Abstract
In Candida albicans-infected resident peritoneal macrophages, activation of group IVA cytosolic phospholipase A2 (cPLA2α) by calcium- and mitogen-activated protein kinases triggers the rapid production of prostaglandins I2 and E2 through cyclooxygenase (COX)-1 and regulates gene expression by increasing cAMP. In C. albicans-infected cPLA2α−/− or COX-1−/− macrophages, expression of Il10, Nr4a2, and Ptgs2 was lower, and expression of Tnfα was higher, than in wild type macrophages. Expression was reconstituted with 8-bromo-cAMP, the PKA activator 6-benzoyl-cAMP, and agonists for prostaglandin receptors IP, EP2, and EP4 in infected but not uninfected cPLA2α−/− or COX-1−/− macrophages. In C. albicans-infected cPLA2α+/+ macrophages, COX-2 expression was blocked by IP, EP2, and EP4 receptor antagonists, indicating a role for both prostaglandin I2 and E2. Activation of ERKs and p38, but not JNKs, by C. albicans acted synergistically with prostaglandins to induce expression of Il10, Nr4a2, and Ptgs2. Tnfα expression required activation of ERKs and p38 but was suppressed by cAMP. Results using cAMP analogues that activate PKA or Epacs suggested that cAMP regulates gene expression through PKA. However, phosphorylation of cAMP-response element-binding protein (CREB), the cAMP-regulated transcription factor involved in Il10, Nr4a2, Ptgs2, and Tnfα expression, was not mediated by cAMP/PKA because it was similar in C. albicans-infected wild type and cPLA2α−/− or COX-1−/− macrophages. CREB phosphorylation was blocked by p38 inhibitors and induced by the p38 activator anisomycin but not by the PKA activator 6-benzoyl-cAMP. Therefore, MAPK activation in C. albicans-infected macrophages plays a dual role by promoting the cPLA2α/prostaglandin/cAMP/PKA pathway and CREB phosphorylation that coordinately regulate immediate early gene expression.
Keywords: c-Jun N-terminal kinase (JNK), cAMP-response element-binding protein (CREB), Candida albicans, cyclic AMP (cAMP), extracellular signal-regulated kinase (ERK), mitogen-activated protein kinase (MAPK), p38 MAPK, phospholipase A, prostaglandin
Introduction
In response to microbial infection, macrophages produce oxygenated metabolites of arachidonic acid that act as regulators of inflammation and innate immune responses (1–4). Arachidonic acid is metabolized by 5-lipoxygenase to pro-inflammatory leukotrienes and by COX-12 and inducible COX-2 to prostanoids, which exert both pro- and anti-inflammatory effects (5–8). The differential expression of downstream prostaglandin synthases determines the types of prostaglandins produced by macrophages (9). Eicosanoids are secreted and act locally by engaging specific G-protein-coupled receptors (5). The types of eicosanoid receptors expressed in the local tissue environment are cell type-specific and mediate receptor-specific signaling cascades to elicit their biological effects (10–12).
Group IVA cytosolic phospholipase A2 (cPLA2α) is an important regulatory enzyme that initiates the production of eicosanoids by preferentially releasing arachidonic acid in response to cell activation (13). We have used resident peritoneal macrophages (RPM) as a model to study mechanisms of cPLA2α activation and production of eicosanoids in response to infection by the opportunistic pathogen Candida albicans (14, 15). Resident tissue macrophages are sentinel cells that are important in sensing invading microorganisms and for production of cytokines and chemokines to initiate innate immune responses for host defense (16). In response to microbial infection, macrophages produce and secrete eicosanoids within minutes that can shape innate immunity, particularly by regulating transcriptional responses (3). Cell wall components of C. albicans, particularly β-glucans and mannans, engage the lectin receptors dectin-1 and dectin-2 on RPM and act with MyD88-dependent pathways to provide signals for cPLA2α activation and eicosanoid production in RPM (14, 15). The rapid activation of cPLA2α involves post-translational processes, including increases in intracellular calcium and activation of mitogen-activated protein kinases (MAPKs) (17–20). Calcium promotes the translocation of cPLA2α from the cytosol to intracellular membrane to access phospholipid substrate, and the MAPKs, ERK1/2 and p38, phosphorylate cPLA2α on Ser-505, which enhances catalytic activity (21–23).
We previously reported that cPLA2α activation in C. albicans-infected RPM initiates an autocrine loop involving production of prostaglandins that increase cAMP to globally regulate expression of genes involved in host defense and to dampen inflammation (24). In this study, we demonstrate that cPLA2α activation, production of prostaglandins from the COX-1 pathway, and increases in cAMP act synergistically with mitogen-activated protein kinases to induce expression of the immediate response genes Il10, Ptgs2 (prostaglandin endoperoxide synthase 2; the gene for COX-2), and Nr4a2 (nuclear receptor subfamily 4, group A, member 2) but to suppress MAPK-dependent expression of Tnfα. Although expression of these cAMP-response element-binding protein (CREB)-inducible genes is regulated by activation of protein kinase A (PKA) in C. albicans-infected RPM, results demonstrate that the phosphorylation of CREB is downstream of p38 and not the prostaglandin/cAMP/PKA pathway.
Experimental Procedures
Materials
FBS from Hyclone was heat-inactivated at 56 °C for 30 min before use. DMEM was from Lonza. Penicillin, streptomycin, l-glutamine, Lipofectamine 2000, TaqMan Fast Universal PCR Master Mix, and TaqMan assay probes were from Life Technologies, Inc. Cell culture grade BSA was from Sigma. The MEK1 inhibitor (U0126), p38 inhibitor (SB202190), and c-Jun N-terminal kinase inhibitor (SP600125) were from Millipore. The ERK1/2 inhibitor SCH772984 and the p38 inhibitor LY2228820 were from Selleckchem. The β-actin antibody, phospho-(Ser/Thr) PKA substrate antibody, and XTT cell viability kit were from Cell Signaling. Greiss reagent was obtained from Promega. Polyclonal antibodies to murine COX-1 and COX-2; the IP, EP2, and EP4 receptor antagonists (CAY10441, PF-04418948, and L-161,982 respectively); and receptor agonists (iloprost, butaprost, and CAY10598, respectively) were from Cayman Chemical Co. The stable cAMP analogue 8-Br-cAMP was from Santa Cruz Biotechnology, Inc. The PKA activator 6-Bnz-cAMP, sodium salt, and the exchange protein directly activated by cAMP (Epac) activator 8-pCPT-2-O-Me-cAMP-AM were from Tocris Bioscience. Polyclonal antibody to cPLA2α was raised as described (25). NR4A2 (Nurr1) antibody (N-20) was from Santa Cruz Biotechnology. Phospho-specific antibodies to ERK, p38, JNK, CREB, and cPLA2α were obtained from Cell Signaling Technology, Inc. The Qiagen RNeasy minikit was used for RNA isolation. Halt phosphatase and protease inhibitor mixtures, the Fermentas Maxima First Strand cDNA synthesis kit, and the BCA protein assay kit were from Thermo Scientific.
Mouse Strains
cPLA2α−/− mice were generated using 129 embryonic stem cells in a C57BL/6 strain as described previously (26). The mixed strain was backcrossed onto a BALB/c background and used after 10 generations. BALB/c mice were obtained from NCI, National Institutes of Health (Bethesda, MD) and Charles River. COX-1−/− (B6.129S6-Ptgs1tm1Fun/J), JNK1−/−(B6.129-Mapk8tm1Flv/J, C57BL/6 background), and control mice (B6129SF1/J and C57BL/6J, respectively) were obtained from the Jackson Laboratory. JNK-2−/− (B6.129-Mapk9tm1Flv/J, C57BL/6 background) breeder mice were kindly provided by Dr. Erwin W. Gelfand (National Jewish Health, Denver, CO). Mice were used for macrophage isolation at 8–12 weeks of age.
C. albicans Culture
C. albicans (ATCC 10261) was grown on Sabouraud dextrose agar plates and maintained at 4 °C. The day before the experiment, it was streaked onto a fresh plate and incubated overnight at 37 °C. C. albicans was scraped from the plate and washed twice in endotoxin-free PBS. Live C. albicans at a multiplicity of infection of 2–5 was used. C. albicans expressing green fluorescent protein (GFP) was kindly provided by Dr. Robert Wheeler (University of Maine). It was generated from the wild type SC5314 strain and exhibits similar virulence as the wild type strain (27). C. albicans strains 10261 and SC5314 are both serotype A and have similar effects in RPM.
RPM Isolation
RPM were obtained by peritoneal lavage as described previously (14). Cells were plated at a density of 5 × 105 cells/cm2 (48-well plate) and incubated for 2 h at 37 °C in a humidified atmosphere of 5% CO2 in air. After washing the cultures to remove non-adherent cells, the adherent macrophages were incubated in DMEM containing 10% heat-inactivated FBS, 100 μg/ml streptomycin sulfate, 100 units/ml penicillin G, and 0.29 mg/ml glutamine for 16–18 h at 37 °C. The cells were washed twice with serum-free DMEM containing 0.1% BSA (stimulation medium) and then infected with C. albicans.
Mass Spectrometry Eicosanoid Analysis
The culture medium was collected after macrophage stimulation, centrifuged, and then stored at −80 °C. The media were thawed and mixed with an equal volume of cold methanol prior to analysis. The samples were diluted in water to a final methanol concentration of <15% and then extracted using a solid phase extraction cartridge (Strata Polymeric Reversed Phase, 60 mg/ml, Phenomenex, Torrance, CA). Metabolites were separated by HPLC that directly interfaced into the electrospray ionization source of a triple quadrapole mass spectrometer (Sciex API 3000, PE-Sciex, Thornhill, Canada). Mass spectrometric analyses were performed in the negative ion mode using multiple-reaction monitoring for specific analytes as described previously (24).
RT-PCR
RNA was isolated from RPM and treated with DNase to remove contaminating genomic DNA, and cDNA was generated using 0.5 μg of RNA. PCRs contained, in a final volume of 20 μl, 10 μl of 2× TaqMan fast universal master mix, 1 μl of 20× TaqMan assay/probe, and 9 μl of cDNA (30–70 ng) in RNase-free water. The Thermal Fast cycle program was as follows: 20 s at 95 °C followed by 40 cycles of 1 s at 95 °C and 20 s at 60 °C, using the StepOne Plus RT-PCR system (Applied Biosystems). Duplicate reactions were analyzed for each sample. The following TaqMan assay probes were used: Gusb Mm01197698_ml; Nr4a2 Mm00443060_ml; Il10 Mm00439614_ml; Tnf Mm99999068_ml; Ptgs2 Mm00478364_ml. Threshold cycle values (CT) were determined and used to calculate relative gene expression using 2−ΔCT analysis (28). For normalization of gene expression, the CT values of the gene of interest were related to the CT values of the housekeeping gene Gusb, which is expressed at a constant level in treated and untreated RPM. A relative expression value of 1 indicates that the gene of interest is expressed at the same level as Gusb.
Western Blotting
Cell monolayers were washed twice in ice-cold PBS and then lysed in a modified radioimmune precipitation assay buffer: 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, 1 mm EGTA, 1 mm EDTA, protease and phosphatase inhibitors. After incubation on ice for 30 min, lysates were centrifuged at 15,000 rpm for 15 min, and protein concentration in the supernatant was determined. Lysates were boiled for 5 min after the addition of Laemmli sample buffer, and then proteins were separated on 10% SDS-polyacrylamide gels. After transfer to nitrocellulose membrane, samples were incubated in 20 mm Tris-HCl, pH 7.6, 137 mm NaCl, 0.05% Tween with 5% nonfat milk (blocking buffer) for 1 h and then incubated overnight at 4 °C with primary antibodies in blocking buffer. The membranes were incubated with horseradish peroxidase-conjugated secondary antibody (1:5000) in 20 mm Tris-HCl, pH 7.6, 137 mm NaCl, 0.05% Tween for 1 h at room temperature. The immunoreactive proteins were detected using the Amersham Biosciences ECL system (GE Healthcare).
Macrophage Function Assays
For evaluating bindingand internalization (recognition assay) of GFP-C. albicans, cPLA2α+/+ and cPLA2α−/− RPM (1 × 105) were seeded onto MatTek 35-mm dishes and incubated for 2 h. After washing, cells were incubated overnight followed by infection with live GFP-C. albicans (multiplicity of infection of 2) in phenol red-free stimulation medium for 30 min at 37 °C and 5% CO2. Macrophages were washed, fixed with 4% paraformaldehyde for 15 min, and then stained with DAPI. Images were captured on a Marianas 200 spinning disk confocal microscope using Intelligent Imaging Innovation Inc. software (Slidebook version 6.0) to determine the number of macrophages containing GFP-C. albicans.
For determining C. albicans killing, cPLA2α+/+ and cPLA2α−/− RPM were plated as described above and then incubated with and without IFNγ (200 units/ml) overnight, followed by infection with C. albicans (multiplicity of infection of 2) in stimulation medium with and without IFNγ (200 units/ml) for 4 h. After removing the culture medium, 1% Triton X-100 was added, which lyses RPM but not C. albicans. C. albicans viability was measured using the XTT cell viability kit as described (29). The procedure measures the ability of viable cells to convert XTT to formazan by enzymes that are inactivated in dead cells. Wells containing an equivalent number of C. albicans without RPM were included as a positive control for determining 100% viability.
For measuring nitric oxide production, RPM were cultured as described for the killing assay. After a 4-h incubation of RPM with C. albicans, culture media were removed, centrifuged, and used to measure nitric oxide production using the Griess reagent. To measure NADPH oxidase activity, superoxide anion production was determined by measuring cytochrome c reduction as described previously (20).
Statistics
Statistics were calculated in GraphPad using unpaired t test to obtain two-tailed p values.
Results
Role of COX-1-derived Prostaglandins in Regulating Gene Expression
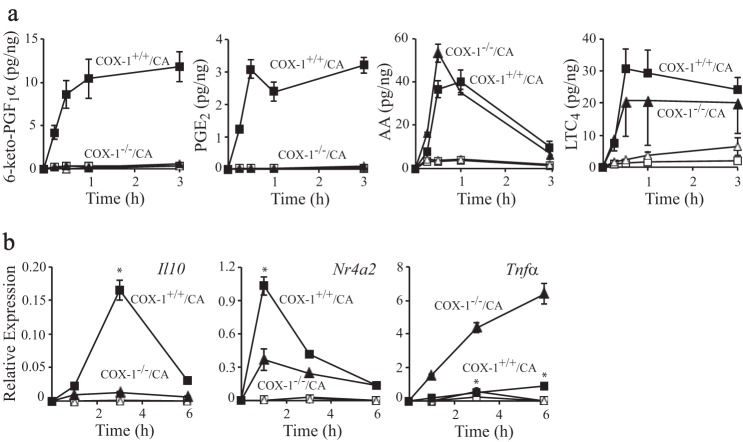
cPLA2α initiates the release of arachidonic acid for production of prostaglandins and leukotrienes in RPM within minutes of infection with C. albicans (24). Prostaglandin I2 (PGI2) and prostaglandin E2 (PGE2) are produced before the expression of COX-2, suggesting that they are derived from the COX-1 pathway. By comparing C. albicans-infected cPLA2α+/+ and cPLA2α−/− RPM, we previously found that cPLA2α-mediated production of prostaglandins regulates gene expression, including immediate early genes, such as Nr4a2, that are expressed within the first 30–60 min of infection (24). To specifically investigate the role of COX-1-derived prostaglandins in regulating gene expression, RPM from wild type and COX-1−/− mice were compared. C. albicans induced rapid production of PGE2 and 6-keto-PGF1α, the stable metabolite of PGI2, in COX-1+/+ RPM, but their levels were reduced to baseline in COX-1−/− RPM infected for up to 3 h (Fig. 1a). In contrast, there was no significant difference in the release of arachidonic acid and leukotriene C4 production.
FIGURE 1.
Prostaglandins from COX-1 mediate gene expression in C. albicans-infected RPM. COX-1+/+ (squares) and COX-1−/− RPM (triangles) were incubated with (solid symbols) and without (open symbols) C. albicans (CA) for the times indicated followed by analysis of the culture media for eicosanoids (a) or RNA isolation to determine relative expression of Il10, Nr4a2, and Tnfα (b) by RT-PCR at 6 h after C. albicans infection (n = 3; *, p < 0.05 compared with COX-1−/−/C. albicans). Error bars, S.E. LTC4, leukotriene C4; AA, arachidonic acid.
cPLA2α-mediated prostaglandin production in response to C. albicans influences expression of genes to dampen inflammation highlighted by an increase in the anti-inflammatory cytokine IL-10 and suppression of the pro-inflammatory cytokine TNFα (24). The expression of these genes and the immediate early gene Nr4a2 was compared in C. albicans-infected COX-1+/+ and COX-1−/− RPM (Fig. 1b). Expression of Il10 and Nr4a2 was significantly lower, and Tnfα expression was higher, in COX-1−/− compared with COX-1+/+ RPM. Expression of Tnfα in COX-1−/− RPM increased from 1–6 h after C. albicans infection, whereas expression of Nr4a2 and Il10 in COX-1+/+ RPM was transient, peaking at 1 and 3 h, respectively.
COX-2 Expression in C. albicans-infected RPM Requires COX-1-derived Prostaglandins
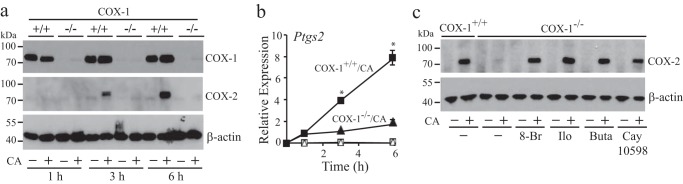
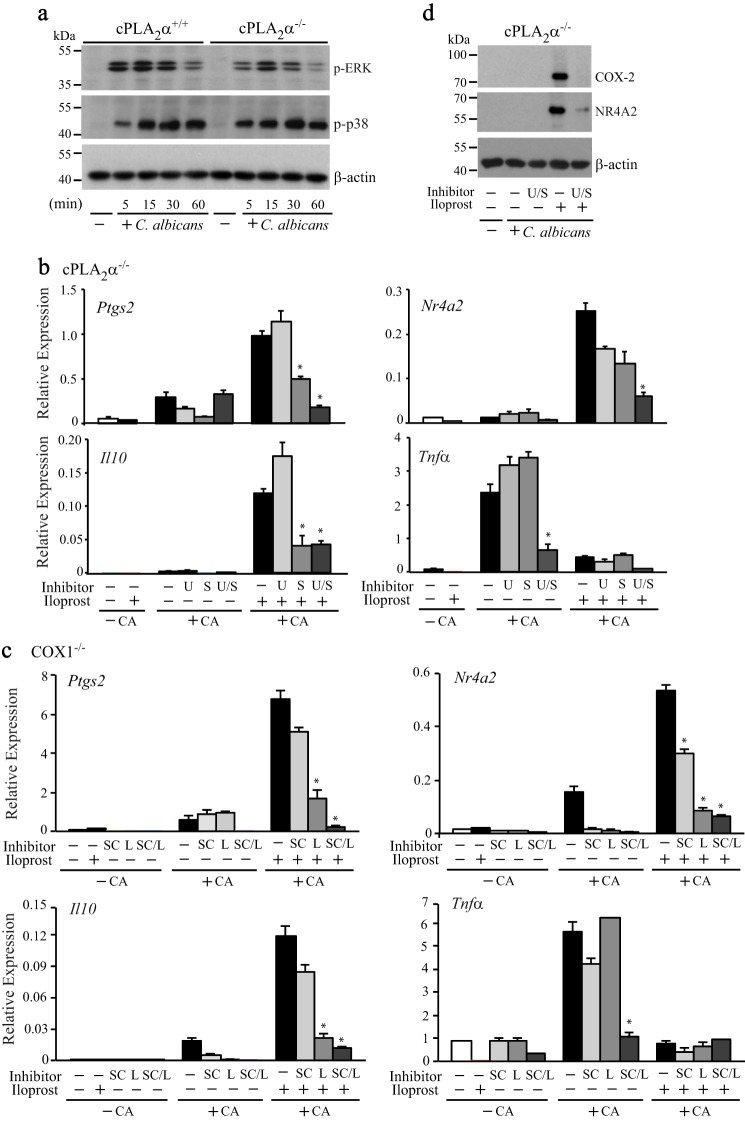
Although results show a role for COX-1-derived prostaglandins in regulating gene expression, most genes that were higher in cPLA2α+/+ RPM compared with cPLA2α−/− RPM peaked 3 h after C. albicans infection (24). Because COX-2 protein can be detected by 3 h in C. albicans-infected cPLA2α+/+ RPM, prostaglandins derived from COX-2 may contribute to regulating gene expression. COX-1−/− RPM are a useful model to investigate a role for COX-2, but it was important to confirm that COX-2 was expressed at similar levels in C. albicans-infected COX-1+/+ and COX-1−/− RPM. In some tissues/cells, COX isoforms compensate for each other when one is ablated (30). Conversely, prostaglandins either added exogenously or produced endogenously can promote COX-2 expression via cAMP-response elements (31–34). In COX-1+/+ RPM, COX-2 was detected on Western blots at 3 h and at higher levels by 6 h but was not detected or was extremely low in COX-1−/− RPM (Fig. 2a). Expression of Ptgs2 mRNA correlated with the Western blotting data showing higher levels in COX-1+/+ than in COX-1−/− RPM after C. albicans infection (Fig. 2b). The results suggest that prostaglandins derived from COX-1 transcriptionally regulate COX-2 expression by increasing cAMP. Consistent with this possibility, COX-2 expression was increased by treating COX-1−/− RPM with 8-Br-cAMP and by agonists for the PGI2 receptor IP (iloprost) and PGE2 receptors EP2 (butaprost) and EP4 (CAY10598) (Fig. 2c). The receptor agonists and 8-Br-cAMP did not increase COX-2 expression in uninfected COX-1−/− RPM, suggesting that cAMP acts synergistically with additional signals triggered by C. albicans.
FIGURE 2.
COX-2 expression is attenuated in C. albicans-infected COX-1−/− RPM. a, Western blot of COX-1, COX-2, and β-actin (loading control) in COX-1+/+ and COX-1−/− RPM incubated with C. albicans (CA) for 1–6 h is shown. b, relative expression of Ptgs2 in COX-1+/+ (squares) and COX-1−/− RPM (triangles) incubated with (solid symbols) and without (open symbols) C. albicans for 1–6 h was determined by RT-PCR (n = 3; *, p < 0.05 compared with COX-1−/−/C. albicans). c, Western blot of COX-2 and β-actin in COX-1+/+ and COX-1−/− RPM preincubated with and without 8-Br-cAMP (8-Br; 2 mm), iloprost (Ilo; 1 μm), butaprost (Buta; 10 μm), and CAY10598 (1 μm) for 30 min followed by incubation with C. albicans for 6 h. a and c, results are representative of three independent experiments. Error bars, S.E.
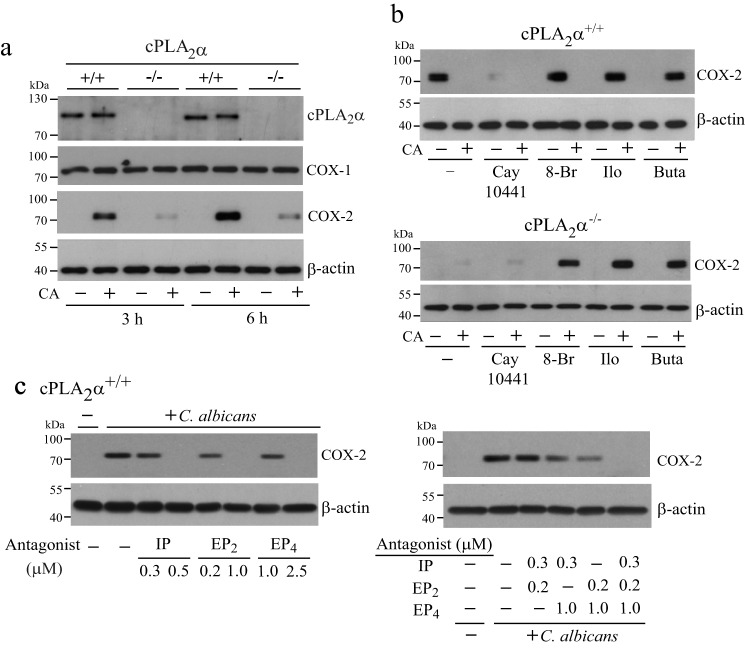
We also confirmed that cPLA2α activation regulates expression of COX-2, which was lower in C. albicans-infected cPLA2α−/− RPM than in cPLA2α+/+ at 3 and 6 h after stimulation (Fig. 3a). As observed in COX-1−/− RPM, the expression of COX-2 was increased by treating cPLA2α−/− RPM with 8-Br-cAMP, iloprost, and butaprost (Fig. 3b). Treatment with the EP4 receptor agonist (CAY10598) also increased COX-2 expression in cPLA2α−/− RPM, as observed in COX-1−/− RPM (data not shown). The expression of COX-2 in C. albicans-infected cPLA2α+/+ RPM was blocked by the IP receptor antagonist CAY10441, suggesting a role for PGI2 (Fig. 3b). We also investigated the role of PGE2 in regulating COX-2 expression by using antagonists for EP2 and EP4 receptors. It was necessary to evaluate different concentrations of the antagonists because their IC50 concentrations differ. At the higher concentration of the receptor antagonists tested, they all blocked COX-2 expression in C. albicans-infected cPLA2α+/+ RPM, suggesting a lack of specificity at this concentration (Fig. 3c). At the lower concentration tested, the receptor antagonists partially blocked COX-2 expression, and this allowed us to evaluate their combined effects (Fig. 3c). When C. albicans-infected cPLA2α+/+ RPM were treated with a combination of two antagonists at the lower concentration, there was partial inhibition of COX-2 expression, but when all three were tested, complete inhibition was observed (Fig. 3c, right). The results suggest that endogenous production of both PGI2 and PGE2 acting through the IP, EP2, and EP4 receptors all contribute to regulating expression of COX-2.
FIGURE 3.
cPLA2α activation promotes COX-2 expression. a, Western blot of cPLA2α, COX-1, COX-2, and β-actin in cPLA2α+/+ and cPLA2α−/− RPM incubated with C. albicans (CA) for 3 and 6 h is shown. b, Western blot of COX-2 and β-actin in cPLA2α+/+ and cPLA2α−/− RPM treated for 30 min with and without Cay10441 (1 μm), 8-Br-cAMP (8-Br; 2 mm), iloprost (Ilo; 1 μm), and butaprost (But; 10 μm) and then incubated with C. albicans for 6 h. c, Western blot of COX-2 and β-actin in cPLA2α+/+ RPM treated for 30 min with the indicated concentrations of the IP (CAY10441), EP2 (PF-04418948), and EP4 (L-161,982) receptor antagonists alone (left) or in combination (right) and then incubated with C. albicans for 6 h. Results are representative of three independent experiments.
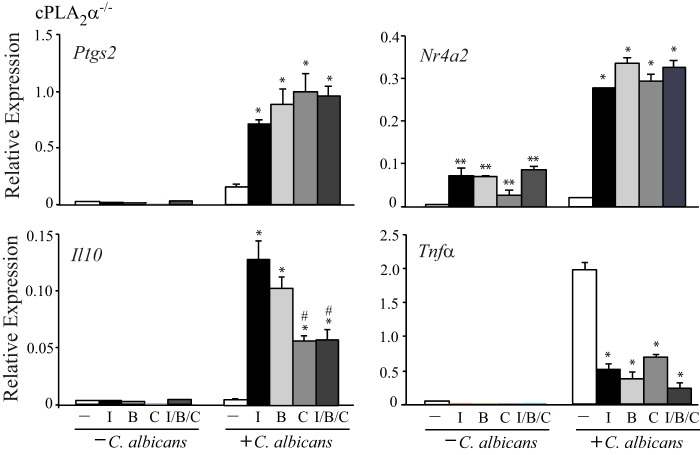
We also investigated the role of prostaglandin receptors in regulating gene expression by using agonists for IP (iloprost), EP2 (Ptgs2), and EP4 (CAY10598). The agonists increased expression of COX-2 (Ptgs2), Nr4a2, and Il10 mRNA and decreased Tnfα expression in C. albicans-infected cPLA2α−/− RPM but not in uninfected RPM except for a small increase in Nr4a2 expression (Fig. 4). The IP, EP2, and EP4 agonists were equally effective in regulating expression of Nr4a2, Ptgs2, and Tnfα when tested alone or in combination. However, Il10 expression was increased to a greater extent by iloprost and butaprost than with the EP4 agonist CAY10598 (Fig. 4). When CAY10598 was tested in combination with IP and EP2 agonists, expression of Il10 was significantly lower than with iloprost and butaprost alone, suggesting that a distinct signaling pathway through EP4 dampens Il10 expression.
FIGURE 4.
Regulation of gene expression by prostaglandin receptor agonists. Relative gene expression cPLA2α−/− RPM treated with and without iloprost (I; 1 μm), butaprost (B; 10 μm), and CAY10598 (C; 1 μm) alone or together (I/B/C) for 30 min and then incubated with C. albicans for 3 h (n = 3; *, p < 0.05 compared with C. albicans-infected RPM not treated with agonists; **, p < 0.05 compared with RPM not treated with C. albicans or agonists; #, p < 0.05 compared with C. albicans-infected RPM treated with iloprost or butaprost). Error bars, S.E.
Role of MAPKs in Regulating Gene Expression
Our next approach was to identify the signals triggered by C. albicans that act synergistically with prostaglandin-induced production of cAMP for regulating expression of Ptgs2, Nr4a2, and Il10. We focused on the role of MAPKs because they are implicated as important regulators of these immediate early genes, although there are cell type-dependent differences in their action (35–38). We previously reported that C. albicans and zymosan stimulate activation of ERK1/2, p38, and JNK1/2 in RPM (15, 20). We first compared ERK1/2 and p38 activation (evaluated using phospho-specific antibodies) in cPLA2α+/+ and cPLA2α−/− RPM to determine whether their activation was influenced by the cPLA2α/prostaglandin/cAMP pathway (Fig. 5a). ERKs and p38 were rapidly activated in both cPLA2α+/+ and cPLA2α−/− RPM in response to C. albicans. The extent of p38 activation was similar in cPLA2α+/+ and cPLA2α−/− RPM and remained elevated from 15 to 60 min. Activation of ERKs, which was slightly lower in cPLA2α−/− RPM, peaked at 15 min and then decreased by 60 min. In some cells, cAMP promotes p38 activation, but our results demonstrate that cPLA2α-mediated prostaglandin production has no effect on p38 activation (39, 40).
FIGURE 5.
Regulation of gene expression by mitogen-activated protein kinases. a, Western blot of phospho-ERKs, phospho-p38, and β-actin in cPLA2α+/+ and cPLA2α−/− RPM treated with and without C. albicans (CA) for 5–60 min. b, relative gene expression in cPLA2α−/− RPM treated with and without U0126 (U; 1 μm), SB20219 (S; 10 μm), or U0126 together with SB20219 (U/S) for 30 min and then incubated with C. albicans for 2 h in the presence or absence of iloprost. c, relative gene expression in COX-1−/− RPM treated with and without the ERK1/2 inhibitor SCH772984 (SC; 1 μm), the p38 inhibitor LY222820 (L; 1 μm), or SCH772984 together with LY222820 (S/L) for 30 min and then incubated with C. albicans for 2 h in the presence or absence of iloprost (n = 3; *, p < 0.05 compared with iloprost-treated C. albicans-infected RPM without inhibitors or for Tnfα compared with C. albicans-infected RPM without either inhibitors or iloprost). d, Western blot of COX-2, NR4A2, and β-actin in cPLA2α−/− RPM treated with and without U0126 together with SB20219 (U/S) for 30 min and then incubated with C. albicans for 6 h in the presence or absence of iloprost. Western blotting results are representative of three independent experiments. Error bars, S.E.
To determine whether activation of MAPKs in response to C. albicans infection regulates gene expression, we tested the effect of inhibitors that block activation of ERKs (U0126) and p38 (SB20219). ERKs and p38 regulate cPLA2α activation; therefore, the MAPK inhibitors were not tested in cPLA2α+/+ RPM because they would inhibit cPLA2α-mediated arachidonic acid release, prostaglandin production, and increases in cAMP. We used C. albicans-infected cPLA2α−/− RPM treated with and without the IP receptor agonist, iloprost, to investigate the contribution of cAMP induced by iloprost and activation of MAPK in regulating gene expression (Fig. 5b). In C. albicans-infected cPLA2α−/− RPM, the increase in expression of Ptgs2, Nr4a2, and Il10 induced by iloprost was significantly inhibited by SB20219 together with U0126. When tested alone, U0126 and SB20219 partially decreased expression of Nr4a2. Expression of Ptgs2 and Il10 was inhibited by SB20219 but not by U0126. The results show that both ERKs and p38 contribute to Nr4a2 expression, but p38 and not ERKs regulate Il10 and Ptgs2. In contrast, increases in cAMP and signals from C. albicans have opposing effects on Tnfα expression, which is lower in cPLA2α−/− RPM treated with iloprost due to inhibition by cAMP (Fig. 5b). The higher level of Tnfα expressed in C. albicans-infected cPLA2α−/− RPM not treated with iloprost was significantly blocked by U0126 together with SB20219, but neither alone had any effect. The results demonstrate that prostaglandins act synergistically with MAPK activation to induce expression of Ptgs2, Nr4a2, and Il10 in response to C. albicans but suppress expression of Tnfα, which is dependent on both ERKs and p38. To provide further support that MAPKs act together with prostaglandins to regulate gene expression, we tested structurally distinct inhibitors of ERKs (SCH772984) and p38 (LY2228820) in C. albicans-infected COX1−/− RPM (Fig. 5c). These inhibitors had effects very similar to those of U0126 and SB20219 on gene expression, and the results additionally show that the effects are consistent in cPLA2α−/− and COX1−/− RPM. Experiments were also carried out to determine whether there was a correlation between the effects of MAPK inhibitors on gene expression and levels of COX2 and NR4A2 protein (Fig. 5d). In cPLA2α−/− RPM, C. albicans and iloprost synergistically induced expression of COX2 and NR4A2 protein, but neither alone was effective. Protein expression of COX-2 and NR4A2 was blocked by inhibition of ERKs and p38, confirming that mRNA expression correlates with protein levels.
Role of JNK1 and JNK2 in Regulating Gene Expression and Eicosanoid Production
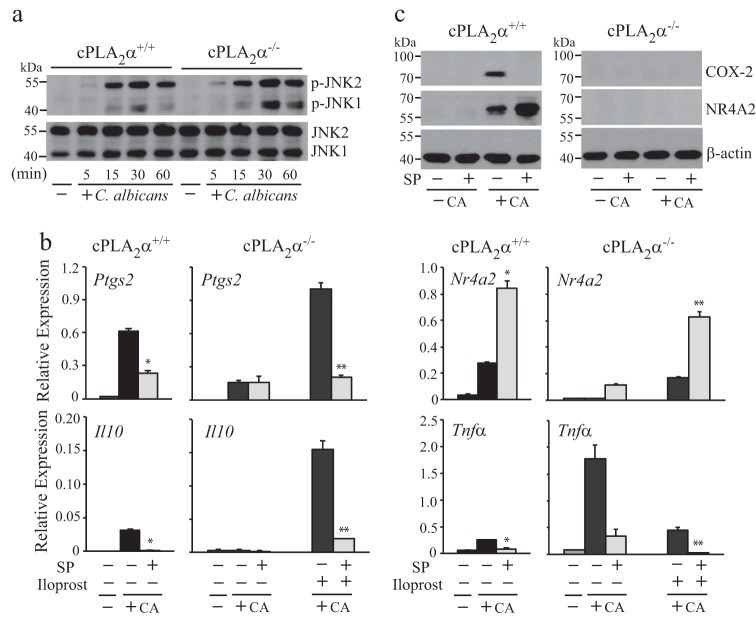
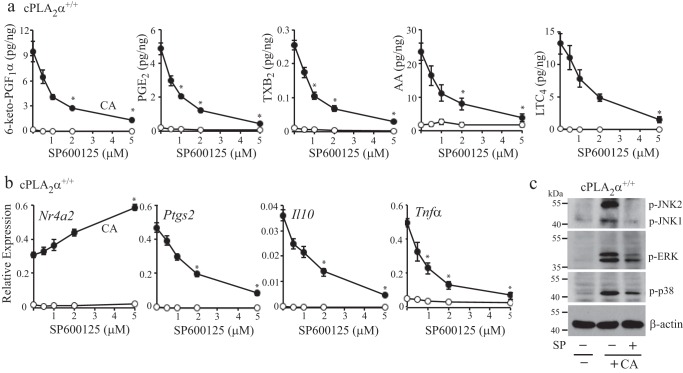
In addition to ERKs and p38, JNKs are also implicated in regulating gene expression, including Ptgs2 (38, 41). As shown in Fig. 6a, C. albicans stimulated activation of JNK1 and JNK2 to a similar extent in cPLA2α+/+ and cPLA2α−/− RPM, which peaked at 30 min. We first tested the effect of the JNK inhibitor SP600125, which inhibits both JNK1 and JNK2. SP600125 significantly inhibited expression of Ptgs2, Il10, and Tnfα in C. albicans-infected cPLA2α+/+ RPM and in cPLA2α−/− RPM treated with and without iloprost (Fig. 6b). In contrast, expression of Nr4a2 was increased by SP600125 (Fig. 6b). Western blotting showed that C. albicans induced an increase in NR4A2 protein in cPLA2α+/+ RPM, and expression was enhanced by the SP600125 inhibitor, consistent with the effect on mRNA levels (Fig. 6c). In contrast, SP600125 inhibited the increase in COX-2 protein in C. albicans-infected cPLA2α+/+ RPM. Expression of NR4A2 and COX2 was not detected in cPLA2α−/− RPM treated with or without the SP600125. A role for JNK in activating cPLA2α has been suggested from studies using SP600125 (42, 43). We tested the effect of SP600125 concentration on gene expression and eicosanoid production (Fig. 7). SP600125 dose-dependently blocked release of arachidonic acid and production of prostaglandins and leukotriene C4 (Fig. 7a). Similar concentrations of SP600125 increased expression of Nr4a2 and decreased expression of Ptgs2, Il10, and Tnfα (Fig. 7b). Although the results suggest that JNKs may play a role in regulating gene expression and cPLA2α activation in C. albicans-infected RPM, SP600125 inhibits several other kinases with equal potency (44, 45). In light of this, we tested the effect of SP600125 on activation of MAPKs and found that in addition to inhibition of JNK1/2 phosphorylation, SP600125 partially inhibited activation of ERKs and p38 in C. albicans-infected cPLA2α+/+ RPM (Fig. 7c).
FIGURE 6.
Effect of SP600125 on gene expression. a, Western blot of phospho-JNK and total JNK in cPLA2α+/+ and cPLA2α−/− RPM treated with and without C. albicans for 5–60 min. b, relative gene expression was determined in PLA2α+/+ and cPLA2α−/− RPM pretreated with and without SP600125 (SP; 5 μm) for 30 min and then incubated with or without iloprost (1 μm) or C. albicans for 2 h (n = 3; *, p < 0.05 compared with C. albicans-infected RPM without inhibitor; **, p < 0.05 compared with C. albicans-infected RPM without inhibitor). c, Western blot of NR4A2, COX-2, and β-actin in cPLA2α+/+ and cPLA2α−/− RPM treated with and without SP600125 (5 μm) and then infected with C. albicans for 4 h. Western blotting results are representative of three independent experiments. Error bars, S.E.
FIGURE 7.
Effect of SP600125 concentration on eicosanoid production and gene expression. a, eicosanoids were analyzed in the culture media of cPLA2α+/+ RPM treated with the indicated concentrations of SP600125 for 30 min and then incubated with (solid circles) or without (open circles) C. albicans (CA) for 1 h. b, RNA was isolated from cPLA2α+/+ RPM treated as described in a, and relative gene expression was determined by RT-PCR. a and b, n = 3; *, p < 0.03 compared with C. albicans-infected cPLA2α+/+ RPM not treated with inhibitor. c, Western blot of phospho-JNK1/2, phospho-ERKs, phospho-p38, and β-actin in cPLA2α+/+ RPM treated with and without SP600125 (5 μm) and then infected with C. albicans for 30 min. Western blotting results are representative of three independent experiments. Error bars, S.E.
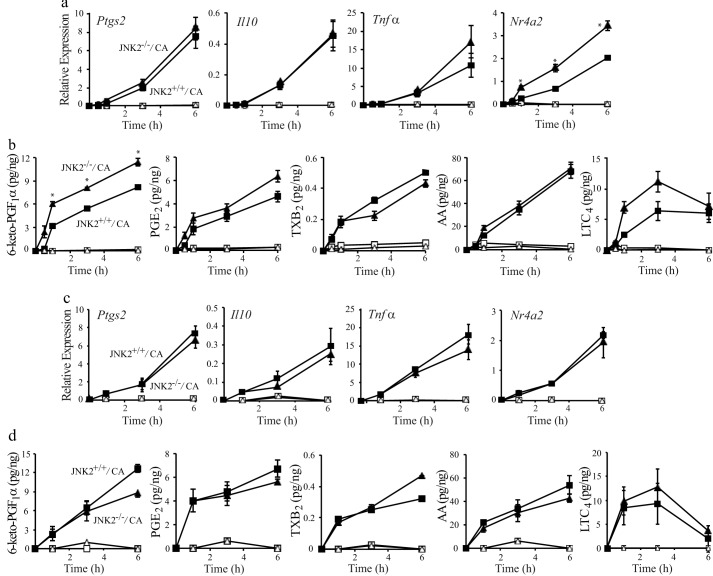
Because the effect of SP600125 on gene expression and eicosanoid production may be due to inhibition of ERKs and p38, another approach was used to investigate the role of JNKs by comparing RPM from WT and JNK1−/− or JNK2−/− mice. As observed with the JNK inhibitor SP600125, the expression of Nr4a2 mRNA was significantly higher in C. albicans-infected JNK2−/− than JNK2+/+ RPM, but there was no difference in expression of Ptgs2, Il10, or Tnfα (Fig. 8a). In contrast to the transient expression of Nr4a2 observed in cPLA2α+/+ RPM (BALB/c) and COX-1+/+ RPM (BL6/129) (see Fig. 1b), Nr4a2 expression in JNK2−/− and JNK2+/+ RPM (C57BL/6) continued to increase from 1 to 6 h, which may reflect strain differences (Fig. 8a). The results suggest that JNK2 activation negatively regulates Nr4a2 expression and that SP600125 enhances expression of Nr4a2 by inhibition of JNK2. The release of arachidonic acid and eicosanoid production in response to C. albicans infection was not significantly different in JNK2+/+ and JNK2−/− RPM with the exception of 6-keto-PGF1α, which was slightly higher in JNK2−/− RPM (Fig. 8b). A comparison of WT and JNK1−/− RPM showed no differences in gene expression (Fig. 8c) or eicosanoid production (Fig. 8d). The results indicate that JNK2 negatively regulates expression of Nr4a2 but that neither JNK1 nor JNK2 plays a role in regulating expression of Ptgs2, Tnfα, or Il10 or activation of cPLA2α in C. albicans-infected RPM. The ability of SP600125 to inhibit gene expression and cPLA2α activation may be due to a nonspecific effect on ERK and p38 activation (see Fig. 7c).
FIGURE 8.
Role of JNK2 and JNK1 in regulating eicosanoid production and gene expression. RPM from JNK2+/+ (squares) and JNK2−/− (triangles) (a and b) and JNK1+/+ (squares) and JNK1−/− (triangles) mice (c and d) were infected with (solid symbols) or without (open symbols) C. albicans (CA) for 1–6 h followed by RNA isolation to determine relative gene expression (a and c) and collection of the culture media for eicosanoid analysis (b and d) (n = 3; *, p < 0.05 compared with C. albicans-infected JNK2+/+ RPM). Error bars, S.E.
Role of MAPKs and cAMP in Regulating Gene Expression and CREB Phosphorylation
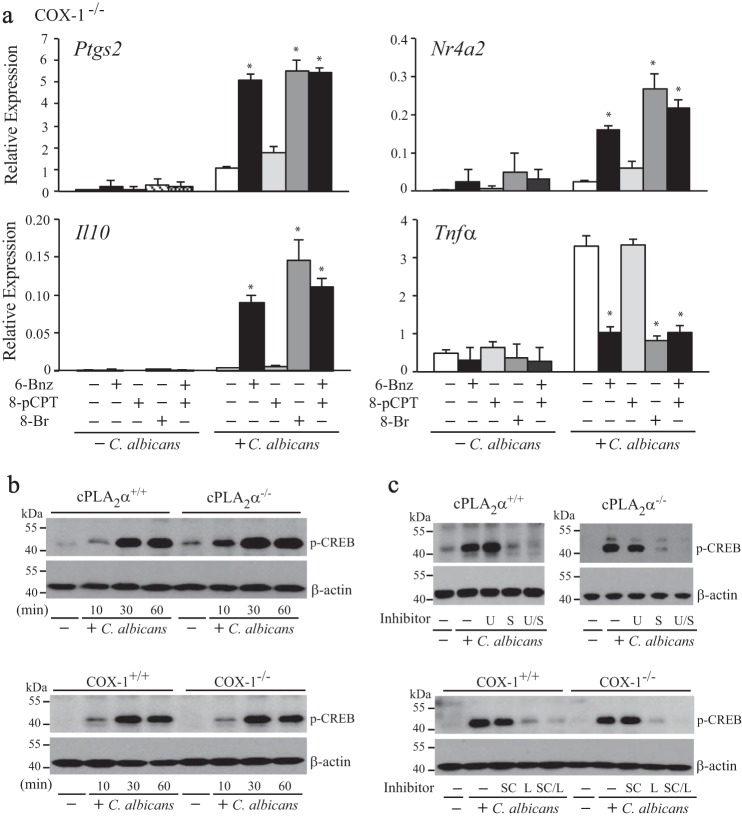
The transcription factor CREB regulates expression of immediate early genes containing cAMP-response element sites in response to cAMP, including Ptgs2, Tnfα, Il10, and Nr4a2 (46–54). One mechanism for CREB activation is phosphorylation of Ser-133 that can be mediated by either PKA or by kinases downstream of MAPKs (55–57). In a previous study, we found that C. albicans stimulates greater production of cAMP in cPLA2α+/+ than in cPLA2α−/− RPM, consistent with an autocrine effect of endogenously produced prostaglandins on gene expression (24). Experiments were carried out to determine whether the regulation of gene expression by the cPLA2α/prostaglandin/cAMP pathway was mediated by PKA or Epacs, the two main cAMP effectors (58). We previously reported that the PKA inhibitor H89 suppressed expression of Il10 and Nr4a2 and enhanced expression of Tnfα in C. albicans-infected cPLA2α+/+ (24). Another approach was used to determine the contribution of PKA and Epac in regulating gene expression that involved treating C. albicans-infected cPLA2α−/− RPM with cAMP analogs that selectively activate PKA (6-Bnz-cAMP) or Epac (8-pCPT-2-O-Me-cAMP-AM). Expression of Ptgs2, Il10, and Nr4a2 was induced, and TNFα expression was suppressed, with 6-Bnz-cAMP and 8-Br-cAMP but not by the Epac activator (Fig. 9a). The results suggest that prostaglandins regulate transcription through cAMP-mediated PKA activation.
FIGURE 9.
Phosphorylation of CREB requires p38 activation but not cPLA2α/COX-1/cAMP/PKA pathway. a, COX-1−/− RPM were treated with or without 6-Bnz-cAMP (6-Bnz; 1 mm), 8-pCPT-2-O-Me-cAMP-AM (8-pCPT; 10 μm), 8-Br-cAMP (8-Br; 2 mm), or 6-Bnz together with 8-pCPT for 30 min and then infected with C. albicans for 3 h. Relative gene expression was determined by RT-PCR (n = 3; *, p < 0.05 compared with C. albicans-infected RPM not treated with cAMP analogues). b, Western blots of phospho-CREB and β-actin in cPLA2α+/+ and cPLA2α−/− RPM or of COX-1+/+ or COX-1−/− RPM treated with or without C. albicans for 10–60 min. c, Western blots of phospho-CREB and β-actin in cPLA2α+/+ and cPLA2α−/− RPM treated with and without U0126 (U; 1 μm), SB20219 (S; 10 μm), or U0126 together with SB20219 (U/S) or in COX-1+/+ and COX-1−/− RPM treated with SCH772984 (SC; 1 μm), LY222820 (L; 1 μm), or SCH772984 together with LY222820 (S/L) for 30 min and then incubated with C. albicans for 30 min. Western blotting results are representative of three independent experiments. Error bars, S.E.
Experiments were then carried out to determine whether CREB phosphorylation was downstream of the prostaglandin/cAMP/PKA or MAPK pathways. The phosphorylation of CREB on Ser-133 occurred to a similar extent in C. albicans-infected cPLA2α+/+ and cPLA2α−/− RPM and COX-1+/+ and COX-1−/− RPM (Fig. 9b). At 10 min, there was a low level of CREB phosphorylation that increased by 30 min and remained elevated for 60 min. The results demonstrate that the prostaglandin/cAMP/PKA pathway does not regulate CREB phosphorylation on Ser-133. We then determined whether CREB phosphorylation is downstream of MAPKs (Fig. 9c). In C. albicans-infected cPLA2α+/+ and cPLA2α−/− RPM, phosphorylation of CREB was blocked by the p38 inhibitor SB20219 but not by blocking ERK activation with the MEK1 inhibitor U0126. The very low level of CREB phosphorylation that remained in RPM treated with the p38 inhibitor was not evident when RPM were treated with both the ERK and p38 inhibitor, suggesting a minor role for ERKs. Similar results were obtained with the structurally distinct ERK (SCH772984) and p38 (LY2228820) inhibitors when tested in COX-1+/+ and COX-1−/− RPM. The results suggest that the role of p38 in regulating gene expression is through phosphorylation of CREB.
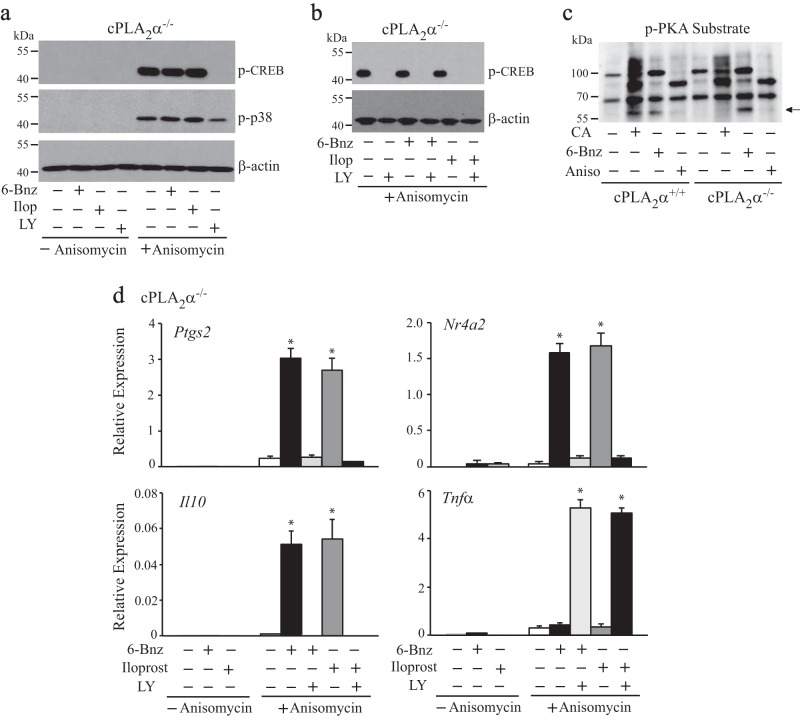
Another approach was used to study the role of p38 MAPK in promoting CREB phosphorylation and gene expression in RPM. This involved treating RPM with anisomycin, a stimulator of cell stress that we previously found preferentially activates p38 but not ERKs in RPM (20). Studies have shown that activation of p38 by anisomycin initiates a kinase cascade leading to the phosphorylation of CREB and regulation of gene expression (59, 60). Treatment of cPLA2α−/− RPM with anisomycin activated p38 and induced phosphorylation of CREB (Fig. 10, a and b). In contrast, activation of PKA with 6-Bnz-cAMP or treatment with the IP receptor agonist iloprost did not induce phosphorylation of CREB or enhance CREB phosphorylation induced by anisomycin (Fig. 10, a and b). The p38 inhibitor LY2228820 blocked CREB phosphorylation in cPLA2α−/− RPM treated with anisomycin. We carried out experiments to evaluate PKA activation in cPLA2α+/+ and cPLA2α−/− RPM using an antibody that detects substrates phosphorylated by AGC family kinases, including PKA (Fig. 10c). Western blots show that the PKA activator 6-Bnz-cAMP induced phosphorylation of a 60-kDa protein in both cPLA2α+/+ and cPLA2α−/− RPM, suggesting that it is a PKA substrate (Fig. 10c). Importantly, Western blots using the phospho-(Ser/Thr) PKA antibody show that C. albicans induces phosphorylation of the 60-kDa protein in cPLA2α+/+ but not in cPLA2α−/− RPM. The results suggest that C. albicans stimulates PKA activation through the cPLA2α/prostaglandin/cAMP pathway in cPLA2α+/+, resulting in phosphorylation of the 60-kDa protein but that C. albicans does not activate PKA in cPLA2α−/− RPM by a mechanism independent of cAMP. In addition, there is little phosphorylation of the 60-kDa protein in RPM treated with anisomycin, suggesting that it poorly activates PKA. The results suggest that phosphorylation of CREB is downstream of p38 activation stimulated by either C. albicans or anisomycin and not by PKA in RPM. As we observed with C. albicans, anisomycin acted synergistically with either 6-Bnz-cAMP or iloprost to induce expression of Ptgs2, Nr4a2, or IL10 but had no effect when tested alone (Fig. 10d). However, unlike C. albicans, anisomycin did not induce TNFα expression in cPLA2α−/− RPM, suggesting the need for additional signaling pathways stimulated by C. albicans. Unexpectedly, TNFα expression was induced by anisomycin together with 6-Bnz-cAMP or iloprost but only when p38 was inhibited. The basis for these results is not known.
FIGURE 10.
The p38 activator anisomycin stimulates CREB phosphorylation and acts synergistically with PKA activators to promote gene expression. a, Western blot of phospho-CREB, phospho-p38, and β-actin in cPLA2α−/− RPM pretreated with LY222820 (LY; 1 μm) for 30 min and then incubated with or without 6-Bnz-cAMP (6-Bnz; 1 mm) or iloprost (Ilop; 1 μm) in the absence or presence of anisomycin (25 ng/ml) for 30 min. b, Western blot of phospho-CREB and β-actin in cPLA2α−/− RPM pretreated with LY222820 (1 μm) for 30 min and then incubated for 30 min with and without 6-Bnz-cAMP (1 mm) or iloprost (1 μm) in the presence of anisomycin. c, Western blot using the phospho-(Ser/Thr)-PKA substrate antibody to evaluate PKA activation in cPLA2α+/+ and cPLA2α−/− RPM treated with C. albicans, 6-Bnz, and anisomycin for 30 min. A protein of ∼60 kDa was phosphorylated in response to the PKA activator 6-Bnz and in C. albicans-infected cPLA2α+/+ RPM but not in cPLA2α−/− RPM or in RPM treated with anisomycin. Western blots are representative of three independent experiments. d, relative gene expression in cPLA2α−/− RPM pretreated with LY222820 (1 μm) for 30 min and then incubated for 3 h with and without 6-Bnz-cAMP (1 mm) or iloprost (1 μm) in the presence or absence of anisomycin (25 ng/ml) (n = 3; *, p < 0.05 compared with anisomycin-treated RPM not treated with 6-Bnz, iloprost, or LY222820). Error bars, S.E.
Role of cPLA2α in Regulating Macrophage Function
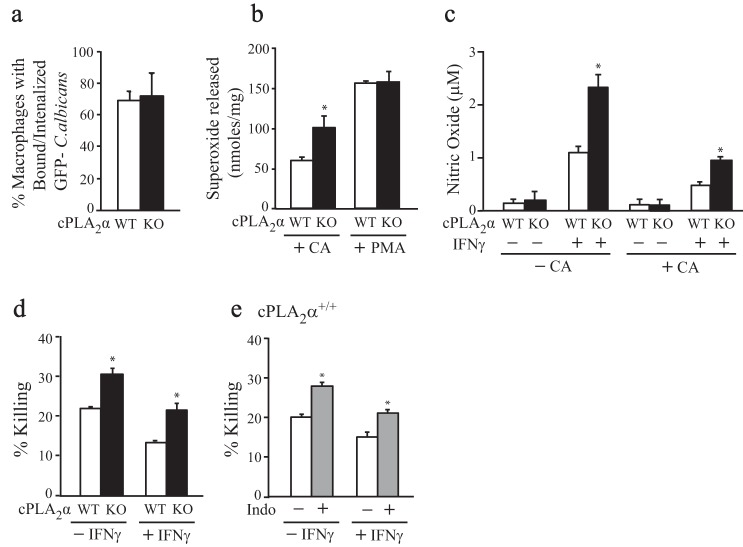
Eicosanoid production can affect innate immune responses by regulating gene expression and macrophage function, including bacterial uptake and killing (61–63). Experiments were carried out to determine the role of cPLA2α in regulating C. albicans recognition and killing and the production of superoxide anion and nitric oxide in response to C. albicans infection. There were no differences in the ability of cPLA2α+/+ and cPLA2α−/− RPM to bind and internalize GFP-C. albicans (recognition assay) (Fig. 11a). C. albicans infection stimulated NADPH oxidase activation (superoxide anion production), which was significantly higher in cPLA2α−/− than in cPLA2α+/+RPM (Fig. 11b). In contrast, superoxide anion production in cPLA2α−/− and cPLA2α+/+ RPM was not significantly different in response to PMA, a known activator of NADPH oxidase. Incubation of RPM overnight with INFγ promoted nitric oxide production that occurred to a greater extent in uninfected RPM. In both uninfected and infected RPM, nitric oxide production was significantly higher in cPLA2α−/− than in cPLA2α+/+ RPM (Fig. 11c). The lower production of nitric oxide in C. albicans-infected RPM is consistent with reports that C. albicans suppresses nitric oxide production that may be due to inhibition of nitric-oxide synthase (64, 65). The killing of C. albicans, tested using an assay that measures C. albicans viability, was found to be higher in cPLA2α−/− than in cPLA2α+/+ RPM, reflecting the increased production of superoxide and nitric oxide (Fig. 11d). Treating cPLA2α+/+ RPM with the cyclooxygenase inhibitor indomethacin enhanced their ability to kill C. albicans to the level observed in cPLA2α−/− RPM, suggesting that killing is suppressed by prostaglandins (Fig. 11e). Prostaglandin E2 has been reported to suppress bacterial killing in macrophages by inhibiting NADPH oxidase (66).
FIGURE 11.
C. albicans killing and production of superoxide and nitric oxide are higher in cPLA2α−/− RPM compared with cPLA2α+/+ RPM. a, the ability of cPLA2α+/+ (WT) and cPLA2α−/− (KO) RPM to bind and internalize GFP-C. albicans (recognition assay) is shown. b, to measure NADPH oxidase activity, superoxide anion production by cPLA2α+/+ and cPLA2α−/− RPM infected with C. albicans or treated with PMA (320 nm) for 90 min was determined. c, nitric oxide production by cPLA2α+/+ and cPLA2α−/− RPM was measured after treatment with and without IFNγ for 18 h, followed by infection with C. albicans for 4 h. d, for measuring C. albicans killing using the XTT viability assay, cPLA2α+/+ and cPLA2α−/− RPM were treated with and without IFNγ for 18 h and then incubated with C. albicans for 4 h. e, for measuring the effect of prostaglandins in regulating C. albicans killing, cPLA2α+/+ RPM were incubated with and without IFNγ for 18 h and then pretreated with indomethacin (Indo; 10 μm) for 30 min prior to incubation with C. albicans for 4 h (n = 3; *, p < 0.05 compared with C. albicans-infected cPLA2α+/+ RPM (b), cPLA2α+/+ RPM treated with IFNγ (c), cPLA2α+/+ RPM incubated with and without IFNγ (d); and cPLA2α+/+ RPM not treated with indomethacin (e)). Error bars, S.E.
Discussion
The results of this study demonstrate how the rapid activation of cPLA2α by C. albicans in RPM and production of prostaglandins from constitutively expressed COX-1 influence the expression of immediate early genes that regulate host defense and the extent of inflammation. Resident tissue macrophages are important first responders to microbial invasion that provide signals for initiating host defense mechanisms (16). Macrophages from different tissues exhibit considerable diversity and distinct properties due to unique transcriptional signatures (67). Recent studies have shown that the specialized phenotype of RPM and ability for self-renewal is determined by the unique expression of GATA6, which is not expressed in other commonly studied primary mouse macrophage populations, such as thioglycollate-elicited peritoneal macrophages and bone marrow-derived macrophages (68, 69). RPM are one of the most highly studied tissue macrophage populations and have been used extensively to investigate the regulation of cPLA2α activation and eicosanoid production, particularly in response to fungal cell walls (zymosan) (18–20, 23, 70–73). They express relatively high levels of COX-1, which contributes to the rapid production of prostaglandins in response to cPLA2α activation (24, 74). Compared with thioglycollate-elicited peritoneal macrophages and bone marrow-derived macrophages, RPM have higher expression of COX-1 and greater capacity for prostanoid production in response to a Toll-like receptor-4 agonist (9). The extent of cPLA2α activation by C. albicans and the level of COX-1 expression contribute to the relatively high levels of prostaglandins produced and secreted within minutes after infection.
The ability of prostaglandins to influence host cell responses is determined by the types of specific G-protein-coupled receptors that are expressed for inducing autocrine or paracrine responses (10). RPM express PGE2 (EP2, EP4) and PGI2 (IP) receptors, and results using antagonists for these receptors suggest that collectively they contribute to the induction or suppression of immediate early genes by increasing cAMP. Endogenously produced PGI2 and PGE2 acting through these receptors suppress production of the pro-inflammatory cytokine TNFα and increase expression of Ptgs2, the transcription factor Nr4a2, and the anti-inflammatory cytokine Il10. These effects are reproduced by treating cPLA2α- or COX-1-deficient RPM with 8-Br-cAMP. The results suggest that prostaglandins/cAMP regulate gene expression through PKA activation because the PKA-selective (6-Bnz-cAMP) but not the Epac-selective (8-pCPT-2-O-Me-cAMP-AM) cAMP analogue induces Ptgs2, Nr4a2, and Il10 and suppresses Tnfα expression in C. albicans-infected COX-1−/− RPM. Agonists for EP2, EP4, and IP increased expression of Ptgs2 and Nr4a2 and suppressed expression of TNFα to a similar extent. This suggests that the autocrine effect of specific prostaglandins on gene expression is in part determined by the amounts produced and the level of the receptor. There is more PGI2 than PGE2 produced in C. albicans-infected RPM, and expression of IP is higher than that of EP2 or EP4 (24). However, the EP4 receptor agonist was less effective at inducing Il10 expression and, when tested together with the IP and EP2 agonists, reduced the levels of Il10 that were expressed compared with using the IP and EP2 agonists alone. Although EP2, EP4, and IP mediate increases in cAMP, there are differences in signaling pathways induced by PGE2 through EP4 and EP2 (10, 75). Differential signaling through EP2 and EP4 has opposing effects in Th17 cells, with EP4 mediating inhibition of Il10 production, as observed in RPM (76). EP4 can couple to pertussis toxin-sensitive G protein Gi, leading to activation of phosphatidylinositol 3-kinase signaling that suppresses cAMP signaling (10). However, the EP4 agonist did not cause a generalized suppression of cAMP signaling in C. albicans-infected RPM because the effect was specific to Il10 and not Ptgs2 or Nr4a2.
The activation of cPLA2α globally affects transcriptional responses, but our focus on immediate early genes in this study illustrates how prostaglandins function as immune modulators to rapidly influence the balance of pro- and anti-inflammatory cytokines for moderating the extent of inflammation during infection. cPLA2α activation and prostaglandin production serve to dampen inflammation by modulating the transcriptional response through cAMP, as exemplified by the induction of the immunosuppressive cytokine IL10 and suppression of TNFα production (24). The transcription factor NR4A2, which is rapidly induced in C. albicans-infected cPLA2α+/+ but not in cPLA2α−/− RPM, has been shown to play a role in dampening inflammation (77, 78). The feedback regulation of COX-2 expression by prostaglandins through increases in cAMP is well documented, but there are cell-specific differences in mechanisms of induction (32, 52). In C. albicans-infected RPM, the rapid production of prostaglandins by COX-1 induces the expression of COX-2, but in thioglycollate-elicited macrophages, the initial induction of COX-2 in response to LPS promotes further COX-2 induction (32). This cAMP-regulated cytokine profile of C. albicans-infected cPLA2α+/+ RPM (increased IL10 and decreased TNFα) and expression of COX-2 are characteristics of resolution phase macrophages that function to restore normal tissue function after injury (79). Our results demonstrate that in response to agonists, such as C. albicans, that strongly activate cPLA2α in cells that constitutively express relatively high amounts of COX-1, such as RPM, the rapid production of prostaglandins can also regulate macrophage responses during the early phases of infection. Stimulation of pro-inflammatory responses in macrophages by microbial pathogens is important for host defense by recruiting myeloid cells to combat infection; however, negative feedback loops are important for dampening excess inflammation to avoid tissue damage (80, 81). Prostaglandin production represents one of the pathways for controlling excess inflammation, but prostaglandins can also negatively influence the ability of macrophages to kill microbial pathogens, as we observed with C. albicans and as others have shown with bacteria (66). This emphasizes the importance of maintaining the balance of pro- and anti-inflammatory pathways for combating infection.
This study focused on understanding how the production of endogenous prostaglandins initiated by cPLA2α in response to infection regulates the expression of immediate early genes. By comparing wild type and both cPLA2α−/− and COX-1−/− RPM, as well as prostaglandin receptor agonists and antagonists, the results demonstrate that prostaglandins act synergistically with activation of MAPKs to regulate gene expression. JNKs are implicated in the regulation of Ptgs2 and Tnfα expression, but we were particularly interested in whether JNKs regulated gene expression through activation of cPLA2α, as reported in studies using the JNK inhibitor SP600125 (36, 42, 43, 82). Our initial screen of the JNK inhibitor SP600125 showed that it inhibited eicosanoid production and gene expression; however, results from JNK1−/− and JNK2−/− RPM demonstrated that JNK1/2 do not regulate these processes. A thorough characterization has shown that SP600125 exhibits poor specificity (44). The ability of SP600125 to block eicosanoid production and gene expression can be explained by our finding that it blocks activation of ERKs and p38, which are required for cPLA2α activation. This highlights the importance of using alternative approaches in determining a role for JNK in cPLA2α activation (43, 82). The only role for JNK that was uncovered in our studies was the finding that JNK2 is a negative regulator of Nr4a2 expression from experiments using SP600125 and JNK2−/− RPM. JNKs play both a positive and negative role in regulating gene expression in a variety of cell types (36). In contrast to SP600125, inhibitors that block ERKs and p38 have well characterized specificity and have been important in defining the role of these MAPKs in activating cPLA2α in RPM by phosphorylating Ser-505 (20, 45, 83). We could not determine the role of MAPKs in regulating gene expression independent of prostaglandin production by treating cPLA2α+/+ RPM with ERK and p38 inhibitors due to inhibition of cPLA2α. The comparison of RPM from WT and either cPLA2α−/− or COX-1−/− mice treated with and without iloprost provided a system to independently interrogate how prostaglandins and MAPKs contribute to gene expression.
The regulation of immediate early gene expression is complex, involving many signal transduction pathways that are cell type- and stimulus-dependent (84, 85). Increases in cAMP by agents that engage G-protein-coupled receptors, such as prostaglandins, and activation of MAPKs are commonly used mechanisms to regulate gene expression and immune responses (37, 86). There is considerable cross-talk between these pathways, and increases in cAMP can promote or inhibit activation of ERKs and p38, depending on the cell type (39, 40). We found that activation of p38 and ERKs is similar in C. albicans-infected wild type and COX-1−/− and cPLA2α−/− RPM, indicating that the cPLA2α/COX-1/prostaglandin/cAMP pathway has little effect on MAPK activation. Our data suggest that MAPKs, particularly activation of p38, contribute to regulation of gene expression by promoting CREB phosphorylation. A comparison of wild type and cPLA2α−/− or COX-1−/− RPM showed no differences in the extent of CREB phosphorylation, indicating that phosphorylation is not downstream of the prostaglandin/cAMP/PKA pathway. In addition, treating COX-1−/− RPM with the PKA activator 6-Bnz-cAMP did not induce CREB phosphorylation, although 6-Bnz-cAMP acted synergistically with C. albicans to induce expression of Ptgs2, Nr4a2, and Il10 and suppress Tnfα. Our results show that CREB phosphorylation stimulated by C. albicans infection and by the stress inducer anisomycin is downstream of p38 activation. A number of kinases downstream of p38, including MSK1/2, MK2, and ribosomal S6 kinases, can mediate CREB phosphorylation (59, 60, 87–89). CREB phosphorylation that occurs downstream of p38 in response to stress signals, such as C. albicans, is not sufficient for transcription of CREB-regulated genes but requires additional co-activators (53, 55, 90). The cPLA2α/prostaglandin/cAMP/PKA pathway may function synergistically with MAPK activation by regulating CREB co-activators, such as transducers of regulated CREB (also called CREB-regulated transcription co-activator), that enhance CREB-mediated transcription of cAMP-responsive genes (49, 53, 58, 91, 92).
As shown in other cell models, the expression of Tnfα in C. albicans-infected RPM involves activation of both ERKs and p38, which regulate transcriptional and post-transcriptional pathways (54). The MAPK isoform p38α plays a major role in regulating Tnfα production by mediating CREB phosphorylation, as we observed in RPM, which primarily express the p38α isoform (24, 93). cPLA2α activation opposes MAPK-dependent expression of Tnfα through prostaglandin-induced increases in cAMP. The ability of cAMP-induced PKA activation to both enhance and suppress gene expression in macrophages involves the binding of PKA subtypes to different scaffold proteins (A kinase-anchoring proteins) that differentially regulate signaling pathways for gene expression (94). The cAMP-mediated suppression of TNFα production has been shown to involve binding of PKA to AKAP95, leading to inhibition of NFκB (94).
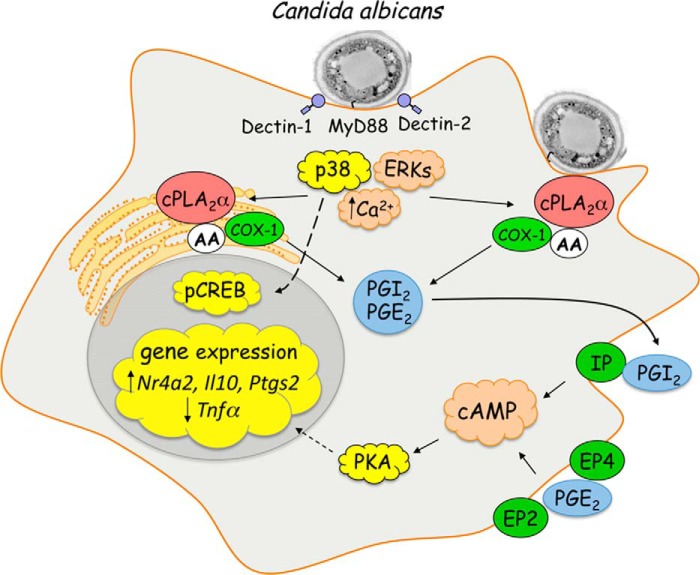
In summary, C. albicans infection of RPM rapidly stimulates activation of MAPKs and calcium increases through MyD88-, Dectin-1-, and Dectin-2-mediated pathways (Fig. 12) (15). ERKs and p38 phosphorylate cPLA2α that enhances catalytic activity, and calcium binds to the C2 domain that promotes translocation to the Golgi and the phagosome (20, 23). In RPM, arachidonic acid released by cPLA2α is metabolized by constitutively expressed COX-1 to PGI2 and PGE2. COX-1 is an integral membrane protein primarily localized in the endoplasmic reticulum but is also found in the Golgi (95, 96). Engagement of IP by PGI2 and of EP2 and EP4 by PGE2 increases cAMP and activation of PKA, which acts synergistically with p38-dependent CREB phosphorylation, resulting in induction of Ptgs2, Nr4a2, and Il10 and suppression of Tnfα. These pathways are important in controlling the balance of cytokines produced early in response to infection for regulating innate immune responses and the extent of inflammation.
FIGURE 12.
Pathways regulating induction of immediate early genes in C. albicans-infected RPM. C. albicans infection of RPM rapidly stimulates activation of MAPKs and calcium increases through MyD88-, Dectin-1-, and Dectin-2-mediated pathways. ERKs and p38 phosphorylate cPLA2α, which enhances catalytic activity, and calcium binds to the C2 domain, which promotes translocation to the Golgi and the phagosome. Arachidonic acid released by cPLA2α is metabolized by constitutively expressed COX-1 to PGI2 and PGE2. COX-1 is an integral membrane protein primarily localized in the endoplasmic reticulum but is also found in the Golgi. Binding of PGI2 to the IP receptor and of PGE2 to the EP2 and EP4 receptors increases cAMP and activation of PKA, which acts synergistically with p38-dependent CREB phosphorylation, resulting in induction of Ptgs2, Nr4a2, and Il10 and suppression of Tnfα. AA, arachidonic acid.
Author Contributions
C. C. L. conceived and coordinated the study and wrote the paper. B. Y. designed, performed, and analyzed experiments. R. C. M. contributed to the design, acquisition of data, and analysis of experiments using mass spectrometry. H. L., S. J., and S. S. provided technical assistance and contributed to the preparation of the figures. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Robert Barkley and Charis Uhlson for mass spectrometry analysis and Dr. Joseph Bonventre for originally providing the cPLA2α−/− mouse breeders.
This work was supported by National Institutes of Health Grant HL34303 (to C. C. L. and R. C. M.). The authors declare that they have no conflicts of interest with the contents of the article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- COX
- cyclooxygenase
- cPLA2
- cytosolic phospholipase A2
- PGI2
- prostaglandin I2
- PGE2
- prostaglandin E2
- RPM
- resident peritoneal macrophage(s)
- 8-Br-cAMP
- 8-bromo-cyclic AMP
- 6-Bnz-cAMP
- 6-benzoyl-cyclic AMP
- CREB
- cAMP-response element-binding protein.
References
- 1. Tam V. C. (2013) Lipidomic profiling of bioactive lipids by mass spectrometry during microbial infections. Semin. Immunol. 25, 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buckley C. D., Gilroy D. W., and Serhan C. N. (2014) Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity 40, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rodríguez M., Domingo E., Municio C., Alvarez Y., Hugo E., Fernández N., and Sanchez Crespo M. (2014) Polarization of the innate immune response by prostaglandin E2: a puzzle of receptors and signals. Mol. Pharmacol. 85, 187–197 [DOI] [PubMed] [Google Scholar]
- 4. Harizi H., Corcuff J. B., and Gualde N. (2008) Arachidonic-acid-derived eicosanoids: roles in biology and immunopathology. Trends Mol. Med. 14, 461–469 [DOI] [PubMed] [Google Scholar]
- 5. Funk C. D. (2001) Prostaglandins and leukotrienes: advances in eicosanoid biology. Science 294, 1871–1875 [DOI] [PubMed] [Google Scholar]
- 6. Haeggström J. Z., and Funk C. D. (2011) Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem. Rev. 111, 5866–5898 [DOI] [PubMed] [Google Scholar]
- 7. Shimizu T. (2009) Lipid mediators in health and disease: enzymes and receptors as therapeutic targets for the regulation of immunity and inflammation. Annu. Rev. Pharmacol. Toxicol. 49, 123–150 [DOI] [PubMed] [Google Scholar]
- 8. Ricciotti E., and FitzGerald G. A. (2011) Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 31, 986–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Norris P. C., Reichart D., Dumlao D. S., Glass C. K., and Dennis E. A. (2011) Specificity of eicosanoid production depends on the TLR-4-stimulated macrophage phenotype. J. Leukoc. Biol. 90, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Woodward D. F., Jones R. L., and Narumiya S. (2011) International Union of Basic and Clinical Pharmacology. LXXXIII: Classification of prostanoid receptors, updating 15 years of progress. Pharmacol. Rev. 63, 471–538 [DOI] [PubMed] [Google Scholar]
- 11. Haeggström J. Z., and Wetterholm A. (2002) Enzymes and receptors in the leukotriene cascade. Cell. Mol. Life Sci. 59, 742–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bäck M., Powell W. S., Dahlén S. E., Drazen J. M., Evans J. F., Serhan C. N., Shimizu T., Yokomizo T., and Rovati G. E. (2014) Update on leukotriene, lipoxin and oxoeicosanoid receptors: IUPHAR Review 7. Br. J. Pharmacol. 171, 3551–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leslie C. C. (2015) Cytosolic phospholipase A2: physiological function and role in disease. J. Lipid Res. 56, 1386–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suram S., Brown G. D., Ghosh M., Gordon S., Loper R., Taylor P. R., Akira S., Uematsu S., Williams D. L., and Leslie C. C. (2006) Regulation of cytosolic phospholipase A2 activation and cyclooxygeanse 2 expression in macrophages by the β-glucan receptor. J. Biol. Chem. 9, 5506–5514 [DOI] [PubMed] [Google Scholar]
- 15. Suram S., Gangelhoff T. A., Taylor P. R., Rosas M., Brown G. D., Bonventre J. V., Akira S., Uematsu S., Williams D. L., Murphy R. C., and Leslie C. C. (2010) Pathways regulating cytosolic phospholipase A2 activation and eicosanoid production in macrophages by Candida albicans. J. Biol. Chem. 285, 30676–30685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davies L. C., Jenkins S. J., Allen J. E., and Taylor P. R. (2013) Tissue-resident macrophages. Nat. Immunol. 14, 986–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ghosh M., Tucker D. E., Burchett S. A., and Leslie C. C. (2006) Properties of the Group IV phospholipase A2 family. Prog. Lipid Res. 45, 487–510 [DOI] [PubMed] [Google Scholar]
- 18. Qiu Z.-H., de Carvalho M. S., and Leslie C. C. (1993) Regulation of phospholipase A2 activation by phosphorylation in mouse peritoneal macrophages. J. Biol. Chem. 268, 24506–24513 [PubMed] [Google Scholar]
- 19. Qiu Z.-H., Gijón M. A., de Carvalho M. S., Spencer D. M., and Leslie C. C. (1998) The role of calcium and phosphorylation of cytosolic phospholipase A2 in regulating arachidonic acid release in macrophages. J. Biol. Chem. 273, 8203–8211 [DOI] [PubMed] [Google Scholar]
- 20. Gijón M. A., Spencer D. M., Siddiqi A. R., Bonventre J. V., and Leslie C. C. (2000) Cytosolic phospholipase A2 is required for macrophage arachidonic acid release by agonists that do and do not mobilize calcium: novel role of mitogen-activated protein kinase pathways in cytosolic phospholipase A2 regulation. J. Biol. Chem. 275, 20146–20156 [DOI] [PubMed] [Google Scholar]
- 21. Evans J. H., Spencer D. M., Zweifach A., and Leslie C. C. (2001) Intracellular calcium signals regulating cytosolic phospholipase A2 translocation to internal membranes. J. Biol. Chem. 276, 30150–30160 [DOI] [PubMed] [Google Scholar]
- 22. Tucker D. E., Ghosh M., Ghomashchi F., Loper R., Suram S., St. John B., Girotti M., Bollinger J. G., Gelb M. H., and Leslie C. C. (2009) Role of phosphorylation and basic residues in the catalytic domain of cytosolic phospholipase A2α in regulating interacial kinetics and binding and cellular function. J. Biol. Chem. 284, 9596–9611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Girotti M., Evans J. H., Burke D., and Leslie C. C. (2004) Cytosolic phospholipase A2 translocates to forming phagosomes during phagocytosis of zymosan in macrophages. J. Biol. Chem. 279, 19113–19121 [DOI] [PubMed] [Google Scholar]
- 24. Suram S., Silveira L. J., Mahaffey S., Brown G. D., Bonventre J. V., Williams D. L., Gow N. A., Bratton D. L., Murphy R. C., and Leslie C. C. (2013) Cytosolic phospholipase A2α and eicosanoids regulate expression of genes in macrophages involved in host defense and inflammation. PLoS One 8, e69002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Carvalho M. S., McCormack F. X., and Leslie C. C. (1993) The 85-kDa, arachidonic acid-specific phospholipase A2 is expressed as an activated phosphoprotein in Sf9 cells. Arch. Biochem. Biophys. 306, 534–540 [DOI] [PubMed] [Google Scholar]
- 26. Bonventre J. V., Huang Z., Taheri M. R., O'Leary E., Li E., Moskowitz M. A., and Sapirstein A. (1997) Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2. Nature 390, 622–625 [DOI] [PubMed] [Google Scholar]
- 27. Wheeler R. T., Kombe D., Agarwala S. D., and Fink G. R. (2008) Dynamic, morphotype-specific Candida albicans β-glucan exposure during infection and drug treatment. PLoS Pathog. 4, e1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 29. Ermert D., Niemiec M. J., Röhm M., Glenthøj A., Borregaard N., and Urban C. F. (2013) Candida albicans escapes from mouse neutrophils. J. Leukoc. Biol. 94, 223–236 [DOI] [PubMed] [Google Scholar]
- 30. Zhang J., Goorha S., Raghow R., and Ballou L. R. (2002) The tissue-specific, compensatory expression of cyclooxygenase-1 and -2 in transgenic mice. Prostaglandins Other Lipid Mediat. 67, 121–135 [DOI] [PubMed] [Google Scholar]
- 31. Sales K. J., Grant V., and Jabbour H. N. (2008) Prostaglandin E2 and F2α activate the FP receptor and up-regulate cyclooxygenase-2 expression via the cyclic AMP response element. Mol. Cell. Endocrinol. 285, 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ishikawa T. O., Jain N., and Herschman H. R. (2009) Feedback regulation of cyclooxygenase-2 transcription ex vivo and in vivo. Biochem. Biophys. Res. Commun. 378, 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bradbury D. A., Newton R., Zhu Y. M., El-Haroun H., Corbett L., and Knox A. J. (2003) Cyclooxygenase-2 induction by bradykinin in human pulmonary artery smooth muscle cells is mediated by the cyclic AMP response element through a novel autocrine loop involving endogenous prostaglandin E2, E-prostanoid 2 (EP2), and EP4 receptors. J. Biol. Chem. 278, 49954–49964 [DOI] [PubMed] [Google Scholar]
- 34. Klein T., Shephard P., Kleinert H., and Kömhoff M. (2007) Regulation of cyclooxygenase-2 expression by cyclic AMP. Biochim. Biophys. Acta 1773, 1605–1618 [DOI] [PubMed] [Google Scholar]
- 35. Darragh J., Soloaga A., Beardmore V. A., Wingate A. D., Wiggin G. R., Peggie M., and Arthur J. S. (2005) MSKs are required for the transcription of the nuclear orphan receptors Nur77, Nurr1 and Nor1 downstream of MAPK signalling. Biochem. J. 390, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rincón M., and Davis R. J. (2009) Regulation of the immune response by stress- activated protein kinases. Immunol. Rev. 228, 212–224 [DOI] [PubMed] [Google Scholar]
- 37. Arthur J. S., and Ley S. C. (2013) Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13, 679–692 [DOI] [PubMed] [Google Scholar]
- 38. Kang Y. J., Mbonye U. R., DeLong C. J., Wada M., and Smith W. L. (2007) Regulation of intracellular cyclooxygenase levels by gene transcription and protein degradation. Prog. Lipid Res. 46, 108–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gerits N., Kostenko S., Shiryaev A., Johannessen M., and Moens U. (2008) Relations between the mitogen-activated protein kinase and the cAMP-dependent protein kinase pathways: comradeship and hostility. Cell. Signal. 20, 1592–1607 [DOI] [PubMed] [Google Scholar]
- 40. Yin F., Wang Y. Y., Du J. H., Li C., Lu Z. Z., Han C., and Zhang Y. Y. (2006) Noncanonical cAMP pathway and p38 MAPK mediate β2-adrenergic receptor-induced IL-6 production in neonatal mouse cardiac fibroblasts. J. Mol. Cell. Cardiol. 40, 384–393 [DOI] [PubMed] [Google Scholar]
- 41. Zhong J., and Kyriakis J. M. (2007) Dissection of a signaling pathway by which pathogen-associated molecular patterns recruit the JNK and p38 MAPKs and trigger cytokine release. J. Biol. Chem. 282, 24246–24254 [DOI] [PubMed] [Google Scholar]
- 42. Casas J., Meana C., Esquinas E., Valdearcos M., Pindado J., Balsinde J., and Balboa M. A. (2009) Requirement of JNK-mediated phosphorylation for translocation of group IVA phospholipase A2 to phagosomes in human macrophages. J. Immunol. 183, 2767–2774 [DOI] [PubMed] [Google Scholar]
- 43. Tang F., Chen Z., Ciszewski C., Setty M., Solus J., Tretiakova M., Ebert E., Han J., Lin A., Guandalini S., Groh V., Spies T., Green P., and Jabri B. (2009) Cytosolic PLA2 is required for CTL-mediated immunopathology of celiac disease via NKG2D and IL-15. J. Exp. Med. 206, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bain J., McLauchlan H., Elliott M., and Cohen P. (2003) The specificities of protein kinase inhibitors: an update. Biochem. J. 371, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S., Alessi D. R., and Cohen P. (2007) The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wen A. Y., Sakamoto K. M., and Miller L. S. (2010) The role of the transcription factor CREB in immune function. J. Immunol. 185, 6413–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. McEvoy A. N., Murphy E. A., Ponnio T., Conneely O. M., Bresnihan B., FitzGerald O., and Murphy E. P. (2002) Activation of nuclear orphan receptor NURR1 transcription by NF-κB and cyclic adenosine 5′-monophosphate response element-binding protein in rheumatoid arthritis synovial tissue. J. Immunol. 168, 2979–2987 [DOI] [PubMed] [Google Scholar]
- 48. Volakakis N., Kadkhodaei B., Joodmardi E., Wallis K., Panman L., Silvaggi J., Spiegelman B. M., and Perlmann T. (2010) NR4A orphan nuclear receptors as mediators of CREB-dependent neuroprotection. Proc. Natl. Acad. Sci. U.S.A. 107, 12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Alvarez Y., Municio C., Alonso S., Sánchez Crespo M., and Fernández N. (2009) The induction of IL-10 by zymosan in dendritic cells depends on CREB activation by the coactivators CREB-binding protein and TORC2 and autocrine PGE2. J. Immunol. 183, 1471–1479 [DOI] [PubMed] [Google Scholar]
- 50. Platzer C., Fritsch E., Elsner T., Lehmann M. H., Volk H. D., and Prösch S. (1999) Cyclic adenosine monophosphate-responsive elements are involved in the transcriptional activation of the human IL-10 gene in monocytic cells. Eur. J. Immunol. 29, 3098–3104 [DOI] [PubMed] [Google Scholar]
- 51. Eliopoulos A. G., Dumitru C. D., Wang C. C., Cho J., and Tsichlis P. N. (2002) Induction of COX-2 by LPS in macrophages is regulated by Tpl2-dependent CREB activation signals. EMBO J. 21, 4831–4840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Díaz-Muñoz M. D., Osma-García I. C., Fresno M., and Iñiguez M. A. (2012) Involvement of PGE2 and the cAMP signalling pathway in the up-regulation of COX-2 and mPGES-1 expression in LPS-activated macrophages. Biochem. J. 443, 451–461 [DOI] [PubMed] [Google Scholar]
- 53. Ravnskjaer K., Kester H., Liu Y., Zhang X., Lee D., Yates J. R. 3rd, and Montminy M. (2007) Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J. 26, 2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sabio G., and Davis R. J. (2014) TNF and MAP kinase signalling pathways. Semin. Immunol. 26, 237–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mayr B., and Montminy M. (2001) Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 56. Naqvi S., Martin K. J., and Arthur J. S. (2014) CREB phosphorylation at Ser133 regulates transcription via distinct mechanisms downstream of cAMP and MAPK signalling. Biochem. J. 458, 469–479 [DOI] [PubMed] [Google Scholar]
- 57. Johannessen M., Delghandi M. P., and Moens U. (2004) What turns CREB on? Cell. Signal. 16, 1211–1227 [DOI] [PubMed] [Google Scholar]
- 58. Sands W. A., and Palmer T. M. (2008) Regulating gene transcription in response to cyclic AMP elevation. Cell. Signal. 20, 460–466 [DOI] [PubMed] [Google Scholar]
- 59. Wiggin G. R., Soloaga A., Foster J. M., Murray-Tait V., Cohen P., and Arthur J. S. (2002) MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 22, 2871–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deak M., Clifton A. D., Lucocq L. M., and Alessi D. R. (1998) Mitogen- and stress- activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17, 4426–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Alvarez Y., Valera I., Municio C., Hugo E., Padron F., Blanco L., Rodriguez M., Fernandez N., and Crespo M. S. (2010) Eicosanoids in the innate immune response: TLR and non-TLR routes. Mediat. Inflamm. 10.1155/2010/201929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Peters-Golden M. (2009) Putting on the brakes: cyclic AMP as a multipronged controller of macrophage function. Sci. Signal. 2, pe37. [DOI] [PubMed] [Google Scholar]
- 63. Kalinski P. (2012) Regulation of immune responses by prostaglandin E2. J. Immunol. 188, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schröppel K., Kryk M., Herrmann M., Leberer E., Röllinghoff M., and Bogdan C. (2001) Suppression of type 2 NO-synthase activity in macrophages by Candida albicans. Int. J. Med. Microbiol. 290, 659–668 [DOI] [PubMed] [Google Scholar]
- 65. Chinen T., Qureshi M. H., Koguchi Y., and Kawakami K. (1999) Candida albicans suppresses nitric oxide (NO) production by interferon-γ (IFN-γ) and lipopolysaccharide (LPS)-stimulated murine peritoneal macrophages. Clin. Exp. Immunol. 115, 491–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Serezani C. H., Chung J., Ballinger M. N., Moore B. B., Aronoff D. M., and Peters-Golden M. (2007) Prostaglandin E2 suppresses bacterial killing in alveolar macrophages by inhibiting NADPH oxidase. Am. J. Respir. Cell Mol. Biol. 37, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gautier E. L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K. G., Gordonov S., Mazloom A. R., Ma'ayan A., Chua W. J., Hansen T. H., Turley S. J., et al. (2012) Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13, 1118–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rosas M., Davies L. C., Giles P. J., Liao C. T., Kharfan B., Stone T. C., O'Donnell V. B., Fraser D. J., Jones S. A., and Taylor P. R. (2014) The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science 344, 645–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Okabe Y., and Medzhitov R. (2014) Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Qiu Z.-H., and Leslie C. C. (1994) Protein kinase C-dependent and -independent pathways of mitogen-activated protein kinase activation in macrophages by stimuli that activate phospholipase A2. J. Biol. Chem. 269, 19480–19487 [PubMed] [Google Scholar]
- 71. Rouzer C. A., Tranguch S., Wang H., Zhang H., Dey S. K., and Marnett L. J. (2006) Zymosan-induced glycerylprostaglandin and prostaglandin synthesis in resident peritoneal macrophages: roles of cyclooxygeanse-1 and -2. Biochem. J. 399, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Scott W. A., Zrike J. M., Hamill A. L., Kempe J., and Cohn Z. A. (1980) Regulation of arachidonic acid metabolites in macrophages. J. Exp. Med. 152, 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bonney R. J., Wightman P. D., Davies P., Sadowski S. J., Kuehl F. A. Jr., and Humes J. L. (1978) Regulation of prostaglandin synthesis and of the selective release of lysosomal hydrolases by mouse peritoneal macrophages. Biochem. J. 176, 433–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rouzer C. A., Kingsley P. J., Wang H., Zhang H., Morrow J. D., Dey S. K., and Marnett L. J. (2004) Cyclooxygenase-1-dependent prostaglandin synthesis modulates tumor necrosis factor-α secretion in lipopolysaccharide-challenged murine resident peritoneal macrophages. J. Biol. Chem. 279, 34256–34268 [DOI] [PubMed] [Google Scholar]
- 75. Konya V., Marsche G., Schuligoi R., and Heinemann A. (2013) E-type prostanoid receptor 4 (EP4) in disease and therapy. Pharmacol. Ther. 138, 485–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Boniface K., Bak-Jensen K. S., Li Y., Blumenschein W. M., McGeachy M. J., McClanahan T. K., McKenzie B. S., Kastelein R. A., Cua D. J., and de Waal Malefyt R. (2009) Prostaglandin E2 regulates Th17 cell differentiation and function through cyclic AMP and EP2/EP4 receptor signaling. J. Exp. Med. 206, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bonta P. I., van Tiel C. M., Vos M., Pols T. W., van Thienen J. V., Ferreira V., Arkenbout E. K., Seppen J., Spek C. A., van der Poll T., Pannekoek H., and de Vries C. J. (2006) Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler. Thromb. Vasc. Biol. 26, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 78. Mahajan S., Saini A., Chandra V., Nanduri R., Kalra R., Bhagyaraj E., Khatri N., and Gupta P. (2015) Nuclear receptor Nr4a2 promotes alternative polarization of macrophages and confers protection in sepsis. J. Biol. Chem. 290, 18304–18314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bystrom J., Evans I., Newson J., Stables M., Toor I., van Rooijen N., Crawford M., Colville-Nash P., Farrow S., and Gilroy D. W. (2008) Resolution-phase macrophages possess a unique inflammatory phenotype that is controlled by cAMP. Blood 112, 4117–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jenner R. G., and Young R. A. (2005) Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 3, 281–294 [DOI] [PubMed] [Google Scholar]
- 81. Ivashkiv L. B. (2011) Inflammatory signaling in macrophages: transitions from acute to tolerant and alternative activation states. Eur. J. Immunol. 41, 2477–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Gubern A., Barceló-Torns M., Barneda D., López J. M., Masgrau R., Picatoste F., Chalfant C. E., Balsinde J., Balboa M. A., and Claro E. (2009) JNK and ceramide kinase govern the biogenesis of lipid droplets through activation of group IVA phospholipase A2. J. Biol. Chem. 284, 32359–32369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Davies S. P., Reddy H., Caivano M., and Cohen P. (2000) Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Healy S., Khan P., and Davie J. R. (2013) Immediate early response genes and cell transformation. Pharmacol. Ther. 137, 64–77 [DOI] [PubMed] [Google Scholar]
- 85. Fowler T., Sen R., and Roy A. L. (2011) Regulation of primary response genes. Mol. Cell 44, 348–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Mosenden R., and Taskén K. (2011) Cyclic AMP-mediated immune regulation: overview of mechanisms of action in T cells. Cell. Signal. 23, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 87. Xing J., Ginty D. D., and Greenberg M. E. (1996) Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273, 959–963 [DOI] [PubMed] [Google Scholar]
- 88. Zaru R., Ronkina N., Gaestel M., Arthur J. S., and Watts C. (2007) The MAPK- activated kinase Rsk controls an acute Toll-like receptor signaling response in dendritic cells and is activated through two distinct pathways. Nat. Immunol. 8, 1227–1235 [DOI] [PubMed] [Google Scholar]
- 89. Tan Y., Rouse J., Zhang A., Cariati S., Cohen P., and Comb M. J. (1996) FGF and stress regulate CREB and ATF-1 via a pathway involving p38 MAP kinase and MAPKAP kinase-2. EMBO J. 15, 4629–4642 [PMC free article] [PubMed] [Google Scholar]
- 90. Xu W., Kasper L. H., Lerach S., Jeevan T., and Brindle P. K. (2007) Individual CREB-target genes dictate usage of distinct cAMP-responsive coactivation mechanisms. EMBO J. 26, 2890–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yao C., Sakata D., Esaki Y., Li Y., Matsuoka T., Kuroiwa K., Sugimoto Y., and Narumiya S. (2009) Prostaglandin E2-EP4 signaling promotes immune inflammation through TH1 cell differentiation and TH17 cell expansion. Nat. Med. 15, 633–640 [DOI] [PubMed] [Google Scholar]
- 92. Altarejos J. Y., and Montminy M. (2011) CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kang Y. J., Chen J., Otsuka M., Mols J., Ren S., Wang Y., and Han J. (2008) Macrophage deletion of p38α partially impairs lipopolysaccharide-induced cellular activation. J. Immunol. 180, 5075–5082 [DOI] [PubMed] [Google Scholar]
- 94. Wall E. A., Zavzavadjian J. R., Chang M. S., Randhawa B., Zhu X., Hsueh R. C., Liu J., Driver A., Bao X. R., Sternweis P. C., Simon M. I., and Fraser I. D. (2009) Suppression of LPS-induced TNF-alpha production in macrophages by cAMP is mediated by PKA-AKAP95-p105. Sci. Signal. 2, ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Smith W. L., Urade Y., and Jakobsson P. J. (2011) Enzymes of the cyclooxygenase pathways of prostanoid biosynthesis. Chem. Rev. 111, 5821–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Grewal S., Ponnambalam S., and Walker J. H. (2003) Association of cPLA2-alpha and COX-1 with the Golgi apparatus of A549 human lung epithelial cells. J. Cell Sci. 116, 2303–2310 [DOI] [PubMed] [Google Scholar]