Abstract
CUL4B ubiquitin ligase belongs to the cullin-RING ubiquitin ligase family. Although sharing many sequence and structural similarities, CUL4B plays distinct roles in spermatogenesis from its homologous protein CUL4A. We previously reported that genetic ablation of Cul4a in mice led to male infertility because of aberrant meiotic progression. In the present study, we generated Cul4b germ cell-specific conditional knock-out (Cul4bVasa),as well as Cul4b global knock-out (Cul4bSox2) mouse, to investigate its roles in spermatogenesis. Germ cell-specific deletion of Cul4b led to male infertility, despite normal testicular morphology and comparable numbers of spermatozoa. Notably, significantly impaired sperm mobility caused by reduced mitochondrial activity and glycolysis level were observed in the majority of the mutant spermatozoa, manifested by low, if any, sperm ATP production. Furthermore, Cul4bVasa spermatozoa exhibited defective arrangement of axonemal microtubules and flagella outer dense fibers. Our mass spectrometry analysis identified INSL6 as a novel CUL4B substrate in male germ cells, evidenced by its direct polyubiquination and degradation by CUL4B E3 ligase. Nevertheless, Cul4b global knock-out males lost their germ cells in an age-dependent manner, implying failure of maintaining the spermatogonial stem cell niche in somatic cells. Taken together, our results show that CUL4B is indispensable to spermatogenesis, and it functions cell autonomously in male germ cells to ensure spermatozoa motility, whereas it functions non-cell-autonomously in somatic cells to maintain spermatogonial stemness. Thus, CUL4B links two distinct spermatogenetic processes to a single E3 ligase, highlighting the significance of ubiquitin modification during spermatogenesis.
Keywords: mouse genetics, protein degradation, spermatogenesis, ubiquitin, ubiquitin ligase
Introduction
Protein degradation via the ubiquitin-proteasome system plays a critical role during mammalian spermatogenesis. Timely removal of outlived proteins is crucial to ensure progression through different phases of spermatogenesis including mitotic, meiotic divisions, and postmeiotic morphogenesis. The ubiquitin-proteasome system selects its targets via a group of E3 ubiquitin ligases, which upon interaction with E2 ubiquitin-conjugating enzymes tag substrates for proteasomal degradation by the 26S proteasome. The largest mammalian E3 ligase family is the CRL (cullin-RING finger ubiquitin ligase) family, which includes eight members. Each cullin family member serves as a molecular scaffold and forms an E3 ligase complex with substrate-recruiting receptors and a linker protein (1). The Cul4b gene, located on the X chromosome, shares extensive sequence homology and functional redundancy with the other CRL4 family member, Cul4a. Both proteins employ DDB1 (DNA damage-binding protein 1) as the linker protein, which in turn recruits common or distinct DDB1-CUL4 associated factors for substrate binding. The DDB1-CUL4-mediated protein modification/degradation is involved in critical cellular processes including DNA replication, DNA repair, cell cycle control, and histone modifications (2).
Male infertility accounts for ∼40–50% of infertility cases in humans (3, 4). Many factors contribute to male infertility; however, the vast majority of male infertility cases are associated with oligozoospermia (decreased sperm number), asthenozoospermia (reduced sperm motility), and/or teratozoospermia (abnormal sperm morphology). In the mammalian testis, a single pluripotent spermatogonial stem cell (SSC)2 undergoes several rounds of mitotic divisions followed by meiotic divisions and complex postmeiotic morphogenesis, eventually giving rise to a cohort of mature spermatozoa. We have previously reported that the CRL4 proteins exhibited complementary expression patterns in adult mouse testis, where CUL4A was predominantly detected in primary spermatocytes, whereas CUL4B was highly expressed in Sertoli cells, spermatogonia, and spermatids (5). The absence of CUL4B in spermatocytes is likely due to meiotic sex chromosome inactivation, a transient X chromosome inactivation caused by sex chromosome condensation and subsequent gene silencing (6). However, the absence of Cul4a gene expression in Cul4b-expressing cells suggests that these two family members play distinct and nonoverlapping/nonredundant roles in specific testicular cell populations. We reported previously that loss of Cul4a in null mutant mice led to male infertility because of meiotic defects (5). Here we investigate the role of Cul4b in murine spermatogenesis. Our previous studies showed that Cul4b mutant mouse embryos died in midgestation because of a G2/M arrest in extraembryonic tissues, whereas epiblast-specific null mutants survived to adulthood with no overt developmental defects (7). To investigate the role of CUL4B protein in mammalian spermatogenesis, we generated and characterized germ cell-specific as well as global Cul4b mutant mice. We demonstrated that CUL4B is required for proper progression of spermiogenesis and spermatozoa motility and uncovered an unexpected non-cell-autonomous role for CUL4B in maintaining the SSC niche.
Experimental Procedures
Mice
Generation of conditional Cul4b mice was described previously (7). The Vasa(Ddx4)-Cre (stock no. 006954), Amh-Cre (stock no. 007915), and Sox2-Cre (stock no. 008454) transgenic mice were purchased from the Jackson Laboratory (Bar Harbor, ME). All mouse colonies were maintained in a barrier facility at Washington University in St. Louis, and all animal experiments were performed in accordance with the institution's regulations under an approved protocol.
Electron Microscopy, Histology, and Immunofluorescence (IF) of the Testis Tissues and Epididymal Spermatozoa
Preparations of testis sections and sperm smears, as well as histological analyses, were performed following protocols described previously (5). Antibodies and dilutions used were: 1:100 for CUL4B (Proteintech Group, Chicago, IL), PLZF (CalBiochem, Gibbstown, NJ), CUL4A (Bethyl Laboratories, Montgomery, TX), and 1:1000 for Alexa 594 goat anti-rabbit and Alexa 488 goat anti-mouse (Life Technologies, Inc.). Images were captured under a Zeiss AxioSkop 2 epifluorescence microscope. Transmission electron microscopy (TEM) was performed on ultrathin cauda epididymal sections as previously described (5).
Sperm Motility Assay
Cauda epididymides were dissected, pierced, and incubated in EmbryoMax® human tubal fluid (EMD Millipore) for 10 min at 37 °C in 5% CO2 to disperse spermatozoa. Sperm motility was determined using a computer-assisted sperm analysis system (Hamilton-Thorne Research, Beverly, MA).
Flow Cytometric Analysis
Single testicular cell suspensions and epididymal spermatozoa were prepared as previously described (5). Live spermatozoa were stained with JC1 dye (Life Technologies, Inc.) according to the manufacturer's instructions, followed by flow cytometry on the BD FACScanTM system (BD Biosciences, San Jose, CA). To prepare splenocyte suspensions, spleen removed from one mouse was meshed through a 70-μm cell strainer (BD Biosciences) with a syringe plunger and washed with cold PBS (calcium- and magnesium-free). After a brief spin, splenocyte pellets were resuspended and incubated in 5 ml of red blood cell lysis buffer (Sigma-Aldrich). The remaining cells were washed once in PBS, centrifuged, and processed for marker staining and FACS analysis. Isolated splenocytes and testicular cells were incubated in blocking solution (PBS with 1% bovine serum albumin and 1% mouse Fc receptor block (BioLegend, San Diego, CA)) for 15 min, followed by incubation with antibodies diluted in blocking solution for 30 min on ice, protected from light. Dilutions for antibodies and fluorescent dyes used were: 1:100 for anti-mouse GR1-FITC, anti-mouse F4/80-PE, anti-mouse CD11b-APC, and anti-mouse CD45-Pacific Blue, and 1:500 for 7AAD (all antibodies were from AnaSpec Inc., Fremont, CA). Following antibody incubation, the cells were fixed in 4% paraformaldehyde with actinomycin (ACROS Organics, Geel, Belgium) overnight at 4 °C and analyzed on a BD LSRFortessaTM cytometer the following day. The data analyses were completed by using FlowJo V10 software (TreeStar Inc., Ashland, OR).
Determination of Sperm ATP Level
Sperm extracts were prepared as previously described (8). The ATP levels were quantified by using the Molecular Probes® ATP determination kit (Life Technologies, Inc.) following the manufacturer's instructions. Bioluminescence was measured on a GloMax® microplate reader (Promega, Madison, WI), and a standard curve for a series of ATP standard solutions with concentrations ranging from 10 pm to 2.5 μm was generated. The ATP concentration of each sample was determined according to the standard curve, and sperm mass (total protein content) was determined by standard Bradford assay. Normalized ATP levels were calculated as ATP (micromoles)/sperm mass (gram). n = 4 for each genotype in two replicates.
Determination of Sperm Phosphoenolpyruvate (PEP) Level
A commercial fluorometric assay kit was used to determine sperm intracellular PEP level (Cayman Chemical Company, Ann Arbor, MI). Epididymal spermatozoa were collected, lysed, and deproteinated in the metaphosphoric acid buffer provided in the kit. Protein pellets were reconstituted, and concentration was determined for normalization purposes. Reaction was set up according to the manufacturer's instructions, and fluorescence was measured on the SynergyTM H4 hybrid microplate reader (Ex = 535 nm, Em = 590 nm). A PEP standard curve was plotted using a series of PEP solutions with concentrations ranging from 0 to 100 μm, and the PEP levels of individual samples were determined accordingly. Normalized PEP level was calculated as PEP concentration (μm)/sperm mass (mg). Four biological replicates were used for each genotype assayed in duplicate.
Affinity Purification and Identification of Ubiquitinated Testicular Proteins
Seminiferous tubules from control or Cul4bVasa testes were minced in PBS, and the suspension was pipetted repetitively for 1 min. Large fragments were allowed to settle for 15 min, and the remaining cell suspension was centrifuged at 600 × g for 5 min. Cell pellets were resuspended in 1 ml of PBS. To enrich for germ cells, cell suspension was carefully layered on top of a Percoll gradient in a 15-ml conical tube, composed of 1 ml each of 45, 30, 22, and 15% Percoll solutions. After centrifugation at 600 × g for 25 min, cell population in the lower 22% Percoll fraction was recovered by slow pipetting. This fraction contained largely round and elongated spermatids, with some remaining spermatocytes. Cell pellets were washed three times in PBS and resuspended in 0.2–0.5 ml immunoprecipitation assay buffer (1% Triton X-100, 150 mm NaCl, 20 mm Tris-Cl, pH 7.4, 1 mm EGTA, 1× protease inhibitor mixture; Roche Applied Science). To affinity-purify ubiquitinated proteins, 10–15 μl of p62-derived ubiquitin-associated (UBA) agarose beads (agarose immobilized recombinant ubiquitin-binding domain of p62/SQSTM1 protein; ENZO, catalog no. UW9010) were added to cell lysates and incubated overnight at 4 °C on a shaking platform. After removing supernatant containing unbound proteins, affinity-purified complexes were pelleted, washed in PBS three times, and submitted to the Proteomics and Mass Spectrometry Program core facility at Washington University School of Medicine for mass spectrometry analysis (detailed method available upon request). Scaffold (Proteome Software, Portland, OR) was used to validate protein identifications using PeptideProphet and to derive corresponding protein ID probability scores (9). The mass spectrometry data processing pipeline and detailed method for peptide quantification were described previously (10).
Cell Culture, Transfection, Immunoprecipitation, and Western Blotting
HEK 293 cells were maintained in DMEM supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. For the His-tagged ubiquitin-expressing plasmid ubiquitination assay, 80% confluent HEK293 cells were transiently transfected with 2 μg of His6-tagged ubiquitin-expressing plasmid (11, 12), HA-CUL4B (generated by Dr. Zhou's lab), and/or MYC-INSL6 (purchased pENTR223-INSL6 plasmid from Harvard PlasmID Database, then subcloned INSL6 ORF into pCS2-Myc vector in Dr. Zhou's lab) as specified, and treated with 20 μm MG132 2 h prior to harvesting. Trypsinized cells were pelleted and washed in PBS, and 1/20 of the cell aliquots were boiled in 2× SDS-PAGE buffer for Western blot analysis. The remaining cells were resuspended in immunoprecipitation buffer (0.1 m NaH2PO4, 10 mm Tris-HCl, pH 8.0, 6 m guanidine HCl, 10 mm imidazol) and sonicated. After brief centrifugation, supernatants were collected and incubated with nickel-nitrilotriacetic acid-agarose beads (Qiagen) for 2 h. nickel-nitrilotriacetic acid-precipitated proteins were released by boiling the agarose beads in 2×SDS-PAGE buffer, and analyzed by Western blotting. For substrate accumulation assay, 80% confluent HEK293 cells were transfected with 6 μg of INSL6-V5-TOPO (purchased pENTR223-INSL6 plasmid from Harvard PlasmID Database then subcloned INSL6 ORF into pCMV-V5 vector by Gateway system in Dr. Zhou's lab) along with 0, 0.5, 1 or 1.5 μg of HA-CUL4B plasmid, respectively. After 48 h, cells were harvested with Nonidet P-40 lysis buffer containing protease inhibitor mixture (Roche), and 15 μg of cell lysates were directly subjected to SDS-PAGE and immunoblotting. HEK293 cells were transfected with siRNA oligonucleotides (GE Healthcare Dharmacon Inc., Lafayette, CO) using Lipofectamine RNAiMAX reagent (Life Technologies, Inc.). The siRNA sequences are as follows: RBX1, 5′-GACTTTCCCTGCTGTTACCTAA-3′ and for DDB1, 5′-CTGTTGATTGCCAAAAAC-3. Co-immunoprecipitation was performed following routine procedure after co-transfection of HEK293 with 5 μg of MYC-INSL6 and 5 μg of HA-DDB1 (generated by Dr. Zhou's lab). Antibody dilutions used are: 1:8000 for anti-α-tubulin (Proteintech), 1:1000 for anti-MYC (Cell Signaling Technology, Danvers, MA), 1:1000 for anti-HA (Cell Signaling Technology), and 1:4000 for anti-V5 (Abcam) 1:500 for β-actin (Santa Cruz).
Results
Germ Cell- but Not Sertoli Cell-specific Cul4b-null Males Are Infertile
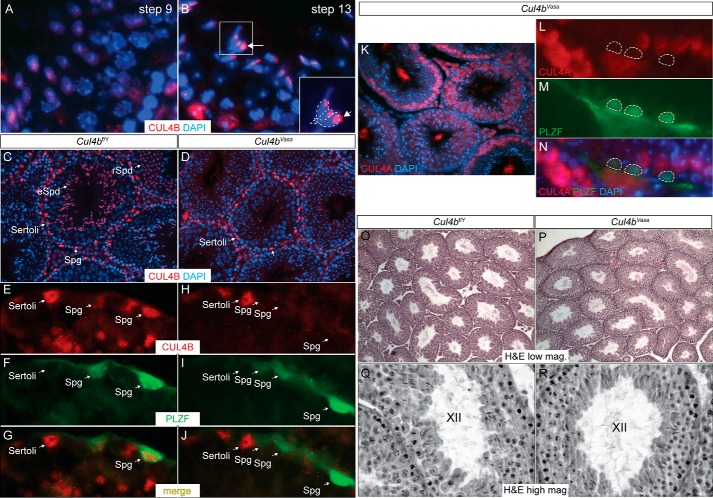
We previously demonstrated dynamic and complimentary expression patterns for the two Cul4 genes in the mouse testis and reported that loss of Cul4a led to male infertility caused by defects in meiotic progression (5). Because Cul4b is highly expressed in both somatic and germ cells in the testis, we first used the well established Vasa-Cre line (13) to generate germ cell-specific Cul4b mutants. These germ cell-specific mutants are hereafter referred to as Cul4bVasa, as compared with their wild type congeners, termed Cul4bf/Y. As shown in Fig. 1C and our previous study (5), CUL4B protein is predominantly detected in Sertoli cells, spermatogonia, and spermatids in adult wild type mouse testes. Close examination of wild type seminiferous tubule sections revealed that as spermiogenesis progresses, CUL4B protein gradually localizes to the cytoplasm adjacent to the postacrosomal segment of elongating spermatids, coinciding with the location of the caudala manchette, a transient microtubule-based spermatid structure (Fig. 1B, compare with Fig. 1A, arrows). In the Cul4bVasa testis, however, CUL4B protein was no longer detectable in germ cells, even though its expression was unaffected in Sertoli cells (Fig. 1D). To further confirm this finding, double immunofluorescence staining was performed using antibodies against CUL4B and an SSC marker, PLZF (14). High magnification epifluorescence microscopy showed complete removal of CUL4B from PLZF-positive spermatogonia, as well as round spermatids residing in the same seminiferous tubules (Fig. 1, E–J), demonstrating successful deletion of Cul4b in male germ cells.
FIGURE 1.
Germ cell-specific deletion of Cul4b does not affect testis morphology. A–D, IF of CUL4B in adult mouse testis. A and B, in wild type testis during spermiogenesis, CUL4B protein is dispersedly distributed in round spermatids (A), whereas more concentrated localization to the cytoplasmic lobe is detected in elongated spermatids (B). The developmental steps of spermatid differentiation were determined according to Russell et al. (47). C, CUL4B protein was detected in Sertoli cells (cells lining the seminiferous tubules with irregularly shaped nuclei and tripartite nucleoli), spermatogonia (Spg, cells lining the seminiferous tubules with round nuclei and often one visible nucleolus), round spermatids (rSpd, haploid cells located closer to the tubular lumen with smaller round nuclei and a large densely stained nucleolus), and elongated spermatids (eSpd, haploid cells located toward the tubular lumen with elongated nuclei) in a 4-month-old control Cul4bf/Y testis. D, Cul4b deletion through Vasa-Cre (Cul4bVasa) showed complete absence of the protein in all germ cell lineages, without affecting its expression in Sertoli cells. E–J, double IF of CUL4B and spermatogonial marker PLZF. CUL4B protein was readily detected in the PLZF-positive spermatogonia in the control testis (E–G) but not in the mutant testis (H–J). K–N, double IF of CUL4A and PLZF in Cul4bVasa testis showing no ectopic CUL4A expression in the absence of Cul4b. O–R, H&E staining showed normal histology of the Cul4bVasa testis, containing seminiferous tubules at all stages. K and L showed stage XII tubules of the two genotypes at higher magnification.
To address whether in the absence of CUL4B, spermatocytes turn on Cul4a expression to compensate for the CRL4 function, we performed double immunofluoroscence staining of CUL4A and PLZF proteins in the Cul4bVasa testis. No CUL4A staining was detected in PLZF-positive spermatogonia or spermatids of the Cul4bVasa testis (Fig. 1, K–N), suggesting that loss of CUL4B protein does not lead to compensatory Cul4a gene expression in these germ cell populations.
Cul4bVasa males exhibit no defects during development and have a normal life span. However, mutant males are completely infertile. Fertility records showed regular and successful copulatory activities of mutant males, evidenced by the presence of vaginal plugs in females housed in the same cages. However, none of the females mated with Cul4bVasa males produced any pups, in contrast to those mated with control males, averaging 11.2 pups per litter (Table 1).
TABLE 1.
Fertility records of Cul4bVasa and control males
| Males (n) | Females (n) | No. vaginal plug observed in 2 months | No. litters | No. pups | Average litter size |
|---|---|---|---|---|---|
| Cul4bf/Y (5) | Cul4bf/f (12) | 18 | 18 | 201 | 11.2 |
| Cul4bVasa (6) | Cul4bf/f (13) | 22 | 0 | 0 | 0 |
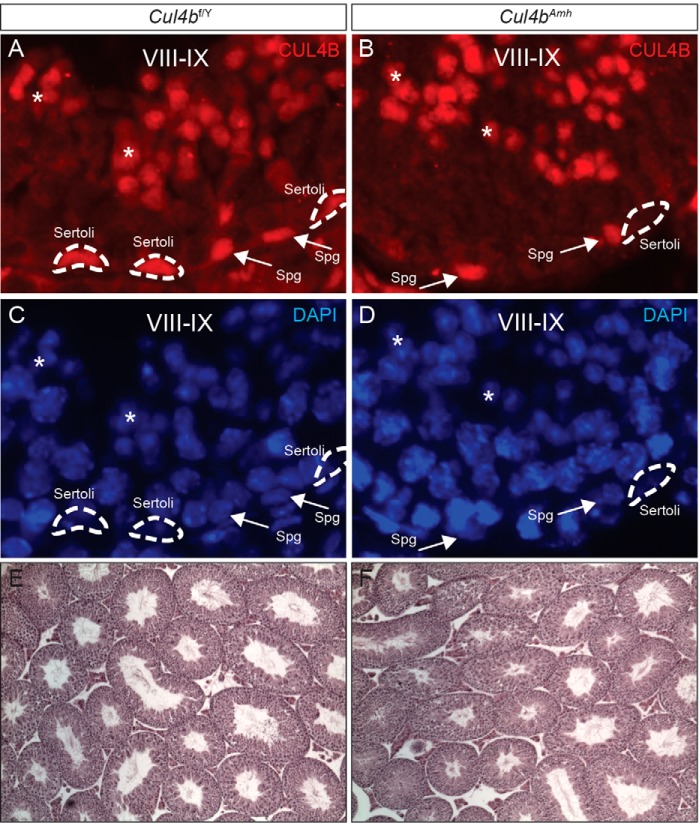
To address the function of CUL4B in Sertoli cells, we generated Sertoli cell-specific Cul4b conditional knock-out mice using the Amh-Cre mouse (15), hereafter referred to as Cul4bAmh. Interestingly, despite its high expression in Sertoli cells (Figs. 1E and 2A), abolishing Cul4b in these somatic cells did not cause any reproductive phenotype. Immunofluorescent staining on Cul4bAmh testis sections showed complete absence of CUL4B in their Sertoli cells (Fig. 2B); however, these males exhibited normal fertility and seminiferous epithelium histology (Fig. 2F). These results suggest that cell autonomous CUL4B function is critical to male germ cell lineage but is dispensable in Sertoli cells.
FIGURE 2.
Sertoli cell-specific deletion of Cul4b has no phenotype. A and B, CUL4B IF showing complete removal of CUL4B protein in the Cul4bAmh Sertoli cells. Dotted lines, Sertoli cells; arrows, spermatogonia; asterisks, spermatids. C and D, DAPI nuclear staining of the same sections shown in A and B. E and F, H&E staining showing normal testis morphology of Cul4bAmh males.
Spermatozoa in Cul4bVasa Testis Are Immotile
Anatomical examination of the Cul4bVasa mice revealed normal internal male reproductive organs, including the testes, epididymides, vas deferens, and seminal vesicles. No significant differences in size, weight, or morphology of the testes were noted between the two genotypes. As shown in Fig. 1 (O–R), seminiferous tubule histology of the two genotypes is indistinguishable, and germ cells of all phases are present in the Cul4bVasa testis.
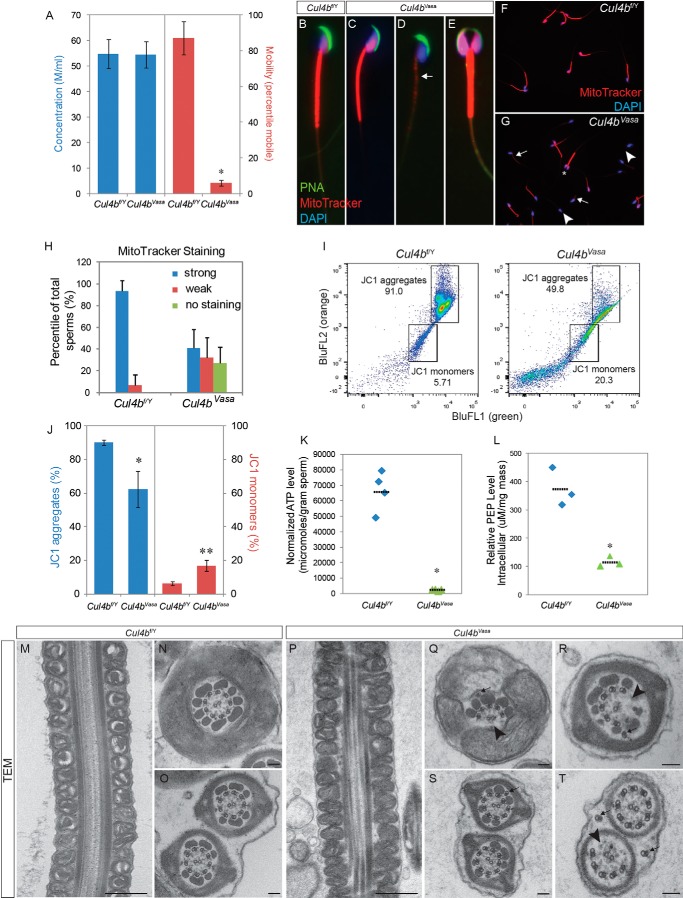
To investigate the cause of infertility in Cul4bVasa males, we harvested epididymal spermatozoa from these mice and used the computer-assisted sperm analysis to evaluate the quality of the mutant spermatozoa. As shown in Fig. 3A, Cul4bVasa produced equivalent numbers of spermatozoa as compared with their wild type counterparts (54.5 ± 5.1 m/ml, n = 4, versus 54.8 ± 5.6 M/ml, n = 5 in controls; p = 0.925). In contrast, sperm motility in the mutants was dramatically compromised: only 6.0 ± 1.4% mutant spermatozoa were progressive motile (n = 4), as compared with 87.2 ± 9.3% in control mice (n = 5, p < 0.0001). We next examined the morphology of epididymal spermatozoa using epifluorescence microscopy after staining with sperm structure-specific dyes. In typical wild type spermatozoa, the sickle-shaped heads contained highly condensed DNA, which stained positive for DAPI (Fig. 3B, blue), and were covered by a crescent-shaped acrosome stained by peanut agglutinin-lectin (green). The midpiece of the spermatozoa is covered by a helical mitochondrial sheath, and active mitochondria were stained bright red after live spermatozoa were incubated with a cell-permeable fluorescent dye, MitoTracker Red CMXRos (Fig. 3B). Most Cul4bVasa sperms exhibited normal morphology when examined under light microscopy; however, more than half of the mutant spermatozoa stained very weakly or negatively for MitoTracker CMXRos (Fig. 3, D, G, and H), suggesting a defect in mitochondrial membrane potential. Intriguingly, a small percentage (6.6 ± 3.4%) of Cul4bVasa spermatozoa exhibited a double-headed phenotype (Fig. 3, E and G, asterisk). The individual heads of these spermatozoa were morphologically normal, with properly condensed nuclei and visible acrosomes. Nevertheless, they were often joined at the connecting piece and shared a common flagellum.
FIGURE 3.
Cul4bVasa mice are infertile because of abnormal sperm morphology and low motility. A, computer-assisted sperm analysis of epididymal spermatozoa revealed normal sperm number but extremely low sperm motility. B–G, morphology of epididymal sperms of control and Cul4bVasa mice stained for acrosome (peanut agglutinin (PNA), green), active mitochondria (MitoTracker, red), and nucleus (DAPI, blue). Weak or no mitochondrial staining was noted in a large fraction of mutant sperms (arrow in D, compared with control in B or a relatively normal mutant sperm in C). E, in addition, 6.6 ± 3.4% mutant sperms showed double heads but joined by a fused flagellum. F and G, representative low magnification images of sperm smear stained with DAPI and MitoTracker. Arrows, weak MitoTracker staining; arrowheads, no MitoTracker staining; asterisk, double-headed sperm. H, quantification of MitoTracker staining. In controls, 93.4 ± 9.8% (n = 8) sperm showed strong staining. In contrast, 32.2 ± 18.1% and 27.0 ± 15.0% (n = 9) of mutant sperm showed either weak or no staining, respectively. I, representative FACS results of JC1-stained spermatozoa showing reduced JC1 aggregate population and increased JC1 monomer population in the mutant (right panel) than control (left panel). J, quantification of JC1 FACS showing significant changes in distribution of the two populations: JC1 aggregates (Cul4bf/Y, 90.2 ± 6.2%, n = 5; Cul4bVasa, 62.3 ± 10.8%, n = 3, p = 0.045) and JC1 monomers (Cul4bf/Y, 6.2 ± 1.1%, n = 5; Cul4bVasa, 16.8 ± 3.3%, n = 3 p = 0.024). K, markedly reduced ATP level in the mutant sperms. L, spermatozoa PEP levels were significantly reduced in the mutant sperms. *, p = 0.017. M–T, transmission electron microscopy revealed abnormal ultrastructure of Cul4bVasa spermatozoa. Mitochondrial sheath around midpiece appeared morphologically normal in Cul4bVasa (P) compared with that of the controls (M). Q and R, a spectrum of axoneme defects, including missing microtubule doublets and associated outer dense fibers (arrowheads), displaced (arrows in Q and T), and extra microtubule doublets (arrow in S). Typical morphology of cross-sections of at the principle piece and end piece were shown in N and O, respectively. Scale bars, 500 nm in M and P, 100 nm in the rest TEM images.
To further characterize the impaired mitochondrial activity observed in the Cul4bVasa spermatozoa, we stained epididymal spermatozoa with a vital carbocyanine dye, JC1, which fluoresces at different wavelengths when binds to mitochondria with differential membrane potentials, and analyzed JC1 staining profiles by FACS (16). Fig. 3I showed the FACS results of JC1-stained spermatozoa from representative control and Cul4bVasa mice. On average, 90.2 ± 6.2% of the control spermatozoa (n = 5) exhibited high mitochondrial membrane potential, evidenced by the orange JC1 staining (Fig. 3, I and J). However, this population in Cul4bVasa mice was significantly reduced (62.3 ± 10.8%, n = 3, p = 0.045), accompanied by an increase in the population of spermatozoa with low mitochondrial membrane potential stained green with JC1 (Cul4bVasa, 16.8 ± 3.3%, n = 3; control, 6.2 ± 1.1%, n = 5; p = 0.024). These results were consistent with our findings with MitoTracker staining and statistically demonstrated that lack of Cul4b in mouse germ cells led to defective mitochondrial activity in some spermatozoa.
ATP is the energy source that is hydrolyzed by the dynein ATPase to generate the propelling and coordinated force along the axoneme that enables proper sperm movement (17, 18). The immotile phenotype of Cul4bVasa prompted us to examine ATP production in the mutant spermatozoa by analyzing intracellular ATP level using a previously described method (8). Not surprisingly, we observed low ATP production in Cul4bVasa spermatozoa (2.4 ± 0.5 × 103 μmoles/g versus 6.7 ± 1.3 × 104 μmoles/g in controls, n = 4, p = 0.002) determined by a bioluminescence assay (Fig. 3K). There is continuous debate on the source of sperm ATP, and two metabolic pathways are believed to be involved in this process. Mitochondria, localized in the sperm tail midpiece, have long been considered to be a major source of ATP production because of their high efficiency of energy production via oxidative respiration (19, 20). Recent evidence, however, suggests that locally produced ATP via less efficient, but high throughput glycolysis in the principle piece of the sperm tail is the major energy source for sperm motility in the mouse (21–23). In our Cul4bVasa mutants, the reduction in mitochondrial membrane potential in spermatozoa may contribute to lowered ATP production. However, loss of ATP production in mutant spermatozoa may also reflect a defective glycolytic pathway. To test this hypothesis, we measured the spermatozoa intracellular PEP level using a fluoremetric assay kit. PEP is the intermediate produced during glycolysis that is metabolized to the final product, pyruvate, by pyruvate kinase. We chose PEP instead of pyruvate as the indicator of glycolysis because pyruvate could also come from other metabolic pathways including amino acid metabolism (serine, alanine, and glycine), as well as conversion from lactate by lactate dehydrogenase. As shown in Fig. 3L, a significant decrease in PEP levels was detected in the mutant spermatozoa (Cul4bf/Y, 374.8 ± 68.0, n = 3; Cul4bVasa, 116.4 ± 18.8, n = 3, p = 0.017), indicating perturbation of glycolysis in the mutant spermatozoa. Taken together, these results suggest that a lack of ATP production resulting from impaired mitochondrial activity and glycolysis was a major cause of low motility/asthenozoospermic phenotype in Cul4bVasa spermatozoa.
We next examined the ultrastructure of mutant spermatozoa by TEM because a loss of sperm motility is often associated with defective axoneme structure. The TEM analysis revealed that Cul4bVasa mutant sperm axonemes frequently displayed missing, supernumerary, or displaced microtubule doublets (Fig. 3, Q–T). These defects were observed mostly in peripheral microtubule doublets, as opposed to the central microtubule doublet, and it was not clear whether the displaced microtubule doublets were still associated with dynein arms or radial spokes. The mitochondrial sheaths of the mutant spermatozoa appeared morphologically normal (Fig. 3P). These data collectively demonstrated that deleting Cul4b from mouse spermatogonia, and subsequent germ cell populations did not affect the mitotic and meiotic phase of spermatogenesis but compromised spermiogenesis and biogenesis of the sperm flagellum in particular. Lack of Cul4b in these cells resulted in abnormal axonemal microtubule structure and reduced mitochondrial activity in the midpiece, which rendered Cul4bVasa spermatozoa immotile and subsequently male infertile.
INSL6 Is a Direct CUL4B Substrate in Mouse Testes
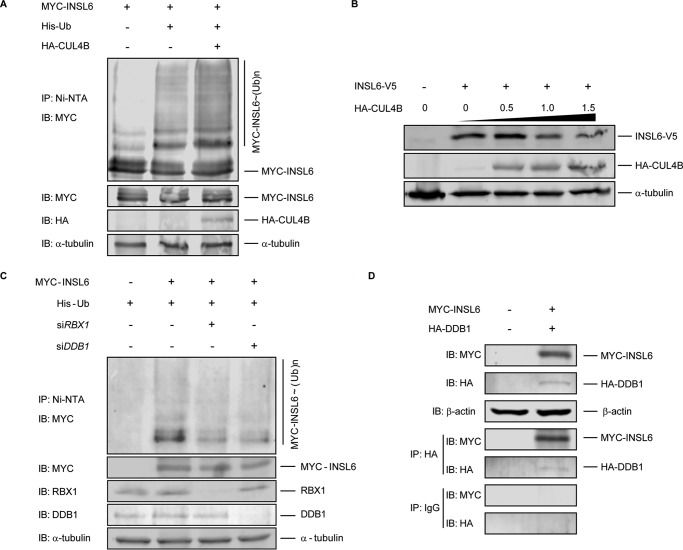
Given the function of CUL4B as an E3 ubiquitin ligase, it is reasonable to hypothesize that in the Cul4b-null germ cells, certain CUL4B substrates may not be efficiently ubiquitinated, which led to defective spermiogenesis. In an attempt to identify potential CUL4B ligase substrates, we performed a mass spectrometry analysis on ubiquitinated proteins purified from enriched spermatids of control and Cul4bVasa mice. Ubiquitinated proteins from pooled spermatid lysates were affinity purified through binding to p62-UBA immobilized agarose beads, and CUL4B target candidates were expected to be under-represented in Cul4bVasa samples. The identity of potential targets were predicted and visualized by Scaffold 4 following mass spectrometry (9). In total, 2504 proteins were identified with a minimum protein threshold of 95.0% and a prophet false discovery rate of 0.6% or lower (supplemental Table S1). Among these, an insulin family member, INSL6, showed high spectrum count in the control testis but was absent in the p62-UBA-enriched testis lysates from Cul4bVasa, suggesting INSL6 as a potential CUL4B substrate in the testis. INSL6 was previously reported to be highly expressed in germ cells, particularly in spermatids of both rodent and human testes (24) (HPA021364 SIGMA), although its function in spermatogenesis remains unknown. To test the ability of CUL4B to directly mediate polyubiquitination of INSL6, a plasmid encoding MYC-labeled full-length INSL6 was co-transfected into HEK293 cells along with His-tagged ubiquitin-expressing plasmid, with or without HA-CUL4B-expressing plasmid. As shown in Fig. 4A, the presence of CUL4B evidently increased polyubiquitinated INSL6, demonstrating that INSL6 is a direct substrate for CUL4B. Next HEK293 cells were co-transfected with INSL6-coding plasmid (INSL6-V5) along with various amount of HA-CUL4B plasmids as indicated in Fig. 4B, and a clear CUL4B dose-dependent decrease in INSL6 protein level was detected, indicating that INSL6 is directly degraded through the CUL4B-mediated ubiquitin-proteasome system mechanism. To further investigate the role of CRL complex in INSL6 polyubiquitination, we tested interactions between INSL6 and two key components of CRL4, the N-terminal binding DDB1 and the C-terminal interacting RING finger protein RBX1 (Rbx1/ROC1/Hrt1) (25). As shown in Fig. 4C, attenuating DDB1 and RBX1 functions, respectively, by RNA silencing in HEK293 cells resulted in reduced levels of INSL6 polyubiquitination. Moreover, DDB1 co-immunoprecipitated with INSL6 after co-transfection of MYC-INSL6 and HA-DDB1 plasmids into HEK293 cells, which indicates direct interactions between the two proteins (Fig. 4D). Together these data demonstrate that CUL4B directly polyubiquitinates INSL6 via the CRL complex in vitro, which leads to its degradation through ubiquitin-proteasome system.
FIGURE 4.
CRL promotes polyubiquitination and degradation of INSL6. A, co-expression of CUL4B resulted in increased amount of multiple slower migrating forms of polyubiquitinated INSL6 in HEK293 cells. Immunoblotting against MYC (Myc-INSL6), HA (HA-CUL4B), and α-tubulin, respectively, were performed on lysates prior to nickel-nitrilotriacetic acid immunoprecipitation. B, immunoblotting against V5 (INSL6-V5), HA (HA-CUL4B), and α-tubulin, respectively, on HEK293 cell lysates, co-transfected with INSL6-V5 and indicated amount of HA-CUL4B plasmid. C, gene silencing by RNAi of DDB1 and RBX1 resulted in reduction of polyubiquitinated INSL6 in HEK293 cells. D, co-immunoprecipitation of DDB1 and INSL6 in HEK293. β-Actin was used as loading control. IB, immunoblotting; IP, immunoprecipitation; Ni-NTA, nickel-nitrilotriacetic acid.
Global Cul4b Deletion Leads to Germ Cell Depletion in Aged Mice
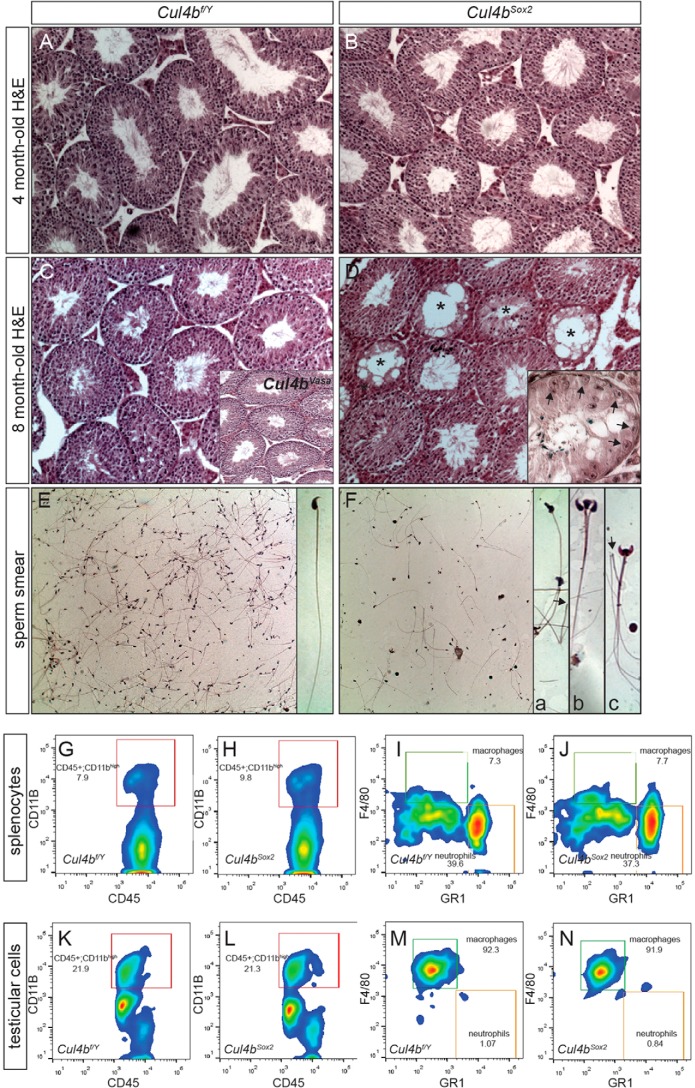
We have previously reported that Cul4b knock-out embryos died soon after implantation because of increased apoptosis in extraembryonic tissues, whereas epiblast-specific deletion of Cul4b via Sox2-Cre gave rise to viable Cul4b-null mice (Cul4bSox2) (7). Because Cul4b is expressed in many somatic cell types, we characterized the fertility phenotype of Cul4bSox2 males and compared with that of Cul4bVasa to determine whether Cul4b also functions in somatic cells to support spermatogenesis. Cul4bSox2 males were completely infertile as expected; however, they exhibited a different and more severe age-dependent testicular/sperm phenotype when compared with their Cul4bVasa counterparts. Younger adult Cul4bSox2 (4-month-old) testes exhibited no distinct defects (Fig. 5B), and their sperm phenotype resembled that in the Cul4bVasa mice (data not shown), suggesting relatively normal mitotic and meiotic progression despite impaired spermiogenesis. However, as Cul4bSox2 mutants aged (8-month-old), their testes shrank, and many seminiferous tubules started to lose their germ cell populations (Fig. 5D). Such a germ cell depletion phenotype varied between individuals (Fig. 5D, inset), but was not detected in older Cul4bVasa mice (Fig. 5C, inset). As a result, oligoasthenozoospermia was often observed in older Cul4bSox2 mutants (Fig. 5F; 54.8 ± 5.6 m/ml, n = 5 in controls; 16.0 ± 13.6 m/ml, n = 4 in Cul4bSox2; p = 0.007). Accompanied by low sperm counts, abnormal morphological phenotypes including deformed sperm heads, double- or triple-headed spermatozoa, and sperm heads detached from the flagella were common in Cul4bSox2 mutants (Fig. 5F, insets). This germ cell depletion phenotype may have resulted from a failure to maintain the SSC niche.
FIGURE 5.
Germ cell depletion in aged Cul4bSox2 testes. A–D, H&E staining of testis sections showing normal morphology at 4 months of age (B), but deteriorated structure by 8 months, with many empty seminiferous tubules (asterisks in D). The phenotype varied among individuals, with some having many tubules composed of Sertoli cells only (arrows, inset in D). Inset in C showed the normal histological organization of the 8-month-old Cul4bVasa testis. E and F, H&E staining of sperm smears of 8-month-old mice, showing reduced number of sperms in the mutant (F) with defective morphology. Inset a, deformed sperm head; inset b, triple head; inset c, double head. Arrows in insets a and c point to severed sperms with only flagella remaining. G–N, cell surface marker staining followed by FACS revealed no distinctive difference in macrophage population between the two genotypes.
Sertoli cells play a crucial role in SSC niche maintenance (26). We tested the hypothesis whether Cul4b expression in Sertoli cells is required to maintain the SSC niche. As mentioned earlier, knocking out Cul4b in Sertoli cells alone did not cause an infertility phenotype (Fig. 2). We followed these mice as they aged (up to 10 months) but did not observe the germ cell depletion phenotype described above. These data argue against a role for Cul4b in Sertoli cells to maintain the SSC niche and suggest that other somatic populations are responsible for this phenotype. One such somatic cell population could be macrophages. It was estimated that macrophages represent as much as one quarter of the interstitial cell population in rodent testes (27, 28), and they play essential roles in regulating the SSC niche (29). In humans, increased number of testicular macrophages was often reported in patients with male infertility (30). Because the germ cell depletion phenotype in aging Cul4bSox2 mutants was often accompanied by splenomegaly (data not shown), we hypothesized that loss of Cul4b in the spleen may have caused a systemic perturbation of macrophages, which may in turn adversely affected spermatogonial maintenance and self-renewal. To address this possibility, surface marker staining followed by FACS analysis was performed on dissociated splenocytes and testicular cells of control and Cul4bSox2 animals at 9 month of age. We employed four-color FACS to distinguish different immunocyte populations: CD45 as a marker for nucleated hematopoietic cells (31), CD11b as a marker for macrophage/microglia (32), F4/80 as a mature macrophage marker (33), and GR1 as a neutrophil marker in peripheral organs (34). The splenocytes isolated from control animals contained 11.05 ± 1.96% CD45+CD11bhigh macrophages/granulocytes of all CD45+ lymphocytes (Fig. 5G), among which 5.81 ± 1.61% were F4/80 highGR1low macrophages (n = 5; Fig. 4I). No statistically significant changes were observed in the Cul4bSox2 splenocytes (CD45+CD11bhigh: 12.02 ± 1.65%, n = 6, p = 0.41; F4/80 highGR1low: 5.50 ± 1.66%, n = 6, p = 0.76; Fig. 5, H and J). The isolated testicular cells from control animals comprised of 21.12 ± 4.54% CD45+CD11bhigh macrophages/granulocytes of all lymphocytes, and the vast majority of them were macrophages (F4/80 highGR1low: 91.84 ± 2.32%, n = 5; Fig. 5, K and M). CD45+CD11bhigh population remained largely unchanged in Cul4bSox2 testes (17.20 ± 5.30%, n = 6, p = 0.22; Fig. 5L); no statistically significant difference in macrophages was noted in these mutants (88.81 ± 3.36%, n = 6, p = 0.11; Fig. 5N). Comparison between the lymphocyte compositions in the spleen and testis of old (10–12 months old) and young (4 months old) adult Cul4bSox2 mice revealed no significant difference. Altogether, these data demonstrated that lack of Cul4b in both germ cells and somatic cells led to male infertility caused by spermiogenesis defects similar to that found in the Cul4bVaas mice and age-dependent germ cell depletion. The cause of the latter phenotype is yet to be determined but is unlikely to be associated with abnormal macrophage infiltration to the mutant testis.
Discussion
In the present study, we assessed the function of the E3 ubiquitin ligase CUL4B in spermatogenesis using genetically modified mouse models. We discovered a male infertility phenotype in both germ cell-specific conditional Cul4b knock-outs, as well as global Cul4b-null mutants.
Abolishing Cul4b in male germ cells did not cause any apparent morphological changes to the testis, and the progression of spermatogenesis appeared to be normal. The seminiferous tubules of Cul4bVasa conditional knock-outs contained male germ cell populations of all phases, and a normal number of spermatozoa were recovered from these animals. The cause of male infertility in the mutants is low sperm motility (93% reduction compared with controls). Several factors could contribute to this decrease in motility. We detected a varied level of decreased mitochondrial activity and loss of mitochondrial membrane potential, evidenced by MitoTracker Red CMXRos and JC1 staining (Fig. 3). The mitochondrial sheath, tightly wrapped around the axoneme of the sperm tail midpiece, may provide energy in the form of ATP required for flagella movement. In humans, numerous cases of idiopathic and varicocele-related asthenozoospermia have been reported to be associated with defective mitochondrial respiratory functionalities, with or without phenotypic abnormalities of the mitochondrial sheath (35–37). Furthermore, a positive correlation was found between high mitochondrial membrane potential and increased sperm motility in a human male infertility study (37). Oxidative phosphorylation relies on the H+ concentration gradient generated and maintained by the electron transport chain, which requires the impermeability of inner mitochondrial membrane. A decrease in mitochondrial membrane potential therefore indicates an inner membrane breach that could potentially lead to impaired ATP production. Indeed, measurement of sperm ATP level revealed an almost complete loss of ATP in the Cul4bVasa spermatozoa. However, the partial reduction in mitochondrial activity cannot fully account for the near total loss of ATP production in mutant spermatozoa. Therefore, the alternative pathway for ATP production, glycolysis, could also be impaired. We measured spermatozoa intracellular levels of PEP, the second to last intermediate of glycolysis, and revealed a significant decrease compared with the controls (Fig. 3). Therefore, loss of CUL4B simultaneously caused reduction of mitochondrial activity in the midpiece and compromised glycolysis, which resulted in lack of energy supply to mutant spermatozoa, ultimately leading to the immotile phenotype. In addition, we observed structural defects in the mutant spermatozoa. Specifically, the axonemes of many mutant spermatozoa showed microtubule anomalies, including supernumerary, missing, or misplaced outer microtubule doublets (Fig. 3). During murine spermiogenesis, the distal centriole nucleates microtubule polymerization to form the axoneme and the proximal centriole organizes the anlage of the capitulum, followed by disintegration of the centrosome in spermatids. Abnormal microtubule arrangements found in Cul4bVasa sperm flagella suggest potential structural defects in the distal centriole or failure, at least in part, of microtubule nucleation.
To uncover the molecular mechanisms leading to these defects, we performed proteomic analysis comparing ubiquitinated proteins isolated from control and Cul4bVasa spermatids by affinity purification using p62-UBA domain-immobilized beads. Among the proteins identified as potential CUL4B targets, INSL6 showed relatively high spectrum count in control samples but was completely absent in Cul4bVasa samples. INSL6 is an insulin-like peptide that shows significant sequence similarity to the relaxin B chain (38). Intriguingly, high INSL6 expression has been reported in both mouse and human male germ cells, specifically in spermatids (24) (HPA021364 SIGMA), and INSL6 mutation in mouse led to compromised fertility in males only (39). Rearrangement of INSL6-containing chromosome 9q24.1 region in humans has been reported to be associated with testicular development failure (40), even though its function in spermatogenesis remains ambiguous. In this study, we performed in vitro experiments and provided solid evidence that CUL4B mediates both INSL6 polyubiquitination, as well as its degradation, implicating INSL6 as a direct substrate of the CUL4B E3 ubiquitin ligase (Fig. 4). Direct interaction between INSL6 and DDB1 was also demonstrated. In addition, knocking down either DDB1 or RBX1 resulted in a reduction in INSL6 polyubiquitination (Fig. 4). It is unfortunate that there is currently no commercially available antibody against mouse INSL6, which prevented us from analyzing INSL6 expression and accumulation in mouse germ cells. Further investigations aiming at revealing the function of INSL6 is underway, which will shed light on understanding the underlying mechanisms of its involvement in spermatogenesis.
Interestingly, Cul4b-null mutation by Sox2-Cre resulted in a different male fertility phenotype, in that the aged mutant mice exhibited a germ cell depletion phenotype. This led us to hypothesize that CUL4B is required to maintain the SSC niche. It is believed that the SSCs are not randomly distributed along the basement membrane of seminiferous tubules but rather preferably located to “SSC niches” that are adjacent to the interstitial tissues rich in blood vessels (41, 42). Several types of somatic cells are responsible for maintaining the SSC niche. Inside the seminiferous tubules, Sertoli cells are thought to provide growth factors regulating SSC self-renewal/proliferation, including FGF2, GDNF (glial cell line-derived neurotrophic factor), and SCF (stem cell factor) (43–45). Because Cul4b was found to be highly expressed in Sertoli cells and loss of Cul4b was not compensated by Cul4a, we first speculated that it played a cell autonomous role in the Sertoli cells to maintain the SSC niche. However, we found no germ cell depletion phenotype in Sertoli-specific Cul4b mutants (Cul4bAmh; Fig. 2). This result suggests that Cul4b is dispensable in Sertoli cells. The interstitial Leydig cells are thought to regulate Sertoli cell function through growth factor production via testosterone signaling. There is a possibility that loss of CUL4B in Leydig cells altered this signaling, causing disturbance in Sertoli cell functions. However, measurements of circulating testosterone in Cul4bSox males revealed no significant difference compared with controls (data not shown). It was also proposed that interstitial macrophages play essential roles in maintaining SSC niche, and infiltration of macrophages in the testis was reported in patients with male infertility (30). High expression of Cul4b was reported in macrophages, and loss of CUL4B protein by shRNA knockdown significantly reduced TNFα protein in the activated macrophages (46). This led us to test whether Cul4b loss caused changes in testicular macrophage populations, which may in turn cause SSC loss. We tested this hypothesis by analyzing immunocyte composition of the spleen and testis tissues of control and Cul4bSox2 animals. We used F4/80 as a macrophage marker for FACS analysis, and no noticeable differences were discovered between the two genotypes or mutants of different ages (Fig. 5). Thus, loss of SSCs in Cul4bSox2 mutant testis is not the result of a change in testicular macrophage number. However, our study does not exclude the possibility that loss of Cul4b may change the properties of testicular macrophages, thereby affecting their ability to maintain the SSC niche. One final cell population that could contribute to the germ cell loss phenotype is the peritubular myoid cell. However, we did not detect either CUL4B or CUL4A expression in this cell type; hence it is unlikely that CUL4B is required in these cells to maintain the stem cell niches. It is noteworthy that our global mutants manifest a male germ cell depletion phenotype with a rather late adult onset, in contrast to most other germ cell depletion mouse models exhibiting early onset. This makes our animals an excellent model for studying long term maintenance of the SSC niche. In conclusion, CUL4B is required in the male germ cells for spermiogenesis and in somatic cells to maintain the SSC population of the testis.
Author Contributions
L. M. and P. Z. designed the study, and Y. Y., L. L., C. Y., C. L., G. M. V., C. W., and P. S. contributed to data acquisition and analysis. Y. Y., L. M., and P. Z. wrote the paper.
Supplementary Material
Acknowledgments
We thank Dr. Indira Mysorekar for providing antibodies for FACS, Drs. Erica Schoeller and Kelle Moley for help with computer-assisted sperm analysis, Dr. Frans van Roy (Universiteit Gent) for kindly gift of α-cateinin antibody, and Jaclynn Lett and the Research Center for Auditory and Vestibular Studies, Department of Otolaryngology at Washington University for technical assistance with electron microscopy. We also thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital (St. Louis, MO) for the use of the High Speed Cell Sorter Core and the Proteomics Core, which provided cell sorting service and mass spectrometry service, respectively.
This work was supported in part by NCI Cancer Center Support Grant P30 CA91842 (to the Siteman Cancer Center). This work was supported by National Institutes of Health Grants ES016597 (to L. M.) and 1R01 CA 159925 and 1R01 CA098210 (to P.Z.) and by U.S. Department of Agriculture National Institute of Food and Agriculture Awards 2011-67015-20025 and 2013-67015-20961 and seed funding from the Food for the 21st Century Program of the University of Missouri (to P. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table S1.
- SSC
- spermatogonial stem cell
- IF
- immunofluorescence
- TEM
- transmission electron microscopy
- H&E
- hematoxylin and eosin
- PEP
- phosphoenolpyruvate
- UBA
- ubiquitin-associated.
References
- 1. Lee J., and Zhou P. (2012) Pathogenic role of the CRL4 ubiquitin ligase in human disease. Front. Oncol. 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sugasawa K. (2009) The CUL4 enigma: culling DNA repair factors. Mol. Cell 34, 403–404 [DOI] [PubMed] [Google Scholar]
- 3. Brugh V. M. 3rd, and Lipshultz L. I. (2004) Male factor infertility: evaluation and management. Med. Clin. N. Am. 88, 367–385 [DOI] [PubMed] [Google Scholar]
- 4. Hirsh A. (2003) Male subfertility. BMJ 327, 669–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin Y., Lin C., Kim S. T., Roig I., Chen H., Liu L., Veith G. M., Jin R. U., Keeney S., Jasin M., Moley K., Zhou P., and Ma L. (2011) The E3 ubiquitin ligase cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 356, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Turner J. M. (2007) Meiotic sex chromosome inactivation. Development 134, 1823–1831 [DOI] [PubMed] [Google Scholar]
- 7. Liu L., Yin Y., Li Y., Prevedel L., Lacy E. H., Ma L., and Zhou P. (2012) Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 22, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miki K., Qu W., Goulding E. H., Willis W. D., Bunch D. O., Strader L. F., Perreault S. D., Eddy E. M., and O'Brien D. A. (2004) Glyceraldehyde 3-phosphate dehydrogenase-S, a sperm-specific glycolytic enzyme, is required for sperm motility and male fertility. Proc. Natl. Acad. Sci. U.S.A. 101, 16501–16506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Searle B. C. (2010) Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 10, 1265–1269 [DOI] [PubMed] [Google Scholar]
- 10. Perrin R. J., Payton J. E., Malone J. P., Gilmore P., Davis A. E., Xiong C., Fagan A. M., Townsend R. R., and Holtzman D. M. (2013) Quantitative label-free proteomics for discovery of biomarkers in cerebrospinal fluid: assessment of technical and inter-individual variation. PLoS One 8, e64314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Treier M., Staszewski L. M., and Bohmann D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78, 787–798 [DOI] [PubMed] [Google Scholar]
- 12. Lee J., Shieh J. H., Zhang J., Liu L., Zhang Y., Eom J. Y., Morrone G., Moore M. A., and Zhou P. (2013) Improved ex vivo expansion of adult hematopoietic stem cells by overcoming CUL4-mediated degradation of HOXB4. Blood 121, 4082–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gallardo T., Shirley L., John G. B., and Castrillon D. H. (2007) Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Costoya J. A., Hobbs R. M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K. E., Wolgemuth D. J., and Pandolfi P. P. (2004) Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 36, 653–659 [DOI] [PubMed] [Google Scholar]
- 15. Holdcraft R. W., and Braun R. E. (2004) Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development 131, 459–467 [DOI] [PubMed] [Google Scholar]
- 16. Gravance C. G., Garner D. L., Baumber J., and Ball B. A. (2000) Assessment of equine sperm mitochondrial function using JC-1. Theriogenology 53, 1691–1703 [DOI] [PubMed] [Google Scholar]
- 17. Storey B. T., and Kayne F. J. (1980) Properties of pyruvate kinase and flagellar ATPase in rabbit spermatozoa: relation to metabolic strategy of the sperm cell. J. Exp. Zool. 211, 361–367 [DOI] [PubMed] [Google Scholar]
- 18. Halangk W., Tröger U., and Bohnensack R. (1990) Quantification of aerobic energy turnover in epididymal bull spermatozoa. Biochim. Biophys. Acta 1015, 243–247 [DOI] [PubMed] [Google Scholar]
- 19. Storey B. T. (1980) Strategy of oxidative metabolism in bull spermatozoa. J. Exp. Zool. 212, 61–67 [DOI] [PubMed] [Google Scholar]
- 20. Storey B. T., and Kayne F. J. (1977) Energy metabolism of spermatozoa. VI. Direct intramitochondrial lactate oxidation by rabbit sperm mitochondria. Biol. Reprod. 16, 549–556 [PubMed] [Google Scholar]
- 21. Krisfalusi M., Miki K., Magyar P. L., and O'Brien D. A. (2006) Multiple glycolytic enzymes are tightly bound to the fibrous sheath of mouse spermatozoa. Biol. Reprod. 75, 270–278 [DOI] [PubMed] [Google Scholar]
- 22. Westhoff D., and Kamp G. (1997) Glyceraldehyde 3-phosphate dehydrogenase is bound to the fibrous sheath of mammalian spermatozoa. J. Cell Sci. 110, 1821–1829 [DOI] [PubMed] [Google Scholar]
- 23. Mukai C., and Okuno M. (2004) Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol. Reprod. 71, 540–547 [DOI] [PubMed] [Google Scholar]
- 24. Lok S., Johnston D. S., Conklin D., Lofton-Day C. E., Adams R. L., Jelmberg A. C., Whitmore T. E., Schrader S., Griswold M. D., and Jaspers S. R. (2000) Identification of INSL6, a new member of the insulin family that is expressed in the testis of the human and rat. Biol. Reprod. 62, 1593–1599 [DOI] [PubMed] [Google Scholar]
- 25. Kamura T., Koepp D. M., Conrad M. N., Skowyra D., Moreland R. J., Iliopoulos O., Lane W. S., Kaelin W. G. Jr., Elledge S. J., Conaway R. C., Harper J. W., and Conaway J. W. (1999) Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661 [DOI] [PubMed] [Google Scholar]
- 26. Oatley M. J., Racicot K. E., and Oatley J. M. (2011) Sertoli cells dictate spermatogonial stem cell niches in the mouse testis. Biol. Reprod. 84, 639–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Niemi M., Sharpe R. M., and Brown W. R. (1986) Macrophages in the interstitial tissue of the rat testis. Cell Tissue Res. 243, 337–344 [DOI] [PubMed] [Google Scholar]
- 28. Giannessi F., Giambelluca M. A., Scavuzzo M. C., and Ruffoli R. (2005) Ultrastructure of testicular macrophages in aging mice. J. Morphol. 263, 39–46 [DOI] [PubMed] [Google Scholar]
- 29. DeFalco T., Potter S. J., Williams A. V., Waller B., Kan M. J., and Capel B. (2015) Macrophages contribute to the spermatogonial niche in the adult testis. Cell Rep. 12, 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frungieri M. B., Calandra R. S., Lustig L., Meineke V., Köhn F. M., Vogt H. J., and Mayerhofer A. (2002) Number, distribution pattern, and identification of macrophages in the testes of infertile men. Fertility Sterility 78, 298–306 [DOI] [PubMed] [Google Scholar]
- 31. Penninger J. M., Irie-Sasaki J., Sasaki T., and Oliveira-dos-Santos A. J. (2001) CD45: new jobs for an old acquaintance. Nat. Immunol. 2, 389–396 [DOI] [PubMed] [Google Scholar]
- 32. Perego C., Fumagalli S., and De Simoni M. G. (2011) Temporal pattern of expression and colocalization of microglia/macrophage phenotype markers following brain ischemic injury in mice. J. Neuroinflammation 8, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Austyn J. M., and Gordon S. (1981) F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur. J. Immunol. 11, 805–815 [DOI] [PubMed] [Google Scholar]
- 34. Miyazaki S., Ishikawa F., Shimizu K., Ubagai T., Edelstein P. H., and Yamaguchi K. (2007) Gr-1high polymorphonuclear leukocytes and NK cells act via IL-15 to clear intracellular Haemophilus influenzae in experimental murine peritonitis and pneumonia. J. Immunol. 179, 5407–5414 [DOI] [PubMed] [Google Scholar]
- 35. Evenson D. P., Darzynkiewicz Z., and Melamed M. R. (1982) Simultaneous measurement by flow cytometry of sperm cell viability and mitochondrial membrane potential related to cell motility. J. Histochem. Cytochem. 30, 279–280 [DOI] [PubMed] [Google Scholar]
- 36. Ruiz-Pesini E., Diez C., Lapeña A. C., Pérez-Martos A., Montoya J., Alvarez E., Arenas J., and López-Pérez M. J. (1998) Correlation of sperm motility with mitochondrial enzymatic activities. Clin. Chem. 44, 1616–1620 [PubMed] [Google Scholar]
- 37. Marchetti C., Obert G., Deffosez A., Formstecher P., and Marchetti P. (2002) Study of mitochondrial membrane potential, reactive oxygen species, DNA fragmentation and cell viability by flow cytometry in human sperm. Hum. Reprod. 17, 1257–1265 [DOI] [PubMed] [Google Scholar]
- 38. Hsu S. Y. (1999) Cloning of two novel mammalian paralogs of relaxin/insulin family proteins and their expression in testis and kidney. Mol. Endocrinol. 13, 2163–2174 [DOI] [PubMed] [Google Scholar]
- 39. Burnicka-Turek O., Shirneshan K., Paprotta I., Grzmil P., Meinhardt A., Engel W., and Adham I. M. (2009) Inactivation of insulin-like factor 6 disrupts the progression of spermatogenesis at late meiotic prophase. Endocrinology 150, 4348–4357 [DOI] [PubMed] [Google Scholar]
- 40. Ion R., Telvi L., Chaussain J. L., Barbet J. P., Nunes M., Safar A., Réthoré M. O., Fellous M., and McElreavey K. (1998) Failure of testicular development associated with a rearrangement of 9p24.1 proximal to the SNF2 gene. Hum. Genet. 102, 151–156 [DOI] [PubMed] [Google Scholar]
- 41. Shetty G., and Meistrich M. L. (2007) The missing niche for spermatogonial stem cells: do blood vessels point the way? Cell Stem Cell 1, 361–363 [DOI] [PubMed] [Google Scholar]
- 42. Yoshida S., Sukeno M., and Nabeshima Y. (2007) A vasculature-associated niche for undifferentiated spermatogonia in the mouse testis. Science 317, 1722–1726 [DOI] [PubMed] [Google Scholar]
- 43. Goriely A., McVean G. A., van Pelt A. M., O'Rourke A. W., Wall S. A., de Rooij D. G., and Wilkie A. O. (2005) Gain-of-function amino acid substitutions drive positive selection of FGFR2 mutations in human spermatogonia. Proc. Natl. Acad. Sci. U.S.A. 102, 6051–6056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tadokoro Y., Yomogida K., Ohta H., Tohda A., and Nishimune Y. (2002) Homeostatic regulation of germinal stem cell proliferation by the GDNF/FSH pathway. Mech. Dev. 113, 29–39 [DOI] [PubMed] [Google Scholar]
- 45. de Rooij D. G., Okabe M., and Nishimune Y. (1999) Arrest of spermatogonial differentiation in jsd/jsd, Sl17H/Sl17H, and cryptorchid mice. Biol. Reprod. 61, 842–847 [DOI] [PubMed] [Google Scholar]
- 46. Pfeiffer J. R., and Brooks S. A. (2012) Cullin 4B is recruited to tristetraprolin-containing messenger ribonucleoproteins and regulates TNF-alpha mRNA polysome loading. J. Immunol. 188, 1828–1839 [DOI] [PubMed] [Google Scholar]
- 47. Russell L. D., Ettlin R. A., Sinha Hikim A. P., and Clegg E. D. (1990) Histological & Histopathological Evaluation of the Testis, Cache River Press, St. Louis, MO [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.