Abstract
Type IV pili (T4P) are ubiquitous bacterial cell surface structures, involved in processes such as twitching motility, biofilm formation, bacteriophage infection, surface attachment, virulence, and natural transformation. T4P are assembled by machinery that can be divided into the outer membrane pore complex, the alignment complex that connects components in the inner and outer membrane, and the motor complex in the inner membrane and cytoplasm. Here, we characterize the inner membrane platform protein PilC, the cytosolic assembly ATPase PilB of the motor complex, and the cytosolic nucleotide-binding protein PilM of the alignment complex of the T4P machinery of Myxococcus xanthus. PilC was purified as a dimer and reconstituted into liposomes. PilB was isolated as a monomer and bound ATP in a non-cooperative manner, but PilB fused to Hcp1 of Pseudomonas aeruginosa formed a hexamer and bound ATP in a cooperative manner. Hexameric but not monomeric PilB bound to PilC reconstituted in liposomes, and this binding stimulated PilB ATPase activity. PilM could only be purified when it was stabilized by a fusion with a peptide corresponding to the first 16 amino acids of PilN, supporting an interaction between PilM and PilN(1–16). PilM-N(1–16) was isolated as a monomer that bound but did not hydrolyze ATP. PilM interacted directly with PilB, but only with PilC in the presence of PilB, suggesting an indirect interaction. We propose that PilB interacts with PilC and with PilM, thus establishing the connection between the alignment and the motor complex.
Keywords: ATPase, cell motility, membrane reconstitution, membrane transport, secretion, type IV pili, Myxococcus xanthus
Introduction
Type IV pili (T4P)3 are versatile surface structures that are important for various processes, including twitching motility, biofilm formation, bacteriophage infection, surface attachment, virulence, and natural transformation. T4P are found on the surfaces of a wide variety of Gram-positive and Gram-negative bacteria and archaea (1–3). T4P systems (T4PS) are related to type II secretion systems (T2SS), which are responsible for the secretion of proteins across the outer membrane of Gram-negative bacteria (4, 5), bacterial competence systems that are involved in the uptake of DNA (6), and systems that are involved in the assembly of archaeal surface structures (7).
T4P are highly dynamic structures that undergo cycles of extension and retraction (8, 9). During extensions, the T4P assembly ATPase PilB stimulates the extraction of pilin monomers from the inner membrane (IM) and their incorporation at the base of the pilus fiber. The fiber has a diameter of ∼6 nm and can extend up to several micrometers in length (10). During retractions, the T4P disassembly ATPase PilT stimulates the removal of pilin monomers from the base of the pilus and their reinsertion into the IM (9, 11). T4P retraction generates forces up to 150 piconewtons (12, 13), pulling a cell forward, and making T4P systems the strongest molecular motors known.
The rod-shaped cells of the δ-proteobacterium Myxococcus xanthus, assemble 5–10 T4P at the leading cell pole that extend and retract to generate cell movement in the direction of the long axis of cells (14). Occasionally, cells reverse their direction of movement (15). During a reversal, the T4P disassemble at the old leading pole and reassemble at the new leading cell pole (16–18). Differences in the timing between reversals result in net movement (19). M. xanthus is the model system for this type of T4P-dependent directional movement, but the biochemistry of the T4P assembly system in this organism has not been studied in detail.
Similarly to other T4PS in Gram-negative bacteria, the T4PS of M. xanthus consists of 12 conserved proteins. The nomenclature for proteins of T4PS varies widely between organisms, even for highly conserved proteins. Here, we use the nomenclature used for the M. xanthus T4PS. The pilin PilA is added onto the pilus base of the T4PS after cleavage of the class III signal sequence of the pre-pilin by the pre-pilin peptidase (PilD) (20). The remaining 10 proteins of the T4PS form three inter-connected subcomplexes. The outer membrane (OM) subcomplex that serves as a conduit for the pilus across the OM and consists of the secretin (PilQ) (21), the peptidoglycan-binding protein (TsaP) (22), and a pilotin (Tgl), which stimulates insertion of PilQ into the OM and/or PilQ oligomerization (23, 24). The alignment subcomplex consists of the cytosolic actin-like ATP-binding protein (PilM) (25), two bitopic IM proteins with large periplasmic domains and short cytosolic N termini (PilN and PilO), and an IM lipoprotein with a large periplasmic domain (PilP) (26–30). Cytosolic PilM interacts with the short N terminus of PilN (25, 26, 31). PilN and PilO interact directly and likely form heterodimers (27, 31). The PilNO complex interacts with PilP (30), and PilP interacts with PilQ (30–32), thus putatively connecting components in the cytosol and IM to components in the OM. The IM motor subcomplex consists of the IM protein PilC (33) and the associated cytosolic ATPases, which provide the energy for extension (PilB) and retraction (PilT), respectively (34). The T4PS in Gram-negative bacteria span both the IM and OM, whereas homologous systems in Gram-positive bacteria and in archaea only span the cytoplasmic membrane. Consistently, only the IM motor subcomplex is conserved in all T4PS. Moreover, the retraction ATPase PilT is only present in bacterial T4PS systems.
Here, we investigate the IM protein PilC, the actin-like protein PilM, and the assembly ATPase PilB and analyze their function and association in the T4PS of M. xanthus. PilC is a polytopic IM protein that belongs to the GspF/PilC superfamily of IM proteins of T2SS and T4PS. Sequence analysis suggests that proteins of this family consist of two cytoplasmic domains with high sequence similarity, separated by two transmembrane domains and followed by a short C-terminal periplasmic extension. Here, we refer to M. xanthus PilC and homologs thereof in T4PS and T2SS as the IM platform protein (33). To date, no structural data are available on a full-length protein of the GspF/PilC superfamily. The structure of the cytosolic N-terminal domain of Thermus thermophilus PilC revealed a dimeric helical bundle structure with the dimer formed by interactions between the fifth and sixth α-helix (35). The N-terminal domain of EpsF from the T2SS of Vibrio cholerae also crystallized as a helix bundle but showed a different dimer interface than T. thermophilus PilC (36). Moreover, the Neisseria meningitidis PilC homolog has been described by analytical ultracentrifugation and single particle analysis to form dimers and tetramers in solution (36). The exact function of PilC remains under discussion. Although a pilG mutant, the pilC ortholog in N. meningitidis, does not assemble pili, the pilG/pilT double mutant assembles pili (37). By contrast, both the pilC and the pilC/pilT double mutants in Pseudomonas aeruginosa are non-piliated (33).
The PilB ATPase is a member of the secretion ATPase superfamily, a subgroup of RecA/Rad51-like motors (39–41). Crystal structures of T2SS assembly ATPases (42–45), T4P retraction ATPases (46, 47), and the motor ATPase of the archaellum (the archaeal T4P-like motility system) (48) revealed that these proteins consist of a conserved C-terminal ATPase domain and a much more variable N-terminal domain. These crystal structures and other data also revealed that PilB-like ATPases function as hexamers and that conformational changes upon hydrolysis of ATP most likely drive the extrusion or retraction of the pilus (48, 49). Several studies have suggested an interaction between PilB and PilC homologs. Localization and stability of the assembly ATPase PilB of P. aeruginosa have been shown to depend on PilC. Co-purification and co-localization experiments revealed an interaction between PilC and PilB (33, 50). Yeast two-hybrid analysis of the T2SS of Erwinia chrysanthemi revealed interactions between GspF (PilC homolog), GspE (PilB homolog), and GspL (PilM-N homolog), but co-immunoprecipitation of GspF with GspE required the presence of GspL (51). In the T2SS of V. cholerae, structural data suggest that the ATPase EpsE interacts and forms a complex with the N-terminal cytosolic domain of the PilM-N homolog EpsL (42), resulting in a stimulation of the ATPase activity (52).
The structure of T. thermophilus PilM has been solved and is most similar to the actin-like ATPase FtsA (25), BfpC (a PilM homolog) of the T4PS of Escherichia coli, and the cytoplasmic domains of GspL (a PilM-N homolog) of the T2SS of V. cholerae (53, 54). Despite the actin-like structure of PilM, the protein was purified and crystallized as a monomer, and no polymerization or ATP hydrolysis has been observed (25). However, PilM has been reported to bind ATP (25). The short and highly conserved cytosolic N terminus of PilN has been reported to interact with PilM (26), and indeed crystallization of T. thermophilus PilM was only successful in the presence of the N terminus of PilN, demonstrating that PilM binds to the N terminus of PilN (25). In addition to the interaction with PilN, PilM has also been suggested to interact with PilC and PilB. Similar to T2SS of E. chrysanthemi (51), in the T2SS of P. aeruginosa, co-expression of XcpY (PilM-N homolog) and XcpR (PilB homolog) is stabilized by XcpS (PilC homolog) suggesting the formation of a ternary complex via direct or indirect association of the three proteins (55, 56).
To examine how PilC, PilB, and PilM interact to stimulate T4P assembly, we overexpressed and purified PilC, PilB, and PilM of the T4PS of M. xanthus and characterized the proteins and their interactions with each other.
Experimental Procedures
Plasmids and Strains
Primers, plasmids, and strains used in this study are listed in Tables 1–3, respectively. Cloning was performed in E. coli DH5α. If no template is named, PCRs were performed on chromosomal DNA of M. xanthus strain DK1622 (14). To generate a plasmid that expresses PilB with an N-terminal His10 tag, a PCR fragment created using primers 1138 and 1139 was digested with NcoI and BamHI and ligated into pETDuet-1 resulting in plasmid pLB5. To generate pLB8, which encodes a fusion of M. xanthus PilB and P. aeruginosa Hcp1, a PCR fragment of pilB generated using primers 1143 and 1176 and a PCR fragment of hcp1 generated using primers 1154 and 1155 on genomic DNA of P. aeruginosa, were digested with BamHI and NsiI and BamHI and HindIII, respectively, and ligated into pLB5 digested with NsiI and HindIII. To create the plasmid pAH94, which encodes a fusion of M. xanthus PilC to E. coli YbeL (also called β), a PCR fragment generated using primers AH75 and AH76, was digested with EcoRI and XhoI and ligated into plasmid β-pET28a(+). To enable removal of the β domain from PilC, a tobacco etch virus (TEV) protease cleavage site was introduced by circular PCR using primers 1152 and 1153 on pAH94 resulting in pLB7. An additional StrepII affinity tag was introduced after the TEV cleavage site by circular PCR using primers 1167 and 1168 on plasmid pLB7, resulting in plasmid pLB11. Plasmid pSC111, which encodes a fusion of the first 16 amino acids of M. xanthus PilN to the C terminus of M. xanthus PilM, was generated by ligating two annealed primers (opilN-29 and opilN-30) into pET45b(+) digested with HindIII and XhoI followed by ligation of a fragment encoding pilM, amplified using primers opilM-49 and opilM-50 in the BamHI and HindIII sites of the same vector. To co-express PilM and PilB from compatible vectors, pilM-N(1–16) was amplified by PCR from pSC111 using primers 1188 and 1189 and ligated into the NdeI and BglII sites of pCDFDuet-1, and PilB:hcp1 was amplified from pLB8 using primers 1186 and 1187 and ligated into the NcoI and HindIII sites of pACYCDuet-1, resulting in pLB15 and pLB16, respectively.
TABLE 1.
Primers used in this study
| Name | Sequence (5′ to 3′; restriction sites in italics) |
|---|---|
| 1138 | GTAATTGGATCCCTAGAAGCGGTCCGGGGCGGTGTTTC |
| 1139 | GCAGTCCCATGGATCATCACCATCACCACCATCATCACCATCACATGTCCGGTCGACTCGGTGAAC |
| 1152 | CGATGGATCCGAAAACCTGTATTTTCAGGGAGAATTCATGGCAGCCCCAGCAGTG |
| 1153 | TTCGGATCCCCCGACCACTTCTC |
| 1154 | GCGCGGATCCAAACTGGCGAGCGGCGCGGGTCACATGGCTGTTGATATGTTCATCAAG |
| 1155 | GCGCAAGCTTTTATTTTTCGAACTGCGGGTGGCTCCAGGCCTGCACGTTCTGG |
| 1143 | CGACGCGCAGCCGCTGGCCTGGTTCAAG |
| 1176 | TTTAAGGATCCGCGGAAGCGGTCCGGGG |
| 1167 | TGGAGCCACCCGCAGTTCGAAAAAGAATTCATGGCAGCCCCAGC |
| 1168 | TTTTTCGAACTGCGGGTGGCTCCATCCCTGAAAATACAGGTTTT |
| 1888 | ATCGCATATGGTGCGAGGCTCCCGTC |
| 1189 | ACTCAGATCTTCAACGCTTCTTCACCGC |
| 1186 | ACGTGCCATGGGTATGTCCGGTCGACTC |
| 1187 | GCACTGAAGCTTTCAATGGTGATGGTGATGATGGTGGTGATGATGGGCCTGCACGTTCTG |
| opilN-29 | AGCTTGGCTCCGCCATGATGATTCGCATCAACCTGCTGCCCGTCCGGGCGGTGAAGAAGCGTTGAC |
| opilN-30 | TCGAGTCAACGCTTCTTCACCGCCCGGACGGGCAGCAGGTTGATGCGAATCATCATGGCGGAGCCA |
| opilM-49 | ATCGGGGATCCGATGGTGCGAGGCTCCCGT |
| opilM-50 | ATCGGAAGCTTGGCGGAGCCGGCCAGCTTGTCGCCCGG |
| pAH75 | GCGAATTCATGGCAGCCCCAGCAGTGAAGTCG |
| pAH76 | CCGCTCGAGCTACTGGATGGCGCCGGCAAGCG |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Source |
|---|---|---|
| pETDuet-1 | Expression vector, AmpR | Novagen |
| β-pET28a(+) | pET28a (+) vector containing His6-YbeL, KanR | 62 |
| pET45b(+) | Expression vector, N-terminal His6 tag; AmpR | Merck Millipore |
| pACYCDuet-1 | Expression vector, CmR | Novagen |
| pCDFDuet1 | Expression vector, SmR | Novagen |
| pSL5 | His6-PilB expression vector, AmpR | 34 |
| pLB5 | pETDuet-1 + His10-PilB, AmpR | This study |
| pLB8 | pETDuet-1 + His10-PilB:Hcp1, AmpR | This study |
| pAH94 | pET28a (+) + His6-β:PilC; KanR | This study |
| pLB7 | pET28a (+) + His6-β:TEV-PilC, KanR | This study |
| pLB11 | pET28a (+) + His6-β:TEV-StrepII-PilC, KanR | This study |
| pSC111 | pET45b (+) + His6PilM-PilN (1–16), AmpR | This study |
| pLB15 | pCDFDuet-1 + PilM-N (1–16); SmR | This study |
| pLB16 | pACYCDuet-1 + PilB:Hcp1-His10; CmR | This study |
TABLE 3.
Strains used in the study
| Strain | Relevant characteristics | Source |
|---|---|---|
| M. xanthus DK1622 | Wild type | 14 |
| E. coli BL21 star | F− ompT hsdSB (rB− mB−) gal dcm rne131 (DE3) | Novagen |
| E. coli Rosetta 2 DE(3) | F− ompT hsdSB(rB− mB−) gal dcm (DE3) pRARE2 (CamR) | Novagen |
| E. coli DH5α | F− endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG ϕ80flacZΔM15Δ (lacZYA-argF)U169 hsdR17 (rK− mk+) Л- | Invitrogen |
Overexpression of PilB, PilB:Hcp1, PilM-N(1–16), and β:PilC
E. coli strains were grown at 37 °C in either Lysogeny Broth (LB) or on plates containing LB and 1.5% agar. When required, ampicillin (100 μg/ml), kanamycin (50 μg/ml), chloramphenicol (34 μg/ml), or spectinomycin (25 μg/ml) were added. Production of PilB, PilB:Hcp1, and PilM-N(1–16) was performed in E. coli BL21 Star transformed with the respective plasmids. Strains were grown at 30 °C and induced with 0.5 mm IPTG at an A600 of 0.5. Growth was continued for 12 h at 18 °C. Production of β:PilC was performed in E. coli Rosetta 2 (DE3) transformed with pLB11, at 25 °C in autoinduction medium (57). Growth was continued until an A600 of 3.0 was reached. Cells were harvested by centrifugation (17,000 × g, 4 °C, 10 min), frozen in liquid nitrogen, and stored at −20 °C. To analyze the amount of overexpressed protein, strains were grown with and without induction with IPTG, and equal amounts (A600) of whole cells were dissolved in SDS-sample buffer and analyzed by SDS-PAGE.
Purification, Reconstitution, and Analysis of PilC
For the isolation of membranes, cells producing β:PilC were thawed on ice and resuspended (1 g cells/10 ml) in buffer C (150 mm NaCl, 20 mm Tris/HCl, pH 7.5) supplemented with EDTA-free cOmplete protease inhibitor mixture (Roche Applied Science) and DNase I (Thermo Fisher Scientific). Cell disruption was performed by two passages through a microfluidizer at 1500 bar. Cell debris was removed by low speed centrifugation (17,000 × g, 4 °C, 20 min). Membranes were pelleted by ultracentrifugation (400,000 × g, 4 °C, 1 h), subsequently homogenized in buffer C, frozen in liquid nitrogen, and stored at −80 °C. To remove the β-fusion protein, membranes were thawed on ice and incubated with TEV protease (0.1 mg/ml TEV protease per 1 mg/ml membranes) for 2 h at 4 °C in buffer C supplemented with 0.5 mm EDTA and 1 mm DTT. After ultracentrifugation (400,000 × g, 4 °C, 1 h), membranes were homogenized in buffer C. Solubilization of PilC from TEV-treated or non-treated membranes was performed by diluting PilC to a concentration of 5 mg/ml in buffer C supplemented with 2% n-decyl-β-d-maltopyranoside (Glycon) and 10% glycerol (buffer D). After 2 h at 4 °C, the sample was subjected to ultracentrifugation (400,000 × g, 4 °C, 1 h). β:PilC was purified using 1 ml of His-Select nickel affinity gel resin (Sigma) equilibrated with buffer D. After incubation of the resin with the solubilized protein, the sample was applied on a gravity column, and the flow-through was collected. The resin was washed with 2 column volumes of buffer E (300 mm NaCl, 0.04% n-decyl-β-d-maltopyranoside, 10% glycerol, 20 mm Tris/HCl, pH 7.5) containing 20 mm imidazole, and elution of the protein was achieved by washing the column with 3 column volumes of buffer E containing 200 mm imidazole. PilC was purified using 0.5 ml of Strep-Tactin Superflow resin (IBA) equilibrated with buffer D. After loading the solubilized protein and the resin on a gravity column, the flow-through was collected, and the resin was washed with 2 column volumes of buffer F (150 mm NaCl, 0.5% n-decyl-β-d-maltopyranoside, 1 mm EDTA, 20 mm Tris/HCl, pH 7.5) followed by elution using 5 column volumes of buffer F supplemented with 2.5 mm desthiobiotin. The protein was frozen in liquid nitrogen and stored at −80 °C. To analyze the oligomeric state of purified PilC, Blue-Native PAGE analyses were performed as described (58) using NativeMarkTM unstained protein standards (Life Technologies, Inc.). Reconstitution of purified PilC into liposomes was performed essentially as described (59). To confirm the reconstitution of PilC into the liposomes, purified PilC or proteoliposomes containing PilC were loaded on sucrose density gradients consisting of steps containing 0, 6.5, 13, 19.5, 26, and 65% (w/v) sucrose in buffer C. The presence of PilC and lipids in the different sucrose fractions were determined by Western blotting and by determining the amount of phosphate in the different fractions after heat/acid destruction of the phospholipids, previously as described by Rouser et al. (60). After ultracentrifugation (400,000 × g, 4 °C, 1 h), samples from each interface were taken, and after the addition of 300 μl of perchloric acid and heating for 3 h at 180 °C, the samples were resuspended in 1.4 ml of water. 200 μl of 2.5% (w/v) ammonium/molybdate solution and 200 μl of 10% (w/v) ascorbic acid were added, and the absorbance at 797 nm was determined. Phosphate standard solution (Merck Millipore) in a concentration range from 0 to 10 mm served as standard. To enable us to compare liposomes and StrepII-PilC-containing proteoliposomes, the amount of phospholipids in both liposomes and proteoliposomes was determined by the method of Rouser et al. (60), and liposomes were adjusted to an equal concentration of phospholipids as found in the StrepII-PilC-containing proteoliposomes. To analyze the orientation of PilC in liposomes, 10 μg/ml PilC were incubated with 0 to 1000 μg/ml proteinase K (Sigma, Steinheim, Germany) in a total final volume of 50 μl of buffer C supplemented with 1 mm CaCl2 or buffer C supplemented with 1 mm CaCl2 and 0.5% n-decyl-β-d-maltopyranoside for 30 min on ice. 1 mm PMSF and 6 μl of 10× SDS-loading dye were added, and samples were incubated at 100 °C for 10 min before being subjected to Western blot analysis using α-PilC antibodies.
Purification of PilB, PilB:Hcp1, and PilM-N(1–16)
To purify PilB:Hcp1 or PilM-N(1–16), cells expressing PilB:Hcp1 or PilM-N(1–16) were thawed and resuspended in buffer A (50 mm Tris/HCl, pH 7.5, 100 mm NaCl, 10 mm imidazole) containing cOmplete protease inhibitor mixture (Roche Applied Science) and 10 μg/ml DNase I (Thermo Fisher Scientific) and lysed by two passages through a microfluidizer (Microfluidics) at 1500 bar. The suspension was centrifuged (17,000 × g, 4 °C, 10 min) to remove inclusion bodies and unbroken cells followed by another centrifugation to remove membranes and aggregated protein (400,000 × g, 4 °C, 1 h). The supernatant was loaded on a HiTrap Chelating HP column (GE Healthcare) equilibrated with buffer A. After washing the column with buffer A containing 40 mm imidazole, protein was eluted using a gradient from 40 to 800 mm imidazole in buffer A, and fractions were collected. After pooling, PilB:Hcp1 or PilM-N(1–16)-containing fractions were concentrated using ultrafiltration (30-kDa cutoff, Merck Millipore) and injected on a size exclusion column (Superdex 200 16/60 for PilB:Hcp1, S.D. 75 16/60 for PilM-N(1–16), GE Healthcare) equilibrated with buffer B (100 mm NaCl, 5% glycerol, 50 mm Tris/HCl, pH 7.5). Thyroglobulin (669 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), and ovalbumin (44 kDa) served as marker proteins. Proteins were collected, frozen in liquid nitrogen, and stored at −80 °C. PilB was purified as described previously (34).
Western Blotting
Western blot analyses were performed following standard procedures. The proteins were transferred to PVDF membranes. Specific α-PilB (34) and α-PilC and α-PilM (17) antibodies were used for protein detection combined with horseradish peroxidase-conjugated secondary anti-rabbit antibodies and Clarity ECL Western blotting substrate (Bio-Rad). Blots were developed using a LAS-4000 imager (GE Healthcare).
Fluorescent Nucleotide Binding Assay
Nucleotide binding assays were performed either in a 120- or 60-μl quartz cuvette at 8 °C using a PC1 photon-counting spectrofluorometer (ISS) or a FluoroMax 4 spectrofluorometer (Horiba Scientific). To monitor 2′(3′)-O-(N-methylanthraniloyl) adenosine 5′-triphosphate (MANT-ATP, Molecular Probes) binding to PilB or PilB:Hcp1, fluorescence spectra (excitation 350 nm; emission 400–500 nm; 5-nm slit width) of 10 μm PilB or PilB:Hcp1 were recorded in the presence of increasing amounts of MANT-ATP. Spectra were corrected for the fluorescence of MANT-ATP in the absence of protein. To monitor 2′,3′-O-(2,4,6-trinitrophenyl)-adenosine 5′-triphosphate (TNP-ATP, Molecular Probes) binding to PilM-N(1–16), fluorescence spectra (excitation 409 nm; emission 500 to 600 nm; 2-nm slit width) of 1 μm TNP-ATP in buffer B were recorded in the presence of increasing amounts of PilM-N(1–16). For the competition assay, fluorescence spectra were recorded of 1 μm TNP-ATP and 1.275 μm PilM-N(1–16) in buffer B in the presence of increasing amounts of ATP.
PilB, PilB:Hcp1, and PilM-N(1–16) Binding to PilC-containing Proteoliposomes
9 μg of PilB:Hcp1 or 9 μg of PilB and/or 8 μg of PilM-N(1–16) were incubated at 4 °C with 400 μg of liposomes or proteoliposomes (containing 100 μg PilC) in a total volume of 200 μl of buffer G (150 mm NaCl, 5 mm MgCl2,50 mm Tris, pH 7.5). After 20 min, samples were submitted to ultracentrifugation in a Beckman Airfuge at 20,000 p.s.i. for 20 min. Pellet and supernatant fractions were separated, and the pellet was resuspended in an equal amount of buffer G. Pellet and supernatant fractions were analyzed by immunoblotting.
Co-purification of PilB:Hcp1 and PilM-N(1–16)
0.5 g of cells expressing either tag-less PilM-N(1–16) alone or co-expressing tag-less PilM-N(1–16) and His10-tagged PilB:Hcp1 were resuspended in 5 ml of buffer C, lysed by sonication (three cycles of 15 s on/30 s off), and centrifuged (400,000 × g, 4 °C). 500 μl of the supernatants were applied to Rainin PureSpeed immobilized metal affinity chromatography tips on a Rainin E4 XLS automated multichannel pipette according to the manufacturer's (Mettler Toledo) specifications. The resin was washed three times with 300 μl at medium speed using buffer C supplemented with 10 mm imidazole, and proteins were eluted by washing three times with 100 μl of buffer C supplemented with 500 mm imidazole. Fractions were analyzed by immunoblotting using specific antibodies.
ATP Hydrolysis
ATP hydrolysis activity was determined in a total volume of 50 μl, containing 2.4 μm PilB:Hcp1 or PilB, 1 mm ATP, 150 mm NaCl, 5 mm MgCl2, and 50 mm Tris/HCl, pH 8.0. The stimulation of PilB:Hcp1 or PilB ATPase activity by solubilized PilC was determined by including 0–10 μl of purified StrepII-PilC (final concentration 0–60 μg of StrepII-PilC/ml (0–1.2 μm)) or buffer F as a control. The stimulation of PilB:Hcp1 or PilB ATPase activity by liposomes or PilC-containing proteoliposomes was determined by including 0–10 μl of either liposomes or StrepII-PilC-containing proteoliposomes (0–240 μg (proteo)liposomes/ml; final concentration 0–60 μg PilC/ml (0–0.6 μm)). The amount of released phosphate was determined using the Malachite Green assay (61) using the phosphate standard solution (Merck Millipore) in a concentration ranging from 0 to 100 μm as standard.
Results
Inner Membrane Protein PilC Forms Dimers and Can Be Reconstituted into Liposomes
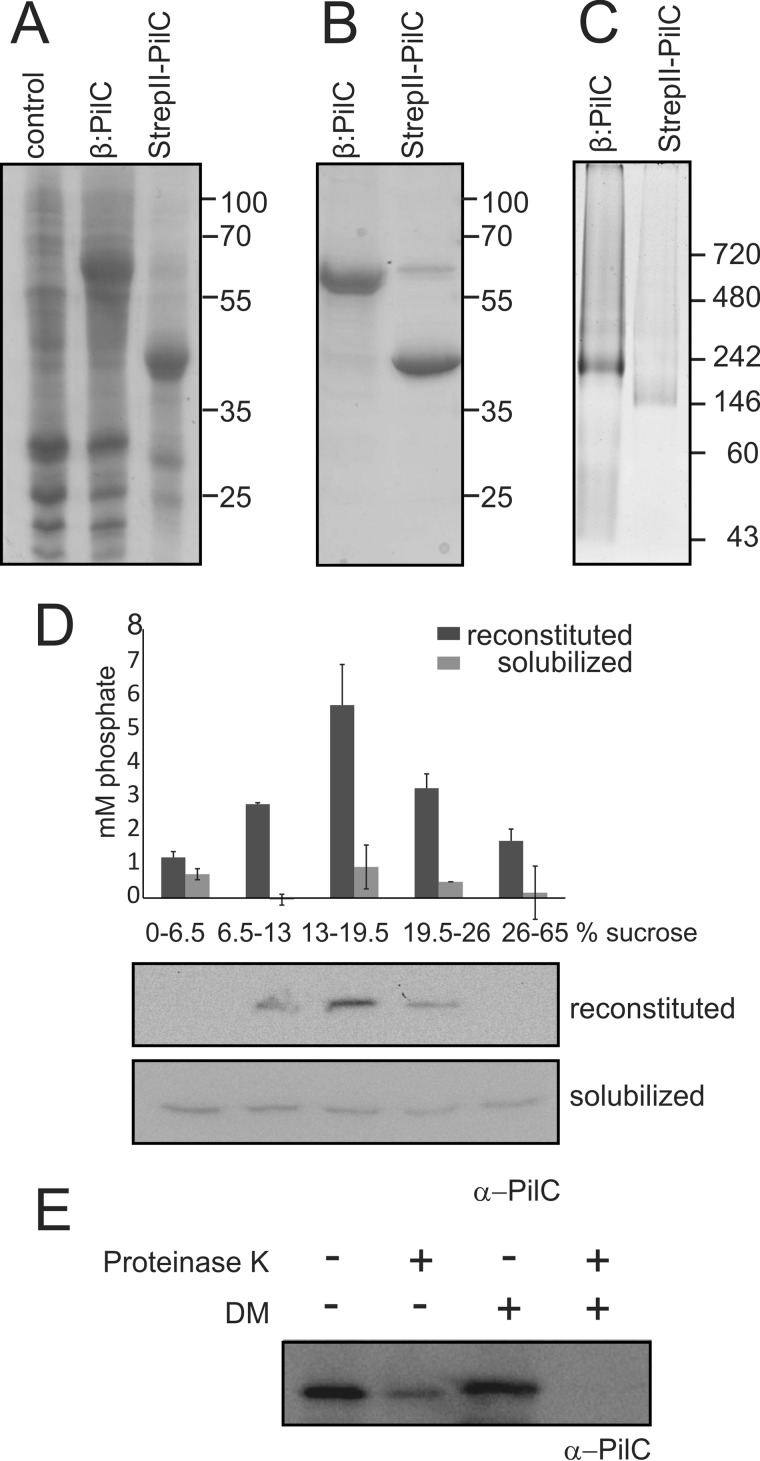
To characterize PilC of M. xanthus, we set out to optimize its overexpression, purification, and reconstitution in liposomes. Optimal overexpression conditions were found when expression was performed in autoinduction medium (57) and when PilC was fused to the C terminus of E. coli His6-tagged YbeL (also called β), a protein previously demonstrated to facilitate the expression of membrane proteins (62). To enable removal of the β domain from PilC, a TEV protease cleavage site followed by a StrepII tag were introduced between YbeL and PilC, resulting in His6-YbeL-TEV-StrepII-PilC (β:PilC). Indeed SDS-PAGE analysis of isolated membranes showed significant levels of overexpressed β:PilC (Fig. 1A). Incubation of isolated membranes with TEV protease resulted in removal of the His6-tagged YbeL giving rise to StrepII-PilC (Fig. 1A). Best solubilization and purification (Fig. 1B) of β:PilC or StrepII-PilC was obtained using n-decyl-β-d-maltopyranoside as detergent. 2 and 1 mg of β:PilC and StrepII-PilC, respectively, could be reproducibly obtained from 4 g wet weight of cells. To determine the oligomeric state of purified β:PilC and StrepII-PilC, Blue-Native PAGE analysis was performed (Fig. 1C). β:PilC and StrepII-PilC migrated corresponding to ∼230 and 170 kDa of the marker consisting of soluble standards. Heuberger et al. (63) showed that the mass of membrane proteins calculated from Blue-Native PAGE using soluble marker proteins overestimates the mass by ∼80%. Taking this observation into account, molecular masses of 125 and 94 kDa were determined for β:PilC and StrepII-PilC, respectively. This demonstrates that purified solubilized β:PilC (63 kDa) and StrepII-PilC (47 kDa) are present in a single oligomeric state and suggests that they are dimeric membrane proteins. As a final step, purified solubilized StrepII-PilC was reconstituted into liposomes derived from E. coli phospholipids. Reconstitution was performed using liposomes that were not destabilized with detergents to promote insertion of StrepII-PilC into the liposomes with the cytosolic domains on the outside of the liposomes (64). Analysis of the resulting proteoliposomes using sucrose density gradient centrifugation showed that StrepII-PilC co-localized with the liposomes, suggesting efficient incorporation of StrepII-PilC (Fig. 1D). Thus, the IM platform protein PilC of M. xanthus can be efficiently overexpressed, purified, and reconstituted into liposomes. To determine the orientation of reconstituted PilC, a protease accessibility assay was performed. Therefore, PilC-containing proteoliposomes were incubated with increasing amounts of proteinase K, either in the presence or absence of detergent, and samples were analyzed by Western blotting using α-PilC, which is directed against the first 185 amino acids (N-terminal cytoplasmic loop). In the presence of 1 μg/ml proteinase K, the N-terminal cytoplasmic loop is fully degraded in the presence of detergent, whereas in the absence of detergent 95% of reconstituted PilC was degraded. Approximately 5% of reconstituted PilC was protected from degradation by proteinase K at concentrations up to 1 mg/ml (Fig. 1E), demonstrating that 95% of PilC was reconstituted in an orientation with its N-terminal cytoplasmic domain on the outside of the proteoliposomes.
FIGURE 1.
Expression, purification, and reconstitution of PilC. A, Coomassie-stained SDS-PAGE of membranes isolated from E. coli Rosetta 2 (DE3) strains expressing no PilC (control), His6-YbeL-TEV-StrepII-PilC (β:PilC), and His6-YbeL-TEV-StrepII-PilC-expressing membranes treated with TEV protease (StrepII-PilC). Representative figure is of at least 10 biological replicates. B, Coomassie-stained SDS-PAGE of purified His6-YbeL-TEV-StrepII-PilC (β:PilC) and StrepII-PilC. Representative figure is of at least 10 biological replicates. C, Blue-Native PAGE analysis of purified His6-YbeL-TEV-StrepII-PilC (β:PilC) and StrepII-PilC. Representative figure is of three biological replicates. Non-native (A and B) and native (C) molecular mass markers are indicated. D, reconstitution of purified StrepII-PilC was analyzed by sucrose density gradient centrifugation. Solubilized and reconstituted PilCs were loaded onto a sucrose step gradient. Samples from each layer were analyzed for the amount of phospholipid-derived phosphate (upper graph) and the presence of StrepII-PilC by immunoblot analyses using α-PilC antibody (lower lanes). The indicated error bars represent the average of two biological replicates containing two technical replicates. E, orientation of PilC in proteoliposomes was analyzed by incubation of PilC-containing proteoliposomes with (+) or without (−) 1 mg/ml proteinase K in the presence (+) and absence (−) of 0.5% n-decyl-β-d-maltopyranoside (DM), followed by Western blotting using an α-PilC antibody, directed against the 185 N-terminal cytoplasmic amino acids. Representative figure is of two biological replicates with two technical replicates.
Assembly ATPase PilB Forms Hexamers and Binds MANT-ATP in a Cooperative Manner
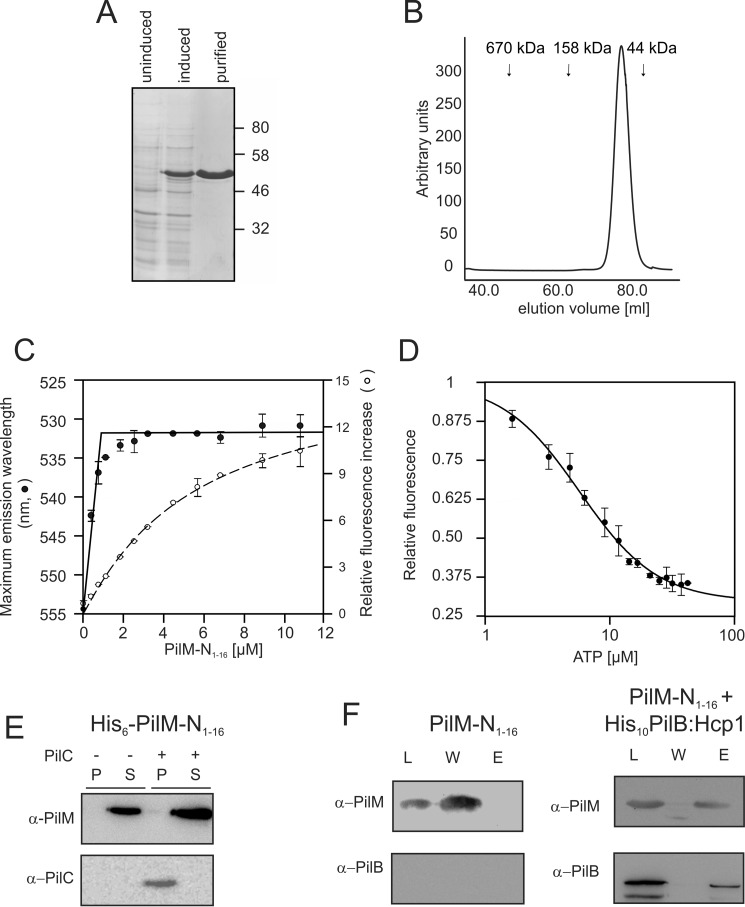
The ATPase PilB is required for assembly of T4P in M. xanthus (34). Purified M. xanthus PilB had a very low ATPase activity (34). The assembly ATPases of T4PS have been reported to function as hexamers, but under all conditions tested, the purified His6-PilB eluted as a sharp peak corresponding to the size of the His6-PilB monomer (34). The hexameric form of GspE, the ATPase of the T2SS of V. cholerae, could be stabilized by a C-terminal fusion to highly stable hexameric Hcp1 of P. aeruginosa (49). Therefore, to stabilize M. xanthus PilB in its hexameric form, a similar fusion of His10-tagged M. xanthus PilB with P. aeruginosa Hcp1 was created. The His10-PilB:Hcp1 fusion protein was expressed at high levels and could be purified using nickel-affinity and size exclusion chromatography (Fig. 2A). The His10-PilB:Hcp1 fusion (84.5 kDa) eluted at a volume corresponding to a molecular mass of ∼510 kDa, demonstrating that the fusion protein formed a stable hexamer (Fig. 2B). To compare His6-PilB with His10-PilB:Hcp1, His6-PilB was purified as described previously (34), and both proteins were tested for ATP binding and hydrolysis. ATP binding was studied by determining the fluorescence increase of the fluorescent ATP homolog MANT-ATP upon binding to His6-PilB or His10-PilB:Hcp1 (Fig. 2C). His6-PilB and His10-PilB:Hcp1 bound MANT-ATP with similar affinities of ∼25 μm. However, although His6-PilB showed non-cooperative binding of MANT-ATP, binding of MANT-ATP to His10-PilB:Hcp1 was highly cooperative (Fig. 2C). This demonstrates that binding of the nucleotide to the hexamer differs from binding to the monomer and suggests a correct hexameric assembly for His10-PilB:Hcp1.
FIGURE 2.
PilB:Hcp1 fusion forms hexamers and binds MANT-ATP in a cooperative manner. A, Coomassie-stained SDS-PAGE of cell lysates of E. coli BL21 Star expressing PilB:Hcp1 with (induced) and without (uninduced) induction with IPTG. PilB:Hcp1 was further purified from induced cells using nickel-affinity chromatography and size exclusion chromatography (purified). Representative figure is of at least 10 biological replicates. B, size exclusion chromatography of purified His10-PilB:Hcp1. Elution volumes of molecular mass markers and the void volume are indicated. Representative figure is of at least 10 biological replicates. C, relative increase in fluorescence of increasing MANT-ATP concentrations upon addition of 10 μm PilB:Hcp1 (closed dots) or 10 μm PilB (open dots). Data were fitted to the Hill equation y = (axn/(bn + xn)) resulting in PilB:Hcp1 (continuous curve) and PilB (dashed curve) in best fits with Hill coefficients (n) of 2.8 and 0.9, respectively. Representative figure of two biological replicates with three technical replicates. Standard deviation was calculated from the technical replicates. D, His6-PilB (left panel) or His10-PilB:Hcp1 (right panel) was incubated with either liposomes (−) or proteoliposomes containing PilC (+). After 20 min, samples were subjected to ultracentrifugation, and the pellet (P) and supernatant (S) fractions were analyzed by Western blotting using the antibodies indicated. Representative figure is of at least two biological replicates with two technical replicates. E, ATP hydrolysis activity of PilB:Hcp1 (200 ng/ml, circles) and PilB (150 ng/ml, triangles) was determined in the presence of increasing concentrations of detergent-solubilized StrepII-PilC (open circles and triangles) or equivalent concentrations of the detergent-containing buffer without StrepII-PilC (closed circles and triangles). F, similar as in E, but now PilB:Hcp1 (200 ng/ml, circles) and PilB (150 ng/ml, triangles) were incubated with increasing concentrations of StrepII-PilC reconstituted into liposomes (open circles and triangles) and empty liposomes (closed circles and triangles) as a control. E and F, representative figures of two biological replicates with three technical replicates. Standard deviation was calculated from three technical replicates.
PilC-dependent ATPase Activity of Hexameric PilB
To determine whether PilB interacts with PilC, binding experiments of His6-PilB and His10-PilB:Hcp1 to liposomes or StrepII-PilC-containing proteoliposomes were performed (Fig. 2D). Notably, His10-PilB:Hcp1 but not His6-PilB bound to StrepII-Pil-containing proteoliposomes, suggesting that hexameric PilB binds with a higher affinity to PilC than monomeric PilB. In line with the observed lack of interaction between His6-PilB and StrepII-PilC, we did not detect stimulation of the ATPase activity of His6-PilB by detergent-stabilized StrepII-PilC or StrepII-PilC-containing proteoliposomes (Fig. 2, E and F). Remarkably, incubation of His10-PilB:Hcp1 with either detergent-stabilized StrepII-PilC (Fig. 2E) or StrepII-PilC-containing proteoliposomes (Fig. 2F) stimulated the ATPase activity of His10-PilB:Hcp1. No stimulation of the ATPase activity of His10-PilB:Hcp1 was observed when His10-PilB:Hcp1 was incubated with the detergent containing StrepII-PilC buffer (Fig. 2E) or with liposomes (Fig. 2F). Thus, the ATPase activity of His10-PilB:Hcp1 is stimulated by StrepII-PilC in micellar solution and by StrepII-PilC reconstituted in proteoliposomes. This is the first demonstration of a functional interaction between PilB and PilC.
PilM Interacts Directly with PilB but Not with PilC
To study whether the actin-like protein PilM interacts with either PilC or PilB, we set out to produce and purify M. xanthus PilM. Different initial production attempts resulted in instable PilM. Because PilM from T. thermophilus formed crystals and diffracted to a much higher resolution in the presence of a peptide corresponding to the cytosolic N terminus of PilN (25) and PilM in M. xanthus is unstable in the absence of PilN (31), we speculated that M. xanthus PilM might also be unstable in E. coli in the absence of PilN. Examination of the crystal structure of T. thermophilus PilM containing the N-terminal PilN peptide suggested that fusion of the first 16 amino acids of M. xanthus PilN (PilN(1–16)) to the C terminus of M. xanthus PilM might orient the PilN(1–16) peptide exactly in its peptide binding pocket. Indeed, this fusion protein (His6-PilM-N(1–16)) showed increased stability, suggesting that, similarly to PilM from T. thermophilus, M. xanthus PilM interacts with the N terminus of PilN. The His6-PilM-N(1–16) protein could be produced and purified as a stable, soluble protein (Fig. 3A). Purified His6-PilM-N(1–16) behaved as a monomer, when analyzed by size exclusion chromatography (Fig. 3B). No significant ATPase activity was observed when His6-PilM-N(1–16) was incubated with ATP (data not shown). His6-PilM-N(1–16) bound MANT-ATP with a low affinity (>20 μm, data not shown), but bound TNP-ATP with high affinity (Fig. 3C). Notably, when TNP-ATP was incubated with increasing amounts of His6-PilM-N(1–16), two clearly different phases of TNP-ATP binding were observed (Fig. 3C). In the first phase, a strong shift in the maximum emission wavelength of TNP-ATP was observed, indicating an affinity better than 1 μm, and in a second phase a further increase in the fluorescence, with an affinity of 4 μm, was observed. The further increase in TNP-ATP fluorescence after initial binding of all TNP-ATP, in a His6-PilM-N(1–16) concentration-dependent manner, might indicate the formation of higher oligomeric His6-PilM-N(1–16) complexes. Competition of TNP-ATP binding with ATP showed that TNP-ATP binding to His6-PilM-N(1–16) can be competed with an IC50 of around 4 μm (Fig. 3D). To determine whether PilM interacts with PilC, binding experiments of His6-PilM-N(1–16) to liposomes or StrepII-PilC-containing proteoliposomes were performed, but no binding of His6-PilM-N(1–16) to liposomes or StrepII-PilC-containing proteoliposomes was detected (Fig. 3E). To determine whether PilM interacts with PilB, a construct was created that expressed tag-less PilM-N(1–16). Tag-less PilM-N(1–16) eluted in the wash fraction when applied to a nickel-immobilized metal affinity chromatography tip (Fig. 3F, left panel). When lysate of a strain co-expressing tag-less PilM-N(1–16) and His10-PilB:Hcp1 was applied to an immobilized metal affinity chromatography tip (Fig. 3F, right panel), tag-less PilM-N(1–16) eluted together with His10-PilB:Hcp1 in the elution fractions. Thus, M. xanthus PilM interacts with PilB. Contrary to what was observed for PilC, PilM did not influence the ATPase activity of PilB (data not shown), suggesting that especially the binding of PilB to PilC is important to stimulate ATPase activity.
FIGURE 3.
PilM-N(1–16) binds to PilB but not to PilC. A, Coomassie-stained SDS-PAGE of cell lysates of E. coli BL21 Star expressing PilM-N(1–16) with (induced) and without (uninduced) induction with IPTG. PilM-N(1–16) was further purified from induced cells using nickel-affinity chromatography and size exclusion chromatography (purified). Representative figure is of five biological replicates. B, size exclusion chromatography of purified PilM-N(1–16). Elution volumes of molecular mass markers and the void volume are indicated. Representative figure is of five biological replicates. C, fluorescence of 1 μm TNP-ATP was determined as a function of increasing concentrations of PilM-N(1–16). The wavelength of the maximum emission (closed dots) and the relative fluorescence increase at 545 nm (open dots) are depicted. The data for the relative fluorescence increase was fitted to the Hill equation y = (axn/(bn + xn)) resulting in a Hill coefficient (n) of 1.06 (dashed line). Representative figure is of two biological replicates with three technical replicates. Standard deviation was calculated from the technical replicates. D, relative fluorescence emission at 445 nm of 1 μm TNP-ATP and 1.2 μm PilM-N(1–16) was determined in the presence of increasing concentrations of ATP. Standard deviation was calculated from three independent experiments. The curve was fitted to a sigmoidal dose-response curve F = 1/(1 + 10̂(log(IC50)-[ATP])). Representative figure is of two biological replicates with three technical replicates. Standard deviation was calculated from the technical replicates. E, PilM-N(1–16) was incubated with either liposomes (−) or proteoliposomes containing PilC (+). After 20 min, samples were subjected to ultracentrifugation, and the pellet (P) and supernatant (S) fractions were analyzed by Western blotting using the antibodies indicated. Representative figure is of two biological replicates with three technical replicates. F, tagless PilM-N(1–16) was either expressed alone (left panels) or together with His10-tagged PilB:Hcp1 (right panels). Cell lysates were loaded onto nickel-immobilized metal affinity chromatography tips (load, L), washed (wash, W) and eluted with 500 mm imidazole (elution, E). Fractions were analyzed by immunoblotting using antibodies indicated. Representative figure is of two biological replicates with three technical replicates.
M. xanthus PilM Interacts with PilC via PilB
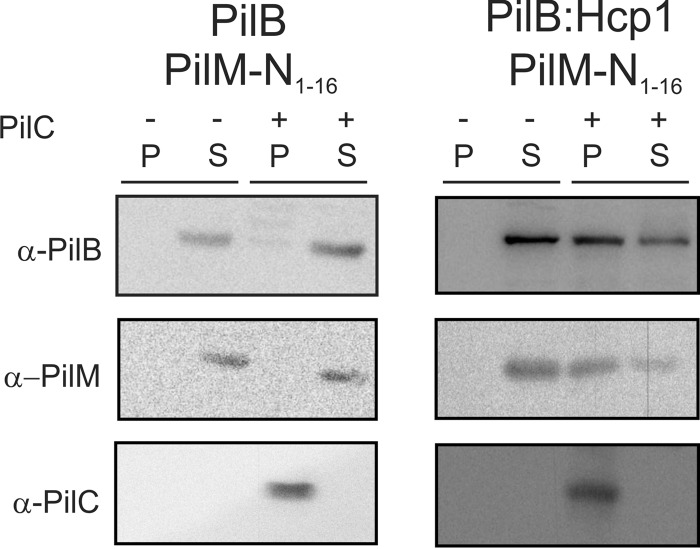
To study whether PilM might interact with PilC via PilB, binding experiments of His6-PilB and His10-PilB:Hcp1 to liposomes or StrepII-PilC-containing proteoliposomes were performed in the presence of PilM-N(1–16) (Fig. 4). As described above, His10-PilB:Hcp1 but not His6-PilB, bound to StrepII-PilC-containing proteoliposomes. Remarkably, PilM-N(1–16) was found only in the StrepII-PilC-containing pellet fraction in the presence of His10-PilB:Hcp1 but not in the presence of His6-PilB. This demonstrates that PilM interacts with PilC via PilB and that hexamer formation by PilB is important for this interaction to occur. In a final step, ATPase assays with PilC and PilB and increasing concentrations of PilM were performed. No influence of PilM on the PilC-stimulated ATPase of PilB was observed. This demonstrated that under these conditions an interaction between the three components was observed, but only the interaction with PilC stimulates the activity of PilB.
FIGURE 4.
Interaction between PilB, PilM-N(1–16), and PilC. PilM-N(1–16) was incubated with PilB (left panel) or PilB:Hcp1 (right panel) with either liposomes (−) or proteoliposomes containing PilC (+). After 20 min, the sample was subjected to ultracentrifugation, and pellet (P) and supernatant (S) were analyzed by Western blotting using the antibodies indicated. Representative figure is of two biological replicates with three technical replicates.
Discussion
We set out to characterize the IM complex of the T4PS of M. xanthus. A previous characterization of PilB, the assembly ATPase of this system, showed that purified M. xanthus PilB had a very low ATPase activity (34). A strong increase in ATPase activity was observed when GspE, the secretion ATPase of the T2SS of V. cholerae, was stabilized in its hexameric form by a C-terminal fusion to Hcp1 of P. aeruginosa (49). Purified M. xanthus PilB behaved as a monomer (34), and indeed fusion of PilB to Hcp1 resulted in formation of the hexamer. Nucleotide binding assays demonstrated that the hexameric His10-PilB:Hcp1 was able to bind nucleotides with a similar affinity as monomeric His6-PilB. Importantly, His10-PilB:Hcp1 bound the nucleotides in a cooperative manner, whereas His6-PilB did not, suggesting that a functional hexamer was formed. However, no significant ATPase activity was observed for His10-PilB:Hcp1. We therefore looked for other proteins that might stimulate ATPase activity of PilB. Studies defining the assembly pathway of the T4PS in M. xanthus revealed that this system is assembled from the OM to the IM and that polar localization of M. xanthus PilB is not dependent on other known components of T4PSs (31). However, T4P and T2S ATPases have been proposed to interact with the IM platform protein, especially their N-terminal cytoplasmic loop (33, 50). Therefore, we set out to overexpress, purify, and reconstitute the IM platform protein PilC from M. xanthus. PilC could be overexpressed as a C-terminal fusion to YbeL, a protein demonstrated to facilitate the functional expression of membrane proteins (62). After cleavage and removal of YbeL, StrepII-PilC was purified to homogeneity. PilG, the PilC ortholog of N. meningitidis, was mainly present as a dimer, but here also a tetrameric species was identified (35). Both the YbeL fusion and StrepII-PilC migrated as single bands on Blue-Native PAGE, demonstrating that they are present on Blue-Native PAGE in a single oligomeric state. Taking into account that the mass of many membrane transporters calculated from Blue-Native PAGE using soluble marker proteins is overestimated by ∼80% suggests that the YbeL-fusion and StrepII-PilC are present as dimers. However, because PilC contains less transmembrane domains than most transporter proteins for which the overestimation of the molecular mass with 80% was determined, and the observation that the oligomeric state of membrane proteins is also dependent on the kind and concentration of detergent used, we cannot fully exclude that PilC is present as a higher oligomeric assembly. To test whether PilB interacts with PilC, pulldown experiments were performed with StrepII-PilC-containing proteoliposomes. Remarkably, His10-PilB:Hcp1, but not His6-PilB, bound to the StrepII-PilC-containing proteoliposomes, demonstrating that the hexameric form of PilB interacts with a higher affinity with PilC than monomeric PilB. This further confirms that these ATPases are functional in the hexameric state and that assembly of the PilB hexamer is an important step in stimulation of T4P assembly. Importantly, the ATPase activity of hexameric His10-PilB:Hcp1 was stimulated by PilC. The stimulation of ATPase activity was observed for the reconstituted PilC in proteoliposomes as well as for solubilized PilC, demonstrating that this interaction also occurs in the solubilized state. To our knowledge this is the first time that a stimulation of the ATPase activity of a T4P or T2S ATPase by its IM platform protein is observed, demonstrating a functional interaction between the ATPase and the IM protein. Assuming that PilC is functional as a dimer in the complex with PilB and that all PilC is functionally reconstituted, the stimulated ATPase activity of the complex can be calculated from the slopes of Fig. 2, E and F, and is ∼1 Pi/min. This value is still relatively low, e.g. the Hcp1-fused GspE has an activity of ∼18 Pi/min in the absence of its platform protein (49). A further stimulation would be expected if hydrolysis of ATP is the driving force for pilus extension.
Another candidate to stimulate the ATPase activity of PilB was PilM. The T2S ATPase GspE interacts with the PilM-N homolog GspL (42, 55, 51, 65–67), and hexamer formation by GspE might be an important step in the interaction (67). To our knowledge, such an interaction has not yet been demonstrated for T4P assembly systems, and therefore we set out to characterize PilM of M. xanthus. As observed for T. thermophilus PilM (25), PilM was stabilized by the N-terminal cytosolic extension of PilN. Notably, contrary to other studies where a peptide was added separately to PilM, we were able to stabilize PilM by creating a fusion with this small peptide. Purified PilM-N(1–16) behaved in size exclusion chromatography as a monomer and bound nucleotides with a high affinity. However, no significant ATP hydrolysis could be detected. Remarkably, binding assays with fluorescent nucleotides showed that after initial nucleotide binding (observed by a decrease in the maximal emission value of the fluorescent nucleotide), the fluorescence of the already bound nucleotide further increased strongly upon increasing the PilM concentration suggesting a higher oligomeric assembly of PilM. Formation of “rods” of the cytoplasmic domain of GspL were recently observed in several crystal structures (68), suggesting that similar to GspL a higher oligomeric form of PilM is assembled in the presence of nucleotides and may play a role in the assembly or the function of the T4P system. In archaea, this role might be fulfilled by either the FlaH ATPase that interacts with the FlaI secretion ATPase (69) or FlaX (38), a membrane protein with a cytosolic domain that forms a ring-like structure around the FlaI secretion ATPase. Importantly, in our pulldown and co-purification experiments, we observed that PilM and hexameric PilB interact directly, whereas an interaction was neither observed between PilM and solubilized PilC nor between PilM and PilC in proteoliposomes. By contrast, in the presence of PilB, a heteromeric complex was formed containing PilM, hexameric PilB, and PilC. We speculate that PilM may form a ring surrounding the hexameric PilB ATPase as has been proposed for FlaX in the case of FlaI secretion ATPase in archaea (38).
Thus, based on our data, we propose a model for T4P assembly, in which the dimeric PilC IM platform protein interacts via its two N-terminal cytoplasmic domains with hexameric PilB. PilB, in turn, also interacts with PilM, and PilM also interacts with the N-terminal cytoplasmic domain of PilN and possibly also with itself. Moreover, we speculate that PilM may form a ring structure around PilB; this ring structure might form a stator during the insertion or retraction of the pili. In this complex, ATP hydrolysis by PilB, stimulated by PilC, would drive the incorporation of PilA pilin subunits at the base of the pilus.
It is interesting to note that also in the T2SSs, the PilB, PilC, and PilM homologs form a complex (51, 55, 56). Similar to what we have observed, formation of a complex, including PilC and PilM, requires all three components, and no direct interaction between the PilC and PilM homologs was detected. Both in T2SS and in T4PS, a direct interaction between the PilB and PilM homologs is found, but although this interaction in T2SS stimulates the ATPase activity of the PilB homolog (52), we did not detect such stimulation in the T4PS of M. xanthus. In T2SSs, a direct interaction between the PilC homolog and the PilB homolog was only identified using yeast two-hybrid analysis in the T2SS of E. chrysanthemi, but complex formation, including the PilC and PilB homologs, generally required the presence of the PilM homolog (51). Contrary to this observation for T2SS, we observed a stable interaction between PilC and PilB, and this interaction stimulated the ATPase activity. Thus in both T2SS and T4PS, a complex seems to be formed in which the platform protein interacts with the secretion ATPase and in which the secretion ATPase interacts with the PilM (homolog). In T2SS, a stimulation of the secretion ATPase by the PilM homolog, but not by the platform protein, was observed, although in our study a stimulation of the secretion ATPase by the platform protein, but not by the PilM homolog, was observed. Thus, although these three homologous proteins form a complex in both systems, the assembly of the components of these complexes and the functional consequences of these interactions are different. We speculate that these differences may relate to the mechanism of these two machines, i.e. T2SS function depends on one ATPase only (GspE) and the T4PS depends on two ATPases, PilB for extension and PilT for retraction, that associate dynamically with the rest of the machine. To address these differences, the dynamic interactions between these components during T4P assembly and retraction as well as during secretion of a substrate should be studied in the future.
Author Contributions
L. F. B. designed, performed, and analyzed the experiments in Figs. 1–4 and wrote the paper. C. F. designed, performed, and analyzed the experiments in Fig. 3. A. H. contributed to Fig. 1. L. S. A. conceived the study and wrote the paper. C. v. d. D. conceived and coordinated the study, analyzed the experiments, and wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Shani Leviatan-Ben-Arye and Nathan Nelson for the constructs that led to overexpression β:PilC. Jan-Hendrik Heilers is thanked for carefully reading the manuscript. We thank Sonja-Verena Albers for allowing Lisa Franziska Bischof to continue this work within the Molecular Biology of Archaea Group in Freiburg, Germany.
This work was supported in part by the German Research Council within the framework of the Collaborative Research Center SFB987 “Microbial Diversity in Environmental Signal Response,” intramural funding of the Max Planck Society, and by a fellowship from the International Max Planck Research School for Environmental, Cellular, and Molecular Microbiology (to C. F.). The authors declare that they have no conflicts of interest with the contents of this article.
- T4P
- type IV pili
- T4PS
- T4P system
- T2SS
- type II secretion system
- IM
- inner membrane
- OM
- outer membrane
- TEV
- tobacco etch virus
- MANT
- 2′(3′)-O-(N-methylanthraniloyl) adenosine 5′-triphosphate
- TNP-ATP
- (2′-(or-3′)-O-(trinitrophenyl) adenosine 5′-triphosphate
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
References
- 1. Berry J.-L., and Pelicic V. (2015) Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 39, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pohlschroder M., Ghosh A., Tripepi M., and Albers S.-V. (2011) Archaeal type IV pilus-like structures—evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363 [DOI] [PubMed] [Google Scholar]
- 3. Melville S., and Craig L. (2013) Type IV pili in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 77, 323–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayers M., Howell P. L., and Burrows L. L. (2010) Architecture of the type II secretion and type IV pilus machineries. Future Microbiol. 5, 1203–1218 [DOI] [PubMed] [Google Scholar]
- 5. Hobbs M., and Mattick J. S. (1993) Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol. Microbiol. 10, 233–243 [DOI] [PubMed] [Google Scholar]
- 6. Chen I., and Dubnau D. (2004) DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2, 241–249 [DOI] [PubMed] [Google Scholar]
- 7. Albers S.-V., and Jarrell K. F. (2015) The archaellum: how archaea swim. Front. Microbiol. 6, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Merz A. J., So M., and Sheetz M. P. (2000) Pilus retraction powers bacterial twitching motility. Nature 407, 98–102 [DOI] [PubMed] [Google Scholar]
- 9. Skerker J. M., and Berg H. C. (2001) Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 98, 6901–6904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Craig L., and Li J. (2008) Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 18, 267–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Morand P. C., Bille E., Morelle S., Eugène E., Beretti J.-L., Wolfgang M., Meyer T. F., Koomey M., and Nassif X. (2004) Type IV pilus retraction in pathogenic Neisseria is regulated by the PilC proteins. EMBO J. 23, 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maier B., Potter L., So M., Long C. D., Seifert H. S., and Sheetz M. P. (2002) Single pilus motor forces exceed 100 pN. Proc. Natl. Acad. Sci. U.S.A. 99, 16012–16017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clausen M., Jakovljevic V., Søgaard-Andersen L., and Maier B. (2009) High-force generation is a conserved property of type IV pilus systems. J. Bacteriol. 191, 4633–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kaiser D. (1979) Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc. Natl. Acad. Sci. U.S.A. 76, 5952–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blackhart B. D., and Zusman D. R. (1985) “Frizzy” genes of Myxococcus xanthus are involved in control of frequency of reversal of gliding motility. Proc. Natl. Acad. Sci. U.S.A. 82, 8767–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun H., Zusman D. R., and Shi W. (2000) Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr. Biol. 10, 1143–1146 [DOI] [PubMed] [Google Scholar]
- 17. Bulyha I., Schmidt C., Lenz P., Jakovljevic V., Höne A., Maier B., Hoppert M., and Søgaard-Andersen L. (2009) Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Mol. Microbiol. 74, 691–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mignot T., Merlie J. P. Jr., and Zusman D. R. (2005) Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science 310, 855–857 [DOI] [PubMed] [Google Scholar]
- 19. Spormann A. M., and Kaiser A. D. (1995) Gliding movements in Myxococcus xanthus. J. Bacteriol. 177, 5846–5852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nunn D. N., and Lory S. (1991) Product of the Pseudomonas aeruginosa gene pilD is a pre-pilin leader peptidase. Proc. Natl. Acad. Sci. U.S.A. 88, 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burkhardt J., Vonck J., and Averhoff B. (2011) Structure and function of PilQ, a secretin of the DNA transporter from the thermophilic bacterium Thermus thermophilus HB27. J. Biol. Chem. 286, 9977–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Siewering K., Jain S., Friedrich C., Webber-Birungi M. T., Semchonok D. A., Binzen I., Wagner A., Huntley S., Kahnt J., Klingl A., Boekema E. J., Søgaard-Andersen L., and van der Does C. (2014) Peptidoglycan-binding protein TsaP functions in surface assembly of type IV pili. Proc. Natl. Acad. Sci. U.S.A. 111, E953–E961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nudleman E., Wall D., and Kaiser D. (2006) Polar assembly of the type IV pilus secretin in Myxococcus xanthus. Mol. Microbiol. 60, 16–29 [DOI] [PubMed] [Google Scholar]
- 24. Koo J., Tammam S., Ku S.-Y., Sampaleanu L. M., Burrows L. L., and Howell P. L. (2008) PilF is an outer membrane lipoprotein required for multimerization and localization of the Pseudomonas aeruginosa type IV pilus secretin. J. Bacteriol. 190, 6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karuppiah V., and Derrick J. P. (2011) Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J. Biol. Chem. 286, 24434–24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Georgiadou M., Castagnini M., Karimova G., Ladant D., and Pelicic V. (2012) Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol. Microbiol. 84, 857–873 [DOI] [PubMed] [Google Scholar]
- 27. Sampaleanu L. M., Bonanno J. B., Ayers M., Koo J., Tammam S., Burley S. K., Almo S. C., Burrows L. L., and Howell P. L. (2009) Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. J. Mol. Biol. 394, 143–159 [DOI] [PubMed] [Google Scholar]
- 28. Li C., Wallace R. A., Black W. P., Li Y. Z., and Yang Z. (2013) Type IV pilus proteins form an integrated structure extending from the cytoplasm to the outer membrane. PLoS ONE 8, e70144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tammam S., Sampaleanu L. M., Koo J., Sundaram P., Ayers M., Chong P. A., Forman-Kay J. D., Burrows L. L., and Howell P. L. (2011) Characterization of the PilN, PilO and PilP type IVa pilus subcomplex. Mol. Microbiol. 82, 1496–1514 [DOI] [PubMed] [Google Scholar]
- 30. Tammam S., Sampaleanu L. M., Koo J., Manoharan K., Daubaras M., Burrows L. L., and Howell P. L. (2013) PilMNOPQ from the Pseudomonas aeruginosa type IV pilus system form a transenvelope protein interaction network that interacts with PilA. J. Bacteriol. 195, 2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Friedrich C., Bulyha I., and Søgaard-Andersen L. (2014) Outside-in assembly pathway of the type IV pilus system in Myxococcus xanthus. J. Bacteriol. 196, 378–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balasingham S. V., Collins R. F., Assalkhou R., Homberset H., Frye S. A., Derrick J. P., and Tønjum T. (2007) Interactions between the lipoprotein PilP and the secretin PilQ in Neisseria meningitidis. J. Bacteriol. 189, 5716–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takhar H. K., Kemp K., Kim M., Howell P. L., and Burrows L. L. (2013) The platform protein is essential for type IV pilus biogenesis. J. Biol. Chem. 288, 9721–9728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jakovljevic V., Leonardy S., Hoppert M., and Søgaard-Andersen L. (2008) PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190, 2411–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Karuppiah V., Hassan D., Saleem M., and Derrick J. P. (2010) Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins 78, 2049–2057 [DOI] [PubMed] [Google Scholar]
- 36. Collins R. F., Saleem M., and Derrick J. P. (2007) Purification and three-dimensional electron microscopy structure of the Neisseria meningitidis type IV Pilus biogenesis protein PilG. J. Bacteriol. 189, 6389–6396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carbonnelle E., Helaine S., Nassif X., and Pelicic V. (2006) A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Mol. Microbiol. 61, 1510–1522 [DOI] [PubMed] [Google Scholar]
- 38. Banerjee A., Ghosh A., Mills D. J., Kahnt J., Vonck J., and Albers S.-V. (2012) FlaX, a unique component of the crenarchaeal archaellum, forms oligomeric ring-shaped structures and interacts with the motor ATPase FlaI. J. Biol. Chem. 287, 43322–43330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iyer L. M., Leipe D. D., Koonin E. V., and Aravind L. (2004) Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 146, 11–31 [DOI] [PubMed] [Google Scholar]
- 40. Planet P. J., Kachlany S. C., DeSalle R., and Figurski D. H. (2001) Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. U.S.A. 98, 2503–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shin D. S., Pellegrini L., Daniels D. S., Yelent B., Craig L., Bates D., Yu D. S., Shivji M. K., Hitomi C., Arvai A. S., Volkmann N., Tsuruta H., Blundell T. L., Venkitaraman A. R., and Tainer J. A. (2003) Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. EMBO J. 22, 4566–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abendroth J., Murphy P., Sandkvist M., Bagdasarian M., and Hol W. G. (2005) The x-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J. Mol. Biol. 348, 845–855 [DOI] [PubMed] [Google Scholar]
- 43. Robien M. A., Krumm B. E., Sandkvist M., and Hol W. G. (2003) Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J. Mol. Biol. 333, 657–674 [DOI] [PubMed] [Google Scholar]
- 44. Yamagata A., and Tainer J. A. (2007) Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J. 26, 878–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen Y., Shiue S.-J., Huang C.-W., Chang J.-L., Chien Y.-L., Hu N.-T., and Chan N.-L. (2005) Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. J. Biol. Chem. 280, 42356–42363 [DOI] [PubMed] [Google Scholar]
- 46. Misic A. M., Satyshur K. A., and Forest K. T. (2010) P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type Iv pilus retraction motor. J. Mol. Biol. 400, 1011–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Satyshur K. A., Worzalla G. A., Meyer L. S., Heiniger E. K., Aukema K. G., Misic A. M., and Forest K. T. (2007) Crystal structures of the Pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure 15, 363–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reindl S., Ghosh A., Williams G. J., Lassak K., Neiner T., Henche A.-L., Albers S.-V., and Tainer J. A. (2013) Insights on FlaI functions in archaeal motor assembly and motilify from structures, conformations and genetics. Mol. Cell 49, 1069–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu C., Turley S., Marionni S. T., Park Y.-J., Lee K. K., Patrick M., Shah R., Sandkvist M., Bush M. F., and Hol W. G. (2013) Hexamers of the type II secretion ATPase GspE from Vibrio cholerae with increased ATPase activity. Structure 21, 1707–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chiang P., Habash M., and Burrows L. L. (2005) Disparate subcellular localization patterns of Pseudomonas aeruginosa type IV pilus ATPases involved in twitching motility. J. Bacteriol. 187, 829–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Py B., Loiseau L., and Barras F. (2001) An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2, 244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Camberg J. L., Johnson T. L., Patrick M., Abendroth J., Hol W. G., and Sandkvist M. (2007) Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. EMBO J. 26, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yamagata A., Milgotina E., Scanlon K., Craig L., Tainer J. A., and Donnenberg M. S. (2012) Structure of an essential type IV pilus biogenesis protein provides insights into pilus and type II secretion systems. J. Mol. Biol. 419, 110–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Abendroth J., Bagdasarian M., Sandkvist M., and Hol W. G. (2004) The structure of the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae: an unusual member of the actin-like ATPase superfamily. J. Mol. Biol. 344, 619–633 [DOI] [PubMed] [Google Scholar]
- 55. Robert V., Filloux A., and Michel G. P. (2005) Subcomplexes from the Xcp secretion system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 252, 43–50 [DOI] [PubMed] [Google Scholar]
- 56. Arts J., de Groot A., Ball G., Durand E., El Khattabi M., Filloux A., Tommassen J., and Koster M. (2007) Interaction domains in the Pseudomonas aeruginosa type II secretory apparatus component XcpS (GspF). Microbiology 153, 1582–1592 [DOI] [PubMed] [Google Scholar]
- 57. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 58. Claeys D., Geering K., and Meyer B. J. (2005) Two-dimensional blue native/sodium dodecyl sulfate gel electrophoresis for analysis of multimeric proteins in platelets. Electrophoresis 26, 1189–1199 [DOI] [PubMed] [Google Scholar]
- 59. van der Laan M., Zerbes R. M., and van der Does C. (2013) Reconstitution of mitochondrial presequence translocase into proteoliposomes. Methods Mol. Biol. 1033, 325–344 [DOI] [PubMed] [Google Scholar]
- 60. Rouser G., Fkeischer S., and Yamamoto A. (1970) Two dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 61. van der Does C., de Keyzer J., van der Laan M., and Driessen A. J. (2003) Reconstitution of purified bacterial preprotein translocase in liposomes. Methods Enzymol. 372, 86–98 [DOI] [PubMed] [Google Scholar]
- 62. Leviatan S., Sawada K., Moriyama Y., and Nelson N. (2010) Combinatorial method for overexpression of membrane proteins in Escherichia coli. J. Biol. Chem. 285, 23548–23556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Heuberger E. H., Veenhoff L. M., Duurkens R. H., Friesen R. H., and Poolman B. (2002) Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317, 591–600 [DOI] [PubMed] [Google Scholar]
- 64. Knol J., Veenhoff L., Liang W. J., Henderson P. J., Leblanc G., and Poolman B. (1996) Unidirectional reconstitution into detergent-destabilized liposomes of the purified lactose transport system of Streptococcus thermophilus. J. Biol. Chem. 271, 15358–15366 [DOI] [PubMed] [Google Scholar]
- 65. Sandkvist M., Bagdasarian M., Howard S. P., and DiRita V. J. (1995) Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14, 1664–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Possot O. M., Vignon G., Bomchil N., Ebel F., and Pugsley A. P. (2000) Multiple interactions between pullulanase secreton components involved in stabilization and cytoplasmic membrane association of PulE. J. Bacteriol. 182, 2142–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shiue S.-J., Kao K.-M., Leu W.-M., Chen L.-Y., Chan N.-L., and Hu N.-T. (2006) XpsE oligomerization triggered by ATP binding, not hydrolysis, leads to its association with XpsL. EMBO J. 25, 1426–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lu C., Korotkov K. V., and Hol W. G. (2014) Crystal structure of the full-length ATPase GspE from the Vibrio vulnificus type II secretion system in complex with the cytoplasmic domain of GspL. J. Struct. Biol. 187, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Banerjee A., Neiner T., Tripp P., and Albers S.-V. (2013) Insights into subunit interactions in the Sulfolobus acidocaldarius archaellum cytoplasmic complex. FEBS J. 280, 6141–6149 [DOI] [PubMed] [Google Scholar]