Abstract
The Hedgehog (HH) signaling pathway is essential for the maintenance and response of several types of stem cells. To study the transcriptional response of stem cells to HH signaling, we searched for proteins binding to GLI proteins, the transcriptional effectors of the HH pathway in mouse embryonic stem (ES) cells. We found that both GLI3 and GLI1 bind to the pluripotency factor NANOG. The ectopic expression of NANOG inhibits GLI1-mediated transcriptional responses in a dose-dependent fashion. In differentiating ES cells, the presence of NANOG reduces the transcriptional response of cells to HH. Finally, we found that Gli1 and Nanog are co-expressed in ES cells at high levels. We propose that NANOG acts as a negative feedback component that provides stem cell-specific regulation of the HH pathway.
Keywords: differentiation, embryonic stem cell, Hedgehog signaling pathway, stem cells, transcription repressor, GLI proteins, Nanog, repressor
Introduction
The HH3 pathway is essential for regulating biological processes in a diverse set of cells. HH ligands, including Sonic hedgehog, bind to the Patched1 (PTCH1) receptor, which activates the transmembrane protein Smoothened, resulting in pathway activation (for a review, see Ref. 1). The transcriptional response to HH ligands is mediated by the GLI family of transcription factors (GLI1–3), which can act as both transcriptional activators and repressors in a context-dependent fashion (for a review, see Ref. 2). The presence of HH ligand causes the maturation of full-length GLI proteins into their transcriptional activator forms (GLI-A), whereas in the absence of ligand GLI proteins undergo C-terminal truncation and then act as transcriptional repressors (GLI-R). Although GLI2 and GLI3 exist in both activator and repressor forms, GLI2 is the major activator, whereas GLI3 is the major repressor (3–5). In contrast, GLI1 only exists as a full-length activator form (5–7). Gli1 is a direct HH target gene, thereby participating in a positive feedback loop (8–11).
The HH pathway was initially characterized for its role in regulating embryonic development, but it also has critical roles in regulating the homeostasis of several adult tissues (for a review, see Ref. 12). In particular, HH regulates two major neural stem cell populations in the brain, the ventral subventricular zone and subgerminal zone, as well as quiescent hair follicle stem cells (13). In the absence or inhibition of the HH pathway, these tissues undergo a marked reduction in the number of proliferating cells, indicating that the pathway is required for normal proliferation (14). Conversely, hyperactivation of the HH pathway results in an expanded population of neural stem cells. In this context, the progeny of neural stem cells is shifted so that they preferentially give rise to two daughter stem cells instead of producing transient amplifying cells capable of generating differentiated progeny (15). Together these results indicate that the levels of HH perceived by neural stem cells regulate the balance between generating stem cells and differentiated progenitors. In addition to regulating normal neural development, various studies have suggested that populations of stem cells play key roles in cancer. In particular, GLI proteins have been shown to activate the transcription of the pluripotency factor Nanog in glioblastoma and medulloblastoma cancer models (16, 17). NANOG in turn is critical for maintaining tumorigenic cell populations, suggesting positive feedback between these factors (16, 17).
Although HH signaling, via GLI transcription factors, is critical for regulating neural stem cells, the underlying transcriptional mechanisms remain poorly understood. In part, this is because it is difficult to isolate large numbers of these stem cells. In an effort to understand this process, we performed a mass spectrometry-based screen to identify GLI-binding proteins in mouse embryonic stem (ES) cells that might act as stem cell-specific cofactors. Here, we report that GLI1 and GLI3 bind to the pluripotency factor NANOG. The presence of NANOG inhibits GLI transcriptional responses, therefore inhibiting HH signaling. We show that Gli1 is expressed at high levels in ES cells along with Nanog. Interestingly, previous studies have shown that NANOG also binds to and inhibits the transcriptional effectors of both the BMP and NF-κB pathways in ES cells (18, 19). Collectively, these results suggest that, by binding to multiple transcriptional effectors, NANOG may help to buffer ES cells from external signals.
Experimental Procedures
Unless specified otherwise, statistical significance was measured using a paired t test with a two-tailed p value.
Tissue Culture and Cell Lines
NIH3T3 and HEK293Tcells were cultured with 10% calf serum in DMEM. P19 cells were cultured with 2.5% fetal bovine serum (FBS) and 7.5% calf serum in α minimum essential medium Eagle (Sigma, M8042). ES cell lines containing a tamoxifen-inducible Cre (CreER) and FLAG-tagged GLI1 or GLI3T driven by the Rosa26 promoter (9) were grown on mouse embryo fibroblast feeder cells. Expression of FLAG-tagged GLI1 and GLI3T was induced by adding 1 μm 4-OH-tamoxifen (Sigma, H7904) for at least 48 h. The feeder-free J1 ES cells (ATCC, SCRC 1010) and J1 biotinylated NANOG ES cells (FB-NANOG) (20) were cultured on gelatinized plates. ES cells were cultured in medium containing 15% FBS with leukemia inhibitory factor (LIF) at a final concentration of 1,000 units/ml. In vitro differentiation of ES cells was induced by removing LIF from the ES cell medium.
shRNA Lentivirus Infection
1,200 ng of Nanog shRNA lentiviral plasmid (shNG; Sigma Mission RNAi TRCN0000075333) or control (shCtrl; pLKO.1-puro vector containing 1.9 kb of inert DNA) was co-transfected with 400 ng of vesicular stomatitis virus G and 800 ng of Δ8.9 into HEK293T cells in 6-well plates using Lipofectamine 2000 (Life Technologies). After 1 day, the medium was changed to ES cell medium without LIF to obtain LIF-free supernatant for ES cell infection. After an additional 24 h, the supernatant containing the viruses was harvested. Immediately before infection, the undiluted supernatant was mixed with Polybrene (Sigma) to a final concentration of 4 μg/ml and then mixed with 5 × 105 resuspended J1 ES cells. The ES cells were then incubated overnight before providing fresh medium on the 2nd day. The ES cells were split on day 3 into ES cell medium containing 5 μg/ml puromycin, and HH signaling was stimulated by treatment with 5 μm purmorphamine or 0.05% dimethyl sulfoxide (vehicle control) for 2 days.
Immunoprecipitation and Mass Spectrometry
A single 15-cm plate (containing ∼3 × 108 ES cells expressing FLAG-tagged GLI1/3R or control cells) was harvested with cell scrapers in cold Dulbecco's PBS. Cells were spun down at 300 × g for 5 min and resuspended in 1 ml of Lysis Buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 0.5 m NaCl, 0.2% Triton X-100, 10 mm KCl, 10% glycerol, 0.5 mm DTT, Complete Mini protease inhibitor mixture (Roche Applied Science))/0.3 ml of cell pellet. The cells were incubated with the Lysis Buffer at 4 °C for 30 min and centrifuged at 20,000 × g for 30 min. 1 ml of supernatant was transferred into a fresh tube, and 0.3 volume of Buffer A (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 10% glycerol) was added to dilute the salt concentration of cell lysate to a final concentration of ∼0.3 m. 50 μl of anti-FLAG M2 affinity gel (Sigma, A2220) was mixed with 1.3 ml of cell lysate, rotated at 4 °C for 2 h, and then washed three times with Washing Buffer (10 mm HEPES, pH 7.9, 1.5 mm MgCl2, 300 mm NaCl, 10 mm KCl, 0.2% Triton X-100, Complete Mini protease inhibitor mixture) and then with Elution Base Buffer (10 mm HEPES, pH 7.9, 0.1 m NaCl, 1.5 mm MgCl2, 0.05% Triton X-100, Complete Mini protease inhibitor mixture, no FLAG peptides) once. This was centrifuged at 2,500 × g for 30 s, and the pellet was resuspended in 30 μl of Complete Elution Buffer (200 μg/ml 3XFLAG peptides (Sigma, F4799) in Elution Base Buffer), and the mixture was incubated at 4 °C for 30 min with frequent vortexing. The supernatant was collected by spinning at 2,500 × g for 30 s, loaded on a 4–20% gradient SDS-polyacrylamide gel, and minimally resolved by electrophoresis for 10 min at 120 V. The gel was subsequently stained with Coomassie Blue for 1 h and destained for 30 min. The stained part of the gel containing proteins was excised and digested with trypsin. Peptides were sequenced by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Proteins were identified as described previously (21, 22) using either the Proxeon Easy-nLC II coupled to the Thermo Velos Pro or the Dionex Ultimate 3000 RSLCnano LC coupled to the Thermo Orbitrap Elite. Briefly, the digested peptides were desalted using Millipore U-C18 ZipTip pipette tips following the manufacturer's protocol. A 2-cm-long × 100-μm-inner diameter C18 5-μm trap column (Proxeon EASY column) was followed by a 75-μm-inner diameter × 15-cm-long analytical column packed with C18 3-μm material (Dionex Acclaim PepMap 100). Peptides were separated by a 60-min 5–45% B gradient using Buffer A (0.1% formic acid in water) and Buffer B (0.1% formic acid in acetonitrile). On the Orbitrap Elite, the Fourier transformed MS resolution was set to 120,000, and the top 20 MS/MS spectra were acquired by collision-induced dissociation using the ion trap. For the Velos Pro, the ion trap was used for MS, and the top 20 MS/MS spectra were collected using the data-dependent acquisition method. Raw data were processed using SEQUEST embedded in Proteome Discoverer v1.3, searching the mouse reference proteome from UniProt (March 2012 containing 54,201 entries). A decoy database was used for calculating peptide and protein probabilities. X! Tandem database searches were performed embedded in Scaffold 4 Q+ (Proteome Software) using the same search parameters as SEQUEST. Scaffold was used for validation of peptide and protein identifications with filtering to achieve 99% protein confidence with two peptides at 95% confidence. The mass spectrometry proteomics data were deposited to the ProteomeXchange Consortium (23) via the PRIDE partner repository with the data set identifier PXD002494.
Western Blotting
Protein samples were resolved by 9% SDS-PAGE and transferred onto nitrocellulose membranes (GE Healthcare). Membranes were blocked with 10% nonfat milk in TBS-Tween 20 buffer for 30 min and incubated with the primary antibodies anti-FLAG antibody (Sigma, FLAG M2 antibody, F1804; 1:4,000), anti-HA (Thermo Scientific, 26183; 1:4,000), anti-NANOG (Calbiochem, SC1000; 1:2,000), anti-actin (Sigma, A2066; 1:2,000), and anti-POU5F1 (OCT4) (Santa Cruz Biotechnology, SC5279; 1:1,000) in 3% nonfat milk at 4 °C overnight. After washing with TBS-Tween 20 for 5 min, membranes were then incubated with secondary antibodies HRP-conjugated rabbit anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories; 1:5,000) and HRP-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories; 1:5,000) at room temperature for 1 h. After washing with TBS-Tween 20 three times, membranes were developed by using ECL Prime Western blotting detection reagent (GE Healthcare, RPN2232) and visualized by exposure to CL-XPosure films (Thermo Scientific, 34091). To determine -fold change, Western blots were quantified using ImageJ, and NANOG intensities were normalized to actin intensities.
Luciferase Assays
P19 cells were seeded at 1.5 × 105 cells/well in 24-well plates and co-transfected using Lipofectamine 2000 (Life Technologies, 11668019) with 300 ng of luciferase reporter plasmid ptcΔ136-pGL3 (24), 100 ng of pCIG-GLI1 (9), 200 ng of pSV-β-galactosidase expression plasmid (24), and 0–100 ng of pCIG-NANOG or pCIG-OCT4 expression constructs. In samples with less than 100 ng of NANOG or OCT4, pCIG vector was added to a total of 100 ng. pBluescript DNA was then added as filler DNA so that each sample had a total of 800 ng of transfected DNA. The serum levels in the medium were reduced to 0.5% at the time of transfection. The cells were harvested 2 days after transfection and assayed for activity using the One-Glo Luciferase Assay lit (Promega, E6120). All luciferase activities were normalized with β-galactosidase activity levels (quantified using the BetaFluor β-gal assay kit (G-Biosciences, 786-654)).
Quantitative RT-PCR
RNA was extracted from ES cells by TRIzol reagent (Life Technologies), and cDNAs were synthesized by SuperScript II reverse transcriptase (Invitrogen) using 1 μg of total RNA. Quantitative PCR was performed on a ViiA7 platform using 2× SensiFast SYBR mixture (Bioline, BIO-94020). PCR primers were as follows: GAPDH: F, GGTGAAGGTCGGTGTGAACG; R, CTCGCTCCTGGAAGATGGTG; Gli1: F, CCCAGCTCGCTCCGCAAACA; R, CTGCTGCGGCATGGCACTCT; Ptch1: F, GACCGGCCTTGCCTCAACCC; R, CAGGGCGTGAGCGCTGACAA; Nanog: F, AGGGTCTGCTACTGAGATGCTCTG; R, CAACCACTGGTTTTTCTGCCACCG; Oct4: F, TCTGGAGACCATGTTTCTGAAGT; R, TACAGAACCATACTCGAACCACAT; Sox2: F, GCGGAGTGGAAACTTTTGTC; R, TATTTATAATCCGGGTGCTCCT; Gata4: F, TTCTCAGAAGGCAGAGAGTGTGT; R, ATGCCGTTCATCTTGTGATAGAG; Gata6: F, GACGGCACCGGTCATTACC; R, ACAGTTGGCACAGGACAGTCC; Gsc: F, AGAAGGTGGAGGTCTGGTTTAAG; R, GAGGACGTCTTGTTCCACTTCT; T: F, CTTCAAGGAGCTAACTAACGAGATG; R, GTCCAGCAAGAAAGAGTACATGG; Gata2: F, GCCTCTACCACAAGATGAATGG; R, GTCTGACAATTTGCACAACAGG; Bmp2: F, CGCTTCTTCTTCAATTTAAGTTCTG; R, AACTACTGTTTCCCAAAGCTTCCT; Nes: F, AGGACCAGGTGCTTGAGAGA; R, TTCGAGAGATTCGAGGGAGA; Fgf5: F, GGATTGTAGGAATACGAGGAGTTTT; R, AACTTACAGTCATCCGTAAATTTGG; Pitx2: F, CTTGGACTCCTCCAAACATAGACT; R, CACATCCTCATTCTTTCCTTGCT; Hand1: F, CCTTCAAGGCTGAACTCAAAAA; R, GCGCCCTTTAATCCTCTTCT; Cdx2: F, GCGAAACCTGTGCGAGTGGATG; R, CGGTATTTGTCTTTTGTCCTGGTTTTCA; Gata3: F, TGGGCTGTACTACAAGCTTCATAA; R, CTTTTTCGATTTGCTAGACATCTTC.
Chromatin Immunoprecipitation
FB-NANOG cells (20) were differentiated over a 4-day period with 5 μm purmorphamine or 0.05% dimethyl sulfoxide (vehicle control) added after 48 h. Chromatin immunoprecipitation (ChIP) was performed as described previously (25) except that samples were sonicated using a Bioruptor for three 10-min sessions (30 s on, 1 min off) at high voltage. After ChIP, enrichment at loci was determined by the ΔΔct method using the following primers: Gfi1b (for normalization): F, CGCCAGATTTTGACACAAATAA; R, CTGCACAGACAGACACTTCTCC; C1-1 (negative control): F, GCCAGAATTCCATCCCACTA; R, CCAATAACCTGCCCTGACAT; Gli1 enhancer: F, GGACAAAGAGACCTGGGACA; R, AGGAGATGCTCTGACGCCTA; Ptch1 enhancer: F, AGGCCTGCACCAATAATGAC; R, TCCTTGCTCGCCTCTTTAAC.
Analysis of Nanog and Gli Co-expression
We searched Barcode-annotated samples (26, 27) for biological contexts associated with three expression patterns: 1) high expression in Gli1 (frozen robust multiarray analysis-normalized and Barcode-standardized Gli1 expression ≥5) and high expression in Nanog (Nanog expression ≥10), 2) high expression in Gli1 (Gli1 expression ≥5) and medium expression in Nanog (1 ≤ Nanog expression < 10), and 3) high expression in Gli1 (Gli1 expression ≥5) and low expression in Nanog (Nanog expression <1). In Barcode, samples were processed to facilitate cross-data set comparisons by minimizing unwanted variation such as laboratory or batch effects. Because of this, expression levels of a gene can be meaningfully compared across heterogeneous samples in the Barcode compendium as shown previously (28). After identifying samples for each expression pattern, statistically enriched biological contexts were identified using the Gene Set Context Analysis package in R/Bioconductor (29), which implements the ChIP-PED method (28). To identify biological contexts associated with a specific gene expression pattern of Gli1 and Nanog, we counted the total number of samples (N), number of total samples with the specified expression pattern (K), total number of samples Nc for each biological context (c), and number of samples of biological context (c) with the specific expression pattern Kc. A Fisher's exact test was then performed for each context (c) to see whether it is enriched in the specified expression pattern. The p values were corrected using the Bonferroni procedure by multiplying the total number of tested contexts (c) to adjust for multiple testing. For gene expression analysis in differentiating ES cells, we obtained expression data from a previous study (30) (GSE3749), which generated triplicate time course measurements during differentiation of J1 mouse ES cells. The CEL files were imported into dChIP software (31) for data normalization and extraction of expression values.
Results
NANOG Binds to GLI Proteins
To identify GLI-associated proteins in ES cells, we used a line that contained a tamoxifen-inducible Cre as well as a Cre-activatable 3XFLAG-tagged GLI3 repressor (GLI3R) allele driven by the ubiquitous Rosa26 promoter (10). We performed mass spectrometry-based protein identification on FLAG-immunoprecipitated lysates from tamoxifen-induced ES cells (GLI3RFLAG) and control, parental ES cells that do not express GLI3RFLAG. We sorted the resulting list based on the Z score enrichment. The most enriched protein in this list was the bait protein, GLI3. This list also contained two proteins, suppressor of fused (SUFU) and 14-3-3 (Table 1), that were previously shown to bind GLI proteins (32–34). We set a cutoff Z-score value of 1.5, which allowed for the inclusion of both of these proteins (Table 1). The remaining 19 proteins have not previously been associated with GLI proteins. Interestingly, NANOG, a well established core pluripotency factor, and its cofactor NR0B1 (hereafter referred to by its alternative designation DAX1) (20, 35) were both present on this list (Table 1). We focused our subsequent efforts on characterizing this interaction.
TABLE 1.
GLI3-interacting proteins identified by mass spectrometry
Proteins identified by mass spectrometry from three independent biological samples and sorted by combined Z scores with a cutoff value of 1.5 are shown. The GLI3 bait protein is bold and underlined along with the previously identified GLI-interacting proteins SUFU and 14-3-3.
| Protein | Accession no. | Z score |
|---|---|---|
| HSPD1, 60-kDa heat shock protein | P63038 | 28.25 |
| GLI3 (bait protein) | Q61602 | 15.34 |
| RUVBL1, RuvB-like 1a | P60122 | 3.65 |
| ATP5A1, ATP synthase subunit α | Q03265 | 3.57 |
| HNRNPM, isoform 2 of heterogeneous nuclear ribonucleoprotein M | Q9D0E1-2 | 3.5 |
| ALDOA, fructose-bisphosphate aldolase A | P05064 | 3.11 |
| DSP, desmoplakina | E9Q557 | 3.04 |
| DHX15, putative pre-mRNA-splicing factor ATP-dependent RNA helicase | O35286 | 2.47 |
| FLNB, filamin-B | Q80X90 | 2.24 |
| JUP, junction plakoglobina | Q02257 | 2.19 |
| CCT2, T-complex protein 1 subunit β | P80314 | 1.97 |
| NR0B1 (DAX1), nuclear receptor subfamily 0 group B member 1 | Q61066 | 1.79 |
| SNRPD2, small nuclear ribonucleoprotein Sm D2 | P62317 | 1.78 |
| CCT3, T-complex protein 1 subunit γ | P80318 | 1.73 |
| HNRNPF, isoform 2 of heterogeneous nuclear ribonucleoprotein Fa | Q9Z2X1-2 | 1.65 |
| LMNA, isoform C of prelamin-A/C | P48678-2 | 1.62 |
| PAICS, multifunctional protein ADE2 | Q9DCL9 | 1.61 |
| NANOG | Q80Z64-2 | 1.61 |
| MYL6, myosin light polypeptide 6 | Q60605-2 | 1.61 |
| SNRPD1, small nuclear ribonucleoprotein Sm D1 | P62315 | 1.61 |
| SON | Q9QX47-4 | 1.61 |
| VCP, transitional endoplasmic reticulum ATPase | Q01853 | 1.6 |
| RPL7, 60S ribosomal protein L7 | P14148 | 1.59 |
| SUFU, suppressor of fused | Q9Z0P7-2 | 1.57 |
| ACIN1, apoptotic chromatin condensation inducer in the nucleus | Q9JIX8 | 1.51 |
| 14-3-3 | O70456 | 1.51 |
a Previously reported as a background protein obtained in mass spectrometry of ES cells (35).
NANOG is a core regulator of stem cells and acts in conjunction with SOX2 and POU5F1 (hereafter referred to by its alternative designation OCT4) to maintain ES cell self-renewal and pluripotency (36, 37). As NANOG and OCT4 physically interact with each other (35, 38–40), we asked whether GLI3 might also associate with OCT4. NANOG efficiently co-immunoprecipitated with GLI3RFLAG, confirming the mass spectrometry interaction. However, OCT4 was not co-immunoprecipitated with GLI3RFLAG (Fig. 1A).
FIGURE 1.
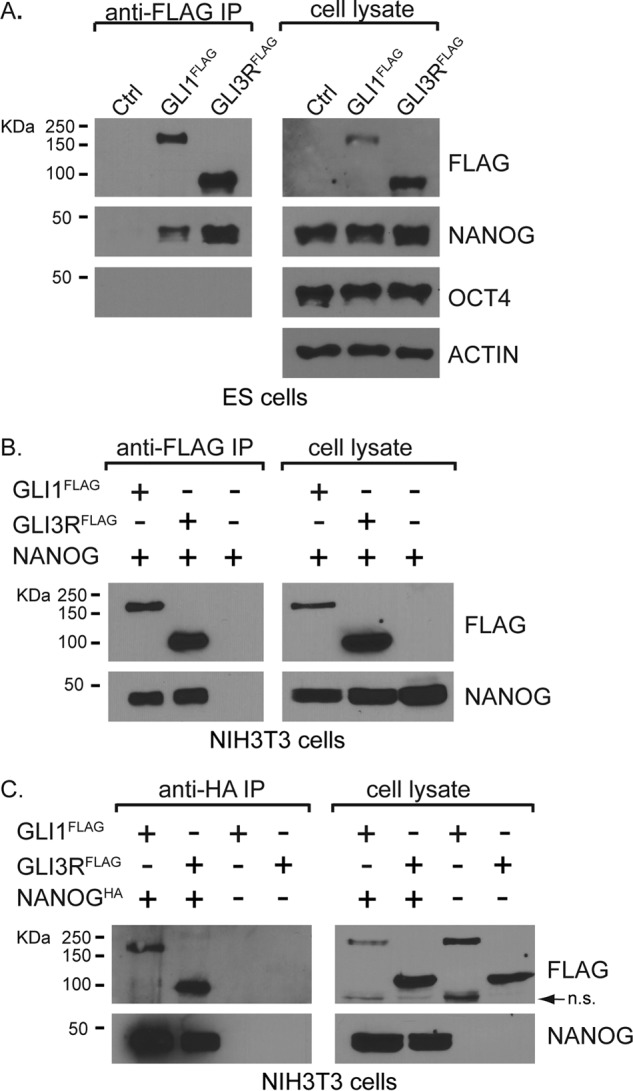
GLI1 and GLI3 bind to NANOG. A, endogenous NANOG (∼42k Da), but not OCT4 (∼38kDa), co-immunoprecipitated with both FLAG-tagged GLI activator (GLI1FLAG; ∼122 kDa) and GLI repressor (GLI3RFLAG; ∼84 kDa) using GLI1FLAG- and GLI3RFLAG-expressing mouse embryonic cell lines (n = 3 biological replicates). B and C, when GLI and NANOG proteins are co-transfected in NIH3T3 cells, they can each immunoprecipitate the other protein (n = 3 biological replicates). The arrow in C indicates a nonspecific (n.s.) band. IP, immunoprecipitation; Ctrl, control.
The previous experiments demonstrated that NANOG binds to the transcriptional repressor, GLI3R. To determine whether NANOG might also be able to interact with additional GLI proteins, we used an ES cell line containing a Cre-inducible, 3XFLAG-tagged full-length GLI1 (GLI1FLAG) to perform additional immunoprecipitations. Unlike GLI2 and GLI3, GLI1 exists only as a full-length protein that acts as a transcriptional activator (5, 6). Similar to GLI3R, GLI1FLAG also bound to NANOG but not to OCT4 (Fig. 1A). To determine whether NANOG interacts with both GLI1 and GLI3R in other cell types, we co-transfected HH-responsive NIH3T3 cells with constructs encoding HA-tagged NANOG and either GLI1FLAG or GLI3RFLAG. Consistent with the interactions shown in ES cells, NANOG binds to both GLI3R and GLI1 in NIH3T3 cells (Fig. 1, B and C).
NANOG Represses GLI1-mediated Transcriptional Activation
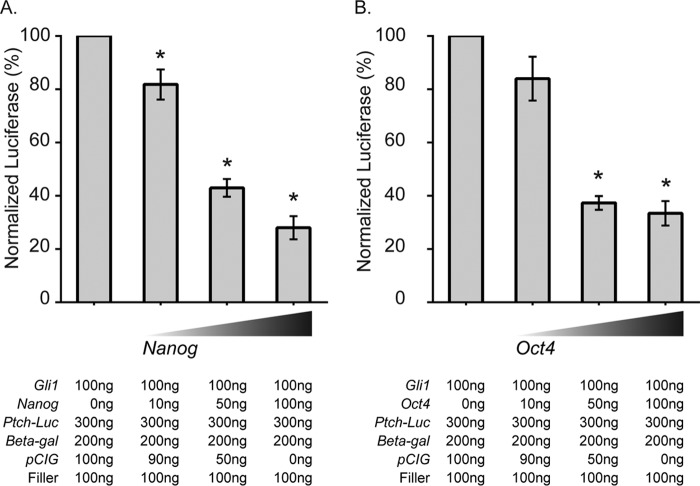
NANOG helps to maintain ES cell self-renewal and pluripotency in part by repressing the transcription of key lineage-specific regulatory genes (39, 40). To determine whether NANOG influences GLI-mediated transcription, we first utilized a GLI-responsive luciferase assay in P19 embryonal carcinoma cells, which express pluripotency markers, including NANOG (41). Compared with cells transfected with Gli1 alone, cells transfected with both Nanog and Gli1 had a dose-dependent reduction in luciferase activity (Fig. 2A).
FIGURE 2.
NANOG inhibits GLI1-mediated transcription. GLI1-responsive luciferase activity was inhibited by co-transfecting with increasing amounts of a Nanog-expressing vector (n = 4 biological replicates) (A) or an Oct4-expressing vector (n = 3 biological replicates) (B) in P19 embryonal carcinoma cells. Error bars indicate S.E. Significantly reduced values are indicated by an asterisk (p < 0.05). The corresponding amounts of transfected DNA are indicated below the figure.
Because NANOG and OCT4 are recursively regulated, we asked whether OCT4 might also be able to repress GLI-mediated responses. P19 cells transfected with Oct4 have a dose-dependent reduction in GLI1-mediated activation that is comparable with cells transfected with Nanog (Fig. 2B). Because GLI1 does not bind to OCT4, a known binding partner of NANOG, under our experimental conditions (Fig. 1A), it is presently unclear whether this reduction occurs because of the presence of OCT4 in a GLI-NANOG complex that was undetectable in our experimental conditions or by indirect mechanisms (see “Discussion”).
NANOG Binds to GLI1 through Its C-terminal Domains
NANOG is a 305-amino acid protein containing a conserved homeodomain (amino acids 95–155) and a tryptophan repeat (WR) domain (amino acids 197–244) (42, 43). To identify the protein-binding domain on NANOG, we generated a series of HA-tagged NANOG truncations (Fig. 3A) and co-transfected them with FLAG-tagged-GLI1 (Fig. 3B). Consistent with our previous results, full-length NANOG co-immunoprecipitated with GLI1. Constructs lacking the C-terminal half of NANOG (amino acids 156–305) did not interact with GLI1, indicating that the C-terminal half is essential for the interaction. To determine whether the C-terminal half of NANOG could bind GLI1, we generated additional constructs C1, C2, and C3 (Fig. 3B). The C1 fragment (amino acids 155–305) bound GLI1 at levels that were comparable with full-length NANOG (Fig. 3B), indicating that the C-terminal half of the NANOG is sufficient for binding GLI1. The C2 construct (amino acids 197–244) bound only minimally to GLI1, whereas C3 (amino acids 155–197) did not bind at all. These results suggest that an extensive region of the C-terminal half of NANOG is involved in the interaction with GLI1.
FIGURE 3.
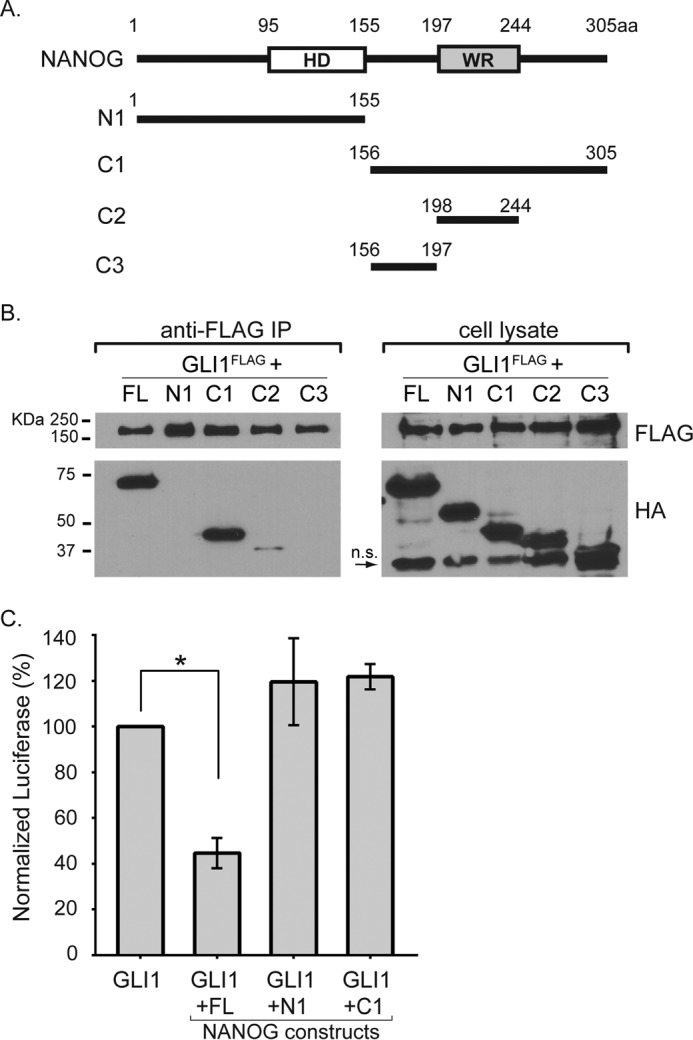
The C-terminal half of NANOG binds GLI1, but the entire protein is required for inhibiting transcriptional response. A, schematic diagram of full-length NANOG and N- or C-terminal deletion constructs that were tested in this study. The numbers indicate the amino acids corresponding to those in full-length NANOG. B, anti-FLAG immunoprecipitation with cell lysates from NIH3T3 cells co-transfected with FLAG-tagged GLI1 and HA-tagged NANOG deletion constructs (n = 3 biological replicates). C, only full-length NANOG is capable of repressing GLI-mediated transcription in P19 embryonal carcinoma cells (n = 3 biological replicates). Error bars indicate S.E. The asterisk indicates statistically significant changes (p < 0.05). FL, full-length; HD, homeodomain; WR, tryptophan repeat domain; n.s., nonspecific; IP, immunoprecipitation; aa, amino acids.
The N Terminus of NANOG Is Essential for Repressing GLI1-mediated Transcription
The previous results indicated that the C-terminal half of NANOG (construct C1) is sufficient to bind GLI1. We next asked whether this region was also sufficient to inhibit GLI1-mediated transcription. We co-transfected GLI1 with specific NANOG deletion constructs and a GLI-responsive luciferase construct. Although C1 robustly bound to GLI1, it did not repress GLI1-mediated transcription. We also observed that the response to GLI1 was not reduced when the N-terminal half of NANOG (construct N1) was co-transfected with GLI1 (Fig. 3C). This suggests that whereas the C terminus of NANOG mediates binding to GLI1 the N terminus of NANOG is required to inhibit GLI1-mediated transcriptional activation. This is consistent with other studies showing that the N-terminal portion of NANOG contains a transcriptional repressor motif (44).
Hedgehog Signaling Up-regulates NANOG in Differentiating ES Cells
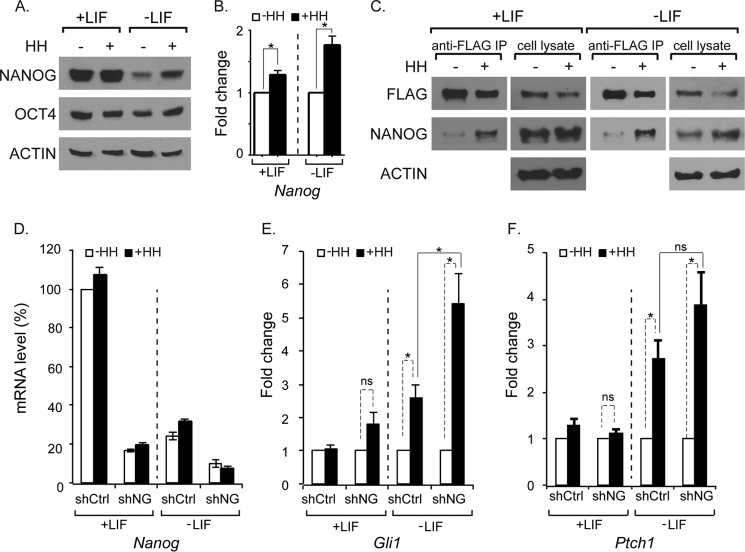
In an effort to examine the significance of NANOG-GLI1 interactions in ES cells, we activated HH signaling with the small molecule purmorphamine under conditions that either maintain stem cells or cause differentiation (by the withdrawal of LIF). In the presence of LIF, NANOG was robustly expressed, and this expression was unaffected by the co-stimulation of HH signaling (Fig. 4A). ES cells began differentiating upon LIF withdrawal and after 4 days expressed markedly lower levels of NANOG. In contrast, differentiating cells stimulated with HH had significantly higher levels of NANOG protein. To determine whether elevated NANOG levels occurred on a transcriptional level, we compared the amount of Nanog mRNA in differentiating cells. Samples in which the HH pathway was activated for 48 h had significantly increased levels of Nanog mRNA compared with those without HH (Fig. 4, A and B). We conclude that HH signaling up-regulates Nanog at the transcriptional level.
FIGURE 4.
NANOG represses GLI target genes. A, NANOG protein levels were significantly up-regulated by HH after 4 days in the absence of LIF (p = 0. 035; -fold change = 4.4; n = 4 biological replicates) but not in the presence of LIF (p = 0.173; -fold change = 1.5; n = 4 biological replicates). B, Nanog mRNA levels were significantly up-regulated by HH after 48 h in either the presence (p = 0.003; n = 4 biological replicates) or absence of LIF (p = 0.0018; n = 4 biological replicates). C, anti-FLAG immunoprecipitations showing that more NANOG protein binds to GLI1FLAG upon HH stimulation either in the presence (n = 3 biological replicates; -fold change = 6; p = 0.0397) or absence of LIF (n = 3 biological replicates; -fold change = 3; p = 0.288). D, transduction of ES cells with a lentivirus producing an shRNA against Nanog (shNG) results in a substantial reduction in Nanog levels compared with a control shRNA (shCtrl) as assayed by quantitative PCR (n = 4 biological replicates). E and F, in the absence of LIF, Gli1 and Ptch1 mRNA levels were up-regulated by HH stimulation (Gli1, p = 0.0282; Ptch1, p = 0.0232). Note the absence of their up-regulation in the presence of LIF. When Nanog levels are reduced by lentiviral shRNA (shNG), there is a significant increase (p = 0.036) in the expression of Gli1 in the absence of LIF compared with a negative control shRNA. There is also an increase, although not significant (p = 0.1242), in Ptch1 in the absence of LIF compared with negative control shRNA (n = 4 biological replicates). Error bars indicate S.E. The asterisk indicates a significant reduction (p < 0.05). PM, purmorphamine; ns, not significant; IP, immunoprecipitation.
We next compared the ability of GLI to interact with NANOG in the presence and absence of HH signaling. Although NANOG was not up-regulated by HH stimulation under conditions that maintained proliferation (LIF-containing medium) (Fig. 4A), there was a marked increase in the amount of NANOG pulled down by GLI1 (Fig. 4C). Unlike endogenous Gli1, which is transcriptionally activated by HH signaling, the Gli1FLAG transgene expressed in these cells is driven by the constitutively active Rosa26 promoter (9). The levels of GLI1FLAG protein are unchanged by HH signaling (Fig. 4C), and the increased amount of NANOG pulled down by GLI1 is therefore not caused by HH-mediated up-regulation of Gli1. A similar enrichment of the GLI1-NANOG complex is also present in differentiating ES cells after LIF withdrawal (Fig. 4C). These results are consistent with a scenario in which HH signaling promotes the formation of GLI1-NANOG complexes in ES cells.
NANOG Represses GLI-mediated Transcription in Differentiating ES Cells
Our results indicated that NANOG acts as a repressor of GLI-mediated transcription. If NANOG represses GLI-mediated transcription in ES cells, then reducing NANOG levels should result in enhanced GLI-mediated transcriptional responses. To test this, we infected ES cells with lentiviral shRNA constructs targeting Nanog (shNG). Under pluripotent conditions with LIF, shNG expression resulted in an ∼75% reduction in Nanog mRNA levels compared with cells infected with a control shRNA construct (shCtrl) (Fig. 4D). As expected, Nanog levels were strongly down-regulated as ES cells began differentiating (24% of undifferentiated levels), and compared with these already down-regulated levels, Nanog was reduced a further 58% in shNG-infected cells (Fig. 4D). To determine whether the reduced levels of Nanog affected HH-mediated transcription, we examined the expression of Gli1 and Ptch1, which are direct HH pathway transcriptional targets in a variety of different tissue types (5, 45). To establish a baseline for comparison, we first assessed levels of Gli1 and Ptch1 mRNA upon HH pathway stimulation. Under pluripotent ES cell culture conditions, we did not observe a difference in Gli1 or Ptch1 in HH-stimulated cells as compared with unstimulated cells. However, when ES cells were cultured in medium that promotes differentiation, there was a significant increase in both Gli1 and Ptch1 upon HH stimulation.
We then examined the levels of Gli1 and Ptch1 when Nanog levels were knocked down. When shNG-expressing cells are stimulated by HH signaling under pluripotent culture conditions (medium containing LIF), they do not significantly up-regulate Gli1 or Ptch1. In contrast, under differentiating conditions (medium not containing LIF), both Gli1 and Ptch1 were up-regulated (Fig. 4, E and F). When compared with controls, shNG-infected cells had significantly higher levels of Gli1 induction in differentiating ES cells (Fig. 4D). The levels of Ptch1, although elevated, were not significantly different from controls (Fig. 4F). We conclude that reduced levels of NANOG result in amplified responses to HH signaling in differentiating ES cells.
NANOG and GLI Are Co-expressed in Stem Cells
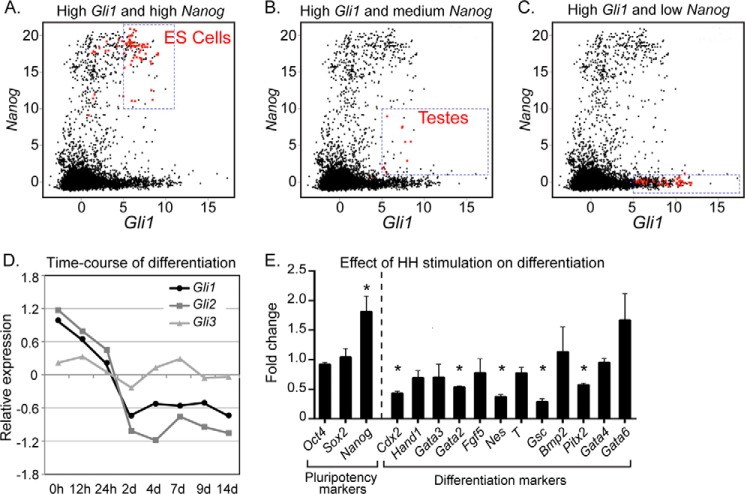
To systematically identify biological contexts where GLI1 and NANOG might function, we examined their expression levels in a compendium of 9,444 gene expression microarray samples generated using Affymetrix Mouse 430 2.0 (GPL1261) arrays. These samples were compiled by the Barcode project and normalized using frozen robust multiarray analysis to ensure that expression levels of each gene can be meaningfully compared across these heterogenous samples (26, 27). The biological context of each sample was annotated and curated by Barcode. In total, these samples represent over 3,000 different biological contexts. We first searched these samples for biological contexts that were enriched for high expression of both Gli1 and Nanog. ES cells were substantially and significantly enriched for co-expression of both of these genes (Fig. 5A and Table 2). We also searched for biological contexts where Gli1 was expressed at high levels along with medium expression of Nanog, uncovering significant enrichment in the embryonic testes (Fig. 5B and Table 3). Finally, we searched for contexts where Gli1 was expressed at high levels and Nanog was expressed at low levels. This pattern of co-expression was enriched in a variety of embryonic tissues as well as in several medulloblastoma samples (Fig. 5C and Table 4).
FIGURE 5.
Nanog and HH signaling are associated with pluripotent cell states. A meta-analysis of mouse microarrays identified tissue types in which there was co-expression of high expression levels of both Gli1 and Nanog (A), high expression levels of Gli1 and medium levels of Nanog (B), and high expression levels of Gli1 and low levels of Nanog (C). The area examined in each graph is indicated by the dashed blue box. The top 13 most enriched regions, all corresponding to different ES cell categories, are indicated in red in A. The top enriched regions in B and C are depicted as red data points within their respective blue boxes. Each data point corresponds to a microarray. Both the x and y axes are in log2 scale. D, relative mRNA expression levels of Gli1, Gli2, and Gli3 upon time course differentiation of mouse J1 ES cells (n = 4 biological replicates). E, expression of genes associated with pluripotency and differentiation in differentiating ES cells treated in the presence of activated HH signaling. The expression values are shown relative to differentiating ES cells without HH stimulation (n = 4 biological replicates). Error bars indicate S.E. The asterisk indicates a significant decrease (p < 0.05). h, hours; d, days.
TABLE 2.
Biological categories enriched for high expression of both Gli1 and Nanog
The table shows the top 15 most enriched categories. The brief description summarizes a description of the biological samples enriched in each category. The experiment ID refers to the corresponding Gene Expression Omnibus (GEO) accession number for the series. Adj. p value, Bonferroni-corrected p values. esiRNA, endoribonuclease-prepared siRNA.
| Rank | Active (Kc) | Total (Nc) | Adj. p value | Sample type | Experiment ID |
|---|---|---|---|---|---|
| 1 | 8 | 8 | 7.42E − 13 | e14tg2a cells: normal | GSE4308, GSE4309 |
| 2 | 9 | 12 | 1.86E − 12 | Embryonic stem cells: r1, undifferentiated mouse embryonic stem cells; fractionated polysomal RNA | GSE9563 |
| 3 | 6 | 6 | 5.15E − 09 | ES cells: ezh2-null ES cells at day 0 (undifferentiated) | GSE12982 |
| 4 | 6 | 13 | 8.22E − 06 | Embryonic stem cells: normal | GSE10476, GSE10573, GSE10553, GSE10610, GSE10806 |
| 5 | 4 | 4 | 3.40E − 05 | ES cell control | GSE14012 |
| 6 | 4 | 4 | 3.40E − 05 | ES cells: ctr9 esiRNA day 4 | GSE12078 |
| 7 | 4 | 4 | 3.40E − 05 | ES cells: luc esiRNA day 4 | GSE12078 |
| 8 | 4 | 8 | 0.00229 | e14tg2a cells: 10 pg amplified | GSE4308, GSE4309 |
| 9 | 3 | 3 | 0.00271 | Embryonic stem cells: ES cells heterozygous for dicer; date of analysis, August 16, 2006 | GSE7141 |
| 10 | 3 | 3 | 0.00271 | Embryonic stem cells: embryonic stem cells transfected with mir290 cluster | GSE8503 |
| 11 | 3 | 3 | 0.00271 | Embryonic stem cells: embryonic stem cells transfected with sirl | GSE8503 |
| 12 | 3 | 3 | 0.00271 | Embryonic stem cells: r1, undifferentiated mouse embryonic stem cells | GSE9563 |
| 13 | 3 | 3 | 0.00271 | ES cells: e14tg1 wild type ES cells at day 0 (undifferentiated) | GSE12982 |
| 14 | 3 | 3 | 0.00271 | Induced pluripotent cells: iPS cells, 4 factors | GSE10806 |
| 15 | 5 | 20 | 0.0056 | Single cell from blimp KO blimp1-null transcript positive, oct4 + cells at ls0b: gene expression data from blimp KO blimp1-null transcript-positive cells (ls0b) | GSE11128 |
TABLE 3.
Biological categories enriched for high expression of Gli1 and medium expression of Nanog
The table shows all enriched categories with an adjusted p value greater than 0.05. The brief description summarizes a description of the biological samples enriched in each category. The experiment ID refers to the corresponding Gene Expression Omnibus (GEO) accession number for the series. Adj. p value, Bonferroni-corrected p values. m, male; na, not applicable; gd, gestational day.
| Rank | Active (Kc) | Total (Nc) | Adj. p value | Sample type | Experiment ID |
|---|---|---|---|---|---|
| 1 | 4 | 5 | 9.80E − 07 | Testes: strain, cd1; pool size, 2; sex, m; developmental stage, gd 11; Theiler stage, 18; somite count, na; developmental landmark, na | GSE4818 |
| 2 | 3 | 3 | 5.97E − 05 | Testes: strain, cd1; pool size, 5; sex, m; developmental stage, gd 12; Theiler stage, 20; somite count, na; developmental landmark, na | GSE4818 |
| 3 | 3 | 3 | 5.97E − 05 | Testes: strain, cd1; pool size, na; sex, m; developmental stage, gd 14; Theiler stage, 22; somite count, na; developmental landmark, na | GSE4818 |
TABLE 4.
Biological categories enriched for high expression of Gli1 and low Nanog
The brief description summarizes a description of the biological samples enriched in each category. The experiment ID refers to the corresponding Gene Expression Omnibus (GEO) accession number for the series. Adj. p value, Bonferroni-corrected p values. rosi, rosiglitazone; e, embryonic day.
| Rank | Active (Kc) | Total (Nc) | Adj. p value | Sample type | Experiment ID |
|---|---|---|---|---|---|
| 1 | 20 | 21 | 6.36E − 25 | Cerebellum: primary tumor cells isolated from conventional patched medulloblastomas | GSE12430 |
| 2 | 7 | 7 | 1.49E − 07 | Cerebellum: cerebellar tumor from olig2tvacre:smom2 mice | GSE11859 |
| 3 | 7 | 7 | 1.49E − 07 | Frontonasal prominence tissue e120: normal | GSE7759 |
| 4 | 7 | 7 | 1.49E − 07 | Frontonasal prominence tissue e125: normal | GSE7759 |
| 5 | 7 | 7 | 1.49E − 07 | Mandibular prominence tissue e100: normal | GSE7759 |
| 6 | 7 | 7 | 1.49E − 07 | Mandibular prominence tissue e110: normal | GSE7759 |
| 7 | 7 | 7 | 1.49E − 07 | Mandibular prominence tissue e115: normal | GSE7759 |
| 8 | 7 | 7 | 1.49E − 07 | Mandibular prominence tissue e120: normal | GSE7759 |
| 9 | 7 | 7 | 1.49E − 07 | Mandibular prominence tissue e125: normal | GSE7759 |
| 10 | 7 | 7 | 1.49E − 07 | Maxillary prominence tissue e115: normal | GSE7759 |
| 11 | 7 | 7 | 1.49E − 07 | Maxillary prominence tissue e120: normal | GSE7759 |
| 12 | 7 | 7 | 1.49E − 07 | Maxillary prominence tissue e125: normal | GSE7759 |
| 13 | 5 | 5 | 0.000105 | Sciatic nerve: mouse sciatic nerve, control rosi | GSE11343 |
| 14 | 5 | 5 | 0.000105 | Sciatic nerve: mouse sciatic nerve, diabetic rosi | GSE11343 |
| 15 | 5 | 5 | 0.000105 | Sciatic nerve: mouse sciatic nerve, diabetic vehicle | GSE11343 |
| 16 | 5 | 5 | 0.000105 | Skeletal muscle: tumor from myf5 cressm2 mouse | GSE6461 |
| 17 | 5 | 6 | 0.000608 | Cerebellum: cerebellar tumor from hgfapcre: smom2 mice | GSE11859 |
| 18 | 4 | 4 | 0.00275 | Medulloblastoma: mouse medulloblastoma | GSE7212 |
| 19 | 4 | 4 | 0.00275 | Palatal shelf tissue e145: normal | GSE11400 |
| 20 | 4 | 4 | 0.00275 | Palatal shelf tissue e145: r26pax3pax3 | GSE11400 |
| 21 | 4 | 4 | 0.00275 | Sciatic nerve: mouse sciatic nerve, nondiabetic vehicle | GSE11343 |
To determine how GLI expression was affected as ES cells start to differentiate, we examined the levels of Gli1, Gli2, and Gli3 upon time course differentiation of ES cells using publicly available microarray data (30). Consistent with the above analysis, Gli1 and Gli2 were robustly expressed in mouse ES cells. Their expression gradually diminishes during differentiation in a fashion similar to ES cell core pluripotency factors such as Oct4 and Nanog (Fig. 5D). Coupled with the previously reported co-expression of Gli1 and Nanog in neural stem cells (16, 17, 46), we conclude that Gli1 is co-expressed with Nanog in several different types of stem/progenitor cells.
To evaluate the effect of HH stimulation on differentiation, we analyzed the expression of several pluripotency and differentiation markers in differentiating ES cells exposed to purmorphamine. Consistent with our previous results (Fig. 4, A and B), HH stimulation resulted in increased Nanog expression during differentiation (Fig. 5E). HH activation also had a modest inhibitory effect on the expression of multiple cell lineage markers, perhaps as a result of increased Nanog expression (Fig. 5E).
Discussion
In this study, we have identified a previously unknown protein-protein interaction between NANOG and GLI proteins. We show that expression of NANOG inhibits the ability of GLI1 to activate transcription. This suggests that NANOG represses the transcriptional activity of GLI proteins. Consistent with this, reduced levels of NANOG increase the level of transcriptional response to HH signaling in differentiating ES cells. At the same time, HH signaling maintains high levels of NANOG in differentiating ES cells, thereby generating a negative feedback loop (Fig. 6A). As NANOG is expressed in a variety of different stem cell populations, this interaction suggests a stem cell-specific mechanism for dampening transcriptional responses to HH signaling.
FIGURE 6.
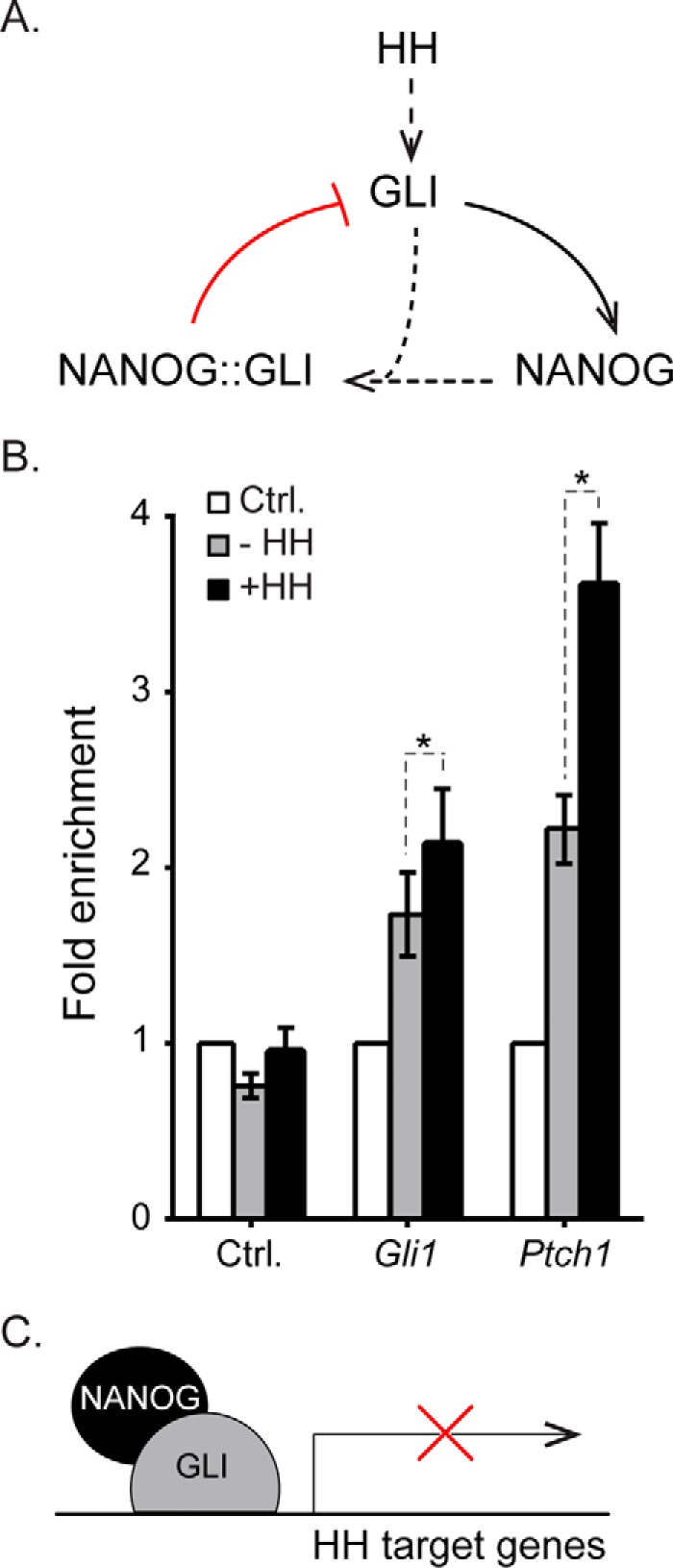
NANOG acts in a negative feedback loop for HH signaling in ES cells. A, GLI is activated in response to HH stimulation. HH signaling up-regulates Nanog via transcriptional mechanisms. NANOG then complexes with GLI1 to inhibit its activity, thereby reducing transcriptional responses to HH signaling. B, enrichment of NANOG at GLI binding regions in differentiating J1 ES cells with biotinylated NANOG or BirA control cells (no biotinylated NANOG) using chromatin immunoprecipitation. Cells were differentiated for 4 days with HH stimulant added after the first 2 days of differentiation. NANOG binding is significantly increased upon HH stimulation at GLI-bound enhancers of the HH target genes Ptch1 (p = 0.003) and Gli1 (p = 0.0063) (n = 5 biological replicates). Error bars indicate S.E. C, schematic representation of NANOG binding to GLI at enhancers of HH target genes to repress transcription. Ctrl, control.
Although HH signaling has well established roles in adult stem cell homeostasis, it is not clear whether HH signaling has a biological role in mouse ES cells. As ES cells lacking the essential HH pathway component Smoothened are capable of contributing to a range of different tissues in chimeric mice, it is not required for survival (47). Nonetheless, Gli1 is strongly expressed in embryonic stem cells (Fig. 5A), and levels are reduced upon ES cell differentiation (Fig. 5D). This pattern of expression mirrors that of ES cell pluripotency factors, and the interaction with NANOG suggests that GLI1 may have an unappreciated role in interacting with the pluripotency network in ES cells. In neural stem cells, the overactivation of HH signaling (by Ptch1 deletion) results in reduced numbers of differentiated neuronal progeny and increased numbers of neuronal stem cells (15). However, neuronal stem cells also require HH signaling to generate differentiated neuronal progeny (for a review, see Ref. 12). Hence, the levels of HH signaling perceived by neuronal stem cells must be tightly regulated to ensure a balance between stem cell renewal and differentiation. By regulating GLI levels, NANOG could provide another level of feedback to moderate a balance between stem cell maintenance and differentiation.
GLI1 and NANOG Participate in a Negative Feedback Loop
As noted above, GLI1 and NANOG have previously been shown to participate in a positive feedback loop in neural stem cells. In particular, GLI1 acts as a transcriptional activator of NANOG in glioma stem cells and cerebellar neurospheres (16, 17). Consistent with this, we found that HH signaling delays the down-regulation of NANOG during ES cell differentiation via transcriptional activation of Nanog (Fig. 4B). HH signaling ultimately causes increased amounts of NANOG protein, thereby acting as a positive regulator. Up-regulated NANOG levels would then be able to form complexes with GLI-A proteins that would inhibit subsequent GLI-mediated transcriptional activation. In doing so, this is acting as a negative feedback loop (Fig. 6A).
This negative feedback loop contrasts with the positive feedback loops by which GLI and NANOG have been proposed to function in human glioblastoma stem cells and cerebellar neurospheres (16, 17). It is also unexpected in light of the high levels of Gli1 that are co-expressed with Nanog in ES cells (Fig. 5A). Because Gli1 is activated by HH signaling, the co-expression of Nanog should ultimately dampen its expression. A possible explanation for this apparent discrepancy might be that NANOG inhibited but did not block HH responses and that high levels of Gli1 observed in these systems might nonetheless represent a dampened response. Alternatively, because Nanog expression fluctuates within a population of ES cells (48–51), a NANOG-GLI negative feedback loop could potentially influence these fluctuations on a single cell level. Our studies did not explore NANOG-GLI-A interactions in other types of stem cells, but given their co-expression, it is feasible that NANOG similarly inhibits GLI1 activation in other stem cells as well.
Mechanism of NANOG-mediated Repression of GLI Target Genes
NANOG binding could repress activation of GLI target genes either through sequestration of GLI proteins or by associating with GLI proteins as they bind to DNA. We used ChIP to test whether NANOG could bind to enhancer-bound GLI proteins. We used ES cells expressing biotinylated NANOG that have previously been used for defining NANOG binding regions in ES cells (20, 25). These cells were differentiated for a total of 4 days with addition of the HH stimulant purmorphamine for the final 2 days. NANOG binding was assessed at enhancers for Gli1 and Ptch1 that are bound by multiple GLI proteins in a range of different tissues (9, 10, 52). Upon HH activation, there was a significant increase in NANOG binding to both enhancers (Fig. 6B). These results are consistent with the model in which NANOG binds to enhancer-bound GLI proteins (Fig. 6C). Our finding that the C-terminal half of NANOG robustly binds GLI1 whereas the N-terminal half, which does not bind GLI1, is required for its repression (Fig. 3, A–C) suggests the presence of a repressor domain that is also consistent with this model.
NANOG has previously been shown to associate with several repressor complexes that also contain OCT4 (35, 53). Because GLI proteins do not bind to OCT4 (Fig. 1A) but OCT4 nonetheless inhibits GLI-mediated transcriptional assays (Fig. 2B), further experiments will be required to determine the composition of the GLI-NANOG repressor complex. One candidate for this complex is DAX1, which was also identified as a GLI3-interacting protein in the mass spectrometry data set (Table 1). DAX1 is a transcriptional co-repressor in several contexts, including ES cells (54–56). Like NANOG, DAX1 is a core member of the ES cell pluripotency network and has previously been identified as a binding partner for both NANOG and OCT4 (20, 35, 38, 56–58). In addition to binding GLI1, NANOG also binds to a truncated, repressor-specific form of GLI3 (Fig. 1). Although assays for GLI transcriptional activation are straightforward, genetic approaches are currently the only meaningful way of determining loss of GLI-R without concomitant GLI activation. In future studies, it will be interesting to determine whether NANOG binding also influences the ability of GLI3R to act as a transcriptional repressor.
NANOG Inhibits Extrinsic Signaling Factors by Binding to Their Transcriptional Effectors
NANOG has previously been shown to interact with SMAD1, a transcriptional mediator of BMP signaling. In this study, NANOG was found to bind to SMAD1 and inhibit BMP-mediated responses that would normally drive ES cells to differentiate (19). NANOG has also been shown to prevent NF-κB-induced differentiation by binding to NF-κB family transcription factors (18). Together with our results, these studies indicate a common mechanism by which NANOG inhibits transcription. Interestingly, the C-terminal half of NANOG that binds GLI proteins also mediates the interaction between NANOG and the NF-κB family transcription factor RELA (18). It remains to be determined whether NANOG binds to these different proteins through an adapter protein or via direct interactions, perhaps through a common protein motif.
Author Contributions
Q. L., S. A. V., H. J., and J. K. conceived and coordinated the study. Q. L., S. A. V., R. K. L., Z. J., and H. J. wrote the manuscript. Q. L., R. K. L., H. C., S. M. G., M. D. P., and Z. J. performed experiments. All authors edited the manuscript, reviewed the results, and approved the final version of this manuscript.
Acknowledgments
We thank Dr. Edward Marcotte and Dr. Daniel Boutz for helpful advice. We thank Dr. Kristin Falkenstein for comments on the manuscript.
This work was supported by National Institutes of Health Grants R01HG006282 (to H. J.) and R01HD073151 (to S. A. V.) and Cancer Prevention Research Institute of Texas Grants RP120343 (to S. A. V.) and RP110782 (to M. D. P.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HH
- Hedgehog
- PTCH1
- Patched1
- BMP
- bone morphogenetic protein
- LIF
- leukemia inhibitory factor
- F
- forward
- R
- reverse
- GLI-A
- GLI activator
- GLI-R
- GLI repressor
- SUFU
- suppressor of fused.
References
- 1. Briscoe J., and Thérond P. P. (2013) The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429 [DOI] [PubMed] [Google Scholar]
- 2. Falkenstein K. N., and Vokes S. A. (2014) Transcriptional regulation of graded Hedgehog signaling. Semin. Cell Dev. Biol. 33, 73–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sasaki H., Nishizaki Y., Hui C., Nakafuku M., and Kondoh H. (1999) Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126, 3915–3924 [DOI] [PubMed] [Google Scholar]
- 4. Wang B., Fallon J. F., and Beachy P. A. (2000) Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423–434 [DOI] [PubMed] [Google Scholar]
- 5. Dai P., Akimaru H., Tanaka Y., Maekawa T., Nakafuku M., and Ishii S. (1999) Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J. Biol. Chem. 274, 8143–8152 [DOI] [PubMed] [Google Scholar]
- 6. Park H. L., Bai C., Platt K. A., Matise M. P., Beeghly A., Hui C. C., Nakashima M., and Joyner A. L. (2000) Mouse Gli1 mutants are viable but have defects in SHH signaling in combination with a Gli2 mutation. Development 127, 1593–1605 [DOI] [PubMed] [Google Scholar]
- 7. Bai C. B., Stephen D., and Joyner A. L. (2004) All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6, 103–115 [DOI] [PubMed] [Google Scholar]
- 8. Bai C. B., Auerbach W., Lee J. S., Stephen D., and Joyner A. L. (2002) Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development 129, 4753–4761 [DOI] [PubMed] [Google Scholar]
- 9. Vokes S. A., Ji H., McCuine S., Tenzen T., Giles S., Zhong S., Longabaugh W. J., Davidson E. H., Wong W. H., and McMahon A. P. (2007) Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development 134, 1977–1989 [DOI] [PubMed] [Google Scholar]
- 10. Vokes S. A., Ji H., Wong W. H., and McMahon A. P. (2008) A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 22, 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Platt K. A., Michaud J., and Joyner A. L. (1997) Expression of the mouse Gli and Ptc genes is adjacent to embryonic sources of hedgehog signals suggesting a conservation of pathways between flies and mice. Mech. Dev. 62, 121–135 [DOI] [PubMed] [Google Scholar]
- 12. Petrova R., and Joyner A. L. (2014) Roles for Hedgehog signaling in adult organ homeostasis and repair. Development 141, 3445–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsu Y. C., Li L., and Fuchs E. (2014) Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 157, 935–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Álvarez-Buylla A., and Ihrie R. A. (2014) Sonic hedgehog signaling in the postnatal brain. Semin. Cell Dev. Biol. 33, 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ferent J., Cochard L., Faure H., Taddei M., Hahn H., Ruat M., and Traiffort E. (2014) Genetic activation of hedgehog signaling unbalances the rate of neural stem cell renewal by increasing symmetric divisions. Stem Cell Rep. 3, 312–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Po A., Ferretti E., Miele E., De Smaele E., Paganelli A., Canettieri G., Coni S., Di Marcotullio L., Biffoni M., Massimi L., Di Rocco C., Screpanti I., and Gulino A. (2010) Hedgehog controls neural stem cells through p53-independent regulation of Nanog. EMBO J. 29, 2646–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zbinden M., Duquet A., Lorente-Trigos A., Ngwabyt S. N., Borges I., and Ruiz i Altaba A. (2010) NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 29, 2659–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Torres J., and Watt F. M. (2008) Nanog maintains pluripotency of mouse embryonic stem cells by inhibiting NFκB and cooperating with Stat3. Nat. Cell Biol. 10, 194–201 [DOI] [PubMed] [Google Scholar]
- 19. Suzuki A., Raya A., Kawakami Y., Morita M., Matsui T., Nakashima K., Gage F. H., Rodríguez-Esteban C., and Izpisúa Belmonte J. C. (2006) Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 103, 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim J., Chu J., Shen X., Wang J., and Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takata K., Reh S., Tomida J., Person M. D., and Wood R. D. (2013) Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nat. Commun. 4, 2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee C. F., Paull T. T., and Person M. D. (2013) Proteome-wide detection and quantitative analysis of irreversible cysteine oxidation using long column UPLC-pSRM. J. Proteome Res. 12, 4302–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vizcaíno J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nybakken K., Vokes S. A., Lin T. Y., McMahon A. P., and Perrimon N. (2005) A genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. Nat. Genet. 37, 1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim J., Cantor A. B., Orkin S. H., and Wang J. (2009) Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat. Protoc. 4, 506–517 [DOI] [PubMed] [Google Scholar]
- 26. McCall M. N., Bolstad B. M., and Irizarry R. A. (2010) Frozen robust multiarray analysis (fRMA). Biostatistics 11, 242–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCall M. N., Uppal K., Jaffee H. A., Zilliox M. J., and Irizarry R. A. (2011) The Gene Expression Barcode: leveraging public data repositories to begin cataloging the human and murine transcriptomes. Nucleic Acids Res. 39, D1011–D1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu G., Yustein J. T., McCall M. N., Zilliox M., Irizarry R. A., Zeller K., Dang C. V., and Ji H. (2013) ChIP-PED enhances the analysis of ChIP-seq and ChIP-chip data. Bioinformatics 29, 1182–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ji Z., Vokes S. A., Dang C. V., and Ji H. (2016) Turning publicly available gene expression data into discoveries using gene set context analysis. Nucleic Acids Res. 44, e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hailesellasse Sene K., Porter C. J., Palidwor G., Perez-Iratxeta C., Muro E. M., Campbell P. A., Rudnicki M. A., and Andrade-Navarro M. A. (2007) Gene function in early mouse embryonic stem cell differentiation. BMC Genomics 8, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schadt E. E., Li C., Ellis B., and Wong W. H. (2001) Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data. J. Cell. Biochem. Suppl. 37, 120–125 [DOI] [PubMed] [Google Scholar]
- 32. Humke E. W., Dorn K. V., Milenkovic L., Scott M. P., and Rohatgi R. (2010) The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kogerman P., Grimm T., Kogerman L., Krause D., Undén A. B., Sandstedt B., Toftgård R., and Zaphiropoulos P. G. (1999) Mammalian suppressor-of-fused modulates nuclear-cytoplasmic shuttling of Gli-1. Nat. Cell Biol. 1, 312–319 [DOI] [PubMed] [Google Scholar]
- 34. Asaoka Y., Kanai F., Ichimura T., Tateishi K., Tanaka Y., Ohta M., Seto M., Tada M., Ijichi H., Ikenoue T., Kawabe T., Isobe T., Yaffe M. B., and Omata M. (2010) Identification of a suppressive mechanism for Hedgehog signaling through a novel interaction of Gli with 14-3-3. J. Biol. Chem. 285, 4185–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., and Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 36. Martello G., and Smith A. (2014) The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 30, 647–675 [DOI] [PubMed] [Google Scholar]
- 37. Young R. A. (2011) Control of the embryonic stem cell state. Cell 144, 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., and Poot R. A. (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loh Y. H., Wu Q., Chew J. L., Vega V. B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K. Y., Sung K. W., Lee C. W., Zhao X. D., Chiu K. P., et al. (2006) The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431–440 [DOI] [PubMed] [Google Scholar]
- 40. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., and Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi S. C., Choi J. H., Park C. Y., Ahn C. M., Hong S. J., and Lim D. S. (2012) Nanog regulates molecules involved in stemness and cell cycle-signaling pathway for maintenance of pluripotency of P19 embryonal carcinoma stem cells. J. Cell. Physiol. 227, 3678–3692 [DOI] [PubMed] [Google Scholar]
- 42. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., and Yamanaka S. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 43. Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., and Smith A. (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 44. Chang D. F., Tsai S. C., Wang X. C., Xia P., Senadheera D., and Lutzko C. (2009) Molecular characterization of the human NANOG protein. Stem Cells 27, 812–821 [DOI] [PubMed] [Google Scholar]
- 45. Agren M., Kogerman P., Kleman M. I., Wessling M., and Toftgård R. (2004) Expression of the PTCH1 tumor suppressor gene is regulated by alternative promoters and a single functional Gli-binding site. Gene 330, 101–114 [DOI] [PubMed] [Google Scholar]
- 46. Clement V., Sanchez P., de Tribolet N., Radovanovic I., and Ruiz i Altaba A. (2007) HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 17, 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wijgerde M., McMahon J. A., Rule M., and McMahon A. P. (2002) A direct requirement for Hedgehog signaling for normal specification of all ventral progenitor domains in the presumptive mammalian spinal cord. Genes Dev. 16, 2849–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., and Smith A. (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- 49. Kalmar T., Lim C., Hayward P., Muñoz-Descalzo S., Nichols J., Garcia-Ojalvo J., and Martinez Arias A. (2009) Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS Biol. 7, e1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Singer Z. S., Yong J., Tischler J., Hackett J. A., Altinok A., Surani M. A., Cai L., and Elowitz M. B. (2014) Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol. Cell 55, 319–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumar R. M., Cahan P., Shalek A. K., Satija R., DaleyKeyser A. J., Li H., Zhang J., Pardee K., Gennert D., Trombetta J. J., Ferrante T. C., Regev A., Daley G. Q., and Collins J. J. (2014) Deconstructing transcriptional heterogeneity in pluripotent stem cells. Nature 516, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee E. Y., Ji H., Ouyang Z., Zhou B., Ma W., Vokes S. A., McMahon A. P., Wong W. H., and Scott M. P. (2010) Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 107, 9736–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., and Songyang Z. (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 54. Li J., Lu Y., Liu R., Xiong X., Zhang Z., Zhang X., Ning G., and Li X. (2011) DAX1 suppresses FXR transactivity as a novel co-repressor. Biochem. Biophys. Res. Commun. 412, 660–666 [DOI] [PubMed] [Google Scholar]
- 55. Uranishi K., Akagi T., Sun C., Koide H., and Yokota T. (2013) Dax1 associates with Esrrb and regulates its function in embryonic stem cells. Mol. Cell. Biol. 33, 2056–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sun C., Nakatake Y., Akagi T., Ura H., Matsuda T., Nishiyama A., Koide H., Ko M. S., Niwa H., and Yokota T. (2009) Dax1 binds to Oct3/4 and inhibits its transcriptional activity in embryonic stem cells. Mol. Cell. Biol. 29, 4574–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang J., Liu G., Ruan Y., Wang J., Zhao K., Wan Y., Liu B., Zheng H., Peng T., Wu W., He P., Hu F. Q., and Jian R. (2014) Dax1 and Nanog act in parallel to stabilize mouse embryonic stem cells and induced pluripotency. Nat. Commun. 5, 5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J., Levasseur D. N., and Orkin S. H. (2008) Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 105, 6326–6331 [DOI] [PMC free article] [PubMed] [Google Scholar]