ABSTRACT
While significant protection from pneumococcal disease has been achieved by the use of polysaccharide and polysaccharide-protein conjugate vaccines, capsule-independent protection has been limited by serotype replacement along with disease caused by nonencapsulated Streptococcus pneumoniae (NESp). NESp strains compose approximately 3% to 19% of asymptomatic carriage isolates and harbor multiple antibiotic resistance genes. Surface proteins unique to NESp enhance colonization and virulence despite the lack of a capsule even though the capsule has been thought to be required for pneumococcal pathogenesis. Genes for pneumococcal surface proteins replace the capsular polysaccharide (cps) locus in some NESp isolates, and these proteins aid in pneumococcal colonization and otitis media (OM). NESp strains have been isolated from patients with invasive and noninvasive pneumococcal disease, but noninvasive diseases, specifically, conjunctivitis (85%) and OM (8%), are of higher prevalence. Conjunctival strains are commonly of the so-called classical NESp lineages defined by multilocus sequence types (STs) ST344 and ST448, while sporadic NESp lineages such as ST1106 are more commonly isolated from patients with other diseases. Interestingly, sporadic lineages have significantly higher rates of recombination than classical lineages. Higher rates of recombination can lead to increased acquisition of antibiotic resistance and virulence factors, increasing the risk of disease and hindering treatment. NESp strains are a significant proportion of the pneumococcal population, can cause disease, and may be increasing in prevalence in the population due to effects on the pneumococcal niche caused by pneumococcal vaccines. Current vaccines are ineffective against NESp, and further research is necessary to develop vaccines effective against both encapsulated and nonencapsulated pneumococci.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a significant human pathogen. Humans are the main reservoir for the pneumococcus, and asymptomatic carriage in the nasopharynx typically occurs at least once by the age of 2 years in the United States (1). However, when the pneumococcus gains access to normally sterile body sites, immune dysregulation and disease can occur. The pneumococcus is a common etiological agent of several diseases such as bacterial meningitis, pneumonia, otitis media (OM), sinusitis, and conjunctivitis (2–4). According to the World Health Organization, the pneumococcus is responsible for 1 million deaths yearly (5). In 2000, over 14 million children worldwide under the age of 5 years were diagnosed with an invasive pneumococcal disease, with the highest incidence seen in Africa (5). Bacterial OM is most commonly caused by the pneumococcus, Haemophilus influenzae, and Moraxella catarrhalis (3). In the United States, OM is the number 1 reason for pediatric clinical visits and antibiotic prescriptions. Currently, an estimated cost of 1.5 to 3 billion dollars is associated with OM despite high rates of childhood vaccination (6–8), while the cost was as high as 5 billion dollars before routine vaccinations (9). Due to high rates of OM despite pneumococcal vaccination, further investigation and increased insight into how these versatile pathogens continue to cause disease are necessary to produce widespread and effective prevention methods.
CLASSIFICATION
One of the most important pneumococcal virulence factors is the polysaccharide capsule, of which there are at least 97 antigenically distinct serotypes (10). The pneumococcal capsule is necessary for virulence of encapsulated pneumococci as it not only protects against opsonophagocytosis but also allows nasopharyngeal (NP) colonization (11, 12). NP colonization is required for progression to pneumococcal disease, and the capsule has been implicated in reducing mucosal clearance from the NP, allowing effective colonization (13). Specific pneumococcal serotypes are associated with invasive pneumococcal disease (IPD), as some serotypes cause more IPD than others even after accounting for the more frequent colonization of some serotypes (14–16). The pneumococcal conjugate vaccines (PCVs) have significantly reduced the rates of IPD while reducing rates of noninvasive disease such as OM, along with NP colonization, only slightly (17–22). Smaller reductions in rates of noninvasive disease were observed because of serotype replacement, which was characterized by increased levels of some pneumococcal nonvaccine serotypes (NVT) (23).
The capsular polysaccharide synthesis (cps) locus, which is required for capsule production, is situated between the highly conserved dexB and aliA pneumococcal genes. The dexB and aliA genes are not known to be used for capsule production (24). Homologous recombination at the cps locus can occur because of these highly conserved flanking regions within the chromosome (25, 26). There have been reports of increased isolation of nonencapsulated Streptococcus pneumoniae (NESp) from patients with NP colonization and noninvasive disease and occasionally from patients with IPD (27–36). NESp can be classified into either group I or group II isolates on the basis of the composition of the cps locus (28). Group I NESp isolates have cps genes, but they do not produce capsule due to mutations or deletions of specific cps genes (28). Group II NESp isolates have novel genes in place of the cps genes. Two aliB-like homologues, called aliC and aliD (also called aliB ORF1 and ORF2, respectively), as well as the gene coding for pneumococcal surface protein K (pspK; also called nspA), replace the cps locus of group II isolates (28, 31, 37). These different variants of group II NESp have been subdivided into so-called null capsule clades (NCCs) based on which genes are located within the cps locus (31). NCC1 strains are pspK-positive (pspK+), aliC negative, and aliD negative strains. NCC2 strains are pspK negative, aliC+, and aliD+ strains and are classified as either NCC2a or NCC2b on the basis of the intergenic length polymorphism between a remnant capN gene and the flanking aliA gene (31). To date, strains containing only the aliD gene (NCC3 [pspK negative, aliC negative, and aliD+ strains]) have been identified as closely related streptococcal species and not true pneumococci. Interestingly, aliD, but not aliC, is also found within the cps locus of serogroups 25 and 38 and a truncated version is found within 66 other serotypes, while pspK has not been shown to occur with any other cps genes (24).
Speculation on the increase in NESp isolation has been based on two thoughts: (i) the introduction of PCVs as mentioned above and (ii) an increase in awareness and more stringent typing of collected samples. Pneumococcal colony morphology on agar plates varies based on capsule status. NESp colonies have a classical “rough” morphology that contrasts with the “smooth,” and sometimes “mucoid,” morphology of encapsulated pneumococci. Also, NESp colonies may be smaller than those of encapsulated pneumococci, making identification and collection more challenging. Together, these characteristics may cause NESp to be overlooked in laboratories and underreported (38, 39). NESp strains produce a null result when typed with capsular serotype-specific antisera and are termed nontypeable (38–40). In general, nontypeable S. pneumoniae strains include both NESp strains, which do not produce capsule, and strains that express novel serotypes or amounts of capsule below the limit of detection. This distinction is important and necessary for understanding the true prevalence of NESp in contrast to the emergence of novel pathogenic serotypes. The U.S. introduction of the PCV in the year 2000 and in other countries shortly after preceded many of the reports about NESp in the population. To draw a connection to the general prevalences of NESp before and after the introduction of the PCV is challenging due to the refinement of and greater sensitivity in typing methods that developed over the years. Despite this, some studies have been able to more directly address pre- and postvaccine NESp prevalences (27, 29, 32, 35, 36). In general, these studies have reported differing results, with some showing no change in NESp prevalence and others showing greater or lesser prevalences.
It is also important to differentiate NESp from other closely related nonencapsulated streptococcal species, such as S. pseudopneumoniae (41) and S. tigurinus (42), which are often confused with S. pneumoniae. Molecular approaches can be used to confirm both species and capsule status with greater precision and accuracy to demonstrate true prevalence (43, 44). These approaches include multilocus sequence typing (MLST), which has allowed reliable species identification of streptococcal isolates (45). Analysis of the cps locus can be used for verification of nonencapsulated status. For example, the first and essential gene of the cps locus, cpsA (also called wzg), is detected by several PCR assays and can differentiate NESp strains as group I or group II (46).
PREVALENCE AND ANTIBIOTIC RESISTANCE
Prevalence.
The human population most susceptible to pneumococcal infections is that of children due to their immature immune system (47). Children within day care centers are in close contact, which promotes pneumococcal transmission (48). Therefore, many studies that document NESp colonization involve children (Table 1). One study in Portugal found that 7% of carriage isolates collected in one day care center were NESp. Interestingly, the only examples of concurrent colonization by multiple strains in that study were of colonization with a NESp and either an encapsulated 23F or 16F strain (49). In another study of Portuguese children attending a day care center, it was found that 12% of pneumococcal isolates were NESp (50). Within a Spanish population of children, 6.2% of carriage isolates were NESp (51), while a study of pneumococcal carriage isolates from Brazilian children found that almost 19% of isolates were NESp (52). Over 13% of the strains isolated from their sampling of children colonized with pneumococcus in another Brazilian study were NESp strains (53). A population of Italian children contained 5.4% NESp carriage isolates (54), and in southeastern France, children had a NESp carriage rate of 9.4% (55). In Israeli pneumococcal carriage isolates, NESp consisted of over 5% of the total number of collected samples (56), while a study in an unvaccinated population of children in an orphanage in Poland found that 3.9% of the pneumococcal strains isolated were NESp (57). In a cohort of 770 infants and mothers in Thailand that had not received any pneumococcal vaccines, up to 16% of carriage isolates were found through genome sequencing to be NESp (58).
TABLE 1 .
Prevalence of NESp within carriage isolates along with rates of resistance
| Country | Total no. of isolatesa | No. of NESp isolates | % NESp isolates | % resistant NESp isolatesb | Typing method(s) | Reference |
|---|---|---|---|---|---|---|
| Portugal | 254 | 18 | 7.1 | 100 | Quelling | 49 |
| Portugal | 202 | 25 | 12.4 | 100 | Quelling | 50 |
| Spain | 194 | 12 | 6.2 | 58 | Quelling/dot blot | 51 |
| Brazil | 253 | 48 | 19 | 100 | Quelling/PCR | 52 |
| Brazil | 166 | 22 | 13.3 | 47.4 (MDR) | Quelling/latex beads/PCR | 53 |
| Italy | 184 | 10 | 5.4 | 80 | Quelling/latex beads | 54 |
| France | 170 | 16 | 9.4 | 94 | Quelling | 55 |
| Israel | 1,763 | 90 | 5.1 | 30 (MDR) | Quelling | 56 |
| Poland | 139 | 5 | 3.9 | 100 (MDR) | Quelling/PCR | 57 |
| Thailand | 3,085 | 512 | 16.6 | 83.6 | Genome sequencing | 58 |
Data represent numbers of isolates that were serotyped in the study.
Data represent percentages of resistance to at least one antibiotic tested. Multiple drug resistance (MDR) data represent resistance to at least three classes of antibiotics and are included where individual rates of resistance were not available.
Resistance.
Resistance profiles for the collected NESp isolates from the studies described above were also reported in most cases and showed that high rates of resistance are common. For example, in the study from Portugal described above, all 18 NESp isolates were resistant to at least one antibiotic and 16 were resistant to multiple drugs (49). Among these isolates, all of the NESp strains of the NNN clonal cluster and 14 of 18 strains of the BE clonal cluster, as defined by pulsed-field gel electrophoresis (PFGE), were resistant to erythromycin, clindamycin, tetracycline, and sulfamethoxazole-trimethoprim, and some were resistant to penicillin. All isolates of the NNN and BE clusters were multilocus sequence type (ST) ST344 or double-locus variants (DLV) of ST344. Interestingly, in a previous study by this group, the NNN cluster was found in only one day care center, while this study isolated NNN clones from four different day care centers (59). Sánchez-Tatay (51) found that 50% of NESp isolates were resistant to penicillin and erythromycin (6 of 12 isolates) and that all of the resistant isolates were ST344. One other NESp isolate was resistant to only penicillin and was ST942 (51). One of the studies of Brazilian children found that all of the NESp isolates were resistant to penicillin and were ST2315 or a single-locus variant (SLV) of ST2315 (ST4746) (52). Another study of Brazilian children did not provide data for all resistance profiles determined, but the NESp isolates had higher rates of multiple drug resistance (MDR [resistance to at least three antibiotic classes]) than other isolates, with 19 of 22 isolates being ST2315 (53). NESp isolates from Italian children were all ST344, and 80% were resistant to at least two antibiotic classes (54). Sixteen NESp isolates were collected from children in southeastern France, and 15 were ST344 or ST344 SLVs, with the remaining NESp isolate being ST63. Of these 16 NESp isolates, 15 were resistant to erythromycin and intermediately susceptible to penicillin, but it was not stated if all 15 resistant NESp isolates were ST344 (55). NESp isolates collected from Israel were divided into 6 clusters (clusters A to F) based on PFGE results, with clusters A and B containing 44% of all 148 isolates. MLST of clusters A and B was done and showed an ST448 SLV for cluster A and a new ST for cluster B that varied from ST344 by three alleles. Resistance profiles for these clusters indicate that at least 70% of cluster A isolates were resistant to a least one antibiotic, while 96% of cluster B isolates were MDR. Rates of resistance were also high in the other clusters (clusters C to F), with the exception of cluster F, which was susceptible to all antibiotics tested. NESp isolates from Poland were all ST344, and all were MDR (57). The genomes of the NESp isolates from Thailand were analyzed for known resistance genes for β-lactam antibiotics or for allelic forms conferring resistance to co-trimoxazole (58). Over 83% of the NESp isolates were predicted on the basis of their sequences to be resistant to either of these antibiotic classes but included no isolates from the ST344 cluster. Of the highly recombinogenic NESp strains contained within a polyphyletic sequence cluster (SC) called BC 3-NT, 86% of isolates were ST4133; of these isolates, 30% had no genes or single nucleotide polymorphisms (SNPs) that would confer resistance to the tested antibiotics. This is in contrast to the ST448 cluster, all isolates of which contained genes or SNPs for resistance to both antibiotic classes. Interestingly, the genes for β-lactam antibiotics and co-trimoxazole resistance occurred together more often than not despite not being closely linked in the genome. Of the NESp isolates, 16.6% were sensitive to both antibiotics compared to the 1.17% that were sensitive to only one antibiotic, showing that multiple drug resistances were more common than resistance to a single antibiotic.
These studies indicated that a significant proportion of colonizing pneumococcal isolates are NESp, and this rate could be higher in vaccinated individuals, as increases in NESp levels have been observed following the introduction of the PCV (36). Also, the results of these various studies show that the global distribution of NESp varies according to geographic location. This is not surprising since the global distribution of pneumococcal serotypes also varies based on geographic location (60, 61). While the currently available data demonstrate that the highest rates of NESp carriage are in Asia and South America, this may simply reflect an increased awareness of NESp isolates in these regions. Additionally, high rates of antibiotic resistance are seen for NESp, specifically, within the ST344 cluster (49, 54, 55, 57). Together, these results indicate that multiple antibiotic resistances may be a common feature of NESp.
COLONIZATION
Colonization is requisite for pneumococcal disease (62, 63). Effective colonization within the NP allows the necessary dissemination into sterile tissue sites leading to disease (62, 63). The loss of capsule from strains that normally express capsule abolishes colonization and virulence (12, 13). Current vaccines induce a strong antibody response to specific capsule serotypes, allowing efficient clearance from the host and reduced colonization of the targeted capsule types, but these vaccines are ineffective against NESp strains (64).
The phase-variable “transparent variants” of encapsulated strains have increased epithelial cell adherence due to reduced capsule expression (65, 66). This allows pneumococcal surface proteins greater access to host cell receptors for adherence and potential immunomodulatory effects (67, 68). Also, protein expression profiles alter depending upon the pneumococcal phase phenotype (69). Increased capsule expression is observed when encapsulated pneumococcal strains cause systemic infection (66, 70). It has been shown that the pneumococcus increases mucosal secretions, leading to dysregulation of mucosal clearance by host ciliary action (71, 72). While capsule is needed for encapsulated pneumococcal colonization, strains that naturally lack capsule, i.e., NESp strains, compensate for the absence of capsule and can still effectively colonize. A potential mechanism of colonization was found within group II NESp strains. Group II NESp isolates with the pspK gene showed significantly greater NP colonization of the mouse than an isogenic pspK mutant (31). Also, extrachromosomal expression of PspK in a PspK− NESp strain increased NP colonization in a mouse (73). Furthermore, it was found that PspK+ NESp strains were able to colonize the NP as effectively as an encapsulated strain and that both colonize at greater densities than PspK− NESp strains (31). The ability of PspK to increase adhesion to epithelial cells may explain how the loss of capsule has been compensated for in some NESp strains (73).
Despite the lack of capsule and PspK, group II NESp strains that contain aliC and aliD within the cps locus are also able to colonize the nasopharynx (28, 31, 58, 74). Of 512 sequenced NESp isolates from Thailand, 197 contained aliC and aliD, while 258 contained pspK (58). Recent work led to the conclusion that AliC increased pneumococcal competence and AliD enhanced early NP colonization of the mouse (74). The oligopeptide transporter Ami-AliA/AliB was also found to be important for NP colonization (75). Thus, results of current studies indicate that both PspK and AliD have a role in colonization.
An important consideration during colonization and disease is evasion of the innate immune response. Despite lacking some important colonization and immune evasion proteins (see Virulence Factors below), NESp strains do colonize and are isolated from the nasopharynx. Pneumococcal clearance is mediated through several host factors, including neutrophil recruitment, macrophage opsonophagocytosis, and CD4+ T cells (76, 77). Interestingly, results from studies of available genome sequences (78, 79) indicate that NESp strains contain DltA (80, 81) and EndA (82), which have been shown to inhibit neutrophil extracellular traps, an important method of bacterial clearance (83).
BIOFILM FORMATION
Pneumococcal biofilm is important for colonization and persistence within the NP (84, 85). Bacterial populations are almost exclusively contained within these surface-associated communities when they are part of the nasopharyngeal community (86–88). The ability to produce biofilm and interact with different bacterial species or different pneumococcal strains is vital for colonization (89). It has been shown that NESp strains produce larger amounts of biofilm than their encapsulated counterparts (90–92). This observation might be attributed to increased exposure to surface proteins that allow greater attachment and, subsequently, enhanced biofilm formation. Therefore, the very nature of NESp may allow increased biofilm formation, which not only enables greater colonization but also reduces susceptibility to antibiotics and the host response (93). NP colonization and biofilm formation allow the pneumococcus to disseminate to other body sites and cause disease (62, 63). In these sites, biofilm formation has been implicated in increased probability of disease, especially during OM (94–97).
VIRULENCE FACTORS
Pneumococcal virulence factors tend to fall within one of three groups: capsule, toxin, and surface proteins (11, 98, 99). The capsule is not necessary for NESp strains as, by definition, they lack this virulence factor. All pneumococci contain the cytolytic toxin pneumolysin, and, in addition, some NESp strains have been recently reported to contain a second pneumolysin gene that is closely related to other streptococcal cytolysins (100, 101). As no other toxins have been described for any pneumococcus, NESp virulence is mainly mediated through surface proteins and other surface structures. Surface proteins are attached to the bacterial cell through various mechanisms, which include attachment to choline on the pneumococcal surface by choline binding repeats, lipid attachment via recognition of lipobox motif (LxxC) by Lgt enzyme (102), and peptidoglycan anchoring through sortase-mediated attachment via the LPxTG motif and other pilus-specific sortases (103). The structure of pneumococcal surface proteins allows anchoring to the pneumococcal surface with an extracellular portion that interacts with the environment, typically to promote survival (98, 104). NESp strains, like encapsulated pneumococci, reside in the NP, and surface proteins can aid in colonization through attachment, immune modulation, or enhanced biofilm formation (67, 105, 106).
Interestingly, the first genome sequences of five genetically diverse NESp isolates lacked genes for common pneumococcal surface proteins such as PspA, PspC (also called CbpA), and PcpA (78). The absence of genes for PspA and PspC was also observed with comparative genomic hybridization of 34 other NESp isolates from diverse genetic backgrounds (107). The surface proteins PspA and PspC have been studied as vaccine candidates (108–111), but these proteins would not be effective immunogens for NESp since they lack these proteins. Other potential protein-based vaccine targets are the pneumococcal histidine triad proteins (Pht). PhtD has been found in all encapsulated strains regardless of serotype and shows a high degree of sequence identity between strains. Despite this, PhtD is absent in the sequenced PspK− NESp strains but is present in the sequenced PspK+ strains (78, 112). Genome sequences of hundreds of NESp isolates were published during 2013 and 2014 (58, 78, 79, 113) and will provide more information about the distribution of NESp virulence factors. However, the genes for some virulence factors, such as PspA and PspC, are difficult to assemble with short-read sequence data, as noted previously (114), so molecular confirmation of their presence or absence may be needed.
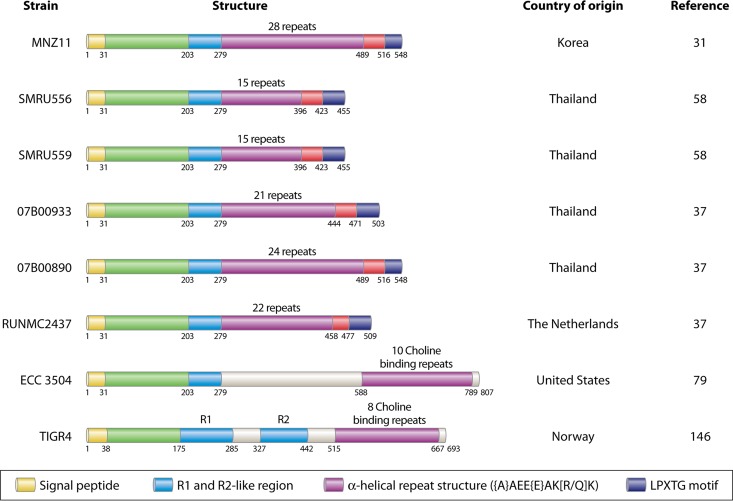
Current results indicate that NESp strains lack surface proteins that are common in encapsulated pneumococci. Thus, it is reasonable to propose that the bacterial surface of NESp differs from that of encapsulated strains. In fact, several novel surface proteins were recently discovered in a genomic study of 21 NESp isolates from patients with conjunctivitis (79). Some of these proteins were reported to be divergent forms of choline-binding and LPxTG-attached proteins (Fig. 1 [ECC_3504]), and others were reported to have glycan and gp-340 binding domains that may contribute to bacterial aggregation (79). As shown in Fig. 1, the PspC (CbpA) of strain EC_3504, which was named CpbAC (79), is divergent from the PspC protein of strain TIGR4. It is interesting that the first 279 amino acids of CbpAC have 77% identity to PspK of MNZ11 but show only 55% identity to the R1 region of the PspC protein of TIGR4. We have shown that PspK enhances epithelial cell attachment and binds secretory IgA (sIgA) (73), while it was predicted that CbpAC binds dimeric IgA based on homology to the beta antigen of S. agalactiae (79). PspK and CbpAC may perform redundant roles, which could explain why CpbAC-containing NESp strains have aliC and aliD in the cps locus and not pspK. This may also indicate that the aliC and aliD genes, which conjunctival isolates often contain, perform a function during conjunctivitis (see Disease below). Another possible explanation for the fact that PspK+ NESp strains have not been observed to cause conjunctivitis is their potentially recent emergence, with the first known PspK+ isolate collected in 2008 (31). While the answer has yet to be experimentally verified, it is important to determine what NESp virulence factors or combinations of virulence factors can lead to various pneumococcal diseases. Therefore, in-depth studies of the way NESp interacts with the host are necessary because these strains have virulence gene repertoires different from those of encapsulated pneumococci.
FIG 1 .
Schematic of PspK from several geographic locations, highlighting homology to PspC and newly described CbpAC1. Representations of various PspK proteins at the amino acid level, CbpAC1 from ECC_3504, and PspC from TIGR4 are shown. Countries of strain isolation and strain references are provided adjacent to the representations. All representative structures have a highly conserved amino terminus for the first 279 amino acids. Variations in amino acids 203 to 279 have been observed in other strains, but the number of repeats after amino acid 279 is what most often varies between PspK sequences. The protein from strain ECC_3504 was isolated from a conjunctival sample and was encoded at the pspC (also known as cbpA) locus instead of at the cps locus as PspK is. Despite identical sequences for the first 279 amino acids, the location on the genome and the presence of choline binding repeats led to this structure being referred to as a divergent PspC called CbpAC1. CbpAC1 also has an additional ~300 amino acids after the conserved region that are not seen in any known PspK sequences. The final protein is PspC from TIGR4 (146) for homology comparison with PspK and variant PspC. Sequence data were obtained from an NCBI database query. Each color in the color-coded key at the bottom of the figure corresponds to regions of the same color on the protein structures except where indicated.
DISEASE
Conjunctivitis.
The best-documented disease caused by NESp is conjunctivitis, which is associated with sequence type 448 (ST448) and ST344 (56, 115, 116). There were reports of conjunctivitis caused by NESp as early as 1980, indicating the presence of NESp in the population for at least several decades (117). Of 271 conjunctival isolates from across the United States, over 90% were NESp and over 85% were represented by 4 STs, ST448, ST344, ST1186, and ST2315, all of which are nonencapsulated (79). An earlier Spanish study found that only 23.2% of conjunctival isolates were nonencapsulated, all of which were represented by 4 STs, but only ST344 was common to that study and the study described above (118). Another Spanish study reported that 34.8% of conjunctival isolates were NESp, with the majority related to ST344, ST448, or ST941. Rates of antibiotic resistance were in agreement with the resistance rates of carriage isolates as outlined above. ST344 were commonly MDR, with ST941 being resistant to at least two antibiotics and ST448 being susceptible to the tested drugs (119). In general, ST344 and ST448 NESp strains commonly cause conjunctivitis outbreaks, while sporadic cases have shown greater variation in both capsule status and ST. Among those involved in outbreaks of pneumococcal conjunctivitis, the majority are of the ST448 clonal cluster, in contrast to reports of the high rates of carriage by the ST344 cluster (119–122). While it is unknown why NESp strains, and specifically the ST448 cluster, cause conjunctivitis at high rates, Valentino et al. reported that all epidemic conjunctival isolates contain the aliC and aliD genes within the capsule locus, indicating a potential role in the virulence of NESp (79). Recent work has elucidated several differences between the gene contents of encapsulated and nonencapsulated S. pneumoniae strains. Some of these differences increase colonization and virulence, but the majority of these differences are uncharacterized, necessitating increased research on these emerging pathogens (73). Further information about conjunctivitis can be found in a review article by Tarabishy and Jeng (123).
Otitis media.
NESp strains have been isolated from children with otitis media (OM) (27, 29, 124). In fact, one study found that as many as 8% of OM isolates were NESp (27), and, more recently, NESp strains were found in 6.4% of OM isolates (125). This is interesting because complement hinders pneumococcal infections in the middle ear (126) and NESp strains lack the protection from complement afforded by the capsule and PspC (78, 107). Other mechanisms to avoid complement-mediated clearance must be used, and several other pneumococcal factors that reduce the effect of complement were found in several NESp genomes. These complement evasion proteins that NESp strains contain include a phosphoglycerate kinase (PGK) (127) that inhibits membrane attack complex formation; endopeptidase O (128), which binds C1q; and Tuf (129), which binds various proteins in the complement-regulating factor H family (130), including factor H. It is currently unknown if these proteins serve the same function in NESp as they do in encapsulated strains.
While there have been almost no studies that have examined the mechanisms that allow NESp to persist and cause disease, the chinchilla has emerged as an effective model of OM caused by NESp (124, 131–133). In this model, the replacement of the cps locus with pspK allowed efficient colonization in the NP along with increased virulence in the middle ear (31, 132, 134). Our study examined the ability of NESp to cause OM in the chinchilla along with the function of PspK during infection. All strains that contained pspK produced OM (132). In contrast, only half of the NESp isolates tested that did not naturally contain pspK were able to cause OM. Additionally, the PspK− NESp isolates that caused disease had higher relative epithelial cell adhesion to nasopharyngeal epithelial cell line Detroit 562 than avirulent PspK− NESp (132). While there was no correlation between epithelial cell adherence and levels of recovered bacteria in PspK-containing strains, it was postulated that PspK allows sufficient attachment for other factors to then increase virulence (132). A recent study determined the prevalence of pspK in NESp isolates from 12 Asian countries (135). That study found that over 42% of the NESp isolates from Thailand contained pspK and that over 15% of the NESp isolates from Asia had the pspK gene (135). Interestingly, the most common ST found was ST1106, which has been previously associated with serotype 14 isolates, and all of the ST1106 isolates were multiple drug resistant (135).
Invasive pneumococcal disease.
NESp strains are able to cause IPD, but the frequency is much lower than that caused by encapsulated strains (28, 30, 37, 136). This can potentially be explained by the innate immune response, specifically, the complement system. Activation of the host complement system by the pneumococcus is the main mechanism of clearance during infection (137, 138). The classical complement pathway is responsible for initiating complement deposition and ultimately leading to clearance through opsonophagocytosis (139). The presence of capsule hinders the recognition of common pneumococcal antigenic sites on the bacterial surface, while the absence of capsule allows rapid recognition and efficient complement activation (140). This lack of capsule reduces the ability of NESp to produce relevant infection in sterile sites that are inundated with complement. Despite this, a review of South East Asian cases of IPD found that anywhere from 0% to 15% were caused by NESp, with some variation by country (141).
The largest study to date on IPD isolates from the United States found that 88 (0.61%) of 14,328 isolates tested were NESp, with the majority being group I isolates (30). Of the 79 group I isolates, 22 were serotype 8 strains that expressed a dysfunctional cpsA gene that stopped capsule production (30). It was suggested that opsonization of capsule-bearing pneumococci would deplete complement levels in the microenvironment to allow transient survival of the cpsA escape mutants. Of the nine group II isolates found in this survey, six contained aliB-like homologues aliC and aliD, and the remaining three isolates were either unclassified or lacked all genes in the cps locus (30). Another study of nearly 5,000 IPD isolates from Chile found that 3.43% of pneumonia cases were caused by NESp, while only 1.5% of bacteremia cases were caused by NESp (142). The first IPD study by Park et al. (30) described above did not differentiate between types of IPD. Since, as reported by Lagos et al. (142), NESp strains are more likely to cause pneumonia than bacteremia, the study by Park et al. (30) may have included higher numbers of isolates from patients with bacteremia than from patients with pneumonia. This could result in the observed lower prevalence of NESp IPD cases than that reported from the earlier Chilean study. Alternatively, the differences between these studies in reported rates of IPD caused by NESp could have been due to geographic differences in the host or the pathogen (30).
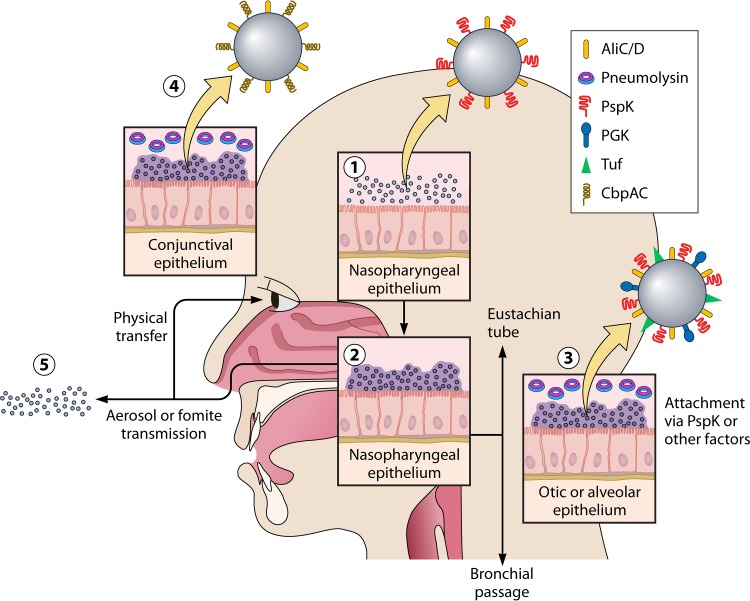
These studies indicate that group II isolates may be specialized for NP colonization. Thus, these upper respiratory commensals may be important pathogens of noninvasive diseases such as otitis media, sinusitis, and nonbacteremic pneumonia. A general model of NESp pathogenesis is presented in Fig. 2.
FIG 2 .
Proposed model for NESp colonization and disease. Initial attachment and colonization is mediated through surface proteins (1), including unknown methods of immune evasion. NESp biofilm formation and persistence are aided by enhanced adhesive properties (2). Planktonic bacteria dissociate from the biofilm matrix and ascend into the middle ear through the Eustachian tube or descend to the lung through the pharynx (3). Disruption of surface-associated communities is followed by physical transmission to eye (4) or transmission to other hosts through aerosol droplets or fomite intermediates (5).
GENETIC EXCHANGE
The pneumococcus has several mechanisms to mediate horizontal gene transfer and recombination (here referred to simply as “recombination”), including transformation, integrative conjugative elements (ICEs), and transducing phage. Several studies have shown, in vitro, that capsule-negative mutants acquire genes through recombination at a higher frequency than their encapsulated progenitors (143–145). These results may indicate that capsule is an anatomical barrier that impedes the receipt of DNA or possibly that bacterial physiology changes with loss of capsule in a way that promotes the receipt of DNA. However, inferences of the frequency of recombination in natural populations of pneumococci are affected by many additional factors, including population sizes, selection for or against the incoming DNA, and the power of recombination tests to detect the recombinant DNA. Several recent genomic studies have provided a nuanced view of the frequency of recombination in natural populations of NESp compared to encapsulated pneumococci, which we discuss below.
A study of the genomes of NP colonization isolates from Massachusetts classified ST448 and ST344 NESp into an early-branching, monophyletic sequence cluster (SC) called SC12 (114). Various other NESp strains were classified into a polyphyletic sequence cluster called SC16 and were not further examined. The per-site recombination-to-mutation ratio (r/m; the ratio of the numbers of single nucleotide polymorphisms accumulated by recombination versus point mutation) of SC12 was 12.3, which ranked it only 7th among 15 SC’s in the order of highest per-site r/m (114). These results indicate that the ST448 and ST344 NESp strains are not particularly recombinant compared to most encapsulated pneumococcal strains isolated in Massachusetts. This conclusion is supported by a study of the genomes of globally sampled ST448 and ST344 NESp strains, where the range of per-event r/m (ratio of the number of recombination events to the number of point mutation events) was 0.15 to 0.18 and was comparable to the range of 0.13 to 0.23 determined for many encapsulated lineages (58, 113). On the other hand, there is evidence that the sporadic, polyphyletic NESp strains are much more recombinant than other pneumococcal strains. The previously mentioned study of colonization isolates from Thailand classified many NESp strains into a polyphyletic sequence cluster called BC 3-NT (58). The per-event r/m of BC 3-NT was 0.32, significantly higher than that for all other sequence clusters. More impressively, the 128 NESp isolates of BC 3-NT, which do not contain NESp of the so-called classical lineages of ST448 and ST334, were significantly more recombinant than the 74 capsular serotype 14 isolates of BC 3-NT (per-event r/m of 0.34 versus 0.20, respectively) (58).
The classical NESp lineages of ST448 and ST344 have been noted to have relatively large genomes with novel gene content in comparison to other pneumococci (77, 112), including an abundance of ICEs and novel regions of difference/genomic islets in addition to the previously mentioned virulence factors (77, 112). Apparently, the presence of multiple ICEs in the ST448 and ST344 NESp strains has not resulted in increased recombination frequency within these lineages or in export of their novel genes to other pneumococci. These classical NESp lineages contain 116 unique clusters of orthologous groups (COGs), including 33 COGs within two ICEs, ICE1 S. pneumoniae ST344 (ICE1SpST344) and ICE2SpST344, and two COGs within pneumococcal pathogenicity island 1 (113). ICE1SpST344 was found within both the classical and sporadic NESp lineages and was shown to aid in epithelial cell adhesion but had reduced growth rates compared to NESp lineages lacking this ICE (113). The remaining COGs unique to NESp include mobilization and conjugation proteins, a zinc ABC transporter, a zinc metalloprotease, a lipoprotein, and numerous hypothetical proteins of unknown function. As discussed earlier, NESp strains are also distinguished by novel gene content that includes variants of surface proteins found in the epidemic conjunctivitis cluster (79) and an altered cps locus (31, 37, 73).
Despite potential colonization advantages conferred by some of these novel elements, the large genomes of classical NESp may hinder growth and result in reduced fitness (113). NESp strains harbor multiple unique mechanisms to aid in colonization, and a balance between growth, colonization, and unique gene content must be maintained. In contrast to classical lineages of NESp, sporadic lineages of NESp have higher rates of recombination than other pneumococci (58, 113). Despite these differences, penicillin nonsusceptibility levels of 0.06 µg/ml to 2.0 µg/ml are common in both classic and sporadic lineages. Moreover, resistance to tetracycline and erythromycin is prevalent regardless of lineage. ST448 bacteria were generally susceptible to trimethoprim-sulfamethoxazole, but this was not true with ST344 (58, 113). Thus, NESp could conceivably represent a growing reservoir of antibiotic resistance genes that could be exported to encapsulated strains, reducing treatment options and survival outcomes.
In summary, these results indicate that the early-branching, monophyletic, classical lineages that include ST448 and ST344 NESp are not unusually recombinant, but they carry novel genes and genetic elements that may aid survival. The polyphyletic, sporadic lineages that include other NESp sequence types such as ST1106 are highly recombinant and also carry novel genes such as pspK that may aid survival (58, 113). Therefore, both sorts of NESp may be poised to increase in prevalence in the wake of PCV.
CONCLUSIONS
A century of research has led to the view that pneumococcal capsule is required for colonization and virulence (12, 13); however, a growing body of evidence is showing that this is not always the case. The loss of the polysaccharide capsule does significantly decrease the chance of IPD but does not eliminate the possibility (30, 142). More commonly, noninvasive diseases such as OM and conjunctivitis are caused by NESp (27, 79). The total number of clinical cases caused by NESp is probably underestimated because serotyping is not routinely performed and serotype-nontypeable strains may not be further classified. Also, the numbers of pneumococcal carriage isolates that lack capsule have been underestimated, with prevalences ranging from 3% to 19% of the pneumococcal population (52, 57). NESp strains have been isolated from the human population for several decades and have most likely been present for much longer (117). Indeed, some of these NESp strains appear to represent early-branching lineages of pneumococcus whereas other NESp strains are highly recombinant, but more work is needed to understand their evolutionary history (58, 113, 114). Increasing pressure against a specific subset of pneumococcal isolates, namely, capsule types within the PCV, affects the environmental niche that NESp strains are able to exploit. This allows increasing numbers of NESp strains to occur within the human population, increasing the risk of pneumococcal disease caused by NESp. In addition, NESp strains have novel gene content that is just beginning to be cataloged and that has been rarely studied from the functional and pathogenic perspectives. Understanding how these unique organisms colonize and have survived within the human population despite the lack of an antiphagocytic capsule will lead to greater insights into bacterial methods of persistence and evasion of the host defense.
Footnotes
Citation Keller LE, Robinson DA, McDaniel LS. 2016. Nonencapsulated Streptococcus pneumoniae: emergence and pathogenesis. mBio 7(2):e01792-15. doi:10.1128/mBio.01792-15.
REFERENCES
- 1.Gray BM, Converse GM, Dillon HC. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 24 months of life. J Infect Dis 142:923–933. doi: 10.1093/infdis/142.6.923. [DOI] [PubMed] [Google Scholar]
- 2.Musher DM. 1992. Infections caused by Streptococcus pneumoniae: clinical spectrum, pathogenesis, immunity, and treatment. Clin Infect Dis 14:801–807. doi: 10.1093/clinids/14.4.801. [DOI] [PubMed] [Google Scholar]
- 3.Lieberthal AS, Carroll AE, Chonmaitree T, Ganiats TG, Hoberman A, Jackson MA, Joffe MD, Miller DT, Rosenfeld RM, Sevilla XD, Schwartz RH, Thomas PA, Tunkel DE. 2013. The diagnosis and management of acute otitis media. Pediatrics 131:e964–e999. doi: 10.1542/peds.2012-3488. [DOI] [PubMed] [Google Scholar]
- 4.Klein JO. 1981. The epidemiology of pneumococcal disease in infants and children. Rev Infect Dis 3:246–253. doi: 10.1093/clinids/3.2.246. [DOI] [PubMed] [Google Scholar]
- 5.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team . 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien MA, Prosser LA, Paradise JL, Ray GT, Kulldorff M, Kurs-Lasky M, Hinrichsen VL, Mehta J, Colborn DK, Lieu TA. 2009. New vaccines against otitis media: projected benefits and cost-effectiveness. Pediatrics 123:1452–1463. doi: 10.1542/peds.2008-1482. [DOI] [PubMed] [Google Scholar]
- 7.Ahmed S, Shapiro NL, Bhattacharyya N. 2014. Incremental health care utilization and costs for acute otitis media in children. Laryngoscope 124:301–305. doi: 10.1002/lary.24190. [DOI] [PubMed] [Google Scholar]
- 8.Huang SS, Johnson KM, Ray GT, Wroe P, Lieu TA, Moore MR, Zell ER, Linder JA, Grijalva CG, Metlay JP, Finkelstein JA. 2011. Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 29:3398–3412. doi: 10.1016/j.vaccine.2011.02.088. [DOI] [PubMed] [Google Scholar]
- 9.Gates GA. 1996. Cost-effectiveness considerations in otitis media treatment. Otolarngal Head Neck Surg 114:525–530. [DOI] [PubMed] [Google Scholar]
- 10.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, Konradsen HB, Nahm MH. 2015. Pneumococcal capsules and their types: past, present, and future. Clin Microbiol Rev 28:871–899. doi: 10.1128/CMR.00024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6:288–301. doi: 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 12.Magee AD, Yother J. 2001. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun 69:3755–3761. doi: 10.1128/IAI.69.6.3755-3761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson AL, Roche AM, Gould JM, Chim K, Ratner AJ, Weiser JN. 2007. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect Immun 75:83–90. doi: 10.1128/IAI.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis 187:1424–1432. doi: 10.1086/374624. [DOI] [PubMed] [Google Scholar]
- 15.Brueggemann AB, Peto TE, Crook DW, Butler JC, Kristinsson KG, Spratt BG. 2004. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J Infect Dis 190:1203–1211. doi: 10.1086/423820. [DOI] [PubMed] [Google Scholar]
- 16.Sleeman KL, Griffiths D, Shackley F, Diggle L, Gupta S, Maiden MC, Moxon ER, Crook DW, Peto TE. 2006. Capsular serotype–specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J Infect Dis 194:682–688. doi: 10.1086/505710. [DOI] [PubMed] [Google Scholar]
- 17.Black S, Shinefield H, Fireman B, Lewis E, Ray P, Hansen JR, Elvin L, Ensor KM, Hackell J, Siber G, Malinoski F, Madore D, Chang I, Kohberger R, Watson W, Austrian R, Edwards K. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Pediatr Infect Dis J 19:187–195. doi: 10.1097/00006454-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Eskola J, Kilpi T, Palmu A, Jokinen J, Eerola M, Haapakoski J, Herva E, Takala A, Käyhty H, Karma P, Kayhty H, Karma P, Kohberger R, Siber G, Makela PH, Finnish Otitis Media Study Group . 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N Engl J Med 344:403–409. doi: 10.1056/NEJM200102083440602. [DOI] [PubMed] [Google Scholar]
- 19.Buznach N, Dagan R, Greenberg D. 2005. Clinical and bacterial characteristics of acute bacterial conjunctivitis in children in the antibiotic resistance era. Pediatr Infect Dis J 24:823–828. doi: 10.1097/01.inf.0000178066.24569.98. [DOI] [PubMed] [Google Scholar]
- 20.Jódar L, Butler J, Carlone G, Dagan R, Goldblatt D, Käyhty H, Klugman K, Plikaytis B, Siber G, Kohberger R, Chang I, Cherian T. 2003. Serological criteria for evaluation and licensure of new pneumococcal conjugate vaccine formulations for use in infants. Vaccine 21:3265–3272. doi: 10.1016/S0264-410X(03)00230-5. [DOI] [PubMed] [Google Scholar]
- 21.Whitney CG, Farley MM, Hadler J, Harrison LH, Bennett NM, Lynfield R, Reingold A, Cieslak PR, Pilishvili T, Jackson D, Facklam RR, Jorgensen JH, Schuchat A, Active Bacterial Core Surveillance of the Emerging Infections Program Network . 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N Engl J Med 348:1737–1746. doi: 10.1056/NEJMoa022823. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien KL, David AB, Chandran A, Moulton LH, Reid R, Weatherholtz R, Santosham M. 2008. Randomized, controlled, trail efficacy of pneumococcal conjugate vaccine against otitis media among Navajo and White Mountain Apache infants. Pediatr Infect Dis J 27:71–73. doi: 10.1097/INF.0b013e318159228f. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien KL, Millar EV, Zell ER, Bronsdon M, Weatherholtz R, Reid R, Becenti J, Kvamme S, Whitney CG, Santosham M. 2007. Effect of pneumococcal conjugate vaccine on nasopharyngeal colonization among immunized and unimmunized children in a community-randomized trial. J Infect Dis 196:1211–1220. doi: 10.1086/521833. [DOI] [PubMed] [Google Scholar]
- 24.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffey TJ, Enright MC, Daniels M, Morona JK, Morona R, Hryniewicz W, Paton JC, Spratt BG. 1998. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol 27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 26.Brueggemann AB, Pai R, Crook DW, Beall B. 2007. Vaccine escape recombinants emerge after pneumococcal vaccination in the United States. PLoS Pathog 3:e168. doi: 10.1371/journal.ppat.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croney CM, Nahm MH, Juhn SK, Briles DE, Crain MJ. 2013. Invasive and noninvasive Streptococcus pneumoniae capsule and surface protein diversity following use of conjugate vaccine. Clin Vaccine Immunol 20:1711–1718. doi: 10.1128/CVI.00381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hathaway LJ, Stutzmann Meier P, Bättig P, Aebi S, Mühlemann K. 2004. A homologue of aliB is found in the capsule region of nonencapsulated Streptococcus pneumoniae. J Bacteriol 186:3721–3729. doi: 10.1128/JB.186.12.3721-3729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ing J, Mason EO, Kaplan SL, Lamberth LB, Revell PA, Luna RA, Hulten KG. 2012. Characterization of non-typeable and atypical Streptococcus pneumoniae pediatric isolates from 1994–2010. J Clin Microbiol 50:1326–1330. doi: 10.1128/JCM.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park IH, Geno KA, Sherwood LK, Nahm MH, Beall B. 2014. Population-based analysis of invasive nontypeable pneumococci reveals that most have defective capsule synthesis genes. PLoS One 9:e97825. doi: 10.1371/journal.pone.0097825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park IH, Kim KH, Andrade AL, Briles DE, McDaniel LS, Nahm MH. 2012. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. mBio 3:e00035-12. doi: 10.1128/mBio.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacapa R, Bliss SJ, Larzelere-Hinton F, Eagle KJ, McGinty DJ, Parkinson AJ, Santosham M, Craig MJ, O’Brien KL. 2008. Changing epidemiology of invasive pneumococcal disease among White Mountain Apache persons in the era of the pneumococcal conjugate vaccine. Clin Infect Dis 47:476–484. doi: 10.1086/590001. [DOI] [PubMed] [Google Scholar]
- 33.Melchiorre S, Camilli R, Pietrantoni A, Moschioni M, Berti F, Del Grosso M, Superti F, Barocchi MA, Pantosti A. 2012. Point mutations in wchA are responsible for the non-typability of two invasive Streptococcus pneumoniae isolates. Microbiology 158:338–344. doi: 10.1099/mic.0.054270-0. [DOI] [PubMed] [Google Scholar]
- 34.Norcross EW, Tullos NA, Taylor SD, Sanders ME, Marquart ME. 2010. Assessment of Streptococcus pneumoniae capsule in conjunctivitis and keratitis in vivo neuraminidase activity increases in nonencapsulated pneumococci following conjunctival infection. Curr Eye Res 35:787–798. doi: 10.3109/02713683.2010.492462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okade H, Funatsu T, Eto M, Furuya Y, Mizunaga S, Nomura N, Mitsuyama J, Yamagishi Y, Mikamo H. 2014. Impact of the pneumococcal conjugate vaccine on serotype distribution and susceptibility trends of pediatric non-invasive Streptococcus pneumoniae isolates in Tokai, Japan over a 5-year period. J Infect Chemother 20:423–428. doi: 10.1016/j.jiac.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Sá-Leão R, Nunes S, Brito-Avô A, Frazão N, Simões AS, Crisóstomo MI, Paulo AC, Saldanha J, Santos-Sanches I, de Lencastre H. 2009. Changes in pneumococcal serotypes and antibiotypes carried by vaccinated and unvaccinated day-care centre attendees in Portugal, a country with widespread use of the seven-valent pneumococcal conjugate vaccine. Clin Microbiol Infect 15:1002–1007. doi: 10.1111/j.1469-0691.2009.02775.x. [DOI] [PubMed] [Google Scholar]
- 37.Salter SJ, Hinds J, Gould KA, Lambertsen L, Hanage WP, Antonio M, Turner P, Hermans PW, Bootsma HJ, O’Brien KL, Bentley SD. 2012. Variation at the capsule locus, cps, of mistyped and non-typable Streptococcus pneumoniae isolates. Microbiology 158:1560–1569. doi: 10.1099/mic.0.056580-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carvalho MG, Steigerwalt AG, Thompson T, Jackson D, Facklam RR. 2003. Confirmation of nontypeable Streptococcus pneumoniae-like organisms isolated from outbreaks of epidemic conjunctivitis as Streptococcus pneumoniae. J Clin Microbiol 41:4415–4417. doi: 10.1128/JCM.41.9.4415-4417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whatmore AM, Efstratiou A, Pickerill AP, Broughton K, Woodard G, Sturgeon D, George R, Dowson CG. 2000. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniae virulence factor-encoding genes. Infect Immun 68:1374–1382. doi: 10.1128/IAI.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter SS, Heilmann KP, Dohrn CL, Riahi F, Beekmann SE, Doern GV. 2008. Accuracy of phenotypic methods for identification of Streptococcus pneumoniae isolates included in surveillance programs. J Clin Microbiol 46:2184–2188. doi: 10.1128/JCM.00461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahinas D, Thornton CS, Tamber GS, Arya G, Wong A, Jamieson FB, Ma JH, Alexander DC, Low DE, Pillai DR. 2013. Comparative genomic analyses of Streptococcus pseudopneumoniae provide insight into virulence and commensalism dynamics. PLoS One 8:e65670. doi: 10.1371/journal.pone.0065670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veloso TR, Zbinden A, Andreoni F, Giddey M, Vouillamoz J, Moreillon P, Zinkernagel AS, Entenza JM. 2013. Streptococcus tigurinus is highly virulent in a rat model of experimental endocarditis. Int J Med Microbiol 303:498–504. doi: 10.1016/j.ijmm.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 43.John J, Gopalkrishnan S, Marie MAM, Gowda KL. 2014. Historical development of typing methods for Streptococcus pneumoniae. Rev Med Microbiol 25:27–33. doi: 10.1097/MRM.0000000000000001. [DOI] [Google Scholar]
- 44.Enright MC, Spratt BG. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 45.Ip M, Chi F, Chau SS, Hui M, Tang J, Chan PK. 2006. Use of the housekeeping genes, gdh (zwf) and gki, in multilocus sequence typing to differentiate Streptococcus pneumoniae from Streptococcus mitis and Streptococcus oralis. Diagn Microbiol Infect Dis 56:321–324. doi: 10.1016/j.diagmicrobio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 46.García E, López R. 1997. Molecular biology of the capsular genes of Streptococcus pneumoniae. FEMS Microbiol Lett 149:1–10. doi: 10.1016/S0378-1097(97)00026-8. [DOI] [PubMed] [Google Scholar]
- 47.McIntosh K. 2002. Community-acquired pneumonia in children. N Engl J Med 346:429–437. doi: 10.1056/NEJMra011994. [DOI] [PubMed] [Google Scholar]
- 48.García-Rodríguez JA, Fresnadillo Martínez MJ. 2002. Dynamics of nasopharyngeal colonization by potential respiratory pathogens. J Antimicrob Chemother 50(Suppl S2):59–74. doi: 10.1093/jac/dkf506. [DOI] [PubMed] [Google Scholar]
- 49.Sá-Leão R, Nunes S, Brito-Avô A, Alves CR, Carriço JA, Saldanha J, Almeida JS, Santos-Sanches I, de Lencastre H. 2008. High rates of transmission of and colonization by Streptococcus pneumoniae and Haemophilus influenzae within a day care center revealed in a longitudinal study. J Clin Microbiol 46:225–234. doi: 10.1128/JCM.01551-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunes S, Sá-Leão R, Carriço J, Alves CR, Mato R, Avô AB, Saldanha J, Almeida JS, Sanches IS, de Lencastre H. 2005. Trends in drug resistance, serotypes, and molecular types of Streptococcus pneumoniae colonizing preschool-age children attending day care centers in Lisbon, Portugal: a summary of 4 years of annual surveillance. J Clin Microbiol 43:1285–1293. doi: 10.1128/JCM.43.3.1285-1293.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sánchez-Tatay D, Arroyo LA, Tarragó D, Lirola MJ, Porras A, Fenoll A, Hausdorff WP, Brueggemann AB, Obando I. 2008. Antibiotic susceptibility and molecular epidemiology of nasopharyngeal pneumococci from Spanish children. Clin Microbiol Infect 14:797–801. doi: 10.1111/j.1469-0691.2008.02025.x. [DOI] [PubMed] [Google Scholar]
- 52.Pimenta FC, Carvalho MDGS, Gertz RE Jr, Bastos-Rocha CGB, Oliveira L, Lacerda Pigosso L, Lima JA, Marquez Franco C, Andrade AL, Beall BW. 2011. Serotype and genotype distributions of pneumococcal carriage isolates recovered from Brazilian children attending day-care centres. J Med Microbiol 60:1455–1459. doi: 10.1099/jmm.0.031450-0. [DOI] [PubMed] [Google Scholar]
- 53.Andrade ALS, Franco CM, Lamaro-Cardoso J, André MCDPB, Oliveira LLG, Kipnis A, Rocha CGBB, Andrade JG, la Alves S, Park IH, Nahm MH, Almeida SG, Brandileone MCC. 2010. Non-typeable Streptococcus pneumoniae carriage isolates genetically similar to invasive and carriage isolates expressing capsular type 14 in Brazilian infants. J Infect 61:314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Camilli R, Daprai L, Cavrini F, Lombardo D, D’Ambrosio F, Del Grosso M, Vescio MF, Landini MP, Pascucci MG, Torresani E, Garlaschi ML, Sambri V, Pantosti A. 2013. Pneumococcal carriage in young children one year after introduction of the 13-valent conjugate vaccine in Italy. PLoS One 8:e76309. doi: 10.1371/journal.pone.0076309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunais B, Bruno P, Touboul P, Degand N, Sakarovitch C, Fontas E, Haas H, Girard-Pipau F, Ruimy R, Pradier C. 2015. Impact of the 13-valent pneumococcal conjugate vaccine on nasopharyngeal carriage of Streptococcus pneumoniae among children attending group daycare in southeastern France. Pediatr Infect Dis J 34:286–288. doi: 10.1097/INF.0000000000000559. [DOI] [PubMed] [Google Scholar]
- 56.Porat N, Greenberg D, Givon-Lavi N, Shuval DS, Trefler R, Segev O, Hanage WP, Dagan R. 2006. The important role of nontypable Streptococcus pneumoniae International clones in acute conjunctivitis. J Infect Dis 194:689–696. doi: 10.1086/506453. [DOI] [PubMed] [Google Scholar]
- 57.Sulikowska A, Grzesiowski P, Sadowy E, Fiett J, Hryniewicz W. 2004. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis isolated from the nasopharynges of asymptomatic children and molecular analysis of S. pneumoniae and H. influenzae strain replacement in the nasopharynx. J Clin Microbiol 42:3942–3949. doi: 10.1128/JCM.42.9.3942-3949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rodgers GL, Arguedas A, Cohen R, Dagan R. 2009. Global serotype distribution among Streptococcus pneumoniae isolates causing otitis media in children: potential implications for pneumococcal conjugate vaccines. Vaccine 27:3802–3810. doi: 10.1016/j.vaccine.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 60.Sá-Leão R, Tomasz A, Sanches IS, Nunes S, Alves CR, Avô AB, Saldanha J, Kristinsson KG, de Lencastre H. 2000. Genetic diversity and clonal patterns among antibiotic-susceptible and -resistant Streptococcus pneumoniae colonizing children: day care centers as autonomous epidemiological units. J Clin Microbiol 38:4137–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sniadack DH, Schwartz B, Lipman H, Bogaerts J, Butler JC, Dagan R, Echaniz-Aviles G, Lloyd-Evans N, Fenoll A, Girgis NI, Henrichsen J, Klugman K, Lehmann D, Takala AK, Vandepitte J, Gove S, Breiman RF. 1995. Potential interventions for the prevention of childhood pneumonia: geographic and temporal differences in serotype and serogroup distribution of sterile site pneumococcal isolates from children—implications for vaccine strategies. Pediatr Infect Dis J 14:503–509. doi: 10.1097/00006454-199506000-00007. [DOI] [PubMed] [Google Scholar]
- 62.Bogaert D, de Groot R, Hermans PW. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis 4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 63.Simell B, Auranen K, Käyhty H, Goldblatt D, Dagan R, O’Brien KL, Pneumococcal Carriage Group . 2012. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 11:841–855. doi: 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 64.Dagan R, Givon-Lavi N, Fraser D, Lipsitch M, Siber GR, Kohberger R. 2005. Serum serotype-specific pneumococcal anticapsular immunoglobulin G concentrations after immunization with a 9-valent conjugate pneumococcal vaccine correlate with nasopharyngeal acquisition of pneumococcus. J Infect Dis 192:367–376. doi: 10.1086/431679. [DOI] [PubMed] [Google Scholar]
- 65.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis 177:368–377. doi: 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- 67.Graham RM, Paton JC. 2006. Differential role of CbpA and PspA in modulation of in vitro CXC chemokine responses of respiratory epithelial cells to infection with Streptococcus pneumoniae. Infect Immun 74:6739–6749. doi: 10.1128/IAI.00954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cundell DR, Weiser JN, Shen J, Young A, Tuomanen EI. 1995. Relationship between colonial morphology and adherence of Streptococcus pneumoniae. Infect Immun 63:757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Overweg K, Pericone CD, Verhoef GG, Weiser JN, Meiring HD, De Jong AP, De Groot R, Hermans PW. 2000. Differential protein expression in phenotypic variants of Streptococcus pneumoniae. Infect Immun 68:4604–4610. doi: 10.1128/IAI.68.8.4604-4610.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim JO, Romero-Steiner S, Sørensen UB, Blom J, Carvalho M, Barnard S, Carlone G, Weiser JN. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect Immun 67:2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ha U, Lim JH, Jono H, Koga T, Srivastava A, Malley R, Pagès G, Pouysségur J, Li J-D. 2007. A novel role for IκB kinase (IKK) α and IKKβ in ERK-dependent up-regulation of MUC5AC mucin transcription by Streptococcus pneumoniae. J Immunol 178:1736–1747. [DOI] [PubMed] [Google Scholar]
- 72.Ha U-H, Lim JH, Kim H-J, Wu W, Jin S, Xu H, Li J-D. 2008. MKP1 regulates the induction of MUC5AC mucin by Streptococcus pneumoniae pneumolysin by inhibiting the PAK4-JNK signaling pathway. J Biol Chem 283:30624–30631. doi: 10.1074/jbc.M802519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keller LE, Jones CV, Thornton JA, Sanders ME, Swiatlo E, Nahm MH, Park IH, McDaniel LS. 2013. PspK of Streptococcus pneumoniae increases adherence to epithelial cells and enhances nasopharyngeal colonization. Infect Immun 81:173–181. doi: 10.1128/IAI.00755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hathaway LJ, Bättig P, Reber S, Rotzetter JU, Aebi S, Hauser C, Heller M, Kadioglu A, Mühlemann K. 2014. Streptococcus pneumoniae detects and responds to foreign bacterial peptide fragments in its environment. Open Biol 4:130244. doi: 10.1098/rsob.130224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kerr AR, Adrian PV, Estevão S, de Groot R, Alloing G, Claverys J-P, Mitchell TJ, Hermans PW. 2004. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect Immun 72:3902–3906. doi: 10.1128/IAI.72.7.3902-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang Z, Clarke TB, Weiser JN. 2009. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119:1899–1909. doi: 10.1172/JCI36731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Van Rossum AM, Lysenko ES, Weiser JN. 2005. Host and bacterial factors contributing to the clearance of colonization by Streptococcus pneumoniae in a murine model. Infect Immun 73:7718–7726. doi: 10.1128/IAI.73.11.7718-7726.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keller LE, Thomas JC, Luo X, Nahm MH, McDaniel LS, Robinson DA. 2013. Draft genome sequences of five multilocus sequence types of nonencapsulated Streptococcus pneumoniae. Genome Announc 1:e00520-13. doi: 10.1128/genomeA.00520-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Valentino MD, McGuire AM, Rosch JW, Bispo PJM, Burnham C, Sanfilippo CM, Carter RA, Zegans ME, Beall B, Earl AM, Tuomanen EI, Morris TW, Haas W, Gilmore MS. 2014. Unencapsulated Streptococcus pneumoniae from conjunctivitis encode variant traits and belong to a distinct phylogenetic cluster. Nat Commun 5:5411. doi: 10.1038/ncomms6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kovács M, Halfmann A, Fedtke I, Heintz M, Peschel A, Vollmer W, Hakenbeck R, Brückner R. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in Gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J Bacteriol 188:5797–5805. doi: 10.1128/JB.00336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wartha F, Beiter K, Albiger B, Fernebro J, Zychlinsky A, Normark S, Henriques-Normark B. 2007. Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell Microbiol 9:1162–1171. doi: 10.1111/j.1462-5822.2006.00857.x. [DOI] [PubMed] [Google Scholar]
- 82.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. 2006. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr Biol 16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 83.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 84.Marks LR, Parameswaran GI, Hakansson AP. 2012. Pneumococcal interactions with epithelial cells are crucial for optimal biofilm formation and colonization in vitro and in vivo. Infect Immun 80:2744–2760. doi: 10.1128/IAI.00488-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolcott RD, Ehrlich GD. 2008. Biofilms and chronic infections. JAMA 299:2682–2684. doi: 10.1001/jama.299.22.2682. [DOI] [PubMed] [Google Scholar]
- 86.Donlan RM, Costerton JW. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 88.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 89.Shak JR, Vidal JE, Klugman KP. 2013. Influence of bacterial interactions on pneumococcal colonization of the nasopharynx. Trends Microbiol 21:129–135. doi: 10.1016/j.tim.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Allegrucci M, Sauer K. 2007. Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol 189:2030–2038. doi: 10.1128/JB.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moscoso M, García E, López R. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol 188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hiller NL, Ahmed A, Powell E, Martin DP, Eutsey R, Earl J, Janto B, Boissy RJ, Hogg J, Barbadora K, Sampath R, Lonergan S, Post JC, Hu FZ, Ehrlich GD. 2010. Generation of genic diversity among Streptococcus pneumoniae strains via horizontal gene transfer during a chronic polyclonal pediatric infection. PLoS Pathog 6:e1001108. doi: 10.1371/journal.ppat.1001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.De Beer D, Stoodley P, Lewandowski Z. 1997. Measurement of local diffusion coefficients in biofilms by microinjection and confocal microscopy. Biotechnol Bioeng 53:151–158. doi:. [DOI] [PubMed] [Google Scholar]
- 94.Oggioni MR, Trappetti C, Kadioglu A, Cassone M, Iannelli F, Ricci S, Andrew PW, Pozzi G. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol Microbiol 61:1196–1210. doi: 10.1111/j.1365-2958.2006.05310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. 2006. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA 296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trappetti C, Ogunniyi AD, Oggioni MR, Paton JC. 2011. Extracellular matrix formation enhances the ability of Streptococcus pneumoniae to cause invasive disease. PLoS One 6:e19844. doi: 10.1371/journal.pone.0019844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bakaletz LO. 2007. Bacterial biofilms in otitis media: evidence and relevance. Pediatr Infect Dis J 26:S17–S19. doi: 10.1097/INF.0b013e318154b273. [DOI] [PubMed] [Google Scholar]
- 98.Jedrzejas MJ. 2001. Pneumococcal virulence factors: structure and function. Microbiol Mol Biol Rev 65:187–207. doi: 10.1128/MMBR.65.2.187-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.AlonsoDeVelasco E, Verheul AF, Verhoef J, Snippe H. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev 59:591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cockeran R, Anderson R, Feldman C. 2002. The role of pneumolysin in the pathogenesis of Streptococcus pneumoniae infection. Curr Opin Infect Dis 15:235–239. doi: 10.1097/00001432-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 101.Morales M, Martín-Galiano AJ, Domenech M, García E. 2015. Insights into the evolutionary relationships of LytA autolysin and ply pneumolysin-like genes in Streptococcus pneumoniae and related streptococci. Genome Biol Evol 7:2747–2761. doi: 10.1093/gbe/evv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect Immun 79:548–561. doi: 10.1128/IAI.00682-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bergmann S, Hammerschmidt S. 2006. Versatility of pneumococcal surface proteins. Microbiology 152:295–303. doi: 10.1099/mic.0.28610-0. [DOI] [PubMed] [Google Scholar]
- 104.Sutcliffe IC, Russell RR. 1995. Lipoproteins of gram-positive bacteria. J Bacteriol 177:1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pracht D, Elm C, Gerber J, Bergmann S, Rohde M, Seiler M, Kim KS, Jenkinson HF, Nau R, Hammerschmidt S. 2005. PavA of Streptococcus pneumoniae modulates adherence, invasion, and meningeal inflammation. Infect Immun 73:2680–2689. doi: 10.1128/IAI.73.5.2680-2689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hammerschmidt S. 2006. Adherence molecules of pathogenic pneumococci. Curr Opin Microbiol 9:12–20. doi: 10.1016/j.mib.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 107.Tavares DA, Simões AS, Bootsma HJ, Hermans PW, de Lencastre H, Sá-Leão R. 2014. Non-typeable pneumococci circulating in Portugal are of cps type NCC2 and have genomic features typical of encapsulated isolates. BMC Genomics 15:863. doi: 10.1186/1471-2164-15-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Briles DE, Hollingshead SK, King J, Swift A, Braun PA, Park MK, Ferguson LM, Nahm MH, Nabors GS. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis 182:1694–1701. doi: 10.1086/317602. [DOI] [PubMed] [Google Scholar]
- 109.Wu H-Y, Nahm MH, Guo Y, Russell MW, Briles DE. 1997. Intranasal immunization of mice with PspA (pneumococcal surface protein A) can prevent intranasal carriage, pulmonary infection, and sepsis with Streptococcus pneumoniae. J Infect Dis 175:839–846. doi: 10.1086/513980. [DOI] [PubMed] [Google Scholar]
- 110.Glover DT, Hollingshead SK, Briles DE. 2008. Streptococcus pneumoniae surface protein PcpA elicits protection against lung infection and fatal sepsis. Infect Immun 76:2767–2776. doi: 10.1128/IAI.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Balachandran P, Brooks-Walter A, Virolainen-Julkunen A, Hollingshead SK, Briles DE. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect Immun 70:2526–2534. doi: 10.1128/IAI.70.5.2526-2534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Godfroid F, Hermand P, Verlant V, Denoël P, Poolman JT. 2011. Preclinical evaluation of the Pht proteins as potential cross-protective pneumococcal vaccine antigens. Infect Immun 79:238–245. doi: 10.1128/IAI.00378-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hilty M, Wüthrich D, Salter SJ, Engel H, Campbell S, Sá-Leão R, de Lencastre H, Hermans P, Sadowy E, Turner P, Chewapreecha C, Diggle M, Pluschke G, McGee L, Köseoğlu Eser Ö, Low DE, Smith-Vaughan H, Endimiani A, Küffer M, Dupasquier M, Beaudoing E, Weber J, Bruggmann R, Hanage WP, Parkhill J, Hathaway LJ, Mühlemann K, Bentley SD. 2014. Global phylogenomic analysis of nonencapsulated Streptococcus pneumoniae reveals a deep-branching classic lineage that is distinct from multiple sporadic lineages. Genome Biol Evol 6:3281–3294. doi: 10.1093/gbe/evu263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Croucher NJ, Finkelstein JA, Pelton SI, Mitchell PK, Lee GM, Parkhill J, Bentley SD, Hanage WP, Lipsitch M. 2013. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat Genet 45:656–663. doi: 10.1038/ng.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hanage WP, Kaijalainen T, Saukkoriipi A, Rickcord JL, Spratt BG. 2006. A successful, diverse disease-associated lineage of nontypeable pneumococci that has lost the capsular biosynthesis locus. J Clin Microbiol 44:743–749. doi: 10.1128/JCM.44.3.743-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zegans ME, Sanchez PA, Likosky DS, Allar RT, Martin M, Schwartzman JD, Pryor JH, Turco JH, Whitney CG. 2009. Clinical features, outcomes, and costs of a conjunctivitis outbreak caused by the ST448 strain of Streptococcus pneumoniae. Cornea 28:503–509. doi: 10.1097/ICO.0b013e3181909362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shayegani M, Parsons LM, Gibbons WE, Campbell D. 1982. Characterization of nontypable Streptococcus pneumoniae-like organisms isolated from outbreaks of conjunctivitis. J Clin Microbiol 16:8–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Berrón S, Fenoll A, Ortega M, Arellano N, Casal J. 2005. Analysis of the genetic structure of nontypeable pneumococcal strains isolated from conjunctiva. J Clin Microbiol 43:1694–1698. doi: 10.1128/JCM.43.4.1694-1698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marimon JM, Ercibengoa M, García-Arenzana JM, Alonso M, Pérez-Trallero E. 2013. Streptococcus pneumoniae ocular infections, prominent role of unencapsulated isolates in conjunctivitis. Clin Microbiol Infect 19:E298–E305. doi: 10.1111/1469-0691.12196. [DOI] [PubMed] [Google Scholar]
- 120.Buck JM, Lexau C, Shapiro M, Glennen A, Boxrud DJ, Koziol B, Whitney CG, Beall B, Danila R, Lynfield R. 2006. A community outbreak of conjunctivitis caused by nontypeable Streptococcus pneumoniae in Minnesota. Pediatr Infect Dis J 25:906–911. doi: 10.1097/01.inf.0000238143.96607.ec. [DOI] [PubMed] [Google Scholar]
- 121.Martin M, Turco JH, Zegans ME, Facklam RR, Sodha S, Elliott JA, Pryor JH, Beall B, Erdman DD, Baumgartner YY, Sanchez PA, Schwartzman JD, Montero J, Schuchat A, Whitney CG. 2003. An outbreak of conjunctivitis due to atypical Streptococcus pneumoniae. N Engl J Med 348:1112–1121. doi: 10.1056/NEJMoa022521. [DOI] [PubMed] [Google Scholar]
- 122.Hennink M, Abbas Z, McDonald RR, Nagle E, Montgomery KL, Diener T, Horsman GB, Levett PN. 2006. Streptococcus pneumoniae outbreak in a rural Régina community. Can Commun Dis Rep 32:181–186. [PubMed] [Google Scholar]
- 123.Tarabishy AB, Jeng BH. 2008. Bacterial conjunctivitis: a review for internists. Cleve Clin J Med 75:507–512. doi: 10.3949/ccjm.75.7.507. [DOI] [PubMed] [Google Scholar]
- 124.Xu Q, Kaur R, Casey JR, Sabharwal V, Pelton S, Pichichero ME. 2011. Nontypeable Streptococcus pneumoniae as an otopathogen. Diagn Microbiol Infect Dis 69:200–204. doi: 10.1016/j.diagmicrobio.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hotomi M, Nakajima K, Hiraoka M, Nahm MH, Yamanaka N. 2016. Molecular epidemiology of nonencapsulated Streptococcus pneumoniae among Japanese children with acute otitis media. J Infect Chemother 22:72–77. doi: 10.1016/j.jiac.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 126.Sabharwal V, Ram S, Figueira M, Park IH, Pelton SI. 2009. Role of complement in host defense against pneumococcal otitis media. Infect Immun 77:1121–1127. doi: 10.1128/IAI.01148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blom AM, Bergmann S, Fulde M, Riesbeck K, Agarwal V. 2014. Streptococcus pneumoniae phosphoglycerate kinase is a novel complement inhibitor affecting the membrane attack complex formation. J Biol Chem 289:32499–32511. doi: 10.1074/jbc.M114.610212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Agarwal V, Sroka M, Fulde M, Bergmann S, Riesbeck K, Blom AM. 2014. Binding of Streptococcus pneumoniae endopeptidase O (PepO) to complement component C1q modulates the complement attack and promotes host cell adherence. J Biol Chem 289:15833–15844. doi: 10.1074/jbc.M113.530212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mohan S, Hertweck C, Dudda A, Hammerschmidt S, Skerka C, Hallström T, Zipfel PF. 2014. Tuf of Streptococcus pneumoniae is a surface displayed human complement regulator binding protein. Mol Immunol 62:249–264. doi: 10.1016/j.molimm.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 130.Józsi M, Zipfel PF. 2008. Factor H family proteins and human diseases. Trends Immunol 29:380–387. doi: 10.1016/j.it.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 131.Giebink GS, Payne EE, Mills EL, Juhn SK, Quie PG. 1976. Experimental otitis media due to Streptococcus pneumoniae: immunopathogenic response in the chinchilla. J Infect Dis 134:595–604. doi: 10.1093/infdis/134.6.595. [DOI] [PubMed] [Google Scholar]
- 132.Keller LE, Friley J, Dixit C, Nahm MH, McDaniel LS. 2014. Nonencapsulated Streptococcus pneumoniae cause acute otitis media in the chinchilla that is enhanced by pneumococcal surface protein K. Open Forum Infect Dis 1:ofu037. doi: 10.1093/ofid/ofu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reid SD, Hong W, Dew KE, Winn DR, Pang B, Watt J, Glover DT, Hollingshead SK, Swords WE. 2009. Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J Infect Dis 199:786–794. doi: 10.1086/597042. [DOI] [PubMed] [Google Scholar]
- 134.Keller LE, Luo X, Thornton JA, Seo K-S, Moon BY, Robinson DA, McDaniel LS. 2015. Immunization with pneumococcal surface protein K of nonencapsulated Streptococcus pneumoniae provides protection in a mouse model of colonization. Clin Vaccine Immunol 22:1146–1153. doi: 10.1128/CVI.00456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Na IY, Baek JY, Park IH, Kim DH, Song J-H, Ko KS. 2015. pspK gene prevalence and characterization of non-typeable Streptococcus pneumoniae isolates from Asian countries. Microbiology 161:973–979. doi: 10.1099/mic.0.000073. [DOI] [PubMed] [Google Scholar]
- 136.Scott JR, Hinds J, Gould KA, Millar EV, Reid R, Santosham M, O’Brien KL, Hanage WP. 2012. Nontypeable pneumococcal isolates among Navajo and White Mountain Apache communities: are these really a cause of invasive disease? J Infect Dis 206:73–80. doi: 10.1093/infdis/jis307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brown EJ. 1985. Interaction of Gram-positive microorganisms with complement, p 159–187. In Loos M (ed), Bacteria and complement. Springer, Berlin, Germany. [DOI] [PubMed] [Google Scholar]
- 138.Winkelstein JA. 1981. The role of complement in the host’s defense against Streptococcus pneumoniae. Rev Infect Dis 3:289–298. doi: 10.1093/clinids/3.2.289. [DOI] [PubMed] [Google Scholar]
- 139.Brown JS, Hussell T, Gilliland SM, Holden DW, Paton JC, Ehrenstein MR, Walport MJ, Botto M. 2002. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc Natl Acad Sci U S A 99:16969–16974. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. 2010. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun 78:704–715. doi: 10.1128/IAI.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jauneikaite E, Jefferies JM, Hibberd ML, Clarke SC. 2012. Prevalence of Streptococcus pneumoniae serotypes causing invasive and non-invasive disease in South East Asia: a review. Vaccine 30:3503–3514. doi: 10.1016/j.vaccine.2012.03.066. [DOI] [PubMed] [Google Scholar]
- 142.Lagos R, Muñoz A, San Martin O, Maldonado A, Hormazabal JC, Blackwelder WC, Levine MM. 2008. Age- and serotype-specific pediatric invasive pneumococcal disease: insights from systematic surveillance in Santiago, Chile, 1994–2007. J Infect Dis 198:1809–1817. doi: 10.1086/593334. [DOI] [PubMed] [Google Scholar]
- 143.Ravin AW. 1959. Reciprocal capsular transformations of pneumococci. J Bacteriol 77:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Taylor HE. 1949. Additive effects of certain transforming agents from some variants of pneumococcus. J Exp Med 89:399–424. doi: 10.1084/jem.89.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Marks LR, Reddinger RM, Hakansson AP. 2012. High levels of genetic recombination during nasopharyngeal carriage and biofilm formation in Streptococcus pneumoniae. mBio 3:e00200-12. doi: 10.1128/mBio.00200-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]