Abstract
Iron is an important biological catalyst and is critical for DNA synthesis during cell proliferation. Cellular iron uptake is enhanced in tumor cells to support increased DNA synthesis. Circadian variations in DNA synthesis and proliferation have been identified in tumor cells, but their relationship with intracellular iron levels is unclear. In this study, we identified a 24-h rhythm in iron regulatory protein 2 (IRP2) levels in colon-26 tumors implanted in mice. Our findings suggest that IRP2 regulates the 24-h rhythm of transferrin receptor 1 (Tfr1) mRNA expression post-transcriptionally, by binding to RNA stem-loop structures known as iron-response elements. We also found that Irp2 mRNA transcription is promoted by circadian clock genes, including brain and muscle Arnt-like 1 (BMAL1) and the circadian locomotor output cycles kaput (CLOCK) heterodimer. Moreover, growth in colon-26(Δ19) tumors expressing the clock-mutant protein (CLOCKΔ19) was low compared with that in wild-type colon-26 tumor. The time-dependent variation of cellular iron levels, and the proliferation rate in wild-type colon-26 tumor was decreased by CLOCKΔ19 expression. Our findings suggest that circadian organization contributes to tumor cell proliferation by regulating iron metabolism in the tumor.
Keywords: circadian, circadian rhythm, iron, iron metabolism, pharmacology
Introduction
Circadian rhythms affect blood pressure, locomotor activity, core body temperature, and the sleep-wake cycle. These circadian controls of physiology and behavior are driven by a master pacemaker located in the suprachiasmatic nucleus of the hypothalamus, which relies on the interplay of interconnected transcriptional and translational feedback loops (1). The brain and muscle Arnt-like 1 (BMAL1)3 and circadian locomotor output cycles kaput (CLOCK) heterodimer drive the transcription of cryptochrome (CRY) and period (PER) genes by binding to the E-box in their promoter regions. CRY and PER homodimers and heterodimers inhibit the function of BMAL1/CLOCK, thus decreasing their own expression (2–5). This core loop creates a 24-h rhythmic oscillation of clock-controlled gene expression in normal and tumor cells.
Iron is an important metal for cell proliferation, metabolism, respiration, and DNA synthesis (6–8). Iron homeostasis is dependent on the expression of transferrin receptor 1 (TfR1) (9, 10). TfR1 is a membrane receptor responsible for iron uptake and ferritin is an intracellular protein that stores iron. When present in excess, cellular iron is toxic (11–13). Therefore, iron concentration must be tightly regulated. In general, the expression of TfR1 is regulated by iron-regulatory proteins (IRPs), which are sensors of intracellular iron levels. IRPs control TfR1 and divalent metal transporter 1 (Dmt1) at the post-transcriptional level by binding to RNA stem-loop structures known as iron-responsive elements (IREs) in the 3′-untranslated region (UTR) of mRNA. IRP binding stabilizes transcripts in these genes (14–16).
Iron metabolism is critical for the rapid growth of tumors and iron is also required for DNA synthesis in tumor cells. Therefore, the cellular iron level is higher in tumor cells. TfR1 expression in tumor cells is higher than that in normal cells (17). Additionally, recent studies have demonstrated diurnal variation in DNA synthesis and cell proliferation in tumor masses and that such variation is important for tumor growth (18, 19). However, the mechanism by which iron metabolism relates to these rhythms is unclear.
We previously reported that TfR1 mRNA expression in tumors exhibits a circadian rhythm (20). Therefore, we hypothesized that iron levels in tumor masses also follow a circadian rhythm. Moreover, because cellular iron metabolism is controlled by IRPs (14–16), IRP mRNA expression may also exhibit a circadian rhythm in tumor masses.
In this study, we identified a 24-h cycle in IRP2 expression in tumor masses. In addition, we found that circadian clock genes controlled the 24-h oscillation of IRP2 mRNA. Furthermore, circadian expression of IRP2 affected the stability of mRNA including that of IRE in colon-26 tumors. Our findings suggest that circadian organization contributes to cell proliferation by regulating iron metabolism.
Experimental Procedures
Animals and Cells
Seven-week-old male BALB/c mice (Charles River Japan) were housed with lights on from 7:00 a.m. to 7:00 p.m. at an ambient temperature of 24 ± 1 °C and a humidity of 60 ± 10%, with food and water provided ad libitum. The colon-26 cell line was purchased from Cell Bank Riken BioResource Center. Cells were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified 5% CO2 atmosphere. Colon-26(Δ19) expressing the stable clock mutant protein was generated by transfection of the clock mutant protein (CLOCKΔ19) expression vector and the cells were clonally selected by treatment of G418 (Wako). Expression of CLOCKΔ19 was confirmed by polymerase chain reaction (PCR) products (Fig. 3c). Colon-26 (irp2 knock down) expressing stable Irp2 microRNA was generated by transfection of Irp2 microRNA expression vector (BLOCK-iT Pol II miR RNAi Expression Vector, Invitrogen) and the cells was clonally selected by treatment of G418 (Wako). We confirmed that the cell line was authenticated by the cell bank using short tandem repeat-PCR analysis, and used within 3 months from frozen stock. Tumor model mice were euthanized after the tumor size reached ∼200 mm3. The tumor volume was estimated using the formula: tumor volume (mm3) = 4πxyz/3, where 2x, 2y, and 2z are the three perpendicular diameters of the tumor. All experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals distributed by the United States National Institutes of Health.
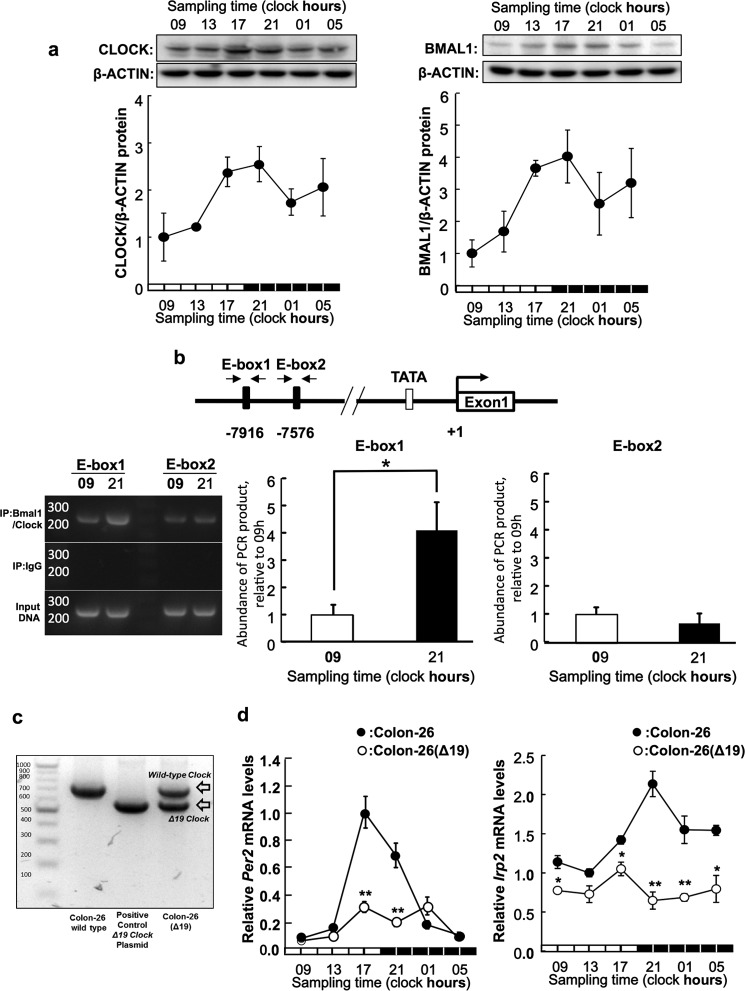
FIGURE 3.
BMAL1/CLOCK regulates the transcription of the IRP2 gene in a time-dependent manner. a, temporal expression profiles of CLOCK (left panel) and BMAL1 (right panel) protein in implanted colon-26 tumor masses. The photographs are shown as representative data. The quantitative data were normalized using β-ACTIN as a control. Values are the mean ± S.E. (n = 3–4). There were significant time-dependent variations in CLOCK and BMAL1 protein levels in implanted colon-26 tumor masses (p < 0.05, ANOVA). b, quantification of temporal changes in the binding of BMAL1/CLOCK to E-box in the 5′-flanking region of IRP2 gene in colon-26 cells implanted in mice. Bindings of Bmal1/CLOCK proteins to each E-box, E-box1 (−7916 bp) and E-box2 (−7576 bp), are detected by ChIP assay. The photographs are shown as representative data of ChIP assay, which detects bindings of Bmal1/CLOCK proteins on each E-box, E-box1 (−7916 bp) and E-box2 (−7576 bp). Data were normalized to the input control, which consisted of PCR from cross-linked chromatin before immunoprecipitation (IP). The quantitative data were calculated as the ratio to input DNA. The mean value of each assay at 9:00 a.m. was set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.05 when the two groups were compared. c, the confirming of the expression of Δclock mRNA. Photograph was PCR bands of wild clock mRNA (761 bp) and Δclock mRNA (608 bp) in wild-type colon-26 and colon-26(Δ19) tumor mass. The expression of clock mutant mRNA was demonstrated in colon-26(Δ19). Primer sequences are shown in Tables 1–3. d, 24-h rhythm of Per2 (left panel) and Irp2 (right panel) mRNA in implanted wild-type colon-26 and colon-26(Δ19) tumor masses. The data were normalized using β-actin as a control. There were significant time-dependent variations in Per2 and IRP2 mRNA levels in implanted colon-26 tumor masses (p < 0.05, ANOVA). Values are shown as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 when compared with wild-type colon-26 group at each time.
Experimental Design
To assess the temporal IRP2 mRNA and protein expression profiles in tumor cells and normal cells, the right tumor mass or left normal footpad was removed from individual tumor-bearing mice at 6 different time points (09:00, 13:00, 17:00, 21:00, 01:00, and 05:00 h) on day 7 post-implantation. The levels of IRP2 mRNA and protein were measured by real-time reverse transcriptase (RT)-PCR and Western blotting analysis, respectively. To determine whether the molecular components of the circadian clock regulate the expression of IRP2, the effect of clock gene products on the transcriptional activity of IRP2 was assessed using a luciferase reporter containing lengths of the IRP2 5′-flanking region. To assess the temporal CLOCK and BMAL1 protein expression profiles in tumor cells, the levels of each protein were measured by Western blotting analysis. To analyze the temporal binding of endogenous BMAL1/CLOCK to the Irp2 promoter in colon-26 tumors, chromatin immunoprecipitation (ChIP) assays were performed using samples isolated at 09:00 and 21:00 h. To assess the relationship between oscillations in IRP2 and clock gene expression, colon-26(Δ19) mutant cells, which overexpress a CLOCK mutant that lacks transcriptional activity (CLOCKΔ19), were used. To assess the temporal Per2 and Irp2 mRNA expression profiles in wild-type colon-26 and colon-26(Δ19) tumor cells, the levels of each mRNA were measured by real-time RT-PCR.
To determine the effect of IRP2 abundance on TfR1 mRNA stability, mRNA was extracted from colon-26 cells treated with actinomycin D (Act D) to inhibit transcription 24 h after IRP2 expression had been induced by deferoxamine (DFO) treatment. The levels of TfR1 mRNA were measured by real-time RT-PCR. Wild-type colon-26 or colon-26(Δ19) tumor masses were removed at 6 different time points and the temporal Tfr1 and Dmt1 mRNA expression profiles were assessed. To assess the time-dependent changes in TfR1 mRNA stability, tumor masses were removed at 09:00 and 21:00 h. The temporal binding of endogenous IRP2 to the IREs of murine TfR1 mRNA in individual tumor masses at 09:00 and 21:00 h was assessed by immunoprecipitation analysis.
To assess the importance of clock genes in tumor cell proliferation, tumor volumes were measured on day 15 post-implantation. To assess the temporal iron concentration profiles in tumor cells, tumor masses were removed from individual wild-type colon-26 or colon-26(Δ19) tumor-bearing mice at 6 different time points on day 7 post-implantation. The iron levels were determined using an atomic absorption photometer. To investigate the influence of iron on cell growth, cell viability of colon-26 cells, and colon-26(Δ19) cells after treatment of apo-transferrin was analyzed with an ATP assay.
Atomic Absorption Spectrophotometry
The samples were added to ∼2 ml of HNO3 (60%) and then heated for 1 h at 70 °C. Organic compounds were completely oxidized and iron was ionized. Samples were condensed by further heating at 150 °C for 3 h. Finally, sample volumes were adjusted to 4 ml using Milli-Q water. Iron levels were quantified by inductively coupled plasma mass spectrometry using the Agilent 7500c system (Agilent, Tokyo, Japan).
RT-PCR Analysis
Total RNA was extracted using RNAiso (TaKaRa, Otsu, Japan). Mouse IRP1 (NM_007386), IRP2 (NM_022655), TfR1 (NM_011638), Dmt1 (NM_008732), Per2 (NM_011066), and β-actin (NM_007393) cDNAs were synthesized using the ReverTra Ace quantitative RT-PCR Kit (Toyobo, Kita, Japan). Real-time PCR analysis was performed with diluted cDNA samples using THUNDERBIRD SYBR qPCR Mix (Toyobo) with the 7500 Real-Time PCR System (Applied Biosystems). Primer sequences are shown in Tables 1–3.
TABLE 1.
Primer list for real-time PCR
| Gene Symbol | GenBankTM accession No. | Sequence | |
|---|---|---|---|
| Irp1 | NM_007386 | Fw | 5′-CAAACACCAGCAATCCATCC-3 |
| Re | 5′-TAGCCCACCACATCAAACC-3′ | ||
| Irp2 | NM_022655 | Fw | 5′-ATGGACTCCCCAAGTGC-3′ |
| Re | 5′-CTATAAGAATTTTCGAGCCAAAG-3′ | ||
| Tfr1 | NM_011638 | Fw | 5′-GGCTAGTGTCAGAAAACCCA-3′ |
| Re | 5′-GGTCTGCCCAATATAAGCGA-3′ | ||
| Dmt1 | NM_008732 | Fw | 5′-ATGAGCATTGCCTACCTAGA-3′ |
| Re | 5′-GGTGACATACTTCAGCAAGA-3′ | ||
| Per2 | NM_011066 | Fw | 5′-ATCTCCAGGCGGTGTTGAAG-3′ |
| Re | 5′-AGGGTTACGTCTGGGCCTCT-3′ | ||
| β-actin | NM_007393 | Fw | 5′-GACGGCCAGGTCATCACTATT-3′ |
| Re | 5′-TACCACCAGACAGCACTGTGTT-3′ |
TABLE 2.
Primer list for RNA RIPAs
| Gene symbol | GenBankTM accession No. | Sequence | |
|---|---|---|---|
| Irp2 | NM_022655 | Fw | 5′-AGAGTTCACTATAAATAGTGTTGTT-3′ |
| Re | 5′-CTCAATGACCACAAATGTTT-3′ |
TABLE 3.
Primer list for Δclock mRNA
| Gene symbol | GenBankTM accession No. | Sequence | |
|---|---|---|---|
| Δclock | NM_007715 | Fw | 5′-GCGAGAACTTGGCATTGAAGAG-3′ |
| Re | 5′-CTGTGTCCACTCATTACACTCTGTTG-3′ |
Western Blotting Analysis
Cytoplasmic proteins in tumor masses were extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce). The protein concentrations were determined using the BCA Protein Assay kit (Pierce Biotechnology). Lysates were separated using 6 or 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes. The membranes were incubated with antibodies against IRP2 (Santa Cruz Biotechnology, Dallas, TX; SC-33682) and β-ACTIN (Santa Cruz Biotechnology; SC-4778HRP). The immunocomplexes were incubated with horseradish peroxidase-conjugated secondary antibodies (Abcam; ab6728) and visualized using SuperSignal Chemiluminescent Substrate (Pierce Biotechnology). The membranes were photographed and the density of each band was analyzed using ImageQuant LAS-3000 (Fuji Film).
Construction of Vectors
The 5′ flanking region of the murine IRP2 gene (from −8328 to −7272 and from −376 to +70; +1 indicates exon1 of IRP2) was amplified from colon-26 DNA using Elongase Enzyme Mix (Invitrogen). PCR was performed using the forward primer 5′-CATGAGCTCGTCCCTTCGATTATCAGCTG-3′ (Sac1) and the reverse primer 5′-GTACTCGAGGAAGCAACAGACTTTCAGCC-3′ (Xho1), as well as the forward primer 5′-ATACTCGAGGCTTTACCATGCAGGGAGAAC-3′ (Xho1) and the reverse primer 5′-GTCAGATCTTCGCCGGAGACCATATTATCC-3′ (Bgl2). PCR products were purified and ligated into the pGL4.12 basic vector (IRP2-Luc).
To construct CLOCKΔ19 expression vector, total RNA was obtained from clock mutant mouse liver. Total RNA was converted to cDNA using ReverTra Ace (TOYOBO). The exon region of the mouse clock gene was amplified from liver cDNA using the Elongase Enzyme mix (Invitrogen). PCR was performed using the forward primer, 5′-ATAGATATC(EcoRV)ATGGTGTTTACCGTAAGCTGTA-3′ and reverse primer, 5′-ATACTCGAG(XhoI)ACCCTTCCAAGGTCCAGCCACAGTAG-3′. The PCR products were purified and ligated into a pcDNA3.1 expression vector (Life Technologies) (CLOCKΔ19). Expression vectors for mouse CLOCK, BMAL1, PER2, and CRY1 were constructed as previously described (21).
BLOCK-iT Pol II miR Irp2 RNAi Expression Vector (Invitrogen) was constructed following the material method. Irp2 microRNA oligo sequence were shown the sense oligo, 5′-GCUGCUGUCUUUGUGGUCCUCUUCA-3′ and the antisense oligo, 5′-UGAAGAGGACCACAAAGACAGCAGC-3′.
Cell Transcription and Luciferase Assays
The day before transfection, colon-26 cells were seeded at 3 × 105 cells/well into 6-well plates containing RPMI 1640 medium supplemented with 10% FBS. The cultured cells were transfected with 100 ng of reporter constructs and 2 μg (total) of clock gene expression constructs using Lipofectamine LTX reagent (Invitrogen) according to the manufacturer's instructions. To correct for fluctuations in transfection efficiency, 0.5 ng of pRL-SV40 (Promega) was co-transfected in all experiments. The total amount of DNA per well was adjusted by adding pcDNA3.1 vector (Invitrogen). At 24 h post-transfection, the cells were disrupted with 500 μl of passive lysis buffer (Promega) and luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega). For each sample, the measured luciferase activity was corrected for transfection efficiency variation by dividing the firefly luciferase activity (expressed from the reporter construct) by the Renilla luciferase activity (expressed from pRL-SV40). The transcription activity in individual experiments was normalized to the corresponding control that was adjusted to 1.
ChIP Assay
Tumors were cross-linked with 10% formaldehyde in phosphate-buffered saline (PBS) at 25 °C for 10 min. Each cross-linked sample was sonicated by Bioruptor (Diagenode) on ice and then incubated with antibodies against BMAL1 and CLOCK (1:1) (Santa Cruz Biotechnology, Inc.). Chromatin-antibody complexes were extracted following the manufacturer's instructions for Chip-IT Express (Active motif). DNA was isolated from the immunoprecipitates and subjected to PCR using the following primer pairs: for the E-box in the IRP2 promoter region, 5′-GTTCTGAACTGCTCAGGAAG-3′ and 5′-CTGAACTGGAGAGAATGTCC-3′, and for the E-box in the IRP2 intron region, 5′-GGCACAGTAACCCTAAGTTC-3′ and 5′-CAAGGAGCTGGAGAATGTAC-3′. Products were separated on 3% agarose gel electrophoresis, stained with ethidium bromide, and photographed with ImageQuant LAS 3000 mini (Fuji film).
RNA Immunoprecipitation Assays (RIPA)
RIPA was performed as previously described (22, 23), using IRP2 or IgG (Santa Cruz Biotechnology) antibody. Each experiment was independently performed 3 times. PCR was performed with diluted cDNA samples using the Gotaq Green Master Mix (Promega). PCR products were run on 2% agarose gels. After staining with ethidium bromide, the gels were photographed using Polaroid-type film or ImageQuant LAS-3000 (Fujifilm) and the density of each band was analyzed using ImageJ software (National Institutes of Health, Bethesda, MD). Primer sequences are shown in Tables 1–3.
Cell Viability Assays
Colon-26 and colon-26(Δ19) cells were seeded at a density of 1 × 105 cells/well in a 6-well culture plate. After 48 and 72 h, intracellular ATP was measured as an indicator of cell viability using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega).
Incorporation of 5-Bromouridine into DNA
Tumor masses were collected from wild-type or colon-26(Δ19) cell-bearing mice at 09:00 or 21:00 h. Tumor mass was chopped by scissors. The tumor pieces were treated with 5-bromouridine (5-BrU)/DMEM solution (100 μg/ml) for 60 min at 37 °C and later fixed in 8% paraformaldehyde in PBS for 15 min at room temperature. The tumor pieces were embedded into gelatin. The tumor-gelatin masses were cut in 20-μm thick slices by Leica CM1100 (Leica). Slices were blocked in 3% bovine serum albumin (BSA), 0.2% Tween (TBS) for 1 h. Then, slices were incubated in anti-5-bromodeoxyuridine-fluorescein isothiocyanate (anti-BrdU-FITC) antibody (Santa Cruz Biotechnology)/T-TBS (1:2000) for 24 h. The stained cells were observed using a fluorescence microscope (BZ-9000, KEYENCE). The images were taken in 3 regions of culture sections at a magnification of ×100.
Statistical Analysis
Analysis of variance (ANOVA) was used for multiple comparisons, and Scheffé's test was used for comparison between two groups. A p value of < 0.05 was considered significant.
Results
The 24-h Rhythm of IRP2 Expression in Colon-26 Tumors
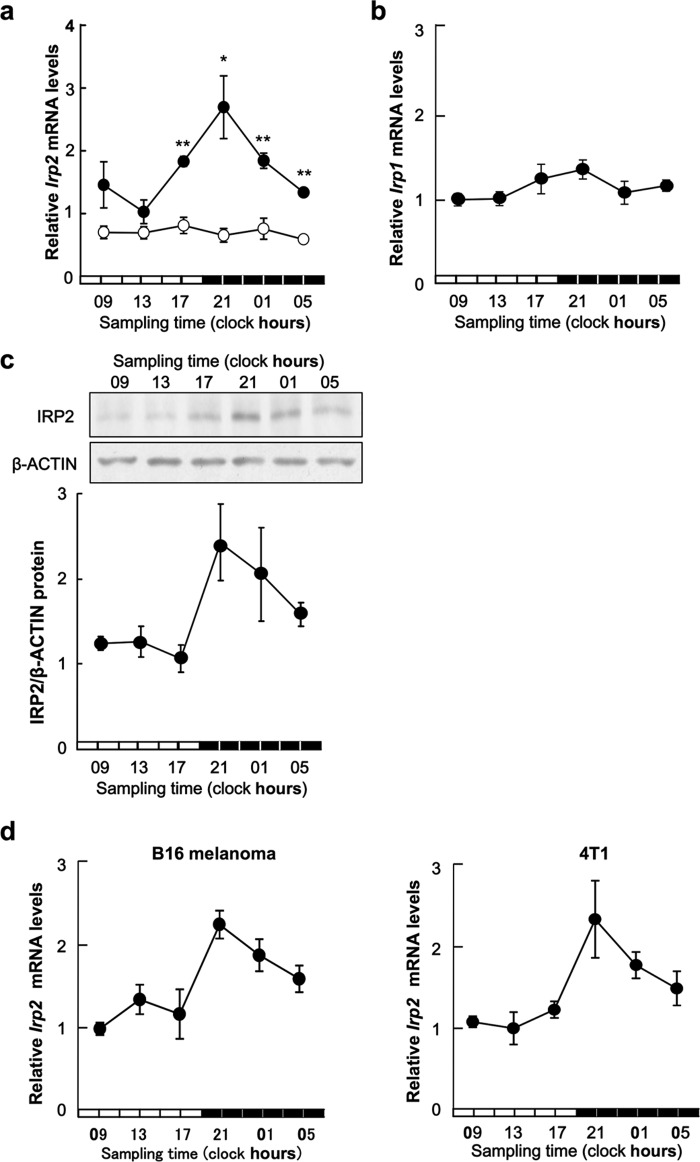
Two IRP isoforms were identified: IRP1 and IRP2. A 24-h cycle was noted in mRNA expression of IRP2, but not IRP1, in mice right footpad bearing colon-26 tumor cells (Fig. 1, a and c). The expression of IRP2 mRNA in the left footpad without tumor cells showed no significant rhythm. IRP2 protein expression also displayed a 24-h rhythm, with higher levels observed during the early dark phase (Fig. 1b). The increase and decrease in IRP2 mRNA levels seemed to underlie the rhythm in IRP2 protein expression in colon-26 tumors. Therefore, we checked how with other cancer cells. The 24-h cycle was noted in mRNA expression of IRP2 in B16 melanoma and 4T1 tumor masses (Fig. 1c).
FIGURE 1.
Twenty-four hour rhythm of IRP2 in colon-26 tumor masses. a, temporal expression profiles of Irp2 mRNA in implanted colon-26 tumor masses (right footpad: ●) or normal tissue (left footpad: ○). The data were normalized using β-actin as a control. Values are the mean ± S.E. (n = 6). There was a significant time-dependent variation in mRNA levels in implanted colon-26 tumor masses (right footpad: ●) (p < 0.05, ANOVA). *, p < 0.05; **, p < 0.01 when compared with normal tissues at each time. b, 24-h rhythm of IRP1 in colon-26 tumor masses. Temporal expression profiles of IRP1 mRNA in implanted colon-26 tumor masses. The data were normalized using β-actin as a control. Values are the mean ± S.E. (n = 6). There was no significant time-dependent variation in mRNA levels in implanted colon-26 tumor masses. c, temporal expression profiles of IRP2 protein in implanted colon-26 tumor masses (representative data, upper panel, quantitative data, lower panel). The data were normalized using β-ACTIN as a control. Values are the mean ± S.E. (n = 3–4). There was a significant time-dependent variation in IRP2 protein levels (p < 0.05, ANOVA). d, 24-h rhythm of IRP2 in B16 melanoma and 4T1 tumor masses. Temporal expression profiles of IRP2 mRNA in implanted B16 melanoma and 4T1 tumor masses. The data were normalized using β-actin as a control. Values are the mean ± S.E. (n = 6). There was a significant time-dependent variation in mRNA levels in implanted B16 melanoma and 4T1 tumor masses (p < 0.05, ANOVA).
Regulation of IRP2 Gene Expression by BMAL1/CLOCK
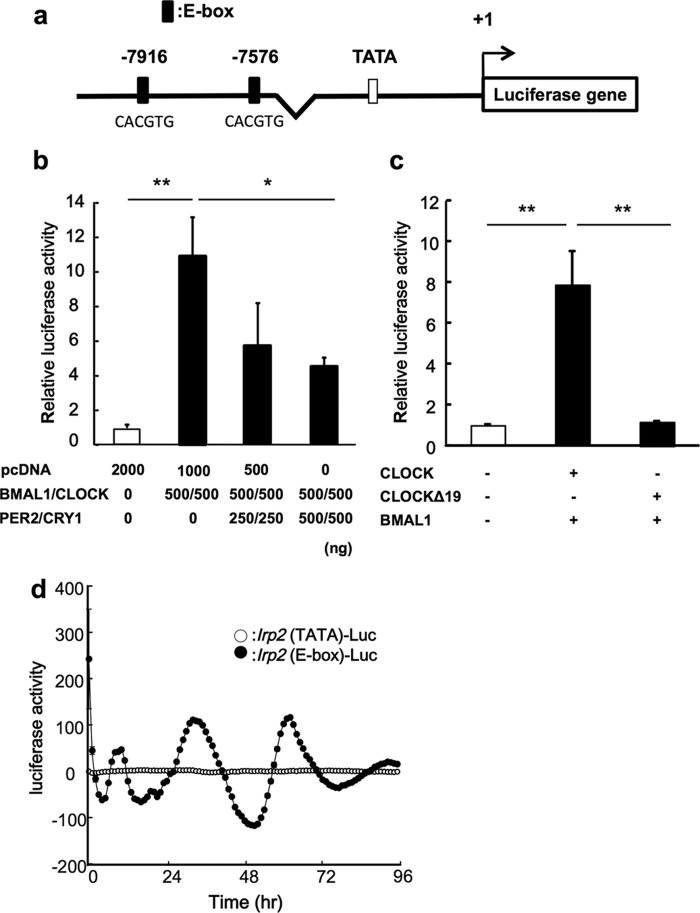
To determine whether clock gene products affect the expression of IRP2 mRNA, we looked for consensus sequences within the promoter region of the IRP2 gene (Fig. 2a). Next, we tested whether BMAL1 and CLOCK transcription factors regulated the rhythmic expression of the IRP2 gene in colon-26 cells. The IRP2-Luc reporter vector contains two BMAL1/CLOCK response site E-boxes. Co-transfection of IRP2-Luc with BMAL1/CLOCK expression constructs resulted in a 10-fold increase in promoter activity. We also assessed whether BMAL1/CLOCK activity was inhibited by transfection with PER2 and CRY1, the main proteins involved in the negative feedback loop. BMAL1/CLOCK-induced luciferase activity was suppressed in cells transfected with PER2 and CRY1 expression plasmids (Fig. 2b). However, there was no significant increase in transcriptional activity when cells were transfected with both, CLOCKΔ19 (CLOCK that lacks transcriptional activity) and BMAL1 (Fig. 2c). Therefore, the region of the Irp2 promoter was investigated by measuring real-time bioluminescence in the colon-26-cultured cell (Fig. 2d). The time-dependent variation of activity in the Irp2 promoter region including of E-box was revealed, but not TATA box region in Irp2.
FIGURE 2.
Regulation of IRP2 gene expression by BMAL1/CLOCK. a, the 5′-upstream DNA fragments of murine IRP2 were incorporated into the pGL4.12-basic vector to construct the reporter plasmids. The distances, in bases, from the putative transcription start site (marked as +1) are shown. b, transcriptional regulation of the IRP2 gene by BMAL1/CLOCK. Luciferase activity induced by BMAL1/CLOCK was inhibited by transfection with PER and CRY. The mean value of the control (pcDNA transfection) was set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 when the two groups were compared. c, transcriptional activity was not induced when the CLOCKΔ19 was expressed. Wild-type CLOCK or CLOCKΔ19 were cotransfected with BMAL1 expression vector (500 ng, respectively). The mean value of the control (pcDNA transfection) was set at 1.0. Values are shown as the mean ± S.E. (n = 3). **, p < 0.01 when the two groups were compared. d, representative traces of bioluminescence oscillations driven by Irp2 (E-box)-luciferase vector (closed circle) or Ipr2 (TATA)-luciferase vector (opened circle) in colon-26-cultured cells after incubation with 10 nm DEX for 2 h.
BMAL1/CLOCK Regulates Transcription of the IRP2 Gene in a Time-dependent Manner
To investigate the critical regions responsible for the time-dependent, BMAL1/CLOCK-mediated transactivation of IRP2, we investigated temporal expression of BMAL1 and CLOCK protein in the tumor mass (Fig. 3a). The BMAL1 and CLOCK protein expression was higher at 21:00 and lower at 09:00 h. Subsequently, we investigated the temporal binding of endogenous BMAL1/CLOCK to the IRP2 promoter by ChIP analysis (Fig. 3b). The results showed that the binding quantity of endogenous BMAL1/CLOCK in implanted colon-26 tumors was significantly higher at 21:00 than at 09:00 h. This result suggests that time-dependent binding of BMAL1/CLOCK to the IRP2 E-box controls its rhythmic expression in the intron region of the IRP2 gene at E-box1 (−7916) compared with E-box2 (−7576).
To examine the relationship between the clock genes and IRP2 expression, we analyzed IRP2 mRNA expression in implanted colon-26 and colon-26(Δ19) (cells overexpressing CLOCKΔ19) tumor masses (Fig. 3c). The ClockΔ19, which carries a deletion of exon 19 in the Clk locus, produces a protein that has been characterized as dominant-negative of E-box mediating transcription by BMAL1/CLOCK (3). The expression of Per2 mRNA was lower in colon-26(Δ19) tumors than in wild-type colon-26 tumors (Fig. 3c). IRP2 mRNA levels showed no significant 24-h rhythm in implanted colon-26(Δ19) tumors (Fig. 3d).
Time-dependent Stabilization of TfR1 mRNA in Implanted Colon-26 Cells
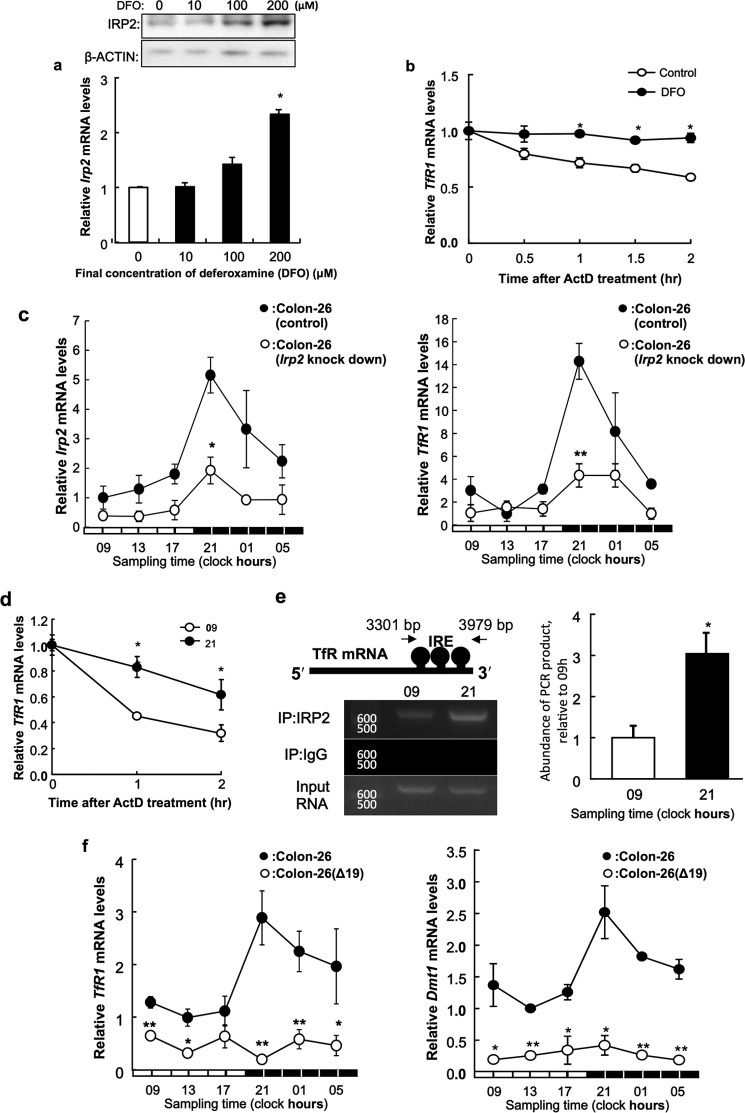
DFO stabilize IRP2 (24). Treatment with DFO, an iron-chelating agent, elicited significant increasing of IRP2 mRNA expression when compared with control treatment (Fig. 4a). Next, we tested whether TfR1 mRNA was stabilized by the high expression of IRP2 in cultured colon-26 cells. After a 24-h DFO treatment (200 μm), transcription was inhibited by actinomycin D, and TfR1 mRNA levels were measured by RT-PCR. Degradation of TfR1 mRNA decreased when IRP2 expression was induced with DFO (Fig. 4b). This result suggests that IRP2 stabilizes TfR1 mRNA in colon-26 cells.
FIGURE 4.
Stabilization of TfR1 mRNA by DFO in cultured colon-26 cells. a, analysis of IRP2 mRNA and protein levels in colon-26 cells grown for 24 h with or without DFO (0, 10, 100, and 200 μm). Upper panel shows the IRP2 protein levels or β-actin protein levels as a control protein. The IRP2 mRNA levels are indicated under the graph. The data were normalized using β-actin as a control. The mean value of the control group was set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.01 when compared with the control. b, colon-26 cells were treated with DFO (200 μm) for 24 h, and the stability of TfR1 mRNA extracted from actinomycin D-treated cells was assessed. The data were normalized using β-actin as a control. The mean value of the control group was set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.01 when compared with the control. c, 24-h rhythm of Irp2 (left panel) and TfR1 (right panel) mRNA in implanted wild-type colon-26 and colon-26 (Irp2 knockdown) tumor masses. The data were normalized using β-actin as a control. There were significant time-dependent variations in Irp2 and TfR1 mRNA levels in implanted wild-type colon-26 and colon-26 (Irp2 knockdown) tumor (p < 0.05, ANOVA). Values are the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 when compared with wild-type colon-26 group at each time. d, quantification of temporal changes in the stability of TfR1 mRNA extracted from actinomycin D-treated implanted colon-26 tumor masses. The data were normalized using β-actin as a control. The value at start time in each assay was set at 1.0. Values are shown as the mean ± S.E. (n = 3). **, p < 0.01 and *, p < 0.05 when the two groups were compared. e, quantification of temporal changes in the binding of IRP2 to the IRE located in the 3′-UTR of TfR1 in implanted colon-26 tumors. Data were normalized to the input control, which consisted of PCR from cross-linked total RNA before immunoprecipitation. The mean value at 9:00 a.m. was set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.05 when the two groups were compared. f, 24-h rhythm of TfR1 (left panel) and Dmt1 (right panel) mRNA in implanted wild-type colon-26 and colon-26(Δ19) tumor masses. The data were normalized using β-actin as a control. There were significant time-dependent variations in TfR1 and Dmt1 mRNA levels in implanted wild-type colon-26 tumor (p < 0.05, ANOVA). Values are the mean ± S.E. (n = 3). *, p < 0.05; **, p < 0.01 when compared with wild-type colon-26 group at each time.
We analyzed the 24-h rhythm of Irp2 or TfR1 mRNA levels in implanted colon-26 (Irp2 knockdown) tumors or wild-type colon-26 (Fig. 4c). Amplitude of 24-h rhythm in Irp2 and TfR1 mRNA levels were decreased significantly in implanted colon-26 Irp2 knockdown tumors compared with wild-type colon-26.
Therefore, there was less degradation of TfR1 mRNA at 21:00 than at 09:00 h, and the expression of TfR1 mRNA after a 2-h actinomycin D treatment was significantly higher at 21:00 than at 09:00 h (Fig. 4d). Therefore, we hypothesized that IRP2 stabilized TfR1 mRNA by binding to IREs located in the 3′-UTR of TfR1 mRNA. We analyzed temporal changes in IRP2 binding to IREs by RIPA. The results showed that a higher quantity of endogenous IRP2 bound to the IREs at 21:00 than at 09:00 h (Fig. 4e). We analyzed temporal changes in TfR1 and Dmt1 mRNA expression in colon-26 tumors. There was a significant 24-h cycle in the levels of TfR1 and Dmt1 mRNA expression in implanted colon-26 tumors, with higher levels occurring during the early dark phase (Fig. 4c). TfR1 and Dmt1 mRNA levels showed no significant 24-h rhythm in implanted colon-26(Δ19) tumors (Fig. 4f).
Clock Genes Regulate Cell Proliferation via Iron Metabolism
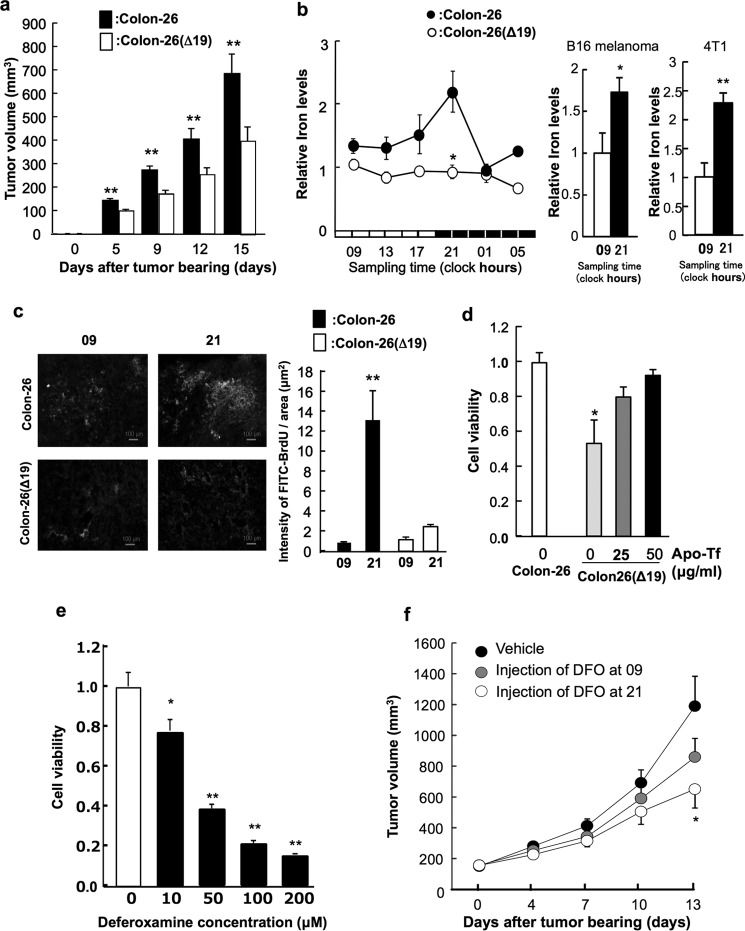
To demonstrate the effect of CLOCKΔ19 expression on tumor cell growth, we measured the tumor volume in mice implanted with colon-26 or colon-26(Δ19) cells (Fig. 5a). The tumor volume in colon-26(Δ19)-implanted mice was significantly lower than that in colon-26-implanted mice.
FIGURE 5.
Clock genes regulate cell proliferation through iron metabolism. a, the tumor volume in implanted wild-type colon-26 and colon-26(Δ19) tumors. Values are shown as the mean ± S.E. (n = 8). **, p < 0.01 when the two groups were compared. b, left panel, the temporal changes in iron levels in implanted wild-type colon-26 and colon-26(Δ19) tumor masses. The data were normalized by weight. There was a significant time-dependent variation in iron level in implanted colon-26 tumor masses (p < 0.05, ANOVA). Values are the mean ± S.E. (n = 6). Right panel, time-dependent difference of intracellular iron levels in implanted B16 melanoma and 4T1 tumor masses. The data were normalized using β-actin as a control. Values are the mean ± S.E. (n = 3–4). There was a significant time-dependent variation in intracellular iron levels in implanted B16 melanoma and 4T1 tumor masses (**, p < 0.01; *, p < 0.05, Student's t test). c, incorporation of 5-BrU in implanted wild-type colon-26 and colon-26(Δ19) tumors. 5-BrU is changed to 5-BrdU by ribonucleotide reductase including of cofactor iron. Photographs indicates that green colors were indicated the cells incorporating of 5-BrdU into DNA using FITC-conjugated anti-BrdU antibody. Right panel indicates the quantification data of luminance in FITC. **, p < 0.01 when the four groups were compared. Values are shown as the mean ± S.E. (n = 4). d, cell viability of wild type colon-26 and colon-26(Δ19) cells after apo-transferrin (Apo-Tf) treatment. Apo-transferrin binds to Fe2+ in medium. The mean value of the control group (wild-type colon-26) was set at 1.0. Values are shown as the mean ± S.E. (n = 3). *, p < 0.05 when compared with the control group. e, influence of dosing time on antitumor activities of DFO. Cell viability of colon-26 cells after DFO treatment. The mean value of the control group was set at 1.0. Values are shown as the mean ± S.E. (n = 3). **, p < 0.01; *, p < 0.05 when compared with the control. f, influence of DFO dosing time on tumor growth. Colon-26 cells were inoculated into ICR male mice. DFO (300 mg/kg, intraperitoneally, 9:00 or 21:00) or vehicle (9:00) were administered five times a week. Each point is the mean ± S.E. (n = 9–10).
We analyzed temporal changes in iron concentration in the colon-26 tumors. There was a significant 24-h cycle in the levels of iron in implanted colon-26 tumors, with higher levels occurring during the early dark phase (Fig. 5b). By contrast, the 24-h rhythm of iron levels were not present in implanted colon-26(Δ19) tumors (Fig. 5b).
Iron is a cofactor of ribonucleotide reductase. 5-BrU is converted to 5-BrdU by ribonucleotide reductase, which is then incorporated into DNA. Tumor masses were collected from mice transplanted with colon-26 or colon-26(Δ19) cells at 09:00 and 21:00 h. These tumors were treated with medium containing 5-BrU. In wild-type colon-26 cells, 5-BrdU incorporation into DNA at 21:00 was higher compared with that at 09:00 h (Fig. 5c). This time-dependent difference of 5-BrdU incorporation was not observed in colon-26(Δ19) cells.
To investigate the importance of iron in cell proliferation, we measured the influence of iron on viability of colon-26(Δ19) cells using apo-transferrin that binds to iron in culture media (Fig. 5d). Cell viability of colon-26(Δ19) was lower than that of colon-26 cells. On the other hand, the inhibition of cell viability by overexpression of CLOCKΔ19 was ameliorated by exposure to apo-transferrin (Fig. 5d). Then, influence of dosing time on the ability of DFO to inhibit tumor growth in vitro and in vivo was studied. The dose-dependent inhibition in administration of DFO was shown (Fig. 5e). Therefore, colon-26 tumor-bearing mice were injected intraperitoneally with 300 mg/kg of DFO or vehicle (saline) 5 times a week at 09:00 or 21:00 h (Fig. 5f). The growth rate of tumor cells in mice injected with DFO at 21:00 h was significantly lower than that in mice injected with saline.
Discussion
Iron is essential for biosynthesis and dysregulation of iron metabolism can cause disease (11–13). Iron is fundamental for many biological processes, including DNA synthesis, electron transport, and cell cycle regulation (6–8). However, excess iron is toxic (11–13) and therefore, cellular iron metabolism must be tightly regulated. Iron metabolism is enhanced in various tumor cells to support increased rates of DNA synthesis and cell proliferation. Thus, iron metabolism is important for tumor growth (22–25). Recent studies have shown that DNA synthesis and cell proliferation exhibit circadian rhythms in tumor cells and that these rhythms are essential for tumor cell proliferation (6–9). Several factors that contribute to this rhythm have been reported, but the relevance of iron is unclear. In this study, we hypothesized that iron levels in tumor cells display circadian rhythms and that these rhythms affect the proliferation of tumor cells.
Because cellular iron levels are regulated by cellular iron metabolism, we hypothesized that the iron rhythm in tumor cells was generated by the regulation of IRPs, which are key factors in iron metabolism (15–17). Therefore, we analyzed the expression of IRPs. Although no marked rhythmic oscillation of Irp1 mRNA levels was observed in implanted colon-26 tumor cells, Irp2 mRNA and protein levels in colon-26 tumor cells implanted in mice showed a clear 24-h oscillation (Fig. 1a).
Moreover, Irp2 mRNA levels in murine breast cancer 4T1 tumor cells or murine B16 melanoma implanted in mice showed a clear 24-h oscillation (Fig. 1d). These results may suggest that Irp2 show rhythmic expression in various tumors that have the higher proliferating ability than normal cell and tissue (15–17). IRP has two isoforms, IRP1 and IRP2. Both isoforms stabilize TfR1 and Dmt1 mRNA by binding to 3′-UTR. Our results established that IRP2 may be more important than IRP1 in inducing circadian expression in implanted tumor cells.
To identify the relationship between circadian clock genes and IRP2, we analyzed whether clock gene products affected the expression of Irp2. The luciferase reporter assay and ChIP assay revealed that Irp2 expression was regulated by BMAL1 and CLOCK (Figs. 2 and 3).
Furthermore, to clear the relationship between circadian clock gene activity and Irp2 expression in vivo, we established a mutant colon-26 cell line, colon-26(Δ19), in which CLOCKΔ19 (CLOCK protein lacking transcriptional activity) was overexpressed. clock-mutant mice are a powerful tool for the analysis of molecular clocks (3). These mice have a point mutation in exon 19 of the clock gene and exhibit low-amplitude rhythms in the expression of clock-controlled genes. Thus, the colon-26(Δ19) cell line may clarify the relationship between the biological clock and iron metabolism within tumors. In implanted colon-26(Δ19) tumor tissue, oscillations in Irp2 mRNA were no longer apparent, and Irp2 mRNA levels were decreased (Fig. 3c). These results suggest that circadian clock regulation of transcription is important for Irp2 rhythmic expression in tumor cells.
Because cellular iron levels are regulated by cellular iron metabolism, we hypothesized that the iron rhythm was controlled by regulation of IRPs, TfR1, and DMT1, which are key factors in iron metabolism (15–17). A previous study showed that the circadian rhythm in Tfr1 mRNA expression is regulated by c-Myc (20). However, it was unclear if the stability of TfR1 was related to IRP2 activity. In this study, temporal analysis of mRNA levels in implanted colon-26 cells suggested that IRP2 was involved in the circadian regulation of TfR1 and Dmt1 mRNA stability. Degradation of TfR1 mRNA at 21:00 h was lower than that at 09:00 h in vivo (Fig. 4d). Moreover, the rhythm of IRP2 protein abundance in colon-26 cells correlated with the time dependence of its binding to the 3′-UTR of TfR1 mRNA (Fig. 4e). These results suggest that oscillation in IRP2 protein levels control the 24-h rhythm of TfR1 mRNA stability. Thus, these mechanisms may affect the stability of Dmt1 mRNA in vivo. Therefore, the amplitude in the 24-h rhythm of TfR1 mRNA in Irp2 knockdown colon-26 tumor mass was decreased (Fig. 4f).
Iron is an essential element for various biological functions (26). Finally, the results of this study suggest that the circadian clock system controls tumor cell proliferation by regulating iron metabolism as one of various function of iron. In vivo, colon-26(Δ19) cell proliferation was lower than that of colon-26 cells (Fig. 5a). Iron levels exhibited a 24-h rhythm in colon-26 tumors (Fig. 5b), with higher levels observed during the early dark phase. Moreover, the time-dependent difference of iron levels in mice implanted with murine breast cancer 4T1 tumor cells or murine B16 melanoma were revealed (Fig. 5b). On the other hand, oscillations in iron were not apparent in implanted colon-26(Δ19) tumor tissues.
Conversion of 5-BrU to 5-BrdU, which is then incorporated into DNA, requires activity of ribonucleotide reductase, of which iron is a cofactor. DNA incorporation of 5-BrdU was low in colon-26(Δ19) cells in a time-dependent manner, which was not observed in colon-26(Δ19) cells (Fig. 5c). In vitro, the inhibition of colon-26(Δ19) cell proliferation was ameliorated by exposure to apo-transferrin. These results indicate that disruption of the circadian clock system reduces DNA synthesis levels and decreases cell proliferation via homeostasis of iron in the tumor. Influence of dosing time on the ability of DFO to inhibit tumor growth in mice was studied (Fig. 5f). Colon-26 tumor-bearing mice were injected intraperitoneally with 300 mg/kg of DFO or vehicle (saline) 5 times a week at 09:00 or 21:00 h. The growth rate of tumor cells in mice injected with DFO at 21:00 h was significantly lower than in mice injected with saline. This result concurs with a previous study, where DNA synthesis was enhanced in the dark phase (9). Because of the iron concentration and tumor growth, it was suggested that the circadian rhythm in iron concentration influences proliferation in the tumor.
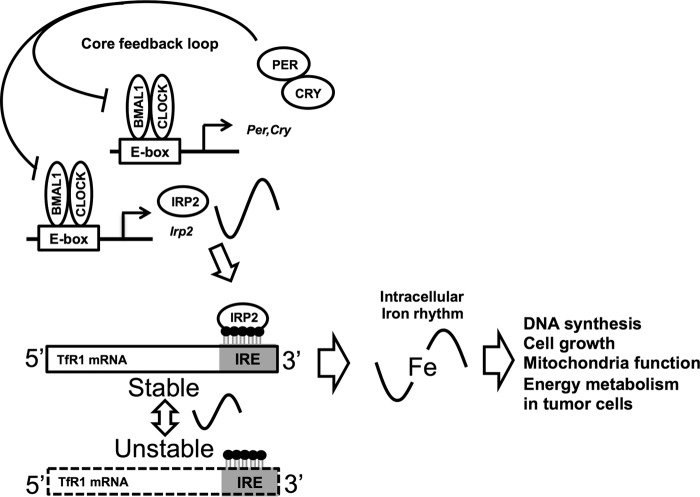
Our results suggest that the post-transcriptional mechanisms for the control of mRNA degradation can regulate circadian expression in tumor cells (Fig. 6). Previously, regulation of the molecular circadian clock by epigenetic regulation, histone acetylation, ubiquitination, and phosphorylation was demonstrated (27–29). We expect that this study will contribute to the elucidation of a novel molecular circadian clock mechanism. Thus, circadian rhythms of biological metals in tumors may be important for time-dependent biological function and tumor development.
FIGURE 6.
The molecular mechanism underlying the 24-h rhythm in iron metabolism. The CLOCK/BMAL1 heterodimer promotes IRP2 transcription in a time-dependent manner. IRP2 regulates the 24-h rhythm of TfR1 expression post-transcriptionally by binding to RNA stem-loop structures known as IREs. This rhythms lead to a 24-h rhythm in iron content in tumor cells. In turn, the 24-h rhythm of iron affects the rhythm of cell proliferation in tumor.
Author Contributions
F. O., N. M., H. O., H. A., S. K., and S. O. conceived and coordinated the study and wrote the paper. F. O., A. T., H. O., H. A., N. M., H. K., H. A., S. K., and S. O. designed, performed, and analyzed the experiments shown in all figures and tables. H. K., Y. T., T. O., Y. H., H. T., T. S., K. H., and H. I. provided technical assistance and contributed to the preparation of all the figures. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University, for technical support.
This work was supported in part by Grants-in-Aid for Scientific Research (A; 25253038) (to S. O.), Scientific Research on Innovative Areas (25136716) (to S. O.), and Challenging Exploratory Research (25670079) (to S. O.), the Uehara Memorial Foundation (to S. O.), Grant-in-Aid for Scientific Research (C; 15K08098) (to N. M.) and Grant-in-Aid for Young Scientists (B; 26860097) (to F. O.), Grant-in-Aid for Young Scientists (B; 15K19167) (to H. O.), and Grant-in-Aid for Young Scientists (Start-up; 15H06795K) (to K. H.), the Japan Research Foundation for Clinical Pharmacology (to N. M.), a Grant-in-Aid for JSPS Fellows (25-4175) (to K. H.) from the Japan Society for the Promotion of Science (JSPS), the Fukuoka Foundation for Sound Health, and the Platform for Drug Discovery, Informatics, and Structural Life Science of the Ministry of Education, Culture, Sports, Science and Technology, Japan. The authors declare that they have no conflicts of interest with the contents of this article.
- BMAL1
- brain and muscle Arnt-like 1
- CLOCK
- circadian locomotor output cycles kaput
- 5-BrU
- 5-bromouridine
- CRY
- cryptochrome
- DFO
- deferoxamine
- Dmt1
- divalent metal transporter 1
- IRE
- iron-responsive element
- IRP
- iron-regulatory protein
- PER
- period
- RIPA
- RNA immunoprecipitation assay
- TBS
- BSA containing Tween
- TfR1
- transferrin receptor 1
- ANOVA
- analysis of variance.
References
- 1. Stephan F. K., and Zucker I. (1972) Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U.S.A. 69, 1583–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez J. D., and Sehgal A. (2002) Circadian rhythms: finer clock control. Nature 419, 798–799 [DOI] [PubMed] [Google Scholar]
- 3. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., and Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 4. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., and Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 5. Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., and Schibler U. (2002) The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110, 251–260 [DOI] [PubMed] [Google Scholar]
- 6. Arredondo M., and Núñez M. T. (2005) Iron and copper metabolism. Mol. Aspects Med. 26, 313–327 [DOI] [PubMed] [Google Scholar]
- 7. Eisenstein R. S. (2000) Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu. Rev. Nutr. 20, 627–662 [DOI] [PubMed] [Google Scholar]
- 8. Fairweather-Tait S. J. (2004) Iron nutrition in the UK: getting the balance right. Proc. Nutr. Soc. 63, 519–528 [DOI] [PubMed] [Google Scholar]
- 9. Nakagawa H., Koyanagi S., Kuramoto Y., Yoshizumi A., Matsunaga N., Shimeno H., Soeda S., and Ohdo S. (2008) Modulation of circadian rhythm of DNA synthesis in tumor cells by inhibiting platelet-derived growth factor signaling. J. Pharmacol. Sci. 107, 401–407 [DOI] [PubMed] [Google Scholar]
- 10. Daniels T. R., Delgado T., Rodriguez J. A., Helguera G., and Penichet M. L. (2006) The transferrin receptor part I: biology and targeting with cytotoxic antibodies for the treatment of cancer. Clin. Immunol. 121, 144–158 [DOI] [PubMed] [Google Scholar]
- 11. Huang X. (2003) Iron overload and its association with cancer risk in humans: evidence for iron as a carcinogenic metal. Mutat. Res. 533, 153–171 [DOI] [PubMed] [Google Scholar]
- 12. Fenton H. J. (1894) Oxidation of tartaric acid in the presence of iron. J. Chem. Soc. Trans. 65, 899–910 [Google Scholar]
- 13. Hentze M. W., Muckenthaler M. U., and Andrews N. C. (2004) Balancing acts: molecular control of mammalian iron metabolism. Cell 117, 285–297 [DOI] [PubMed] [Google Scholar]
- 14. Henderson B. R. (1996) Iron regulatory proteins 1 and 2. Bioessays 18, 739–746 [DOI] [PubMed] [Google Scholar]
- 15. Pantopoulos K. (2004) Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N.Y. Acad. Sci. 1012, 1–13 [DOI] [PubMed] [Google Scholar]
- 16. Wallander M. L., Leibold E. A., and Eisenstein R. S. (2006) Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim. Biophys. Acta 1763, 668–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Niitsu Y., Kohgo Y., Nishisato T., Kondo H., Kato J., Urushizaki Y., and Urushizaki I. (1987) Transferrin receptors in human cancerous tissues. Tohoku J. Exp. Med. 153, 239–243 [DOI] [PubMed] [Google Scholar]
- 18. Smaaland R., Laerum O. D., Lote K., Sletvold O., Sothern R. B., and Bjerknes R. (1991) DNA synthesis in human bone marrow is circadian stage dependent. Blood 77, 2603–2611 [PubMed] [Google Scholar]
- 19. Wood P. A., Du-Quiton J., You S., and Hrushesky W. J. (2006) Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol. Cancer Ther. 5, 2023–2033 [DOI] [PubMed] [Google Scholar]
- 20. Okazaki F., Matsunaga N., Okazaki H., Utoguchi N., Suzuki R., Maruyama K., Koyanagi S., and Ohdo S. (2010) Circadian rhythm of transferrin receptor 1 gene expression controlled by c-Myc in colon cancer-bearing mice. Cancer Res. 70, 6238–6246 [DOI] [PubMed] [Google Scholar]
- 21. Koyanagi S., Kuramoto Y., Nakagawa H., Aramaki H., Ohdo S., Soeda S., and Shimeno H. (2003) A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 63, 7277–7283 [PubMed] [Google Scholar]
- 22. Trowbridge I. S., and Lopez F. (1982) Monoclonal antibody to transferrin receptor blocks transferrin binding and inhibits human tumor cell growth in vitro. Proc. Natl. Acad. Sci. U.S.A. 79, 1175–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levy J. E., Jin O., Fujiwara Y., Kuo F., and Andrews N. C. (1999) Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat. Genet. 21, 396–399 [DOI] [PubMed] [Google Scholar]
- 24. Hanson E. S., Foot L. M., Leibold E. A. (1999) Hypoxia post-translationally activates iron-regulatory protein 2. J. Biol. Chem. 274, 5047–5052 [DOI] [PubMed] [Google Scholar]
- 25. Richardson D. R., and Baker E. (1990) The uptake of iron and transferrin by the human malignant melanoma cell. Biochim. Biophys. Acta 1053, 1–12 [DOI] [PubMed] [Google Scholar]
- 26. Abbaspour N., Hurrell R., Kelishadi R. (2014) Review on iron and its importance for human health. J. Res. Med. Sci. 19, 164–174 [PMC free article] [PubMed] [Google Scholar]
- 27. Harada Y., Sakai M., Kurabayashi N., Hirota T., and Fukada Y. (2005) Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3β. J. Biol. Chem. 280, 31714–31721 [DOI] [PubMed] [Google Scholar]
- 28. Hirano A., Yumimoto K., Tsunematsu R., Matsumoto M., Oyama M., Kozuka-Hata H, Nakagawa T., Lanjakornsiripan D., Nakayama K. I., and Fukada Y. (2013) FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118 [DOI] [PubMed] [Google Scholar]
- 29. Feng D., Liu T., Sun Z., Bugge A., Mullican S. E., Alenghat T., Liu X. S., and Lazar M. A. (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]