ABSTRACT
We describe a novel symbiotic association between a kinetoplastid protist, Novymonas esmeraldas gen. nov., sp. nov., and an intracytoplasmic bacterium, “Candidatus Pandoraea novymonadis” sp. nov., discovered as a result of a broad-scale survey of insect trypanosomatid biodiversity in Ecuador. We characterize this association by describing the morphology of both organisms, as well as their interactions, and by establishing their phylogenetic affinities. Importantly, neither partner is closely related to other known organisms previously implicated in eukaryote-bacterial symbiosis. This symbiotic association seems to be relatively recent, as the host does not exert a stringent control over the number of bacteria harbored in its cytoplasm. We argue that this unique relationship may represent a suitable model for studying the initial stages of establishment of endosymbiosis between a single-cellular eukaryote and a prokaryote. Based on phylogenetic analyses, Novymonas could be considered a proxy for the insect-only ancestor of the dixenous genus Leishmania and shed light on the origin of the two-host life cycle within the subfamily Leishmaniinae.
IMPORTANCE
The parasitic trypanosomatid protist Novymonas esmeraldas gen. nov., sp. nov. entered into endosymbiosis with the bacterium “Ca. Pandoraea novymonadis” sp. nov. This novel and rather unstable interaction shows several signs of relatively recent establishment, qualifying it as a potentially unique transient stage in the increasingly complex range of eukaryotic-prokaryotic relationships.
INTRODUCTION
For at least 1.5 billion years, prokaryotes and eukaryotes coevolved, and they have established numerous symbiotic associations. Even the very origin of eukaryotes would most likely be impossible without the acquisition of an endosymbiotic bacterium which became the omnipresent mitochondria of extant eukaryotic cells (1). Similarly, the rise of algae was associated with the incorporation of a cyanobacterium that eventually transformed into the photosynthetic organelle—the plastid. At the later stages of eukaryogenesis, various primary, secondary, and even tertiary endosymbiotic events added additional levels of complexity to eukaryotic cells (2).
More recent intracellular associations involving prokaryotes are relatively widespread among eukaryotic taxa and have considerable impacts on the ecology, physiology, and metabolism of both participants. The particular effects of such endosymbiotic associations can vary depending on whether they evolved toward mutualism, parasitism, or a range of intermediate forms. In the case of mutualism, endosymbionts provide some advantages to the host, from which they receive a sheltered environment rich in nutrients. However, if they are parasites, host defense mechanisms have to be overcome, and only one partner benefits from such a relationship. The host either benefits from the expanded metabolic capabilities derived from the endosymbiont, potentially allowing it to colonize new ecological niches, or experiences stress due to destructive effects of unwanted dwellers and the necessity to feed them.
The studies of endosymbiotic prokaryotes were for a long time impeded by the failure to cultivate most of them. Indeed, it was proposed recently that the majority of bacteria are uncultivable, as in nature they are part of complex communities, members of which depend on mutually exchanged metabolites (3). The study of uncultured bacteria has been aided by the advent of genomics and bioinformatics. These approaches have allowed a closer look at interactions between the partners of symbiotic associations and have uncovered common trends in the evolution of endosymbiont genomes (4–6). Recently, it was shown that some bacterial endosymbionts have reached an extreme level of genome reduction that is compensated by the balancing effect of collaborative networks among them (7).
While the most intricate relationships seem to have evolved between different bacteria and sap-feeding insects (8), unicellular eukaryotes have also engaged in such associations, leading to profound changes in their lifestyle and ensuing important evolutionary and ecological implications. The best known examples include cyanobacteria in the cercozoan Paulinella chromatophora, nitrogen-fixing bacteria in parabasalids of termites, and methanogenic archaea in anaerobic ciliates and pelobiontids (9, 10). However, the findings of endosymbiotic bacteria in parasitic protists are frequently confined to mere descriptions, as is the case for cytoplasmic bacteria in the apicomplexan Gregarina garnhami (11), the dinoflagellate Hematodinium sp. (12), the heterokont Blastocystis sp. (13), or the ciliate Balantidium jocularum (14). More attention has been given to the recently discovered facultative symbiosis between two sexually transmitted agents, the flagellate Trichomonas vaginalis and the bacterium Mycoplasma hominis (15). This association is of medical importance, as it likely leads to a more severe disease manifestation (16).
The most extensively studied endosymbiont-containing protists belong to the family Trypanosomatidae (Euglenozoa, Kinetoplastea), a group of obligatory parasites found in a wide range of arthropods, vertebrates, and plants (17). The best-known representatives are dixenous species (i.e., with two alternating hosts in the life cycle) of the genera Trypanosoma and Leishmania that cause severe diseases in humans and domestic animals, whereas the widest segment of this group’s diversity is represented by monoxenous insect parasites (18, 19). Among those, members of the subfamily Strigomonadinae (genera Strigomonas, Angomonas, and Kentomonas) harbor obligatory symbiotic bacteria of the genus “Ca. Kinetoplastibacterium” (20–22). A common ancestor of this group acquired a betaproteobacterium of the family Alcaligenaceae (22, 23). The ensuing long-term coevolution led to significant changes in the morphology, metabolism, and physiology of both partners of the association. Thus, each trypanosomatid cell bears a single bacterial cell in its cytoplasm, which undergoes a synchronous division with the host cell and is vertically transmitted (24, 25). The endosymbionts lack the cell wall, presumably to ensure intense metabolic exchange with the host cell (26). In endosymbiont-containing trypanosomatids, the corset of subpellicular microtubules gets reorganized in comparison to those of other groups, possibly as a consequence of the extensive branching that is evident in the mitochondrion of those cells. The enlarged mitochondrion might be a consequence of an increased energy consumption by these flagellates compared to the energy consumption of their asymbiotic kin (20, 27). Another characteristic feature is a reduction of the paraflagellar rod (28). It was proposed that the close association of the bacterium with glycosomes in the host cell cytoplasm ensures provision with ATP from the trypanosomatid host (29), which also supplies its partner with phosphatidylcholine required for the endosymbiont’s envelope (30). In return, the bacterium provides enzymes for completing the metabolic pathways for biosynthesis of heme, vitamins, coenzymes, lipids, and essential amino acids within the host cell (31–34). Moreover, the endosymbiont also supplies its host with purines and boosts the production of polyamines, leading to accelerated host cell division (35–37). Trypanosomatids artificially deprived of bacteria can survive in culture, and yet, they are unable to colonize insect hosts (38), likely due to the altered expression of surface glycoconjugates and gp63-like protease (39, 40).
The endosymbiotic association described above was so far considered a singular event in the evolutionary history of trypanosomatids. However, in the course of a broad-scale survey of biodiversity in Ecuador (41), we have isolated and cultured a new species of trypanosomatid possessing intracytoplasmic bacteria. Neither the eukaryotic host nor the bacterial endosymbiont has close relatives involved in similar endosymbiotic consortia, thus confirming an independent origin of this novel association. Furthermore, the phylogenetic positions of both the trypanosomatid and the bacterial partner of this newly discovered endosymbiotic system suggest that their relationship has been established relatively recently. Here, we characterize this association by describing the morphology and phylogenetic affinities of both organisms, as well as details of their interactions and phylogenetic affinities. We argue that this symbiotic consortium represents a very good model for studying the initial stages of endosymbiosis between a bacterium and a protist.
RESULTS
Isolation, light microscopy, and cultivation.
A specimen of Niesthrea vincentii (Hemiptera: Rhopalidae) collected in July 2008 in the vicinity of Atacames (Esmeraldas Province, Ecuador) was found to be positive for trypanosomatids. The primary culture, labeled E262AT, was established and passaged in brain heart infusion (BHI) medium supplemented with hemin and antibiotics. Next, the trypanosomatids in the primary culture were compared to the corresponding original environmental isolate 262AT (41) by sequencing the spliced leader (SL) RNA gene from both sources. Their sequences exhibited 99% similarity (GenBank accession number KP717858), confirming the identity of the cultured isolate. The clonal axenic culture E262AT.01 was obtained using the limiting dilution method and shown to carry an 18S rRNA sequence (GenBank accession number KT944309) identical to that of the primary culture. Both primary and clonal cultures could be propagated in hemin-free BHI or M199 medium, with the cell division rates being similar regardless of the presence of hemin. Cultured cells could not withstand an elevated temperature (37°C) but, similar to Leishmania, grew faster in the medium with an acidic pH of 5.5 (data not shown).
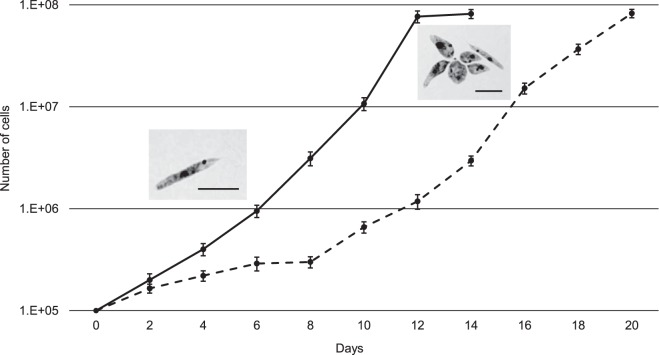
Light microscopic examination of E262AT.01 revealed the presence of three distinct morphotypes: promastigotes, choanomastigotes with various flagellum lengths, and rarely occurring The amastigotelike cells. Cell measurements are presented in Table S1 in the supplemental material. The proportions of individual morphotypes varied throughout cultivation. Promastigotes prevailed in the early- and mid-log-phase stages, while choanomastigotes dominated in the stationary phase. Cells were also observed forming multicellular rosettes firmly attached to the plastic surface of the cultivation flask (Fig. 1). Occasionally, those rosettes reached a few millimeters in size and contained thousands of cells arranged in multiple layers. Promastigotes divided significantly faster than choanomastigotes. When a stationary-phase culture, composed predominantly of choanomastigotes, was diluted to the same density as a promastigote-dominated culture, it took 14 days to reach the mid-log phase, whereas promastigotes achieved that level in just 8 days (Fig. 1). Alternatively, this lag can be explained by morphotype switching: only promastigotes can divide, and some time is needed for the choanomastigote-promastigote transformation. The addition of antibiotics into the culture medium showed that even at the highest concentrations tested (see Materials and Methods), elimination of the intracellular bacteria from trypanosomatids did not occur. However, under these conditions, the cells divided considerably more slowly and amassed conspicuously more bacteria than in the absence of antibiotics (data not shown). Importantly, no bacterium-free cells were observed under such growth conditions.
FIG 1 .
Growth curves of Novymonas esmeraldas (isolate E262AT.01). Prevailing morphotypes at the early/mid-log (promastigotes) and stationary (choanomastigotes in rosettes) stages are shown in insets. Scale bars are 10 µm. Stationary-phase (dotted line) and early-log-phase (solid line) cultures were diluted to the same initial cell density and compared side by side. Data are from three independent biological replicates. The error bars indicate standard deviations.
Detection of bacterial endosymbionts by FISH.
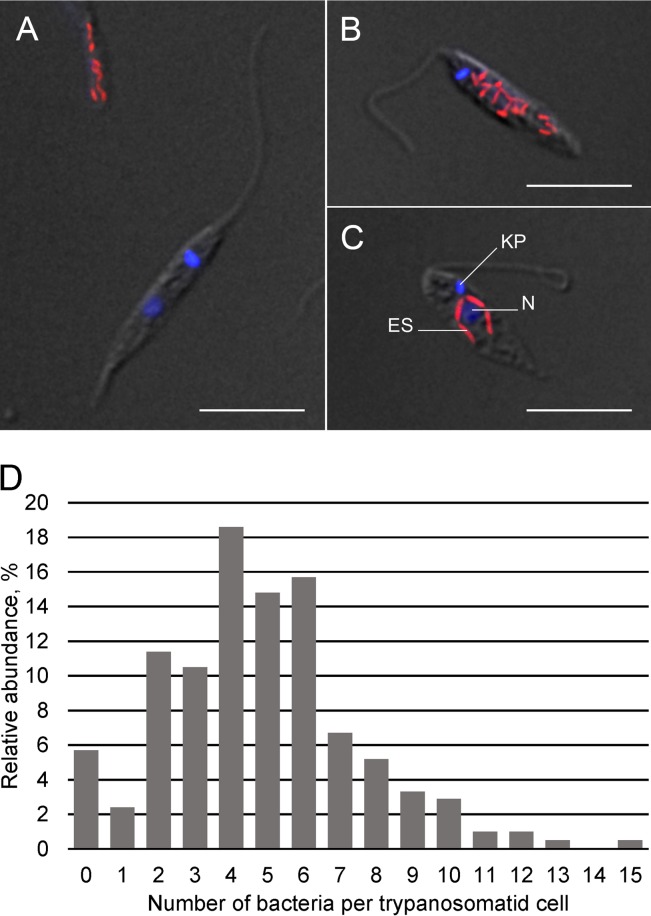
Giemsa and 4′,6-diamidino-2-phenylindole (DAPI) staining allowed the detection of rod-shaped structures in most E262AT.01 cells. Similar bodies were previously observed in members of the subfamily Strigomonadinae and identified as bacterial endosymbionts (20). In order to confirm the nature of the Giemsa- and DAPI-positive structures, we employed fluorescent in situ hybridization (FISH) using probe Eub338, which recognizes bacterial 16S rRNA (42). An absolute majority of trypanosomatid cells was positive, pointing to their identification as bacterial endosymbionts (Fig. 2A to C). Strikingly, and in contrast to the representatives of the Strigomonadinae flagellates studied so far (24), in the E262AT.01 culture, the number of endosymbionts per host cell varied drastically. It ranged from 0 to 15, with about 6% of the cells lacking an endosymbiont (Fig. 2D). In about 70% of the flagellates, two to six bacteria were usually randomly distributed throughout the cytoplasm (Fig. 2A to C). In some cases, especially in cells with low numbers of bacteria, the latter tended to be located in the vicinity of the nucleus (Fig. 2C). With increasing numbers of bacteria per cell, the proportions of such hosts declined steeply (Fig. 2D).
FIG 2 .
Bacterial endosymbionts in Novymonas esmeraldas (isolate E262AT.01) cells were detected by FISH. (A to C) DAPI- and Eub338-probed cells were analyzed by differential interference contrast (DIC) combined with fluorescence microscopy. Scale bars are 10 µm. (D) Relative abundances of the endosymbionts in host cells (n = 210). ES, endosymbiont; KP, kinetoplast; N, nucleus.
Isolation of axenic bacterial culture.
Upon lysis of the host cells, the bacterial endosymbionts released could be cultured on Trypticase soy agar and propagated in liquid BHI without supplements. The identity of the isolated bacterial culture was confirmed by 16S rRNA gene sequencing (see below). The growth of the bacterial culture was halted by ampicillin and kanamycin at 100 µg/ml and by chloramphenicol at 64 µg/ml (data not shown).
Subcloning of E262AT.01.
In order to determine whether it is possible to obtain an endosymbiont-free culture of the E262AT.01, we performed several experiments with cloning by limiting dilution in different media independently in the laboratories in Ostrava, Prague, and České Budějovice (see Table S2 in the supplemental material). None of the 41 subclones obtained, which were analyzed in detail, was free of bacteria. Similarly to the original E262AT.01 culture, a fraction of cells lacked bacteria. Different clones exhibited different rates of cell division, but there was no correlation between their growth and the number of endosymbiotic bacteria per host cell.
Phylogenetic analyses.
Since individual phylogenetic trees for small subunit (18S) and large subunit (28S) rRNAs and heat shock protein 83 (Hsp83) genes were highly congruent, a concatenated alignment was made to infer the phylogenetic position of the new trypanosomatid. Both maximum-likelihood and Bayesian analyses showed the same topology, positioning it within the subfamily Leishmaniinae (Fig. 3). In fact, the isolate E262AT.01 (hereinafter called Novymonas esmeraldas gen. nov., sp. nov.; GenBank accession numbers KT944293, KT944303, and KT944309 for Hsp83, 28S rRNA, and 18S rRNA, respectively) proved to be the closest known relative of the genus Leishmania. The phylogenetic position was supported by the highest posterior probability and only a moderate bootstrap percentage. The latter was higher when different subsets of taxa were tested; in particular, the removal of Endotrypanum monterogeii led to an increase of this value to 85 (data not shown).
FIG 3 .
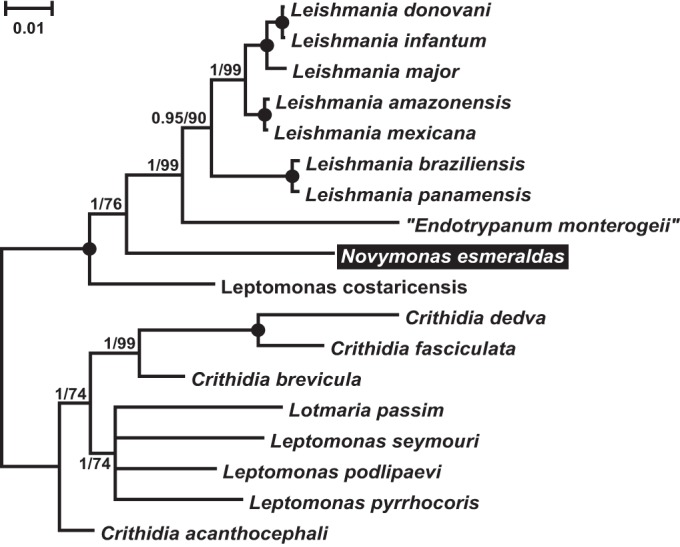
Phylogenetic tree for the trypanosomatid isolate studied in this work, inferred by the maximum-likelihood method using an 18S SSU rRNA, 28S LSU-α rRNA, and Hsp83 gene concatenated sequence set. Numbers at nodes indicate the posterior probability/bootstrap percentage. Nodes having 1.0 posterior probability and 100% bootstrap support are marked with black circles. The bar represents the number of substitutions per site. The name “Endotrypanum monterogeii” is enclosed in quotation marks since it is considered to be a misidentified member of the genus Leishmania. It is traditionally used for the corresponding culture. The species under study is highlighted.
Analysis of the SL RNA gene sequences (GenBank accession numbers KT944298 and KT944299) revealed that the new species belongs to typing unit 42 (TU42) (19, 41). It also includes environmental DNA isolates 104SI and CAR-B7 (GenBank accession number DQ864304), isolated in Ecuador and Central African Republic from the predatory true bug Zelus sp. (Heteroptera) and the biting midge Culicoides cf. fulvithorax (Diptera), respectively (41, 43). Moreover, the 18S rRNA gene sequences of isolates CAR-B7, E262AT, and GAB3 (the latter collected from Culicoides cf. distinctipennis in Gabon) were identical (GenBank accession numbers KT944308 and KT944309).
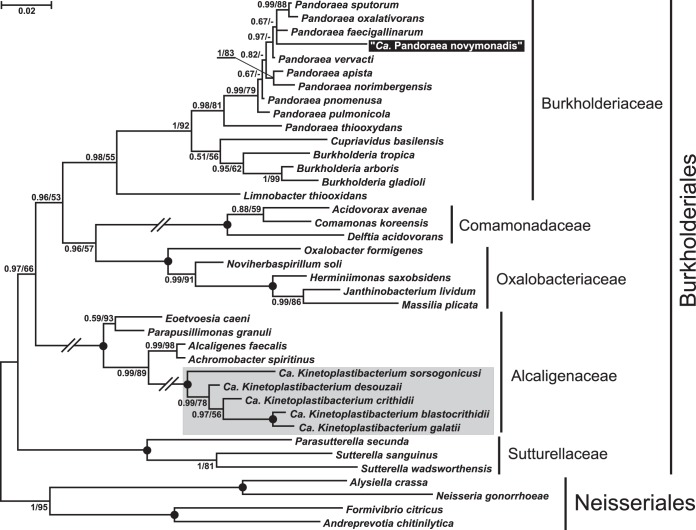
Bayesian and maximum-likelihood trees of the bacterial sequences were mostly consistent, with just minor differences in the branching order of clades with low bootstrap support (Fig. 4). The trypanosomatid endosymbiont analyzed herein (referred to as “Candidatus Pandoraea novymonadis”) is located at the very crown of the genus Pandoraea Coenye et al. 2000, being part of the family Burkholderiaceae (order Burkholderiales, class Betaproteobacteria). The affiliation to this genus was supported by high posterior probability and notably high bootstrap values. The exact position of the bacterium within this taxon could not be determined, as the phylogenetic relationships were poorly resolved. Its 16S rRNA gene sequence (GenBank accession number KT944310) differed by 3.3 to 4.8% from those of other Pandoraea spp. The branch that it formed on the phylogenetic tree proved to be much longer than those of the previously described members of this genus. Meanwhile, other known endosymbiotic bacteria of trypanosomatids (“Candidatus Kinetoplastibacterium” spp., family Alcaligenaceae) were only distantly related (Fig. 4).
FIG 4 .
16S rRNA-based Bayesian phylogenetic tree of bacterial endosymbionts of trypanosomatids. Names of species for sequences retrieved from GenBank are indicated. The species newly described in this work is highlighted. “Candidatus Kinetoplastibacterium” spp. (family Alcaligenaceae) are boxed and shaded. Bayesian posterior probabilities and bootstrap percentages for maximum likelihood analysis are shown at the nodes. Slashed branches are at 50% of their original lengths. Dashes indicate bootstrap support below 50% or different maximum-likelihood topology. Black dots represent 100% bootstrap support and Bayesian posterior probability of 1.0. The tree was rooted with sequences of four species of the order Neisseriales. The scale bar denotes the number of substitutions per site.
Ultrastructural characterization of the trypanosomatid-bacteria association.
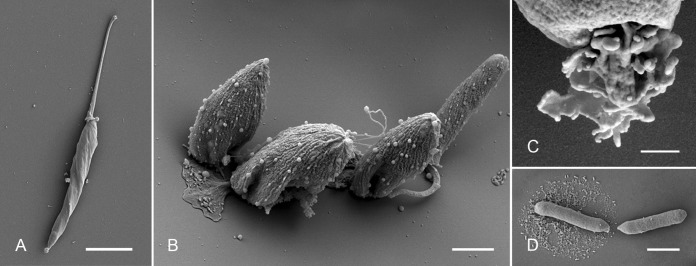
The Novymonas cells were further analyzed by scanning electron microscopy (SEM) and high-pressure freezing followed by transmission electron microscopy (HPF-TEM) (Fig. 5 and 6). SEM analysis confirmed the presence of all of the main morphotypes (promastigotes, choanomastigotes, and amastigote-like cells) identified by light microscopy (Fig. 5A and B; also data not shown). Upon prolonged cultivation, the prevailing choanomastigotes were found firmly attached to the plastic surface (Fig. 5B). This attachment was mediated by a modified flagellum, which was shortened and widened, forming an attachment pad, along with a gluelike substance cementing cells onto the plastic (Fig. 5B). On the occasional detached cells, we observed that this modified flagellum had a multilobe structure (Fig. 5C). Interestingly, some promastigotes were also found to be attached (Fig. 5B); although in this case, the flagellum was not modified, the gluelike substance was present (data not shown). Examination of the axenic culture of endosymbiotic “Candidatus Pandoraea novymonadis” by SEM documented uniformly sausage-shaped bacilli (Fig. 5D), which measured 0.4 to 0.7 µm in diameter and 1.5 to 3.0 µm in length (N = 50).
FIG 5 .
Scanning electron microscopy (SEM) images of Novymonas esmeraldas sp. nov. and “Candidatus Pandoraea novymonadis” sp. nov. (A) Free-swimming promastigote; (B) sessile forms (both pro- and choanomastigote-shaped, with or without modified flagellum); (C) prominently modified flagellum of a sessile choanomastigote; (D) bacilli in the axenic culture of “Ca. Pandoraea novymonadis.” Scale bars are 5 µm (A), 2 µm (B), 400 nm (C), and 1 µm (D).
FIG 6 .
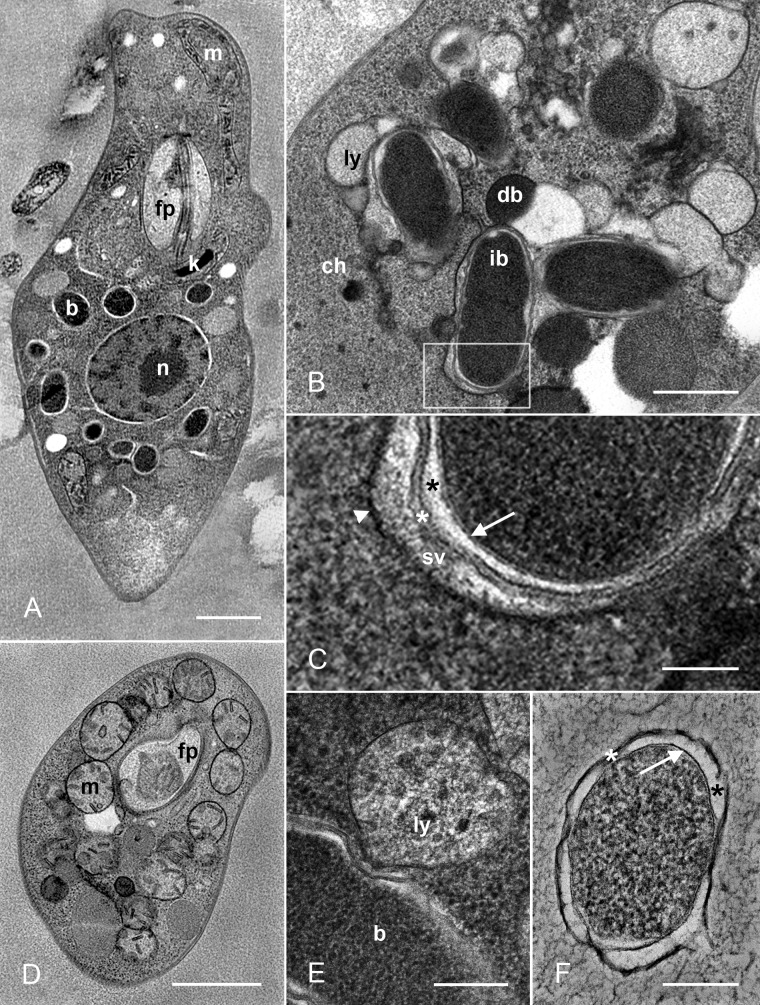
Transmission electron microscopy (TEM) images of Novymonas esmeraldas sp. nov. and “Candidatus Pandoraea novymonadis” sp. nov. (A) General view of Novymonas cell showing typical features of trypanosomatids such as the nucleus (n), kinetoplast (k), mitochondrion (m), and flagellar pocket (fp), as well as the bacterial symbionts (b). (B) Interaction between the bacteria and the trypanosomatid cell demonstrating fusion of lysosomes (ly) with bacterium-containing vacuoles in the cytoplasm of the host (ch). Intact and degrading bacteria are labeled ib and db, respectively. (C) Magnification of boxed part of panel B showing the membrane (arrowhead) of the symbiontophorous vacuole (sv), bacterial cell wall (white asterisk), periplasmic space (black asterisk), and internal membrane (arrow). (D) Cross section of Novymonas cell showing mitochondrial hypertrophy. (E) The early stage of the fusion between a bacterium and a lysosome. (F) Endosymbiotic bacillus in the axenic culture of “Ca. Pandoraea novymonadis” with the same structure of cell covering as is seen in panel C. Scale bars are 1 µm (A, D), 500 nm (B), 100 nm (C), and 200 nm (E, F).
HPF-TEM revealed typical morphological features of trypanosomatids: an oval nucleus located in the posterior half of the cell, an elongated kinetoplast disk positioned perpendicular to the basal body of the flagellum, and an extensively branched single mitochondrion. In addition, some distinctive traits were observed, such as the hypertrophied mitochondrion and multiple electron-dense bacteria within the cytoplasm (Fig. 6A to D), which were enclosed in the symbiontophorous vacuoles either individually (Fig. 6B and C) or in pairs, occasionally dividing (data not shown). Quite frequently, vacuoles containing bacteria were accompanied by lysosomes of the host cell (Fig. 6B). Several phases of interaction between the lysosomes, symbiontophorous vacuoles, and bacteria were observed. They ranged from early membrane contacts (Fig. 6E) to complete fusion of organelles and subsequent degradation of the bacterium (Fig. 6B). Intact endosymbionts had an envelope typical for the Gram-negative bacteria: an inner cytoplasmic membrane and a relatively thin cell wall with periplasmic space in between (Fig. 6C). The same structure was found in the free bacteria obtained from the axenic culture, although their periplasmic space was somewhat wider (Fig. 6F).
Taxonomic summary
Trypanosomatid host
Class Kinetoplastea (Honigberg 1963) Vickerman 1976
Subclass Metakinetoplastina Vickerman 2004
Order Trypanosomatida Kent 1880
Family Trypanosomatidae (Doflein 1901) Grobben 1905
Genus Novymonas gen. nov. Kostygov and Yurchenko 2015
Diagnosis: The genus is defined by a unique position on the 18S rRNA-28S rRNA-Hsp83-based phylogenetic tree(s) within the clade Leishmaniinae. It does not cluster within either the Leishmania clade or the Leptomonas-Lotmaria-Crithidia group. The main morphotypes are promastigotes and choanomastigotes.
Etymology: The generic name honors Frederick George Novy, an American bacteriologist and parasitologist who pioneered studies of insect trypanosomatids. He was the first to document structures (“diplosomes”) (44) that were later proved to be bacterial endosymbionts in Strigomonas culicis. The name also relates to the word nový (“new” in many Slavic languages), reflecting the novelty of the discovered trypanosomatid-bacterium association.
Novymonas esmeraldas sp. nov. Votýpka, Kostygov, Maslov, and Lukeš (Fig. 2 and 5)
Species diagnosis and description: The species is identified by its distinct phylogenetic position on the 18S rRNA and other gene trees, as well as by its unique SL RNA gene sequences. It forms promastigotes and choanomastigotes in culture, with free-swimming promastigotes and attached choanomastigotes in rosettes dominating in log and stationary phases, respectively. Cells in the culture range from 10.9 to 18 µm in length and from 1.3 to 4.8 µm in width. The length of the flagellum varies from 7.8 to 19.5 µm for elongated promastigotes. Spherical choanomastigotes are 4.5 to 9.7 µm long and 2.8 to 6.4 µm wide, with the flagellum ranging between 8.6 and 20.4 µm. The kinetoplast disk is compactly packed and varies between 553 and 938 nm in diameter and 114 to 213 nm in cross section (measured in HPF-TEM pictures). Cells can propagate at low pH but cannot withstand elevated temperature.
Type host: Niesthrea vincentii (Hemiptera: Rhopalidae).
Site: Intestine: hindgut. Only short choanomastigotelike cells have been observed in situ.
Type locality: Vicinity of Atacames (Esmeraldas Province, Ecuador, 00°52′31″S; 79°50′32″W).
Type material: The name-bearing type, a hapantotype, is a Giemsa-stained slide of the clonal isolate E262AT.01, deposited in the research collection of the Life Science Research Centre, Ostrava, Czech Republic (accession code 2015/E262AT.01/S). Axenic cultures of the primary (E262AT) and clonal (E262AT.01) isolates are deposited in the research collections of the Life Science Research Centre of the University of Ostrava, Department of Parasitology at Charles University, Prague, and Institute of Parasitology, České Budějovice, Czech Republic, and the Department of Biology, University of California at Riverside, United States.
Etymology: The species name (esmeraldas) is derived from the name of the province in Ecuador where the host of this parasite was collected.
Gene sequences: GenBank accession numbers KT944309 (18S rRNA), KT944303 (28S rRNA), KT944298, KT944299 (SL RNA), KT944300 (glycosomal glyceraldehyde-3-phosphate dehydrogenase [gGAPDH]), and KT944293 (Hsp83).
Remarks: Two environmental DNA isolates from biting midges—CAR-B7, collected in September 2012 from Culicoides cf. fulvithorax in Dzanga-Sangha Protected Areas, Central African Republic (2°13′N, 16°11′E), and GAB3, collected in June 2014 from Culicoides cf. distinctipennis in Loango National Park, Gabon (02º20′S, 09°35′E)—as well as the DNA isolate 104SI, sampled in March 2005 from the reduviid Zelus sp. in Casanga, Ecuador (00°35′S, 77°53′W), belong to the same species according to 18S rRNA and SL RNA gene sequences.
Bacterial endosymbiont
Class Betaproteobacteria Garrity et al. 2006
Order Burkholderiales Garrity et al. 2006
Family Burkholderiaceae Garrity et al. 2006
Genus Pandoraea Coenye et al. 2000
“Candidatus Pandoraea novymonadis” sp. nov. Kostygov, Grybchuk-Ieremenko, and Yurchenko 2015
Species diagnosis and description: Cells are Gram-negative, nonsporulating, rodlike in shape, measuring between 0.4 and 0.7 in length by 1.5 to 3.0 µm in width, fitting the genus description (45). They are cultivable axenically and motile. The species is identified by its unique position on the 16S rRNA-based phylogenetic tree.
Type host: Novymonas esmeraldas (Trypanosomatidae).
Type material: The name-bearing type, a hapantotype, is a Giemsa-stained slide of the axenic culture of “Ca. Pandoraea novymonadis,” deposited in the research collection of the Life Science Research Centre in Ostrava and the Institute of Parasitology, České Budějovice, Czech Republic (accession code 2015/E262AT.01/Pandoraea).
Etymology: The species name (novymonadis) refers to the specific trypanosomatid host.
Gene sequences: GenBank accession number KT944310 (16S rRNA).
DISCUSSION
In this work, we have characterized a new endosymbiont-bearing species of the family Trypanosomatidae. In contrast to the previously known bacterium-harboring flagellates of the subfamily Strigomonadinae, which constitute a separate clade (20), this monoxenous species is the closest known relative of the dixenous genus Leishmania and qualifies as a representative of the newly established genus Novymonas. Similarly to its relatives, it was isolated in the Neotropics, a region from which all the leishmanias might have radiated (46, 47). Therefore, Novymonas may share some preadaptations to dixeny with its sister group, although it is clearly incapable of withstanding elevated temperature, thus proving its monoxenous status. Novymonas could be considered a proxy for the monoxenous ancestor of Leishmania, and hence, scrutiny of its genetics and biochemistry might shed light on the origin of the two-host life cycle within the Leishmaniinae.
However, the new species is even more interesting since it harbors a bacterial endosymbiont in what appears to be an unstable relationship. The endosymbiotic bacterium of Novymonas belongs to the genus Pandoraea within the family Burkholderiaceae and, therefore, is only distantly related to the other known bacterial endosymbionts of trypanosomatids (“Ca. Kinetoplastibacterium” spp.) that belong to the family Alcaligenaceae (22). Consequently, it represents a separate lineage of intracellular symbionts and may have quite different adaptations to such a lifestyle. Indeed, it appears that the Novymonas-Pandoraea endosymbiosis was established relatively recently. The following features favor a late origin of this relationship: (i) the endosymbiont preserves its cell wall; (ii) in contrast to Strigomonadinae (24, 25), the division of “Ca. Pandoraea novymonadis” is not coordinated with the division of the host, resulting in various numbers of bacteria per cell and, thus, an overall instability of this association; (iii) the host employs lysosomes to exercise control over the bacteria; (iv) unlike “Ca. Kinetoplastibacterium” spp. (5, 48), the symbiont of Novymonas can be axenically cultivated; and finally (v), other known Pandoraea spp. are either emerging opportunistic human pathogens or free-living organisms.
Despite the eventual loss of the bacterial symbionts from a fraction of cells in culture, the endosymbiosis is likely to be obligatory for the trypanosomatid, which cannot be cultivated without Pandoraea. Moreover, cultivation under high concentrations of antibiotics leads to the elimination of the endosymbiont-free cells, demonstrating that the bacteria are indispensable under these conditions. It is reasonable to assume that Novymonas farms bacteria inside its cytoplasm, using them as a source of some essential nutrients. Hence, the aposymbiotic cells are ultimately doomed. Nevertheless, they emerge at a relatively high frequency in the culture, since mechanisms ensuring synchronization between the cohabitants and a proper segregation of bacteria during host cell division seem to be missing. Under high concentrations of antibiotics, cells divide more slowly and accumulate more endosymbionts. In clonal cultures originating from single cells with different numbers of endosymbionts, the whole spectrum of bacterial load can be observed. All our efforts to obtain an endosymbiont-free clonal culture of Novymonas failed, apparently due to the reduced viability of such cells. Counterintuitively, the addition of antibiotics at concentrations affecting trypanosomatids did not trigger the loss of bacteria, implying that the host cells actively protect their endosymbionts.
In the relatively closely related Strigomonadinae-“Ca. Kinetoplastibacterium” system, the division of the endosymbiont was proposed to depend on host factors, as the gene implicated in cell division has been lost from the “Ca. Kinetoplastibacterium” genome (37). Moreover, inhibition of protein synthesis in the protist host blocks the symbiont’s cytokinesis (49). Importantly, all other members of the genus Pandoraea are free living, and yet, many of them were isolated as opportunistic agents from cystic fibrosis patients (45, 50, 51). This suggests that representatives of this genus actively explore new evolutionary niches and adapt to host-associated lifestyles.
A similar situation was observed in the ciliate Euplotes aediculatus, harboring the bacterium Polynucleobacter necessarius, which is from the same family as Pandoraea (52). This symbiosis was demonstrated to be obligatory for both the ciliate and the bacterium (53). Nevertheless, free-living strains of presumably the same species, as judged by their 16S rRNA gene sequences, have also been discovered (54). However, the Euplotes-Polynucleobacter system is quite different from the Novymonas-Pandoraea association, since no lysosome-mediated digestion has been detected in the former partnership. Moreover, another interesting aspect of the association described herein is the fact that the eukaryotic partner is a parasite. The impact of endosymbiosis on a host-parasitic lifestyle is largely unknown. Parasites are usually supplied with essential nutrients by their hosts (55), and it is therefore counterintuitive that some of them may need an additional source. To understand why the Novymonas trypanosomatid entered into a lasting, although still unstable and rather unique partnership with Pandoraea, the whole genomes of both partners will have to be sequenced, and ideally, Novymonas should be modified into a genetically tractable organism. Both aims are now among our priorities, as we are convinced that this symbiotic relationship may serve as a model to study the evolution of early endosymbiosis in general and in parasitic protists in particular.
MATERIALS AND METHODS
Field work, establishment of primary cultures, cloning, and cultivation.
Niesthrea vincentii Westwood 1842 (Hemiptera: Rhopalidae) was collected in the vicinity of Atacames (Esmeraldas Province, Ecuador, 00º52′31″S; 79°50′32″W) in July 2008. The insects were dissected and examined under a light microscope as described previously (56). The primary isolate E262AT was cultivated in brain heart infusion (BHI) medium (Sigma-Aldrich, St. Louis, MO) supplemented with 10 µg/ml hemin (Jena Biosciences, Jena, Germany), pH 7.6, and antibiotics as reported previously (20, 57). The clonal isolate E262AT.01 was obtained using the limiting dilution method as described previously (58). The identity of the clonal line was confirmed by sequencing its 18S rRNA gene. The primary culture and clonal line thus obtained were deposited in the collections of the Department of Parasitology, Charles University, Prague, in the Life Science Research Centre of the University of Ostrava, and in the Institute of Parasitology, České Budějovice, Czech Republic. Of note, cells also grew well in BHI medium without hemin or in M199 medium supplemented with 10% fetal bovine serum (FBS) (both from Life Technologies, Grand Island, NY) and antibiotics as described above.
For growth curves, cells were seeded at a density of 1 × 105 cells per ml in BHI medium, pH 7.6, and incubated at 23°C or 37°C for 20 days with counting every other day.
In order to eliminate symbionts from cultured trypanosomatid cells, we tested a number of antibiotics at different concentrations. Culture were grown in the presence of either ampicillin or kanamycin at 100, 200, 400, and 800 µg/ml or chloramphenicol at 64, 128, and 256 µg/ml.
For the same reason, we also performed a large-scale experiment with subcloning by limiting dilution. The work was done independently in three laboratories (Ostrava, Prague, and České Budějovice). The following media were employed: (i) M199 with additives as described above, pH 7.4; (ii) M199 medium with additives as described above, pH 5.5; (iii) preconditioned supplemented M199 medium, pH 7.4; (iv) BHI-RPMI (1:1) medium with 10% FBS and 200 µg/ml amikacin; and (v) RPMI medium with 10% FBS. In total, we analyzed 11 96-well plates and obtained 41 subclones. Fifty to eighty cells of each subclone were examined for the presence of endosymbionts (see Table S2 in the supplemental material).
Isolation of axenic endosymbiont culture.
An amount of 5 × 107 cells was spun down and then lysed in 1 ml of distilled water for 3 days until no moving trypanosomatid cells could be observed under the light microscope. The suspension was divided into 100-µl aliquots, which were plated on Trypticase soy agar, LB agar (both from Sigma-Aldrich), or blood agar plates and incubated at 37°C. Colonies grown on Trypticase soy agar were propagated in liquid BHI without supplements, and experiments with antibiotics were performed as described above.
Light and electron microscopy.
Light microscopy of Giemsa or 4′,6-diamidino-2-phenylindole (DAPI)-stained smears on poly-l-lysine-coated slides was done as described elsewhere (46, 59) using an Olympus BX51 microscope equipped with a DP70 charge-coupled device (CCD) camera (Olympus, Tokyo, Japan). Standard measurements were performed for 50 cells of each morphotype on Giemsa-stained smears and expressed in micrometers (19). For scanning electron microscopy (SEM), cultured cells were fixed in 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) and processed as described previously (58, 60). Samples were observed using a JEOL JSM-7401-F microscope (JEOL, Tokyo, Japan) with an accelerating voltage of 4 kV. High-pressure freezing followed by transmission electron microscopy (HPF-TEM) was performed essentially as described elsewhere (61). Images were captured on a JEOL JEM-1010 microscope (JEOL) using a Mega View III camera (EMSIS GmbH, Münster, Germany). Kinetoplasts were measured after HPF-TEM as described previously (62).
FISH.
Bacterial endosymbionts were visualized by fluorescent in situ hybridization (FISH) using the bacterium-specific probe Eub338 (5′-GCTGCCTCCCGTAGGAGT-3′) labeled on the 5′ end with Cy3 fluorescent dye (63). E262AT.01 cells were fixed for 30 min in 4% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature and processed as described elsewhere (64, 65). Slides were mounted in ProLong gold antifade reagent with DAPI (Life Technologies) and observed with the Axioplan 2 fluorescence microscope (Carl Zeiss Microscopy GmbH, Jena, Germany), and images were captured using cellSens imaging software version 1.11 (Olympus Life Science, Tokyo, Japan). The numbers of endosymbiotic bacteria were counted for 210 randomly selected trypanosomatid cells.
DNA isolation, PCR amplification, cloning, and sequencing.
Total genomic DNA of the trypanosomatids or bacteria was isolated from the axenically grown culture (5 ml for trypanosomatids and 1 ml for bacteria) using the DNeasy blood & tissue kit (Qiagen, Hilden, Germany) or GenElute bacterial genomic DNA kit (Sigma-Aldrich), respectively, according to the manufacturers’ protocols. The 18S rRNA gene was amplified using primers S762 and S763 and sequenced directly, as described previously (66, 67). Hsp83 gene amplification was performed using the primers 100XF (5′-CAGCTGATGTCCCTGATCATYAAYACNTTYTA-3′) and 970XR (5′-TCGAGGGAGAGRCCNARCTTRATC-3′) as described elsewhere (68). The PCR products were sequenced directly with the amplification primers, as well as with two internal oligonucleotides, XF2 (5′-AAGAAGCGCAACAACATCAAGC-3′) and XR2 (5′-GCACAGRTCCTCRCAGTTGTCCA-3′). The LSU α-segment of the 28S rRNA gene was amplified using primers LSF (5′-ACAGACCTGAGTGTGGCAGGACTAC-3′) and LMR (5′-CCCACATGCAATTTCTTTTTGGA-3′) and sequenced with oligonucleotides LSIF (5′-CGAAAGGTGGTGAACTATGCCTGAACA-3′) and LSIR (5′-CCAGCTACTAGGTGGTTCGATGAGTC-3′). For amplification of the spliced leader (SL) RNA gene, primers M167 and M168 were used (69). The resulting PCR products were cloned using the InstTA PCR cloning kit (Thermo Fischer Scientific, Waltham, MA) and sequenced as described previously (57, 70). To amplify the complete 16S rRNA sequence of the bacterial endosymbiont, we used the primers P1seq and 1486R (23). The internal transcribed spacer (ITS) region between the 16S and 23S rRNA genes was amplified with primers P3Seq and P23sRev (22). The PCR products were sequenced directly. We also amplified and sequenced the glycosomal glyceraldehyde-3-phosphate dehydrogenase (gGAPDH) gene, which is widely used as a phylogenetic marker (62, 67, 71). However, we did not apply it to phylogenetic inference since it has been demonstrated to produce serious artefacts (20).
Phylogenetic analyses.
The 18S small subunit (SSU) rRNA, 28S LSU-α rRNA, and Hsp83 gene sequences of 18 species of trypanosomatids (including isolate E262AT.01) were aligned using Muscle version 3.8.3.1 (72). The resulting alignments were refined manually using BioEdit version 7.2.5 (73), and ambiguously aligned positions from 18S and 28S sequences were removed prior to concatenation using Gblocks software (74) as described previously (75). The resulting data set, containing 5,782 (2,125 + 1,749 + 1,908) positions, was used for phylogenetic inference under a partitioned model with maximum-likelihood criterion and a Bayesian approach in Treefinder version 03.2011 (http://www.treefinder.de) and MrBayes version 3.2.5 (76). Analysis in Treefinder was performed with the following parameters: the TN + G model for the 18S rRNA gene, GTR + G for the 28S rRNA gene, and J3 + GI, GTR + GI, and J3 + G, respectively, for the three codon positions of the Hsp83 gene (as selected by the built-in model selector of Treefinder using the Akaike information criterion); 5 gamma categories; and optimized substitution rates, nucleotide frequencies, and partition rates. Edge support was estimated by the bootstrap method with 1,000 replicates. Bayesian inference of phylogeny was accomplished with an analysis run for 5 million generations under the GTR + I + G substitution model (5 gamma categories) with the nucleotide frequencies, substitution rates, partition rates, and parameters of rate heterogeneity among sites unlinked for all 4 partitions (defined as described above). Other analysis parameters were left at their default states. Rooting of the tree obtained was done according to the previously published phylogenetic reconstructions that demonstrate subdivision of the subfamily Leishmaniinae into two clades (60, 77).
Reconstruction of the bacterial phylogeny was performed in a similar way, with a few alterations as specified below. Since the alignment of the 16S rRNA gene sequences was more accurate than the sequence alignment described above, no positions were removed from the alignment (except for end trimming). The data set contained 39 taxa and 1,446 nucleotide positions. The phylogenetic inference under the maximum-likelihood criterion was done in RAxML version 8.0 (78) under the GTR + G + I model as selected in jModelTest 2.1.4 (79). Edge reliability was estimated with 1,000 “slow” (as defined by the software) replicas of bootstrap resampling. No partitioning was applied in either maximum-likelihood or Bayesian analysis.
The accession numbers of the sequences used in all of the analyses described above are available from the authors upon request.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the new sequences determined in this work are KT944291 (Crithidia dedva Hsp83), KT944292 (Crithidia brevicula Hsp83), KT944293 (Novymonas esmeraldas Hsp83), KT944294 (Leptomonas costaricensis Hsp83), KT944295 (Leptomonas podlipaevi Hsp83), KT944296 (Leptomonas pyrrhocoris Hsp83), KT944297 (Leptomonas seymouri Hsp83), KT944298 and KT944299 (N. esmeraldas SL RNA), KT944300 (N. esmeraldas gGAPDH), KT944301 (C. brevicula 28S rRNA), KT944302 (C. dedva 28S rRNA), KT944303 (N. esmeraldas 28S rRNA), KT944304 (L. costaricensis 28S rRNA), KT944305 (L. podlipaevi 28S rRNA), KT944306 (L. pyrrhocoris 28S rRNA), KT944307 (L. seymouri 28S rRNA), KT944308 (Trypanosomatidae sp. CAR-B7 18S rRNA), KT944309 (N. esmeraldas 18S rRNA), and KT944310 (“Candidatus Pandoraea novymonadis” 16S rRNA).
SUPPLEMENTAL MATERIAL
Morphometric characteristics of different morphotypes of Novymonas esmeraldas. All measurements (µm) were done on 50 (promastigotes and choanomastigotes) or 20 (amastigotelike) cells.
Subcloning of Novymonas esmeraldas. Forty-one subclones were cultivated in different media (see Materials and Methods) and analyzed for the presence of endosymbionts.
ACKNOWLEDGMENTS
We thank all the members of our laboratories for helpful and stimulating discussions and Eva Nowack and Jorge Morales (Heinrich Heine Universität, Düsseldorf, Germany) for helpful advice and for providing us with the Eub338 FISH probe.
Funding Statement
This work, including the efforts of Julius Lukeš, was funded by Czech Science Foundation (14-23986S), COST action (CM 1307), and Seventh Framework Programme (FP7) (316304). This work, including the efforts of Vyacheslav Yurchenko and Alexei Y. Kostygov, was funded by Moravskoslezsky Kraj Research Initiative (DT1/RRC/2013-2014). This work, including the efforts of Vyacheslav Yurchenko, was funded by Moravskoslezsky Kraj Research Initiative (00955/RRC/2015). This work, including the efforts of Anastasiia Grybchuk-Ieremenko, was funded by Grant from University of Ostrava (SGS27/PrF/2015). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Kostygov A, Dobáková E, Grybchuk-Ieremenko A, Váhala D, Maslov DA, Votýpka J, Lukeš J, Yurchenko V. 2016. Novel trypanosomatid-bacterium association: evolution of endosymbiosis in action. mBio 7(2):e01985-15. doi:10.1128/mBio.01985-15.
REFERENCES
- 1.Gray MW. 2012. Mitochondrial evolution. Cold Spring Harb Perspect Biol 4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keeling PJ. 2013. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- 3.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. 2015. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun 6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 4.Wernegreen JJ. 2015. Endosymbiont evolution: predictions from theory and surprises from genomes. Ann N Y Acad Sci 1360:16–35 doi: 10.1111/nyas.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alves JM, Klein CC, da Silva FM, Costa-Martins AG, Serrano MG, Buck GA, Vasconcelos AT, Sagot MF, Teixeira MM, Motta MC, Camargo EP. 2013. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol 13:190. doi: 10.1186/1471-2148-13-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moya A, Peretó J, Gil R, Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat Rev Genet 9:218–229. doi: 10.1038/nrg2319. [DOI] [PubMed] [Google Scholar]
- 7.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 8.Van Leuven JT, Meister RC, Simon C, McCutcheon JP. 2014. Sympatric speciation in a bacterial endosymbiont results in two genomes with the functionality of one. Cell 158:1270–1280. doi: 10.1016/j.cell.2014.07.047. [DOI] [PubMed] [Google Scholar]
- 9.Nowack EC, Melkonian M. 2010. Endosymbiotic associations within protists. Philos Trans R Soc Lond B Biol Sci 365:699–712. doi: 10.1098/rstb.2009.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ossipov DV, Karpov SA, Smirnov AV, Rautian MS. 1997. Peculiarities of the symbiotic systems of protists with diverse patterns of cellular organisation. Acta Protozool 36:3–21. [Google Scholar]
- 11.Mackenzie C, Walker MH. 1979. Bacteria-like structures in the endoplasm of Gregarina garnhami (Eugregarinida, Protozoa). Cell Tissue Res 202:33–39. doi: 10.1007/BF00239219. [DOI] [PubMed] [Google Scholar]
- 12.Stentiford GD, Bateman KS, Small HJ, Pond M, Ungfors A. 2012. Hematodinium sp. and its bacteria-like endosymbiont in European brown shrimp (Crangon crangon). Aquat Biosyst 8:24. doi: 10.1186/2046-9063-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stenzel DJ, Boreham PF. 1994. Bacteria-like endosymbionts in Blastocystis sp. Int J Parasitol 24:147–149. doi: 10.1016/0020-7519(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 14.Grim JN. 1993. Endonuclear symbionts within a symbiont—the surgeonfish intestinal symbiont, Balantidium jocularum (Ciliophora) is host to a Gram-positive macronuclear inhabiting bacterium. Endocytobiosis Cell Res 9:209–214. [Google Scholar]
- 15.Dessì D, Rappelli P, Diaz N, Cappuccinelli P, Fiori PL. 2006. Mycoplasma hominis and Trichomonas vaginalis: a unique case of symbiotic relationship between two obligate human parasites. Front Biosci 11:2028–2034. doi: 10.2741/1944. [DOI] [PubMed] [Google Scholar]
- 16.Hirt RP. 2013. Trichomonas vaginalis virulence factors: an integrative overview. Sex Transm Infect 89:439–443. doi: 10.1136/sextrans-2013-051105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V. 2014. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol 195:115–122. doi: 10.1016/j.molbiopara.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Maslov DA, Votýpka J, Yurchenko V, Lukeš J. 2013. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol 29:43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Votýpka J, d’Avila-Levy CM, Grellier P, Maslov DA, Lukeš J, Yurchenko V. 2015. New approaches to systematics of Trypanosomatidae: criteria for taxonomic (re)description. Trends Parasitol 31:460–469. doi: 10.1016/j.pt.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 20.Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, Grybchuk D, Lukeš J, Yurchenko V. 2014. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist 165:825–838. doi: 10.1016/j.protis.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Alves JM, Serrano MG, Maia da Silva F, Voegtly LJ, Matveyev AV, Teixeira MM, Camargo EP, Buck GA. 2013. Genome evolution and phylogenomic analysis of Candidatus Kinetoplastibacterium, the betaproteobacterial endosymbionts of Strigomonas and Angomonas. Genome Biol Evol 5:338–350. doi: 10.1093/gbe/evt012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Y, Maslov DA, Chang KP. 1994. Monophyletic origin of beta-division proteobacterial endosymbionts and their coevolution with insect trypanosomatid protozoa Blastocrithidia culicis and Crithidia spp. Proc Natl Acad Sci U S A 91:8437–8441. doi: 10.1073/pnas.91.18.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teixeira MM, Borghesan TC, Ferreira RC, Santos MA, Takata CS, Campaner M, Nunes VL, Milder RV, de Souza W, Camargo EP. 2011. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist 162:503–524. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Motta MC, Catta-Preta CM, Schenkman S, de Azevedo Martins AC, Miranda K, de Souza W, Elias MC. 2010. The bacterium endosymbiont of Crithidia deanei undergoes coordinated division with the host cell nucleus. PLoS One 5:e12415. doi: 10.1371/journal.pone.0012415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catta-Preta CM, Nascimento MT, Garcia MC, Saraiva EM, Motta MC, Meyer-Fernandes JR. 2013. The presence of a symbiotic bacterium in Strigomonas culicis is related to differential ecto-phosphatase activity and influences the mosquito-protozoa interaction. Int J Parasitol 43:571–577. doi: 10.1016/j.ijpara.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Chang KP. 1974. Ultrastructure of symbiotic bacteria in normal and antibiotic-treated Blastocrithidia culicis and Crithidia oncopelti. J Protozool 21:699–707. doi: 10.1111/j.1550-7408.1974.tb03733.x. [DOI] [PubMed] [Google Scholar]
- 27.Freymuller E, Camargo EP. 1981. Ultrastructural differences between species of trypanosomatids with and without endosymbionts. J Protozool 28:175–182. doi: 10.1111/j.1550-7408.1981.tb02829.x. [DOI] [PubMed] [Google Scholar]
- 28.Gadelha C, Wickstead B, de Souza W, Gull K, Cunha-e-Silva N. 2005. Cryptic paraflagellar rod in endosymbiont-containing kinetoplastid protozoa. Eukaryot Cell 4:516–525. doi: 10.1128/EC.4.3.516-525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motta MC, Soares MJ, Attias M, Morgado J, Lemos AP, Saad-Nehme J, Meyer-Fernandes JR, De Souza W. 1997. Ultrastructural and biochemical analysis of the relationship of Crithidia deanei with its endosymbiont. Eur J Cell Biol 72:370–377. [PubMed] [Google Scholar]
- 30.de Azevedo-Martins AC, Frossard ML, de Souza W, Einicker-Lamas M, Motta MC. 2007. Phosphatidylcholine synthesis in Crithidia deanei: the influence of the endosymbiont. FEMS Microbiol Lett 275:229–236. doi: 10.1111/j.1574-6968.2007.00892.x. [DOI] [PubMed] [Google Scholar]
- 31.Chang KP, Chang CS, Sassa S. 1975. Heme biosynthesis in bacterium-protozoon symbioses: enzymic defects in host hemoflagellates and complemental role of their intracellular symbiotes. Proc Natl Acad Sci U S A 72:2979–2983. doi: 10.1073/pnas.72.8.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camargo EP, Freymuller E. 1977. Endosymbiont as supplier of ornithine carbamoyltransferase in a trypanosomatid. Nature 270:52–53. doi: 10.1038/270052a0. [DOI] [PubMed] [Google Scholar]
- 33.Galinari S, Camargo EP. 1978. Trypanosomatid protozoa: survey of acetylornithinase and ornithine acetyltransferase. Exp Parasitol 46:277–282. doi: 10.1016/0014-4894(78)90141-8. [DOI] [PubMed] [Google Scholar]
- 34.Salzman TA, Batlle AM, Angluster J, de Souza W. 1985. Heme synthesis in Crithidia deanei: influence of the endosymbiote. Int J Biochem 17:1343–1347. doi: 10.1016/0020-711X(85)90058-8. [DOI] [PubMed] [Google Scholar]
- 35.Faria e Silva PM, Fiorini JE, Soares MJ, Alviano CS, de Souza W, Angluster J. 1994. Membrane-associated polysaccharides composition, nutritional requirements and cell differentiation in herpetomonas roitmani: influence of the endosymbiont. J Eukaryot Microbiol 41:55–59. doi: 10.1111/j.1550-7408.1994.tb05934.x. [DOI] [PubMed] [Google Scholar]
- 36.Frossard ML, Seabra SH, DaMatta RA, de Souza W, de Mello FG, Motta MC. 2006. An endosymbiont positively modulates ornithine decarboxylase in host trypanosomatids. Biochem Biophys Res Commun 343:443–449. doi: 10.1016/j.bbrc.2006.02.168. [DOI] [PubMed] [Google Scholar]
- 37.Motta MC, Martins AC, de Souza SS, Catta-Preta CM, Silva R, Klein CC, de Almeida LG, de Lima Cunha O, Ciapina LP, Brocchi M, Colabardini AC, de Araujo Lima B, Machado CR, de Almeida Soares CM, Probst CM, de Menezes CB, Thompson CE, Bartholomeu DC, Gradia DF, Pavoni DP, Grisard EC, Fantinatti-Garboggini F, Marchini FK, Rodrigues-Luiz GF, Wagner G, Goldman GH, Fietto JL, Elias MC, Goldman MH, Sagot MF, Pereira M, Stoco PH, de Mendonca-Neto RP, Teixeira SM, Maciel TE, de Oliveira Mendes TA, Urmenyi TP, de Souza W, Schenkman S, de Vasconcelos AT. 2013. Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the Trypanosomatidae family. PLoS One 8:e60209. doi: 10.1371/journal.pone.0060209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fampa P, Corrêa-da-Silva MS, Lima DC, Oliveira SM, Motta MC, Saraiva EM. 2003. Interaction of insect trypanosomatids with mosquitoes, sand fly and the respective insect cell lines. Int J Parasitol 33:1019–1026. doi: 10.1016/S0020-7519(03)00124-3. [DOI] [PubMed] [Google Scholar]
- 39.d’Avila-Levy CM, Santos LO, Marinho FA, Matteoli FP, Lopes AH, Motta MC, Santos AL, Branquinha MH. 2008. Crithidia deanei: influence of parasite gp63 homologue on the interaction of endosymbiont-harboring and aposymbiotic strains with Aedes aegypti midgut. Exp Parasitol 118:345–353. doi: 10.1016/j.exppara.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 40.d’Avila-Levy CM, Silva BA, Hayashi EA, Vermelho AB, Alviano CS, Saraiva EM, Branquinha MH, Santos AL. 2005. Influence of the endosymbiont of Blastocrithidia culicis and Crithidia deanei on the glycoconjugate expression and on Aedes aegypti interaction. FEMS Microbiol Lett 252:279–286. doi: 10.1016/j.femsle.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 41.Kozminsky E, Kraeva N, Ishemgulova A, Dobáková E, Lukeš J, Kment P, Yurchenko V, Votýpka J, Maslov DA. 2015. Host-specificity of monoxenous trypanosomatids: statistical analysis of the distribution and transmission patterns of the parasites from neotropical Heteroptera. Protist 166:551–568. doi: 10.1016/j.protis.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 42.Amann R, Snaidr J, Wagner M, Ludwig W, Schleifer KH. 1996. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol 178:3496–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maslov DA, Westenberger SJ, Xu X, Campbell DA, Sturm NR. 2007. Discovery and bar coding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J Eukaryot Microbiol 54:57–65. doi: 10.1111/j.1550-7408.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- 44.Novy FG, MacNeal WJ, Torrey HN. 1907. The trypanosomes of mosquitoes and other insects. J Infect Dis 4:223–276. doi: 10.1093/infdis/4.2.223. [DOI] [Google Scholar]
- 45.Coenye T, Falsen E, Hoste B, Ohlén M, Goris J, Govan JR, Gillis M, Vandamme P. 2000. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int J Syst Evol Microbiol 50(Pt 2):887–899. doi: 10.1099/00207713-50-2-887. [DOI] [PubMed] [Google Scholar]
- 46.Yurchenko VY, Lukeš J, Jirků M, Zeledón R, Maslov DA. 2006. Leptomonas costaricensis sp. n. (Kinetoplastea: Trypanosomatidae), a member of the novel phylogenetic group of insect trypanosomatids closely related to the genus Leishmania. Parasitology 133:537–546. doi: 10.1017/S0031182006000746. [DOI] [PubMed] [Google Scholar]
- 47.Lukeš J, Mauricio IL, Schönian G, Dujardin JC, Soteriadou K, Dedet JP, Kuhls K, Tintaya KW, Jirků M, Chocholová E, Haralambous C, Pratlong F, Oborník M, Horák A, Ayala FJ, Miles MA. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A 104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Souza W, Motta MC. 1999. Endosymbiosis in protozoa of the Trypanosomatidae family. FEMS Microbiol Lett 173:1–8. doi: 10.1016/S0378-1097(99)00005-1. [DOI] [PubMed] [Google Scholar]
- 49.Catta-Preta CM, Brum FL, da Silva CC, Zuma AA, Elias MC, de Souza W, Schenkman S, Motta MC. 2015. Endosymbiosis in trypanosomatid protozoa: the bacterium division is controlled during the host cell cycle. Front Microbiol 6:520. doi: 10.3389/fmicb.2015.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anandham R, Indiragandhi P, Kwon SW, Sa TM, Jeon CO, Kim YK, Jee HJ. 2010. Pandoraea thiooxydans sp. nov., a facultatively chemolithotrophic, thiosulfate-oxidizing bacterium isolated from rhizosphere soils of sesame (Sesamum indicum L.). Int J Syst Evol Microbiol 60:21–26. doi: 10.1099/ijs.0.012823-0. [DOI] [PubMed] [Google Scholar]
- 51.Sahin N, Tani A, Kotan R, Sedlácek I, Kimbara K, Tamer AU. 2011. Pandoraea oxalativorans sp. nov., Pandoraea faecigallinarum sp. nov. and Pandoraea vervacti sp. nov., isolated from oxalate-enriched culture. Int J Syst Evol Microbiol 61:2247–2253. doi: 10.1099/ijs.0.026138-0. [DOI] [PubMed] [Google Scholar]
- 52.Heckmann K, Schmidt HJ. 1987. Polynucleobacter necessarius gen. nov., sp. nov., an obligately endosymbiotic bacterium living in the cytoplasm of Euplotes aediculatus. Int J Syst Bacteriol 37:456–457. doi: 10.1099/00207713-37-4-456. [DOI] [Google Scholar]
- 53.Fujishima M, Heckmann K. 1984. Intraspecies and interspecies transfer of endosymbionts in Euplotes. J Exp Zool 230:339–345. doi: 10.1002/jez.1402300302. [DOI] [Google Scholar]
- 54.Jezbera J, Jezberová J, Brandt U, Hahn MW. 2011. Ubiquity of Polynucleobacter necessarius subspecies asymbioticus results from ecological diversification. Environ Microbiol 13:922–931. doi: 10.1111/j.1462-2920.2010.02396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gunn A, Pitt SJ. 2012. Parasitology: an integrated approach. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 56.Yurchenko VY, Lukeš J, Jirků M, Maslov DA. 2009. Selective recovery of the cultivation-prone components from mixed trypanosomatid infections: a case of several novel species isolated from neotropical Heteroptera. Int J Syst Evol Microbiol 59:893–909. doi: 10.1099/ijs.0.001149-0. [DOI] [PubMed] [Google Scholar]
- 57.Yurchenko VY, Lukeš J, Tesařová M, Jirků M, Maslov DA. 2008. Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus Crithidia. Protist 159:99–114. doi: 10.1016/j.protis.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Hamilton PT, Votýpka J, Dostálová A, Yurchenko V, Bird NH, Lukeš J, Lemaitre B, Perlman SJ. 2015. Infection dynamics and immune response in a newly described Drosophila-trypanosomatid association. mBio 6:e01356-15. doi: 10.1128/mBio.01356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maslov DA, Yurchenko VY, Jirků M, Lukeš J. 2010. Two new species of trypanosomatid parasites isolated from Heteroptera in Costa Rica. J Eukaryot Microbiol 57:177–188. doi: 10.1111/j.1550-7408.2009.00464.x. [DOI] [PubMed] [Google Scholar]
- 60.Jirků M, Yurchenko VY, Lukeš J, Maslov DA. 2012. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J Eukaryot Microbiol 59:537–547. doi: 10.1111/j.1550-7408.2012.00636.x. [DOI] [PubMed] [Google Scholar]
- 61.Yurchenko V, Votýpka J, Tesařová M, Klepetková H, Kraeva N, Jirků M, Lukeš J. 2014. Ultrastructure and molecular phylogeny of four new species of monoxenous trypanosomatids from flies (Diptera: Brachycera) with redefinition of the genus Wallaceina. Folia Parasitol 61:97–112. doi: 10.14411/fp.2014.023. [DOI] [PubMed] [Google Scholar]
- 62.Yurchenko V, Kostygov A, Havlová J, Grybchuk-Ieremenko A, Ševčíková T, Lukeš J, Ševčík J, Votýpka J 5 October 2015. Diversity of trypanosomatids in cockroaches and the description of Herpetomonas tarakana sp. n. J Eukaryot Microbiol doi: 10.1111/jeu.12268. [DOI] [PubMed] [Google Scholar]
- 63.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cottrell MT, Kirchman DL. 2000. Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol 66:5116–5122. doi: 10.1128/AEM.66.12.5116-5122.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fuchs BM, Pernthaler J, Amann R. 2007. Single cell identification by fluorescence in situ hybridization, p 886–896. In Reddy CA, Beveridge TJ, Breznak JA, Marzluf G, Schmidt TM, Snyder LR (ed), Methods for general and molecular microbiology, 3rd ed. ASM Press, Washington, DC. [Google Scholar]
- 66.Maslov DA, Lukeš J, Jirků M, Simpson L. 1996. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: implications for the evolution of parasitism in the trypanosomatid protozoa. Mol Biochem Parasitol 75:197–205. doi: 10.1016/0166-6851(95)02526-X. [DOI] [PubMed] [Google Scholar]
- 67.Kostygov AY, Grybchuk-Ieremenko A, Malysheva MN, Frolov AO, Yurchenko V. 2014. Molecular revision of the genus Wallaceina. Protist 165:594–604. doi: 10.1016/j.protis.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Simpson AG, Roger AJ. 2004. Protein phylogenies robustly resolve the deep-level relationships within Euglenozoa. Mol Phylogenet Evol 30:201–212. doi: 10.1016/S1055-7903(03)00177-5. [DOI] [PubMed] [Google Scholar]
- 69.Westenberger SJ, Sturm NR, Yanega D, Podlipaev SA, Zeledón R, Campbell DA, Maslov DA. 2004. Trypanosomatid biodiversity in Costa Rica: genotyping of parasites from Heteroptera using the spliced leader RNA gene. Parasitology 129:537–547. doi: 10.1017/S003118200400592X. [DOI] [PubMed] [Google Scholar]
- 70.Votýpka J, Maslov DA, Yurchenko V, Jirků M, Kment P, Lun ZR, Lukeš J. 2010. Probing into the diversity of trypanosomatid flagellates parasitizing insect hosts in South-West China reveals both endemism and global dispersal. Mol Phylogenet Evol 54:243–253. doi: 10.1016/j.ympev.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 71.Yurchenko V, Lukeš J, Xu X, Maslov DA. 2006. An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n. sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae). J Eukaryot Microbiol 53:103–111. doi: 10.1111/j.1550-7408.2005.00078.x. [DOI] [PubMed] [Google Scholar]
- 72.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 74.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 75.Chistyakova LV, Kostygov AY, Kornilova OA, Yurchenko V. 2014. Reisolation and redescription of Balantidium duodeni Stein, 1867 (Litostomatea, Trichostomatia). Parasitol Res 113:4207–4215. doi: 10.1007/s00436-014-4096-1. [DOI] [PubMed] [Google Scholar]
- 76.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Votýpka J, Klepetková H, Yurchenko VY, Horák A, Lukeš J, Maslov DA. 2012. Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist 163:616–631. doi: 10.1016/j.protis.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 78.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphometric characteristics of different morphotypes of Novymonas esmeraldas. All measurements (µm) were done on 50 (promastigotes and choanomastigotes) or 20 (amastigotelike) cells.
Subcloning of Novymonas esmeraldas. Forty-one subclones were cultivated in different media (see Materials and Methods) and analyzed for the presence of endosymbionts.