Abstract
Regulators of G protein Signaling (RGS) promote deactivation of heterotrimeric G proteins thus controlling the magnitude and kinetics of responses mediated by G protein-coupled receptors (GPCR). In the nervous system, RGS7 and RGS9–2 play essential role in vision, reward processing, and movement control. Both RGS7 and RGS9–2 belong to the R7 subfamily of RGS proteins that form macromolecular complexes with R7-binding protein (R7BP). R7BP targets RGS proteins to the plasma membrane and augments their GTPase-accelerating protein (GAP) activity, ultimately accelerating deactivation of G protein signaling. However, it remains unclear if R7BP serves exclusively as a membrane anchoring subunit or further modulates RGS proteins to increase their GAP activity. To directly answer this question, we utilized a rapidly reversible chemically induced protein dimerization system that enabled us to control RGS localization independent from R7BP in living cells. To monitor kinetics of Gα deactivation, we coupled this strategy with measuring changes in the GAP activity by bioluminescence resonance energy transfer-based assay in a cellular system containing μ-opioid receptor. This approach was used to correlate changes in RGS localization and activity in the presence or absence of R7BP. Strikingly, we observed that RGS activity is augmented by membrane recruitment, in an orientation independent manner with no additional contributions provided by R7BP. These findings argue that the association of R7 RGS proteins with the membrane environment provides a major direct contribution to modulation of their GAP activity.
Keywords: allosteric regulation, G protein, G protein-coupled receptor (GPCR), GTPase-activating protein (GAP), regulator of G protein signaling (RGS), signal transduction
Introduction
In the nervous system, signaling through transmembrane G protein-coupled receptors (GPCRs)2 plays a crucial role in a number of fundamental processes including differentiation, neurotransmission, and synaptic plasticity (1). Upon binding to an extracellular ligand, GPCRs undergo a conformational change that leads to activation of intracellular heterotrimeric Gαβγ proteins. This process involves dissociation of G proteins into GαGTP and Gβγ subunits freeing them for the interaction with downstream effectors (2). The magnitude of the response depends on the amount of time G proteins spend in their activated state. Therefore, timely deactivation of G proteins is crucial for determining the overall extent of signaling.
Deactivation of G proteins is controlled by Regulators of G protein signaling (RGS) proteins which act as GTPase-activating proteins (GAPs) accelerating the rate of GTP hydrolysis on the Gα subunit thereby promoting return of G protein to its inactive Gαβγ heterotrimeric state. It is currently well accepted that the RGS-catalyzed G protein deactivation is essential for determining timing, extent and sensitivity of G protein signaling in a number of GPCR pathways (3, 4). Among more than 30 known RGS proteins in mammalian genomes, critical roles for the signaling in the nervous system has been attributed to the R7 family that includes RGS6, RGS7, RGS9, and RGS11. Two members of the R7 family in particular, RGS7 and long splice isoform of RGS9: RGS9–2, ensure timely Gα deactivation required in key neuronal processes including vision, motor control, reward behavior, and nociception (5). For example, the broadly expressed RGS7 has been shown to play key role in synaptic communication of ON-bipolar neurons in the retina (6), regulation GABA neurotransmission in hippocampus (7), and control of morphine reward through μ-opioid receptor (MOR) signaling in the nucleus accumbens (8). RGS9–2 expression is restricted to the striatum where it also regulates signaling through MOR (9) and D2 dopamine receptors (10) and is involved in reward signaling, motor control and neuropathic pain (11–16).
In neurons, the function of RGS7 and RGS9–2 is regulated by their association with several binding partners. Both RGS proteins form an obligatory heterodimer with type 5 G protein β-subunit (Gβ5), which facilitates their folding and provides proteolytic stability (17–19). In addition, both RGS7/Gβ5 and RGS9–2/Gβ5 form complexes with small palmitoylated protein R7-binding protein (R7BP), which plays essential role in controlling their membrane localization and stability (20–25). Another binding partner, the orphan receptor GPR158, is a selective membrane anchor for RGS7, which is similarly critical for RGS7 expression and membrane localization in the nervous system (26, 27).
Targeting to the plasma membrane is a consistent theme in regulation of RGS7/Gβ5 and RGS9–2/Gβ5 function. Both RGS7/Gβ5 and RGS9–2/Gβ5 are largely cytoplasmic when expressed in heterologous system (28–31), although a fraction of RGS7 has been shown to undergo palmitoylation thought to promote its membrane localization (32, 33). The RGS7/Gβ5 and RGS9–2/Gβ5 complexes can be effectively recruited to the plasma membrane via interaction with their substrate, activated Gαo (33) or membrane anchors R7BP (20, 24) and GPR158 (27). In the case of R7BP, this membrane recruitment has been reported to result in augmentation of RGS9–2 and RGS7 GAP activity toward its substrates Gαi and Gαo, accelerating termination of the G protein driven responses (21, 34). The mechanism behind this stimulatory effect is unclear. On the one hand, membrane recruitment of RGS proteins may facilitate G protein deactivation by increasing the proximity to of RGS proteins to their membrane-bound substrate Gα-GTP (21, 27, 34). On the other hand, R7BP has been also proposed to act as an allosteric modulator of RGS proteins (35, 36). However, the impact of membrane association relative to possible direct effects of R7BP is unknown.
In this study we examined the contribution of membrane localization of RGS7 and RGS9–2 in regulation of MOR receptor signaling. We implemented a reversible-chemical dimerization system to control RGS localization independent of binding partners and evaluated real-time changes in GAP activity with and without R7BP in live cells. Our results show that membrane recruitment of RGS is sufficient for attaining the full potentiation of their GAP activity. This finding suggests that augmentation of RGS activity by R7BP may result from positioning R7 RGS proteins in the context of lipid environment.
Experimental Procedures
DNA Constructs
The cloning of R7BP, Gβ5S, and RGS9–2 has been described (20, 28). RGS7 was obtained from Missouri S&T cDNA Resource Center. The KRas membrane targeting construct (37) as well as the BRET biosensors Venus155–239-Gβ1, Venus1–155-Gγ2, were provided by Nevin A. Lambert (Medical College of Georgia, Augusta, GA) (38). Cloning of masGRK3ctNluc reporter has been described (7). The In-Fusion HD Cloning Kit (Clontech, Mountain View, CA) was used to generate the following constructs used in this study (all in pcDNA3.1): mSNAPf, RGS7-P2A-Gβ5, mCherry-RGS7-P2A-Gβ5, FKBP-mCherry-RGS7-P2A-Gβ5, mCherry-RGS7-FKBP-P2A-Gβ5, RGS9–2-P2A-Gβ5, mCherry-RGS9–2-P2A-Gβ5, FKBP-mCherry-RGS9–2-P2A-Gβ5, mCherry-RGS9–2-FKBP-P2A-Gβ5, sR7BP, sR7BP-FKBP, Venus-sR7BP-FKBP, and Venus-R7BP. mCherry was cloned from pmCherry-N1 (Clontech), SNAPf was cloned from pENTR4-SNAPf (provided by Eric Campeau/Addgene plasmid # 29652), and FKBP was cloned from Lyn-FKBP-FKBP-CFP (provided by Tobias Meyer/Addgene plasmid # 20149). Detailed cloning strategies and a list of primer sequences are available upon request.
Cell Culture
NG108–15 cells were cultured in DMEM supplemented with 10% fetal bovine serum, sodium hypoxanthine (0.1 mm), aminopterin (0.4 μm), thymidine (16 μm), penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a 5% CO2 humidified incubator. Cells were transfected at ∼80% confluence in 35-mm plates using Lipofectamine PLUS (2.5 μl) and LTX (4 μl) reagents for Bioluminescence Resonance Energy Transfer assay as previously described with modification (34). MOR, GαoA, Venus155–239-Gβ1, Venus1–155-Gγ2, masGRK3ct-Nluc, and SNAPf-KRas constructs were transfected at a 1:2:1:1:1:1 ratio. Experiments using RGS7, RGS9–2, or R7BP constructs were used at a 1:0.5:1 ratio relative to MOR. A total of 2.5 μg was transfected in each experiment using an empty vector to normalize the total amount of plasmid DNA. rCD1 and FK506 were purchased from Sirius Fine Chemicals (Bremen, Germany) and used at a final concentration of 1 μm.
Confocal Microscopy
Cells were grown on poly-l-lysine coated coverslips, handled as stated above, fixed in a 4% paraformaldehyde/2% glucose solution for 10 min at room temperature, and mounted on slides with DAPI Fluoromount-G (SouthernBiotech). Confocal images were obtained using a Zeiss LSM 880 under a 20× objective (Carl Zeiss). DAPI and mCherry channels were overlaid using ImageJ software.
Fast Kinetic BRET Assay
As we have previously described (34), cells were detached in 5 mm EDTA in PBS at room temperature, centrifuged at 750 × g for 5 min, and resuspended in PBS containing 0.5 mm MgCl2 and 0.1% glucose. Approximately 75,000 cells were added to a 96-well plate followed by an equal volume of Nano-Glo Luciferase Assay Substrate (Promega, Madison, WI). BRET measurements were recorded at room temperature on a POLARstar Omega micro plate reader (BMG Labtech, Cary, NC) utilizing two emission photomultiplier tubes enabling simultaneous detection of light from masGRK3ct-Nluc (475 nm) and Gβ1γ2-Venus (535 nm) with a resolution of 50 milliseconds for every data point. BRET signal was calculated as the ratio of raw 535 nm intensity divided by the raw 475 nm intensity, which was then normalized by subtracting the baseline BRET ratio prior to agonist application and expressed as a percent of maximal BRET signal.
Western Blotting
Following BRET assay samples were centrifuged and resuspended in PBS supplemented with 150 mm NaCl, 1% Triton X-100, and Complete protease inhibitor mixture (Roche, Indianapolis, IN). Cells were lysed by sonication, centrifuged at 14,000 rpm for 20 min at 4 °C, and total protein concentration of the supernatant was determined using the Pierce 660 nm Protein Assay Reagent. Samples were denatured in SDS/urea sample buffer, resolved on PAGEr Gold Gels (Lonza, Basel, Switzerland), and transferred to PVDF membranes for detection with the following primary antibodies: sheep anti-RGS9–2 (20), rabbit anti-RGS7 (39), rabbit anti-R7BP (40), mouse anti-GAPDH (Millipore; AB2302). Species-specific HRP-conjugated secondary antibodies and SuperSignal West Pico ECL (Pierce) was used to capture the signal on film.
Statistical Analysis
Western blots were quantified using ImageJ software. The relative expression of RGS was determined by subtracting the band density from cells only expressing endogenous RGS proteins. For each BRET experiment a single exponential fit, 1/τ (s−1), was obtained from the deactivation phase of the curve. A kGAP rate constant was determined by subtracting the basal deactivation rate of cells only expressing endogenous RGS proteins. To compare GAP activity across experiments, kGAP values were then normalized to RGS expression from Western analysis and presented as kGAP/expression (s−1). A minimum of three biological replicates were performed for each experiment.
Results
Real-time Reversible System for Rapid Membrane Recruitment and Release of RGS/Gβ5 Complexes in Living Cells
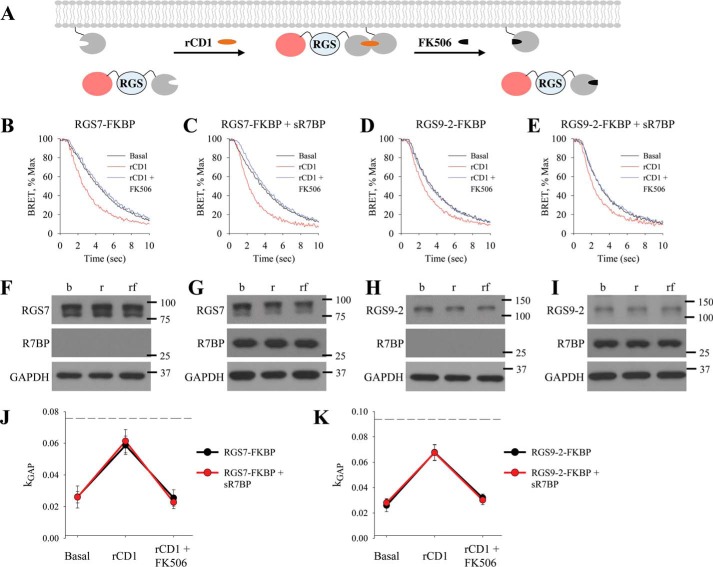
To separate the contribution of protein-protein interactions from the effects of changes in the subcellular localization we sought to develop a system where we can measure RGS GAP activity as we change its association with the plasma membrane. To achieve this, we applied a recently described chemically induced dimerization system that allows controlling protein-protein interactions by a set of small molecule drugs in a rapid and completely reversible manner (41). The system utilizes interaction between two proteins, SNAPf and FKBP, which dimerize upon binding to a cell permeable small molecule rCD1. This interaction is rapidly disrupted by addition of a second small molecule FK506 that serves as a competitive antagonist.
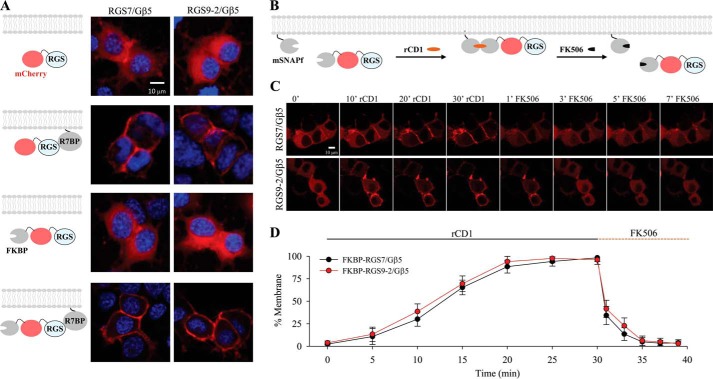
We positioned SNAPf to the plasma membrane, by appending it to the C-terminal membrane-targeting domain of K-Ras, which contains a polybasic cluster of amino acids followed by a site for prenylation, calling the resulting construct mSNAPf. The matching counterpart, FKBP was fused with the fluorescent protein mCherry and added to the N terminus of RGS7 and RGS9–2 (Fig. 1, A and B). When expressed in NG108–15 together with Gβ5 both FKBP-mCherry-RGS7 and FKBP-mCherry-RGS9–2 localized exclusively to the cytoplasm (Fig. 1A). This localization was no different than distribution of RGS7/Gβ5 and RGS9–2/Gβ5 complexes tagged only with mCherry but not FKBP (Fig. 1A) and matched previously documented localization of these proteins in transfected cells (28, 29, 31). Furthermore, FKBP-mCherry-RGS7 or FKBP-mCherry-RGS9–2 complexes with Gβ5 retained their ability to be completely recruited to the plasma membrane upon co-expression with R7BP (Fig. 1A).
FIGURE 1.
Reversible chemical dimerization system controls RGS/Gβ5 subcellular localization on a minute timescale. A, NG108–15 cells were transfected with mCherry-RGS7/Gβ5 and mCherry-RGS9–2/Gβ5, which revealed that these proteins were predominantly in the cytosol. Co-transfection with R7BP localized both RGS7/Gβ5 and RGS9–2/Gβ5 to the plasma membrane. Localization of FKBP-mCherry-RGS/Gβ5 was also cytosolic. Co-transfection of FKBP-mCherry-RGS/Gβ5 with R7BP localized the complex on the membrane and therefore recapitulated conditions of non FKBP-tagged RGS/Gβ5. DAPI stain (blue) was used to visualize the nucleus. B, schematic representation of the reversible chemically-induced dimerization system used to manipulate protein localization. C, representative confocal imaging of living NG108–15 cells co-transfected with mSNAPf and FKBP-mCherry-RGS7/Gβ5 or FKBP-mCherry-RGS9–2/Gβ5. Time course reveals localization dynamics after addition of rCD1 and FK506. All treatments are at 1 μm final concentration. D, quantitation was performed by co-transfection with Venus-tagged membrane anchor (not shown) to compare percentage fluorescence intensity of mCherry-RGS co-localization with Venus. (mean ± S.E.; n = 3 independent experiments. *, p < 0.05; Student's paired t test).
Co-expression of FKBP-mCherry-RGS7 or FKBP-mCherry-RGS9–2 with mSNAPf at the baseline untreated conditions did not change their cytoplasmic localization (Fig. 1, B and C). We next applied rCD1 to the cells co-expressing mSNAPf with either FKBP-mCherry-RGS7 or FKBP-mCherry-RGS9–2 while monitoring changes in protein localization (Fig. 1C). We observed progressive recruitment of fluorescence to the plasma membrane that resulted in ∼90% membrane localization in 20 min (RGS7: 88.5% ± 7.1, RGS9–2: 94.1% ± 5.9) and achieved complete translocation after 30 min (RGS7: 98.3% ± 4.2, RGS9–2: 96.1% ± 5.1) (Fig. 1, C and D). Conversely, addition of FK506 caused extensive and rapid loss of the fluorescence from the plasma membrane and its even redistribution in the cytosol, which quantitatively matched basal conditions within 5 min (RGS7: 4.9% ± 4.8, RGS9–2: 6.2% ± 5.2). Notably the translocation kinetics were similar for both RGS7 and RGS9–2 complexes and were consistent with published applications of this dimerization system (41–43) ultimately enabling us to modulate RGS localization in living cells independent of native membrane anchors. In all subsequent experiments, we chose 30 min rCD1 and 5 min FK506 incubation conditions to achieve complete translocation and reversal of RGS localization back to the cytosol.
Direct Recruitment of RGS7 and RGS9–2 to the Plasma Membrane Potentiates GAP Activity, in the Absence of Membrane Anchors
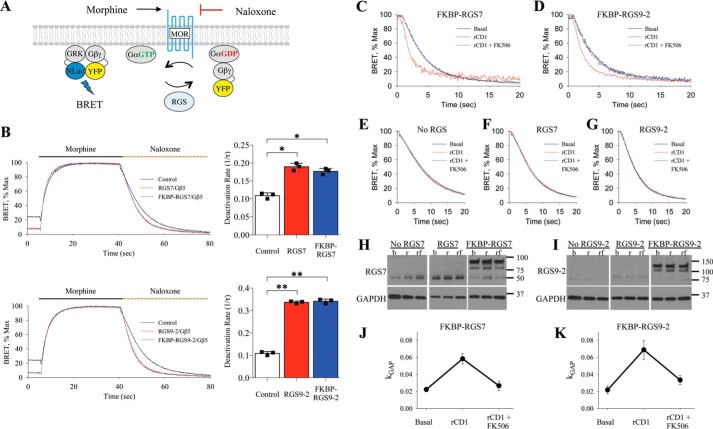
To examine the impact of changes in subcellular localization on the activity of RGS/Gβ5 complexes we used a real-time Bioluminescence Resonance Energy Transfer (BRET)-based assay to monitor kinetics of G protein deactivation in living cells while inducing and reversing recruitment of RGS proteins on the membrane. In this assay (34, 38), NG108–15 cells are transfected with μ-opioid receptor (MOR) together with Gαo, a preferred substrate of RGS7 and RGS9–2, and reporter constructs Gβγ-Venus, and GRK3ct-Nluc (Fig. 2A). Activation of MOR with agonist morphine, initiates signaling during which Venus-tagged Gβγ interacts with the NLuc-tagged effector GRK3ct, generating the BRET signal. Subsequent application of the antagonist naloxone leads to response deactivation, which is rate limited by the GTP hydrolysis on the Gαo (34). Subsequent re-association of Gαo-GDP with the Gβγ quenches the BRET signal. RGS proteins speed up the rate of GTP hydrolysis on the Gα subunits and thereby accelerate the deactivation phase of the response. Consistent with this notion and our previous work (27, 34, 44), we observed that FKBP-mCherry-RGS7 or FKBP-mCherry-RGS9–2 co-expressed with Gβ5 significantly accelerated the deactivation phase of the response (Fig. 2B). Thus, monitoring kinetics of the response deactivation provides a convenient real-time measure of changes in RGS catalytic activity toward Gαo.
FIGURE 2.
Characterization of RGS/Gβ5 subcellular localization on MOR-Gαo signal transduction. A, schematic representation of the fast kinetic BRET assay used to monitor G protein activation and deactivation in live cells. B, representative trace of BRET ratio demonstrating the RGS-dependent deactivation of G proteins in this assay. A single exponential curve fit, 1/τ, was obtained from the deactivation phase of the curve and plotted as a bar graph (mean ± S.E.; n = 3; *, p < 0.005; **, p < 0.001; Student's paired t test). C, in cells co-transfected with FKBP-RGS7/Gβ5, plot of deactivation phase of experiment shows kinetics during basal conditions, addition of rCD1, and addition of rCD1 followed by FK506 treatment. D, deactivation kinetics in cells co-transfected with FKBP-RGS9–2/Gβ5. E–G, negligible effect of rCD1 and FK506 in the absence of FKBP-tagged proteins. H, RGS7 expression determined by Western blotting during basal (b), 30 min rCD1 treatment (r), and 30 min 1 μm rCD1 followed by 5 min 1 μm FK506 treatment (rf). I, RGS9–2 expression determined by Western blotting during basal (b), rCD1 (r), and rCD1 followed by FK506 (rf) treatment. J, kGAP quantification was calculated by subtracting FKBP-RGS7/Gβ5 deactivation rate (1/τ) from the deactivation rate of cells expressing only endogenous RGS, which was then normalized to RGS7 expression levels obtained from Western blot analysis. This was performed from basal, rCD1, and rCD1 followed by FK506 conditions. K, kGAP quantification for FKBP-RGS9–2/Gβ5. Each BRET trace and Western blot is from a single experiment representative of three independent experiments.
Using this system we evaluated the activity of FKBP-mCherry-RGS7/Gβ5 or FKBP-mCherry-RGS9–2/Gβ5 proteins co-expressed with the mSNAPf membrane recruiting module at three time points: basal (cytosolic RGS/Gβ5), 30 min after rCD1 treatment (RGS/Gβ5 recruited the plasma membrane), and 5 min after the addition of FK506 (reversal of RGS/Gβ5 to the cytosol). Treatment with rCD1 substantially accelerated the deactivation kinetics of the response mediated by FKBP-mCherry-RGS7/Gβ5, whereas application of FK506 completely reverted this activity back to basal levels (Fig. 2C). Similar behavior was also observed with FKBP-mCherry-RGS9–2/Gβ5 (Fig. 2D). We next performed a series of controls to validate this approach. We observed no effects of rCD1 or FK506 on response deactivation kinetics when they were added in the absence of RGS expression (Fig. 2E) or when wild-type RGS7 or RGS9–2 were used instead of FKBP tagged constructs (Fig. 2, F and G). Western blot analysis further revealed that rCD1 and FK506 did not impact RGS7 or RGS9–2 expression levels (Fig. 2, H and I). We next quantified RGS activity by calculating kGAP parameter that reports acceleration in Gαo deactivation kinetics induced by RGS proteins, following normalization to protein expression levels. Comparison of kGAP revealed that membrane rCD1 membrane recruitment similarly augmented catalytic activity of both RGS7 and RGS9–2, to about 3-fold relative to baseline levels and the effect was completely reversible by the FK506 (Fig. 2, J and K). Thus, direct recruitment of RGS7 and RGS9–2 enhances their GAP activity toward Gαo.
R7BP-mediated Recruitment of RGS/Gβ5 to the Plasma Membrane Potentiates GAP Activity
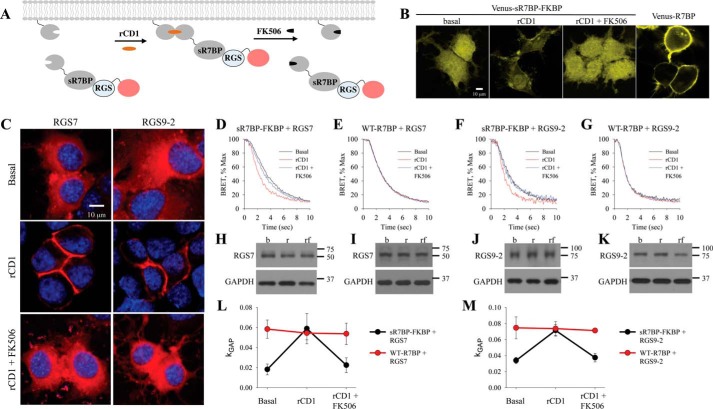
Both RGS7 and RGS9–2 can also be targeted to the plasma membrane by the association with their membrane anchor R7BP and this binding has also been reported to augment the GAP activity of the RGS proteins (21, 34). Therefore, we next examined the impact of controlled localization of RGS7 and RGS9–2 via R7BP. To reversibly control its cellular localization, we made R7BP soluble by replacing its constitutive membrane targeting sequence located at the C terminus with FKBP resulting in a “soluble” R7BP construct (sR7BP-FKBP) compatible with the chemical dimerization system (Fig. 3A). To verify this approach cells were transfected with an N-terminal Venus-tagged sR7BP-FKBP, which localized in the cytosol (Fig. 3B). Membrane translocation kinetics and reversal following rCD1 and FK506 treatment were then examined and found to be similar to FKBP-RGS experiments. When co-expressed in mSNAPf containing cells with mCherry-tagged RGS7/Gβ5 or RGS9–2/Gβ5, sR7BP-FKBP was able to translocate RGS to the plasma membrane within 30 min of rCD1 application followed by complete reversal to the cytosol after 5 min of FK506 treatment (Fig. 3C), recapitulating the behavior of RGS proteins in direct recruitment experiments.
FIGURE 3.
R7BP-mediated localization of RGS/Gβ5 increases GAP activity. A, schematic representation of reversible chemically-induced dimerization system where soluble R7BP contains the FKBP-binding partner (sR7BP-FKBP). B, visualization of R7BP reversible dimerization in NG108–15 cells co-transfected with Venus-tagged sR7BP-FKBP and mSNAPf for comparison with cells transfected with Venus-tagged full-length R7BP. C, representative confocal images of NG108–15 cells co-transfected with mSNAPf and mCherry-RGS7/Gβ5 or mCherry-RGS9–2/Gβ5 under basal, 30 min of rCD1 (1 μm), and 30 min of rCD1 (1 μm) followed by 5 min of FK506 (1 μm) treatment. DAPI stain (blue) was used to visualize the nucleus. D–G, cells were co-transfected with RGS7/Gβ5 or RGS9–2/Gβ5 in the presence of sR7BP-FKBP and WT-R7BP. BRET assay was performed and the deactivation phase during basal, rCD1, and rCD1 followed by FK506 treatment was plotted. H–K, Western blot analysis was used to determine RGS expression level. L–M, kGAP quantification was calculated as described above. Each BRET trace and Western blot is from a single experiment representative of three independent experiments.
Importantly, the addition of rCD1 accelerated the Gαo deactivation kinetics of the response in the system containing sR7BP-FKBP upon termination of MOR signaling (Fig. 3, D and F). This effect was completely reversed after addition of FK506, which released the R7BP-RGS complexes back to the cytosol. In contrast, when the same experiment was performed with wild-type R7BP, we saw no influence of rCD1 or FK506 on deactivation kinetics (Fig. 3, E and G). The treatments had no significant effect on the expression level of RGS proteins (Fig. 3, H–K) as assessed by Western blotting. The relative RGS content was used to normalize the data while calculating the kGAP values (Fig. 3, L and M). Comparison of kGAP between treatments and conditions revealed that rCD1 increases the catalytic activity of both RGS7 and RGS9–2 ∼3-fold and brings it to the level seen with wild-type R7BP. These data indicate that that membrane targeting through sR7BP-FKBP or wild type R7BP has the same impact on augmenting RGS/Gβ5 GAP activity.
The GAP Activity of RGS/Gβ5 Localized on the Membrane Is Not Further Potentiated by R7BP
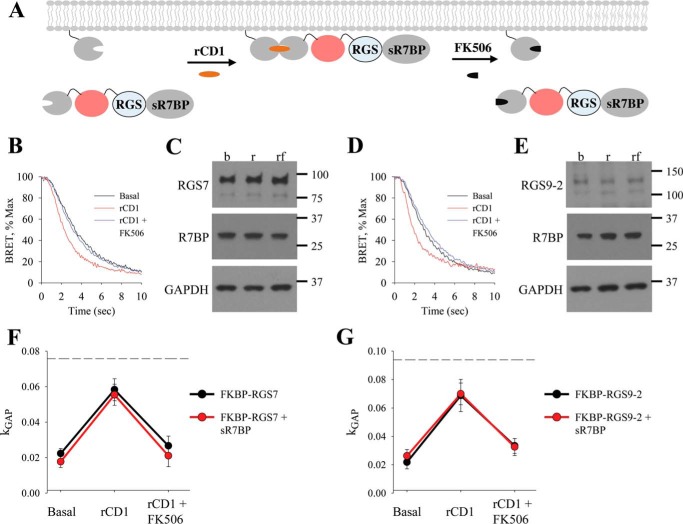
To test whether interaction with R7BP may lead to further potentiation of RGS/Gβ5 activity, we studied the effect of soluble R7BP devoid of FKBP tag (sR7BP) on the activity of FKBP-RGS/Gβ5 complexes before and after their membrane recruitment to the membrane via mSNAPf. (Fig. 4A). Similar to prior experiments, treatment with rCD1 accelerated the deactivation of the response catalyzed by FKBP-mCherry-RGS7 and FKBP-mCherry-RGS9–2 in the presence of sR7BP (Fig. 4, B and D). Again, this effect was completely reversible by the application of FK506. Western blot analysis revealed that no changes in levels of RGS or sR7BP were induced by treatments, and respective band density values were used to normalize kGAP values (Fig. 4, C and E). The resulting data were then plotted together with data from Fig. 2 to compare the effect of FKBP-RGS/Gβ5S when bound or unbound to R7BP (Fig. 4, F and G). Analysis of these data reveals that both baseline and membrane-potentiated GAP activity of RGS7/Gβ5 and RGS9–2/Gβ5 complexes are quantitatively identical regardless of R7BP presence. Additional experiments with higher amounts of transfected RGS proteins have revealed that observable kGAP values do not reach saturation under these conditions ruling out that R7BP fails to further augment RGS activity due to the ceiling effect.
FIGURE 4.
Direct recruitment of RGS/Gβ5 to the membrane increases GAP activity which is not further potentiated by R7BP. A, schematic representation of reversible chemically-induced dimerization system with FKBP-mCherry-RGS in the presence of sR7BP. B, cells were co-transfected with sR7BP and FKBP-mCherry-RGS7/Gβ5. BRET assay was performed and the deactivation phase during basal, rCD1, and rCD1 followed by FK506 treatment was plotted. C, Western blot analysis was used to determine expression levels of RGS7 and R7BP. D, cells were co-transfected with sR7BP and FKBP-mCherry-RGS9–2/Gβ5. BRET assay was performed, and the deactivation phase during basal, rCD1, and rCD1 followed by FK506 treatment was plotted. E, Western blot analysis was used to determine expression levels of RGS9–2 and R7BP. F–G, kGAP quantification was calculated for FKBP-RGS/Gβ5 in the presence of sR7BP and plotted with kGAP for FKBP-RGS/Gβ5 in the absence of R7BP (data from Fig. 2). Dotted line shows maximum kGAP value observed in this system when higher amount of RGS were used in the transfection. Each BRET trace and Western blot is from a single experiment representative of three independent experiments.
Membrane-mediated Increase in Catalytic Activity of RGS/Gβ5 Is Insensitive to their Orientation on the Plasma Membrane
We next examined the role of the RGS/Gβ5 complex orientation relative to the plasma membrane in facilitating their catalytic activity. RGS/Gβ5 proteins bind R7BP via their N-terminal DEP domain. Thus by fusing FKBP to the N terminus of RGS we mimicked the native orientation of the complex during recruitment to the membrane. To understand the relevance of this orientation to modulation of catalytic activity, we next inverted the orientation of RGS/Gβ5 at the plasma membrane. This was achieved by fusing FKBP to the C terminus of mCherry-tagged RGS7 and RGS9–2 (Fig. 5A). Surprisingly, addition of rCD1 accelerated deactivation kinetics in the presence of both mCherry-RGS7-FKBP and mCherry-RGS9–2-FKBP (Fig. 5, B and D). This effect was completely reversible by FK506, indicating that it was caused by changes in protein localization (Fig. 5, C and E). Western blotting analysis did not reveal any significant changes in protein expression levels (Fig. 5, F–I). Analysis of the kGAP values indicated significant enhancement in catalytic activity of both RGS7 and RGS9–2 upon membrane recruitment. The changes in the activity were indistinguishable between the absence and presence of sR7BP (Fig. 5, J and K) indicating that R7BP had no effect on either baseline or membrane-augmented activity of RGS complexes.
FIGURE 5.
Membrane orientation of RGS/Gβ5 does not impact GAP activity. A, schematic representation of reversible chemically-induced dimerization system with FKBP-binding partner on the C terminus of RGS (mCherry-RGS-FKBP). BRET assay was performed, and the deactivation phase during basal, rCD1, and rCD1 followed by FK506 treatment was plotted for: B, mCherry-RGS7-FKBP/Gβ5; C, mCherry-RGS7-FKBP/Gβ5 in the presence of sR7BP; D, mCherry-RGS9–2-FKBP/Gβ5; E, mCherry-RGS9–2-FKBP/Gβ5 in the presence of sR7BP. F–I, Western blot analysis was used to determine expression levels of RGS and R7BP. J–K, kGAP quantification was calculated as described above. Dotted line shows maximum kGAP value observed in this system when higher amount of RGS were used in the transfection. Each BRET trace and Western blot is from a single experiment representative of three independent experiments.
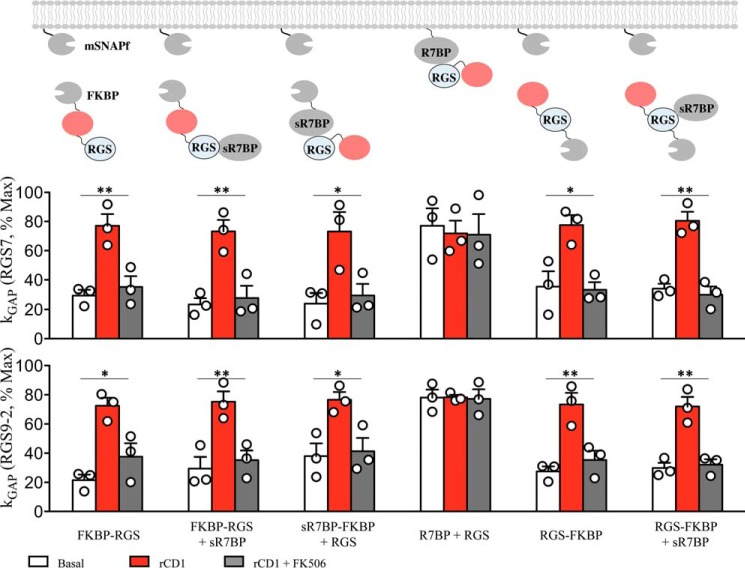
RGS GAP Activity Is Equivalently Potentiated by Membrane Recruitment Regardless of Molecular Environment
Having established the operational range within which the GAP activity of RGS proteins does not reach saturation, we sought to directly compare the effects of various membrane recruitment modes. This was achieved by normalizing the levels of GAP activity to the maximal levels and comparing the resulting values across conditions (Fig. 6). This analysis reveals that the activity of both RGS7 and RGS9–2 complexes were similarly potentiated by membrane recruitment by ∼3-fold from ∼ 20–30% to 70–80% relative to maximal activity levels. Importantly, the GAP activity values on the membrane reached the same values regardless of the mechanism of recruitment, membrane orientation, and the identity of the RGS complex. Furthermore, in each instance the potentiation was fully reversible by the dissociation of the complexes from the membrane. These data suggest that membrane recruitment potentiates the activity of RGS/Gβ5 complexes regardless of their orientation on the membrane or association with R7BP.
FIGURE 6.
Comparative analysis of RGS/Gβ5 kGAP activity. Normalization of kGAP values as a percentage of the maximum kGAP in this system (100%), which was obtained by experiments with higher RGS transfection ratios (mean ± S.E.; n = 3; *, p < 0.05; **, p < 0.02; Student's paired t test).
Discussion
In this study we found that RGS7 and RGS9–2 association with the plasma membrane is sufficient for increasing their catalytic activity of deactivating Gα signaling. We first developed a reversible chemically induced protein interaction scheme to manipulate RGS subcellular localization on a minute timescale. Using this system in combination with highly sensitive cell-based BRET assay to measure RGS activity, we found that RGS plasma membrane recruitment directly correlates with increases in the GAP activity toward Gαo in a signaling system based on MOR. Curiously, addition of the R7BP did not result in further potentiation of the GAP activity of R7 RGS complexes. These results suggest R7BP serves purely as a membrane targeting element that does not directly contribute to the GAP activity of RGS/Gβ5 complexes.
In general, GPCR signaling components are tightly integrated with the plasma membrane to efficiently receive and propagate external stimuli. GPCRs are transmembrane proteins, the Gαβγ subunits are subject to post-translational lipid modifications for membrane attachment, and many effector molecules are either transmembrane (adenylate cyclases, GIRK channels, etc.) or membrane associated (GRKs, Phospholipases, etc.) proteins. Therefore, targeting RGS proteins to the membrane is essential to regulate GPCR signaling. Although a fraction of RGS7 has been reported to undergo palmitoylation (32) or be recruited to the membrane through Gαo (33), both RGS7 and RGS9–2 are predominantly cytosolic proteins which rely on membrane anchors for membrane recruitment (28–31). Thus, it appears natural that the activity of RGS proteins is modulated by membrane-associated binding partners. RGS7 is localized to the plasma membrane by direct binding to R7BP or GPR158 and in either case this has been shown to accelerate Gα deactivation in transfected cells (27, 34). Similarly, RGS9-2 is targeted to the membrane by association with R7BP, an event that also accelerates Gα signal termination in transfected cells (34).
In the context of the membrane, several factors may play a role in the regulation of RGS GAP activity. The placement of RGS on the membrane where Gα is co-localized may augment the activity of RGS proteins through increased likelihood for interaction simply due to proximity. The membrane further increases the frequency of collision between molecules by restricting diffusion to a two-dimensional plane as opposed to a three-dimensional cytosolic space. There are also biochemical principles to consider. For example, different lipid modifications may result in a non-random distribution throughout the membrane. This makes it plausible for palmitoylated R7BP to exist in such lipid raft microdomains as other palmitoylated proteins (i.e. Gαo). Lipid raft compartmentalization may further impact signaling due to G protein subunit diversity (16 Gα, 5 Gβ, 12 Gγ) providing hundreds of possible heterotrimeric combinations (45, 46), each of which with a differing intrinsic lipid modification footprint (i.e. Gαo is palmitoylated and myristoylated whereas Gγ1 is farnesylated) (47).
In addition to diffusional/proximity effects, membrane association is increasingly documented to impact the activity of the signaling molecules by allosteric effects. Such mechanism where the membrane acts directly as an allosteric modulator over protein function was documented for RhoGEFs (48, 49) whose catalytic activity is enhanced by tethering to phospholipid vesicles and for phospholipase C-β (PLC-β) whose ability to hydrolyze phosphatidylinositol 4,5-bisphosphate requires electrostatic displacement of the inhibitory “cap” by charged lipid heads of the membrane (50). Similarly, the results of our study show that the primary role in modulating the activity of RGS/Gβ5 complexes belongs to the plasma membrane. Although the exact mechanisms and structural basis of this effect remain to be fully elucidated, we think that the lack of the orientation effect suggests that the enhancement of RGS/Gβ5 GAP activity toward Gαo is likely achieved by passive proximity mechanism rather than active membrane induced allosterism.
Apart from the effects of the membrane, the activity of RGS/Gβ5 complex have been suggested to be regulated allosterically by interaction with their membrane-anchoring proteins. Retina specific membrane anchor R9AP is capable of facilitating the GAP activity of RGS11/Gβ5 toward Gαo (51). Because RGS11/Gβ5 efficiently associates with the membranes independently from R9AP binding, it was concluded that this potentiation resulted from additional allosteric effects in the context of the membrane. Outside of the membrane association, R9AP/R7BP-like domain in orphan receptor GPR158 was shown to facilitate GAP activity of RGS7 toward Gαo in solution (26). Because it was observed in a purified recombinant system, this augmentation is also likely to result from allosteric affects directly associated with RGS7/Gβ5 binding to GPR158 fragment. Allosterism was also implicated in the action of R7BP in transfected cell system, where it facilitated complex formation of RGS7/Gβ5 and RGS9–2/Gβ5 with the effector channel GIRK (35). However, in that study the effects of R7BP on the catalytic activity of RGS complexes were not assessed directly, and hence the contribution of the membrane recruitment versus protein-protein interaction to the action of R7BP remained unknown.
Surprisingly, our investigation concludes that association of RGS7/Gβ5 and RGS9–2/Gβ5 with the plasma membrane is sufficient to fully potentiate their catalytic activity. This observation is consistent with the lack of the functional effect produced by R7BP in solution when tested in recombinant system with purified components (26), which indicates that just binding to R7BP does not alter GAP activity of RGS/Gβ5 complexes. Considering these results together, the most parsimonious explanation of our findings is that the main role of R7BP in regulating the activity of RGS complexes may be in bringing them to the membrane environment, which in turn directly potentiates the GAP activity by increasing the proximity of RGS proteins to their substrates, activated Gα-GTP.
While the present study did not detect significant effects of R7BP on the GAP activity of Gαo, binding to R7BP has been documented to result in conformational changes in RGS7/Gβ5 and that altering the dynamics of these changes affects the ability of RGS7/Gβ5 to regulate signaling via M3 muscarinic receptor (36). Thus it is possible that that R7BP still exerts allosteric effects on RGS/Gβ5 complexes, which affect their interactions with GPCRs instead of directly influencing the catalytic GAP activity of the RGS domain.
In summary, our study highlights the importance of RGS localization to the membrane in providing timely termination of GPCR signaling. Regulation of the membrane association of R7 RGS proteins with multiple binding partners and membrane environment thus serves as a key event that can powerfully shape cellular and behavioral responses to neurotransmitters.
Author Contributions
B. S. M. designed and executed all of the experiments in this study and wrote the paper. K. A. M. designed the experiments and wrote the paper.
Acknowledgments
We thank Samantha Cocco and Alexander Crain for technical assistance and Dr. Cesare Orlandi for assistance with cloning RGS9–2-P2A-Gβ5 construct. Rabbit anti-R7BP was a generous gift from Dr. William Simonds (NIDDK, National Institutes of Health, Bethesda, MD). BRET biosensors Venus155-239-Gβ1 and Venus1-155-Gγ2 were a generous gift from Dr. Nevin A. Lambert (Medical College of Georgia, Augusta, GA).
This work was supported by National Institutes of Health Grants DA036596 and DA026405 (to K. A. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- GPCR
- G protein-coupled receptor
- GAP
- GTPase-activating protein
- RGS
- regulator of G protein signaling
- R7BP
- R7-binding protein
- MOR
- μ-opioid receptor.
References
- 1. Wettschureck N., and Offermanns S. (2005) Mammalian G proteins and their cell type specific functions. Physiol. Rev. 85, 1159–1204 [DOI] [PubMed] [Google Scholar]
- 2. Neer E. J. (1995) Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80, 249–257 [DOI] [PubMed] [Google Scholar]
- 3. Ross E. M., and Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 4. Hollinger S., and Hepler J. R. (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54, 527–559 [DOI] [PubMed] [Google Scholar]
- 5. Anderson G. R., Posokhova E., and Martemyanov K. A. (2009) The R7 RGS protein family: multi-subunit regulators of neuronal G protein signaling. Cell Biochem. Biophys. 54, 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cao Y., Pahlberg J., Sarria I., Kamasawa N., Sampath A. P., and Martemyanov K. A. (2012) Regulators of G protein signaling RGS7 and RGS11 determine the onset of the light response in ON bipolar neurons. Proc. Natl. Acad. Sci. U.S.A. 109, 7905–7910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ostrovskaya O., Xie K., Masuho I., Fajardo-Serrano A., Lujan R., Wickman K., and Martemyanov K. A. (2014) RGS7/Gbeta5/R7BP complex regulates synaptic plasticity and memory by modulating hippocampal GABABR-GIRK signaling. eLife 3, e02053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sutton L. P., Ostrovskaya O., Dao M., Xie K., Orlandi C., Smith R., Wee S., and Martemyanov K. A. (2015) Regulator of G-Protein Signaling 7 Regulates Reward Behavior by Controlling Opioid Signaling in the Striatum. Biol. Psychiatry, pii: S0006–3223(15)00653-8, 10.1016/j.biopsych.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Psigfogeorgou K., Terzi D., Papachatzaki M. M., Varidaki A., Ferguson D., Gold S. J., and Zachariou V. (2011) A unique role of RGS9–2 in the striatum as a positive or negative regulator of opiate analgesia. J. Neurosci. 31, 5617–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cabrera-Vera T. M., Hernandez S., Earls L. R., Medkova M., Sundgren-Andersson A. K., Surmeier D. J., and Hamm H. E. (2004) RGS9–2 modulates D2 dopamine receptor-mediated Ca2+ channel inhibition in rat striatal cholinergic interneurons. Proc. Natl. Acad. Sci. U.S.A. 101, 16339–16344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mitsi V., Terzi D., Purushothaman I., Manouras L., Gaspari S., Neve R. L., Stratinaki M., Feng J., Shen L., and Zachariou V. (2015) RGS9–2-controlled adaptations in the striatum determine the onset of action and efficacy of antidepressants in neuropathic pain states. Proc. Natl. Acad. Sci. U.S.A. 112, E5088–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gold S. J., Hoang C. V., Potts B. W., Porras G., Pioli E., Kim K. W., Nadjar A., Qin C., LaHoste G. J., Li Q., Bioulac B. H., Waugh J. L., Gurevich E., Neve R. L., and Bezard E. (2007) RGS9–2 negatively modulates L-3,4-dihydroxyphenylalanine-induced dyskinesia in experimental Parkinson's disease. J. Neurosci. 27, 14338–14348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovoor A., Seyffarth P., Ebert J., Barghshoon S., Chen C. K., Schwarz S., Axelrod J. D., Cheyette B. N., Simon M. I., Lester H. A., and Schwarz J. (2005) D2 dopamine receptors colocalize regulator of G-protein signaling 9–2 (RGS9–2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J. Neurosci. 25, 2157–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rahman Z., Schwarz J., Gold S. J., Zachariou V., Wein M. N., Choi K. H., Kovoor A., Chen C. K., DiLeone R. J., Schwarz S. C., Selley D. E., Sim-Selley L. J., Barrot M., Luedtke R. R., Self D., Neve R. L., Lester H. A., Simon M. I., and Nestler E. J. (2003) RGS9 modulates dopamine signaling in the basal ganglia. Neuron 38, 941–952 [DOI] [PubMed] [Google Scholar]
- 15. Zachariou V., Georgescu D., Sanchez N., Rahman Z., DiLeone R., Berton O., Neve R. L., Sim-Selley L. J., Selley D. E., Gold S. J., and Nestler E. J. (2003) Essential role for RGS9 in opiate action. Proc. Natl. Acad. Sci. U.S.A. 100, 13656–13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson G. R., Cao Y., Davidson S., Truong H. V., Pravetoni M., Thomas M. J., Wickman K., Giesler G. J. Jr., and Martemyanov K. A. (2010) R7BP complexes with RGS9–2 and RGS7 in the striatum differentially control motor learning and locomotor responses to cocaine. Neuropsychopharmacology 35, 1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen C. K., Eversole-Cire P., Zhang H., Mancino V., Chen Y. J., He W., Wensel T. G., and Simon M. I. (2003) Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc. Natl. Acad. Sci. U.S.A. 100, 6604–6609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cabrera J. L., De Freitas F., Satpaev D. K., and Slepak V. Z. (1998) Identification of the G*5-RGS7 protein complex in the retina. Biochem. Biophys. Res. Commun. 249, 898–902 [DOI] [PubMed] [Google Scholar]
- 19. Makino E. R., Handy J. W., Li T. S., and Arshavsky V. Y. (1999) The GTPase activating factor for transducin in rod photoreceptors is the complex between RGS9 and type 5 G protein * subunit. Proc. Natl. Acad. Sci. U.S.A. 96, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martemyanov K. A., Yoo P. J., Skiba N. P., and Arshavsky V. Y. (2005) R7BP, a novel neuronal protein interacting with RGS proteins of the R7 family. J. Biol. Chem. 280, 5133–5136 [DOI] [PubMed] [Google Scholar]
- 21. Drenan R. M., Doupnik C. A., Jayaraman M., Buchwalter A. L., Kaltenbronn K. M., Huettner J. E., Linder M. E., and Blumer K. J. (2006) R7BP augments the function of RGS7*Gβ5 complexes by a plasma membrane-targeting mechanism. J. Biol. Chem. 281, 28222–28231 [DOI] [PubMed] [Google Scholar]
- 22. Anderson G. R., Lujan R., Semenov A., Pravetoni M., Posokhova E. N., Song J. H., Uversky V., Chen C. K., Wickman K., and Martemyanov K. A. (2007) Expression and localization of RGS9–2/G 5/R7BP complex in vivo is set by dynamic control of its constitutive degradation by cellular cysteine proteases. J. Neurosci. 27, 14117–14127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Posokhova E., Uversky V., and Martemyanov K. A. (2010) Proteomic identification of Hsc70 as a mediator of RGS9–2 degradation by in vivo interactome analysis. J. Proteome Res. 9, 1510–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drenan R. M., Doupnik C. A., Boyle M. P., Muglia L. J., Huettner J. E., Linder M. E., and Blumer K. J. (2005) Palmitoylation regulates plasma membrane-nuclear shuttling of R7BP, a novel membrane anchor for the RGS7 family. J. Cell Biol. 169, 623–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson G. R., Semenov A., Song J. H., and Martemyanov K. A. (2007) The membrane anchor R7BP controls the proteolytic stability of the striatal specific RGS protein, RGS9–2. J. Biol. Chem. 282, 4772–4781 [DOI] [PubMed] [Google Scholar]
- 26. Orlandi C., Xie K., Masuho I., Fajardo-Serrano A., Lujan R., and Martemyanov K. A. (2015) Orphan receptor GPR158 is an allosteric modulator of RGS7 catalytic activity with an essential role in dictating its expression and localization in the brain. J. Biol. Chem. 290, 13622–13639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Orlandi C., Posokhova E., Masuho I., Ray T. A., Hasan N., Gregg R. G., and Martemyanov K. A. (2012) GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J. Cell Biol. 197, 711–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Song J. H., Waataja J. J., and Martemyanov K. A. (2006) Subcellular targeting of RGS9–2 is controlled by multiple molecular determinants on its membrane anchor, R7BP. J. Biol. Chem. 281, 15361–15369 [DOI] [PubMed] [Google Scholar]
- 29. Liapis E., Sandiford S., Wang Q., Gaidosh G., Motti D., Levay K., and Slepak V. Z. (2012) Subcellular localization of regulator of G protein signaling RGS7 complex in neurons and transfected cells. J. Neurochem. 122, 568–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang J. H., Barr V. A., Mo Y., Rojkova A. M., Liu S., and Simonds W. F. (2001) Nuclear localization of G protein β5 and regulator of G protein signaling 7 in neurons and brain. J. Biol. Chem. 276, 10284–10289 [DOI] [PubMed] [Google Scholar]
- 31. Bouhamdan M., Michelhaugh S. K., Calin-Jageman I., Ahern-Djamali S., and Bannon M. J. (2004) Brain-specific RGS9–2 is localized to the nucleus via its unique proline-rich domain. Biochim. Biophys. Acta 1691, 141–150 [DOI] [PubMed] [Google Scholar]
- 32. Rose J. J., Taylor J. B., Shi J., Cockett M. I., Jones P. G., and Hepler J. R. (2000) RGS7 is palmitoylated and exists as biochemically distinct forms. J. Neurochem. 75, 2103–2112 [DOI] [PubMed] [Google Scholar]
- 33. Takida S., Fischer C. C., and Wedegaertner P. B. (2005) Palmitoylation and plasma membrane targeting of RGS7 are promoted by alpha o. Mol. Pharmacol. 67, 132–139 [DOI] [PubMed] [Google Scholar]
- 34. Masuho I., Xie K., and Martemyanov K. A. (2013) Macromolecular composition dictates receptor and G protein selectivity of regulator of G protein signaling (RGS) 7 and 9–2 protein complexes in living cells. J. Biol. Chem. 288, 25129–25142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhou H., Chisari M., Raehal K. M., Kaltenbronn K. M., Bohn L. M., Mennerick S. J., and Blumer K. J. (2012) GIRK channel modulation by assembly with allosterically regulated RGS proteins. Proc. Natl. Acad. Sci. U.S.A. 109, 19977–19982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Narayanan V., Sandiford S. L., Wang Q., Keren-Raifman T., Levay K., and Slepak V. Z. (2007) Intramolecular interaction between the DEP domain of RGS7 and the Gβ5 subunit. Biochemistry 46, 6859–6870 [DOI] [PubMed] [Google Scholar]
- 37. Lan T. H., Liu Q., Li C., Wu G., and Lambert N. A. (2012) Sensitive and high resolution localization and tracking of membrane proteins in live cells with BRET. Traffic 13, 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hollins B., Kuravi S., Digby G. J., and Lambert N. A. (2009) The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signal. 21, 1015–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rojkova A. M., Woodard G. E., Huang T. C., Combs C. A., Zhang J. H., and Simonds W. F. (2003) Gγ subunit-selective G protein β5 mutant defines regulators of G protein signaling protein binding requirement for nuclear localization. J. Biol. Chem. 278, 12507–12512 [DOI] [PubMed] [Google Scholar]
- 40. Nini L., Waheed A. A., Panicker L. M., Czapiga M., Zhang J. H., and Simonds W. F. (2007) R7-binding protein targets the G protein β 5/R7-regulator of G protein signaling complex to lipid rafts in neuronal cells and brain. BMC Biochem. 8, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feng S., Laketa V., Stein F., Rutkowska A., MacNamara A., Depner S., Klingmüller U., Saez-Rodriguez J., and Schultz C. (2014) A rapidly reversible chemical dimerizer system to study lipid signaling in living cells. Angew Chem. Int. Ed Engl. 53, 6720–6723 [DOI] [PubMed] [Google Scholar]
- 42. MacNamara A., Stein F., Feng S., Schultz C., and Saez-Rodriguez J. (2015) A single-cell model of PIP3 dynamics using chemical dimerization. Bioorg. Med. Chem. 23, 2868–2876 [DOI] [PubMed] [Google Scholar]
- 43. Schifferer M., Feng S., Stein F., Tischer C., and Schultz C. (2015) Reversible chemical dimerizer-induced recovery of PIP2 levels moves clathrin to the plasma membrane. Bioorg. Med. Chem. 23, 2862–2867 [DOI] [PubMed] [Google Scholar]
- 44. Xie K., Masuho I., Brand C., Dessauer C. W., and Martemyanov K. A. (2012) The complex of G protein regulator RGS9–2 and Gβ(5) controls sensitization and signaling kinetics of type 5 adenylyl cyclase in the striatum. Sci. Signal 5, ra63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hillenbrand M., Schori C., Schöppe J., and Plückthun A. (2015) Comprehensive analysis of heterotrimeric G-protein complex diversity and their interactions with GPCRs in solution. Proc. Natl. Acad. Sci. U.S.A. 112, E1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Masuho I., Ostrovskaya O., Kramer G. M., Jones C. D., Xie K., and Martemyanov K. A. (2015) Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Sci. Signal 8, ra123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wedegaertner P. B., Wilson P. T., and Bourne H. R. (1995) Lipid modifications of trimeric G proteins. J. Biol. Chem. 270, 503–506 [DOI] [PubMed] [Google Scholar]
- 48. Medina F., Carter A. M., Dada O., Gutowski S., Hadas J., Chen Z., and Sternweis P. C. (2013) Activated RhoA is a positive feedback regulator of the Lbc family of Rho guanine nucleotide exchange factor proteins. J. Biol. Chem. 288, 11325–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carter A. M., Gutowski S., and Sternweis P. C. (2014) Regulated localization is sufficient for hormonal control of regulator of G protein signaling homology Rho guanine nucleotide exchange factors (RH-RhoGEFs). J. Biol. Chem. 289, 19737–19746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Charpentier T. H., Waldo G. L., Barrett M. O., Huang W., Zhang Q., Harden T. K., and Sondek J. (2014) Membrane-induced allosteric control of phospholipase C-beta isozymes. J. Biol. Chem. 289, 29545–29557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masuho I., Celver J., Kovoor A., and Martemyanov K. A. (2010) Membrane anchor R9AP potentiates GTPase-accelerating protein activity of RGS11 x Gbeta5 complex and accelerates inactivation of the mGluR6-G(o) signaling. J. Biol. Chem. 285, 4781–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]