Abstract
The structural similarity between defensins and scorpion neurotoxins suggests that they might have evolved from a common ancestor. However, there is no direct experimental evidence demonstrating a functional link between scorpion neurotoxins and defensins. The scorpion defensin BmKDfsin4 from Mesobuthus martensii Karsch contains 37 amino acid residues and a conserved cystine-stabilized α/β structural fold. The recombinant BmKDfsin4, a classical defensin, has been found to have inhibitory activity against Gram-positive bacteria such as Staphylococcus aureus, Bacillus subtilis, and Micrococcus luteus as well as methicillin-resistant Staphylococcus aureus. Interestingly, electrophysiological experiments showed that BmKDfsin4,like scorpion potassium channel neurotoxins, could effectively inhibit Kv1.1, Kv1.2, and Kv1.3 channel currents, and its IC50 value for the Kv1.3 channel was 510.2 nm. Similar to the structure-function relationships of classical scorpion potassium channel-blocking toxins, basic residues (Lys-13 and Arg-19) of BmKDfsin4 play critical roles in peptide-Kv1.3 channel interactions. Furthermore, mutagenesis and electrophysiological experiments demonstrated that the channel extracellular pore region is the binding site of BmKDfsin4, indicating that BmKDfsin4adopts the same mechanism for blocking potassium channel currents as classical scorpion toxins. Taken together, our work identifies scorpion BmKDfsin4 as the first invertebrate defensin to block potassium channels. These findings not only demonstrate that defensins from invertebrate animals are a novel type of potassium channel blockers but also provide evidence of a functional link between defensins and neurotoxins.
Keywords: bacteria, defensin, neurotoxin, potassium channel, structure-function
Introduction
Over the course of long-term evolution, venomous scorpions have gradually developed a powerful venom system as a primary weapon for capturing prey and defending against predators (1, 2). Genomic, transcriptomic, and proteomic analyses have indicated that venoms contain diverse types of cysteine-rich neurotoxins that block or modulate different types of ion channels that open and close to generate electrical signals for nerve cell communication (3–6). The vast majority of scorpion toxins contain a core topology comprising α helices connected to an antiparallel β sheet stabilized by three or four disulfide bonds, termed the cystine-stabilized α/β motif, including ChTX4 from the scorpion Leiurus quinquestriatus hebraeus (7) and BmKTX from the scorpion Mesobuthus martensii (8). In addition to producing these classical toxins, scorpions also produce defensins, which are also synthesized by fungi (9), plants (10, 11), and other invertebrate (12) and vertebrate animals (13). Many defensins are also cysteine-rich cationic peptides with cystine-stabilized α/β motifs. On the basis of the similarity between scorpion defensins and venom neurotoxins in gene organization, protein sequence, and three-dimensional structures, it has been proposed that neurotoxins likely originated from defensins (6, 14–17). However, there is no direct experimental evidence demonstrating a functional link between scorpion neurotoxins and defensins.
Here we performed an in-depth investigation into the potential of scorpion defensins to function as neurotoxin-like potassium channel blockers on the basis of diverse neurotoxin-potassium channel interactions (8, 18–22). As expected, BmKDfsin4, an ancient antimicrobial defensin from the scorpion M. martensii Karsch (6), effectively blocked the voltage-gated potassium channels Kv1.1, Kv1.2, and Kv1.3 in the same manner as classical scorpion potassium channel toxins. These findings are the first to experimentally demonstrate that a venomous animal defensin is a potassium channel blocker, revealing that venomous animal defensins are a novel type of potassium channel blocker.
Materials and Methods
Peptide and Peptide Mutants
Recently, BmKDfsin4 has been identified from the scorpion M. martensii Karsch by our group, and BmKDfsin4 has been expressed using a prokaryotic expression technique on the basis of our previous strategy for expressing recombinant potassium channel-blocking scorpion toxins (19–21). The KOD-Plus-Neo kit was used to produce BmKDfsin4 mutants on the basis of the wild-type pGEX-6p-1-BmKDfsin4 plasmid. All plasmids were verified by DNA sequencing before expression. The mutants were also expressed and purified using the same method.
Sequence and Structure Analysis of BmKDfsin4
Multiple sequence alignments of BmKDfsin4, Lqh def, charybdotoxin (ChTX), and BmKTX were performed using the GeneDoc program. The secondary structures of peptides were measured using CD spectroscopy. The three-dimensional structure of BmKDfsin4 was modeled using micasin as a template (PDB code 2LR5) through the SWISS-MODEL server as described preciously (19). The solution structures of BmKTX and ChTX were retrieved from the PDB (BmKTX PDB code, 1BKT; ChTX PDB code, 2CRD).
Bacterial Strains
Staphylococcus aureus AB94004, S. aureusATCC25923, Micrococcus luteus AB93113, Bacillus subtilis AB91021, Bacillus thuringiensis AB92037, Escherichia coli AB94012, and E. coli ATCC25922 were purchased from the China Center of Type Culture Collection. Methicillin-resistant S. aureus P1381 and penicillin-resistant Staphylococcus epidermidis P1389 were obtained from the 302nd Military Hospital (Beijing, China).
Antimicrobial Assays
Antimicrobial activity was determined using the broth microdilution assay according to the procedure recommended by the Clinical and Laboratory Standards Institute, with some modifications. Briefly, bacteria were cultured in LB medium to A600 = 0.6 at 37 °C and then diluted to 105-106 cfu/ml in LB medium. The peptide was serially diluted in 0.9% saline. An 80-μl sample of the bacterial suspension and 20 μl of the diluted peptide at varying concentrations were added to 96-well plates and then incubated for 16 h at 37 °C with continuous shaking at 250 rpm. The minimum inhibitory concentration (MIC) was determined as the lowest peptide concentration at which no bacterial growth was observed.
Time-kill Kinetics
S. aureus AB94004 was cultured in LB medium to the exponential phase (A600 = 0.5) and then diluted to 107 cfu/ml in LB medium. The cells were treated with BmKDfsin4 and ampicillin at 5 × MIC. The negative control cells were treated with 0.9% saline. Aliquots were taken at defined intervals, washed with 0.9% saline, and then diluted appropriately in saline and plated on LB agar. The plates were incubated at 37 °C for 24 h, and the colony-forming units were counted.
Fluorescence Measurements
S. aureus AB94004 cells were cultured to the exponential phase (107 cfu/ml), and then the fluorescent dye SYTOX Green (Invitrogen) was added to a final concentration of 5 μm. After incubation for 10 min, BmKDfsin4 was added to final concentrations of 1 × MIC, 2 × MIC, 5 × MIC, and 10 × MIC. MSI-78 and 0.9% saline were used as positive and negative controls, respectively. Fluorescence was measured using the excitation and emission wavelengths of 488 and 525 nm, respectively.
Transmission electron microscopy
Exponential phase S. aureusAB94004 (107 cfu/ml) cells were treated with BmKDfsin4 at 10 × MIC for 30 min. Negative control cells were treated with 0.9% saline. Samples were harvested by centrifugation and then washed with 0.9% saline. The bacteria were fixed with 2% glutaraldehyde in 0.1 m PBS for 1 h and then with 1% osmium tetroxide at 4 °C for another 1 h. After fixation, the bacteria were stained with 1% uranyl acetate and then dehydrated using a graded series of ethyl alcohol. After the steps described above, the samples were embedded in epoxy resin and stained with 1% uranyl acetate and lead citrate. Semithin sections of the samples were then prepared and examined using a Hitachi H-8100 transmission electron microscope.
Expression of Potassium Channels
The pRc/CMV-hKv1.3 vector was provided by Prof. Stephan Grissmer (University of Ulm, Ulm, Germany) and Prof. Olaf Pongs (Zentrum für Molekulare Neurobiologie der Universität Hamburg, Hamburg, Germany). The pRc/CMV-hKv1.3 vector was used directly. The cDNAs encoding the channels mKv1.1, hKv1.2, hIK, hSKCa3, hERG, and hKCNQ1 (from Prof. Stephan Grissmer and Prof. George Chandy, University of California, Irvine, CA) were subcloned into the vector pIRES2-EGFP (Clontech) for coexpression with enhanced GFP. All of the potassium channels are from humans, except the Kv1.1 channel, which is from mice. The QuikChange site-directed mutagenesis kit (Stratagene) was used to produce the mutants on the basis of the wild-type hKv1.3 plasmid. All plasmids and mutants were verified by DNA sequencing before expression.
Cell Culture
HEK293 cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a humidified 5% CO2 incubator at 37 °C. Cells were transfected using FuGENE transfection reagent (Roche Diagnostics) following the instructions of the manufacturer and used for electrophysiology 24–48 h after transfection.
Electrophysiological Recordings
Electrophysiological experiments were conducted using the patch clamp whole-cell recording mode at room temperature. Current measurements and data acquisition were performed with an EPC 10 patch clamp amplifier (HEKA Elektronik) controlled by Patchmaster software (HEKA Elektronik). For measurements of mKv1.1, hKv1.2, hKv1.3, hERG, and hKCNQ1 channel currents, the internal pipette solution contained 140 mm KCl, 1 mm MgCl2, 1 mm EGTA, 1 mm Na2ATP, and 5 mm HEPES (pH 7.2 with KOH, and the external solution contained 5 mm KCl, 140 mm NaCl, 10 mm HEPES, 2 mm CaCl2, 1 mm MgCl2, and 10 mm d-glucose (pH 7.4 with NaOH). For measurements of hIK and hSKCa3 channel currents, the internal pipette solution contained 145 mm potassium aspartate, 8.7 mm CaCl2, 2 mm MgCl2, 10 mm EGTA, and 10 mm HEPES (pH 7.2 with KOH) to achieve an intracellular free Ca2+ concentration of 1 μm, and the external solution contained 130 mm sodium aspartate, 30 mm potassium aspartate, 2 mm CaCl2, 1 mm MgCl2, and 10 mm HEPES (pH 7.4 with NaOH). Each channel current was elicited using an individual recording protocol. BmKDfsin4 was dissolved in the external solution containing 0.01% BSA for the electrophysiological experiments. A multichannel microperfusion system (MPS-2, INBIO Inc., Wuhan, China) was used to exchange the external recording bath solution.
Results
Structural Similarity between a Scorpion Defensin and Potassium Channel-blocking Toxins
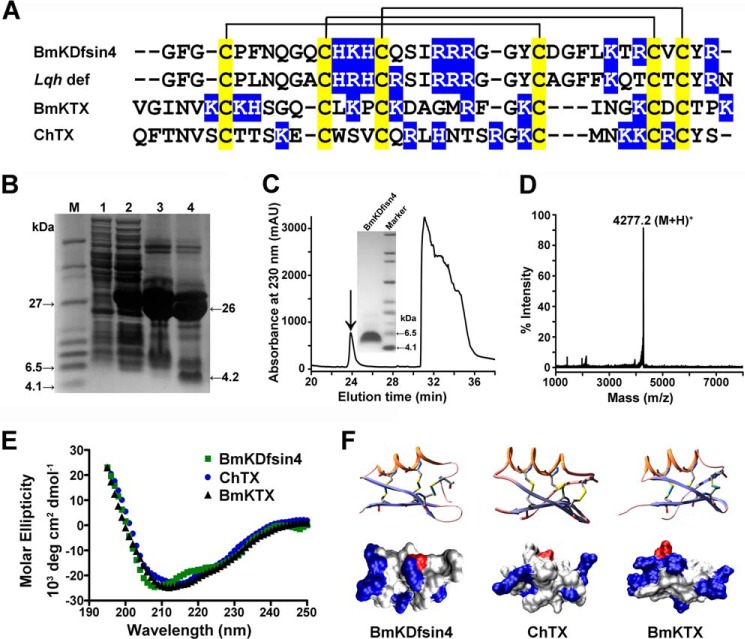
BmKDfsin4 is a representative scorpion defensin that was found in the M. martensii genome (6). This peptide is highly homologous to the defensin Lqh def isolated from scorpion hemolymph (23) (Fig. 1A). Through our previous strategy for expressing recombinant potassium channel-blocking scorpion toxins (19–21), BmKDfsin4 was expressed by a prokaryotic expression technique, and the recombinant BmKDfsin4 (rBmKDfsin4) with the correct molecular weight was obtained after a series of purification steps (Fig. 1, B–D). Subsequently, its secondary structure was determined using CD spectroscopy. The similar CD spectra of rBmKDfsin4 and the potassium channel-sensitive BmKTX toxin indicated that BmKDfsin4 likely adopted the cystine-stabilized α/β fold and that the CD spectrum of another potassium channel-blocking toxin, ChTX, is also comparable (Fig. 1, E and F). Charged residue analyses showed that one acidic residue of BmKDfsin4 was distributed regionally and that nine basic residues were distributed widely throughout the whole peptide, which was also similar to the classical potassium channel toxins ChTX and BmKTX (8). These common structural features and charged residue distributions of the defensin BmKDfsin4 and the neurotoxins ChTX and BmKTX suggest that BmKDfsin4 might be an antimicrobial defensin with neurotoxic activity.
FIGURE 1.
Sequence alignment, production, and structural analysis of BmKDfsin4. A, multiple amino acid sequence alignment of BmKDfsin4, Lqh def (a scorpion defensin from L. quinquestriatus), ChTX (a toxin from L. quinquestriatus hebraeus), and BmKTX (a native toxin from M. martensii Karsch). Cysteines are shadowed in yellow, and basic residues are shown in blue. B, Tricine SDS-PAGE analysis of the expression of the GST-BmKDfsin4 fusion protein and split products. Lane M, molecular mass markers; lane 1, total cell-free extract of E. coli carrying pGEX-6p-1-BmKDfsin4 without isopropyl 1-thio-β-d-galactopyranoside induction; lane 2, total cell-free extract of E. coli carrying pGEX-6p-1-BmKDfsin4 induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside; lane 3, the purified GST-BmKDfsin4 fusion protein after desalting and concentration; lane 4, the fusion protein cleaved by enterokinase. C, the HPLC profile of the GST-BmKDfsin4 fusion protein cleaved by enterokinase. The fraction containing BmKDfsin4 is indicated by the arrow and was analyzed by Tricine SDS-PAGE. D, mass spectrum of the BmKDfsin4 peptide measured by MALDI-TOF/MS. The measured value is 4277.2 Da, and the calculated value is 4276.9 Da. E, the circular dichroism spectra of BmKDfsin4, measured at 25 °C from 195–250 nm. Data were collected at 1-nm intervals with a continuous scan rate of 200 nm/min. F, three-dimensional structures of BmKDfsin4, ChTX, and BmKTX (top row). The disulfide bonds are shown. Also shown are space-filling molecular surfaces of BmKDfsin4, ChTX, and BmKTX (bottom row) showing the locations of different types of amino acids (blue, basic residues; red, acidic residues).
BmKDfsin4 Inhibits Bacteria without Destroying Bacterial Membrane Integrity
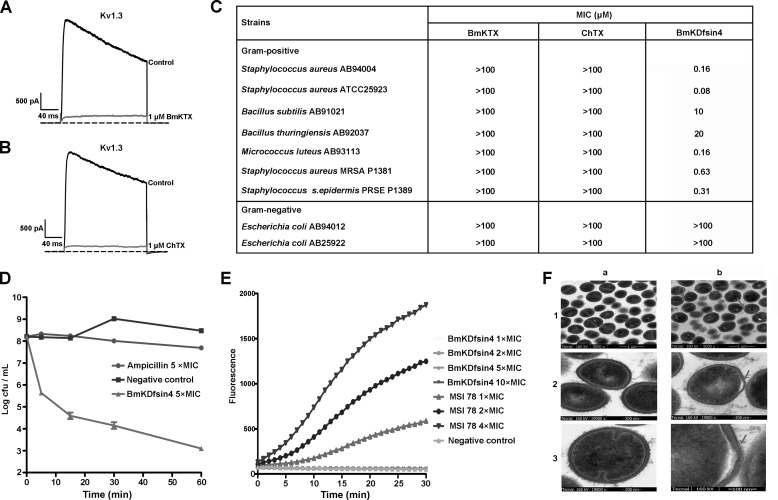
Defensin, as an effector in innate immunity, plays an important role in preventing microbial infection. Before measuring the antibacterial activity of rBmKDfsin4, we investigated the antibacterial activity of the classicalBmKDfsin4-like scorpion toxins BmKTX and ChTX (Fig. 1). As shown in Fig. 2, A–C, 1 μm BmKTX and 1 μm ChTX blocked about 94.9% and 93.0% of Kv1.3 channel currents, respectively, but they could not inhibit the growth of either Gram-positive or Gram-negative bacteria. However, the scorpion defensin BmKDfsin4 exhibited its inherent inhibitory activity against bacteria. As shown in Fig. 2C, rBmKDfsin4 inhibited the growth of Gram-positive bacteria, including standard strains and antibiotic-resistant bacteria (e.g. methicillin-resistant S. aureus and penicillin-resistant S. epidermidis) with MICs of 0.08–20 μm, but, even at concentrations up to 100 μm, it could not effectively inhibit the growth of Gram-negative bacteria.
FIGURE 2.
The antibacterial activity and mechanism of BmKDfsin4. A, representative current traces in the absence (control) or presence of 1 μm BmKTX on the currents of the Kv1.3 channel. 1 μm BmKTX blocked 94.9% ± 1.1% of Kv1.3 channel currents. B, representative current traces in the absence (control) or presence of 1 μm ChTX on the currents of the Kv1.3 channel. 1 μm ChTX blocks 93.0% ± 3.1% of Kv1.3 channel currents. Each experiment of channel current blockage was performed at least three times (n ≥ 3). C, MICs of BmKTX, ChTX, and BmKDfsin4 against bacteria. Every concentration of peptides was set three parallel duplicates, and only the concentration for all three parallel samples without bacterial growth was used as the MIC of the peptide against bacteria. MIC determination of peptides was repeated three times (n = 3), and the determined MIC values were the same. D, killing kinetics of BmKDfsin4 against S. aureus AB94004. The bacteria were treated with BmKDfsin4 or ampicillin at 5 × MIC. The experiment was repeated with similar results. E, the effect of BmKDfsin4 on bacterial membrane integrity. BmKDfsin4 was used at 1 × MIC, 2 × MIC, 5 × MIC, and 10 × MIC. The negative and positive controls were 0.9% saline and MSI-78 (an amphipathic α-helical peptide with an antibacterial mechanism of disrupting the membrane), respectively. The MIC of MSI-78 against S. aureus AB94004 was 3.53 μm. The experiment was repeated with similar results. F, transmission electron microscopy observation of S. aureus AB94004 in the absence or presence of BmKDfsin4. a1–3, negative control. b1–3, treatment with BmKDfsin4 at a concentration of 10 × MIC for 30 min. Cells showing plasmolysis are indicated by arrows. Each experiment was performed in duplicate (n = 3).
To explore the mechanism by which BmKDfsin4 inhibits bacterial growth, S. aureus AB94004 was selected as a standard model bacterium. The killing kinetics of BmKDfsin4 against S. aureus showed that it killed this bacterium more rapidly than ampicillin, an antibiotic inhibiting bacterial cell wall biosynthesis (Fig. 2D). The capacity of BmKDfsin4 to permeabilize the bacterial membrane was also assessed with the fluorescent nucleic acid stain SYTOX. The pore-forming antimicrobial peptide MSI-78 caused an immediate increase in fluorescence in a dose-dependent manner when S. aureus was exposed to this peptide (Fig. 2E). In contrast to the fluorescence changes caused by MSI-78, no increase in fluorescence was observed over 30 min after S. aureus cells were exposed to BmKDfsin4 at 1 × MIC, 2 × MIC, 5 × MIC, or 10 × MIC, similar to the results for the negative control, 0.9% saline. These results showed that the bacterial membrane integrity was not destroyed after rBmKDfsin4 treatment, indicating that the BmKDfsin4 peptide does not disrupt membranes. To further elucidate the antibacterial mechanism of BmKDfsin4, electron microscopy was used to visualize the potential damage to S. aureus caused by the scorpion defensin. There were obvious morphological changes in S. aureus treated with BmKDfsin4 at 10 × MIC for 30 min compared with S. aureus in the absence of BmKDfsin4. Although the cell outline was clear, the bacteria appeared irregularly fractured after BmKDfsin4 treatment. In addition, plasmolysis was observed using electron microscopy (Fig. 2F, b1–3). Therefore, BmKDfsin4 selectively inhibited Gram-positive bacteria but did not disrupt bacterial membrane integrity.
BmKDfsin4 Is a New Blocker of Potassium Channels
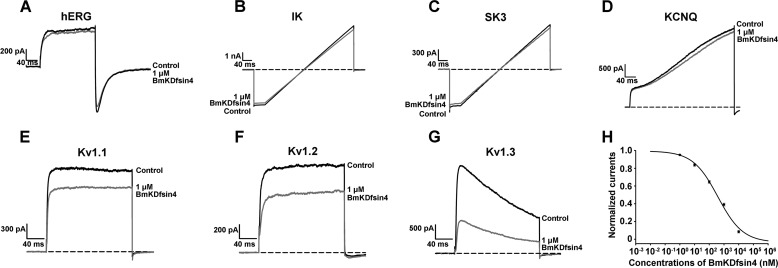
On the basis of the similarity between BmKDfsin4 and potassium channel-blocking scorpion neurotoxins, we evaluated the activity of BmKDfsin4 on various potassium channels. After transfection and expression of various potassium channels in HEK293 cells, electrophysiological experiments indicated that 1 μm rBmKDfsin4 could inhibit about 11.6%, 3.9%, 7.0%, 5.9%, 25.2%, 30.5%, and 61.0% of the potassium currents mediated by the hERG, IK, SK3, KCNQ, Kv1.1, Kv1.2, and Kv1.3 channels, respectively (Fig. 3, A–G). On the basis of the potassium channel-blocking profiles of rBmKDfsin4, concentration-dependent experiments were conducted. As shown in Fig. 3H, rBmKDfsin4 blocked hKv1.3 channel currents with an IC50 value of 510.2 ± 161.2 nm. These data not only demonstrate that BmKDfsin4 is the first potassium channel blocker from scorpion defensins but also show that this defensin can target different potassium channel subtypes.
FIGURE 3.
The BmKDfsin4 interaction with potassium channels. A–G, representative current traces in the absence (control) or presence of 1 μm BmKDfsin4 on the currents from the potassium channels hERG, IK, SK3, KCNQ, Kv1.1, Kv1.2, and Kv1.3. At 1 μm, BmKDfsin4 blocked 11.6% ± 1.2% of ERG, 3.9% ± 0.3% of IK, 7.0% ± 0.6% of SK3, 5.9% ± 0.5% of KCNQ, 25.2% ± 1.9% of Kv1.1, 30.5% ± 1.1% of Kv1.2, and 61.0% ± 1.6% of Kv1.3 currents. The Kv1.1 channel is from mice, and the other channels are from humans. The black and red lines represent the currents measured in the absence (control) and presence of BmKDfsin4, respectively. H, average normalized hKv1.3 channel current inhibition by various concentrations of BmKDfsin4. The IC50 was 510.2 ± 161.2 nm. Each channel was tested at least three times (n ≥ 3). The results are shown as mean ± S.E.
Basic Residues Play Critical Roles in BmKDfsin4 Affinity
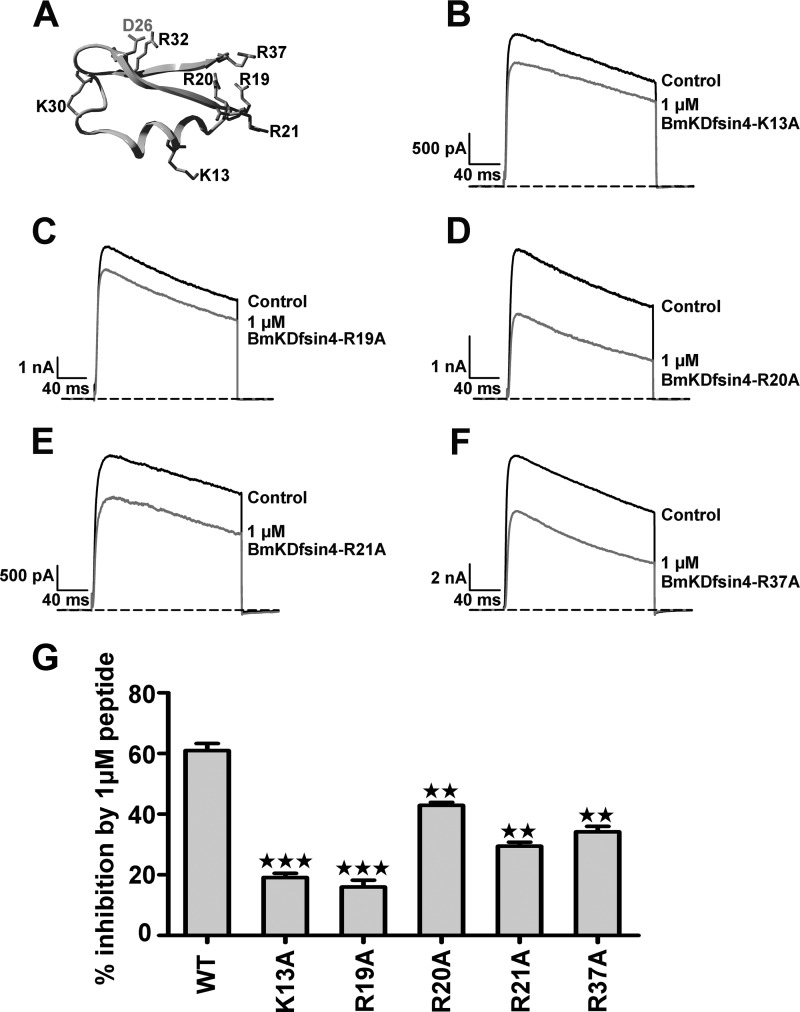
It is well known that basic residues are essential for animal toxin affinities because of the dominant electrostatic interactions between the basic residues in scorpion toxins and acidic residues in potassium channels (18, 19, 24). On the basis of the evolutionary function of acidic residues in orienting the toxin binding interface (8, 25), the channel affinity effects of five basic residues (Lys-13, Arg-19, Arg-20, Arg-21, and Arg-37), which are physically far from Asp-26 of BmKDfsin4, were investigated (Fig. 4A). As shown in Fig. 4, B–G, mutagenesis and electrophysiological experiments indicated that these basic residues played different roles in inhibiting Kv1.3 channel currents. In comparison with the ∼61% inhibition of wild-type Kv1.3 channel currents by 1 μm rBmKDfsin4, 1 μm rBmKDfsin4-K13A and rBmKDfsin4-R19A mutants blocked about 19.1% and 16.0% of Kv1.3 channel currents, respectively, demonstrating the necessity of Lys-13 and Arg-19 for the BmKDfsin4 affinity for Kv1.3 (Fig. 4, B, C, and G). In addition to Lys-13 and Arg-19 in BmKDfsin4, the Arg-20, Arg-21, and Arg-37 residues moderately affected rBmKDfsin4 activity. The corresponding mutants caused inhibitions of about 42.9%, 29.4%, and 34.2%, respectively, at identical concentrations (Fig. 4, D–G). These results clearly reveal that the positively charged molecular surface of BmKDfsin4, similar to classical animal toxins, is used to recognize the potassium channel.
FIGURE 4.
The effect of basic residues on the binding affinity of BmKDfsin4. A, the basic residue distribution in the three-dimensional structure of BmKDfsin4. B–F, at 1 μm, BmKDfsin4-K13A, R19A, R20A, R21A, and R37A cause inhibitions of 19.1% ± 1.4%, 16.0% ± 2.3%, 42.9% ± 1.0%, 29.4% ± 1.4%, and 34.2% ± 1.8%, respectively. The black and red lines represent the currents measured in the absence (control) and presence of BmKDfsin4, respectively. G, average inhibition of Kv1.3 channel currents by 1 μm wild-type or mutant BmKDfsin4. The control current amplitude in each experiment was fixed as 1 for the normalized currents, and the inhibition rates were compared. Each channel was tested at least three times (n ≥ 3). The results are shown as mean ± S.E. ★★★, p < 0.001; ★★, p < 0.01).
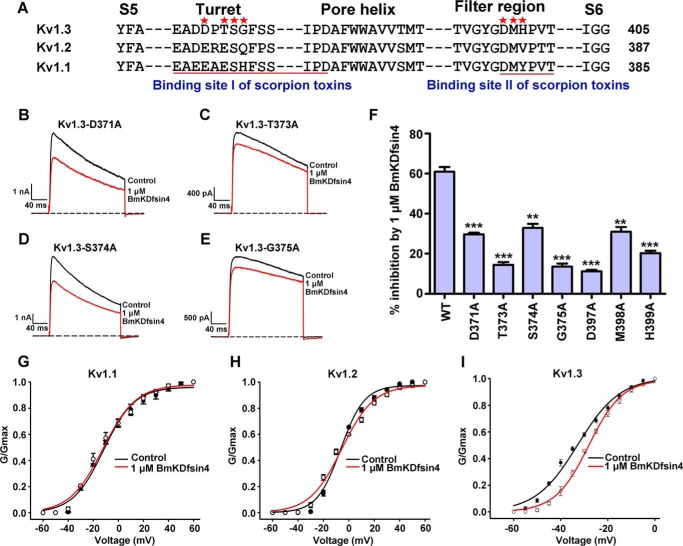
The Extracellular Pore Region of the Kv1.3 Channel Serves as the BmKDfsin4-interacting Interface
Given that the potassium channel vestibule acts as the binding site of scorpion toxin blockers (18, 19, 24) (Fig. 5A), we further investigated whether the Kv1.3 channel extracellular pore region is the BmKDfsin4-interacting interface. As expected, the outer vestibule of the Kv1.3 channel markedly affected BmKDfsin4 binding. In the Kv1.3 channel turret domain, the alanine substitutions of Asp-371, Thr-373, Ser-374 and Gly-375 differentially decreased rBmKDfsin4 affinity. In contrast to the 61% inhibition of wild-type Kv1.3 channel currents by 1 μm rBmKDfsin4, about 29.6%, 14.4%, 32.8%, and 13.5% inhibitions of potassium currents for the Kv1.3-D371A, Kv1.3-T373A, Kv1.3-S374A, and Kv1.3-G375A mutant channels, respectively, were observed for equal rBmKDfsin4 concentrations (Fig. 5, B–F). In addition to the channel turret, the selectivity filter-S6 transmembrane helix linker of the Kv1.3 channel also played an important role in BmKDfsin4 binding. As shown in Fig. 5F, 11.2%, 30.9%, and 20.2% inhibition of potassium currents for Kv1.3-D397A, Kv1.3-M398A, and Kv1.3-H399A, respectively, were observed for treatment with 1 μm rBmKDfsin4. These effects were much smaller than the corresponding inhibition of wild-type Kv1.3 channel currents. In line with the findings of the mutagenesis experiments, there were no significant shifts of the conductance-voltage curves of Kv1.1, Kv1.2, and Kv1.3 channels between the absence and presence of rBmKDfsin4 (Fig. 5, G–I), further indicating that the potassium channel pore region is the binding site of defensin BmKDfsin4. The critical effect of the Kv1.3 channel vestibule on BmKDfsin4 binding affinity clearly demonstrates that BmKDfsin4 and potassium channel-blocking animal toxins have similar binding modes for recognizing potassium channels.
FIGURE 5.
The Kv1.3 channel outer vestibule is responsible for BmKDfsin4 binding. A, multiple sequence alignment of the outer vestibules of the hKv1.3, hKv1.2, and mKv1.1 channels. B–E, representative current traces of the Kv1.3 channel with mutations in the pore region in response to 1 μm BmKDfsin4. At 1 μm, BmKDfsin4 inhibited potassium currents as follows: 29.6% ± 0.8% for the Kv1.3-D371A channel, 14.4% ± 1.4% for the Kv1.3-T373A channel, 32.8% ± 2.0% for the Kv1.3-S374A channel, and 13.5% ± 1.5% for the Kv1.3-G375A channel. The black and red lines represent the currents measured in the absence (control) and presence of BmKDfsin4, respectively. F, average inhibition of wild-type and mutant Kv1.3 channel currents by 1 μm BmKDfsin4. The control current amplitude in each experiment was fixed as 1 for the normalized currents, and the inhibition rates were compared. Each channel was tested at least three times (n ≥ 3). The results are shown as mean ± S.E. (***, p < 0.001; **, p < 0.01. G–I, the conductance-voltage curves from the peak currents were plotted for Kv1.1, Kv1.2, and Kv1.3 in the absence and presence of 1 μm BmKDfsin4. No significant G-V curve shift was observed. The detailed ΔV50 values before and after BmKDfsin4 interacting with Kv1.1, Kv1.2, and Kv1.3 channels were −0.61, 0.82, and 4.62 mV, respectively.
Discussion
The origin of animal toxins through the molecular evolution of defensins has been presumed on the basis of their similarities in gene organization and in protein primary and three-dimensional structures (12–16, 23, 26). Recently, a truncated insect defensin has been reported to block potassium channels (17), but no wild-type potassium channel-blocking defensin from venomous animals has been reported. Recently, our group first reported two kinds of wild-type defensins that could directly inhibit potassium channels. One kind of wild-type defensin is from mammals, such as human β-defensin 2 (hBD2) (27, 28), α-defensin 1 (HNP1), and α-defensin 5 (HD5) (29), and the other kind of wild-type defensin is from fungi, such as plectasin (30). These findings encouraged us to explore whether venomous animal defensins can block potassium channels. Through extensive experiments, we found that the scorpion defensin BmKDfsin4 is a dual-function peptide with antimicrobial and potassium channel-blocking activities. Our work first indicated that a defensin from a venomous animal could block potassium channels. The dual function of scorpion defensin BmKDfsin4 provides a functional link between scorpion neurotoxins and defensins.
BmKDfsin4 Is a typical Antimicrobial Defensin
The scorpion defensin BmKDfsin4 is highly homologous to two scorpion defensins (Lqh def and Androctonus defensin) isolated from scorpion hemolymph (23, 31) (Fig. 1A). Similar to many known defensins, such as Micasin (32), Cg-def (33), and defensin A (34, 35), BmKDfsin4 could effectively and selectively inhibit the growth of Gram-positive bacteria, including some standard and antibiotic-resistant strains, without destroying the integrity of the cell wall (Fig. 2). This antimicrobial mechanism is similar to that of many known ancient invertebrate-type defensins from scorpions, dragonflies, spiders, fungi, and mussels. For example, the fungal defensin plectasin achieves antibacterial activity by targeting the bacterial cell wall precursor Lipid II (36). These common functional and mechanistic features revealed the essence of BmKDfsin4 as a typical defensin against microbes.
BmKDfsin4 Is a Novel Blocker Interacting with the Potassium Channel with a Neurotoxin-like Pore-blocking Mode
In line with the structural similarity between BmKDfsin4 and potassium channel-blocking animal toxins (Fig. 1), BmKDfsin4 is a novel potassium channel blocker with IC50 values of 510.2 ± 161.2 nm for the channel Kv1.3 (Fig. 3). Unlike hBD2 and HNP1 as both blockers and gating modifiers (28, 29), which bind to the Kv1.3 channel more selectively and affect Kv1.3 channel activation, rBmKDfsin4 at a 1 μm concentration could block about 25.2%, 30.5%, and 61.0% of the potassium currents mediated by Kv1.1, Kv1.2, and Kv1.3 channels, respectively (Fig. 3, E–H). Meanwhile, rBmKDfsin4 has less of an effect on the activation of Kv1.1, Kv1.2, and Kv1.3 channels (Fig. 5, G–I), showing a function similar to those of defensin HD5 and plectasin as only potassium channel blockers (29, 30). More interestingly, many scorpion toxins, such as Kaliotoxin (37, 38), Agitoxin 2 (39, 40) and OSK1 (41), simultaneously bind multiple potassium channel subtypes. Such similar potassium channel-blocking affinity profiles in BmKDfsin4 and scorpion toxins experimentally highlight the potential for animal defensins to evolve into neurotoxins targeting various potassium channel subtypes.
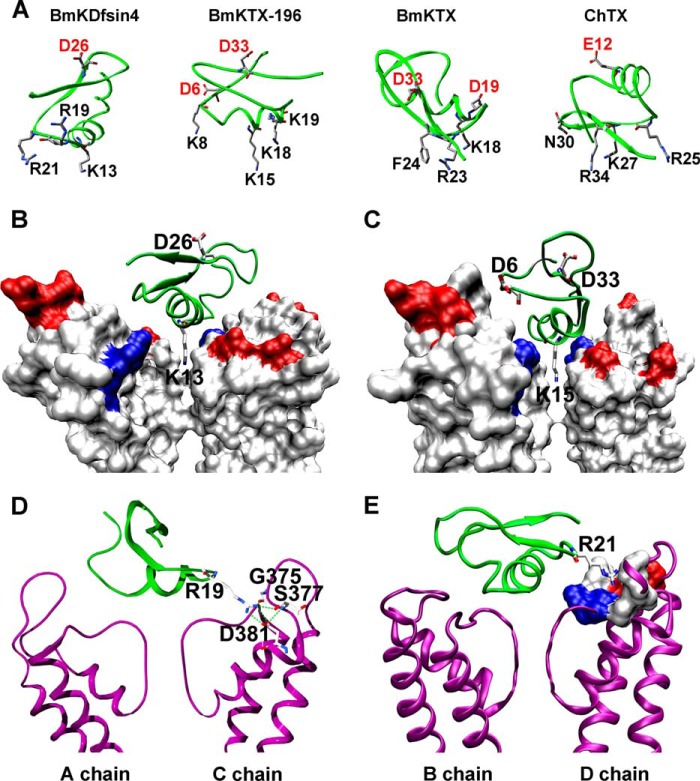
In addition to the similar potassium channel-blocking affinity profiles of BmKDfsin4 and animal toxins (Figs. 3 and 5), BmKDfsin4 was also found to adopt an animal toxin-like mechanism to bind to potassium channels. During the scorpion toxin-potassium channel interactions, the toxin basic residues usually play important roles in binding to the negatively charged potassium channel pore region. As shown in Fig. 6A, the basic residues in the potassium channel-interacting interfaces of the scorpion toxins BmKTX196, BmKTX and ChTX were found to be responsible for their affinities (8, 22). Similar to the functional importance of basic residues in the classical animal toxins, the basic residues in BmKDfsin4 were also used to recognize the negatively charged vestibule of the potassium channel (19, 27) (Figs. 4, 5, and 6A). Such common interaction models are also suggested by their predicted complex structures, which were constructed using our previous computational simulations (Fig. 6, B–E) (8, 18–21, 28) The classical scorpion toxins usually have a critical potassium channel pore-blocking residue (8, 19–21). BmKDfsin4 mainly uses its α helix domain to recognize the Kv1.3 channel. Therefore, the critical Lys-13 in the middle of the defensin α helix domain was predicted to be the channel pore-blocking residue (Fig. 6B). Such an interaction model has also been observed in the interaction between the scorpion toxin analog BmKTX196 and the channel Kv1.3 (22) (Fig. 6C). In addition to Lys-13 in BmKDfsin4, another critical residue, Arg-19, formed strong electrostatic interactions with Gly-375, Ser-377, and Asp-381 through hydrogen bonds and salt bridges (Fig. 6D). In addition, the functional residue Arg-21 was found to be surrounded by a “pocket” formed by many nonpolar and polar residues from the turret and selectivity filter-S6 transmembrane helix linker of the Kv1.3 channel (Fig. 6E). These features, like those of classical scorpion toxins, revealed that BmKDfsin4 is a novel potassium channel blocker.
FIGURE 6.
The interaction model between BmKDfsin4 and the tetrameric Kv1. 3 channel. A, the distribution of acidic residues and main functional residues of BmKDfsin4, BmKTX-196, BmKTX, and ChTX. B, Lys-13 of BmKDfsin4 interacts with residues around the selectivity filter of the Kv1.3 channel. C, Lys-15 of BmKTX196 interacts with residues around the selectivity filter of the Kv1.3 channel (22) D, Arg-19 of BmKDfsin4 interacts with the Kv1.3 channel through hydrogen bonds and salt bridges. E, Arg-21 of BmKDfsin4 interacts with polar and nonpolar residues in the Kv1.3 channel.
Based on the genome of M. martensii (6), evolutionary analysis comparing defensins and neurotoxins indicated that the defensin family and potassium channel-blocking toxin family formed one group, suggesting that they originated from a common ancestor. In view of the longer evolutionary history of defensins that are widely present in fungi, plants, and invertebrate and vertebrate animals (42) and the shorter evolutionary history of the scorpion venom system, which has only emerged from the oldest scorpion fossils dated from the late Silurian period (Proscorpius osborni, 418 million years before present), the dual function of BmKDfsin4 with both antimicrobial and neurotoxin-like activities not only highlights the functional link between scorpion defensins and neurotoxins but also indicates that venomous animal defensins are a new source of potassium channel blockers.
Author Contributions
L. M., Z. X., Q. Z., Y. L., F. Y., Z. Chen, W. L., Z. Cao, and Y. W. read and approved the final manuscript. Y. W., Z. Cao, and Z. Chen conceived and designed the experiments. L. M., Z.X., Q. Z., Y. L., and F. Y. performed the experiments. Z. Cao, W. L., Z. Chen, and Y. W. analyzed the data. Z. Cao and Y. W. wrote the paper.
This work was supported by National Natural Sciences Foundation of China Grants 31470812 and 31170789, National Science Fund for Excellent Young Scholars Grant 31422049, Hubei Science Fund for Excellent Scholars Grant 2015CFA042, and Wuhan University Natural Sciences Foundation Grant 2042015kf0168. The authors declare that they have no conflicts of interest with the contents of this article.
- ChTX
- charybdotoxin
- MIC
- minimum inhibitory concentration
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1. Dunlop J. A., Kamenz C., and Scholtz G. (2007) Reinterpreting the morphology of the Jurassic scorpion Liassoscorpionides. Arthropod Struct. Dev. 36, 245–252 [DOI] [PubMed] [Google Scholar]
- 2. Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., and de la Vega R. C. (2009) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511 [DOI] [PubMed] [Google Scholar]
- 3. Batista C. V., D'Suze G., Gómez-Lagunas F., Zamudio F. Z., Encarnación S., Sevcik C., and Possani L. D. (2006) Proteomic analysis of Tityus discrepans scorpion venom and amino acid sequence of novel toxins. Proteomics 6, 3718–3727 [DOI] [PubMed] [Google Scholar]
- 4. de Oliveira U. C., Candido D. M., Coronado Dorce V. A., and Junqueira-de-Azevedo I. L. (2015) The transcriptome recipe for the venom cocktail of Tityus bahiensis scorpion. Toxicon 95, 52–61 [DOI] [PubMed] [Google Scholar]
- 5. Xu X., Duan Z., Di Z., He Y., Li J., Li Z., Xie C., Zeng X., Cao Z., Wu Y., Liang S., and Li W. (2014) Proteomic analysis of the venom from the scorpion Mesobuthus martensii. J. Proteomics 106, 162–180 [DOI] [PubMed] [Google Scholar]
- 6. Cao Z., Yu Y., Wu Y., Hao P., Di Z., He Y., Chen Z., Yang W., Shen Z., He X., Sheng J., Xu X., Pan B., Feng J., Yang X., Hong W., Zhao W., Li Z., Huang K., Li T., Kong Y., Liu H., Jiang D., Zhang B., Hu J., Hu Y., Wang B., Dai J., Yuan B., Feng Y., Huang W., Xing X., Zhao G., Li X., Li Y., and Li W. (2013) The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 4, 2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bontems F., Roumestand C., Boyot P., Gilquin B., Doljansky Y., Menez A., and Toma F. (1991) Three-dimensional structure of natural charybdotoxin in aqueous solution by 1H-NMR: charybdotoxin possesses a structural motif found in other scorpion toxins. Eur. J. Biochem. 196, 19–28 [DOI] [PubMed] [Google Scholar]
- 8. Chen Z., Hu Y., Hu J., Yang W., Sabatier J. M., De Waard M., Cao Z., Li W., Han S., and Wu Y. (2014) Unusual binding mode of scorpion toxin BmKTX onto potassium channels relies on its distribution of acidic residues. Biochem. Biophys. Res. Commun. 447, 70–76 [DOI] [PubMed] [Google Scholar]
- 9. Mygind P. H., Fischer R. L., Schnorr K. M., Hansen M. T., Sönksen C. P., Ludvigsen S., Raventós D., Buskov S., Christensen B., De Maria L., Taboureau O., Yaver D., Elvig-Jørgensen S. G., Sørensen M. V., Christensen B. E., Kjaerulff S., Frimodt-Moller N., Lehrer R. I., Zasloff M., and Kristensen H. H. (2005) Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 437, 975–980 [DOI] [PubMed] [Google Scholar]
- 10. Thomma B. P., Cammue B. P., and Thevissen K. (2002) Plant defensins. Planta 216, 193–202 [DOI] [PubMed] [Google Scholar]
- 11. Lay F. T., and Anderson M. A. (2005) Defensins: components of the innate immune system in plants. Curr. Protein Pept. Sci. 6, 85–101 [DOI] [PubMed] [Google Scholar]
- 12. Rodríguez de la Vega R. C., and Possani L. D. (2005) On the evolution of invertebrate defensins. Trends Genet. 21, 330–332 [DOI] [PubMed] [Google Scholar]
- 13. Lehrer R. I., and Ganz T. (2002) Defensins of vertebrate animals. Curr. Opin. Immunol. 14, 96–102 [DOI] [PubMed] [Google Scholar]
- 14. Froy O., and Gurevitz M. (2004) Arthropod defensins illuminate the divergence of scorpion neurotoxins. J. Pept. Sci. 10, 714–718 [DOI] [PubMed] [Google Scholar]
- 15. Zhu S., and Tytgat J. (2004) The scorpine family of defensins: gene structure, alternative polyadenylation and fold recognition. Cell. Mol. Life Sci. 61, 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu S., Gao B., and Tytgat J. (2005) Phylogenetic distribution, functional epitopes and evolution of the CSαβ superfamily. Cell. Mol. Life Sci. 62, 2257–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu S., Peigneur S., Gao B., Umetsu Y., Ohki S., and Tytgat J. (2014) Experimental conversion of a defensin into a neurotoxin: implications for origin of toxic function. Mol. Biol. Evol. 31, 546–559 [DOI] [PubMed] [Google Scholar]
- 18. Yi H., Qiu S., Cao Z., Wu Y., and Li W. (2008) Molecular basis of inhibitory peptide maurotoxin recognizing Kv1.2 channel explored by ZDOCK and molecular dynamic simulations. Proteins 70, 844–854 [DOI] [PubMed] [Google Scholar]
- 19. Han S., Yi H., Yin S. J., Chen Z. Y., Liu H., Cao Z. J., Wu Y. L., and Li W. X. (2008) Structural basis of a potent peptide inhibitor designed for Kv1.3 channel, a therapeutic target of autoimmune disease. J. Biol. Chem. 283, 19058–19065 [DOI] [PubMed] [Google Scholar]
- 20. Chen Z. Y., Hu Y. T., Yang W. S., He Y. W., Feng J., Wang B., Zhao R. M., Ding J. P., Cao Z. J., Li W. X., and Wu Y. L. (2012) Hg1, novel peptide inhibitor specific for Kv1.3 channels from first scorpion Kunitz-type potassium channel toxin family. J. Biol. Chem. 287, 13813–13821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Feng J., Hu Y., Yi H., Yin S., Han S., Hu J., Chen Z., Yang W., Cao Z., De Waard M., Sabatier J. M., Li W., and Wu Y. (2013) Two conserved arginine residues from the SK3 potassium channel outer vestibule control selectivity of recognition by scorpion toxins. J. Biol. Chem. 288, 12544–12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Z., Hu Y., Hong J., Hu J., Yang W., Xiang F., Yang F., Xie Z., Cao Z., Li W., Lin D., and Wu Y. (2015) Toxin acidic residue evolutionary function-guided design of de novo peptide drugs for the immunotherapeutic target, the Kv1.3 channel. Sci. Rep. 5, 9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cociancich S., Goyffon M., Bontems F., Bulet P., Bouet F., Menez A., and Hoffmann J. (1993) Purification and characterization of a scorpion defensin, a 4 kDa antibacterial peptide presenting structural similarities with insect defensins and scorpion toxins. Biochem. Biophys. Res. Commun. 194, 17–22 [DOI] [PubMed] [Google Scholar]
- 24. Gan G., Yi H., Chen M., Sun L., Li W., Wu Y., and Ding J. (2008) Structural basis for toxin resistance of β4-associated calcium-activated potassium (BK) channels. J. Biol. Chem. 283, 24177–24184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Han S., Yin S., Yi H., Mouhat S., Qiu S., Cao Z., Sabatier J. M., Wu Y., and Li W. (2010) Protein-protein recognition control by modulating electrostatic interactions. J. Proteome Res. 9, 3118–3125 [DOI] [PubMed] [Google Scholar]
- 26. Goyffon M., and Landon C. (1998) Scorpion toxins and defensins. C. R. Seances Soc. Biol. Fil. 192, 445–462 [PubMed] [Google Scholar]
- 27. Yang W., Feng J., Xiang F., Xie Z., Zhang G., Sabatier J. M., Cao Z., Li W., Chen Z., and Wu Y. (2015) Endogenous animal toxin-like human β-defensin 2 inhibits own K+ channels through interaction with channel extracellular pore region. Cell. Mol. Life Sci. 72, 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feng J., Yang W., Xie Z., Xiang F., Cao Z., Li W., Hu H., Chen Z., and Wu Y. (2015) Kv channel S1-S2 linker working as a binding site of human β-Defensin 2 for channel activation modulation. J. Biol. Chem. 290, 15487–15495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xie Z., Feng J., Yang W., Xiang F., Yang F., Zhao Y., Cao Z., Li W., Chen Z., and Wu Y. (2015) Human α-defensins are immune-related Kv1.3 channel inhibitors: new support for their roles in adaptive immunity. FASEB J. 29, 4324–4333 [DOI] [PubMed] [Google Scholar]
- 30. Xiang F., Xie Z., Feng J., Yang W., Cao Z., Li W., Chen Z., and Wu Y. (2015) Plectasin, first animal toxin-like fungal defensin blocking potassium channels through recognizing channel pore region. Toxins 7, 34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ehret-Sabatier L., Loew D., Goyffon M., Fehlbaum P., Hoffmann J. A., van Dorsselaer A., and Bulet P. (1996) Characterization of novel cysteine-rich antimicrobial peptides from scorpion blood. J. Biol. Chem. 271, 29537–29544 [DOI] [PubMed] [Google Scholar]
- 32. Zhu S., Gao B., Harvey P. J., and Craik D. J. (2012) Dermatophytic defensin with antiinfective potential. Proc. Natl. Acad. Sci. U.S.A. 109, 8495–8500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gueguen Y., Herpin A., Aumelas A., Garnier J., Fievet J., Escoubas J. M., Bulet P., Gonzalez M., Lelong C., Favrel P., and Bachère E. (2006) Characterization of a defensin from the oyster Crassostrea gigas: recombinant production, folding, solution structure, antimicrobial activities, and gene expression. J. Biol. Chem. 281, 313–323 [DOI] [PubMed] [Google Scholar]
- 34. Chalk R., Albuquerque C. M., Ham P. J., and Townson H. (1995) Full sequence and characterization of two insect defensins: immune peptides from the mosquito Aedes aegypti. Proc Biol Sci 261, 217–221 [DOI] [PubMed] [Google Scholar]
- 35. Maget-Dana R., and Ptak M. (1997) Penetration of the insect defensin A into phospholipid monolayers and formation of defensin A-lipid complexes. Biophys. J. 73, 2527–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider T., Kruse T., Wimmer R., Wiedemann I., Sass V., Pag U., Jansen A., Nielsen A. K., Mygind P. H., Raventós D. S., Neve S., Ravn B., Bonvin A. M., De Maria L., Andersen A. S., Gammelgaard L. K., Sahl H. G., and Kristensen H. H. (2010) Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328, 1168–1172 [DOI] [PubMed] [Google Scholar]
- 37. Crest M., Jacquet G., Gola M., Zerrouk H., Benslimane A., Rochat H., Mansuelle P., and Martin-Eauclaire M. F. (1992) Kaliotoxin, a novel peptidyl inhibitor of neuronal BK-type Ca2+-activated K+ channels characterized from Androctonus mauretanicus mauretanicus venom. J. Biol. Chem. 267, 1640–1647 [PubMed] [Google Scholar]
- 38. Romi R., Crest M., Gola M., Sampieri F., Jacquet G., Zerrouk H., Mansuelle P., Sorokine O., Van Dorsselaer A., and Rochat H. (1993) Synthesis and characterization of kaliotoxin: is the 26–32 sequence essential for potassium channel recognition? J. Biol. Chem. 268, 26302–26309 [PubMed] [Google Scholar]
- 39. Garcia M. L., Garcia-Calvo M., Hidalgo P., Lee A., and MacKinnon R. (1994) Purification and characterization of three inhibitors of voltage-dependent K+ channels from Leiurus quinquestriatus var. hebraeus venom. Biochemistry 33, 6834–6839 [DOI] [PubMed] [Google Scholar]
- 40. Felix J. P., Bugianesi R. M., Schmalhofer W. A., Borris R., Goetz M. A., Hensens O. D., Bao J. M., Kayser F., Parsons W. H., Rupprecht K., Garcia M. L., Kaczorowski G. J., and Slaughter R. S. (1999) Identification and biochemical characterization of a novel nortriterpene inhibitor of the human lymphocyte voltage-gated potassium channel, Kv1.3. Biochemistry 38, 4922–4930 [DOI] [PubMed] [Google Scholar]
- 41. Mouhat S., Visan V., Ananthakrishnan S., Wulff H., Andreotti N., Grissmer S., Darbon H., De Waard M., and Sabatier J. M. (2005) K+ channel types targeted by synthetic OSK1, a toxin from Orthochirus scrobiculosus scorpion venom. Biochem. J. 385, 95–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ng T. B., Cheung R. C., Wong J. H., and Ye X. J. (2013) Antimicrobial activity of defensins and defensin-like peptides with special emphasis on those from fungi and invertebrate animals. Curr. Protein Pept. Sci. 14, 515–531 [DOI] [PubMed] [Google Scholar]