to determine their dissociation constant (Kd) (Fig. 3). The results show that the full-length (two-site model; Kd1 = 0.2 ± 0.4 μm and Kd2 = 3.0 ± 0.9 μm), N-terminal (one-site model; Kd = 1.4 ± 0.1 μm), and C-terminal domains (one-site model; Kd = 1.47 ± 0.07 μm) could each bind F420 in solution. Surprisingly, the full-length and C-terminal constructs could also bind FMN although with a lower affinity (one-site model; Kd = 14.7 ± 0.9 μm). Although binding with 10-fold lower affinity to the C-terminal domain of FbiB, FMN was subsequently included in the functional studies of the different domain constructs to assess a possible role in catalytic function or inhibition/regulation.
FIGURE 3.
Ligand binding by various FbiB constructs. The intrinsic fluorescence signal of F420 and FMN were used to monitor ligand binding and also to calculate dissociation constants (Kd) against the full-length protein and both of the individual domains, N- and C-terminal, as indicated. The F420 dissociation constants derived from these binding curves are Kd1 = 0.2 ± 0.4 μm and Kd2 = 3.0 ± 0.9 μm (full-length, two-site model), Kd = 1.4 ± 0.1 μm (N-terminal), and Kd = 1.47 ± 0.07 μm (C-terminal). FMN binding to the C-terminal domain of FbiB is also included, as labeled, and gives a dissociation constant of 14.7 ± 0.9 μm.
Structure Determination
The requirement of the C-terminal domain for full activity and its binding to both F420 and FMN prompted us to pursue x-ray crystal structures of three FbiB constructs. Neither full-length FbiB nor its N-terminal domain could be crystallized. The C-terminal domain of FbiB (FbiB(249–448)), expressed in M. smegmatis cells, was successfully crystallized (26), however, and its structure was determined at 1.9 Å resolution by a multiwavelength anomalous dispersion method using crystals soaked in KBr solution (42) (Table 1). The unliganded FbiB C-terminal domain was crystallized in tetragonal space group P41212 and contains four molecules in the asymmetric unit. F420-0- and FMN-bound structures were produced by soaking ligand into preformed crystals and were solved using the unliganded structure for molecular replacement (Table 3).
Overall Structure
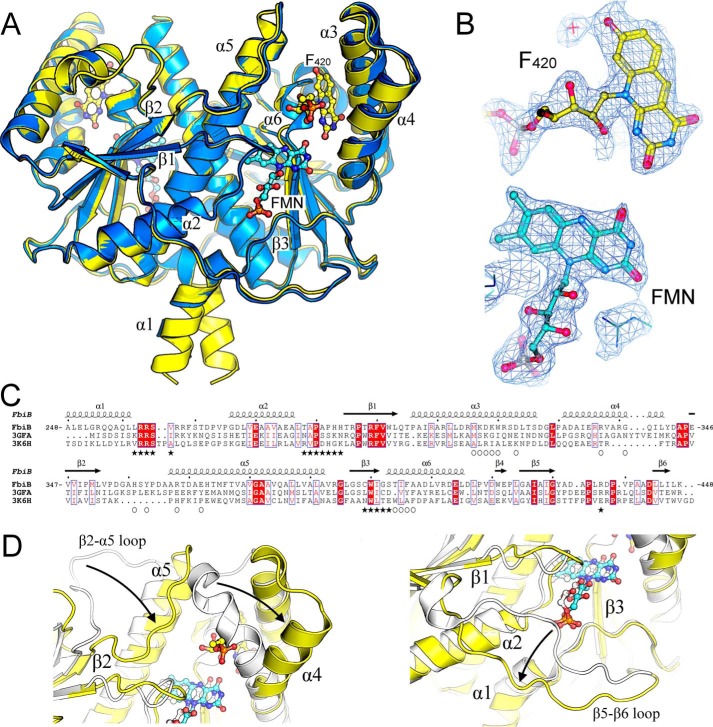
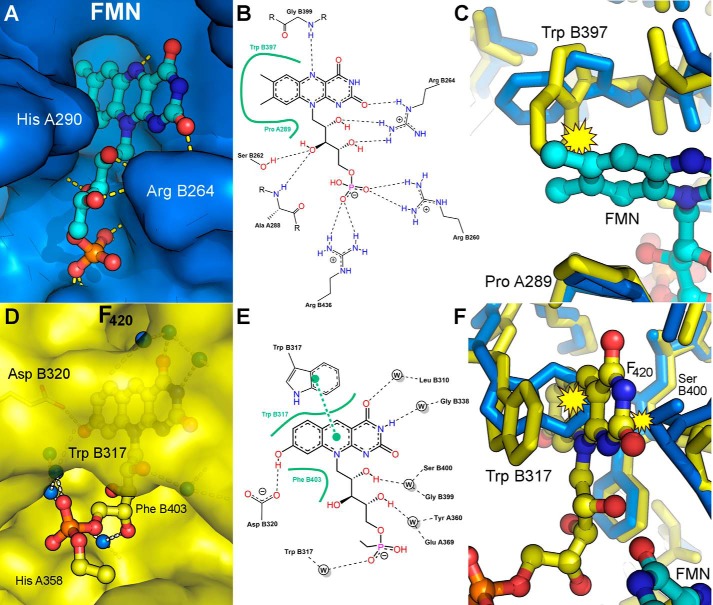
The four molecules in the asymmetric unit are organized as two sets of dimers that are related by a 2-fold non-crystallographic symmetry axis. The more extensive chain A-chain B dimer interface buries an average solvent-accessible surface area of 3247 Å2/monomer, representing 27% of each monomer's surface, as assessed by the PDBePISA server (43), and contains 33 hydrogen bonds, 15 salt bridges, and 204 non-bonded contacts. The protein displays a fold that is typical of the family of FMN-dependent nitroreductases (Pfam PF00881). This fold is based around a central five-stranded β-sheet, made up of four antiparallel β-strands together with a fifth one (parallel to the first) that is contributed by the C-terminal residues of the opposing monomer (Fig. 4A). The β-sheet is flanked by two helices on the internal side, involved in dimerization, and three helices on the external (surface-exposed) side.
FIGURE 4.
Crystal structure of the dimeric C-terminal domain of FbiB. A, overlay of F420-bound (yellow) and FMN-bound (blue) structures. F420 and FMN molecules are shown as ball-and-stick models with carbon atoms colored in yellow and cyan, respectively. Selected secondary structure elements are labeled. B, electron density (2Fo − Fo, omit map) surrounding F420 and FMN molecules in their respective structures. C, multiple sequence alignment of FbiB (C-terminal domain), the putative FMN-dependent nitroreductase from C. difficile (Protein Data Bank entry 3GFA), and a nitroreductase from A. tumefaciens (Protein Data Bank entry 3K6H). Secondary structure elements are shown for the FbiB structure, and residues involved in F420 and FMN binding are indicated by open circles and solid stars, respectively. Many residues involved in FMN binding appear conserved across the three structures, whereas those involved in F420 binding are not conserved. D, overlay of enlarged view of FbiB (yellow) and Protein Data Bank entry 3GFA (white). Incorporation of an F420 binding site (left) produces a large shift in the top of helix α4 and in the β2-α5 loop compared with the C. difficile homolog structure. Another large difference occurs at the bottom of the molecule (right), involving the β5-β6 loop, that opens up in FbiB to produce the binding site for the phosphate group of FMN.
FMN Binding
Although FMN is not a previously known ligand for the reaction catalyzed by FbiB, our results show FMN binding to FbiB in solution. Four molecules of FMN are located in the C-terminal domain crystal structure, one molecule in each protomer, as identified unambiguously in the electron density map (Fig. 4B, bottom). A number of residues within the binding site are conserved in the multiple sequence alignment in Fig. 4C, in which the FMN binding regions are indicated by black stars. The binding site is formed in a pocket at the dimer interface (Fig. 4A) and comprises a relatively non-polar pocket that binds the isoalloxazine chromophore, with a collection of basic residues and hydrogen bond donors arrayed about the ribitol chain (Fig. 5, A and B).
FIGURE 5.
Ligand binding in the C-terminal domain of FbiB. A, surface drawing of the FMN-binding pocket with FMN shown as a ball-and-stick model and hydrogen bonds indicated by dashed yellow lines. Whereas part of the FMN isoalloxazine ring system is buried deeply in a hydrophobic pocket, sandwiched between TrpB397 and ProA289, the remainder of the molecule remains relatively solvent-exposed. B, schematic diagram of protein-FMN contacts. Hydrogen bonds are shown as dashed lines, and extensive non-polar interactions are shown by an annotated, solid green line. C, FMN binding “pushes” TrpB397 aside; if F420 were already bound (yellow structure), then an irreconcilable steric clash (yellow clash symbol) between the FMN and the tryptophan side chain would exist. D, F420 binding pocket; the ring system is fully buried within the protein, with only the tail exposed to solvent space. E, schematic of F420 binding showing hydrogen bonds (dashed lines), extensive non-polar interactions (solid green lines), and a π-stacking interaction as two connected green dots. F420 hydrogen bonding to the protein is almost exclusively indirect and mediated through water molecules. F, F420 binding into an FMN-bound structure (blue structure) would also be precluded due to steric clash at both the front and the back of the binding site.
An overlay of the apo-FbiB structure with the FMN-bound structure shows generally very small differences, with an overall root mean square deviation in 768 aligned Cα atomic positions of 0.43 Å. However, there are some obvious local changes seen upon FMN binding. The FMN isoalloxazine ring system inserts between the polypeptide segments Cys396-Trp397-Ile398 and Ala288-Pro289-His290 and forces these elements apart by ∼1 Å. The side chain of Trp397 of the apo-structure swings 3.5 Å (at its tip) clear of the pocket to allow binding of and provide π-stacking with the FMN ring system, as illustrated in Fig. 5C. The side chain of Ile398 also provides a backstop to this binding site with close contacts to the FMN ring (∼3.8 Å) and also moves noticeably.
F420-0 Binding
Four F420-0 molecules were located unambiguously from the electron density maps (Fig. 4B, top), one in each protomer and binding in a conserved mode. Only two F420-0 molecules (bound to chains A and B), however, have well defined electron density beyond the phosphate group, enabling two atoms of a lactyl moiety to be modeled. Superimposition of the unliganded and F420-bound structures of the C-terminal domain shows little change in the protein on ligand binding, with an average root mean square deviation of 0.31 Å over 782 aligned Cα atomic positions.
The F420-0 molecule binds to the C-terminal domain in a pocket near the dimer interface (Fig. 4A) with the 8-hydroxy-5-deazaalloxazine ring system buried deeply in the protein, whereas the tail of the molecule binds in a solvent-exposed channel (Fig. 5, D and E). The 8-hydroxy-5-deazaalloxazine chromophore is sandwiched between hydrophobic elements, at the back primarily by the side chain of Phe403 and at the front by a π-stacking interaction with the side chain of Trp317 (as illustrated in Fig. 5F). Intriguingly, only a single hydrogen bond is formed directly to the protein with most other polar/charged elements of the ligand binding indirectly to the protein through water-mediated hydrogen bonding.
Structural Comparisons
A search using the PDBeFold server (44) for structural homologs using one FbiB C-terminal dimer produces numerous matches, two of which are shown in multiple sequence alignment in Fig. 4C. The putative nitroreductase from Clostridium difficile in complex with FMN (Protein Data Bank entry 3GFA) shows only 23% sequence identity but a root mean square deviation of 2.1 Å over 338 Cα atoms and a nitroreductase family protein from Agrobacterium tumefaciens (Protein Data Bank entry 3K6H), also in complex with FMN, that displays no detectable sequence similarity by BLAST but has a root mean square deviation of 2.4 Å over 291 Cα atoms. Intriguingly, 3GFA shows a very similar dimer interface to that of the FbiB C-terminal domain with an average solvent-accessible surface area of 3538 Å2 (31% of each monomer's surface). The most obvious difference between the FbiB and the homologous structures is in regions illustrated in Fig. 4D. Movement of the β2-α5 loop and α4 helix produces the F420 binding site; the β5-β6 loop shift opens up the bottom of the FbiB structure near the FMN binding site.
Building a Full-length Model Using SAXS
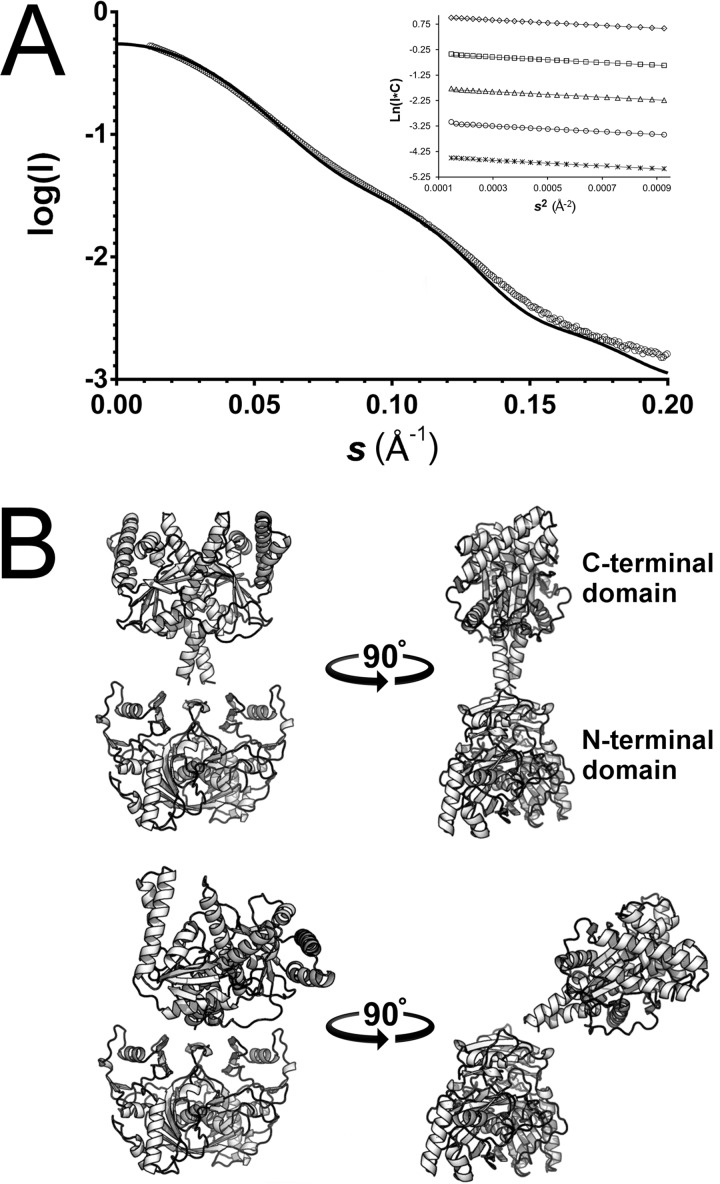
The full-length FbiB protein was subjected to SAXS at the Australian Synchrotron to characterize its structure in solution. Analysis of the SAXS data (Table 4 and Fig. 6A), including the calculation of various analytical curves, Rg, and I(0)/c values from a concentration series, shows that the scattering profile is not concentration-dependent and, with Guinier plots displaying linearity at low angle (Fig. 6A, inset), is indicative of no interparticle interference or aggregation. A P(r) function (not shown) suggests a maximum scattering particle dimension of 130 Å, and Kratky analysis shows a well folded protein.
FIGURE 6.
Full-length FbiB models by SAXS. A, SAXS data plotted against scattering angle (open circles, averaged and solvent-subtracted) and overlaid with the best fit combination of two models of full-length FbiB (χ = 3.92) calculated by the CRYSOL software and indicated by a solid black line. Inset, Guinier plots of a 2-fold dilution series from 5.2 mg/ml (top) to 0.325 mg/ml (bottom). The Guinier calculated radius of gyration (Rg) is consistent at 39.8 Å over the concentration range. B, the two models that in combination best fit the SAXS scattering profile in A. The so-called “straight” model (top) has the N-terminal and C-terminal domains spatially separated and with an approximately 180° angle between each other (top right). The “bent” model (bottom) again has the two domains separated but with the C-terminal domain displaced by an angle of ∼50° relative to the straight model (bottom right). The models were produced by combining a homology model of the N-terminal domain (Phyre2 server) with our x-ray structure of the C-terminal domain and by using rigid body minimization (SASREF) against SAXS data.
A full-length model of FbiB was produced via rigid body modeling by combining our structure of the C-terminal domain of FbiB with an N-terminal homology model and using the SAXS data for minimization. The homology model is based on the structure of an F420:γ-glutamyl ligase from Archaeoglobus fulgidus that shares 40% identity and 55% similarity with the FbiB N-terminal domain over 215 residues.
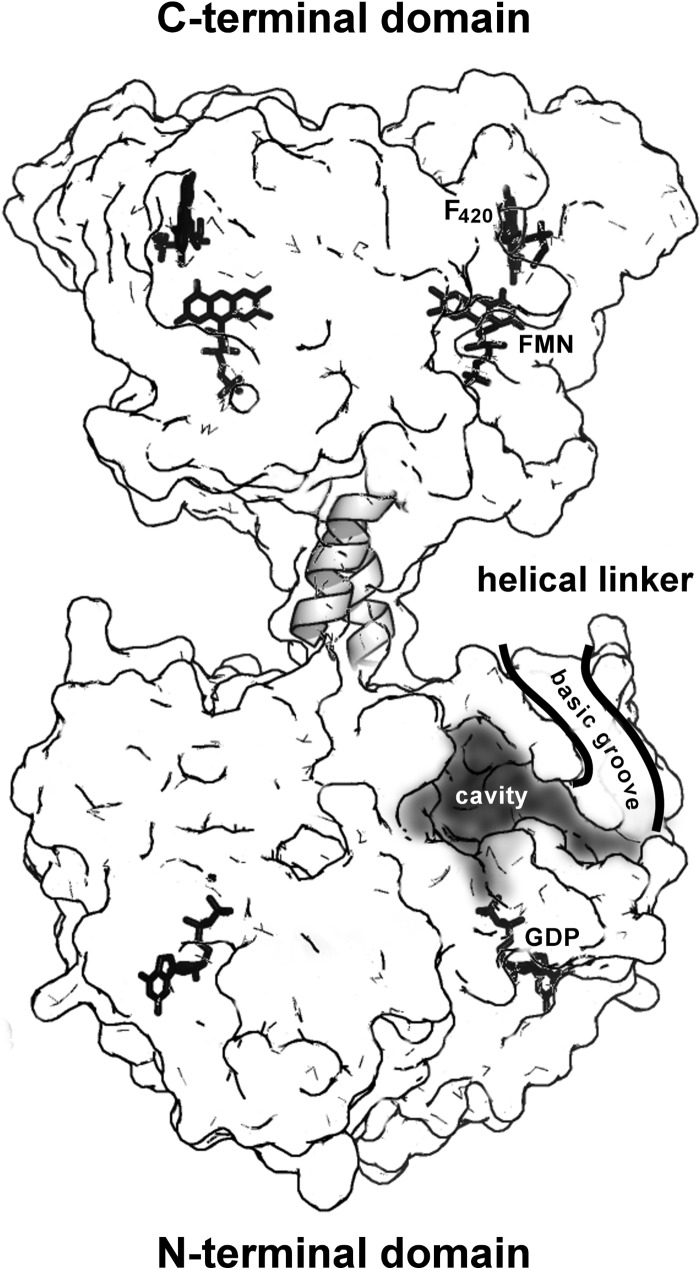
The full-length model shows the two domains as separated and linked by an α-helical segment (Figs. 6B and 7). The SAXS profile (Fig. 6A) is best fit by a combination of two models of the full-length protein, one “straight” (Fig. 6B, top) and one “bent” (Fig. 6B, bottom) and in equal proportions (50:50). A surface representation of the straight model (Fig. 7) shows the relative locations of the putative N-terminal active site cavity and the C-terminal FMN/F420 ligand binding sites. Directly adjacent to the N-terminal active site cavity is a surface groove lined with numerous basic residues that extends toward the C-terminal domain and into solvent space. The implications of this surface groove and the other features of this full two-domain model are discussed below.
FIGURE 7.
Functional implications of a full-length structural model of FbiB. A surface representation of the “straight” model of full-length FbiB showing the separation of N- and C-terminal domains and a postulated α-helical linker. Both domains are dimeric and thus have mirrored features in the diagram, including confirmed locations of FMN and F420 in the C-terminal domain and GDP in the N-terminal domain (by comparison with the homolog CofE structure) as well as a postulated binding cavity for F420 and l-glutamate in the N-terminal domain. The surface is partially transparent, and ligand binding or active sites indicated on the left are located in the back of the model, whereas those on the right are on the front of the model. We hypothesize that electrostatically positive surface grooves (basic grooves) lined with numerous arginine and lysine side chains have a role in binding an elongated and negatively charged F420 poly-γ-glutamate tail and directing it toward solvent space (only the front groove is shown for clarity).
Discussion
FbiB Function
The reaction catalyzed by CofE adds two l-glutamate residues to F420-0 to produce F420-2 in archaea (17). The presence of an additional domain in the mycobacterial FbiB protein suggested that this domain could assist the N-terminal domain in producing F420 molecules with longer poly-γ-glutamate tails in mycobacterial species. Our experimental results indeed demonstrate that the full-length FbiB protein does produce F420 molecules with multiple l-glutamate residues in the poly-γ-glutamate tail (Fig. 1B). The addition of further l-glutamate residues to F420-2 is evidently the rate-limiting step of the process because it takes a much longer time for the F420 species with longer poly-γ-glutamate tails to appear. When F420 is purified from mycobacterial cells, the major species found contain between 5 and 7 l-glutamate residues in their poly-γ-glutamate tails (3, 19). In addition, the full-length FbiB protein expressed in M. smegmatis shows co-purification of F420 molecules with up to 11 l-glutamate residues incorporated, with the major species having 6–9 residues. After 72 h in our in vitro experiments, we observe two major products presumably containing a larger number of l-glutamate residues in the poly-γ-glutamate tail than F420-5, but our inability to characterize these species by mass spectrometry does not allow us to comment further on the identity of these reaction products.
Investigation of the N-terminal and C-terminal domains of FbiB shows that these domains, by themselves, are unable to add l-glutamate residues to F420-0, although the N-terminal domain shows production of F420-1 in a very slow manner. This is a surprising observation because the N-terminal domain is homologous with the CofE protein of archaea (17), and we would have expected it to show γ-glutamyl ligase activity on its own. Our observation that both F420-0 and FMN bind to the C-terminal domain suggests a regulatory role for this domain in the reaction catalyzed by FbiB, perhaps both as a requirement for normal catalysis and as a sensor for the relative concentrations of F420 and FMN, in a feedback regulatory role. We could not, however, observe any effect of FMN on the activity of full-length FbiB over a wide range of concentrations, up to 500 times that of F420-0 in the reaction mixture. It has been suggested that the intracellular concentration of FMN in M. smegmatis is ∼10-fold higher than that of F420 (4).
Comparison of F420 and FMN Binding
Given our observed binding of F420 and FMN to FbiB in solution, we speculated that they would share the same binding site in the C-terminal domain. Surprisingly, this was not the case, as apparent in our crystal structure, although the two binding sites are closely adjacent in the protein structure.
A comparison of the two binding sites and the chemical structures of F420 and FMN explains why “crossover” is not seen in the ligand binding. Although both binding sites have similar electrostatic surface potentials, the F420 chromophore extends a hydroxyl group toward Asp320, forming a hydrogen bond (Fig. 5E). If FMN were placed in this site, the bulk of its dimethyl ring substituents could not be accommodated in the cavity available and additionally would not hydrogen-bond with Asp320 as seen for F420. In contrast, in its own site, FMN projects this same nonpolar dimethyl structure into a larger nonpolar pocket (Fig. 5B); F420 would, if placed in the FMN pocket, extend its hydroxyl group into the nonpolar pocket with no hydrogen bond partner available, effectively producing a buried and unsatisfied hydrogen bonding potential.
The binding of both F420 and FMN in the FbiB C-terminal domain is an interesting feature but not a unique occurrence. A 2012 study of F420-dependent reductases from M. smegmatis demonstrated cofactor promiscuity in three F420-dependent reductases, which can utilize both F420 and FMN to catalyze different chemistries (oxidation and reduction) of the same substrate (4). The F420-dependent reductases catalyze the reduction of aflatoxins and plant-derived furanocoumarins in the presence of F420H2. When FMN replaces F420, three of the F420-dependent reductases tested were found to catalyze FMN-mediated oxidation of two major aflatoxins, AFG1 and AFG2, via dehydrogenation. In another context, an example of competitive FMN/F420 binding is provided by FprA, a di-iron flavoprotein F420H2 oxidase found in methanogenic archaea, which catalyzes the four-electron reduction of O2 to 2H2O with 2 molar equivalents of reduced F420 (45). A recent systematic study of the mycobacteria oxidoreductase superfamily (flavin/deazaflavin oxidoreductases) shows some degree of promiscuity within single proteins, binding F420, FMN, and FAD with micromolar affinity (8). However, whereas the proteins appear promiscuous, they also show selectivity for a single cofactor (∼5–10-fold). Similarly, F420 binding in the C-terminal (oxidoreductase-derived) domain of FbiB shows a 10-fold selectivity over FMN, and both bind at low micromolar affinities (F420 Kd = 1.47 versus FMN Kd = 14.7).
Our crystal structure analysis of FMN and F420 binding, illustrated in Fig. 5, C and F, suggests that although these ligands do not occupy the same site, they cannot bind simultaneously because steric clashes would result; competitive binding in the closely adjacent site cannot, however, be precluded, and we have not examined this. Although it is not entirely unexpected that the C-terminal domain binds F420, potentially in a feedback regulation role, there is no suggestion of why FMN also binds to this domain; could FMN binding in the FbiB C-terminal domain merely represent an evolutionary relic from an ancestral FMN-dependent nitroreductase domain? In this hypothesis, F420 binding evolved in the C-terminal domain after being acquired through a recombination event; the FMN binding site has been retained, although we might have expected it to mutate over an evolutionary time scale to abolish binding unless FMN plays some role in the poly-γ-glutamylation reaction. A more compelling explanation is that the ancestral nitroreductase appropriated by FbiB through recombination was already a promiscuous enzyme, binding both F420 and FMN for oxidoreductase activity potentially toward multiple substrates. Again, this hypothesis does not explain the retention of distinct F420 and FMN binding sites in the C-terminal domain of FbiB nor the potential roles these redox molecules play in the poly-γ-glutamylation reaction.
Reaction Mechanism and the Role of the C-terminal Domain
Our biochemical evidence implies that the catalytic machinery of the FbiB protein is located in the N-terminal domain and functions similarly to the single domain homologs that produce F420-2 as the largest product. The homolog structure from A. fulgidus (Protein Data Bank entry 2PHN), equivalent to the FbiB N-terminal domain, displays metal and GDP binding; glutamate and F420 binding is not observed, but the locations of their binding sites and the catalytic residues involved in glutamate addition have been postulated (46). In relation to the C-terminal domain of FbiB, our biochemical analysis suggests that this domain only provides support for the catalytic activity of the N-terminal domain; both domains are required for catalysis, and either domain alone does not show catalytic activity. We have produced a composite model of the whole FbiB protein by combining our crystal structure of the C-terminal domain with a homology model of the N-terminal domain and by rigid body refinement of these two half-structures against SAXS data (Fig. 6). The two domains are linked via an α-helical segment that we infer is contiguous between the domains; the C terminus of the homolog structure CofE is α-helical, as is the N terminus of our crystal structure. Two models of full-length protein in combination show best agreement with the SAXS data. The first model can be referred to as “straight” with the C-terminal domain located directly over the N-terminal domain with both domains together showing 2-fold symmetry; the two domains are separated by ∼14 Å and, outside of the linker segment, do not interact. Our second model, best referred to as “bent,” shows the C-terminal domain bending ∼50°, toward the N-terminal domain (Fig. 6B). This model brings the putative N-terminal catalytic site and the FMN/F420 sites of the C-terminal domain to within ∼40 Å of each other, still relatively distant.
Either in a catalytic enhancement or a regulatory role, the C-terminal domain may produce an allosteric signal from one or the other of the ligand binding sites to influence catalysis in the N-terminal domain (although we have not observed any effect). We can also hypothesize that the C-terminal domain and perhaps its dynamic behavior in solution, implied by our two-structure model of the SAXS data, apply some mechanical force through the connecting linker sequence to promote or regulate the N-terminal domain function; this is a hypothesis that we have previously explored for another M. tuberculosis enzyme, 2-isopropylmalate synthase, where its C-terminal domain is required for both feedback regulation and catalytic activity in the separate and distant N-terminal domain (47).
One very interesting feature of our N-terminal homology model is the presence of a positively charged surface groove lined by 11 arginine and lysine residues; this is not a feature of the homologous A. fulgidus structure from which the model was produced. If, as we contend, the N-terminal domain contains all of the catalytic machinery for F420 elongation, then this basic surface groove connected to the postulated active site cavity (Fig. 7), could bind to the growing poly-γ-glutamate tail with its negatively charged carboxylate groups and direct the growing chain toward solvent space or toward the adjacent C-terminal domain.
Elongation Mechanism; Insertion or Extension?
Poly-γ-glutamate tails are found on a limited number of biomolecules, including cofactor F420 and folate derivatives. Poly-γ-glutamic acid is also a polymer that is produced by a number of microorganisms, with roles from virulence to promising potential for medical and industrial applications (48). FbiB (CofE in archaea), folylpolyglutamate synthase, and the poly-γ-glutamic acid synthetase complex, catalyze the poly-γ-glutamylation reactions to produce these molecules. The chemical mechanism has been generally assumed to be similar for all of these enzymes, involving activation of the carboxylic acid on the elongated substrate in a nucleotide-dependent manner (GTP in F420 and ATP in folates and poly-γ-glutamic acid), formation of an acyl phosphate intermediate, and finally nucleophilic attack by the incoming l-glutamate (17, 46).
Our functional characterization described in the present work demonstrates the addition of multiple l-glutamate residues to the growing poly-γ-glutamate tail of F420 carried out by FbiB. It is not clear, however, whether each l-glutamate is added to the terminal residue of the growing chain (an extension mechanism) or inserted somewhere into the middle of the chain, perhaps between the phospholactate moiety and the first l-glutamate residue (an insertion mechanism). Elucidation of the full mechanistic details of the elongation mechanism will also have implications for further understanding of the function and mechanism of the folylpolyglutamate synthase and poly-γ-glutamic acid synthetase enzymes that also carry out poly-γ-glutamylation reactions.
Author Contributions
G. B., E. N. B., and C. J. S. designed the study. G. B. and A. M. R. performed the majority of experiments, including protein production and characterization. A. M. R. carried out protein crystallization. G. B. and S. S. performed the functional experiments. Final structure refinement and deposition was carried out by H. M. B. and C. J. S. G. B., E. N. B., and C. J. S. wrote the paper, and all authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Ehab Jirgis for technical assistance and Martin Middleditch for mass spectrometry, performed at the Centre for Genomics, Proteomics, and Metabolomics at the University of Auckland. This research was undertaken on the MX1, MX2, and SAXS/WAXS beamlines at the Australian Synchrotron (Victoria, Australia). We thank Drs. Neil Patterson, Jeremy Keown, David Goldstone, and Shaun Lott for synchrotron data collection.
This work was supported by the Health Research Council of New Zealand, and access to the Australian Synchrotron was supported by the New Zealand Synchrotron Group Ltd. The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 4XOM, 4XOQ, and 4XOO) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FO
- 7,8-didemethyl-8-hydroxy-5-deazariboflavin
- SAXS
- small angle x-ray scattering
- WAXS
- wide angle x-ray scattering
- TEV
- tobacco etch virus.
References
- 1. Selengut J. D., and Haft D. H. (2010) Unexpected abundance of coenzyme F420-dependent enzymes in the genomes of Mycobacterium tuberculosis and other Actinobacteria. J. Bacteriol. 192, 5788–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bashiri G., Perkowski E. F., Turner A. P., Feltcher M. E., Braunstein M., and Baker E. N. (2012) Tat-dependent translocation of an F420-binding protein of Mycobacterium tuberculosis. PLoS One 7, e45003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bashiri G., Squire C. J., Moreland N. J., and Baker E. N. (2008) Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J. Biol. Chem. 283, 17531–17541 [DOI] [PubMed] [Google Scholar]
- 4. Lapalikar G. V., Taylor M. C., Warden A. C., Onagi H., Hennessy J. E., Mulder R. J., Scott C., Brown S. E., Russell R. J., Easton C. J., and Oakeshotta J. G. (2012) Cofactor promiscuity among F420-dependent reductases enables them to catalyse both oxidation and reduction of the same substrate. Catal. Sci. Technol. 2, 1560–1567 [Google Scholar]
- 5. Taylor M. C., Jackson C. J., Tattersall D. B., French N., Peat T. S., Newman J., Briggs L. J., Lapalikar G. V., Campbell P. M., Scott C., Russell R. J., and Oakeshott J. G. (2010) Identification and characterization of two families of F420H2 -dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 78, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mashalidis E. H., Gittis A. G., Tomczak A., Abell C., Barry C. E. 3rd, and Garboczi D. N. (2015) Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis. Protein Sci. 24, 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Purwantini E., and Mukhopadhyay B. (2013) Rv0132c of Mycobacterium tuberculosis encodes a coenzyme F420-dependent hydroxymycolic acid dehydrogenase. PLoS One 8, e81985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmed F. H., Carr P. D., Lee B. M., Afriat-Jurnou L., Mohamed A. E., Hong N. S., Flanagan J., Taylor M. C., Greening C., and Jackson C. J. (2015) Sequence-structure-function classification of a catalytically diverse oxidoreductase superfamily in Mycobacteria. J. Mol. Biol. 427, 3554–3571 [DOI] [PubMed] [Google Scholar]
- 9. Darwin K. H., Ehrt S., Gutierrez-Ramos J.-C., Weich N., and Nathan C. F. (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302, 1963–1966 [DOI] [PubMed] [Google Scholar]
- 10. Gurumurthy M., Rao M., Mukherjee T., Rao S. P., Boshoff H. I., Dick T., Barry C. E. 3rd, and Manjunatha U. H. (2013) A novel F420-dependent anti-oxidant mechanism protects Mycobacterium tuberculosis against oxidative stress and bactericidal agents. Mol. Microbiol. 87, 744–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Purwantini E., and Mukhopadhyay B. (2009) Conversion of NO2 to NO by reduced coenzyme F420 protects mycobacteria from nitrosative damage. Proc. Natl. Acad. Sci. U.S.A. 106, 6333–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DiMarco A. A., Bobik T. A., and Wolfe R. S. (1990) Unusual coenzymes of methanogenesis. Annu. Rev. Biochem. 59, 355–394 [DOI] [PubMed] [Google Scholar]
- 13. Choi K.-P., Kendrick N., and Daniels L. (2002) Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F420 and FO biosynthesis. J. Bacteriol. 184, 2420–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Graham D. E., Xu H., and White R. H. (2003) Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F420 biosynthesis. Arch. Microbiol. 180, 455–464 [DOI] [PubMed] [Google Scholar]
- 15. Choi K. P., Bair T. B., Bae Y. M., and Daniels L. (2001) Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F420 biosynthesis by Mycobacterium bovis BCG. J. Bacteriol. 183, 7058–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Graupner M., Xu H., and White R. H. (2002) Characterization of the 2-phospho-l-lactate transferase enzyme involved in coenzyme F420 biosynthesis in Methanococcus jannaschii. Biochemistry 41, 3754–3761 [DOI] [PubMed] [Google Scholar]
- 17. Li H., Graupner M., Xu H., and White R. H. (2003) CofE catalyzes the addition of two glutamates to F420-0 in F420 coenzyme biosynthesis in Methanococcus jannaschii. Biochemistry 42, 9771–9778 [DOI] [PubMed] [Google Scholar]
- 18. Eirich L. D., Vogels G. D., and Wolfe R. S. (1978) Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry 17, 4583–4593 [DOI] [PubMed] [Google Scholar]
- 19. Bair T. B., Isabelle D. W., and Daniels L. (2001) Structures of coenzyme F420 in Mycobacterium species. Arch. Microbiol. 176, 37–43 [DOI] [PubMed] [Google Scholar]
- 20. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. r., Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 21. Moreland N., Ashton R., Baker H. M., Ivanovic I., Patterson S., Arcus V. L., Baker E. N., and Lott J. S. (2005) A flexible and economical medium-throughput strategy for protein production and crystallization. Acta Crystallogr. D Biol. Crystallogr. 61, 1378–1385 [DOI] [PubMed] [Google Scholar]
- 22. Goldstone R. M., Moreland N. J., Bashiri G., Baker E. N., and Shaun Lott J. (2008) A new Gateway vector and expression protocol for fast and efficient recombinant protein expression in Mycobacterium smegmatis. Protein Expr. Purif. 57, 81–87 [DOI] [PubMed] [Google Scholar]
- 23. Wang F., Jain P., Gulten G., Liu Z., Feng Y., Ganesula K., Motiwala A. S., Ioerger T. R., Alland D., Vilchèze C., Jacobs W. R. Jr., and Sacchettini J. C. (2010) Mycobacterium tuberculosis dihydrofolate reductase is not a target relevant to the antitubercular activity of isoniazid. Antimicrob. Agents Chemother. 54, 3776–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bashiri G., Squire C. J., Baker E. N., and Moreland N. J. (2007) Expression, purification and crystallization of native and selenomethionine labeled Mycobacterium tuberculosis FGD1 (Rv0407) using a Mycobacterium smegmatis expression system. Protein Expr. Purif. 54, 38–44 [DOI] [PubMed] [Google Scholar]
- 25. Blommel P. G., and Fox B. G. (2007) A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 55, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rehan A. M., Bashiri G., Paterson N. G., Baker E. N., and Squire C. J. (2011) Cloning, expression, purification, crystallization and preliminary x-ray studies of the C-terminal domain of Rv3262 (FbiB) from Mycobacterium tuberculosis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 1274–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 29. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., and Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 31. Vonrhein C., Blanc E., Roversi P., and Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 32. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., and Paciorek W. (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 33. Abrahams J. P., and Leslie A. G. W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 [DOI] [PubMed] [Google Scholar]
- 34. Morris R. J., Perrakis A., and Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 35. Emsley P., and Cowtan K. (2004) Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., and Womack T. O. (2011) BUSTER. Global Phasing Ltd., Cambridge, United Kingdom [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kirby N. M., Mudie S. T., Hawley A. M., Cookson D. J., Mertens H. D. T., Cowieson N., and Samardzic-Boban V. (2013) A low-background-intensity focusing small-angle x-ray scattering undulator beamline. J. Appl. Cryst. 46, 1670–1680 [Google Scholar]
- 40. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dauter Z., and Dauter M. (1999) Anomalous signal of solvent bromides used for phasing of lysozyme. J. Mol. Biol. 289, 93–101 [DOI] [PubMed] [Google Scholar]
- 43. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 44. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 45. Seedorf H., Hagemeier C. H., Shima S., Thauer R. K., Warkentin E., and Ermler U. (2007) Structure of coenzyme F420H2 oxidase (FprA), a di-iron flavoprotein from methanogenic Archaea catalyzing the reduction of O2 to H2O. FEBS J. 274, 1588–1599 [DOI] [PubMed] [Google Scholar]
- 46. Nocek B., Evdokimova E., Proudfoot M., Kudritska M., Grochowski L. L., White R. H., Savchenko A., Yakunin A. F., Edwards A., and Joachimiak A. (2007) Structure of an amide bond forming F420:γ-glutamyl ligase from Archaeoglobus fulgidus: a member of a new family of non-ribosomal peptide synthases. J. Mol. Biol. 372, 456–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koon N., Squire C. J., and Baker E. N. (2004) Crystal structure of LeuA from Mycobacterium tuberculosis, a key enzyme in leucine biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 101, 8295–8300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ogunleye A., Bhat A., Irorere V. U., Hill D., Williams C., and Radecka I. (2015) Poly-γ-glutamic acid: production, properties and applications. Microbiology 161, 1–17 [DOI] [PubMed] [Google Scholar]