ABSTRACT
Urinary tract infection (UTI) is a disease of extremely high incidence in both community and nosocomial settings. UTIs cause significant morbidity and mortality, with approximately 150 million cases globally per year. Uropathogenic Escherichia coli (UPEC) is the primary cause of UTI and is generally treated empirically. However, the rapidly increasing incidence of UTIs caused by multidrug-resistant UPEC strains has led to limited available treatment options and highlights the urgent need to develop alternative treatment and prevention strategies. In this study, we performed a comprehensive analysis to define the regulation, structure, function, and immunogenicity of recently identified UPEC vaccine candidate C1275 (here referred to as IrmA). We showed that the irmA gene is highly prevalent in UPEC, is cotranscribed with the biofilm-associated antigen 43 gene, and is regulated by the global oxidative stress response OxyR protein. Localization studies identified IrmA in the UPEC culture supernatant. We determined the structure of IrmA and showed that it adopts a unique domain-swapped dimer architecture. The dimeric structure of IrmA displays similarity to those of human cytokine receptors, including the interleukin-2 receptor (IL-2R), interleukin-4 receptor (IL-4R), and interleukin-10 receptor (IL-10R) binding domains, and we showed that purified IrmA can bind to their cognate cytokines. Finally, we showed that plasma from convalescent urosepsis patients contains high IrmA antibody titers, demonstrating the strong immunogenicity of IrmA. Taken together, our results indicate that IrmA may play an important role during UPEC infection.
IMPORTANCE
Uropathogenic E. coli (UPEC) is the primary cause of urinary tract infection (UTI), a disease of major significance to human health. Globally, the incidence of UPEC-mediated UTI is strongly associated with increasing antibiotic resistance, making this extremely common infection a major public health concern. In this report, we describe the regulatory, structural, functional, and immunogenic properties of a candidate UPEC vaccine antigen, IrmA. We demonstrate that IrmA is a small UPEC protein that forms a unique domain-swapped dimer with structural mimicry to several human cytokine receptors. We also show that IrmA binds to IL-2, IL-4, and IL-10, is strongly immunogenic in urosepsis patients, and is coexpressed with factors associated with biofilm formation. Overall, this work suggests a potential novel contribution for IrmA in UPEC infection.
INTRODUCTION
Urinary tract infections (UTIs) are among the most common infectious diseases of humans and are the most common nosocomial infections in the developed world. UTIs cause significant morbidity and mortality, with approximately 150 million cases globally per year. It is estimated that 40% to 50% of women and 5% of men will develop a UTI in their lifetime, and UTI accounts for more than 1 million hospitalizations and $3.5 billion in medical expenses each year in the United States (1, 2). Community-acquired UTIs are estimated to account for 0.7% of ambulatory care visits (3), and nosocomial UTI is estimated to occur in approximately 7.3% of all hospital admissions (4). UTIs can present as uncomplicated or complicated infections of the bladder (cystitis) or kidney (pyelonephritis), potentially leading to bacteremia and urosepsis (5, 6).
Uropathogenic Escherichia coli (UPEC) is by far the most common cause of community-acquired and nosocomial UTI (2). In women, UPEC is responsible for 75% to 95% of all cases of uncomplicated cystitis and pyelonephritis (5). Antibiotic therapy is the primary treatment, and UTI represents the second-most-common reason for antibiotic prescription worldwide (7). Overall, this has led to an increase in the prevalence of multidrug-resistant (MDR) UPEC strains, increased rates of treatment failure with standard antibiotic therapies, and a rise in the use of second- and third-line therapies, further promoting the emergence of MDR UPEC strains such as the globally disseminated E. coli sequence type 131 (ST131) clone (7–11). Reports of resistance to last-line carbapenem antibiotics among MDR UPEC strains (including ST131 strains) support the classification of carbapenem-resistant Enterobacteriaceae as an urgent threat to human health (12) and reinforce the urgency for new strategies to treat and prevent pan-resistant UTI (13–16).
The paucity of effective treatments for MDR UPEC has prompted the development of novel genomic, proteomic, and structural vaccinology strategies for the design of a broadly protective vaccine. UPEC pathogenesis relies on multiple outer membrane-associated and secreted virulence factors for colonization and infection of the urinary tract, including adhesins, toxins, siderophores, and polysaccharide components (17). Some of these targets have been investigated as antigens in the development of diagnostic, drug, and vaccine applications (18, 19).
Recently, a reverse vaccinology approach led to the identification of several novel vaccine antigens that conferred protection against E. coli in a murine model of sepsis infection (20). Among these antigens, a soluble protein of unknown function (C1275; referred to here as E. coli interleukin [IL] receptor mimic protein A, or IrmA) was identified that conferred significant protection in mouse challenge experiments. In this study, we characterized the prevalence, genetic location, transcriptional regulation, immunogenicity, structure, and function of IrmA. We show that irmA is cotranscribed as part of a newly defined operon with the biofilm-associated antigen 43 (agn43 [flu]) gene and that its transcription is controlled by the global oxidative stress response regulator OxyR. We further demonstrate that IrmA has a fibronectin III (FNIII)-like fold that forms a unique domain-swapped dimer with structural mimicry to the binding domain of the IL-2 receptor (IL-2R), the IL-4 receptor (IL-4R) and, to a lesser extent, the IL-10 receptor (IL-10R). We show that IrmA binds to IL-2, IL-4, and IL-10 with different affinities, highlighting a potential novel role in manipulating inflammation. Finally, evidence of the immunogenicity of IrmA is presented based on the measurement of increased antibody titers in urosepsis patients.
RESULTS
The irmA gene is highly prevalent among UPEC strains.
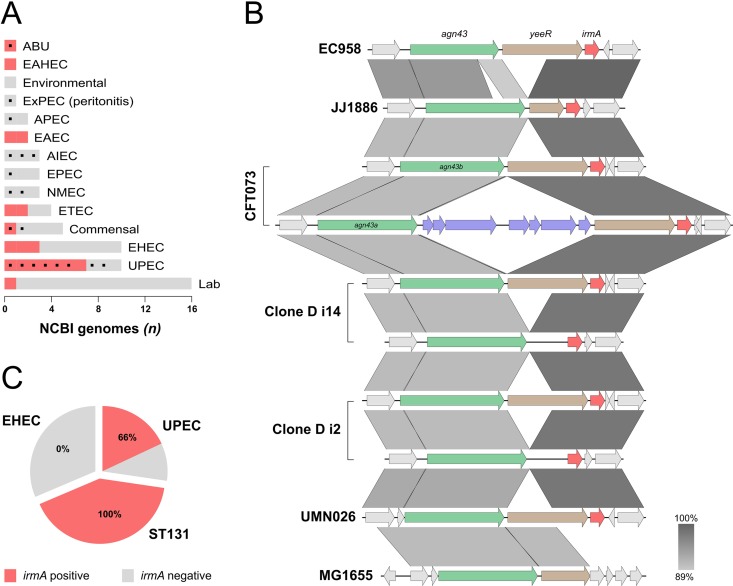
The prevalence of the irmA gene was assessed in 62 completely sequenced E. coli genomes available in the NCBI database (see Table S1 in the supplemental material). The irmA gene was present in 29% (18/62) of the strains, often in more than one copy (Fig. 1A). However, further analysis revealed a much higher prevalence of irmA in UPEC strains, including the globally disseminated MDR ST131 clone, which is represented by the EC958 and JJ1886 strains. irmA was present in 70% (7/10) of completely sequenced UPEC strains (Fig. 1A), 66% (31/47) of urosepsis strains from our own laboratory collection, and all 71 strains from the globally dominant fluoroquinolone-resistant subgroup of ST131 that we reported recently (11). In contrast, irmA was absent in our own non-O157 enterohemorrhagic E. coli (EHEC) collection (Fig. 1C). The IrmA sequences from all of these strains were compared, and the sequence was shown to be highly conserved, with >97% identity at the amino acid level.
FIG 1 .
Genomic context and prevalence of the irmA gene. (A) Prevalence of irmA in the completely sequenced E. coli strains available on the NCBI database according to their pathotype (ABU, asymptomatic bacteriuria; EAHEC, enteroaggregative hemorrhagic E. coli; ExPEC, extraintestinal pathogenic E. coli; APEC, avian-pathogenic E. coli; EAEC, enteroaggregative E. coli; AIEC, adherent invasive E. coli; EPEC, enteropathogenic E. coli; NMEC, neonatal meningitis-associated E. coli; ETEC, enterotoxigenic E. coli; EHEC, enterohemorrhagic E. coli; UPEC; uropathogenic E. coli). Phylogenetic groups of the strains were determined (93), and the B2 phylogeny is indicated (black box). (B) Alignment of the irmA genomic region from completely sequenced irmA-positive UPEC E. coli strains available on the NCBI database. The confirmed genes that comprise the newly defined agn43-yeeR-irmA operon are highlighted (agn43, green; yeeR, brown; irmA, red) as well as an insertion sequence responsible for the partial disruption of the CFT073 operon (purple). Regions are ordered according to their localization in the chromosome. The alignment was generated using Easyfig (94). (C) Prevalence of irmA in our genome-sequenced laboratory collections.
irmA is located downstream of the agn43 and yeeR genes.
The genetic context of irmA was examined by comparing its locations on the chromosomes from six irmA-positive completely sequenced UPEC strains (Fig. 1B). Our analysis revealed that irmA is located downstream of the agn43 and yeeR genes. In CFT073, two identical copies of irmA (c1275 and c3665) are located within pathogenicity islands (PAIs): PAI-CFT073-serX and PAI-CFT073-pheV, respectively. These islands differ in size, content, and associated tRNA (21); however, they share a small subset of genes, including agn43, yeeR, and the irmA alleles, and this pattern is also observed in other UPEC strains (Fig. 1B). The region containing irmA also corresponds to module 5 of AGI-3, a PAI inserted at the selC tRNA in avian-pathogenic E. coli (APEC) strain BEN2908 (22). Further analysis showed that the irmA region has been the subject of acquisition and loss of genetic elements in UPEC, often leading to the disruption of one or more of the components (Fig. 1B). A large region in MG1655 has been deleted, leading to the disruption of yeeR and possibly to deletion of irmA.
The transcription of irmA is regulated by OxyR.
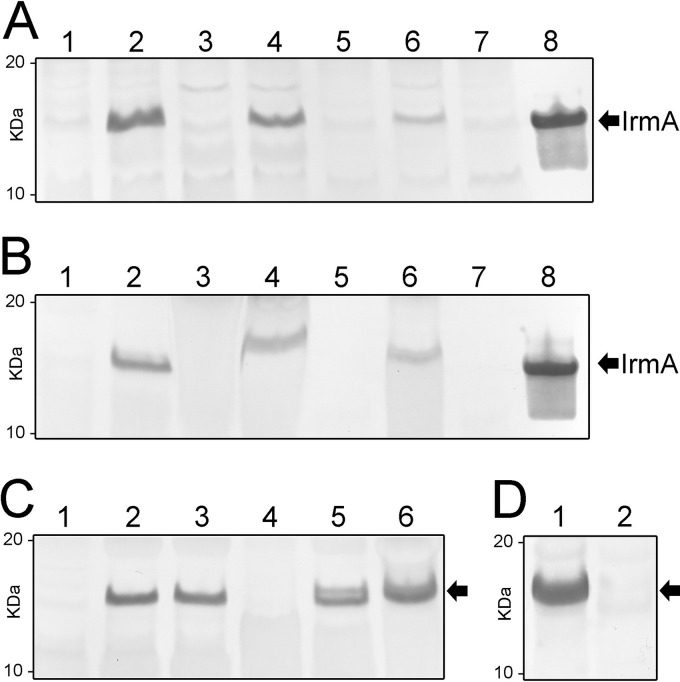
The genetic association of irmA with agn43 led us to examine if these genes are coregulated. The agn43 gene encodes the Ag43 autotransporter protein, which mediates bacterial aggregation, biofilm formation, and long-term bladder colonization (23–28). The expression of Ag43 is phase variable and is controlled by an epigenetic mechanism involving global regulatory protein OxyR and the Dam methyltransferase (29). Mutation of oxyR leads to derepression of agn43 transcription and strong expression of Ag43 (29, 30). Given the colocation of the agn43-yeeR-irmA genes in E. coli, we hypothesized that these three genes could constitute a single transcriptional unit that is regulated by OxyR. To test this, we compared the expression levels of IrmA in total cell lysates prepared from wild-type CFT073 and a CFT073 oxyR mutant (CFT073oxyR) by Western blotting using an IrmA-specific polyclonal antibody (Fig. 2A, lanes 1 and 2). While IrmA expression in CFT073 was barely detectable, we observed a dramatic increase in IrmA expression in CFT073oxyR, indicating that OxyR represses the transcription of irmA. To analyze whether this was strain specific, we also tested the expression of IrmA in MDR ST131 strain EC958. As observed for CFT073, mutation of oxyR (EC958oxyR) resulted in enhanced expression of IrmA (Fig. 2A, lanes 5 and 6). The specificity of the IrmA antibody was confirmed by the lack of a cross-reacting band in an irmA-oxyR double mutant (EC958irmA oxyR; Fig. 2A, lane 7). Interestingly, OxyR also regulated irmA transcription in asymptomatic bacteriuria (ABU) strain 83972 (Fig. 2A, lanes 3 to 4). The regulation of irmA was confirmed by quantitative reverse transcription (qRT)-PCR analysis of wild-type EC958 and EC958oxyR, which revealed 12.9 (±0.98)-fold and 121.4 (±8.43)-fold increases in irmA and agn43 transcription, respectively, in EC958oxyR. Taken together, our data demonstrate that, like the agn43 gene, expression of irmA is repressed by OxyR.
FIG 2 .
Western blot analysis demonstrating detection of IrmA in the culture supernatant and molecular characterization of the agn43-yeeR-irmA operon. (A) Expression of IrmA detected from total cell lysates prepared from E. coli strains CFT073 (lane 1), CFT073oxyR (lane 2), 83972 (lane 3), 83972oxyR (lane 4), EC958 (lane 5), EC958oxyR (lane 6), EC958irmA oxyR (lane 7), and recombinant IrmA (control; lane 8). Mutation of oxyR resulted in the derepression of IrmA. (B) Expression of IrmA detected from the supernatant fraction prepared from E. coli strains CFT073 (lane 1), CFT073oxyR (lane 2), 83972 (lane 3), 83972oxyR (lane 4), EC958 (lane 5), EC958oxyR (lane 6), EC958irmA oxyR (lane 7), and recombinant IrmA (control; lane 8). IrmA was detected from the supernatant fraction and migrated as an ~13-kDa protein. (C) Molecular investigation into the mechanism of IrmA secretion: Western blot analysis of whole-cell lysates prepared from MG1655agn43(pSU2718) (lane 1), MG1655agn43(pSU2718-irmA) (lane 2), and MG1655tamAB(pSU2718-irmA) (lane 3) and Western blot analysis of supernatant fraction prepared from MG1655agn43(pSU2718) (lane 4), MG1655agn43(pSU2718-irmA) (lane 5), and MG1655tamAB(pSU2718-irmA) (lane 6). Secretion of IrmA was independent of the production of Ag43 or the TAM. (D) Expression and release of IrmA is abrogated by an insertion downstream of agn43a in strain CFTfluB+ (lane 2), in contrast to strain CFTfluA+ (lane 1).
IrmA is present in the culture supernatant.
IrmA contains a predicted signal sequence and represents a protective antigen in a sepsis infection model (20); thus, we predicted that it would be either located on the cell surface or secreted. Immunofluorescence microscopy performed using an IrmA-specific polyclonal antibody failed to detect IrmA on the cell surface of CFT073oxyR or EC958oxyR (data not shown). We tested for the presence of IrmA in the culture supernatant of CFT073oxyR, 83972oxyR, and EC958oxyR and observed a strong cross-reacting IrmA-specific band (Fig. 2B, lanes 2, 4, and 6). To investigate if the presence of extracellular IrmA was dependent on the (uncharacterized) YeeR protein and/or Ag43, we used a previously constructed MG1655agn43 mutant (31), which does not contain a functional copy of any of the agn43-yeeR-irmA genes. Introduction of a plasmid containing irmA (pSU2718-irmA) into MG1655agn43 resulted in the detection of IrmA in the culture supernatant, indicating that Ag43 and YeeR are not involved in its secretion (Fig. 2C, lanes 2 and 5). Recently, the transport of Ag43 across the outer membrane has been shown to depend on a newly defined translocation and assembly module (TAM) (32). We therefore tested whether the TAM contributes to the release of IrmA by introducing plasmid pSU2718-irmA into an MG1655 TAM mutant (MG1655tamAB). No effect on IrmA expression and release was observed in MG1655tamAB (Fig. 2C, lanes 3 and 6), indicating that IrmA release occurs independently of the TAM.
The agn43-yeeR-irmA genes are transcribed as a single transcriptional unit.
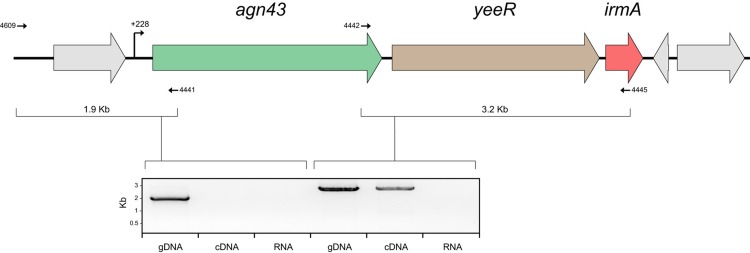
To examine the transcriptional organization of the agn43-yeeR-irmA genes, we utilized a series of primers specific for each individual gene in combination with RT-PCR. RNA was extracted from EC958oxyR (to maximize irmA transcription) and reverse transcribed into cDNA. PCRs using EC958 cDNA yielded specific amplification products demonstrating cotranscription of agn43-yeeR-irmA (Fig. 3). The correct size of the amplicons was confirmed using EC958oxyR genomic DNA, while no amplicons were obtained from EC958oxyR RNA (negative control). The transcription start site of this polycistronic mRNA was determined by 5′ rapid amplification of cDNA ends (RACE) at +228 bp from the agn43 start codon (Fig. 3). Although no evidence for the cotranscription of irmA with other downstream genes was observed, we were unable to determine the termination transcription site by 3′ RACE. To confirm the cotranscription of these genes, we used two previously described CFT073 constructs that constitutively express either c1273 (agn43b; strain CFT073amp, PcLfluB, or CFTfluB+) or c3665 (agn43a; strain CFT073km, PcLfluA, or CFTfluA+) (26). While IrmA was detected in the supernatant fraction of strain CFTfluB+, it could not be detected in the supernatant fraction of strain CFTfluA+ (Fig. 2D), indicating that the insertion downstream of agn43a (Fig. 1B) disrupts the transcription of the yeeR-irmA genes. Taken together, the data demonstrate that the agn43-yeeR-irmA genes are cotranscribed as part of a newly defined operon in E. coli.
FIG 3 .
Structural organization of the agn43-yeeR-irmA operon. A PCR amplification product was obtained using primers overlapping the agn43-yeeR-irmA genes and flanking regions. Primers are represented by arrows, and the expected amplicon size is indicated. The amplified product from EC958 genomic DNA (gDNA; positive control), EC958oxyR cDNA, and EC958oxyR RNA (RNA; negative control) is shown.
IrmA has a fibronectin III (FNIII)-like fold.
The structure of the mature form of IrmA was determined by single-wavelength anomalous diffraction (SAD) and refined to an R-free value of 21.5% (R-factor, 26.1%) at 2.3-Å resolution (see Table S2 in the supplemental material). The crystals belong to the space group P213, with two subunits per asymmetric unit. Although the majority of the electron density could be unambiguously interpreted, N-terminal residues 1 to 7 and C-terminal residues 128 to 131 could not be modeled due to poor density. The overall architecture of IrmA contains a fibronectin III (FNIII)-like domain. This immunoglobulin (Ig)-like domain consists of a β-sandwich framework comprising seven antiparallel β-strands linked by loop regions that form two β-sheets (Fig. 4A). The β-sandwich is stabilized by hydrophobic interactions in the inner core of the FNIII module, and, unlike other FNIII-containing proteins, IrmA incorporates a disulfide bond between residues cys21 and cys80 (Fig. 4B). The electrostatic potential of IrmA shows a negatively charged surface, whereby several acidic residues (Asp7, Asp17, Asp33, Asp38, Glu39, Asp52, Glu64, Glu66, Asp70, Asp78, Glu82, Glu85, Glu88, Glu93, Glu99, Glu100, Asp114, and Asp117) line the surface of the β-strands, forming acidic patches on the surface of the β-sandwich (Fig. 4C).
FIG 4 .
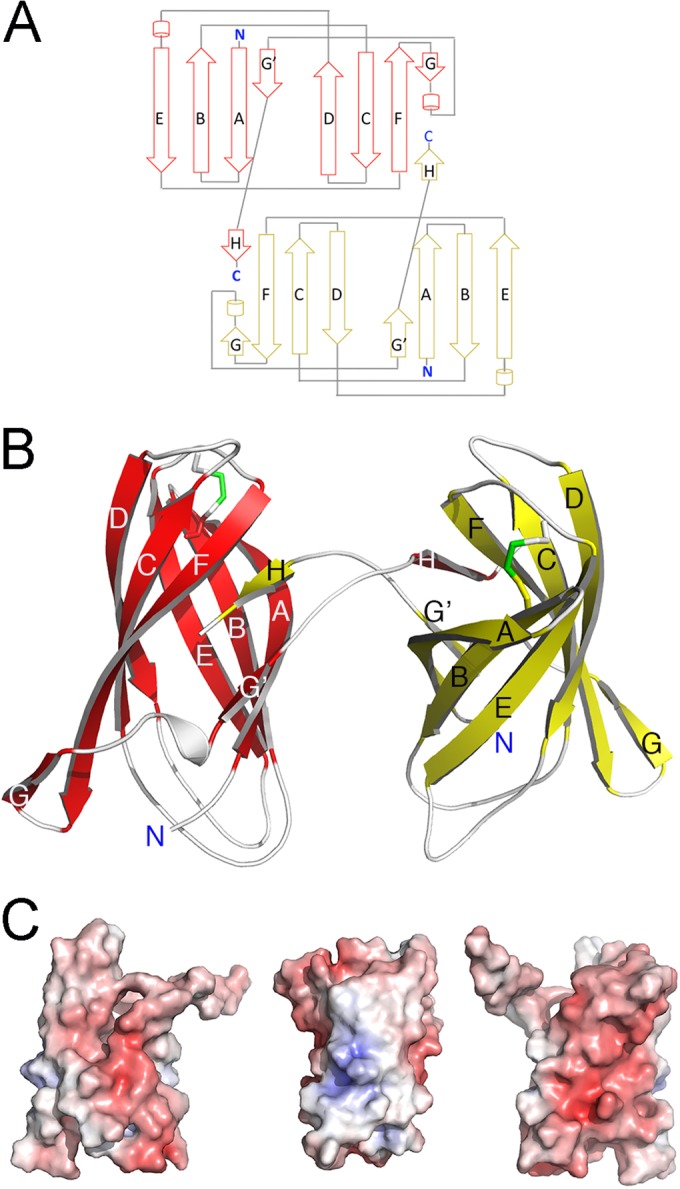
Crystal structure of IrmA. (A) Topology diagram of the IrmA dimer. The monomer conformations are shown in red (top) and yellow (bottom). (B) Ribbon representation of IrmA domain-swapped dimer with the protomers of the dimer shown in red and yellow (β-strands are indicated as arrows and are labeled). (C) Electrostatic surface of the IrmA monomer. Positive and negative electrostatic potentials are shown in blue and red, respectively (saturation at 10 kT/e). The orientation of the left panel corresponds to that in the lower panel in Fig. 4A. The orientations in the middle and left panels correspond to the protein rotated by 90° and 180° along the axis of the FNIII domain (y axis).
IrmA forms a domain-swapped dimer.
Notably, the IrmA monomers in the asymmetric unit form a domain-swapped dimer. The swapped section of the structure localizes in the C-terminal tail (residues 110 to 128) of each IrmA monomer, which protrudes from the FNIII domain and is intertwined with a second identical protein chain. This extended section comprises an unstructured region and a short β-strand, which complements the incomplete FNIII fold present in each of the IrmA monomers that comprise the dimer (Fig. 4B). The two crystallographically independent subunits are very similar (with a root mean square deviation [RMSD] value for 118 Cα atoms of 0.79 Å). The overall buried area at the dimer interface is extensive (approximately 1,481 Å2 per monomer) and is stabilized by 17 hydrogen bonds (33), and the dimer displays an elongated shallow groove between the two subunits (6,773 Å3) (33).
To define whether the IrmA dimer is also present in solution, the protein was recombinantly expressed in BL21pLysS cells and purified using the protocol previously developed for structural studies. IrmA was subjected to size exclusion chromatography (SEC) using a HiLoad 16/60 Superdex 75 column (GE Healthcare) and eluted at the apparent molecular mass of approximately 26 kDa, corresponding to a dimer (see Fig. S1A in the supplemental material). The dimeric state of the protein in aqueous solution was confirmed by blue native polyacrylamide gel electrophoresis (BN-PAGE) and sedimentation velocity analytical ultracentrifugation (SV-AUC) experiments. BN-PAGE analysis showed that IrmA migrates as a dimer (see Fig. S1B). Similarly, SV-AUC studies indicated that IrmA exists as a stable dimer in solution (see Fig. S1C). The absorbance versus radial position profile of IrmA showed a single sedimentation boundary with a standardized sedimentation coefficient (s20,w) of 2.58 S (29 kDa) and a frictional ratio (f/fo) of 1.36 (see Fig. S1C). The complete absence of a species corresponding to the monomer at 0.15 mg/ml indicated that this domain-swapped dimer was tightly associated. The RMSD and run test Z values for the fit were <0.004 and 2.03, respectively. The hydrodynamic properties of IrmA are summarized in Table S3.
We also analyzed the oligomeric state of endogenous IrmA secreted from an E. coli MG1655 strain containing a plasmid encoding irmA (strain MS5667) by SEC and BN-PAGE. Released IrmA was concentrated and purified from the culture supernatant by two consecutive gel filtration steps on a HiLoad 16/60 Superdex 75 column (GE Healthcare). Similarly to the recombinant protein, released IrmA eluted at the apparent molecular mass of 30 kDa and migrated as a dimer on a BN-PAGE (see Fig. S1A and B in the supplemental material).
IrmA shares structural similarity with human interleukin receptors.
IrmA shares low sequence identity with other proteins of known function. A database search for structural homologues using the server Dali (34) revealed that IrmA shows high structural similarity to a number of proteins that are variants of the Ig-like fold, including the C-terminal β-sandwich domain of the SadB autotransporter (PDB 4C47) and the Ig-like domain of aminopeptidases (PDB 3EBG). This structural comparison also identified other related proteins for which a function of the Ig-like domain has been defined (e.g., cytokine receptors). IrmA shares structural similarity to the extracellular domains of human IL-2R and IL-4R. These receptors belong to the largest group in the cytokine receptor family (type I) and contain two covalently linked FNIII modules forming the binding site for their cognate cytokines (35). The FNIII domains in IrmA overlap one FNIII module in both IL-2R and IL-4R (Fig. 5). Pairwise comparison between individual FNIII domains in IrmA with IL-2R (PDB 2ERJ) and IL-4R (PDB 3BPL) revealed that, despite showing very low sequence identity (5% and 9%, respectively), they had considerable structural similarity, with overall RMSD values of 3.4 Å (91 Cα aligned) and 3.7 Å (87 Cα aligned), respectively. This similarity is primarily localized in the β-strand regions, with the loop regions being structurally divergent (Fig. 5). Using the Dali program, it was also observed that IrmA can be superimposed on other cytokine receptors, including IL-10R (PDB 1J7V [amino acid sequence identity of 6%]), but with a lower Z score of 4.5 (see Fig. S2 in the supplemental material).
FIG 5 .
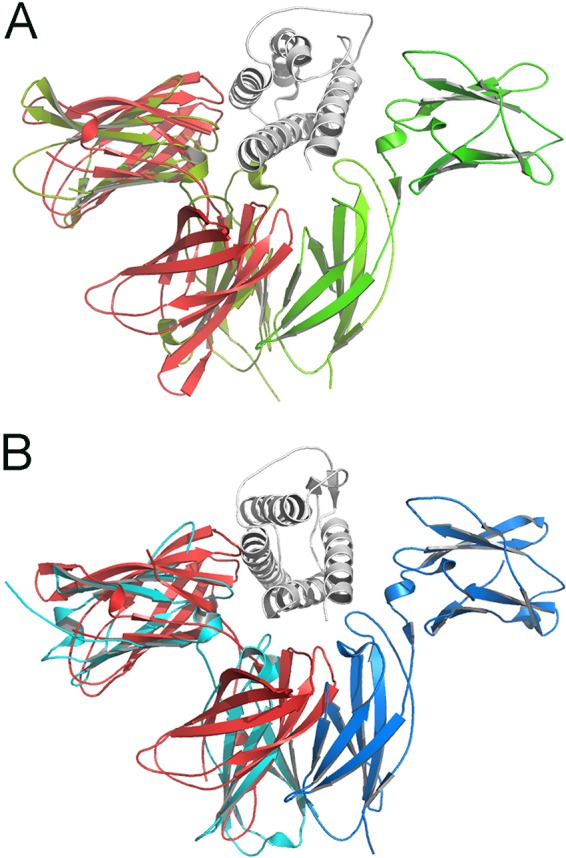
Comparison of IrmA with IL-2R and IL-4R and α-carbon trace superposition of IrmA (shown in red) with the soluble domains of (A) IL-2R (PDB 2ERJ) (green) and (B) IL-4R (PDB 3BPL) (blue). IL-2 and IL-4 bound to their cognate receptors are displayed in gray.
IrmA binds to human cytokines.
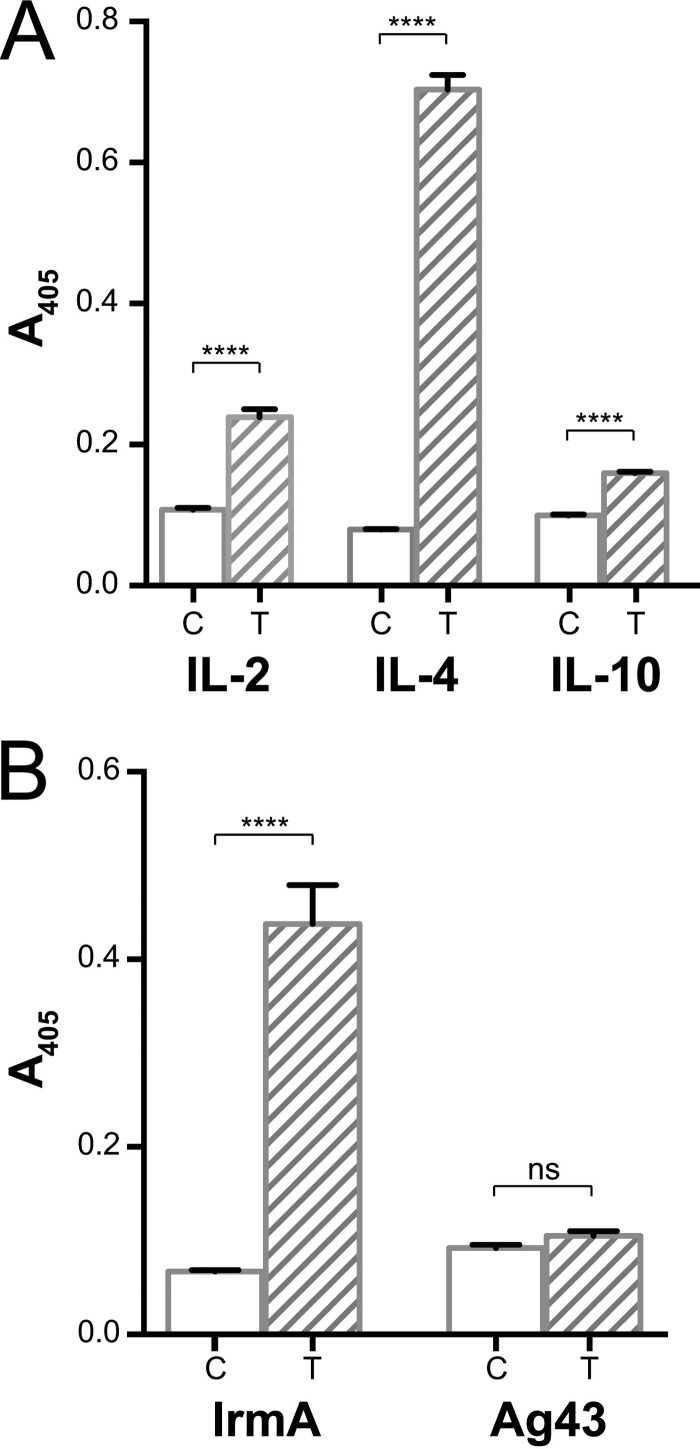
We investigated the structural homology of IrmA with IL-2R, IL-4R, and IL-10R further by examining the ability of IrmA to bind to recombinant IL-2, IL-4, and IL-10, respectively. Using an enzyme-linked immunosorbent assay (ELISA), we demonstrated that IrmA binds to all three immobilized cytokines, with the strongest interaction detected for IL-4 (Fig. 6A). To confirm these results, we also performed a reverse ELISA with IL-4; in these experiments, immobilized IrmA bound strongly to IL-4 (Fig. 6B). We also tested the specificity of the IrmA-IL-4 interaction by examining the ability of recombinant Ag43 to bind to IL-4. Ag43, which is structurally distinct from IrmA (25), did not bind to IL-4 under the experimental conditions tested (Fig. 6B). Finally, we demonstrated that IrmA does not bind to interleukin-17A (IL-17A; see Fig. S3A in the supplemental material), which is structurally distinct from IL-4 (see Fig. S3B).
FIG 6 .
IrmA binding to IL-2, IL-4, and IL-10. (A) ELISA showing the binding of IrmA to IL-2, IL-4, and IL-10, in the absence (C, control) or presence (T, treated) of recombinant IrmA. (B) ELISA results showing the specific interaction of IrmA with IL-4 but not with Ag43. Statistical analysis was performed using an unpaired t test (****, P < 0.0001; ns, not significant). Error bars represent the standard deviations of the results from three replicates.
IrmA is immunogenic in urosepsis patients.
Given the potential of IrmA as an ExPEC vaccine target (20), and our new data demonstrating the presence of IrmA in the supernatant following UPEC culture in LB broth, we assessed the immunogenicity of IrmA using plasma samples from urosepsis patients and healthy individuals. Purified IrmA was used in an ELISA to detect specific IrmA antibodies. In this assay, plasma samples from urosepsis patients infected by IrmA-positive UPEC strains exhibited significantly higher IrmA antibody titers than those from healthy individuals (P < 0.01; see Fig. S4 in the supplemental material).
DISCUSSION
IrmA is a small (~13 kDa) UPEC protein that was originally identified in a large reverse genetic screen as a broadly protective vaccine antigen. Here, we have determined the molecular regulation of the irmA gene and characterized the structural, functional, and immunogenic properties of the IrmA protein.
Comprehensive bioinformatic analysis of the irmA genetic locus revealed its frequent colocation with the Ag43-encoding agn43 gene, and further transcriptional studies demonstrated that the agn43-yeeR-irmA genes are part of a newly defined operon structure. The transcription of agn43 is phase variable and is regulated by the concerted action of Dam (positive regulation) and OxyR (negative regulation) (28–30, 36–39). We showed that the irmA gene is also part of the OxyR regulon. Indeed, mutation of oxyR in two well-characterized UPEC strains (CFT073 and EC958) and ABU strain 83972 led to enhanced expression of IrmA. OxyR is a transcriptional regulator that controls a regulon comprising more than 30 genes involved in the cellular response to oxidative stress (40). In the absence of OxyR, bacteria are hypersensitive to hydrogen peroxide and several other oxidants (41). UPEC encounters reactive oxygen species during UTI, and mutation of oxyR attenuates UPEC colonization of the mouse urinary tract (42) and bloodstream (43). Thus, the regulation of IrmA expression by OxyR and its high prevalence in UPEC strains are consistent with a potential role during human infection.
Analysis of the supernatant fraction from CFT073oxyR, 83972oxyR, and EC958oxyR cultures revealed that IrmA is present in the extracellular milieu. Extracellular IrmA was detected in the absence of both Ag43 and YeeR, demonstrating that these proteins do not contribute to its secretion. We also showed that the TAM, which contributes to Ag43 translocation (32), is not required for IrmA secretion. Thus, the mechanism of IrmA secretion, as well as the function of the uncharacterized YeeR protein, remains to be elucidated.
The crystal structure of IrmA revealed that it forms a domain-swapped dimer where each monomer adopts a FNIII-like fold. Biochemical and biophysical data supported the idea of dimer formation by both the recombinant and endogenous IrmA protein. Notably, although FNIII domains are present in many proteins, they typically do not dimerize; instead, they form string-like structures with other FNIII or modular domains such as fibronectin. Only two examples of FNIII dimerization have been previously described: irisin, which forms an intersubunit β-sheet dimer (44), and the head-to-tail associations of oncofetal fibronectin (45). These dimerization modes are different from the domain-swap mechanism observed in IrmA, which is somewhat reminiscent of chaperone-fimbrial protein assembly, where the chaperone donates a surrogate β-strand to complement the incomplete Ig fold of the fimbrial subunit (46). In itself, domain swapping is also a relatively rare event, with only about 60 structures of domain-swapped proteins present in the Protein Data Bank. The unique structure of IrmA is to the best of our knowledge the first example of a domain-swapped dimer between two FNIII modules and would account for the observed stability of this conformation.
Structural characterization of IrmA revealed that it displays similarity with IL binding receptors IL-2R and IL-4R (and, to a lesser extent, IL-10R), which contain two covalently linked FNIII modules that form the binding partner for their cognate cytokines. Thus, we investigated the ability of purified IrmA to interact with human IL-2, IL-4, and IL-10 cytokines. Our analysis revealed that IrmA binds to all three cytokines, with the greatest affinity observed for IL-4. In contrast, IrmA did not bind to the structurally distinct IL-17A cytokine (47, 48). IL-2 and IL-4 belong to a common family of cytokines that contribute to the differentiation, growth, and proliferation of immune cells (49). IL-2 is required for the development of Treg cells, the stimulation of antibody synthesis, and the proliferation and differentiation of natural killer cells to combat infection (50, 51). IL-4 is a multifunctional pleotropic cytokine that is primarily produced by Th2 polarized T cells and promotes the proliferation and differentiation of B cells, controls allergic conditions, and provides protection against parasitic infection (52, 53). IL-10, on the other hand, is an anti-inflammatory cytokine which inhibits the production of proinflammatory cytokines and chemokines, downregulates major histocompatibility class II and antigen presentation costimulatory markers on macrophages, and suppresses T cell activation (54–56). While a direct role for IL-2 and IL-4 has not been demonstrated in UTI, IL-10 forms part of the early bladder response to acute UPEC-mediated UTI in mice and helps to control infection in the bladder (57). Furthermore, in vitro coculture studies have demonstrated that monocytes and uroepithelial cells produce IL-10 synergistically following UPEC infection (58), and the expression of H4 flagella by EC958 stimulates IL-10 production in monocytes and monocyte/uroepithelial cell cocultures (59). Given the role of Ag43 in biofilm formation, our data invoke an intriguing model whereby expression of IrmA during Ag43-mediated UPEC biofilm growth could compromise the function of IL-10 (and possibly IL-2 and IL-4) in the host, by competing for binding sites and rendering these cytokines less available for immune activation via their cognate host cell receptors.
The ability to modulate and evade the host immune system is a common feature of many microbial pathogens. In some cases, this involves molecular mimicry, where the pathogen produces a molecule with structural similarity to a cytokine or cytokine receptor to inactivate or modulate the immune response (60). For example, several such molecules have been described in herpesviruses, including an IL-10 homologue that possesses anti-inflammatory properties (61–64), an IL-17 homologue that can stimulate T-cell proliferation (65), and an IL-6 homologue that binds directly to cytokine signaling receptor gp130 (66). Some viruses can also produce soluble cytokine receptor mimics that sequester host cytokines and compete with their cognate receptors. Several examples have been described in poxviruses, including a glycoprotein that functions as a soluble IL-1β receptor (67, 68), a gamma interferon (IFN-γ) receptor homologue (69), an IFN-α-binding protein (70, 71), and an IL-18 receptor homologue (72–74). In line with our observations for IrmA, these viral cytokine-binding proteins exhibit structural similarity to their receptor counterparts despite limited sequence homology (60). Further work is now required to determine if IrmA is able to manipulate host immune responses; however, our observation that IrmA is immunogenic in urosepsis patients and the fact that IL-10 plays an important role in the innate immune response to UTI (57) support this hypothesis.
In summary, we have identified a newly defined operon structure comprising the agn43-yeeR-irmA genes that is regulated by OxyR. Elucidation of the structure of IrmA revealed an intriguing mimicry of human cytokine receptors IL-2R, IL-4R, and, to a lesser extent, IL-10R. The cytokine-binding data suggest that IrmA may contribute to manipulation of the innate immune response during UPEC infection.
MATERIALS AND METHODS
Bacteria and growth conditions.
E. coli strains and plasmids used in this study are listed in Table S4 in the supplemental material. The UPEC urosepsis strains and non-O157 EHEC strains were part of our in-house collection. Bacteria were routinely grown at 37°C on solid or in liquid Luria-Bertani (LB) medium (75) supplemented with appropriate antibiotics: chloramphenicol (30 µg/ml), kanamycin (50 µg/ml), and ampicillin (100 µg/ml). For the generation of total cell lysates and supernatant fractions, bacteria were inoculated at a starting optical density at 600 nm (OD600) equal to 0.050 and cultures were grown overnight at 37°C with shaking (180 rpm). Cells were harvested at 10,000 × g for 10 min at 4°C to generate the total cell lysate sample. To generate the supernatant fraction, the culture supernatant was filter sterilized (using a 0.22-µm-pore-size filter) and precipitated overnight at 4°C by adding trichloroacetic acid (TCA) at a final concentration of 10%. After precipitation, supernatant preparations were centrifuged at 18,000 × g for 30 min at 4°C. Pellets were washed with 1 ml 10% TCA and centrifuged at 18,000 × g for 15 min at 4°C. Pellets were resuspended in ice-cold ethanol and centrifuged at same speed. Pellets were dried using a SpeedVac at a low drying rate for 20 min and then resuspended in appropriate resuspension buffer (50 mM ammonium bicarbonate, 3 M urea, 5 mM dithiothreitol [DTT]) to generate the supernatant fraction.
Molecular methods.
Genomic DNA was extracted using an UltraClean microbial DNA isolation kit (Mo Bio Laboratories). Isolation of plasmid DNA was carried out using a QIAprep Spin Miniprep kit (Qiagen). PCRs were performed using Phusion High-Fidelity DNA polymerase (New England Labs); PCR products were gel purified using a QIAquick gel extraction kit (Qiagen) and sequenced using a BigDye Terminator v3.1 cycle sequencing kit (Invitrogen). Primers used in this study are described in Table S5 in the supplemental material. Extraction of RNA was carried out using an RNeasy minikit (Qiagen), and reverse transcription reactions were performed using a SuperScript III first-strand synthesis system (Invitrogen). All protocols were performed following the manufacturer’s specifications. Mutants were obtained using homologous recombination mediated by lambda Red recombinase and a three-step procedure with 600-bp overhangs for recombination (for irmA, primers 5225/5226; for oxyR, primers 5227/5228) (see Table S5) (76–79). The irmA gene was amplified using primers 4350/4351 (see Table S5) and cloned into pSU2718 using XbaI/HindIII restriction sites to generate the plasmid pSU2718-irmA. Quantitative PCR was performed on cDNA generated from EC958 and EC958oxyR RNA using SYBR Green PCR master mix (Applied Biosystems) and a ViiA 7 real-time PCR system (Applied Biosystems). Relative transcript levels of irmA (primers 6623/6624; see Table S5) and agn43 (primers 6625/6626; see Table S5) were normalized against gapA (primers 0820/0821; see Table S5) as the endogenous gene control using the cycle threshold (2−ΔΔCT) method. 5′ RACE was performed using RNA isolated from EC958oxyR following the manufacturer’s specifications (Invitrogen).
Expression and purification of recombinant proteins.
The irmA gene was amplified (primers 2677/2678; see Table S5) and cloned into pMCSG7 using the ligation-independent method (80). The mature form of IrmA was expressed by autoinduction as a His-tagged recombinant soluble protein using BL21pLysS as previously described (81). Soluble recombinant IrmA protein was purified by nickel-chelate chromatography and treated with tobacco etch virus (TEV) protease to cleave the His tag. The protein was purified to homogeneity by a second round of nickel-chelate chromatography followed by gel filtration chromatography on a Superdex S75 column (GE Healthcare). Selenomethionine (SeMet)-labeled IrmA was expressed from E. coli BL21pLysS in minimal medium containing 50 µg/ml SeMet (l/d mixture) using a previously described method (82). Briefly, cultures were grown to exponential phase (OD600 of ~0.6) and induced with isopropyl β-d-thiogalactopyranoside (IPTG; final concentration of 0.5 mM). Cells were harvested at 4 h postinduction, and SeMet IrmA was purified following the same procedures as those described for the native protein. Recombinant Ag43 was obtained as previously described (25).
Generation of polyclonal antiserum and Western blotting.
A rabbit polyclonal antiserum was raised against purified IrmA using four immunizations (400-µg recombinant protein/dose) at the Walter and Eliza Hall Institute of Medical Research Antibody Facility. For immunoblotting, samples were subjected to SDS-PAGE using NuPAGE Novex 4% to 12% bis-Tris precast gels with NuPAGE MES (morpholineethanesulfonic acid) running buffer (Life Technologies) and were subsequently transferred to polyvinylidene difluoride (PVDF) microporous membrane filters using an iBlot dry blotting system as described by the manufacturer (Invitrogen). IrmA antiserum was used as primary serum (1:10,000 dilution), and the secondary antibody was alkaline phosphatase-conjugated anti-rabbit IgG. Sigma Fast BCIP/NBT (5-bromo-4-chloro-3-indolylphosphate/Nitro Blue Tetrazolium) was used as the substrate in the detection process.
Crystallization and structure determination.
Recombinant IrmA was concentrated to 10 mg/ml in 25 mM HEPES–NaOH (pH 6.7)–50 mM NaCl for crystallization experiments. IrmA crystals were obtained from 4 M potassium formate, 100 mM bis-Tris propane (pH 9.0), and 2% (wt/vol) polyethylene glycol (PEG) 2000; 2.5 M sodium malonate was added stepwise to the mother liquor as a cryoprotectant. Diffraction data for native and SeMet IrmA were collected at the UQ ROCX facility (using a Rigaku FR-E SuperBright X-ray generator and a Rigaku Saturn 944 charge-coupled-device [CCD] detector) and at the protein crystallography beamline (MX2 beamline 3ID1) at the Australian Synchrotron, respectively. The crystal-to-detector distances were 50 mm and 300 mm for native and SeMet crystals, respectively; 0.5° oscillation images were collected for a total of 360° for native IrmA and 720° in the case of SeMet IrmA. Diffraction data collected from native IrmA crystals were integrated and scaled with HKL2000 (83) and SeMet derivative data with iMosflm (84) and AIMLESS (85). The crystal structure of IrmA was solved using selenomethionine single-wavelength anomalous dispersion (SAD). Phase calculation, density modification, and initial model building were performed with Auto-Rickshaw (86) and the BUCCANEER program of the CCP4 suite (87). The structure of IrmA was completed by iterative cycles of model building and refinement using Coot (88) and phenix.Refine (89). Molecular figures were generated using PyMOL Molecular Graphics System v. 1.5.0.4 (Schrödinger, LLC). The crystallography, atomic coordinates, and structural features of IrmA have been deposited in the Protein Data Bank.
BN-PAGE.
BN-PAGE was performed using a Novex bis-Tris system (Life Technologies) following the manufacturer’s specifications. Briefly, precast NativePAGE Novex 4% to 16% (vol/vol) bis-Tris gels were run at pH 7.5, 4°C, using 150 V (1 h) followed by 250 V (40 min). Protein samples were prepared by mixing 5 µg of protein solution with NativePAGE sample buffer and water. NativeMark unstained protein standard (Invitrogen) was used to estimate the molecular weight of IrmA after native electrophoresis.
SV-AUC.
SV-AUC experiments were carried out using methods previously described (90) on a Beckman model XL-A analytical ultracentrifuge at a temperature of 20°C. Double-sector quartz cells were loaded with 320 µl of buffer (reference solution) and 300 µl of IrmA at initial concentrations of 0.96 mg/ml, 0.38 mg/ml, and 0.15 mg/ml in 25 mM HEPES–150 mM NaCl (pH 7.0). The cells were mounted in a Beckman 4-hole An60 Ti rotor. Absorbance data at 232 nm was collected at 129,000 × g in continuous mode using a step size of 0.003 cm without averaging. SEDNTERP (91) was used to calculate the partial specific volume of 0.7257 ml/g, solvent density of 1.005 g/ml, and solvent viscosity of 1.023 cp. The resulting sedimentation velocity absorbance data from multiple time points were fitted to a continuous size distribution model using the program SEDFIT (92) with a range of 0 to 10 S, a resolution value of 200, and a P value of 0.6. Frictional ratios (f/fo) and standardized sedimentation coefficients (s20,w) were calculated using SEDNTERP with the measured sedimentation coefficients.
Human plasma samples and ELISA methods.
Human plasma samples and UPEC strains were collected from 47 urosepsis patients at the time of admission to the Princess Alexandra Hospital (Brisbane, Australia). Blood and urine samples were also collected and cultured; in all cases, the blood and urine isolates gave identical results. Human plasma samples were also collected from 59 healthy age- and sex-matched volunteers with no recent history of UTI. Recombinant IrmA protein (10 µg/ml) was coated onto Nunc MaxiSorp flat-bottom 96-well plates in carbonate coating buffer (18 mM Na2CO3, 450 mM NaHCO3, pH 9.3) by incubation at 4°C overnight. Plates were then washed twice with 0.05% Tween 20–phosphate-buffered saline (PBS) (PBST) and blocked with 5% skim milk–PBST (150 µl) for 90 min at 37°C. Plates were washed four times with PBST, and then patient plasma samples were added to the wells at 1:10 dilution. Plates were incubated for 90 min at 37°C and washed four times with PBST. Peroxidase-conjugated anti-human IgG (1:30,000 dilution in 0.5% skim milk) was applied as a secondary antibody, and the reaction mixture was incubated for 90 min at 37°C. Plates were washed four times with PBST before development with 3,3′,5,5′-tetramethylbenzidine was performed. Reactions were stopped with 1 M hydrochloric acid. Streptococcus M1 protein was used as a positive control. The absorbance was measured at 450 nm using a SpectraMax 190 absorbance microplate reader. Statistical analysis was performed using an unpaired t test.
To determine the interaction between recombinant IrmA protein and human IL proteins, recombinant IL-2 (Invitrogen), IL-4 (Sino Biological), IL-10 (Invitrogen), and IL-17A (Invitrogen) proteins were resuspended according to the manufacturer’s specifications and coated onto Nunc MaxiSorp flat-bottom 96-well plates as previously described. After overnight incubation at 4°C, plates were washed twice with PBST and blocked with 5% skim milk–PBST (150 µl) for 90 min at 37°C. After incubation, plates were washed four times with PBST and then incubated with or without recombinant IrmA (100 µg/ml in 0.5% skim milk) for 90 min at 37°C. Plates were then washed four times with PBST and incubated with rabbit polyclonal antiserum raised against purified IrmA (1:50 dilution in 0.5% skim milk) for 90 min at 37°C. Plates were then washed four times with PBST, anti-rabbit IgG alkaline phosphatase (1:10,000 dilution in 0.5% skim milk) was applied as a secondary antibody, and the reaction mixture was incubated for 90 min at 37°C. Plates were washed four times with PBST before development was performed with alkaline phosphatase substrate (pNPP; Sigma). The absorbance was measured at 405 nm using a SpectraMax 190 absorbance microplate reader. Statistical analysis was performed using an unpaired t test. To test binding specificity of IrmA to IL-4, the same protocol was followed using the recombinant Ag43 as a control and rabbit polyclonal antiserum raised against purified Ag43, obtained as previously described (25).
Ethics statement.
This study was performed in accordance with the ethical standards of the University of Queensland, Princess Alexandra Hospital, Griffith University, and the Helsinki Declaration. The study was approved, and the need for informed consent was waived by the institutional review boards of the Princess Alexandra Hospital (research protocol 2008/264) and Griffith University (MSC/18/10/HREC).
Database deposition.
The crystallography, atomic coordinates, experimental data and structure factors of IrmA have been deposited in the Protein Data Bank (PDB ID code 5EK5).
SUPPLEMENTAL MATERIAL
Oligomerization state of IrmA. (A) SEC profile of recombinant IrmA (continuous line) and secreted endogenous IrmA (dashed line) on a Superdex-75 column (GE Healthcare). The two proteins eluted at approximately the same elution volume, with an apparent molecular mass of 30 kDa. (B) (Left panel) SDS polyacrylamide gel electrophoresis (SDS-PAGE) analysis of IrmA SEC samples. The fractions corresponding to native secreted (N) and recombinant (R) protein after SEC were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining. The recombinant sample consisted exclusively of IrmA, while the native sample contained minor high-molecular-mass contaminants. IrmA samples migrate with an apparent molecular mass of about 15 kDa (right panel). BN-PAGE analysis of IrmA SEC samples was performed, and both samples migrated at a molecular mass corresponding to dimer IrmA. (C) SV-AUC analysis of recombinant IrmA sample. Continuous standardized sedimentation coefficient distribution analysis of IrmA was performed at an initial protein concentration of 0.15 mg/ml. The resulting distribution shows that IrmA exists as a dimer (2.58 s20,w). The top panel shows the residuals resulting from the c(s) distribution best fits plotted as a function of radial position from the axis of rotation. Download
Comparison of IrmA with IL-10R1. The figure shows an α-carbon trace superposition of IrmA (shown in red) with the soluble domain of IL-10R1 (PDB 1J7V) (yellow). IL-10 bound to its cognate receptor is displayed in gray. A database search performed using the Dali server showed that IrmA shares structural similarity with different IL binding receptors, including IL-10R1 (PDB 1J7V) (6% sequence identity, Z score of 4.5, with 81 C atoms overlapping with an RMSD of 3 Å). Download
Binding of IrmA to IL-17A and IL-17A structural comparison. (A) ELISA showing the binding of IrmA to IL-4, but not to IL-17A, in the absence (C [control]) or presence (T [treated]) of recombinant IrmA. Statistical analysis was performed using an unpaired t test (****, P < 0.0001; ns, not significant). Error bars represent the standard deviations of results from six replicates. (B) Structural differences between IL-4 and IL-17A cytokines in complex with their cognate receptors (PDB 3BPL and 4HSA, respectively). Download
Reactivity of plasma from urosepsis patients and healthy control subjects. Blood plasma was collected from 30 IrmA-positive urosepsis patients at the time of admittance to hospital (T = 0) and at least 4 days postadmittance (t > 4). The presence of IgG-specific antibodies was determined by ELISA, and the data were compared to results obtained from 59 healthy volunteers with no recent history of UTI (column C). Statistical analysis was performed using an unpaired t test. P values of <0.01 are indicated (**). Download
Completely sequenced NCBI genomes used in this study.
Data collection and refinement statistics (SAD phasing).
Hydrodynamic properties of IrmA.
Bacterial strains and plasmids used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
We thank Matthew Perugini and Gordon King for expert technical advice, and Kate Peters for expert technical assistance. We acknowledge use of the Australian Synchrotron and the University of Queensland Remote Operation Crystallization and X-Ray Diffraction Facility (UQROCX).
Funding Statement
This work was supported by grants from the Australian National Health and Medical Research Council (NHMRC; APP1042651, APP1084889) and the Australian Research Council (ARC; DP150102287). BH and GCU are supported by ARC Future Fellowships (FT130100580 and FT110101048, respectively). SAB is supported by an NHMRC Career Development Fellowship (APP1090456) and MAS is supported by an NHMRC Senior Research Fellowship (APP1106930).
Footnotes
Citation Moriel DG, Heras B, Paxman JJ, Lo AW, Tan L, Sullivan MJ, Dando SJ, Beatson SA, Ulett GC, Schembri MA. 2016. Molecular and structural characterization of a novel Escherichia coli interleukin receptor mimic protein. mBio 7(2):e02046-15. doi:10.1128/mBio.02046-15.
REFERENCES
- 1.Foxman B, Barlow R, D’Arcy H, Gillespie B, Sobel JD. 2000. Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol 10:509–515. doi: 10.1016/S1047-2797(00)00072-7. [DOI] [PubMed] [Google Scholar]
- 2.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284 doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schappert SM, Rechtsteiner EA. 2011. Ambulatory medical care utilization estimates for 2007. Vital Health Stat 13 13:1–38. [PubMed] [Google Scholar]
- 4.Cohn EB, Schaeffer AJ. 2004. Urinary tract infections in adults. Scientificworldjournal 4(Suppl 1):76–88. doi: 10.1100/tsw.2004.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hooton TM. 2012. Clinical practice. Uncomplicated urinary tract infection. N Engl J Med 366:1028–1037. doi: 10.1056/NEJMcp1104429. [DOI] [PubMed] [Google Scholar]
- 6.Johansen TE, Botto H, Cek M, Grabe M, Tenke P, Wagenlehner FM, Naber KG. 2011. Critical review of current definitions of urinary tract infections and proposal of an EAU/ESIU classification system. Int J Antimicrob Agents 38(Suppl):64–70. doi: 10.1016/j.ijantimicag.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Foxman B. 2010. The epidemiology of urinary tract infection. Nat Rev Urol 7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 8.Totsika M, Beatson SA, Sarkar S, Phan MD, Petty NK, Bachmann N, Szubert M, Sidjabat HE, Paterson DL, Upton M, Schembri MA. 2011. Insights into a multidrug resistant Escherichia coli pathogen of the globally disseminated ST131 lineage: genome analysis and virulence mechanisms. PLoS One 6:e26578. doi: 10.1371/journal.pone.0026578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CDC 2013. Antibiotic resistance threats in the United States. CDC, Atlanta, GA. [Google Scholar]
- 13.Tadesse DA, Zhao S, Tong E, Ayers S, Singh A, Bartholomew MJ, McDermott PF. 2012. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg Infect Dis 18:741–749. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zilberberg MD, Shorr AF. 2013. Secular trends in gram-negative resistance among urinary tract infection hospitalizations in the United States, 2000–2009. Infect Control Hosp Epidemiol 34:940–946. doi: 10.1086/671740. [DOI] [PubMed] [Google Scholar]
- 15.Pitout JD. 2012. Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev Anti Infect Ther 10:1165–1176. doi: 10.1586/eri.12.110. [DOI] [PubMed] [Google Scholar]
- 16.WHO 2014. Antimicrobial resistance: global report on surveillance. WHO, Geneva, Switzerland. [Google Scholar]
- 17.Totsika M, Moriel DG, Idris A, Rogers BA, Wurpel DJ, Phan MD, Paterson DL, Schembri MA. 2012. Uropathogenic Escherichia coli mediated urinary tract infection. Curr Drug Targets 13:1386–1399. doi: 10.2174/138945012803530206. [DOI] [PubMed] [Google Scholar]
- 18.Brumbaugh AR, Mobley HL. 2012. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines 11:663–676. doi: 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moriel DG, Schembri MA. 2013. Vaccination approaches for the prevention of urinary tract infection. Curr Pharm Biotechnol 14:967–974. doi: 10.2174/1389201014666131226144824. [DOI] [PubMed] [Google Scholar]
- 20.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VA, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc Natl Acad Sci U S A 107:9072–9077. doi: 10.1073/pnas.0915077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd AL, Rasko DA, Mobley HL. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol 189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chouikha I, Germon P, Brée A, Gilot P, Moulin-Schouleur M, Schouler C. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J Bacteriol 188:977–987. doi: 10.1128/JB.188.3.977-987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diderichsen B. 1980. Flu, a metastable gene controlling surface properties of Escherichia coli. J Bacteriol 141:858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danese PN, Pratt LA, Dove SL, Kolter R. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol Microbiol 37:424–432. doi: 10.1046/j.1365-2958.2000.02008.x. [DOI] [PubMed] [Google Scholar]
- 25.Heras B, Totsika M, Peters KM, Paxman JJ, Gee CL, Jarrott RJ, Perugini MA, Whitten AE, Schembri MA. 2014. The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping. Proc Natl Acad Sci U S A 111:457–462. doi: 10.1073/pnas.1311592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klemm P, Hjerrild L, Gjermansen M, Schembri MA. 2004. Structure-function analysis of the self-recognizing antigen 43 autotransporter protein from Escherichia coli. Mol Microbiol 51:283–296. doi: 10.1046/j.1365-2958.2003.03833.x. [DOI] [PubMed] [Google Scholar]
- 28.Schembri MA, Klemm P. 2001. Coordinate gene regulation by fimbriae-induced signal transduction. EMBO J 20:3074–3081. doi: 10.1093/emboj/20.12.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson IR, Meehan M, Owen P. 1997. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv Exp Med Biol 412:349–355. doi: 10.1007/978-1-4899-1828-4_56. [DOI] [PubMed] [Google Scholar]
- 30.Henderson IR, Owen P. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J Bacteriol 181:2132–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol Microbiol 48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 32.Selkrig J, Mosbahi K, Webb CT, Belousoff MJ, Perry AJ, Wells TJ, Morris F, Leyton DL, Totsika M, Phan MD, Celik N, Kelly M, Oates C, Hartland EL, Robins-Browne RM, Ramarathinam SH, Purcell AW, Schembri MA, Strugnell RA, Henderson IR, Walker D, Lithgow T. 2012. Discovery of an archetypal protein transport system in bacterial outer membranes. Nat Struct Mol Biol 19:506–510. doi: 10.1038/nsmb.2261. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA. 2009. PDBsum new things. Nucleic Acids Res 37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Lupardus P, Laporte SL, Garcia KC. 2009. Structural biology of shared cytokine receptors. Annu Rev Immunol 27:29–60. doi: 10.1146/annurev.immunol.24.021605.090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haagmans W, van der Woude M. 2000. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol 35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- 37.Correnti J, Munster V, Chan T, Woude Mv. 2002. Dam-dependent phase variation of Ag43 in Escherichia coli is altered in a seqA mutant. Mol Microbiol 44:521–532. doi: 10.1046/j.1365-2958.2002.02918.x. [DOI] [PubMed] [Google Scholar]
- 38.Waldron DE, Owen P, Dorman CJ. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol Microbiol 44:509–520. doi: 10.1046/j.1365-2958.2002.02905.x. [DOI] [PubMed] [Google Scholar]
- 39.Schembri MA, Hjerrild L, Gjermansen M, Klemm P. 2003. Differential expression of the Escherichia coli autoaggregation factor antigen 43. J Bacteriol 185:2236–2242. doi: 10.1128/JB.185.7.2236-2242.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiang SM, Schellhorn HE. 2012. Regulators of oxidative stress response genes in Escherichia coli and their functional conservation in bacteria. Arch Biochem Biophys 525:161–169. doi: 10.1016/j.abb.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Christman MF, Morgan RW, Jacobson FS, Ames BN. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753–762. doi: 10.1016/S0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 42.Johnson JR, Clabots C, Rosen H. 2006. Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1:K1:H7 in a mouse model of ascending urinary tract infection. Infect Immun 74:461–468. doi: 10.1128/IAI.74.1.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson JR, Russo TA, Drawz SM, Clabots C, Olson R, Kuskowski MA, Rosen H. 2013. OxyR contributes to the virulence of a clonal group A Escherichia coli strain (O17:K+:H18) in animal models of urinary tract infection, subcutaneous infection, and systemic sepsis. Microb Pathog 64:1–5. doi: 10.1016/j.micpath.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 44.Schumacher MA, Chinnam N, Ohashi T, Shah RS, Erickson HP. 2013. The structure of irisin reveals a novel intersubunit beta-sheet fibronectin type III (FNIII) dimer: implications for receptor activation. J Biol Chem 288:33738–33744. doi: 10.1074/jbc.M113.516641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiefner A, Gebauer M, Skerra A. 2012. Extra-domain B in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein. J Biol Chem 287:17578–17588. doi: 10.1074/jbc.M111.303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rose RJ, Welsh TS, Waksman G, Ashcroft AE, Radford SE, Paci E. 2008. Donor-strand exchange in chaperone-assisted pilus assembly revealed in atomic detail by molecular dynamics. J Mol Biol 375:908–919. doi: 10.1016/j.jmb.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 47.Liu S, Song X, Chrunyk BA, Shanker S, Hoth LR, Marr ES, Griffor MC. 2013. Crystal structures of interleukin 17A and its complex with IL-17 receptor A. Nat Commun 4:1888. doi: 10.1038/ncomms2880. [DOI] [PubMed] [Google Scholar]
- 48.LaPorte SL, Juo ZS, Vaclavikova J, Colf LA, Qi X, Heller NM, Keegan AD, Garcia KC. 2008. Molecular and structural basis of cytokine receptor pleiotropy in the interleukin-4/13 system. Cell 132:259–272. doi: 10.1016/j.cell.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akdis M, Burgler S, Crameri R, Eiwegger T, Fujita H, Gomez E, Klunker S, Meyer N, O’Mahony L, Palomares O, Rhyner C, Ouaked N, Schaffartzik A, Van De Veen W, Zeller S, Zimmermann M, Akdis CA. 2011. Interleukins, from 1 to 37, and interferon-γ: receptors, functions, and roles in diseases. J Allergy Clin Immunol 127:701–721.e70. doi: 10.1016/j.jaci.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 50.Malek TR. 2008. The biology of interleukin-2. Annu Rev Immunol 26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 51.Liao W, Lin JX, Leonard WJ. 2013. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity 38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bao K, Reinhardt RL. 2015. The differential expression of IL-4 and IL-13 and its impact on type-2 immunity. Cytokine 75:25–37. doi: 10.1016/j.cyto.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wynn TA. 2015. Type 2 cytokines: mechanisms and therapeutic strategies. Nat Rev Immunol 15:271–282. doi: 10.1038/nri3831. [DOI] [PubMed] [Google Scholar]
- 54.Duell BL, Tan CK, Carey AJ, Wu F, Cripps AW, Ulett GC. 2012. Recent insights into microbial triggers of interleukin-10 production in the host and the impact on infectious disease pathogenesis. FEMS Immunol Med Microbiol 64:295–313. doi: 10.1111/j.1574-695X.2012.00931.x. [DOI] [PubMed] [Google Scholar]
- 55.Chan LL, Cheung BK, Li JC, Lau AS. 2010. A role for STAT3 and cathepsin S in IL-10 down-regulation of IFN-gamma-induced MHC class II molecule on primary human blood macrophages. J Leukoc Biol 88:303–311. doi: 10.1189/jlb.1009659. [DOI] [PubMed] [Google Scholar]
- 56.Taylor A, Akdis M, Joss A, Akkoç T, Wenig R, Colonna M, Daigle I, Flory E, Blaser K, Akdis CA. 2007. IL-10 inhibits CD28 and ICOS costimulations of T cells via Src homology 2 domain-containing protein tyrosine phosphatase 1. J Allergy Clin Immunol 120:76–83. doi: 10.1016/j.jaci.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Duell BL, Carey AJ, Tan CK, Cui X, Webb RI, Totsika M, Schembri MA, Derrington P, Irving-Rodgers H, Brooks AJ, Cripps AW, Crowley M, Ulett GC. 2012. Innate transcriptional networks activated in bladder in response to uropathogenic Escherichia coli drive diverse biological pathways and rapid synthesis of IL-10 for defense against bacterial urinary tract infection. J Immunol 188:781–792. doi: 10.4049/jimmunol.1101231. [DOI] [PubMed] [Google Scholar]
- 58.Duell BL, Carey AJ, Dando SJ, Schembri MA, Ulett GC. 2013. Human bladder uroepithelial cells synergize with monocytes to promote IL-10 synthesis and other cytokine responses to uropathogenic Escherichia coli. PLoS One 8:e78013. doi: 10.1371/journal.pone.0078013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kakkanat A, Totsika M, Schaale K, Duell BL, Lo AW, Phan MD, Moriel DG, Beatson SA, Sweet MJ, Ulett GC, Schembri MA. 2015. The role of H4 flagella in Escherichia coli ST131 virulence. Sci Rep 5:16149. doi: 10.1038/srep16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alcami A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol 3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 61.Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold ME, Schall TJ. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol 76:1285–1292. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc Natl Acad Sci U S A 97:1695–1700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jenkins C, Abendroth A, Slobedman B. 2004. A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J Virol 78:1440–1447. doi: 10.1128/JVI.78.3.1440-1447.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. 2002. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proc Natl Acad Sci U S A 99:9404–9409. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao Z, Fanslow WC, Seldin MF, Rousseau AM, Painter SL, Comeau MR, Cohen JI, Spriggs MK. 2011. Herpesvirus Saimiri encodes a new cytokine, IL-17, which binds to a novel cytokine receptor. J Immunol 187:4392–4402. doi: 10.1016/1074-7613(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 66.Boulanger MJ, Chow DC, Brevnova E, Martick M, Sandford G, Nicholas J, Garcia KC. 2004. Molecular mechanisms for viral mimicry of a human cytokine: activation of gp130 by HHV-8 interleukin-6. J Mol Biol 335:641–654. doi: 10.1016/j.jmb.2003.10.070. [DOI] [PubMed] [Google Scholar]
- 67.Alcamí A, Smith GL. 1992. A soluble receptor for interleukin-1beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell 71:153–167. doi: 10.1016/0092-8674(92)90274-G. [DOI] [PubMed] [Google Scholar]
- 68.Spriggs MK, Hruby DE, Maliszewski CR, Pickup DJ, Sims JE, Buller RM, VanSlyke J. 1992. Vaccinia and cowpox viruses encode a novel secreted interleukin-1-binding protein. Cell 71:145–152. doi: 10.1016/0092-8674(92)90273-F. [DOI] [PubMed] [Google Scholar]
- 69.Upton C, Mossman K, McFadden G. 1992. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science 258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 70.Symons JA, Alcamí A, Smith GL. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 71.Colamonici OR, Domanski P, Sweitzer SM, Larner A, Buller RM. 1995. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem 270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- 72.Xiang Y, Moss B. 1999. IL-18 binding and inhibition of interferon gamma induction by human poxvirus-encoded proteins. Proc Natl Acad Sci U S A 96:11537–11542. doi: 10.1073/pnas.96.20.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith VP, Bryant NA, Alcamí A. 2000. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. J Gen Virol 81:1223–1230. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- 74.Born TL, Morrison LA, Esteban DJ, VandenBos T, Thebeau LG, Chen N, Spriggs MK, Sims JE, Buller RM. 2000. A poxvirus protein that binds to and inactivates IL-18, and inhibits NK cell response. J Immunol 164:3246–3254. doi: 10.4049/jimmunol.164.6.3246. [DOI] [PubMed] [Google Scholar]
- 75.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watts RE, Totsika M, Challinor VL, Mabbett AN, Ulett GC, De Voss JJ, Schembri MA. 2012. Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect Immun 80:333–344. doi: 10.1128/IAI.05594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect Immun 78:1659–1669. doi: 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo JM, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect Immun 80:321–332. doi: 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Donnelly MI, Zhou M, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. 2006. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif 47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif 41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 82.Heras B, Edeling MA, Schirra HJ, Raina S, Martin JL. 2004. Crystal structures of the DsbG disulfide isomerase reveal an unstable disulfide. Proc Natl Acad Sci U S A 101:8876–8881. doi: 10.1073/pnas.0402769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 84.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Panjikar S, Parthasarathy V, Lamzin VS, Weiss MS, Tucker PA. 2005. Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr D Biol Crystallogr 61:449–457. doi: 10.1107/S0907444905001307. [DOI] [PubMed] [Google Scholar]
- 87.Cowtan K. 2006. The buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 88.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 89.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 90.Burgess BR, Dobson RC, Bailey MF, Atkinson SC, Griffin MD, Jameson GB, Parker MW, Gerrard JA, Perugini MA. 2008. Structure and evolution of a novel dimeric enzyme from a clinically important bacterial pathogen. J Biol Chem 283:27598–27603. doi: 10.1074/jbc.M804231200. [DOI] [PubMed] [Google Scholar]
- 91.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. 1992. Computer-aided interpretation of analytical sedimentation data for proteins, p 90–125. In Harding SE (ed), Analytical ultracentrifugation in biochemistry and polymer science. The Royal Society of Chemistry, Cambridge, United Kingdom. [Google Scholar]
- 92.Brown PH, Schuck P. 2008. A new adaptive grid-size algorithm for the simulation of sedimentation velocity profiles in analytical ultracentrifugation. Comput Phys Commun 178:105–120. doi: 10.1016/j.cpc.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. BioInformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Oligomerization state of IrmA. (A) SEC profile of recombinant IrmA (continuous line) and secreted endogenous IrmA (dashed line) on a Superdex-75 column (GE Healthcare). The two proteins eluted at approximately the same elution volume, with an apparent molecular mass of 30 kDa. (B) (Left panel) SDS polyacrylamide gel electrophoresis (SDS-PAGE) analysis of IrmA SEC samples. The fractions corresponding to native secreted (N) and recombinant (R) protein after SEC were analyzed by SDS-PAGE followed by Coomassie brilliant blue staining. The recombinant sample consisted exclusively of IrmA, while the native sample contained minor high-molecular-mass contaminants. IrmA samples migrate with an apparent molecular mass of about 15 kDa (right panel). BN-PAGE analysis of IrmA SEC samples was performed, and both samples migrated at a molecular mass corresponding to dimer IrmA. (C) SV-AUC analysis of recombinant IrmA sample. Continuous standardized sedimentation coefficient distribution analysis of IrmA was performed at an initial protein concentration of 0.15 mg/ml. The resulting distribution shows that IrmA exists as a dimer (2.58 s20,w). The top panel shows the residuals resulting from the c(s) distribution best fits plotted as a function of radial position from the axis of rotation. Download
Comparison of IrmA with IL-10R1. The figure shows an α-carbon trace superposition of IrmA (shown in red) with the soluble domain of IL-10R1 (PDB 1J7V) (yellow). IL-10 bound to its cognate receptor is displayed in gray. A database search performed using the Dali server showed that IrmA shares structural similarity with different IL binding receptors, including IL-10R1 (PDB 1J7V) (6% sequence identity, Z score of 4.5, with 81 C atoms overlapping with an RMSD of 3 Å). Download
Binding of IrmA to IL-17A and IL-17A structural comparison. (A) ELISA showing the binding of IrmA to IL-4, but not to IL-17A, in the absence (C [control]) or presence (T [treated]) of recombinant IrmA. Statistical analysis was performed using an unpaired t test (****, P < 0.0001; ns, not significant). Error bars represent the standard deviations of results from six replicates. (B) Structural differences between IL-4 and IL-17A cytokines in complex with their cognate receptors (PDB 3BPL and 4HSA, respectively). Download
Reactivity of plasma from urosepsis patients and healthy control subjects. Blood plasma was collected from 30 IrmA-positive urosepsis patients at the time of admittance to hospital (T = 0) and at least 4 days postadmittance (t > 4). The presence of IgG-specific antibodies was determined by ELISA, and the data were compared to results obtained from 59 healthy volunteers with no recent history of UTI (column C). Statistical analysis was performed using an unpaired t test. P values of <0.01 are indicated (**). Download
Completely sequenced NCBI genomes used in this study.
Data collection and refinement statistics (SAD phasing).
Hydrodynamic properties of IrmA.
Bacterial strains and plasmids used in this study.
Primers used in this study.