Abstract
Mammalian Argonaute proteins (AGO1–4), in combination with microRNAs (miRs), bind to target mRNAs to initiate degradation and/or translation repression, but the relationships between these two effects is unclear. Although the AGO isoforms of Drosophila and plants perform different functions, mammalian AGO isoforms are considered to be functionally degenerate in terms of miR loading and downstream silencing effects. However, we found that, in quiescent (G0) rat myoblasts transiting to the G1 phase, cyclin D1 (Ccnd1) mRNA was associated with two functionally distinct AGO-miR complexes. While AGO1-miR-1 down-regulated the mRNA level, AGO2-let-7 delayed the timing of translation. Knockdown (KD) of AGO2, or antisense-mediated depletion of Let-7, caused Ccnd1 translation to occur earlier, but had no significant effect on mRNA abundance. Conversely, down-regulation of either AGO1 or miR-1, resulted in elevated Ccnd1 mRNA levels at early times, but failed to affect the timing of translation. These results show that the two miR-mediated silencing effects, viz. mRNA decay and translation repression, are independent processes induced by individual AGO isoforms in association with specific miRs.
Keywords: Argonaute, cyclin D1, microRNA (miRNA), mRNA, myogenesis
Introduction
The regulation of translation of cellular mRNAs may be complex, involving multiple RNA or protein regulators acting in a cell-specific and/or time-dependent manner. The role of various RNA-binding proteins (RBPs)3 and of microRNAs (miRNAs) as translation regulators has been highlighted (1, 2). In many cases the regulatory region of the mRNA (usually the 3′-untranslated region or 3′ UTR) contains both RBP and miR binding sites, making it difficult to ascertain the relative importance of the individual interactions in a specific physiological context.
The miRs pair with their mRNA targets in association with an Argonaute (AGO) protein present in RNA Induced Silencing Complexes (RISC). In mammalian cells, there are 4 AGO isoforms (AGO1–4) which are considered to be functionally redundant as far as loading of miRs is concerned; thus, immunoprecipitates with antibodies against individual isoforms contain nearly identical spectra of miRs (3–5). However, there are exceptions; miR-451 is exclusively loaded on to, and processed by AGO2 in a Dicer independent manner (6). Moreover, non-miR small RNAs have been found to be specifically associated with AGO1 (7). Given that miR loading on mammalian AGO is generally not isoform specific, the question remains whether the isoforms interact specifically with their mRNA targets. It is unclear whether any mRNA may associate simultaneously with multiple AGO isoforms, and if so, whether individual isoforms have specific roles in post-transcriptional regulation of the target mRNA.
In a variety of experimental systems, miRs have been shown to induce specific effects on mRNA levels (decay), polyadenylation (deadenylation) and translation (repression), but there are conflicting interpretations of the relative contributions of these effects on gene silencing. While ribosome profiling in human cells indicated that the effects of miR-1 and miR-155 could be accounted for by mRNA decay (8), kinetic analyses indicated that translation repression by Let-7 and miR-21 in human cells (9), miR-279, miR-6b, or bantam in Drosophila cells (10), or miR-430 in developing zebrafish embryos (11) precede mRNA decay. It is unknown whether different miRs induce recruitment of different effector proteins for decay or repression through functionally distinct RISC components.
During myogenesis in post-injury skeletal muscle, satellite stem cells are activated by passage from the quiescent state (G0) to the G1 phase, forming myoblasts that undergo rounds of proliferation (12, 13). We previously observed that during this process, cyclin D1 (Ccnd1), a regulator of the G1 phase, is post-transcriptionally regulated by the ATP sensor mTORC1 complex acting through miR-1 and miR-26a (14). In glioblasoma cells (15) and in Respiratory Syncytial Virus (RSV) infected alveolar epithelial cells (16), Ccnd1 is regulated by Let-7 family members. The repression of Ccnd1 protein by miR-1 in mouse myoblasts has also been reported (17). In these cases, the predominant effect of the miR on decay versus repression is unclear. In tumor cells, Ccnd1 is inhibited by the mTORC1-regulated 4EBP1 (18), which binds to the 5′-cap binding protein eIF4E and blocks translation initiation. How these various miRs, as well as 4EBP1, contribute to the abundance and/or translation of Ccnd1 mRNA during passage of myoblasts or other cells through the G1 phase remains unknown.
In this report, we have investigated the kinetics of RBP and miR interaction with Ccnd1 mRNA in quiescent rat myoblasts stimulated by serum. We observed specific associations of AGO1 and AGO2, bound to different miRs, with repressed Ccnd1 mRNA, and showed that these AGO-miR complexes have independent effects on Ccnd1 mRNA levels and on the timing of Ccnd1 translation. Our results highlight the complexity of post-transcriptional effects on a single mRNA target by multiple miRs.
Experimental Procedures
Cell Culture
L6 rat myoblast cells were cultured using Dulbecco's Modified Eagle Medium (DMEM, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen). Cells were always grown up to 70–80% confluency. Cells were starved in DMEM lacking FBS for 24 h, then exposed to DMEM-10% FBS for various times. Where indicated, 50 nm rapamycin (Sigma) was added during serum stimulation.
Bioinformatic Analysis
The rat 3′ UTR sequence was obtained from NCBI (Accession No. 171992). miRNA sequences were obtained from www.mirbase.org. miRNA targets were predicted using miRNA-Target Gene Prediction at EMBL and also verified manually, using the criterion of 7 or more contiguous base pairs starting at position +1 to +4 of the miRNA (19).
RNA Cross-linking Immunoprecipitation (R-CLIP)
R-CLIP assay was done as previously described (20), with modifications. Briefly, harvested cells were cross-linked in phosphate-buffered saline (PBS), pH 7.0 containing 1% formaldehyde (Qualigens) for 10 min. The cross-linking reaction was quenched by adding 0.25 m glycine, pH 7.0. Cells were washed and resuspended in 200 μl of low stringency RIPA buffer (50 mm Tris-Cl, pH 6.5, 1% Nonidet P-40, 0.5% sodium deoxycholate (DOC), 0.05% SDS, 1 mm EDTA, 150 mm NaCl), and sonicated 3 times, 20 s each at 8–9 Watt output. Then RNase Out (20 units, Invitrogen) was added, and the lysate (200 μl) was pre-cleared using protein A-Sepharose beads (Amersham Biosciences) (40 μl of 1:1 suspension containing 20 units of RNase Out) and 100 μg/ml competitor tRNA (Sigma). Antibody-coated beads were prepared separately by rotating protein A-Sepharose beads (50 μl of 1:1 suspension containing 20 units of RNase Out) and 1–2 μl of desired antibody (supplemental Table S1) for 2 h at 4 °C, followed by extensive washing (5–6 times) with low stringency RIPA buffer and resuspension in 25 μl of the same buffer. The pre-cleared lysate (50 μl) and antibody-coated beads (50 μl) were mixed and rotated for 90 min at 4 °C, and the beads collected by centrifugation at 6000 × g for 1 min. The immunoprecipitate was washed 5–6 times at 4 °C with high stringency RIPA buffer (50 mm Tris-Cl, pH 7.4, 1% Nonidet P-40, 1% DOC, 0.1% SDS, 1 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride (PMSF), 1 m NaCl), resuspended in 100 μl of Resuspension buffer (50 mm Tris-Cl, pH 7.4, 5 mm EDTA, 1% SDS, 10 mm DTT), and incubated at 70 °C for 45 min to remove the cross-links, followed by centrifugation to remove the beads. The supernatant (100 μl) was saved for analysis or sequential IP. For sequential IP, the cross-linked lysate was immunoprecipitated with the first antibody as above; the supernatant (100 μl) was then subjected to a second IP with a different antibody. In each case, the solubilized pellet or supernatant was divided into two equal parts for preparation of RNA or protein.
Preparation of RNA
Total RNA was extracted from cells or immunoprecipitates (containing 100 μg/ml tRNA) using Trizol (Invitrogen), as per the manufacturer's instructions. The final RNA pellet was dissolved in 20 μl of nuclease-free water (Invitrogen).
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
The rat Ccnd1 reverse primer (5 pmol, see below) was annealed to RNA (3 μl) in 5 μl containing 1.25 mm of each dNTP at 65 °C for 5 min and then chilled on ice for at least 2 min. Then 10 mm MgCl2, 1× RT buffer, 10 mm dithiothreitol, 20 units of RNase Out, and 100 units of Superscript III (Invitrogen) were added in a total volume of 10 μl and incubated at 50 °C for 50 min. The reaction was stopped by heating at 85 °C for 5 min and chilling on ice. Then 1 unit of RNase H (Invitrogen) was added and incubated at 37 °C for 20 min. The cDNA (2 μl) was PCR-amplified with Platinum Supermix (23 μl, Invitrogen) in 25 μl, containing 12.5 pmol of each of the following Ccnd1 primers (supplemental Table S2): forward O-401 and reverse O-402, spanning positions +982 to +1394, or forward O-390 and reverse O-405, spanning positions +889 to+1119, of the Ccnd1 3′ UTR (supplemental Table S2). PCR was performed in a thermal cycler (model Veriti, Applied Biosystems), for 35 cycles with 30 s denaturation at 94 °C, 30 s annealing at 55 °C, and 30 s extension at 72 °C. Real time PCR was performed in 20 μl using Roche LightCycler 96. Each reaction contained 1 μl of the cDNA as isolated above, 3 pmol each of forward and reverse primer and 10 μl of 2× SYBR Premix Ex TaqII (Takara). The reaction conditions were: Preincubation: 95 °C for 100 s; Amplification: 95 °C for 10 s, 55 °C for 20 s, 72 °C for 25 s; Melting: 95 °C for 10 s, 65 °C for 60 s, 97 °C for 1 s; Cooling: 37 °C for 30 s; Cycles: 40. From the critical threshold (Ct) values of input (In) and immunoprecipitated (IP) samples, the % recovery (i.e. fraction of total mRNA that was bound to the protein) was calculated as 100 × 2Ct(In) − Ct(IP). For quantification of cellular mRNA at different times of serum stimulation, real time PCR was performed in triplicate using Ccnd1 (target) and GAPDH (reference) primers and the Ct values determined. At each time point, ΔCt = Ct(target) − Ct(reference). The fold change of a treatment (with siRNA or LNA) with respect to control is 2ΔΔCt, where ΔΔCt = ΔCt(treated) − ΔCt(control).
Northern Blotting
Northern blots of RNA were probed with 32P-labeled gene specific antisense primers (supplemental Table S2). For quantitative comparisons one-fifth of each RNA sample from a single 6-well plate at 80% confluency (∼1 × 105 cells) was hybridized with 5′-labeled probe preparations of identical specific activity, and the blots were exposed under identical conditions (5 h at −80 °C with intensifying screen). Longer exposures resulted in loss of proportionality of the signals. Band intensities were estimated as mean luminosity of bands in the inverted image using the histogram option of Adobe Photoshop 7 software, after background subtraction. The intensity profiles of control samples obtained thus (Figs. 3 and 4) were highly superimposable (data not shown).
FIGURE 3.
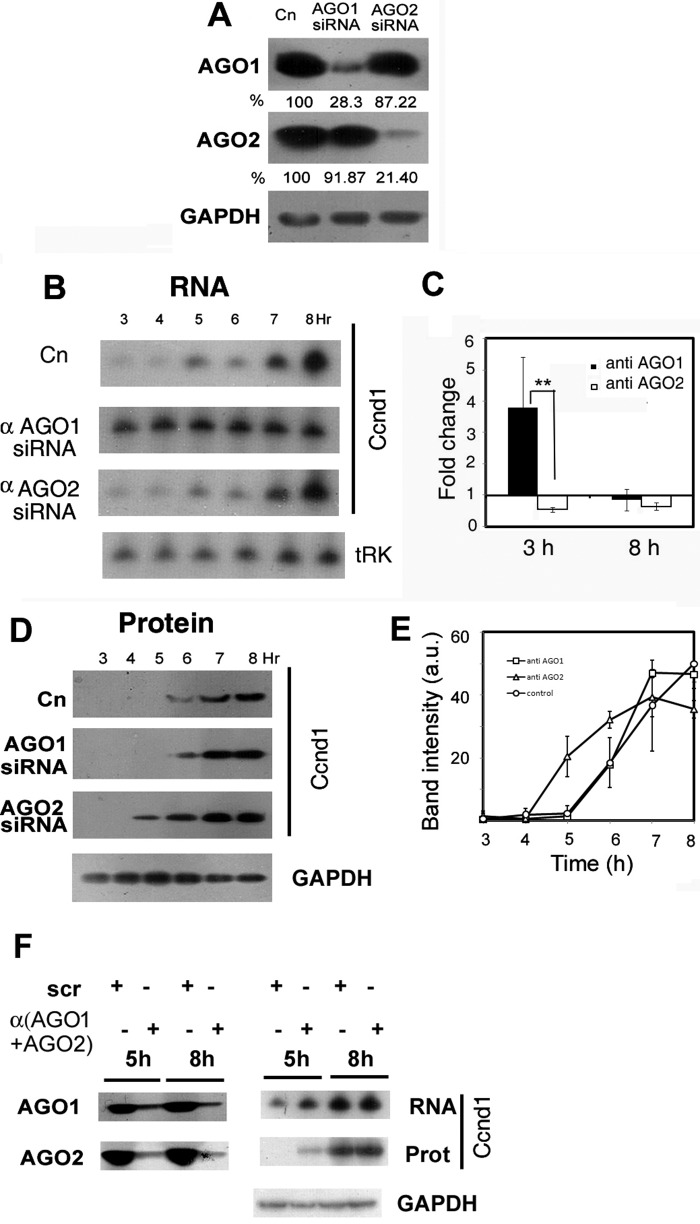
Effect of knockdown of AGO isoforms on Ccnd1 expression. Normal or anti-AGO1 or anti-AGO2 siRNA-treated L6 cells were starved and serum-stimulated for the indicated times. A, specificity and efficiency of siRNA-mediated knockdown. Western blots of 3-h stimulated L6 extracts of untreated or siRNA-treated cells probed with the indicated antibody; numbers represent the AGO1 or AGO2 levels compared with the untreated control (Cn). B, Northern blot of RNA from control or siRNA-treated cells probed for Ccnd1 or tRNALys (tRK). C, fold change of Ccnd1 mRNA in siRNA-treated over untreated cells at 3 h or 8 h of serum stimulation, estimated by real time PCR. D, Western blots analyzing Ccnd1 protein in normal or siRNA-treated cells. E, quantification of the band intensities of triplicate blots as in panel D. F, effect of double knockdown of AGO1 and AGO2 (left) on Ccnd1 mRNA (top right) and protein (middle right) at 5 and 8 h of serum stimulation.
FIGURE 4.
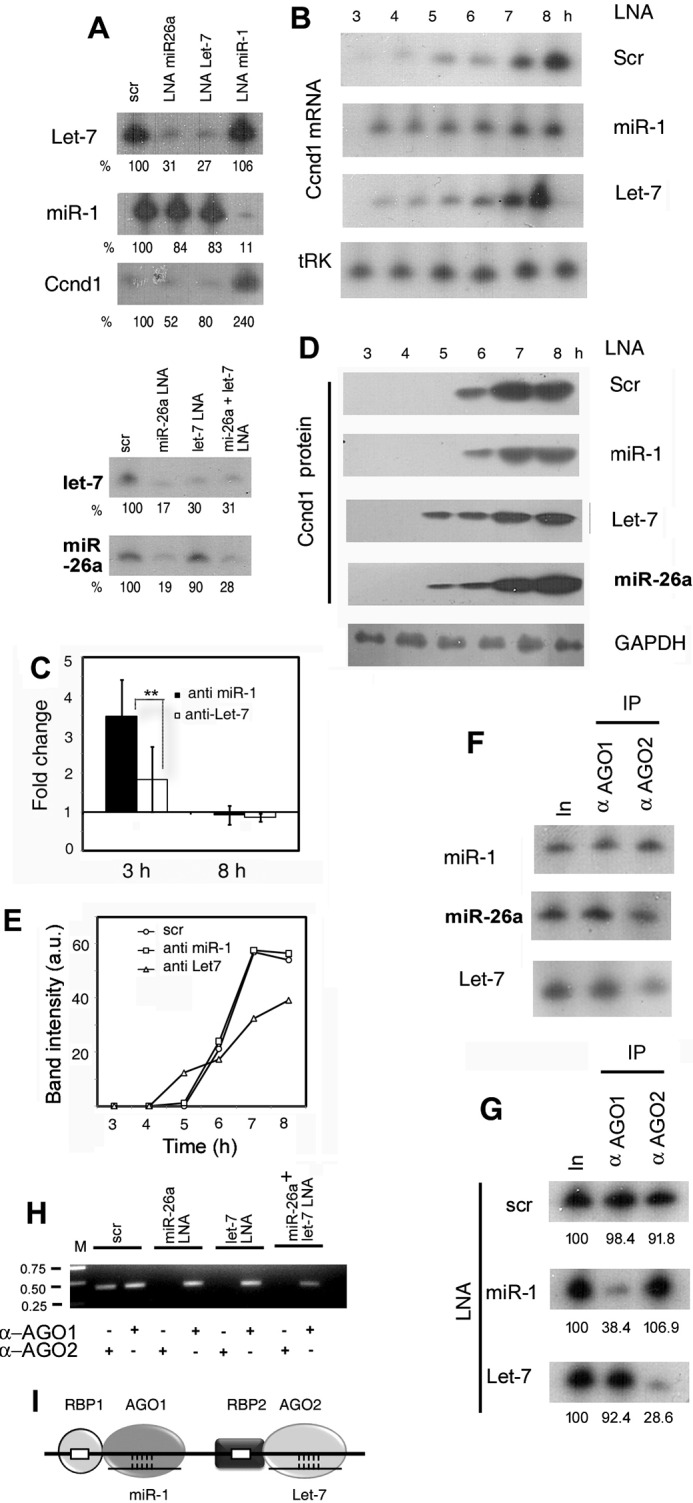
Effect of antagomirs on Ccnd1 expression and AGO-mRNA association. A, specificity and efficiency of antagomir action. Northern blots of 4-h serum stimulated L6 cells treated with LNAs of scrambled-sequence (scr), or targeted against the indicated miR, probed with the specified antisense oligonucleotide. The top and bottom panels are from two independent experiments. B, time course of Ccnd1 RNA in cells treated with control or targeted antagomirs. C, fold change of Ccnd1 mRNA in targeted LNA-treated versus scrambled LNA-treated cells at 3 and 8 h post-serum addition. D and E, time course of Ccnd1 protein expression of LNA-treated cells. The loading control (tRNALys or tRK) for AGO2 siRNA-treated cells is shown at the bottom. F, binding of AGO isoforms to miRs. Immunoprecipitates of AGO1 or AGO2 were probed for indicated miRs by Northern blot. G and H, effect of LNA treatment on AGO-mRNA binding. R-CLIP assay of extracts of L6 cells transfected with indicated LNA antagomirs and serum stimulated for 4 h, using anti-AGO1 or anti-AGO2 antibody. The two panels show independent experiments. In, input RNA. G, Northern blot probed for Ccnd1; (H) RT PCR using Ccnd1 primers. I, postulated role of RBP in positioning of miR-AGO complexes on mRNA. The AGO isoforms interact with distinct RBPs, which have binding sites on the mRNA in close proximity to the miR target site.
Western Blotting
Equal volumes of cell lysate and 2× SDS-PAGE sample loading buffer (100 mm Tris, pH 7.5, 2% SDS, 20% glycerol, 0.02% bromphenol blue, and 1 mm β-mercaptoethanol) were mixed and resolved by 10% SDS- PAGE. Proteins were then transferred to Hybond C membrane (Amersham Biosciences) using a Small Tank transfer unit (Hoefer) at 46 mA for 3 h. The membrane was blocked in Tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% bovine serum albumin (BSA) and incubated with primary antibody (1:1000 in TBS-0,01% Tween 20–0.5% BSA; supplemental Table S1) overnight at 4 °C. The membrane was washed six times in TBS-0.01% Tween 20 (TBST) and incubated with HRP-conjugated secondary antibody (1: 4000 in TBS − 0,01% Tween 20 − 0.5% BSA) for 1 h at room temperature. This was followed by six more washes with TBST. Protein bands were visualized using ECL detection reagent (GE Healthcare).
RNA Interference
All treatments were done as recommended by the manufacturer. Briefly, Accell (set of 4 upgrade) small interfering (si) RNAs against rat AGO1 or AGO2 (Dharmacon) was resuspended in 1× siRNA buffer (Dharmacon). The cell culture medium was replaced with 2 ml of Accell siRNA Delivery Medium (Dharmacon) and siRNA (1 μm) was added to ∼1 × 105 cells in each well of a 6-well plate. Cells were incubated in 37 °C in CO2 incubator for 48 h. This was followed by serum starvation and re-stimulation as described above.
Antagomir Treatment
Lipofectamine-2000 (3 μl of 1 mg/ml, Invitrogen) and 3 μl of 50 nm Locked Nucleic Acid (LNA, Exiqon) (supplemental Table S3) were mixed in serum-free DMEM, incubated at room temperature for 5 min. and added to each well of a 6-well plate containing serum-free DMEM. Cells were incubated for 24 h at 37 °C, followed by serum stimulation as above.
Statistical Analysis
For multiple independent determinations (n ≥ 3, as indicated), the fold change or percent recovery was expressed as mean ± S.D. p values were obtained by paired one-tailed Student's t test using Microsoft Excel.
Results
Delay in Ccnd1 Translation during Serum Stimulation of Rat Myoblasts
Cultured rat myoblasts were serum-starved for 24 h and then cultured for various times in serum-containing medium. Under these conditions, the cells traverse the G1 phase at 8 h, the S phase at 12 h and mitosis at 16 h, and continue into the next cycle with a cycling time (G1-G1) of about 16 h (data not shown). A low level of Ccnd1 mRNA was maintained between 3 and 4 h, then increased linearly between 4 and 8 h (Fig. 1A and C). In contrast, expression of Ccnd1 protein occurred after a lag of 5 h (Fig. 1, B and C). This indicates translation repression of Ccnd1 mRNA between 3 and 5 h of serum stimulation.
FIGURE 1.
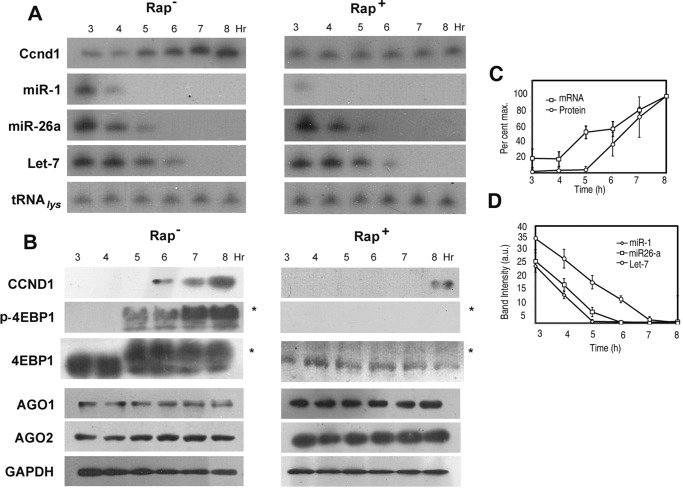
Expression of Ccnd1 and miRNAs during serum stimulation. Serum-starved L6 myoblasts were stimulated with serum for indicated times. A, Northern, and B, Western blots probed with antisense oligonucleotide (A) or antibody (B), respectively, against the indicated RNA (1 × 105 cell-equivalent) or protein (4 × 104 cell-equivalent). *, phospho-form of 4EBP1, migrating slower than the dephospho-form. C, quantified band intensities of Ccnd1 mRNA and protein between 3 and 8 h of serum stimulation, represented as percent of the maximum value. D, parallel down-regulation of miR-1, miR-26a, and Let-7 in serum-stimulated cells. Northern blot band intensities (arbitrary units) were quantified by densitometry.
In serum-stimulated myoblasts, the mTORC1 substrate 4EBP1 was phosphorylated after 4 h, preceding the expression of Ccnd1 (Fig. 1B); the total amount of 4EBP1 was unchanged during this time (Fig. 1B). Ccnd1 protein levels were clearly inhibited by the mTORC1 inhibitor rapamycin (Fig. 1B). Additionally, rapamycin inhibited the up-regulation of Ccnd1 mRNA between 3 and 8 h (Fig. 1A). These results show that mTORC1 controls Ccnd1 expression at multiple post-transcriptional levels.
Interaction Kinetics of AGO Isoforms with Ccnd1 mRNA
Bioinformatic analysis of the 3′ UTR of rat Ccnd1 mRNA revealed the presence of putative binding sites for a number of miRs (Ref. 14; see Fig. 2A). Of these, miR-1, miR-26a, and Let-7 have been shown to affect Ccnd1 expression (8, 13, 16, 17), indicating their functional interaction with the mRNA through AGO proteins.
FIGURE 2.
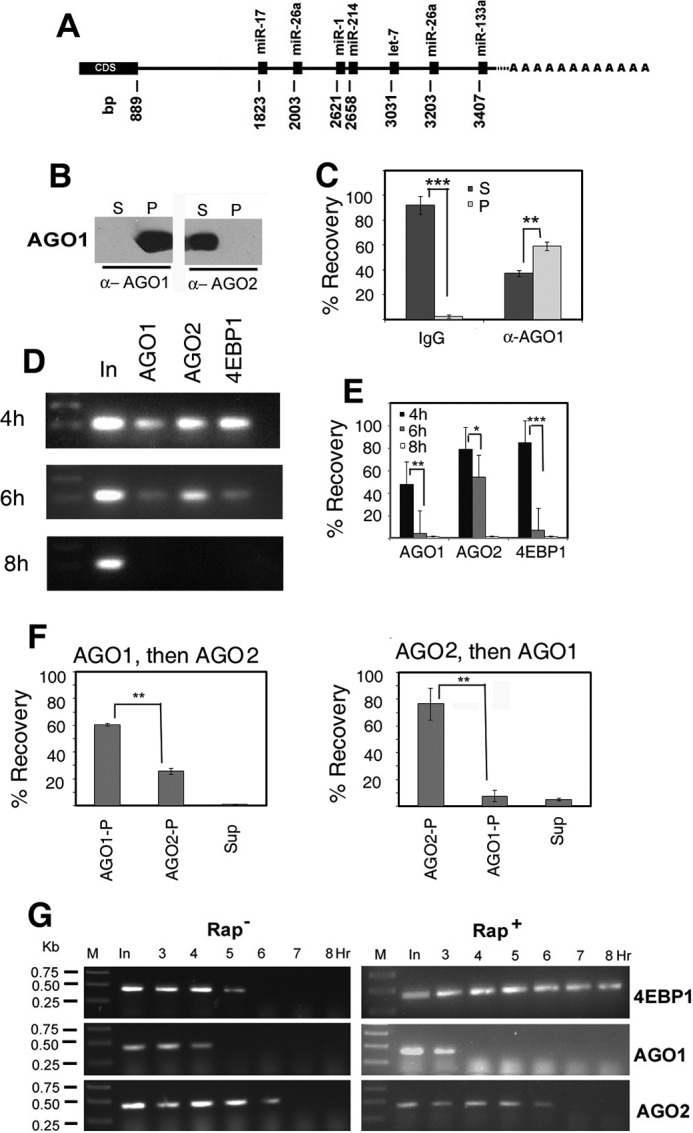
Association of AGO isoforms with Ccnd1 mRNA. A, putative target sights for microRNAs on the 3′ UTR of Ccnd1 mRNA. The positions of the miRNA binding sites (with respect to the AUG start codon at +1) are indicated. cds, coding sequence. B and C, specificity of anti-AGO antibodies. B, an extract from 4-h stimulated L6 cells was treated with protein A-Sepharose beads coated with anti (α) -AGO1 or -AGO2 antibody and after centrifugation, equivalent amounts (4 μl) of the pellet (P) and supernatant (S) were analyzed by Western blot probed with anti-AGO1 or anti-AGO2 antibody. C, 4 h extract was incubated with Sepharose beads coated non-immune IgG (left) or anti-AGO1 antibody (right); RNA was prepared from the pellet (P) or supernatant (S) and equivalent amounts quantified by real time RT PCR as percent input recovered. In this and other figures, p values are indicated by stars: *, p < 0.5; **, p < 0.05; ***, p < 0.005. D and E, cells were serum stimulated for indicated times, and the lysates were subjected to R-CLIP using the specified antibodies. In, input RNA. D, agarose gel electrophoresis of the final PCR products. E, percent recovery calculated on the basis of Ct values by real time PCR (mean +S.D., n = 3–5). F, sequential immunoprecipitation of Ccnd1 mRNA from 4 h lysates and quantification of percent recovery in pellets (P) or final supernatant (S). Left, anti-AGO1 followed by anti-AGO2; right, the order of antibody was reversed. G, RT-PCR amplification of cross-linked and immunoprecipitated (CLIP) RNA from L6 myoblasts serum stimulated for the indicated times in the absence (left) or presence (right) of rapamycin, using antibodies against the proteins shown at the right. In, input RNA from pre-cleared lysate before immunoprecipitation. M, DNA markers of indicated size. Primers O-401 (forward) and O-402 (reverse; supplemental Table S2) were used to yield a 424-bp product.
To detect specific ribonucleoprotein (RNP) complexes formed between Ccnd1 mRNA and AGO isoforms, cross-linked RNPs were subject to immunoprecipitation (R-CLIP) by specific antibodies and the presence of Ccnd1 RNA assayed by real time RT-PCR. The specificities of the antibodies were verified by Western blot analysis of immunoprecipitates; thus, AGO1 was pulled down by anti-AGO1 antibody but not by anti-AGO2 (Fig. 2B). In control experiments, beads coated with non-immune IgG precipitated only 2.67 ± 1.37% of the input Ccnd1 mRNA (Fig. 2C); the remaining 91.9 ± 7.19% was recovered from the supernatant, indicating the absence of significant RNA degradation under the conditions of IP. After 3–4 h of serum stimulation, at which time Ccnd1 is repressed (Fig. 1), Ccnd1 RNPs with the miR-binding proteins AGO1 and AGO2, and the mTORC-regulated repressor 4EBP1, were detected (Fig. 2D). At 4 h, 47.9 + 16.7% (n = 5) of Ccnd1 mRNA was bound to AGO1, while 78 + 11.3% (n = 3) was bound to AGO2 (Fig. 2E). The third protein found to be associated with Ccnd1 mRNA was 4EBP1, which binds to the 5′-cap-binding protein eIF4E but dissociates from it upon mTORC-induced phosphorylation (21). At 4 h of serum stimulation, 84.2 + 11.1% (n = 3) of Ccnd1 mRNA was bound to 4EBP1 (Fig. 2, D and E). By 6 h, both AGO1 and 4EBP1 dissociated almost completely from the mRNA, but AGO2 binding persisted, the high p value indicating a lack of significant difference between 4 and 6 h (Fig. 2E). At 8 h, none of these proteins was significantly bound to Ccnd1 mRNA (Fig. 2E). Thus, at early times, Ccnd1 mRNA was predominantly associated with AGO2 and 4EBP1, and to a lesser extent with AGO1, but the proteins were differentially released from the mRNA at later times. The dissociation of the AGO isofoms was not a result of different abundances of the two proteins, which maintained constant levels during this time period (Fig. 1B), but reflected the abundance of specific miRs (see below). Dissociation of 4EBP1 coincided with its phosphorylation (Fig. 1B).
Since AGO1 and AGO2 are believed to be functionally degenerate (3–5), the association of both these isoforms with Ccnd1 mRNA would imply either that they bind singly but randomly to individual Ccnd1 RNA molecules, or that each mRNA has independent binding sites for each isoform. To distinguish between these possibilities, sequential IPs were performed. When anti-AGO1 was used for the first IP, 60.7 + 0.91% (n = 5) of the RNA was recovered in the pellet; anti-AGO2 pulled down 25.8 + 2.1% (n = 3) from the anti-AGO1 supernatant (Fig. 2F, left panel). When the order of addition of the two antibodies was reversed, 76.6 + 11.9% (n = 3) of the RNA was precipitated by anti-AGO2 antibody, and 7.67 + 4.42% (n = 3) by anti-AGO1 (Fig. 2D, right panel). Taken together, this indicates that at 4 h, ∼50% of Ccnd1 mRNA forms a heteromeric complex with both AGO1 and AGO2, while ∼8 and ∼25% bind to AGO1 or AGO2, respectively. The results are incompatible with random binding of one or two AGO molecules to the mRNA, since in that case the two sequential IPs would yield identical recoveries irrespective of the specificity of the antibody used (62 and 38%, respectively).
Using R-CLIP, the effect of rapamycin on the kinetics of association of the three proteins with Ccnd1 mRNA could be examined (Fig. 2G). In presence of rapamycin, the association of 4EBP1 was prolonged; AGO1 dissociated faster, while the dissociation kinetics of AGO2 was not affected (Fig. 2G).
Specific Roles of AGO1 and AGO2 in Ccnd1 Expression
Since both AGO1 and AGO2 were associated with Ccnd1 mRNA, it was pertinent to ask which one of them was involved in the regulation of Ccnd1 expression after serum addition. Myoblasts were transfected with siRNAs targeting AGO1 or AGO2, then serum-stimulated. The siRNAs induced specific knockdown (KD) of their respective targets, attesting to their specificity (Fig. 3A). Cell cycle parameters such as the G1-specific Ccnd1 cycle (16 h) or PCNA expression in the S phase were not grossly affected by these perturbations (data not shown). KD of AGO1 resulted in a 3.8 + 1.6 (n = 3)-fold up-regulation of Ccnd1 mRNA at early times (3–4 h), and the mRNA was maintained at a constant level thereafter; whereas in the control cells, the mRNA continued to increase to a high level at late times (Fig. 3, B and C). Thus AGO1 destabilizes Ccnd1 mRNA at early times but also has an effect on the rate of mRNA accumulation. Depletion of AGO1 did not affect the onset of Ccnd1 translation: in control as well as in AGO1-depeleted cells, Ccnd1 protein appeared at 6 h (Fig. 3, D and E).
KD of AGO2 did not have any significant effect on the Ccnd1 mRNA profile (Fig. 3, B and C). In AGO2-depleted cells, the time of onset of Ccnd1 translation was reduced from 6 to 5 h following serum addition (Fig. 3, D and E), but the rate of accumulation of Ccnd1 protein was nearly identical to that in control cells, resulting in a higher level of Ccnd1 by 8 h (Fig. 3E). Thus the two AGO isoforms appear to have distinct effects on the abundance and translation of Ccnd1 mRNA.
Double knockdown of AGO1 and AGO2 produced an additive effect: at early times (5 h) the levels of Ccnd1 mRNA as well as protein were elevated with respect to controls (Fig. 3F). This confirmed the independent effects of the two AGO isoforms on Ccnd1 expression.
Distinct Functions of miR-1 and miR-26a/Let7
Since interaction of AGO proteins with mRNAs is critically dependent on the cognate miR, the time-dependent dissociation of the AGO1 and AGO2 complexes from Ccnd1 mRNA could be limited by the intracellular concentrations of specific miRs during serum stimulation. Indeed, several miRs were downregulated in presence of serum, but with different kinetics: miR-1 was reduced earlier than miR-26a or Let-7 (Fig. 1A). In presence of rapamycin, miR-1 was stabilized, but miR-26a and Let-7 were unaffected (Fig. 1A).
To examine the roles of these miRs in regulating Ccnd1 expression, serum-starved myoblasts were transfected with specific LNA-antisense oligonucleotides, followed by serum stimulation. Anti-miR-1 LNA down-regulated endogenous miR-1 by ∼90% but had no significant effect on non-target miRs such as Let-7 (Fig. 4A). Conversely, anti-Let-7 LNA knocked down the target by ∼70% but did not affect miR-1 or miR-26a (Fig. 4A). However, KD of miR-26a resulted in the simultaneous down-regulation of Let-7 by 70–80% (Fig. 4A). It has been shown that miR-26a up-regulates Let-7 through Lin28 and Zcchc11, which repress Let-7 maturation (22). There are putative miR-1, miR-26a, as well as Let-7 target sites on Ccnd1 mRNA (Fig. 2A). Thus any effect of anti-miR-26a LNA on Ccnd1 expression could be direct or indirect (i.e. through Let-7).
KD of miR-1, but not of miR-26a or Let-7, resulted in a 3–4-fold increase in the Ccnd1 mRNA level at early times (3–4 h) of serum stimulation, and its maintenance at a constant level during the experimental period (Fig. 4, A--C). Anti-miR-1 LNA had no effect on the time of onset or rate of Ccnd1 translation (6 h in control and anti-miR-1-treated samples (Fig. 4, D and E). KD of Let-7 did not significantly affect the Ccnd1 mRNA profile (Fig. 4, B and C). In the presence of anti-Let7 or anti-miR26a LNA, translation onset was advanced from 6 to 5 h (Fig. 4D), but the translation rate for anti-Let7 appeared to be slower than in control samples (Fig. 4E). Double KD of miR-1 and Let-7 resulted in stimulation of Ccnd1 mRNA as well as protein expression at early times (data not shown), indicating that the two miRs act independently.
Formation of Specific AGO-miR Complexes on Ccnd1 mRNA
From the above experiments it was clear that either AGO1 or miR-1 modulates the Ccnd1 mRNA level, while either AGO2 or Let-7 affects the time of onset of Ccnd1 translation, suggesting formation of specific AGO-miR complexes on the mRNA. In immunoprecipitates of AGO1 or AGO2, nearly equivalent amounts of each of the three different miRs were present (Fig. 4F), confirming, as shown previously in other cells (3–5), the absence of specificity of binding of the two AGO isoforms for particular miRs in myoblasts. To identify Ccnd1 mRNA-bound complexes, we performed R-CLIP assays in cells treated with specific antagomirs and serum stimulated for 4 h. In control cells containing scrambled antagomir, complexes of Ccnd1 mRNA with either AGO1 or AGO2 were observed, as expected (Fig. 4G). In cells treated with anti-miR-1 antagomir, the AGO1 complex was reduced by 60%, but the AGO2 complex was unaffected. Conversely, on treatment with anti-Let-7 antagomir, the AGO1 complex was unaffected but the AGO2 complex was reduced by over 70% (Fig. 4G). Antagomir-mediated depletion of Let-7 by 70% (Fig. 4A) resulted in reduction of the AGO2 complex by a comparable amount (72%, Fig. 4G), despite the fact that the intracellular level of miR26a was normal (Fig. 4A). In a separate experiment, depletion of miR26a specifically affected the AGO2 complex, as did double depletion of miR26a and Let-7 (Fig. 4H). Thus, Ago1-miR1 and AGO2-Let7 complexes are independently stabilized on Ccnd1 mRNA; moreover, miR26a does not significantly bind to Ccnd1 mRNA-associated AGO1 or AGO2, but exerts its effect through Let-7.
Discussion
Complex regulatory UTRs in mRNAs may contain multiple RNA and protein-interaction sites that are utilized in a time and cell-dependent sequence as the cell traverses the mitotic cycle. The large complex 3′ UTR of Ccnd1 mRNA, with its multiplicity of putative regulatory sites (Fig. 2), provides an ideal platform for determining the molecular events signaling the entry of myoblasts into the cell cycle. The quasi-synchronous passage of serum-starved G0-phase cells to G1 allowed for precise kinetic analyses of RNA-protein complexes in real time. This approach differs from the traditional cell-based assays using asynchronous cultures that yield time-average data. The kinetic analyses made it possible to correlate the association of AGO isoforms with mRNA with the abundance of the corresponding regulatory miRs, and to observe the effects of molecular perturbations on the timing of Ccnd1 translation.
In mammalian cells, the four AGO isoforms are believed to be functionally redundant due to the fact that each isoform may be loaded by a near-identical spectrum of miRs (3–5; see Fig. 4F). If so, any mRNA-AGO complex should contain multiple AGO isoforms in proportions determined by their relative expression in the particular cell. AGO1 and AGO2 were present at comparable levels in myoblasts (Fig. 1); functional degeneracy would predict the random association of either AGO isoform with a specific mRNA, resulting in the formation of mRNA-AGO complexes containing equivalent amounts of the two isoforms. However, we observed Ccnd1 mRNA subpopulations containing Ago1 and AGO2, or AGO2 alone (Fig. 2), indicating non-random association of the AGO isoforms. Second, the AGO1 and AGO2 complexes dissociated with distinct kinetics, and were differentially affected by rapamycin (Fig. 2); this is incompatible with random binding of the AGO isoforms at two sites on the mRNA, since in that case the dissociation kinetics of AGO1 and AGO2 would be identical. Third, the AGO1 complex was dependent on miR-1, whereas the AGO2 complex contained Let-7, and the two complexes appeared to bind independently of each other (Fig. 4). These results indicate the tethering of AGO1 and AGO2 to specific sites on the mRNA.
The various AGO isoforms associate with Dicer, GW182 (TNRC6A-C in mammals) and other proteins to form RNA-induced silencing complexes (miRISC). Multiple forms of miRISC were observed in human (23, 24) and Drosophila (25) cells. In the mammalian system, immunoprecipitated AGO1 and AGO2 containing complexes are broadly identical in protein content, but there are some differences between the AGO1 and AGO2 complexes with respect to the presence of RNA-binding proteins (RBP); e.g. RBM4, a RBP specifically present in the AGO2 complex was shown to be associated with RNA and required for AGO2-mediated gene silencing (23). RBPs such as HuR (26) and FXR1 (27) have been shown to influence miR-mediated post-transcriptional regulation. Thus, the specific binding of AGO1 and AGO2 on Ccnd1 mRNA, as observed here, could be due to the initial tethering of the appropriate miRISC via an associated RBP on the mRNA, followed by its stabilization through the cognate miR-target interaction (Fig. 4I). Compared with the case where RBP binding does not occur, this would result in an increase in the total concentration of the active silencing complex (with miRNA paired to the target). The mammalian AGO1 and AGO2 proteins are highly homologous along most of their length (86–88% in the PAZ, PIWI, and MID domains, but the N-terminal domain is more divergent (70% identity). Moreover, the N-PAZ region of Drosophila AGO1 has been shown to be involved in target RNA binding (28). Therefore, it is possible that specific interactions of the two AGO isoforms through their divergent N-terminal domains with distinct RBPs results in their tethering to specific sites on the target.
We report here for the first time that specific AGO-miR complexes may have different silencing effects on the same mRNA. Depletion of either AGO1 or miR-1 resulted in early elevation of Ccnd1 mRNA but there was no effect on the onset time of Ccnd1 translation (Figs. 3 and 4); conversely, down-regulation of AGO2 or Let-7 affected the onset of Ccnd1 translation but had no effect on mRNA levels (Figs. 3 and 4). The similar phenotypes of AGO and miRs-depleted cells, coupled with the R-CLIP data from antagomir-treated cells (Fig. 4), strongly implicate the corresponding AGO-miR complexes in specific functions. Our results are in agreement with previous reports, which indicated that in human cells, the silencing effect of miR-1 is primarily at the level of target degradation (8), whereas Let-7 induces translation repression prior to mRNA decay (9). Our current results add further distinction between these two primary effects of miRs. Thus, loss of the mir1-AGO1 complex had two distinct effects on Ccnd1 mRNA: it resulted in up-regulation at early times, but it also led to inhibition of the normal rate of accumulation (Figs. 3 and 4); one of the possible explanations of the effect on transcript accumulation is that the miR1-AGO1 complex up-regulates Ccnd1 transcription. Indeed, AGO1 has been reported to be associated with RNA polymerase II and to bind in close proximity to the transcription start site of a number of cell cycle genes including Ccnd1 (29). Depletion of AGO2 did not affect the rate of translation, but reduced the onset time (Fig. 3). An identical reduction of onset time was observed upon down-regulation of Let-7, reflecting the earlier assembly of a functional translation initiation complex on Ccnd1 mRNA, but the translation rate post-onset was lower than normal (Fig. 4); one possible explanation is that Let-7, in combination with an AGO isoform other than AGO2, regulates targets encoding one or more translation factors.
miR-induced target decay is apparently caused by the recruitment of the deadenylase complex CCR4-NOT to GW182 (TNRC) in miRISC, followed by activation of decapping of the 5′ end of the mRNA and exonucleolytic digestion of the template (30). In Drosophila cells, GW182-independent translation repression was observed (31). The mechanism of miRNA-induced repression remains to be worked out, but could involve the recruitment of a translation repressor. The current thinking is that a single miRISC is capable of mediating both mRNA decay and translation repression; our results imply, instead, that specific miRISC complexes containing different AGO isoforms mediate these two effects through interaction with different effector proteins (RBPs) on mRNA.
In serum-stimulated myoblasts, Ccnd1 mRNA was associated at early times with 4EBP1, presumably through eIF4E bound at the 5′-cap site, but was dissociated as it underwent mTORC1-dependent phosphorylation (Fig. 2). However, dissociation of 4EBP1 (Fig. 2) by 4EBP1 phosphorylation at 5 h (Fig. 1) preceded the onset of CCnd1 translation at 6 h (Figs. 1, 3, 4) due to the presence of the AGO2-Let-7 complex. Thus, depletion of the AGO2-Let 7 complex reduced the onset time from 6 to 5 h (Figs. 3 and 4). Prevention of 4EBP1 dissociation by rapamycin did not affect dissociation of the AGO2 complex (Fig. 2), but this was insufficient to initiate Ccnd1 translation (Fig. 1). This observation has two implications: first, that miR-mediated translation repression occurs subsequent to the assembly of eIF4F at the 5′-cap; and second, that the continued mRNA association of 4EBP1, which depends on the presence of the 5′-cap binding protein eIF4E, in presence of rapamycin implies that the AGO-miR complexes do not induce significant decapping of the mRNA, as suggested (30).
Author Contributions
U. G. performed experiments and analyzed data. S. A. designed experiments; analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Ruksana Chowdhury for Real Time PCR and gel documentation, and Tanay Roy for technical and administrative assistance.
This work was supported by Project BEnD (BSC0206) of CSIR-IICB. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Tables S1–S3.
- RBP
- RNA-binding protein
- miR
- microRNA
- AGO
- Argonaute
- RISC
- RNA Induced Silencing Complexes
- RNP
- ribonucleoprotein
- UTR
- untranslated region
- LNA
- locked nucleic acid
- R-CLIP
- RNA cross-linking immunoprecipitation.
References
- 1. Glisovic T., Bachorik J. L., Yong J., and Dreyfuss G. (2008) RNA-binding proteins and posttranscriptional gene regulation. FEBS Lett. 582, 1977–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fabian M. R., Sonenberg N., and Filipowicz W. (2010) Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 79, 351–379 [DOI] [PubMed] [Google Scholar]
- 3. Burroughs A. M., Ando Y., de Hoon M. J. L., Tomaru Y., Suzuki H., Hayashizaki Y., and Daub C. O. (2011) Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol. 8, 158–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dueck A., Ziegler C., Eichner A., Berezikov E., and Meister G. (2012) microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res. 40, 9850–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang D., Zhang Z., O'Loughlin E., Lee T., Houel S., O'Carroll D., Tarakhovsky A., Ahn N. G., and Yi R. (2012) Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang J. S., Maurin T., Robine N., Rasmussen K. D., Jeffrey K. L., Chandwani R., Papapetrou E. P., Sadelain M., O'Carroll D., and Lai E. C. (2010) Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 15163–15168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yamakawa N., Okuyama K., Ogata J., Kanai A., Helwak A., Takamatsu M., Imadome K., Takakura K., Chanda B., Kurosaki N., Yamamoto H., Ando K., Matsui H., Inaba T., and Kotani A. (2014) Novel functional small RNAs are selectively loaded onto mammalian Ago1. Nucleic Acids Res. 42, 5289–5301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo H., Ingolia N. T., Weissman J. S., and Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Béthune J., Artus-Revel C. G., and Filipowicz W. (2012) Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 13, 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Djuranovic S., Nahvi A., and Green R. (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336, 237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bazzini A. A., Lee M. T., and Giraldez A. J. (2012) Ribosome profiling shows that miR-430 reduces translation on before causing mRNA decay in zebrafish. Science 336, 233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Le Grand F., and Rudnicki M. A. (2007) Skeletal muscle satellite cells and adult myogenesis. Curr. Opin. Cell Biol. 19, 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jash S., and Adhya S. (2012) Induction of muscle regeneration by RNA-mediated mitochondrial restoration. FASEB J. 23, 4187–4197 [DOI] [PubMed] [Google Scholar]
- 14. Jash S., Dhar G., Ghosh U., and Adhya S. (2014) Role of the mTORC1 complex in satellite cell activation by RNA induced mitochondrial restoration: Dual control of cyclin D1 through microRNAs. Mol. Cell Biol. 34, 3594–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo Y., Yan K., Fang J., Qu Q., Zhou M., and Chen F. (2013) Let-7b expression determines response to chemotherapy through the regulation of cyclin D1 in glioblastoma. J. Exp. Clin. Cancer Res. 32, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bakre A., Mitchell P., Coleman J. K., Jones L. P., Saavedra G., Teng M., Tompkins S. M., and Tripp R. A. (2012) Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 93, 2346–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang D., Li X., Chen C., Li Y., Zhao L., Jing Y., Liu W., Wang X., Zhang Y., Xia H., Chang Y., Gao X., Yan J., and Ying H. (2012) Attenuation of p38-mediated miR-1/133 expression facilitates myoblast proliferation during the early stage of muscle regeneration. PLoS ONE 7, e41478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Averous J., Fonseca B. D., and Proud C. G. (2008) Regulation of cyclin D1 expression by mTORC1 signaling requires eukaryotic initiation factor 4E-binding protein 1. Oncogene 27, 1106–1113 [DOI] [PubMed] [Google Scholar]
- 19. Ritchie W., and Rasko J. E. J. (2014) Refining microRNA target predictions: sorting the wheat from the chaff. Biochem. Biophys. Res. Comm. 445, 780–784 [DOI] [PubMed] [Google Scholar]
- 20. Niranjanakumari S., Lasda E., Brazas R., and Garcia-Blanco M. A. (2002) Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods 26, 182–190 [DOI] [PubMed] [Google Scholar]
- 21. Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fu X., Meng Z., Liang W., Tian Y., Wang X., Han W., Lou G., Wang X., Lou F., Yen Y., Yu H., Jove R., and Huang W. (2014) miR-26a enhances miRNA biogenesis by targeting Lin28B and Zcchc11 to suppress tumor growth and metastasis. Oncogene 33, 4296–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Höck J., Weinmann L., Ender C., Rüdel S., Kremmer E., Raabe M., Urlaub H., and Meister G. (2007) Proteomic and functional analysis of Argonaute containing mRNA–protein complexes in human cells. EMBO Rep. 8, 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Landthaler M., Gaidatzis D., Rothballer A., Chen P. Y., Soll S. J., Dinic L., Ojo T., Hafner M., Zavolan M., and Tuschl T. (2008) Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14, 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu P.-H., Isaji M., and Carthew R. W. (2013) Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Mol. Cell 52, 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., and Filipowicz W. (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125, 1111–1124 [DOI] [PubMed] [Google Scholar]
- 27. Vasudevan S., and Steitz J. A. (2007) AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128, 1105–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hur J. K., Zinchenko M. K., Djuranovic S., and Green R. (2013) Regulation of Argonaute slicer activity by guide RNA 3′ end interactions with the N-terminal lobe. J. Biol. Chem. 288, 7829–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang V., Zheng J., Qi Z., Wang J., Place R. F., Yu J., Li H., and Li L.-C. (2013) Ago1 interacts with RNA Polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 9, e1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Izaurralde E. (2015) Breakers and blockers—miRNAs at work. Science 349, 380–382 [DOI] [PubMed] [Google Scholar]
- 31. Fukaya T., and Tomari Y. (2012) MicroRNAs mediate gene silencing via multiple different pathways in Drosophila. Mol. Cell 48, 825–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.