ABSTRACT
Members of the ABC-F subfamily of ATP-binding cassette proteins mediate resistance to a broad array of clinically important antibiotic classes that target the ribosome of Gram-positive pathogens. The mechanism by which these proteins act has been a subject of long-standing controversy, with two competing hypotheses each having gained considerable support: antibiotic efflux versus ribosomal protection. Here, we report on studies employing a combination of bacteriological and biochemical techniques to unravel the mechanism of resistance of these proteins, and provide several lines of evidence that together offer clear support to the ribosomal protection hypothesis. Of particular note, we show that addition of purified ABC-F proteins to an in vitro translation assay prompts dose-dependent rescue of translation, and demonstrate that such proteins are capable of displacing antibiotic from the ribosome in vitro. To our knowledge, these experiments constitute the first direct evidence that ABC-F proteins mediate antibiotic resistance through ribosomal protection.
IMPORTANCE
Antimicrobial resistance ranks among the greatest threats currently facing human health. Elucidation of the mechanisms by which microorganisms resist the effect of antibiotics is central to understanding the biology of this phenomenon and has the potential to inform the development of new drugs capable of blocking or circumventing resistance. Members of the ABC-F family, which include lsa(A), msr(A), optr(A), and vga(A), collectively yield resistance to a broader range of clinically significant antibiotic classes than any other family of resistance determinants, although their mechanism of action has been controversial since their discovery 25 years ago. Here we present the first direct evidence that proteins of the ABC-F family act to protect the bacterial ribosome from antibiotic-mediated inhibition.
INTRODUCTION
Antibiotic resistance undermines effective antibacterial chemotherapy and represents a major threat to global public health (1, 2). A comprehensive response to this problem includes gaining a detailed understanding of the mechanisms by which antibiotic resistance is mediated, particularly since such information could inform the development of novel antibacterial agents able to overcome or circumvent extant resistance phenotypes. While the majority of clinically important antibiotic resistance mechanisms are by now well characterized (3), some key gaps in our knowledge remain. One such gap concerns the mechanism by which ABC-F proteins mediate resistance to a broad array of clinically important antibiotic classes that target protein synthesis in Gram-positive pathogens.
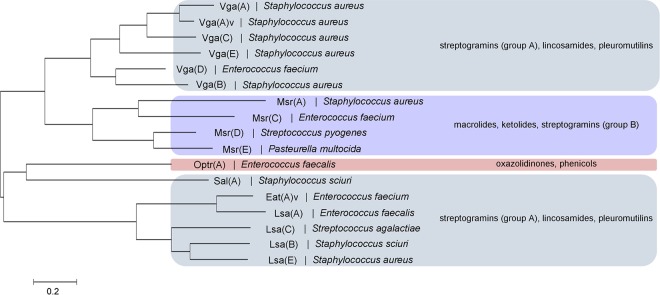
ABC-F proteins are found across all three domains of life, and comprise a single polypeptide containing two ATP-binding cassette (ABC) domains separated by a linker of ~80 amino acids. In contrast to canonical ABC transporters, the ABC portions of ABC-F proteins are not fused to transmembrane domains (TMDs), nor are they genetically associated with TMDs in operons (4). While it may be that some ABC-F proteins associate with TMDs to mediate transport across membranes, it is nevertheless apparent that members of this family participate in biological processes other than transport, including DNA repair, enzyme regulation, and translational control (5). In Gram-positive bacteria, a subgroup of the ABC-F proteins mediates resistance to antibiotics that exert their action on the ribosome. These proteins, referred to here as antibiotic resistance (ARE) ABC-F proteins, are found in both antibiotic-producing bacteria (e.g., the streptomycetes) and in pathogenic bacteria that include the staphylococci, streptococci, and enterococci (4, 6) (Fig. 1). Collectively, the ARE ABC-F family of proteins mediates resistance to the majority of antibiotic classes that bind to the 50S subunit of the ribosome, including the ketolides (7), lincosamides (8, 9), macrolides (10), oxazolidinones (11), phenicols (11), pleuromutilins (12), and streptogramins of groups A (9, 13) and B (10) (Fig. 1). However, no single ARE ABC-F determinant confers resistance to every listed class, and three phenotypic resistance profiles are distinguished in clinical isolates. Combined lincosamide and streptogramin A (and sometimes pleuromutilin) resistance, referred to as the LSA (or LSAP) phenotype, is conferred by vga-, lsa-, and sal-type genes (9, 14, 15), concurrent resistance to macrolides and group B streptogramins (and sometimes ketolides) (MSB phenotype) by the msr-type determinants (7, 10), and resistance to phenicols and oxazolidinones by the recently identified optrA gene (11) (Fig. 1).
FIG 1 .
Phylogenetic tree and antibiotic resistance profiles of the ARE ABC-F proteins found in representative Gram-positive pathogens. The tree was generated using the maximum likelihood method with the MEGA 6.0.6 software package (47). An overview of the antibiotic resistance phenotypes conferred by the different subgroups of determinant is given at the right of the figure, denoted by colored boxes (although variations in individual resistance phenotypes within each subgroup are not shown).
The mechanism by which the ARE ABC-F proteins mediate antibiotic resistance has been a subject of long-standing controversy, with two competing hypotheses having each attracted considerable support (4, 6, 10, 16–20). The efflux hypothesis posits that ARE ABC-F proteins associate with as-yet-unidentified TMDs to form a functional efflux complex capable of exporting antibiotics out of the cell, while the ribosomal protection hypothesis suggests that these resistance proteins act instead to reduce the accessibility or affinity of the antibiotic binding sites on the 50S subunit of the ribosome, thereby directly protecting the translational machinery from antibiotic-mediated inhibition (16). In proposed support of the efflux hypothesis, previous work has reported membrane localization of Vga(A) in Staphylococcus epidermidis (18, 21), and evidence has been obtained for interaction between the streptococcal ARE ABC-F protein Msr(D) and the major facilitator protein Mef(E) when both are heterologously expressed in Escherichia coli (19). Furthermore, studies showing that staphylococci expressing ARE ABC-F resistance determinants exhibit decreased accumulation of antibiotic classes that fall within their phenotypic resistance profile (8, 10, 16, 22, 23) have been considered evidence of efflux. However, subsequent work has refuted this interpretation, demonstrating that such uptake studies are incapable of distinguishing drug efflux from ribosomal protection as decreased accumulation of antibiotics will also result from protection of ribosomes (16), which ordinarily act as an intracellular “sink” to increase drug accumulation (24).
Direct evidence for the ribosomal protection hypothesis is similarly lacking. However, the specificity of the ARE ABC-F resistance mechanism for multiple, structurally unrelated classes of protein synthesis inhibitors is easier to interpret in the context of ribosomal protection, and several ABC proteins not involved in antibiotic resistance have recently been shown to interact directly with the ribosome or with ribosomally associated proteins (4, 25).
In order to clarify the mechanism of the ARE ABC-F proteins, we have studied the action of members of this family using bacteriological and biochemical assays, and thereby provide the first direct evidence for a resistance mechanism involving ribosomal protection.
RESULTS
The antibiotic resistance phenotype conferred by ARE ABC-F proteins is suggestive of ribosomal protection.
In initial experiments using staphylococci expressing vga(A), we sought preliminary support for a mechanism of resistance involving either ribosomal protection or efflux. Previous studies have noted a correlation between the resistance phenotypes mediated by ARE ABC-F proteins and the extent of overlap between binding sites of protein synthesis inhibitors within the PTC and peptide exit tunnel of the 50S subunit (16, 17). A testable prediction is that, if Vga(A) does indeed mediate resistance through ribosomal protection, this protein would likely offer cross-resistance to further classes of structurally unrelated antibiotic that bind the ribosome in close proximity to the group A streptogramins. To examine this, we performed susceptibility determinations with Staphylococcus aureus RN4220(pSEPSA5:vga(A)) using a panel of 50S-targeted antibiotics (see Table S1 in the supplemental material). In line with published data, expression of vga(A) conferred reduced susceptibility to virginiamycin M (64-fold), lincomycin (8-fold), and retapamulin (4-fold) but had no impact on susceptibility to erythromycin, florfenicol, or linezolid. However, we also detected a 4-fold decrease in susceptibility to the 16-member macrolides carbomycin A and leucomycin A1, structurally related antibiotics whose binding site on the ribosome is predicted to partially overlap that of members of the group A streptogramins (26). To our knowledge, this is the first report of a vga-type ARE ABC-F protein mediating any degree of macrolide resistance, and offers further indirect support for a mechanism of resistance involving ribosomal protection.
A common feature of antibiotic resistance mechanisms involving efflux is that, when coresident in a bacterial cell with a determinant conferring resistance to the same antibacterial agent through protection of the drug target, a synergistic or additive increase in resistance is observed. For example, S. aureus strains expressing both the tetracycline ribosome protection protein (RPP), Tet(M), and the tetracycline efflux pump, Tet(K), exhibit a substantial enhancement of tetracycline resistance compared with strains expressing only one of these resistance mechanisms (27). In contrast, no such enhancement in antibiotic resistance may be observed when two resistance determinants, both of which act at the level of the drug target, coexist in a bacterial cell. For example, in fusidic acid-resistant strains of S. aureus carrying both resistance polymorphisms in the drug target (EF-G) and a horizontally acquired fusidic acid resistance gene (fusB), the level of resistance observed does not exceed that of the determinant that alone provides the greatest degree of resistance (28). We therefore reasoned that failure of Vga(A) to exhibit an additive or synergistic effect when coresident in S. aureus with a ribosomal protection mechanism of streptogramin resistance would suggest that Vga(A) does not mediate resistance by efflux. To test this, we created a strain carrying both vga(A) and cfr, the latter of which encodes an rRNA methyltransferase capable of methylating 23S rRNA at position 8 of adenine 2503 (E. coli numbering) and thereby protects ribosomes from inhibition by several antibiotic classes, including those encompassed within the spectrum of resistance of Vga(A) (29). Susceptibility to virginiamycin M, lincomycin, and linezolid was determined for S. aureus RN4220 expressing Vga(A) alone or Vga(A) and Cfr. As expected, expression of Vga(A) alone mediated resistance to virginiamycin M and lincomycin, but not to linezolid, while expression of Cfr alone gave resistance to all three drugs (see Table S2 in the supplemental material). Coexpression of both resistance determinants did not confer a further decrease in susceptibility to any of the drugs beyond that exhibited by the strain solely expressing Cfr, establishing that coexpression of the two resistance proteins does not produce an additive or synergistic effect (Table S2) and further reinforcing the idea that resistance is more likely mediated through ribosomal protection than efflux.
Purified ARE ABC-F proteins mediate specific and dose-dependent protection of staphylococcal translation from antibiotic inhibition in vitro.
Since our bacteriological studies pointed toward ribosomal protection as the likely mechanism of resistance of ARE ABC-F proteins, we sought to directly test the ability of these proteins to protect the translation apparatus from antibiotic-mediated inhibition. We therefore overexpressed and purified recombinant Vga(A) from E. coli for addition into an S. aureus in vitro coupled transcription/translation (T/T) assay. Addition of a C-terminal hexahistidine (6×His) tag to Vga(A) has previously been demonstrated not to perturb the ability of the protein to mediate resistance in whole cells (21), and we therefore expressed Vga(A) fused to this tag to facilitate purification by immobilized-metal affinity chromatography (IMAC). Effective purification of Vga(A) required removal of contaminating nucleic acids (21), which was achieved by the addition of 2 M NaCl during IMAC and subsequent gel filtration purification steps.
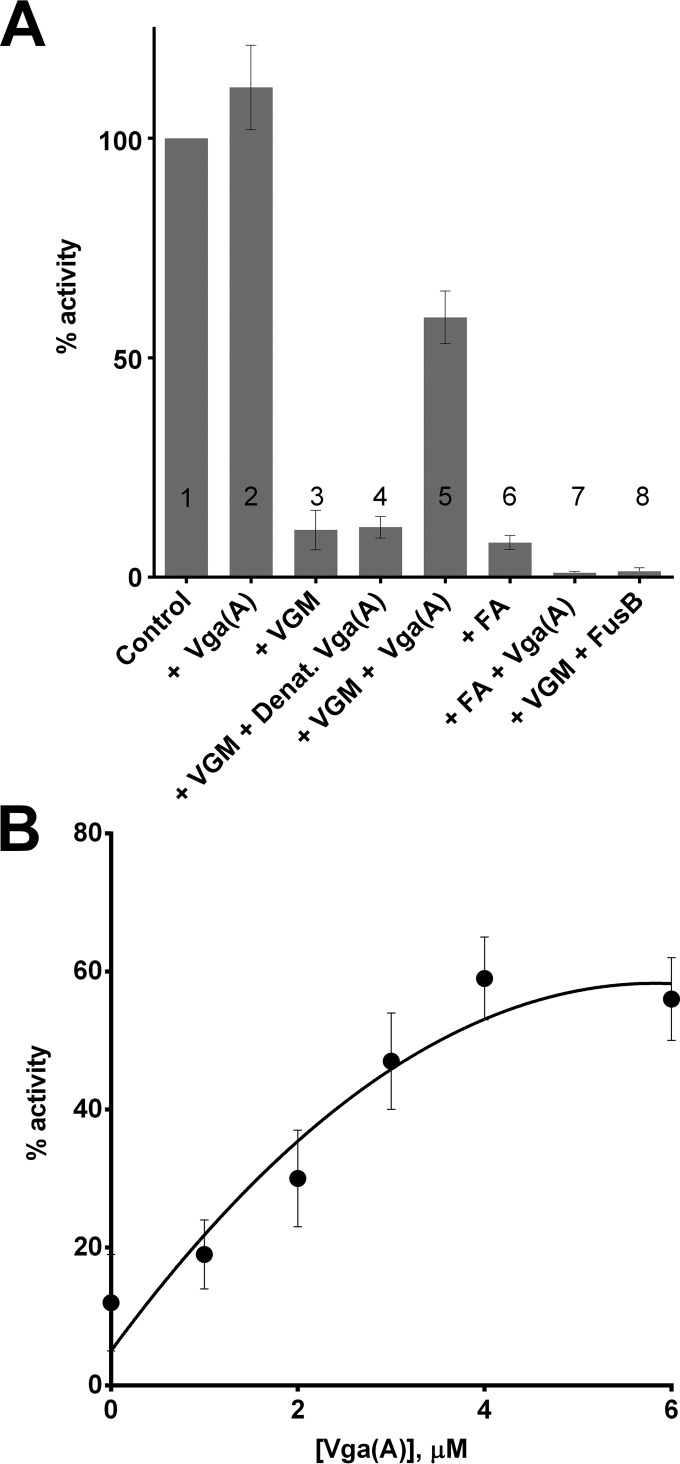
Introduction of 4 µM purified Vga(A) into T/T assays inhibited with a ≥90% inhibitory concentration (IC90) of virginiamycin M resulted in substantial restoration of translation, with activity rising to ~60% of that of the untreated control (Fig. 2A). In contrast, Vga(A) offered no protection against inhibition of translation by a structurally and mechanistically unrelated inhibitor of protein synthesis (fusidic acid) (Fig. 2A). The ability to rescue translation from virginiamycin M was a specific property of Vga(A), since no protection against this antibiotic was observed when the fusidic acid resistance protein FusB (28) was substituted for Vga(A), and protection was also lost upon heat denaturation of Vga(A) (Fig. 2A). Protection of translation from virginiamycin M by Vga(A) exhibited dose dependence in the concentration range between ~1 and 4 µM Vga(A) (Fig. 2A).
FIG 2 .
Vga(A) protects an S. aureus-derived T/T assay from inhibition by virginiamycin M. (A) Column 1 shows an uninhibited T/T assay with no addition of exogenous protein, whilst column 2 shows an uninhibited assay with the addition of 4 µM Vga(A). In columns 3 and 5, 4 µM Vga(A) added to a T/T assay mixture containing ≥IC90 of virginiamycin M (VGM [column 3]) rescued protein synthesis. In columns 3, 4, and 8, addition of 4 µM heat-denatured (Denat.) Vga(A) (column 4) or 4 µM fusidic acid resistance protein FusB (column 8) failed to rescue protein synthesis from inhibition by virginiamycin M (column 3). In columns 6 and 7, addition of 4 µM Vga(A) to a T/T assay mixture containing ≥IC90 of fusidic acid (FA) did not rescue protein synthesis. (B) Dose-dependent rescue of protein synthesis by Vga(A) from inhibition with ≥IC90 of virginiamycin M. Results are means from at least three independent determinations, and error bars show standard deviations.
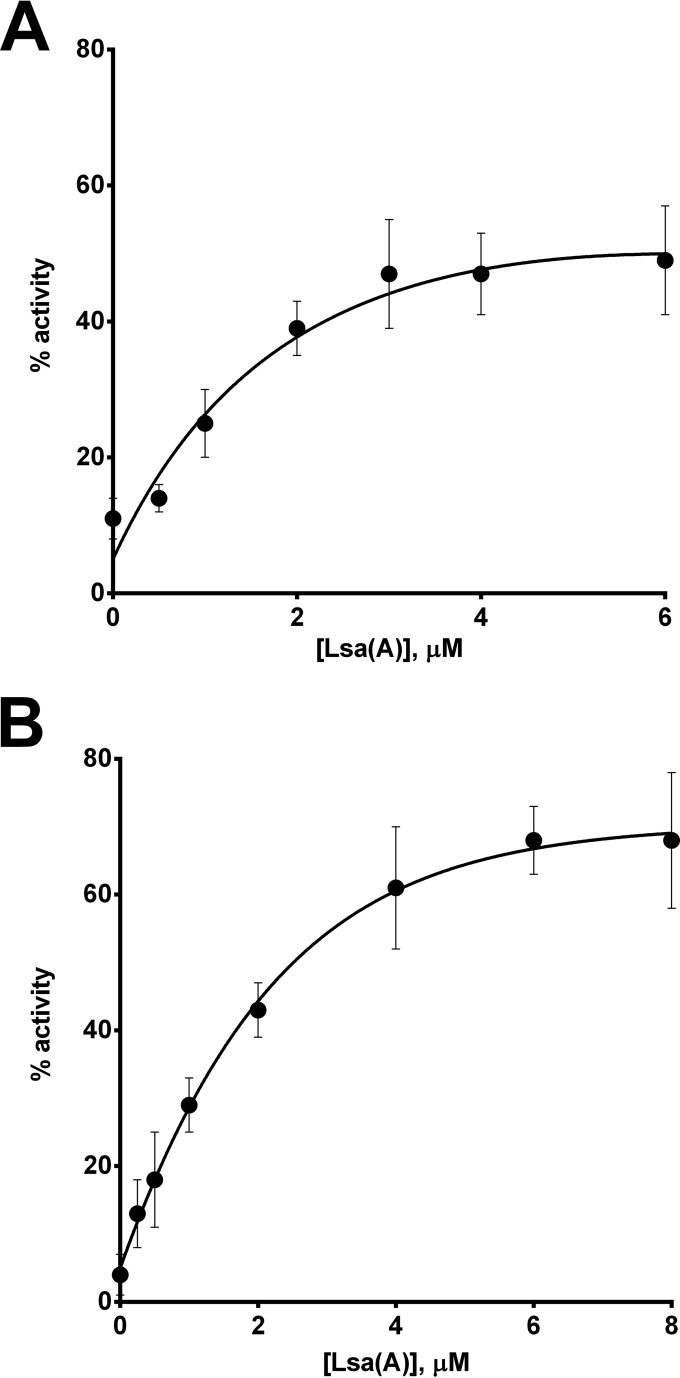
To confirm that the ability to protect the translation apparatus from antibiotic-mediated inhibition is not unique to Vga(A) but is a property shared by other ARE ABC-F proteins, we also purified and tested the ability of a phylogenetically distant member of this family, Lsa(A) (Fig. 1), to rescue translation. Lsa(A) was overexpressed in E. coli with N-terminal His and SUMO tags to maximize protein solubility. Both tags were cleaved following an initial IMAC purification step, yielding untagged Lsa(A) with an additional N-terminal glycine residue, which was subsequently purified to homogeneity by gel filtration. Purified Lsa(A) was titrated into T/T reaction mixtures inhibited with a ≥IC90 of virginiamycin (Fig. 3A) or lincomycin (Fig. 3B). As for Vga(A), rescue of translation was concentration dependent up to ~4 to 6 µM and at the highest protein concentrations tested restored translation activity to at least ~50% in each case (Fig. 3A and B).
FIG 3 .
Lsa(A) mediates dose-dependent protection of a S. aureus-derived transcription/translation assay from inhibition by virginiamycin M (A) and lincomycin (B). Results are means from at least three independent determinations, and error bars show standard deviations.
Recapitulation of resistance phenotypes associated with ARE ABC-F proteins in an in vitro translation assay.
To provide further confirmation that the observed ability of ARE ABC-F proteins to protect an in vitro translation assay from antibiotics reflects the activity of these proteins in whole cells and to further explore the phenomenon of protection, we sought to recapitulate in the T/T assay several phenotypes that have been associated with these proteins in bacteria.
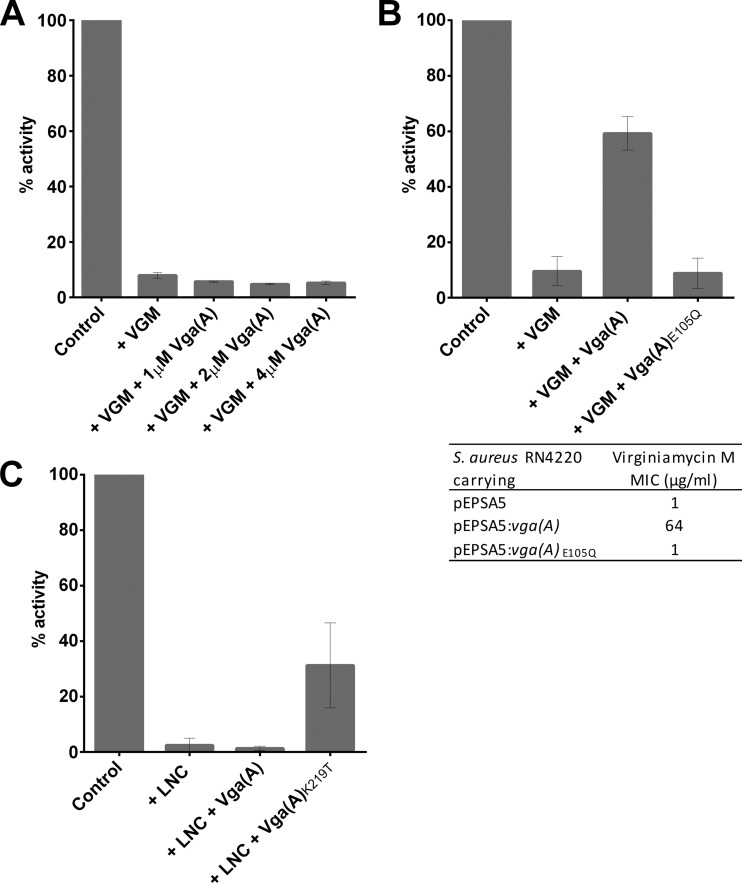
The Vga(A) protein is not functional in E. coli, failing to confer any reduction in virginiamycin M susceptibility even when detectably overexpressed (21; data not shown). This result was mirrored in an in vitro T/T assay using E. coli S30 extract; addition of increasing concentrations of Vga(A) (Fig. 4A) and Lsa(A) (data not shown) to maximum of 4 µM into T/T reactions inhibited with ≥IC90 of virginiamycin M produced no rescue of translation.
FIG 4 .
Recapitulation of resistance phenotypes associated with ARE ABC-F proteins in vitro. (A) When expressed in E. coli, Vga(A) does not confer resistance to virginiamycin M (21); addition of Vga(A) to an E. coli T/T assay containing ≥IC90 of virginiamycin M (VGM) also failed to restore translational activity. (B) ATPase activity is essential for Vga(A) function (21), and abrogation of ATPase activity of the N-terminal ABC domain rendered Vga(A) inactive when expressed in S. aureus RN4220; the purified ATPase-deficient Vga(A)E105Q protein also failed to protect staphylococcal translation from inhibition by virginiamycin M in vitro. (C) A single-amino-acid substitution in the interdomain linker expands the resistance spectrum of Vga(A) to encompass lincomycin (20). Addition of the purified Vga(A)K219T to a staphylococcal T/T assay inhibited with a >IC90 of lincomycin (LNC) restored translational activity, while addition of the wild-type protein did not. Results are means from at least three independent determinations, and error bars show standard deviations.
It has previously been demonstrated that substitution for glutamine of the catalytic glutamate residue following the Walker B motif in either nucleotide binding domain of Vga(A) results in a nonfunctional protein incapable of mediating resistance to virginiamycin M in cells of Staphylococcus epidermidis (21). We confirmed that this also holds true in cells of S. aureus, with expression of Vga(A)E105Q in S. aureus RN4220 having no effect on virginiamycin M susceptibility (Fig. 4B). The same loss of the ability of Vga(A)E105Q to mediate virginiamycin M resistance could also be demonstrated in vitro, with addition of up to 4 µM purified Vga(A)E105Q to a T/T assay employing S. aureus S30 extract producing no restoration of translation activity (Fig. 4B).
A single-amino-acid substitution (K219T) in the linker region between the two nucleotide binding domains of Vga(A) has recently been reported to increase the level of phenotypic resistance to lincosamides from low level (4-fold) to high level (64-fold) (20). This shift in resistance profile was successfully recapitulated in the S. aureus T/T assay; addition of purified Vga(A)K219T to a T/T reaction mixture inhibited with lincomycin resulted in restoration of translation activity to ~30% of that of the uninhibited control, while 4 µM wild-type Vga(A) did not detectably protect translation against lincomycin (Fig. 4C).
Lsa(A) prevents binding of antibiotic to staphylococcal ribosomes, and displaces ribosome-bound antibiotic.
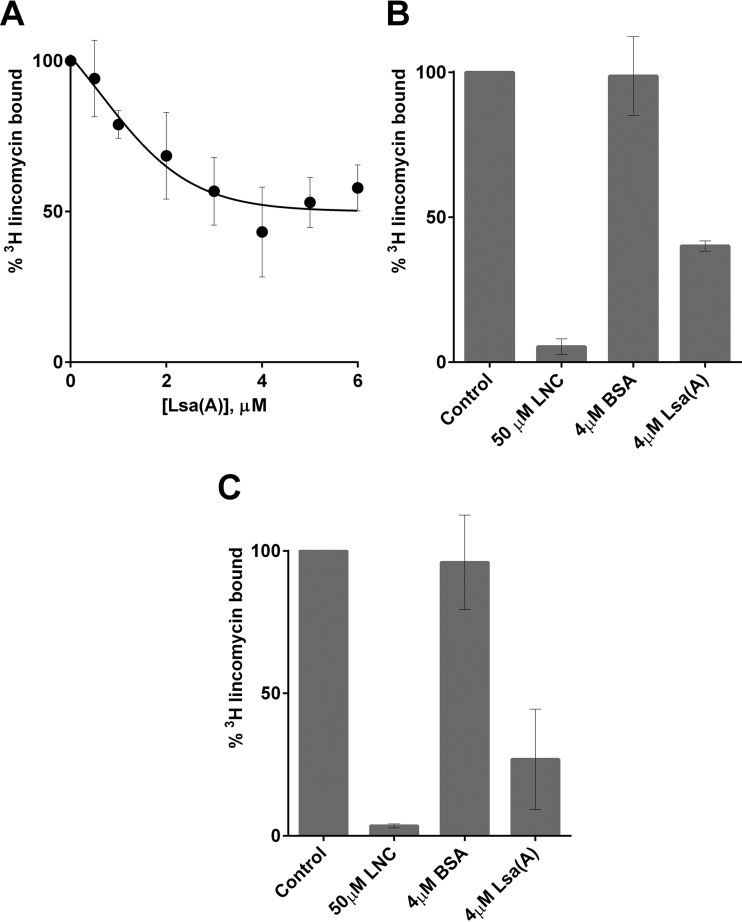
We sought to examine whether the ARE ABC-F proteins protect the translation apparatus from antibiotic-mediated inhibition by directly interfering with the interaction between the antibiotic and the ribosome. In the first instance, we evaluated the ability of the Lsa(A) protein to prevent binding of 3H-radiolabeled lincomycin to purified staphylococcal ribosomes. Preincubation of between 1:1 and 8:1 molar ratios of Lsa(A) to ribosomes prior to the addition of [3H]lincomycin resulted in a dose-dependent decrease in subsequent binding of lincomycin to ribosomes, before reaching a plateau past which addition of Lsa(A) caused no further reduction in lincomycin binding (Fig. 5A). In contrast, preincubation of ribosomes with an 8-fold molar excess of an unrelated control protein (bovine serum albumin [BSA]) did not reduce the level of ribosomally associated [3H]lincomycin (Fig. 5B).
FIG 5 .
Lsa(A) prevents binding of lincomycin to staphylococcal ribosomes, and displaces ribosome-bound lincomycin. (A) Preincubation of increasing concentrations of Lsa(A) with 0.5 µM staphylococcal ribosomes caused a reduction in binding of [3H]lincomycin. (B) Preincubation of 0.5 µM ribosomes with a 50× excess of unlabeled lincomycin (LNC) decreased subsequent binding by [3H]lincomycin (column 2 versus 1). Preincubation with 4 µM BSA did not protect ribosomes from binding by [3H]lincomycin (columns 3 and 1). Addition of 4 µM Lsa(A) resulted in decreased association of [3H]lincomycin with ribosomes (column 4 versus 1). (C) Addition of a 50× excess of unlabeled lincomycin caused dissociation of [3H]lincomycin prebound to staphylococcal ribosomes (column 2 versus 1), as did addition of 4 µM Lsa(A) (column 4 versus 1); however, addition of BSA did not (column 3 versus 1). Results are means from at least three independent determinations, and error bars show standard deviations.
Subsequently, we examined the ability of Lsa(A) to displace prebound [3H]lincomycin from ribosomes. We first confirmed that this assay could demonstrate displacement of radiolabeled lincomycin following addition of a 50-fold excess of the unlabeled drug and that addition of an 8-fold molar excess of BSA had essentially no effect on the level of ribosome-bound drug (Fig. 5C). Addition of an 8-fold molar-excess of Lsa(A) to ribosomes preincubated with a 2-fold molar-excess of drug resulted in a substantial (~73%) reduction in ribosome-associated [3H]lincomycin (Fig. 5C).
DISCUSSION
The ARE ABC-F proteins collectively yield resistance to a broader range of clinically significant antibiotic classes than any other family of resistance determinants. Despite their importance, the mechanism by which they mediate antibiotic resistance has remained obscure since their discovery 25 years ago (10). Here we have provided several lines of indirect and direct evidence that together reveal a mechanism of resistance involving ribosomal protection. The demonstration that more than one member of this family protects the staphylococcal translation apparatus from antibiotic-mediated inhibition in vitro, under conditions where transport cannot occur, coupled with the lack of evidence for drug efflux by the ARE ABC-F proteins, would appear to render the efflux hypothesis redundant.
ARE ABC-F proteins rescue translation from antibiotic-mediated inhibition by driving dissociation of bound antibiotic molecules from the ribosome. The fact that a given ARE ABC-F protein is capable of triggering dissociation of multiple structurally distinct classes of antibiotic, provided these compounds share overlapping binding sites, argues for a simple mechanism of resistance based upon protein-mediated drug displacement. Such a displacement mechanism underlies the only other clinically important example of antibiotic resistance involving ribosomal protection: resistance to tetracyclines mediated by RPPs. These proteins, such as Tet(O) and Tet(M), reach into the tetracycline binding site to directly dislodge the drug (30, 31).
While molecular detail regarding the interaction occurring between ARE ABC-F proteins and the ribosome will be contingent on structural characterization of the complex they form, the recent functional and structural characterization of a non-ARE, but ribosome-binding, ABC-F protein offers a basis for informed speculation. Energy-dependent translational throttle A (EttA) is a bacterial ABC-F protein that binds the ribosome to regulate protein synthesis in response to changing cellular energy levels (32). This protein binds into the E-site of the ribosome, bridging the L1 stalk and P-site, and modulates the conformation of the PTC through contacts with ribosomal proteins, rRNA and P-site fMet-tRNA (33). These interactions occur primarily through the EttA interdomain linker, a well-conserved region of the protein designated the P-site tRNA interaction motif (32). In a recent paper, Lenart et al. (20) noted that this motif is conserved in Vga(A), although extended by an additional 30 amino acids, and demonstrated that single-amino-acid substitutions within this additional region can alter the resistance profile of Vga(A). On this basis, they speculated that the Vga(A) linker may act in an analogous manner to the EttA linker, but with the 30-amino-acid extension allowing further penetration toward the PTC, where it causes dissociation of its target drugs either directly, or through contacts with the P-site tRNA (20). In light of the findings presented here, which collectively confirm that the ARE ABC-F proteins do mediate resistance at the ribosome—and indeed, that these proteins act to displace bound antibiotic from the ribosome—such an explanation for the mechanism of resistance appears compelling. However, it is of note that the interaction of EttA with the ribosomal L1-stalk is mediated by a second functionally important motif, a 44-amino-acid region between the Q-loop and signature motif of the N-terminal ABC domain referred to as the arm region, which is not present in ARE ABC-F proteins. This region is important for determining the specificity of the protein-protein interactions in which other ABC proteins participate (5, 34, 35). Therefore, while the interdomain linker of ARE ABC-F proteins may interact with the P-site tRNA in a similar manner to EttA, the site and mode of binding of the ARE ABC-F proteins to the ribosome may be distinct.
MATERIALS AND METHODS
Bacteria and plasmids.
The bacteria and plasmids used or generated in this study are listed in Table 1.
TABLE 1 .
Bacteria and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. faecalis ATCC 29212 | Source of lsa(A) gene | ATCC (48) |
| S. aureus RN4220 | Restriction-deficient derivative of S. aureus 8325-4, used for routine cloning and antibiotic susceptibility testing | 49 |
| E. coli | ||
| DH5α | For routine cloning procedures | Invitrogen, Paisley, United Kingdom |
| BL21(λDE3) Gold | For expression of Vga(A) | Agilent Technologies |
| BL21-CodonPlus(λDE3) RIL | For expression of Lsa(A) | Agilent Technologies |
| CopyCutter EPI400 | To maintain plasmid pSAluc at low copy number | Epicenter, Madison, WI |
| S. aureus | ||
| RN4220 vga(A)+ | S. aureus RN4220 with vga(A) integrated at ϕ11 attB locus under control of cap1a promoter | This study |
| RN4220 vga(A)+(pEPSA5:cfr) | S. aureus RN4220 vga(A)+, with cfr under control of pT5X promoter on pEPSA5 | This study |
| RN4220(pEPSA5) | RN4220 carrying pEPSA5 | This study |
| RN4220(pEPSA5:cfr) | RN4220 carrying cfr under control of pT5X promoter on pEPSA5 | This study |
| RN4220(pEPSA5:vga(A)) | RN4220 carrying vga(A) under control of pT5X promoter on pEPSA5 | This study |
| Plasmids | ||
| pEPSA5 | S. aureus/E. coli shuttle vector for expression of genes in S. aureus from xylose-inducible promoter pT5X | 37 |
| pLL39 | Single-copy integration vector for integration at L54a attB or ϕ11 attB sites on S. aureus chromosome | 40 |
| pLL2787 | Accessory plasmid carrying ϕ11 int gene | 36 |
| pIVEX2.3d:vga(A) | For expression of Vga(A) with C-terminal 6×His tag in E. coli | 5 Prime GmbH, Düsseldorf, Germany (21, 50) |
| pBEST | Contains firefly luciferase (luc) gene under control of E. coli tac promoter | Promega, Madison, WI |
| pSAluc | Modified pBEST plasmid with luc gene under control of strong staphylococcal cap1A promoter | 44 |
| pEPSA5:vgaA | For expression of Vga(A) in S. aureus | This study |
| pEPSA5:vgaE105Q | For expression of ATPase-deficient mutant of Vga(A) in S. aureus | This study |
| pEPSA5:vgaAK219T | For expression of expanded-phenotype mutant of Vga(A) | This study |
| pEPSA5:cfr | For expression of cfr in S. aureus | This study |
| pLL39:vga(A) | For integration of vga(A) onto S. aureus chromosome under control of cap1a promoter | This study |
| pIVEX2.3d:vga(A)E105Q | For expression of ATPase-deficient mutant of Vga(A) with C-terminal 6×His tag in E. coli | This study |
| pIVEX2.3d:vga(A)K219T | For expression of expanded-phenotype mutant of Vga(A) with C-terminal 6×His tag in E. coli | This study |
| pET28a:SUMO-lsa(A) | Modified pET28a expression vector encoding N-terminal 6×His and SUMO (type 3) tags followed by recognition site for U1p protease, used to express Lsa(A) | This study |
Antibiotics, chemicals, and susceptibility testing.
Antibiotics were from Sigma (Poole, United Kingdom), with the exception of tylosin (Cambridge Bioscience, Cambridge, United Kingdom), spiramycin (Cambridge Bioscience), and sparsomycin (E. Cundliffe, University of Leicester). Tritium (3H)-labeled lincomycin was synthesized by Quotient Bio Research (Nottingham, United Kingdom).
MICs were determined by exposing bacteria to 2-fold serial dilutions of antibacterial agents in Mueller-Hinton broth 2 (Oxoid, Cambridge, United Kingdom) according to the guidelines provided by the CLSI (36). For MIC determinations using strains carrying antibiotic resistance genes on expression plasmid pEPSA5, 1% (wt/vol) xylose was added to cultures following their dilution to a 0.5 McFarland standard to induce expression from the pT5X promoter.
DNA manipulation.
The oligonucleotide primers used for PCR are listed in Table S3 in the supplemental material.
For expression in S. aureus, PCR amplicons corresponding to vga(A) and lsa(A), and a synthesized DNA fragment corresponding to cfr (GenScript, Piscataway, NJ), were ligated into the staphylococcal expression vector pEPSA5 (37) and introduced into S. aureus RN4220 by electroporation (38). In order to assess the activity of Vga(A) in the presence of the cfr determinant, a strain of RN4220 was generated carrying vga(A) on the chromosome and cfr on plasmid pSEPSA5:cfr. To achieve this, a PCR amplicon of vga(A) downstream of the strong, constitutive promoter Pcap1A (39) was ligated into the integrative vector pLL39 (40) and introduced into S. aureus RN4220(pLL2787) (40) by electroporation, whereupon it became integrated into the chromosome at the ϕ11 attB site. A construct for overexpression of Vga(A) with a C-terminal 6×His tag in E. coli was generated as previously described (21). The lsa(A) gene was amplified from Enterococcus faecalis ATCC 29212 (NCBI WP_002365053.1) and ligated into a modified pET28b plasmid (Merck KGaA, Darmstadt, Germany) encoding N-terminal 6×His and SUMO tags (41). The resulting expression constructs were transformed into E. coli BL21(λDE3) Gold (Agilent Technologies, Cheshire, United Kingdom) and BL21-CodonPlus(λDE3) RIL (Agilent Technologies), respectively. Nucleotide replacements to encode amino acid substitutions E105Q and K219T in Vga(A) were independently engineered into constructs carrying vga(A) using the QuikChange Lightning kit (Agilent Technologies).
Heterologous expression and purification of proteins.
Vga(A) and Lsa(A) were overexpressed in E. coli by autoinduction (42) at 25°C for 4 days, and cells were harvested by centrifugation. Cell pellets were resuspended at 3 ml/g in buffer A (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole), incubated with 7,000 U chicken egg-white lysozyme (Sigma), complete EDTA-free protease inhibitor tablets (Roche, Mannheim, Germany), and 17 U Basemuncher endonuclease/ml of suspension (Expedeon, Harston, United Kingdom) for 30 min, and lysed by sonication. Lysates were clarified by centrifugation at 30,000 × g for 30 min. Cleared lysate was loaded onto a 25-ml free-flow gravity column (GeneFlow, Elmhurst, United Kingdom) packed with 5 ml Ni-nitrilotriacetic acid (NTA) agarose (Expedeon), washed with buffer A containing 20 mM imidazole and 2 M NaCl, eluted in buffer A containing 250 mM imidazole and 2 M NaCl, and dialyzed overnight into buffer B (20 mM HEPES [pH 7.5], 2 M NaCl, 1 mM dithiothreitol [DTT]). The C-terminal 6×His tag of Vga(A) was not removed, but the N-terminal 6×His and SUMO tags were removed from Lsa(A) by digestion with SUMO protease (Ulp, Thermo, Fisher Scientific, MA) during dialysis. Dialyzed protein was subjected to gel filtration using a Superdex 200 (16/60) column (GE Healthcare, Buckinghamshire, United Kingdom) preequilibrated with buffer B. Lsa(A) was then exchanged into buffer C (50 mM HEPES [pH 7.4]) containing 300 mM NaCl on HiTrap desalting columns (GE Healthcare) and stored at −80°C. Vga(A) was exchanged into buffer C with 50 mM NaCl and further purified using a resource S column by elution with a 500 mM NaCl gradient. Finally, Vga(A) was also exchanged to buffer C containing 300 mM NaCl prior to storage at −80°C. SDS-PAGE and peptide mass fingerprinting were used to detect and confirm the identity of purified proteins, with the latter performed by the University of Leeds Mass Spectrometry Facility. The FusB protein was expressed and purified as described previously (43).
In vitro transcription/translation assays.
Staphylococcal S30 extract was prepared from S. aureus RN4220 following the protocol of Murray et al. (44), although the preincubation step was omitted. E. coli S30 extract was from Promega (Madison, WI). For T/T assays, an optimized quantity of S30 extract was added into a 25-µl reaction mixture containing 0.1 mM amino acids (Promega), 10 µl S30 premix (Promega), 1 µg of DNA template (pSAluc or pBESTluc for S. aureus and E. coli T/T reactions, respectively), and antibiotics and purified protein as required. Reaction mixtures were incubated for 1 h at 37°C, and the level of transcription/translation was quantified by monitoring the expression of luciferase produced from pSAluc/pBESTluc by the addition of luciferase assay reagent (Promega) and measurement of luminescence.
Purification of staphylococcal ribosomes and use in antibiotic binding assays.
Ribosomes were purified from S30 fractions of S. aureus RN4220 using l-cysteine Sulfolink resin (Thermo Fisher Scientific, Waltham, MA) and ultracentrifugation, as previously described (45). For quantitation, ribosomes were separated on a denaturing agarose gel alongside an RNA standard of known concentration and analyzed by two-dimensional (2D) densitometry using AIDA software (Raytest, Straubenhardt, Germany).
The ability of Lsa(A) to prevent binding of radiolabeled lincomycin to the ribosome was assessed essentially as previously described for tetracycline-ribosome binding studies (46). Ribosomes (500 nM) were preincubated in 50-µl reaction mixtures with Lsa(A), BSA, or unlabeled lincomycin (concentrations described in Results) in assay buffer (10 mM Tris [pH 7.5], 60 mM KCl, 10 mM NH4Cl, 300 mM NaCl, 6 mM MgCl2, 0.1 mM ATP) at 37°C. After 10 min, 1 µM [3H]lincomycin was added, and the reaction mixtures were incubated for a further 10 min before recovering ribosomes on 0.45-µm-pore nitrocellulose filters and two rounds of washing with 200 µl of ice-cold wash buffer (50 mM Tris, 50 mM KCl, 20 mM MgOAc) to remove unbound [3H]lincomycin. Ribosome-associated [3H]lincomycin was then quantified by scintillation counting.
The ability of Lsa(A) to displace prebound lincomycin from ribosomes was investigated using the same assay conditions; however, ribosomes were preincubated with [3H]lincomycin for 10 min at 37°C prior to the addition of Lsa(A), BSA, or unlabeled lincomycin. Reaction mixtures were then incubated for a further 10 min at 37°C, before proceeding to ribosome recovery, washing, and quantitation of ribosome-bound [3H]lincomycin as described above.
SUPPLEMENTAL MATERIAL
MICs of 50S-targeted antibiotics against S. aureus expressing or lacking Vga(A).
MICs of 50S-targeted antibiotics against S. aureus RN4220 expressing chromosomally encoded Vga(A) alone, plasmid-encoded Cfr alone, or both together.
Oligonucleotide primers used in this study. Restriction sites and sequences complementary to the pLL39 vector used for Gibson assembly are italicized, codons targeted for mutagenesis are underlined, and expression signals (promoters and ribosome binding sites) are shown in boldface.
ACKNOWLEDGMENTS
We thank E. Cundliffe (University of Leicester, United Kingdom) for the generous gift of sparsomycin, and C. Y. Lee (University of Arkansas for Medical Sciences) for providing plasmids pLL39 and pLL2787. We are grateful to the following past and present colleagues at the University of Leeds for their advice, assistance, and input into this work: J. Clarke, J. Cove, G. Cox, A. Gupta, and N. Ooi.
Funding Statement
This work was funded by BBSRC-DTG studentship BB/F016603/1 awarded to the University of Leeds. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Sharkey LKR, Edwards TA, O’Neill AJ. 2016. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 7(2):e01975-15. doi:10.1128/mBio.01975-15.
REFERENCES
- 1.WHO 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Review on Antimicrobial Resistance 2014. Antimicrobial resistance: tackling a crisis for the future health and wealth of nations. Review on Antimicrobial Resistance, London, United Kingdom: http://amr-review.org/Publications. [Google Scholar]
- 3.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. 2015. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol 13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 4.Kerr ID. 2004. Sequence analysis of twin ATP binding cassette proteins involved in translational control, antibiotic resistance, and ribonuclease L inhibition. Biochem Biophys Res Commun 315:166–173. doi: 10.1016/j.bbrc.2004.01.044. [DOI] [PubMed] [Google Scholar]
- 5.Davidson AL, Dassa E, Orelle C, Chen J. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorrian JM, Kerr ID. 2009. Can ABC proteins confer drug resistance in microorganisms without being export pumps? p 63–77. In Ponte-Sucre A (ed), ABC transporters in microorganisms: research, innovation and value as targets against drug resistance. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 7.Reynolds ED, Cove JH. 2005. Resistance to telithromycin is conferred by msr(A), msrC and msr(D) in Staphylococcus aureus. J Antimicrob Chemother 56:1179–1180. doi: 10.1093/jac/dki378. [DOI] [PubMed] [Google Scholar]
- 8.Novotna G, Janata J. 2006. A new evolutionary variant of the streptogramin A resistance protein, Vga(A)(LC), from Staphylococcus haemolyticus with shifted substrate specificity towards lincosamides. Antimicrob Agents Chemother 50:4070–4076. doi: 10.1128/AAC.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh KV, Weinstock GM, Murray BE. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 46:1845–1850. doi: 10.1128/AAC.46.6.1845-1850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross JI, Eady EA, Cove JH, Cunliffe WJ, Baumberg S, Wootton JC. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol Microbiol 4:1207–1214. doi: 10.1111/j.1365-2958.1990.tb00696.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, Zhang R, Li J, Zhao Q, He T, Wang D, Wang Z, Shen Y, Li Y, Feßler AT, Wu C, Yu H, Deng X, Xia X, Shen J. 2015. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother 70:2182–2190. doi: 10.1093/jac/dkv116. [DOI] [PubMed] [Google Scholar]
- 12.Gentry DR, McCloskey L, Gwynn MN, Rittenhouse SF, Scangarella N, Shawar R, Holmes DJ. 2008. Genetic characterization of Vga ABC proteins conferring reduced susceptibility to pleuromutilins in Staphylococcus aureus. Antimicrob Agents Chemother 52:4507–4509. doi: 10.1128/AAC.00915-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allignet J, Loncle V, el Sohl N. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45–51. doi: 10.1016/0378-1119(92)90488-B. [DOI] [PubMed] [Google Scholar]
- 14.Tesse S, Trueba F, Berthet N, Hot C, Chesneau O. 2013. Resistance genes underlying the LSA phenotype of French staphylococcal isolates. Antimicrob Agents Chemother 57:4543–4546. doi: 10.1128/AAC.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hot C, Berthet N, Chesneau O. 2014. Characterization of sal(A), a novel gene responsible for lincosamide and streptogramin A resistance in Staphylococcus sciuri. Antimicrob Agents Chemother 58:3335–3341. doi: 10.1128/AAC.02797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynolds E, Ross JI, Cove JH. 2003. Msr(A) and related macrolide/streptogramin resistance determinants: incomplete transporters? Int J Antimicrob Agents 22:228–236. doi: 10.1016/S0924-8579(03)00218-8. [DOI] [PubMed] [Google Scholar]
- 17.Kerr ID, Reynolds ED, Cove JH. 2005. ABC proteins and antibiotic drug resistance: is it all about transport? Biochem Soc Trans 33:1000–1002. doi: 10.1042/BST20051000. [DOI] [PubMed] [Google Scholar]
- 18.Chesneau O, Ligeret H, Hosan-Aghaie N, Morvan A, Dassa E. 2005. Molecular analysis of resistance to streptogramin A compounds conferred by the Vga proteins of staphylococci. Antimicrob Agents Chemother 49:973–980. doi: 10.1128/AAC.49.3.973-980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunez-Samudio V, Chesneau O. 2013. Functional interplay between the ATP binding cassette Msr(D) protein and the membrane facilitator superfamily Mef(E) transporter for macrolide resistance in Escherichia coli. Res Microbiol 164:226–235. doi: 10.1016/j.resmic.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Lenart J, Vimberg V, Vesela L, Janata J, Balikova Novotna G. 2015. Detailed mutational analysis of Vga(A) interdomain linker: implication for antibiotic resistance specificity and mechanism. Antimicrob Agents Chemother 59:1360–1364. doi: 10.1128/AAC.04468-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacquet E, Girard J-M, Ramaen O, Pamlard O, Lévaique H, Betton J-M, Dassa E, Chesneau O. 2008. ATP hydrolysis and pristinamycin IIA inhibition of the Staphylococcus aureus Vga(A), a dual ABC protein involved in streptogramin A resistance. J Biol Chem 283:25332–25339. doi: 10.1074/jbc.M800418200. [DOI] [PubMed] [Google Scholar]
- 22.Matsuoka M, Jánosi L, Endou K, Nakajima Y. 1999. Cloning and sequences of inducible and constitutive macrolide resistance genes in Staphylococcus aureus that correspond to an ABC transporter. FEMS Microbiol Lett 181:91–100. doi: 10.1111/j.1574-6968.1999.tb08830.x. [DOI] [PubMed] [Google Scholar]
- 23.Olano C, Rodríguez AM, Méndez C, Salas JA. 1995. A second ABC transporter is involved in oleandomycin resistance and its secretion by Streptomyces antibioticus. Mol Microbiol 16:333–343. doi: 10.1111/j.1365-2958.1995.tb02305.x. [DOI] [PubMed] [Google Scholar]
- 24.Barre J, Fournet MP, Zini R, Deforges L, Duval J, Tillement JP. 1986. In vitro [3H]-erythromycin binding to Staphylococcus aureus. Biochem Pharmacol 35:1001–1004. doi: 10.1016/0006-2952(86)90090-0. [DOI] [PubMed] [Google Scholar]
- 25.Fredrick K, Ibba M. 2014. The ABCs of the ribosome. Nat Struct Mol Biol 21:115–116. doi: 10.1038/nsmb.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Giambattista M, Engelborghs Y, Nyssen E, Cocito C. 1987. Kinetics of binding of macrolides, lincosamides, and synergimycins to ribosomes. J Biol Chem 262:8591–8597. [PubMed] [Google Scholar]
- 27.Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother 45:763–770. doi: 10.1093/jac/45.6.763. [DOI] [PubMed] [Google Scholar]
- 28.O’Neill AJ, Chopra I. 2006. Molecular basis of fusB-mediated resistance to fusidic acid in Staphylococcus aureus. Mol Microbiol 59:664–676. doi: 10.1111/j.1365-2958.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 29.Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arenz S, Nguyen F, Beckmann R, Wilson DN. 2015. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-A resolution. Proc Natl Acad Sci U S A 112:5401–5406. doi: 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W, Atkinson GC, Thakor NS, Allas U, Lu CC, Chan KY, Tenson T, Schulten K, Wilson KS, Hauryliuk V, Frank J. 2013. Mechanism of tetracycline resistance by ribosomal protection protein Tet(O). Nat Commun 4:1477. doi: 10.1038/ncomms2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boël G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, Gonzalez RL Jr, Hunt JF. 2014. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol 21:143–151. doi: 10.1038/nsmb.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen B, Boël G, Hashem Y, Ning W, Fei J, Wang C, Gonzalez RL Jr, Hunt JF, Frank J. 2014. EttA regulates translation by binding the ribosomal E site and restricting ribosome-tRNA dynamics. Nat Struct Mol Biol 21:152–159. doi: 10.1038/nsmb.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dassa E, Bouige P. 2001. The ABC of ABCS: a phylogenetic and functional classification of ABC systems in living organisms. Res Microbiol 152:211–229. doi: 10.1016/S0923-2508(01)01194-9. [DOI] [PubMed] [Google Scholar]
- 35.Dassa E. 2011. Natural history of ABC systems: not only transporters. Essays Biochem 50:19–42. doi: 10.1042/bse0500019. [DOI] [PubMed] [Google Scholar]
- 36.CLSI 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard M07-A9, 9th ed. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, Kedar GC, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Zy ZY, Carr G, Mosca DA. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 38.Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. doi: 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang S, Lee CY. 1997. Transcriptional analysis of type 1 capsule genes in Staphylococcus aureus. Mol Microbiol 23:473–482. doi: 10.1046/j.1365-2958.1997.d01-1865.x. [DOI] [PubMed] [Google Scholar]
- 40.Luong TT, Lee CY. 2007. Improved single-copy integration vectors for Staphylococcus aureus. J Microbiol Methods 70:186–190. doi: 10.1016/j.mimet.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ariza A, Tanner SJ, Walter CT, Dent KC, Shepherd DA, Wu W, Matthews SV, Hiscox JA, Green TJ, Luo M, Elliott RM, Fooks AR, Ashcroft AE, Stonehouse NJ, Ranson NA, Barr JN, Edwards TA. 2013. Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization. Nucleic Acids Res 41:5912–5926. doi: 10.1093/nar/gkt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Cox G, Thompson GS, Jenkins HT, Peske F, Savelsbergh A, Rodnina MV, Wintermeyer W, Homans SW, Edwards TA, O’Neill AJ. 2012. Ribosome clearance by FusB-type proteins mediates resistance to the antibiotic fusidic acid. Proc Natl Acad Sci U S A 109:2102–2107. doi: 10.1073/pnas.1117275109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray RW, Melchior EP, Hagadorn JC, Marotti KR. 2001. Staphylococcus aureus cell extract transcription-translation assay: firefly luciferase reporter system for evaluating protein translation inhibitors. Antimicrob Agents Chemother 45:1900–1904. doi: 10.1128/AAC.45.6.1900-1904.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maguire BA, Wondrack LM, Contillo LG, Xu Z. 2008. A novel chromatography system to isolate active ribosomes from pathogenic bacteria. RNA 14:188–195. doi: 10.1261/rna.692408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burdett V. 1996. Tet(M)-promoted release of tetracycline from ribosomes is GTP dependent. J Bacteriol 178:3246–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim EB, Kopit LM, Harris LJ, Marco ML. 2012. Draft genome sequence of the quality control strain Enterococcus faecalis ATCC 29212. J Bacteriol 194:6006–6007. doi: 10.1128/JB.01423-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fairweather N, Kennedy S, Foster TJ, Kehoe M, Dougan G. 1983. Expression of a cloned Staphylococcus aureus alpha-hemolysin determinant in Bacillus subtilis and Staphylococcus aureus. Infect Immun 41:1112–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogé J, Betton JM. 2005. Use of pIVEX plasmids for protein overproduction in Escherichia coli. Microb Cell Fact 4:18. doi: 10.1186/1475-2859-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MICs of 50S-targeted antibiotics against S. aureus expressing or lacking Vga(A).
MICs of 50S-targeted antibiotics against S. aureus RN4220 expressing chromosomally encoded Vga(A) alone, plasmid-encoded Cfr alone, or both together.
Oligonucleotide primers used in this study. Restriction sites and sequences complementary to the pLL39 vector used for Gibson assembly are italicized, codons targeted for mutagenesis are underlined, and expression signals (promoters and ribosome binding sites) are shown in boldface.