ABSTRACT
An outbreak of cholera occurred in 1991 in Mexico, where it had not been reported for more than a century and is now endemic. Vibrio cholerae O1 prototype El Tor and classical strains coexist with altered El Tor strains (1991 to 1997). Nontoxigenic (CTX−) V. cholerae El Tor dominated toxigenic (CTX+) strains (2001 to 2003), but V. cholerae CTX+ variant El Tor was isolated during 2004 to 2008, outcompeting CTX− V. cholerae. Genomes of six Mexican V. cholerae O1 strains isolated during 1991 to 2008 were sequenced and compared with both contemporary and archived strains of V. cholerae. Three were CTX+ El Tor, two were CTX− El Tor, and the remaining strain was a CTX+ classical isolate. Whole-genome sequence analysis showed the six isolates belonged to five distinct phylogenetic clades. One CTX− isolate is ancestral to the 6th and 7th pandemic CTX+ V. cholerae isolates. The other CTX− isolate joined with CTX− non-O1/O139 isolates from Haiti and seroconverted O1 isolates from Brazil and Amazonia. One CTX+ isolate was phylogenetically placed with the sixth pandemic classical clade and the V. cholerae O395 classical reference strain. Two CTX+ El Tor isolates possessing intact Vibrio seventh pandemic island II (VSP-II) are related to hybrid El Tor isolates from Mozambique and Bangladesh. The third CTX+ El Tor isolate contained West African-South American (WASA) recombination in VSP-II and showed relatedness to isolates from Peru and Brazil. Except for one isolate, all Mexican isolates lack SXT/R391 integrative conjugative elements (ICEs) and sensitivity to selected antibiotics, with one isolate resistant to streptomycin. No isolates were related to contemporary isolates from Asia, Africa, or Haiti, indicating phylogenetic diversity.
IMPORTANCE
Sequencing of genomes of V. cholerae is critical if genetic changes occurring over time in the circulating population of an area of endemicity are to be understood. Although cholera outbreaks occurred rarely in Mexico prior to the 1990s, genetically diverse V. cholerae O1 strains were isolated between 1991 and 2008. Despite the lack of strong evidence, the notion that cholera was transmitted from Africa to Latin America has been proposed in the literature. In this study, we have applied whole-genome sequence analysis to a set of 124 V. cholerae strains, including six Mexican isolates, to determine their phylogenetic relationships. Phylogenetic analysis indicated the six V. cholerae O1 isolates belong to five phylogenetic clades: i.e., basal, nontoxigenic, classical, El Tor, and hybrid El Tor. Thus, the results of phylogenetic analysis, coupled with CTXϕ array and antibiotic susceptibility, do not support single-source transmission of cholera to Mexico from African countries. The association of indigenous populations of V. cholerae that has been observed in this study suggests it plays a significant role in the dynamics of cholera in Mexico.
INTRODUCTION
Cholera, a deadly waterborne disease, is caused by Vibrio cholerae and continues to be a health hazard for millions around the world, particularly in developing countries. Of more than 200 “O” serogroups, only V. cholerae O1 and O139 have been associated with cholera epidemics. Serogroup O1 has been classified into two biotypes, classical (CL) and El Tor (ET), the latter linked to the ongoing 7th pandemic first reported in 1961 (1, 2). Although cholera has been endemic in the Ganges Delta region of South Asia for centuries, several countries of sub-Saharan Africa and Latin America were severely affected during the 7th pandemic and subsequently are now considered areas of endemicity (1). That is, cholera appeared in Mexico in June 1991, after the Latin American epidemic had begun along the Peruvian coast in January 1991 (3). The disease soon broke out in neighboring countries by 1992, with the exceptions of Uruguay and French Guyana (4). In Mexico, a total of 43,536 cholera cases were reported between 1991 and 1996, with a substantial number of deaths (3). Epidemiological investigations confirmed the association of V. cholerae O1 biotype El Tor with the majority of those cholera cases, although the classical biotype was isolated from some cases in Mexico during subsequent years until 1997 (4–7).
It has long been established that V. cholerae O1 had caused seven pandemics since 1817, of which the 7th pandemic is the largest, considering its longevity and geographical distribution. V. cholerae El Tor replaced the classical biotype of the 6th pandemic and presumably earlier pandemics (1, 8). Variants of El Tor (hybrid El Tor and/or altered El Tor) possessing classical biotype-specific traits have been reported in Asia, Africa, and Latin America (5, 9, 10). Genetic changes (i.e., gain or loss of mobile genetic elements and genomic islands) occur in V. cholerae due to its genomic plasticity (11). An example is the emergence of V. cholerae O139 in late 1992 in India, which is a non-O1 serogroup that caused a massive outbreak in South Asia and beyond (12, 13). Since 2001, variants of El Tor have been associated with cholera epidemics globally, including the recent epidemic in Haiti and previously Zimbabwe (14–16). Although significant advances have been made in the understanding of the genetics, epidemiology, and ecology of V. cholerae over the past two decades, the lack of an extensive genomic database severely limits source attribution for some of the recent outbreaks.
The cholera epidemic in Latin America was hypothesized to have been imported from areas of endemicity since Latin America had not reported cholera for more than 100 years prior to 1991 (17). Three hypotheses have been offered: (i) international trade ships from Asia discharged the pathogen into Peruvian ports in ballast water (18), (ii) immigrants who came from Africa to Latin America in the 1970s brought the pathogen with them (6, 19), and (iii) environmental factors (e.g., El Nino) played a significant role (20, 21). Preliminary analysis using molecular typing indicated V. cholerae strains isolated in Latin America during the 1990s’ epidemic were clonal and represented intrusion of the seventh pandemic El Tor strain into the Western hemisphere (unrelated to the U.S. Gulf Coast clone) (6). However, subsequent genomic analysis of 30 single-nucleotide polymorphisms (SNPs) indicated close relatedness of the Latin American isolates from the 1990s to African strains isolated in the 1970s and 1990s (19). This finding was supported by a recent phylogenetic analysis showing isolates from the Latin American epidemic in the 1990s were related to a V. cholerae strain from Angola, the study that analyzed only seventh pandemic El Tor strains from the Latin American epidemic that carried the ctxB3 genotype (B3 allele) (8). However, V. cholerae altered El Tor has been found to coexist with classical and prototype El Tor in Mexico since the Latin American epidemic began (5). A serious limitation of that retrospective epidemiological study was that the analysis included only a limited number of strains collected spatio-temporally, thereby masking the full genetic diversity of the Mexican V. cholerae population. Phenotypic and genotypic characteristics of 182 V. cholerae O1 strains from Mexico that had been isolated between 1983 and 2008 previously had been reported to have several unique features (5, 7, 22) (see Table S1 in the supplemental material). In this study, six V. cholerae O1 isolates from Mexico were selected (Table 1) based on previously published data (5, 7, 22), for whole-genome sequencing to compare their genomes with genomes of 124 V. cholerae archival and recent isolates to elucidate the evolutionary dynamics of V. cholerae in Mexico.
TABLE 1 .
Characteristics of Vibrio cholerae serogroup O1 strains analyzed in this studya
| Strain | Serotype | Biotype | Source | Yr of isolation |
CTXΦ | Accession no. |
|---|---|---|---|---|---|---|
| CP1032 | Ogawa | El Tor | Human | 1991 | + | ALDA00000000 |
| 95412 | Inaba | Classical | Human | 1997 | + | APFM00000000 |
| CP1033 | Ogawa | El Tor | Human | 2000 | + | AJRL00000000 |
| CP1037 | Ogawa | El Tor | Environment | 2003 | − | ALDB00000000 |
| CP1035 | Ogawa | El Tor | Human | 2004 | − | AJRM00000000 |
| CP1030 | Inaba | El Tor | Environment | 2008 | + | ALCZ00000000 |
Mexico was the country of origin for all strains shown here.
RESULTS AND DISCUSSION
Variations in CTXΦ-RS1.
Four of the six isolates of V. cholerae O1 (95412, CP1030, CP1032, and CP1033) were lysogenic CTXΦ positive, while the remaining two isolates (CP1035 and CP1037) lacked CTXΦ (Table 1). Lysogenic CTXΦ contains two gene clusters, a core region and and RS2 element (23, 24). The core region comprises ctxAB, encoding cholera toxin (CT), and five other genes, namely, psh, cep, orfU, ace, and zot, that are required for phage morphogenesis. The RS2 element encodes proteins associated with CTXΦ replication (RstA), integration (RstB), and regulation (RstR) (23, 24). Satellite phage RS1 carries an additional rstC gene (encoding anti-repressor protein), along with the entire RS2 element that is usually present in the flanking region of CTXΦ in V. cholerae El Tor (24). The chromosomal location of CTXΦ and its orientation and copies of CTXΦ may differ among toxigenic V. cholerae strains (25–27). The CTXΦ-RS1 array of CP1030 has been shown to be unique, lacking RS1 and carrying a truncated CTXΦ instead of RS1 in the upstream region of CTXΦ (B3 allele) in the large chromosome (Chr I) (7). The V. cholerae O1 El Tor strains isolated in Mexico between 2004 and 2008, show the same CTXΦ array (TLC-truncated CTX-CTXΦB3) (7). Moreover, predicted CTXΦ mapping of El Tor isolates associated with the 1990s’ Latin American epidemic in Peru, Mexico, Bolivia, Columbia, and Argentina showed two copies of CTXΦ (B3 allele) together with TLC and RS1 in Chr I (TLC-CTXΦB3-CTXΦB3-RS1) (8). CTXΦ arrays, either TLC-truncated CTX-CTXΦB3, or TLC-CTXΦB3-CTXΦB3-RS1 detected in Latin American isolates was not found in El Tor, altered El Tor, or El Tor variants from Asia, Africa, and Haiti that have been studied to date (8, 25–27). However, an isolate from Sweden was found to contain the latter. Recently, genomic analysis of V. cholerae O1 showed close relatedness between isolates from Latin America and Angola, but the CTXΦ array was different (8).
As shown in Table 2, the rstA and rstB gene sequence of V. cholerae 95412 classical is identical to that of the reference V. cholerae O395 classical isolate, whereas variation was observed in V. cholerae CTX+ El Tor isolates. V. cholerae CP1030, CP1032, and CP1033 contained three unique base substitutions in the rstA gene at 927 (T→C), 933 (C→T), and 942 (G→T), compared to V. cholerae N16961, CIRS101, and the recent Haitian isolate HCO1. In addition, CP1032 had a base substitution at 315 (T→C) in the rstA gene. Interestingly, all point mutations are synonymous for RstA. DNA sequence analysis of CP1030, CP1032, and CP1033 at the rstB gene showed a GTA deletion at positions 77 to 79 and polymorphism at positions 90 (A→T), 96 (T→C), 108 (G→A), and 192 (A→G), unlike V. cholerae El Tor strains, except or the GTA deletion, which had been reported in Haitian isolates (14).
TABLE 2 .
Sites of nucleotide polymorphisms in CTX prophages
| Strain | Country of origin | Yr of isolation | Gene position | rstR type |
rstA polymorphism at position: |
rstB polymorphism at position: |
No. of copies of heptamer in zot-ctxA regiona | ctxB allele type | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 162 | 183 | 258 | 315 | 345 | 516 | 540 | 579 | 609 | 774 | 927 | 933 | 942 | 77–79 | 90 | 96 | 108 | 192 | 288 | 291 | 363 | |||||||
| N16961 | Bangladesh | 1975 | CTXET | ET | C | C | C | G | T | G | G | A | T | T | C | T | C | G | GTA | A | T | G | A | A | C | A | 4 | B3 |
| O395 | India | 1965 | CTXCL | CL | T | T | A | C | * | T | A | G | C | C | T | * | * | * | −c | T | C | * | * | G | T | * | 7 | B1 |
| CIRS101 | Bangladesh | 2002 | CTXHYB | ET | *b | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 3 | B1 |
| HCO1 | Haiti | 2010 | CTXHYB | ET | * | * | * | * | * | * | * | * | * | * | * | * | * | * | − | * | * | * | * | * | * | * | 5 | B1 |
| 95412 | Mexico | 1997 | CTXCL | CL | T | T | A | C | * | T | A | G | C | C | T | * | * | * | − | T | C | * | * | G | T | * | 6 | B1 |
| CP1030 | Mexico | 2008 | CTXET | ET | * | * | * | * | * | * | * | * | * | * | * | C | T | T | − | T | C | A | * | * | * | * | 4 | B3 |
| CP1032 | Mexico | 1991 | CTXHYB | ET and CL | * | * | * | * | C | * | * | * | * | * | * | C | T | T | − | T | C | A | * | * | * | * | 4 | B1 |
| CP1033 | Mexico | 2000 | CTXHYB | ET and CL | * | * | * | * | * | * | * | * | * | * | * | C | T | T | − | T | C | A | G | * | * | G | 4 | B1 |
Shown are the numbers of copies of the TTTTGAT heptamer repeat sequence.
*, indicates sequence identical to that of V. cholerae N16961.
−, GTA deletion.
Virulence gene expression in V. cholerae is regulated by ToxR, a transcriptional regulator that binds with the promoter region (between zot and ctxA) located upstream of ctxAB. The heptamer repeat sequences (TTTTGAT) directly influence the affinity of ToxR binding and promote binding of ToxR, which is followed by activation of the ctxAB promoter (28). As shown in Table 2, V. cholerae CP1030, CP1032, and CP1033 contain four copies of the heptamer repeat, like El Tor, altered El Tor, and hybrid variants from Asia and Africa. However, they differ from the Haitian isolates in having five repeats (14, 29). The V. cholerae 95412 classical isolate contains six copies of the heptamer repeat, unlike the classical V. cholerae reference strain O395, which possesses seven copies of the repeat (Table 2).
Vibrio pathogenicity islands 1 and 2.
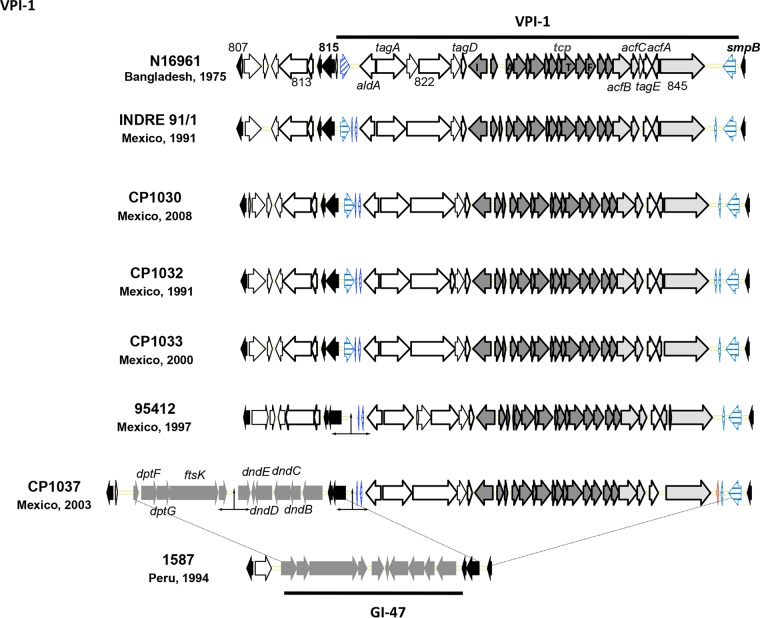
Vibrio pathogenicity island-1 (VPI-1) encodes the toxin-coregulated pilus (TCP) that promotes colonization of intestinal mucosal epithelium, is involved in biofilm formation, and serves as the receptor for the lysogenic CTXΦ (30). Five of the six V. cholerae O1 isolates from Mexico contained VPI-1, but CP1035 lacked this gene cluster. As shown in Fig. 1, V. cholerae CP1030, CP1032, and CP1033 possess VPI-1 of the seventh pandemic V. cholerae El Tor isolates, whereas the genetic organization of VPI-1of CP1037 is homologous to that of V. cholerae 95412 (classical), despite having a genomic island, GI-47, in the upstream region. Interestingly, the tcpA gene, encoding the major pilin subunit (TcpA) of CP1037, is different from the classical and El Tor tcpA genes. The TCP region showed highest level of sequence polymorphism in VPI-1, with tcpA having the most divergence (31). Previous studies reported TcpA had significant differences in the epitope or antigenic structure when classical and El Tor biotype strains were compared (32). Four of the V. cholerae O1 isolates, CP1030, CP1032, CP1033, and 95412, contain the complete VPI-2, whereas the other isolates lack VPI-2. VPI-2 comprises several genes, including those encoding sialidase, the type I restriction modification system, and Mu-like prophage protein genes.
FIG 1 .
Vibrio pathogenicity island 1 (VPI-1) of V. cholerae O1 strains isolated in Mexico and reference El Tor strain N16961. Mexican CTX− V. cholerae O1 strain CP1037 contains GI-47 in the upstream region of VPI-1.
Vibrio seventh pandemic islands.
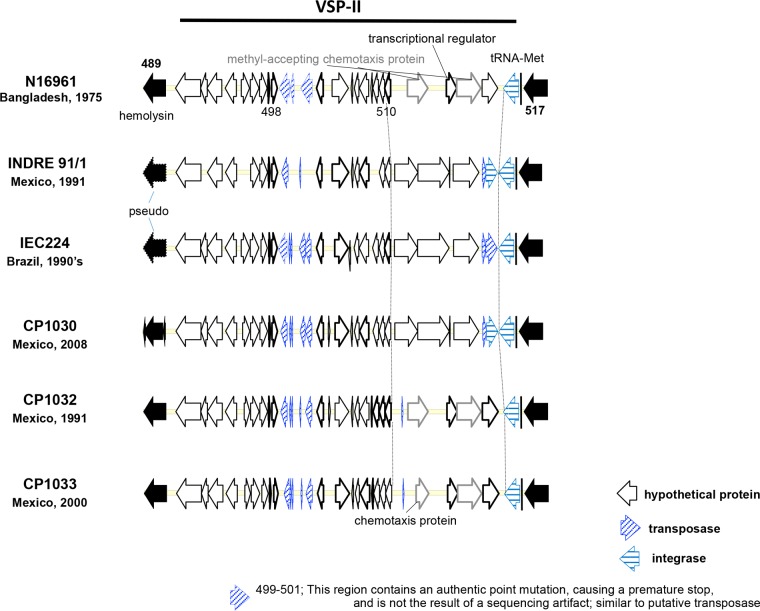
The Vibrio seventh pandemic islands I and II (VSP-I and -II) in V. cholerae are characteristically found in El Tor strains, and they serve as a distinguishing marker from classical strains (33). However, a variant of the VSP-II gene cluster has also been detected in V. cholerae non-O1/O139 strains and in Vibrio mimicus (34, 35). V. cholerae El Tor strains CP1032 and CP1033 from Mexico contained all of the open reading frames (ORFs) in VSP-I and -II, whereas the CTX− isolates CP1035 and CP1037 lack VSP-I and -II, as does the classical strain 95412. CP1030 possesses a variant VSP-II with an insertion between VC0510 and VC0516 (Fig. 2), commonly referred to as the West African and South American (WASA) insertion (8). An identical VSP-II gene cluster has been reported in V. cholerae isolated in Peru and Angola (36, 37). Conversely, the VSP-II gene cluster in contemporary V. cholerae isolates from Asia and Haiti has a 14.4-kb deletion that spans the ORF from VC0495 to VC0512 (CIRS101 type VSP-II) (14, 35, 38). The distribution of the variant VSP-II types among the V. cholerae isolates suggests this island contains hot spots highly prone to genetic rearrangement by recombination (35).
FIG 2 .
Vibrio seventh pandemic island II (VSP-II) of V. cholerae O1 strains isolated in Mexico and Brazil. VSP-II of hybrid V. cholerae O1 El Tor (CP1032 and CP1033) was similar to that of reference El Tor N16961. However, it is different from those of prototype El Tor isolates (CP1030, ICE224, and INDRE 91/1) in Mexico and Brazil.
GIs and ICEs.
V. cholerae O1 isolates from Mexico contain diverse genomic islands (GIs) that differ among the El Tor, classical, and CTX− strains (see Table S2 in the supplemental material). V. cholerae El Tor isolates CP1030, CP1032, and CP1033 uniformly contained GI-1 to GI-10 and GI-85. V. cholerae CP1033 (14), on the other hand, contains GI-15 in the large chromosome, which encodes the putative integrase found in the Mozambique variant of V. cholerae (B-33) and also in hybrid isolates of CP1067 from Bangladesh, that had been isolated in 1991. Moreover, V. cholerae CP1030 contains the WASA1 genomic island, which has been reported previously in West African and South American strains (8). V. cholerae classical strain 95412 has GIs typical of the reference classical strain O395, along with GI-11 and GI-21 in the small chromosome (see Table S2). GI-11 encodes the kappa prophage, whereas the function of GI-21 (~34 kb) has not yet been identified. V. cholerae CP1035 contains genomic islands that are similar to those of V. cholerae non-O1/O139 and differ from classical and El Tor strains. CP1035 contains several previously described genomic islands, including GI-125 and GI-126, encoding a type I restriction modification system and integrase. Interestingly, CP1037 carries GI-36, which has been detected previously in V. cholerae non-O1/O139 TM11079-80 and Amazonia, isolated in Brazil. CP1037 also possesses GI-47 in the upstream region of VPI-1, as previously observed in Peruvian V. cholerae isolated in 1994 (Fig. 1) and a unique genomic island, GI-112, carrying umuCD and a nucleotidyltransferase gene (see Table S2) (11, 14).
The integrating and conjugative elements (ICEs) are self-transmissible mobile genetic elements in bacteria that confer resistance to various antibiotics. SXT is an ~100-kb ICE originally discovered in V. cholerae O139 (39). Since the emergence of V. cholerae O139 on the Indian subcontinent in 1992, the SXT/R391 ICE has been reported to be present in most clinical V. cholerae O1 or O139 strains isolated in Asia and Africa (8). V. cholerae isolates carrying the SXT/R391 ICE are resistant to streptomycin, chloramphenicol, sulfamethoxazole, and trimethoprim (39). Results of recent phylogenetic analysis suggest V. cholerae O1 acquired SXT/R391 ICE sometime between 1978 and 1984, before its discovery in V. cholerae O139, and it is hypothesized that it provides a selective advantage to V. cholerae O1, allowing it to be globally disseminated (8). In the present study, except for CP1035, all of the Mexican isolates lacked the SXT/R391 ICE. The genome sequences of Latin American isolates (INDRE 91/1 [Mexico]; CP1044, CP1046, and CP1047 [Peru]; and IEC224, RC144, and 116059 [Brazil]) are devoid of SXT/R391 ICE. This observation was confirmed by PCR—i.e., except for CP1035, none of the Mexican isolates amplified DNA fragments for primers targeting the SXT integrase gene (intSXT) (40). Lack of the SXT/R391 ICE in epidemic strains isolated in Latin America in the 1990s has been reported (8). Absence of the SXT/R391 ICE among V. cholerae isolates has also been reported in a recent cholera outbreak in the Philippines (41). Antibiotic susceptibility analyses of the five Mexican isolates, CP1030, CP1032, CP1035, CP1037, and 95412, revealed all were sensitive to penicillin, ampicillin, streptomycin, chloramphenicol, trimethoprim-sulfamethoxazole, tetracycline, kanamycin, erythromycin, nalidixic acid, and ciprofloxacin. V. cholerae CP1033 shows resistance only to streptomycin. Despite possessing SXT/R391 ICE, CP1035 was sensitive to antibiotics, suggesting SXT/R391 ICE lacks genes conferring resistance to streptomycin, chloramphenicol, and trimethoprim-sulfamethoxazole. However, V. cholerae O1 strains showed resistance to different antibiotics in Asia and Africa at least a decade earlier than the 1990s’ Latin American epidemic. V. cholerae El Tor strains isolated in 1977 in Africa were resistant to multiple drugs, including tetracycline (42), and classical strains from Bangladesh isolated during 1982 to 1989 were resistant to ampicillin, furazolidone, and trimethoprim-sulfamethoxazole (22).
LPS coding region.
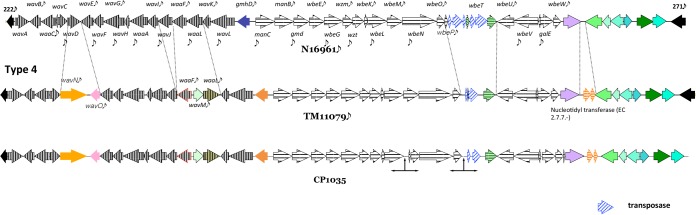
The lipopolysaccharide (LPS) of V. cholerae is comprised of three main regions: lipid A, the core oligosaccharide (OS), and the O antigen. V. cholerae synthesizes the core OS and O antigen using the wav and wb* gene clusters, respectively (43). The wav gene cluster (VC0223 to -240) of the Mexican isolates is similar to that of V. cholerae N16961, except for CP1035, which is different in seven of the ORFs (Fig. 3). V. cholerae CP1035 has a wav gene cluster homologous to V. cholerae TM11079-80, an environmental strain isolated in Brazil in 1980 (Fig. 3). Interestingly, both strains are phenotypically El Tor, but they lack two major virulence-associated genomic islands, CTXΦ encoding CtxAB and Vibrio pathogenicity island VPI-1, which contains the genes for biosynthesis of toxin-coregulated pilus (TCP).
FIG 3 .
O antigen biosynthetic genes of V. cholerae O1 strains CP1035, TM11079, and N16961. The wav and wb* gene clusters of CP1035 are homologous to those of TM11079 and different from those of reference El Tor N16961.
Phylogenetics of the Mexican isolates.
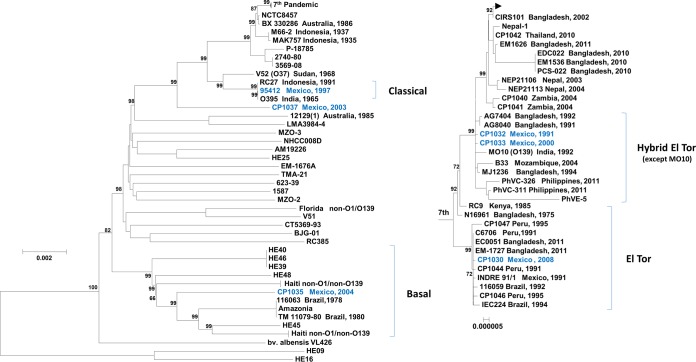
The phylogeny of the V. cholerae strains isolated in Mexico was determined by constructing a genome-relatedness neighbor-joining tree using homologous alignment of 905 orthologous protein-coding genes (~897,461 bp) of 124 V. cholerae genomes (Fig. 4), which placed El Tor, classical, and nontoxigenic V. cholerae isolates from Mexico into distinct phylogenetic clades. CP1035, a CTX− isolate, was placed into a basal clade with other nontoxigenic non-O1/O139 isolates from Haiti and O1 isolates from Brazil and Amazonia. The other CTX− isolate, V. cholerae CP1037, was phylogenetically placed into an independent node ancestral to all sixth and seventh pandemic isolates. The presence of ancestral isolate in the Latin American region is indicative of greater phylogenetic diversity and succession of indigenous V. cholerae populations in that ecosystem. The classical biotype isolate of V. cholerae 95412, isolated from Mexico in 1997, was placed into a monophyletic clade with the other sixth pandemic reference V. cholerae strain, O395, and RC27. Classical biotype strains are considered to have been outcompeted by seventh pandemic V. cholerae El Tor strains in the 1980s and have not been isolated in Asia and Africa after 1990 (22). In contrast, V. cholerae classical strains had been isolated in Mexico until 1997, even though V. cholerae El Tor strains were dominant at the beginning of the Latin American epidemic and during the years following, indicating the Mexican ecosystem to be a reservoir for the classical biotype of V. cholerae (5).
FIG 4 .
Neighbor-joining trees showing phylogenetic relationships of 124 V. cholerae genomes based on 905 orthologs of protein-coding genes (~897,461 bp). The two V. cholerae non-O1/O139 strains (HE09 and HE16) isolated from surface water during the 2010 cholera epidemic in Haiti were used as an outgroup of the tree, and bootstrap values are percentages of 1,000 replications. Mexican V. cholerae O1 strains are shown in blue, indicating the distribution among five distinct phylogenetic clades. The top node represents the genomes of isolates from Haiti, Bangladesh, Nepal, the United States, Cameroon, South Africa, the Russian Federation, Zimbabwe, and the Dominican Republic obtained between 2005 and 2011.
V. cholerae strains CP1032 and CP1033 isolated in Mexico were placed into the paraphyletic hybrid El Tor clade along with Mozambique and Matlab variants of V. cholerae El Tor, namely, B-33 and MJ-1236, together with V. cholerae O139 isolate MO10 (Fig. 4). These isolates also showed close relatedness to 1991 hybrid V. cholerae El Tor strain AG8040 isolated from patients in Bangladesh. Phylogenetic analysis of V. cholerae hybrid strains isolated in Mexico clearly shows a separation from contemporary V. cholerae El Tor and altered El Tor strains from Asia, Africa, and Haiti. The Matlab variant strains, isolated in 1994 in Bangladesh, were the first to have been reported in the literature as “hybrid,” showing classical biotype specific traits in an El Tor genetic background (9). A decade later, genetically similar hybrid variants were isolated in Mozambique during the 2004 cholera outbreak (11, 44). Isolation of V. cholerae CP1032 in 1991 in Mexico suggests hybrid El Tor V. cholerae was present at the same time in two different continents—i.e., Asia and America. V. cholerae CP1030 also belongs to the seventh pandemic clade. However, it clustered tightly into the monophyletic El Tor clade with V. cholerae strains isolated in Mexico, Peru, and Brazil during the Latin American epidemics of the 1990s but distant from recent isolates from Bangladesh, India, Nepal, and Thailand. Furthermore, Zambia, Zimbabwe, and Haiti isolates are also separated from CP1030, suggesting a conserved V. cholerae O1 clone that carries a truncated CTXΦ instead of RS1 in the upstream region of CTXΦ, circulating in the Mexican ecosystem during 2004 to 2008. Since 2000, variants of V. cholerae O1 El Tor have prevailed in areas of Asia and Africa where cholera is endemic, with V. cholerae prototype El Tor strains rarely isolated (45).
Conclusion.
This study provides important insights into the molecular epidemiology of cholera in Mexico. Overall, the results of our study and previous studies show the existence of genetically diverse V. cholerae O1 in Mexico during 1991 to 2008 (5, 7). Considering the global epidemiology of cholera, although the succession of V. cholerae O1 in Mexico remains a mystery, our observations clearly do not support the hypothesis of global transmission of cholera from Africa to Latin America, as proposed elsewhere (8). During the 1990s’ Latin American epidemic, Peru was the first country to have been affected by cholera, and a clonal CTX+ V. cholerae O1 El Tor strain was found to be the etiological agent, which was present on the Peruvian coast for at least several months prior to the onset of the cholera epidemic (21). Furthermore, CTX− V. cholerae O1 El Tor had been isolated from two patients with diarrhea in Lima, Peru, in 1988 (21, 46) and from sewage in Brazil in 1982 (21, 47). The environmental stimulus for V. cholerae (i.e., the increase in the temperature and phytoplankton abundance due to the El Nino phenomenon or changes in salinity and/or nutrient concentrations) may have triggered the existing CTX+ V. cholerae O1 El Tor strains to upsurge rapidly during the 1990s in Peru (21). Molecular typing and phylogenetic analysis of 1990s’ Latin American V. cholerae O1 isolates have been done in several studies, and no significant correlation was found between isolates from Asia and Latin America (8, 19). Phylogenetic analysis of the isolates shows that cholera in Mexico during 1991 to 2008 was caused by genetically diverse V. cholerae O1 strains belonging to distinct phylogenetic clades. Although, Mexican hybrid isolates show close relatedness to one hybrid isolate from Bangladesh, all of which were isolated in 1991, we do not have sufficient metadata to find out the direction of transmission either from Asia to Latin America or vice versa. Additionally, the antibiotic susceptibility patterns and CTX arrangements of the Mexican isolates strongly contradict the notion of a single-source transmission of V. cholerae O1 into Mexico from African countries. The lack of the SXT/R391 ICE in the Latin American CTX+ V. cholerae isolates is yet another interesting observation, which requires further study, since concurrent Asian and African isolates generally possess SXT/R391 ICE. Therefore, genetic events occurring in V. cholerae O1 strains associated with endemic cholera in Mexico are different from those of Asian and African countries (5, 7). Results provided in this study are concordant with those of previous investigations (5, 7, 22) and suggest a likely association of indigenous populations of V. cholerae that play a significant role in the dynamics of cholera in Mexico.
MATERIALS AND METHODS
Bacterial strains.
Vibrio cholerae O1 strains analyzed in the present study (n = 6) are listed in Table S1 in the supplemental material with the source, location, and year of isolation. Vibrio cholerae O1 strains were provided by the Department of Public Health, Faculty of Medicine, National Autonomous University of Mexico (UNAM) and Centro de Investigación Científica y de Educación Superior de Ensenada. The strains were isolated from cholera patients as part of a nationwide cholera surveillance program conducted between 1983 and 2008 in Mexico (5, 7). The bacterial strains were shipped in T1N1 soft agar (1% trypticase, 1% NaCl, 0.7% agar [pH 7.4]), and the identities were confirmed by standard culture methods and biochemical tests, followed by serogroup and biotype determination, as described previously (48, 49).
Sequencing, assembly, and annotation.
Genomic DNA of six V. cholerae strains was subjected to next-generation whole-genome Illumina and hybrid Illumina/454 sequencing and closure strategies, as previously described (11, 14). Libraries were constructed with target insert sizes of 3 kb and paired-end sizes of 100 bp. Hybrid and Illumina sequences were assembled using Celera and Velvet assemblers, respectively (50) and all chromosomes were manually annotated using the Manatee system (http://manatee.sourceforge.net/).
Comparative genomics.
Genome-to-genome comparison was performed by using different approaches because the completeness and quality of the nucleotide sequences varied from strain to strain. First, ORFs of a given pair of genomes were identified and reciprocally compared with each other using the BLASTN, BLASTP, and tBLASTx programs (ORF-dependent comparison). Second, a bioinformatic pipeline was constructed to identify homologous regions of a given query ORF. Initially, a segment on the target contig, which is homologous to a query ORF, was identified using the BLASTN program. This potentially homologous region was expanded in both directions by 2,000 bp. Nucleotide sequences of the query ORF and selected target homologous regions were aligned using a pairwise global alignment algorithm, and the resultant matched region in the subject contig was extracted and saved as a homologue (ORF-independent comparison). Orthologues and paralogs were differentiated by reciprocal comparison, as described previously (11).
Identification and annotation of genomic islands.
We defined genomic islands (GIs) as a continuous array of five or more coding sequences (CDSs) that were discontinuously distributed among genomes of test strains. Correct transfer or insertion of GIs was readily differentiated from a deletion event by comparing the genome-based phylogenetic tree and full matrices showing pairwise detection of orthologous genes between test strains. Identified GIs were designated and annotated using the BLASTP search of its member CDSs against the GenBank NR database, as described elsewhere (11).
Phylogenetics based on genome sequences.
Orthologous regions of V. cholerae N16961 were identified by comparisons based on similarity and were used to generate phylogenetic trees (14). The set of orthologous regions for each CDS of a reference genome was identified according to nucleotide similarity and aligned using CLUSTALW2. The resultant multiple alignments were concatenated to form genome-scale alignments, which were then used to generate the neighbor-joining phylogenetic trees (51).
Nucleotide sequence accession numbers.
Whole-genome sequences of CP1030, CP1032, CP1033, CP1035, CP1037, and 95412 have been deposited in the DDBJ/EMBL/GenBank databases under accession no. ALCZ00000000, ALDA00000000, AJRL00000000, AJRM00000000, ALDB00000000, and APFM00000000, respectively.
SUPPLEMENTAL MATERIAL
Phenotypic and genotypic characteristics of V. cholerae O1 strains (n = 182) isolated from clinical and environmental samples collected in Mexico from 1983 to 2008 (adopted from Alam et al. [5, 7]).
Distribution of genomic islands (GIs) in Vibrio cholerae O1 strains isolated in Mexico from 1991 to 2008.
ACKNOWLEDGMENTS
This project was funded, in part, with federal funds from the National Institute of Allergy and Infectious Diseases, Department of Health and Human Services, under contract no. HHSN2722009000. Partial support was provided by NOAA grant no. SO660009 and NSF-NIH Ecology of Infectious Diseases program grant no. EF-0813066.
We acknowledge the valuable assistance of Jonathan Crabtree, Amy Egan, Naomi Sengamalay, and Lisa Sadzewicz in sample processing, sequencing and logistics. icddr,b is thankful to the Governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support.
Funding Statement
This project was funded, in part, with federal funds from the National Institute of Allergy and Infectious Diseases, Department of Health and Human Services under contract number HHSN2722009000. Partial support was provided by NOAA grant no. SO660009, NSF-NIH Ecology of Infectious Diseases program Grant No. EF-0813066.
Footnotes
Citation Choi SY, Rashed SM, Hasan NA, Alam M, Islam T, Sadique A, Johura F-T, Eppinger M, Ravel J, Huq A, Cravioto A, Colwell RR. 2016. Phylogenetic diversity of Vibrio cholerae associated with endemic cholera in Mexico from 1991 to 2008. mBio 7(2):e02160-15. doi:10.1128/mBio.02160-15.
REFERENCES
- 1.Sack DA, Sack RB, Nair GB, Siddique AK. 2004. Cholera. Lancet 363:223–233. doi: 10.1016/S0140-6736(03)15328-7. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sepúlveda J, Valdespino JL, García-García L. 2006. Cholera in Mexico: the paradoxical benefits of the last pandemic. Int J Infect Dis 10:4–13. doi: 10.1016/j.ijid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Olsvik O. 1992. The cholera epidemic in Latin America. Tidsskr Nor Laegeforen 112:1843–1846. [PubMed] [Google Scholar]
- 5.Alam M, Nusrin S, Islam A, Bhuiyan NA, Rahim N, Delgado G, Morales R, Mendez JL, Navarro A, Gil AI, Watanabe H, Morita M, Nair GB, Cravioto A. 2010. Cholera between 1991 and 1997 in Mexico was associated with infection by classical, El Tor, and El Tor variants of Vibrio cholerae. J Clin Microbiol 48:3666–3674. doi: 10.1128/JCM.00866-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wachsmuth IK, Evins GM, Fields PI, Olsvik O, Popovic T, Bopp CA, Wells JG, Carrillo C, Blake PA. 1993. The molecular epidemiology of cholera in Latin America. J Infect Dis 167:621–626. doi: 10.1093/infdis/167.3.621. [DOI] [PubMed] [Google Scholar]
- 7.Alam M, Rashed SM, Mannan SB, Islam T, Lizarraga-Partida ML, Delgado G, Morales-Espinosa R, Mendez JL, Navarro A, Watanabe H, Ohnishi M, Hasan NA, Huq A, Sack RB, Colwell RR, Cravioto A. 2014. Occurrence in Mexico, 1998–2008, of Vibrio cholerae CTX+ El Tor carrying an additional truncated CTX prophage. Proc Natl Acad Sci U S A 111:9917–9922. doi: 10.1073/pnas.1323408111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J Clin Microbiol 40:3296–3299. doi: 10.1128/JCM.40.9.3296-3299.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safa A, Sultana J, Cam PD, Mwansa JC, Kong RYC. 2008. Vibrio cholerae O1 hybrid El Tor strains, Asia and Africa. Emerg Infect Dis 14:987–988. doi: 10.3201/eid1406.080129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun J, Grim CJ, Hasan NA, Lee JH, Choi SY, Haley BJ, Taviani E, Jeon YS, Kim DW, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Munk AC, Chertkov O, Meincke L, Saunders E, Walters RA, Huq A, Nair GB, Colwell RR. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc Natl Acad Sci U S A 106:15442–15447. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Albert MJ, Nair GB. 2005. Vibrio cholerae O139 Bengal—10 years on. Rev Med Microbiol 16:135–143. doi: 10.1097/01.revmedmi.0000184743.75679.0a. [DOI] [Google Scholar]
- 13.Albert M, Siddique AK, Islam MS, Faruque ASG, Ansaruzzaman M, Faruque SM, Sack R. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. doi: 10.1016/0140-6736(93)90481-U. [DOI] [PubMed] [Google Scholar]
- 14.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci U S A 109:E2010–E2017. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam MS, Mahmud ZH, Ansaruzzaman M, Faruque SM, Talukder KA, Qadri F, Alam M, Islam S, Bardhan PK, Mazumder RN, Khan AI, Ahmed S, Iqbal A, Chitsatso O, Mudzori J, Patel S, Midzi SM, Charimari L, Endtz HP, Cravioto A. 2011. Phenotypic, genotypic, and antibiotic sensitivity patterns of strains isolated from the cholera epidemic in Zimbabwe. J Clin Microbiol 49:2325–2327. doi: 10.1128/JCM.00432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eppinger M, Pearson T, Koenig SS, Pearson O, Hicks N, Agrawal S, Sanjar F, Galens K, Daugherty S, Crabtree J, Hendriksen RS, Price LB, Upadhyay BP, Shakya G, Fraser CM, Ravel J, Keim PS. 2014. Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic. mBio 5:e01721. doi: 10.1128/mBio.01721-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthmann JP. 1995. Epidemic cholera in Latin America: spread and routes of transmission. J Trop Med Hyg 98:419–427. [PubMed] [Google Scholar]
- 18.McCarthy SA, Khambaty FM. 1994. International dissemination of epidemic Vibrio cholerae by cargo ship ballast and other nonpotable waters. Appl Environ Microbiol 60:2597–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam C, Octavia S, Reeves P, Wang L, Lan R. 2010. Evolution of seventh cholera pandemic and origin of 1991 epidemic, Latin America. Emerg Infect Dis 16:1130–1132. doi: 10.3201/eid1607.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mourino-Perez RR. 1998. Oceanography and the seventh cholera pandemic. Epidemiology 9:355–357. doi: 10.1097/00001648-199805000-00024. [DOI] [PubMed] [Google Scholar]
- 21.Seas C, Miranda J, Gil AI, Leon-Barua R, Patz J, Huq A, Colwell RR, Sack RB. 2000. New insights on the emergence of cholera in Latin America during 1991: the Peruvian experience. Am J Trop Med Hyg 62:513–517. [DOI] [PubMed] [Google Scholar]
- 22.Alam M, Islam MT, Rashed SM, Johura FT, Bhuiyan NA, Delgado G, Morales R, Mendez JL, Navarro A, Watanabe H, Hasan NA, Colwell RR, Cravioto A. 2012. Vibrio cholerae classical biotype strains reveal distinct signatures in Mexico. J Clin Microbiol 50:2212–2216. doi: 10.1128/JCM.00189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 24.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Choi SY, Lee JH, Jeon YS, Lee HR, Kim EJ, Ansaruzzaman M, Bhuiyan NA, Endtz HP, Niyogi SK, Sarkar BL, Nair GB, Nguyen BM, Hien NT, Czerkinsky C, Clemens JD, Chun J, Kim DW. 2010. Multilocus variable-number tandem repeat analysis of Vibrio cholerae O1 El Tor strains harbouring classical toxin B. J Med Microbiol 59:763–769. doi: 10.1099/jmm.0.017939-0. [DOI] [PubMed] [Google Scholar]
- 26.Lee JH, Choi SY, Jeon YS, Lee HR, Kim EJ, Nguyen BM, Hien NT, Ansaruzzaman M, Islam MS, Bhuiyan NA, Niyogi SK, Sarkar BL, Nair GB, Kim DS, Lopez AL, Czerkinsky C, Clemens JD, Chun J, Kim DW. 2009. Classification of hybrid and altered Vibrio cholerae strains by CTX prophage and RS1 element structure. J Microbiol 47:783–788. doi: 10.1007/s12275-009-0292-6. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen BM, Lee JH, Cuong NT, Choi SY, Hien NT, Anh DD, Lee HR, Ansaruzzaman M, Endtz HP, Chun J, Lopez AL, Czerkinsky C, Clemens JD, Kim DW. 2009. Cholera outbreaks caused by an altered Vibrio cholerae O1 El Tor biotype strain producing classical cholera toxin B in Vietnam in 2007 to 2008. J Clin Microbiol 47:1568–1571. doi: 10.1128/JCM.02040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfau JD, Taylor RK. 1996. Genetic footprint on the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol 20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. 2014. Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India. PLoS One 9:e112973. doi: 10.1371/journal.pone.0112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyd EF, Moyer KE, Shi L, Waldor MK. 2000. Infectious CTXPhi and the Vibrio pathogenicity island prophage in Vibrio mimicus: evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun 68:1507–1513. doi: 10.1128/IAI.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay CY, Reeves PR, Lan R. 2008. Importation of the major pilin TcpA gene and frequent recombination drive the divergence of the Vibrio pathogenicity island in Vibrio cholerae. FEMS Microbiol Lett 289:210–218. doi: 10.1111/j.1574-6968.2008.01385.x. [DOI] [PubMed] [Google Scholar]
- 32.Jonson G, Holmgren J, Svennerholm AM. 1992. Analysis of expression of toxin-coregulated pili in classical and El Tor Vibrio cholerae O1 in vitro and in vivo. Infect Immun 60:4278–4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dziejman M, Balon E, Boyd D, Fraser CM, Heidelberg JF, Mekalanos JJ. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc Natl Acad Sci U S A 99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, Faruque SM, Rahman MH, Heidelberg JF, Decker J, Li L, Montgomery KT, Grills G, Kucherlapati R, Mekalanos JJ. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci U S A 102:3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taviani E, Grim CJ, Choi J, Chun J, Haley B, Hasan NA, Huq A, Colwell RR. 2010. Discovery of novel Vibrio cholerae VSP-II genomic islands using comparative genomic analysis. FEMS Microbiol Lett 308:130–137. doi: 10.1111/j.1574-6968.2010.02008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nusrin S, Gil AI, Bhuiyan NA, Safa A, Asakura M, Lanata CF, Hall E, Miranda H, Huapaya B, Vargas GC, Luna MA, Sack DA, Yamasaki S, Nair GB. 2009. Peruvian Vibrio cholerae O1 El Tor strains possess a distinct region in the Vibrio seventh pandemic island-II that differentiates them from the prototype seventh pandemic El Tor strains. J Med Microbiol 58:342–354. doi: 10.1099/jmm.0.005397-0. [DOI] [PubMed] [Google Scholar]
- 37.Valia R, Taviani E, Spagnoletti M, Ceccarelli D, Cappuccinelli P, Colombo MM. 2013. Vibrio cholerae O1 epidemic variants in Angola: a retrospective study between 1992 and 2006. Front Microbiol 4:354. doi: 10.3389/fmicb.2013.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N Engl J Med 364:33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waldor MK, Tschäpe H, Mekalanos JJ. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 178:4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother 45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klinzing DC, Choi SY, Hasan NA, Matias RR, Tayag E, Geronimo J, Skowronski E, Rashed SM, Kawashima K, Rosenzweig CN, Gibbons HS, Torres BC, Liles V, Alfon AC, Juan ML, Natividad FF, Cebula TA, Colwell RR. 2015. Hybrid Vibrio cholerae El Tor lacking SXT identified as the cause of a cholera outbreak in the Philippines. mBio 6:e00047-15. doi: 10.1128/mBio.00047-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mhalu FS, Mmari PW, Ijumba J. 1979. Rapid emergence of El Tor Vibrio cholerae resistant to antimicrobial agents during first six months of fourth cholera epidemic in Tanzania. Lancet i:345–347. [DOI] [PubMed] [Google Scholar]
- 43.Nesper J, Kraiss A, Schild S, Blass J, Klose KE, Bockemühl J, Reidl J. 2002. Comparative and genetic analyses of the putative Vibrio cholerae lipopolysaccharide core oligosaccharide biosynthesis (wav) gene cluster. Infect Immun 70:2419–2433. doi: 10.1128/IAI.70.5.2419-2433.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ansaruzzaman M, Bhuiyan NA, Nair GB, Sack DA, Lucas M, Deen JL, Ampuero J, Chaignat C. 2004. Cholera in Mozambique, variant of Vibrio cholerae. Emerg Infect Dis 10:2057–2059. doi: 10.3201/eid1011.040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rashed SM, Iqbal A, Mannan SB, Islam T, Rashid M, Johura F, Watanabe H, Hasan NA, Huq A, Stine OC, Sack RB, Colwell RR, Alam M. 2013. Vibrio cholerae O1 El Tor and O139 Bengal strains carrying ctxB(ET), Bangladesh. Emerg Infect Dis 19:1713–1715. doi: 10.3201/eid1910.130626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Batchelor RA, Wignall FS. 1988. Nontoxigenic 01 Vibrio cholerae in Peru: a report of two cases associated with diarrhea. Diagn Microbiol Infect Dis 10:135–138. doi: 10.1016/0732-8893(88)90031-4. [DOI] [PubMed] [Google Scholar]
- 47.Levine MM, Black RE, Clements ML, Cisneros L, Saah A, Nalin DR, Gill DM, Craig JP, Young CR, Ristaino P. 1982. The pathogenicity of nonenterotoxigenic Vibrio cholerae serogroup O1 biotype El Tor isolated from sewage water in Brazil. J Infect Dis 145:296–299. doi: 10.1093/infdis/145.3.296. [DOI] [PubMed] [Google Scholar]
- 48.Alam M, Hasan NA, Sadique A, Bhuiyan NA, Ahmed KU, Nusrin S, Nair GB, Siddique AK, Sack RB, Sack DA, Huq A, Colwell RR. 2006. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl Environ Microbiol 72:4096–4104. doi: 10.1128/AEM.00066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alam M, Sultana M, Nair GB, Sack RB, Sack DA, Siddique AK, Ali A, Huq A, Colwell RR. 2006. Toxigenic Vibrio cholerae in the aquatic environment of Mathbaria, Bangladesh. Appl Environ Microbiol 72:2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zerbino DR. 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr Protoc Bioinformatics Chapter 11:Unit 11.15. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic and genotypic characteristics of V. cholerae O1 strains (n = 182) isolated from clinical and environmental samples collected in Mexico from 1983 to 2008 (adopted from Alam et al. [5, 7]).
Distribution of genomic islands (GIs) in Vibrio cholerae O1 strains isolated in Mexico from 1991 to 2008.