ABSTRACT
Escherichia coli sequence type 131 (ST131) has emerged globally as the most predominant extraintestinal pathogenic lineage within this clinically important species, and its association with fluoroquinolone and extended-spectrum cephalosporin resistance impacts significantly on treatment. The evolutionary histories of this lineage, and of important antimicrobial resistance elements within it, remain unclearly defined. This study of the largest worldwide collection (n = 215) of sequenced ST131 E. coli isolates to date demonstrates that the clonal expansion of two previously recognized antimicrobial-resistant clades, C1/H30R and C2/H30Rx, started around 25 years ago, consistent with the widespread introduction of fluoroquinolones and extended-spectrum cephalosporins in clinical medicine. These two clades appear to have emerged in the United States, with the expansion of the C2/H30Rx clade driven by the acquisition of a blaCTX-M-15-containing IncFII-like plasmid that has subsequently undergone extensive rearrangement. Several other evolutionary processes influencing the trajectory of this drug-resistant lineage are described, including sporadic acquisitions of CTX-M resistance plasmids and chromosomal integration of blaCTX-M within subclusters followed by vertical evolution. These processes are also occurring for another family of CTX-M gene variants more recently observed among ST131, the blaCTX-M-14/14-like group. The complexity of the evolutionary history of ST131 has important implications for antimicrobial resistance surveillance, epidemiological analysis, and control of emerging clinical lineages of E. coli. These data also highlight the global imperative to reduce specific antibiotic selection pressures and demonstrate the important and varied roles played by plasmids and other mobile genetic elements in the perpetuation of antimicrobial resistance within lineages.
IMPORTANCE
Escherichia coli, perennially a major bacterial pathogen, is becoming increasingly difficult to manage due to emerging resistance to all preferred antimicrobials. Resistance is concentrated within specific E. coli lineages, such as sequence type 131 (ST131). Clarification of the genetic basis for clonally associated resistance is key to devising intervention strategies. We used high-resolution genomic analysis of a large global collection of ST131 isolates to define the evolutionary history of extended-spectrum beta-lactamase production in ST131. We documented diverse contributory genetic processes, including stable chromosomal integrations of resistance genes, persistence and evolution of mobile resistance elements within sublineages, and sporadic acquisition of different resistance elements. Both global distribution and regional segregation were evident. The diversity of resistance element acquisition and propagation within ST131 indicates a need for control and surveillance strategies that target both bacterial strains and mobile genetic elements.
INTRODUCTION
Resistance to extended-spectrum cephalosporins in extraintestinal pathogenic Escherichia coli (ExPEC) represents a major clinical challenge and is commonly caused by the presence of extended-spectrum beta-lactamases (ESBLs). The majority of ESBL-associated E. coli infections are due to a recently emerged, globally distributed ExPEC clone, sequence type 131 (ST131) (1). ST131 predominantly corresponds to serogroup O25b (2, 3) or O16 (4) and belongs to phylogenetic group B2 (5, 6). It remains unclear which features of this clone have resulted in its recent widespread clinical dominance, although antimicrobial resistance and virulence factors are suspected contributors (7).
The blaCTX-M-15 beta-lactamase gene is the dominant ESBL gene in ST131, but other genetically divergent CTX-M genes also occur in this ST, particularly blaCTX-M-14/14-like variants, e.g., in Canada, China, and Spain (8, 9). The almost contemporaneous identification of blaCTX-M in ST131 strains from multiple geographic locations suggests repeated acquisition via multiple horizontal gene transfer events (10). Consistently, both blaCTX-M-15 and blaCTX-M-14/14-like variants occur on conjugative plasmids, especially multireplicon IncFII plasmids additionally harboring FIA/FIB replicons (11).
Other data, however, suggest that the widespread distribution of these genes is mediated by clonal expansion of CTX-M-containing strains and global dissemination (12). This is also a plausible hypothesis, since CTX-M plasmids can be inherited stably and blaCTX-M-15 and blaCTX-M-14 variants can also integrate into the chromosome (13–15). Nevertheless, clonal expansion of E. coli strains with chromosomally integrated blaCTX-M has not yet been demonstrated.
Two recent studies used whole-genome sequence (WGS) data to investigate the population structure of ST131. The first found that ST131 expansion in the United States has been driven by a single sublineage, H30, defined by the presence of a specific fimbrial adhesin allele, fimH30. Within H30, nested clades have emerged: H30R, containing mutations in the chromosomal genes gyrA and parC that confer fluoroquinolone resistance, and H30Rx, containing the same gyrA and parC mutations but additionally associated with blaCTX-M-15 (13). The second study (16), which included samples from six locations around the world, resolved the ST131 population structure into three clades, A, B, and C, with clade C comprising two subgroups, C1 and C2, corresponding to the H30R and H30Rx clades. However, this study included only four isolates from Asia, where ESBL ExPEC prevalence may be highest (17, 18). Furthermore, neither study directly tested the competing hypotheses that ESBL dissemination in ST131 has occurred through multiple horizontal gene transfer events versus clonal expansion.
Here, we used a broader set of ST131 WGS data, including many more isolates from Asia and blaCTX-M-14/14-like-containing strains, alongside a subset of CTX-M plasmid sequences, to estimate the contribution of each potential route of dissemination to the worldwide prevalence of ST131.
RESULTS
The 215 ST131 genome sequences analyzed included 67 strains from various locations in Southeast Asia, 33 from Oxford in the United Kingdom, 11 from a global resistance surveillance program at AstraZeneca, 8 from Canada, and 96 predominantly North American isolates previously reported by Price et al. (13) (details on new isolates are shown in Table S1 in the supplemental material; these strains included both human and animal isolates and clinical and carriage isolates.)
Asian ST131 strains are consistent with the previously described core phylogeny, and the C1/H30 and C2/H30Rx clades emerged from a North American ancestor.
For the 4,717,338 sites in the SE15 ST131 reference genome (19), the mean mapping call rate across the data set was 93.3%. In total, 40,057 (0.85%) sites were variable, with 6,879 (0.15%) representing core single-nucleotide variants (SNVs) called in all 215 isolates. Overall, 611,770 (13%) sites were in recombinant regions, including 4,120 core SNVs, leaving 2,759 core nonrecombinant SNVs for phylogenetic analysis.
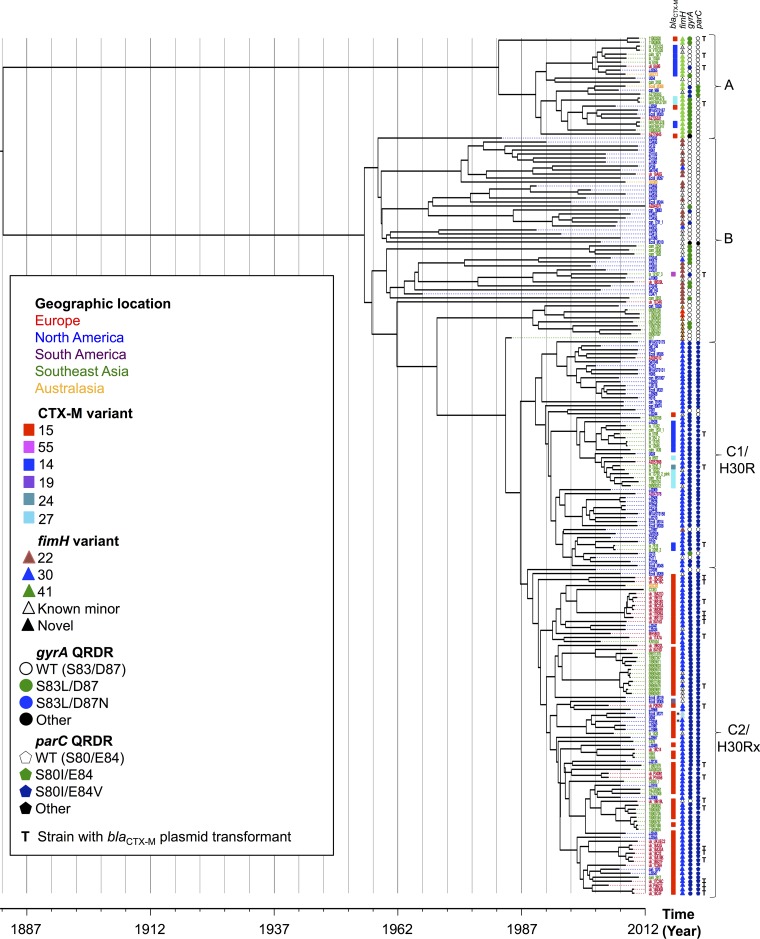
Consistent with the two previous WGS-based ST131 phylogenies (13, 16), the time-scaled phylogeny inferred from this ST131 data set (which included >10 times more Asian isolates than considered previously) comprised three clades (Fig. 1), A (n = 25), B (n = 51), and C (n = 139), with C containing two subclades, C1 (n = 57) and C2 (n = 82), characterized by the presence versus absence, respectively, of blaCTX-M-15 (16). Isolates from all geographic regions were identified within each clade, although there were smaller, geographically restricted clusters within these (Fig. 1, tip color). This supports both global transmission and localized lineage expansion following specific introductions into a geographic locality.
FIG 1 .
Time-scaled phylogeny of ST131 E. coli (n = 215), with associated blaCTX-M/fimH variants, and quinolone resistance-determining region (QRDR) mutations in gyrA (WT, wild-type QRDR). Curly brackets represent ST131 clades as described in the text. Tips are colored by geographic region, per the key. T, blaCTX-M plasmid transformant generated for strain; *, cases with putative deletions in the assembled blaCTX-M-15 gene.
The estimated time to most recent common ancestor (TMRCA) for the whole genomic data set was ~130 years ago, when clade A diverged from clades B and C. Twenty-five years ago, clade C emerged out of the paraphyletic clade B, which was quickly followed by the split between subclades C1 and C2. The number of core SNVs separating the clades was approximately 250 for clade A versus clades B and C, 50 to 60 for clade B versus clades C1 and C2, and 10 to 30 for clade C1 versus clade C2. The evolutionary rate of ST131 was estimated in BEAST (see Materials and Methods) at 2.46 × 10−7 mutations per site per year (95% confidence interval [CI], 2.18 × 10−7 to 2.75 × 10−7), equating to 1.00 (95% CI, 0.89 to 1.12) mutation per genome per year.
All possible geographic origins of the root of the ST131 lineage were inferred to be equally likely since the root is far back in time relative to the estimated migration rates. Clade A was inferred to originate in Southeast Asia with ~70% confidence (78% when the unsampled deme was included in the model [see Materials and Methods]), and the B/C clades were inferred to originate from North America with ~88% confidence. The ancestral origin of C1/H30 and C2/H30Rx was strongly inferred as being in North America (98% confidence; 85% confidence when the unsampled deme was included in the model) with subsequent dissemination to Europe and Asia. Locations of more recent nodes are inferred with high confidence, as expected (20).
blaCTX-M, fimH, and gyrA variants are strongly associated with specific ST131 clades.
Overall, 105 (49%) ST131 isolates harbored blaCTX-M, including 74 isolates (34%) with blaCTX-M-15, 20 (9%) with blaCTX-M-14, 8 (4%) with blaCTX-M-27, and one each (0.4%) with blaCTX-M-19, blaCTX-M-24, and blaCTX-M-55 (Fig. 1). blaCTX-M-15 was almost completely restricted to the C2 clade, as described previously (16), occurring in 69/82 (84%) C2 isolates but only sporadically in other clades (4/133; P < 0.001, Fisher exact test). blaCTX-M-14 and blaCTX-M-27 were also clustered within the two different clades A and C1 and absent from B and C2 (Fig. 1). Overall, the presence of shared blaCTX-M variants within clusters was constrained to those with a TMRCA of less than 25 years, suggestive of the emergence of blaCTX-M within ST131 after the widespread introduction of extended-spectrum cephalosporins in clinical practice.
The most common fimH variant was fimH30 (n = 123; 57%), followed by fimH22 (n = 24; 11%) and fimH41 (n = 21; 10%), whereas 23 isolates had novel fimH variants, and one was fimH null. As observed for blaCTX-M, fimH alleles were strongly associated with clade, with 21/25 (84%) isolates in clade A having fimH41, 23/51 (45%) in clade B having fimH22, and 122/139 (88%) in clade C having fimH30 (P < 0.001; Fisher exact test).
Fluoroquinolone resistance mutations in gyrA and parC were also clade associated, with isolates in clades A and B typically having no or only single mutations in these genes’ quinolone resistance-determining regions (QRDRs) (Fig. 1). In contrast, most clade C isolates had double mutations in both gyrA and parC, shown to confer high-level fluoroquinolone resistance (21) (132/139 [95%]). The seven clade C isolates without these mutations (5 in C1 and 2 in C2) were sporadic, with two having non-fimH30 variants, suggesting intermittent recombination events affecting gyrA, parC, and fimH. The emergence of this double-mutation, high-level fluoroquinolone resistance genotype was dated by our methods to 25 to 40 years ago, consistent with the introduction of fluoroquinolones in clinical practice.
blaCTX-M-15 in clade C2 is present in a consistent but short flanking structure, frequently truncated by IS26 elements and within different genetic backgrounds.
In four of the 74 blaCTX-M-15-containing isolates, blaCTX-M-15 was present on two different contigs (C1353, JJ2643, CD358, and JJ2434). In another isolate (JJ2547), the assembled contig with blaCTX-M-15 contained a series of N’s, suggesting possible uncertainty around the contig assembly or multiple locations of the gene. These five isolates were excluded from further analysis of flanking regions. In the 69 remaining isolates (3 in clade A, 1 in clade C1, and 65 in clade C2), blaCTX-M-15 was found downstream of a homologous tract of 48 bp preceded by an ISEcp1 right-end inverted repeat region (IRR-R) and upstream of a homologous tract of 46 bp followed by ORF477. This is consistent with the introduction of an ISEcp1-blaCTX-M-15-ORF477 unit within ST131 and subsequent rearrangement events affecting this structure.
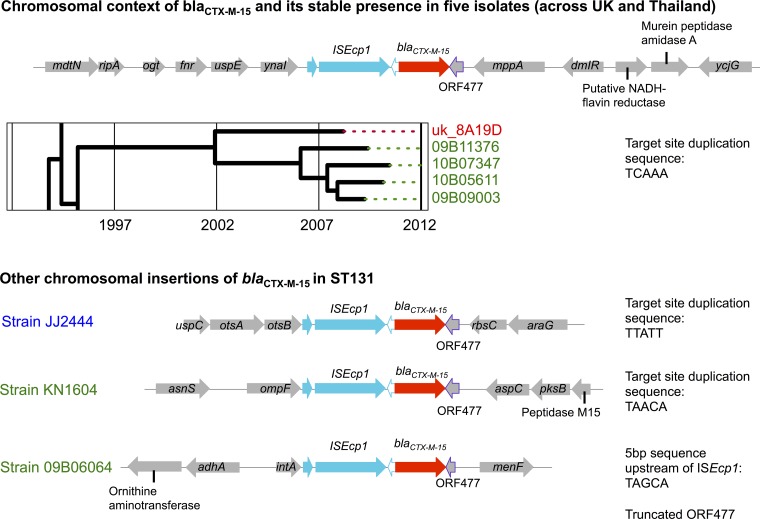
In clade C2, blaCTX-M-15 was integrated into the chromosome of 8/65 (12%) isolates, with four unique integration events, one of which was stably present in a subcluster of five isolates with a common ancestor around 2002 and spread across two geographical regions (Fig. 2). All chromosomal integration events were associated with an intact ISEcp1 upstream of blaCTX-M-15. In three isolates, the ISEcp1-blaCTX-M-15-ORF477 unit was flanked by 5-bp target site duplications consistent with transposition, and in one isolate, the ORF477 was truncated, suggestive of either one-ended transposition (22) or standard transposition followed by a deletion event (Fig. 2).
FIG 2 .
Chromosomal location of ISEcp1-mediated blaCTX-M-15 insertion events and evidence of acquisition and evolution by descent (inset phylogeny) over approximately 8 years across two geographic regions (Oxford, United Kingdom; Mae Sot, Thailand-Myanmar border). Coloring of isolate names represents geographic location (red, Europe; green, Southeast Asia; blue, North America).
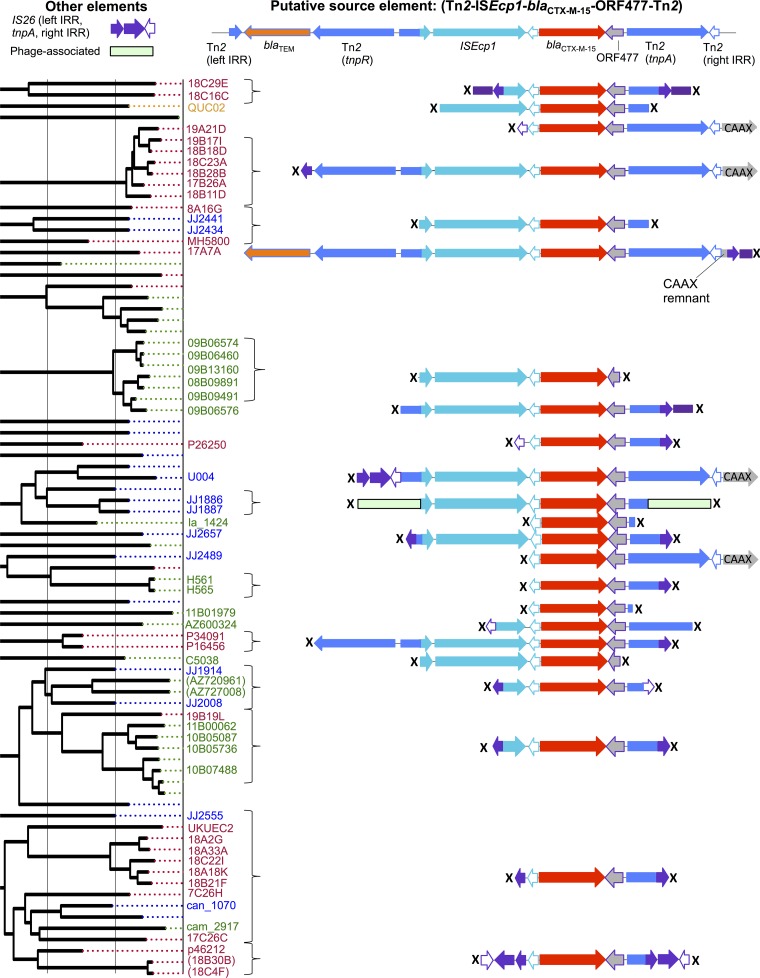
In 27 of the 57 remaining C2 isolates, blaCTX-M-15 appeared plasmid associated, being either present in plasmid transformants (n = 20) or flanked by likely plasmid-associated sequences in the contig assemblies (n = 7). In the remaining 30 isolates, the location of blaCTX-M-15 could not be defined due to limitations of the short-read assemblies. In all 57 isolates, the upstream sequence was either an intact or truncated ISEcp1 sequence, and in 51/57 (89%) isolates, the sequence downstream of ORF477 was either an intact or a truncated Tn2 structure (Fig. 3). In 12 isolates distributed throughout clade C2, a continuation of the Tn2 sequence was also observed upstream of the ISEcp1 sequence, consistent with the ISEcp1 element (flanked by a pair of 5-bp repeats, all TCATA) being nested within a complete or partial Tn2 transposon. In 40/57 (65%) isolates, IS26 repeat regions truncated either or both of these upstream and downstream contexts (Fig. 3).
FIG 3 .
Genetic flanking context of blaCTX-M-15 region for clade C2/H30Rx isolates. (Top) Putative source element. (Left) C2/H30Rx phylogeny. Many contexts are limited by the extent of the assembled region around the blaCTX-M-15 gene (marked with “X”). For all similarly colored, vertically aligned regions below the “Putative source element,” sequence identity is 100%. Curly brackets cluster those isolates with identical flanking sequences. Flanking contexts are not shown and tip labels are omitted for isolates with known chromosomal integration of blaCTX-M, for blaCTX-M-negative isolates in the clade, or for isolates where the flanking sequence was not evaluable (see Results).
blaCTX-M-14 and blaCTX-M-27 are present in diverse genetic backgrounds and within a common ISEcp1-IS903B transposition unit.
For blaCTX-M-14, evidence of chromosomal integration was also found: two related blaCTX-M-14 isolates had ISEcp1-mediated chromosomal integration of blaCTX-M-14 downstream of the gatY gene (clade A, isolates HFMK328 and HFMK347). Six isolates had plasmid-associated blaCTX-M-14 based on annotated flanking sequences/transformants, whereas for the rest (n = 14), the location of blaCTX-M-14 was uncertain due to limitations of the de novo assemblies.
In all these isolates, ISEcp1 was consistently located upstream of blaCTX-M-14, as with blaCTX-M-15, but at only a 43-bp distance, and the downstream flanking sequences were composed of either intact or truncated IS903B elements. In clade A, the genetic flanking sequences surrounding blaCTX-M-14 were consistent with the host strain subcluster and were homologous over the observed contig length within this subcluster (see Fig. S1 in the supplemental material, clade A CTX-M-14 subcluster [i]). This supports a single blaCTX-M-14 plasmid acquisition event followed by either evolution with plasmid inheritance or subsequent transfer of a blaCTX-M-14-containing genetic unit within the subcluster. The flanking sequence for isolate la_5108_T in clade C1 also incorporated an ISEc23 element downstream of IS903B and was homologous to that in clade A CTX-M-14 subcluster [i] (see Fig. S2), suggesting horizontal transfer of this genetic unit between clades.
Six of eight isolates with blaCTX-M-27 were closely related in clade C1, again supporting a single plasmid acquisition event. However, they also all contained bilateral truncation of the ISEcp1-blaCTX-M-IS903B structure by IS26 elements, which occurred in four different contexts, suggesting frequent IS26-mediated blaCTX-M-27 transposition events within this subcluster (see Fig. S2 in the supplemental material).
Plasmid replicon analysis demonstrates a degree of clade-associated plasmid segregation suggestive of ancient IncF plasmid acquisition events.
The predominant replicon family was IncF, identified in 206/215 ST131 isolates (96%). Specific IncF variants differed in frequency, with FII found in 199/215 isolates (93%), FIB in 155/215 (72%), FIA in 145/215 (67%), and FIC in 17/215 (8%). Specific IncF replicons and combinations thereof were clade associated (Table 1). A number of non-F Inc types were also identified; of these, IncH was associated with clade B and IncI was associated with clade C1 (Table 1). Col-like plasmids were also common (189/215 isolates [88%]); however, there was no clear association of any Col type with clade (see Fig. S3 in the supplemental material).
TABLE 1 .
Plasmid replicon families/types by clade
| Inc type | No. of isolates (row %) for clade: |
Total no. of isolates | Difference in replicon prevalence across clades, P | |||
|---|---|---|---|---|---|---|
| A/H41 | B/H22 | C1/H30R | C2/H30Rx | |||
| A/C | 1 (33) | 1 (33) | 1 (33) | 3 | 1 | |
| B/O/K/Z | 1 (10) | 2 (20) | 2 (20) | 5 (50) | 10 | 0.9 |
| FIA only | 1 (50) | 1 (50) | 2 | 1 | ||
| FIA total | 15 (10) | 1 (0.7) | 52 (36) | 77 (53) | 145 | <0.001a |
| FIA-FIB | 3 (100) | 3 | 0.07 | |||
| FIA-FII | 4 (9) | 39 (91) | 43 | <0.001 | ||
| FIA-FIB-FII | 15 (15) | 1 (1) | 44 (45) | 37 (38) | 97 | <0.001 |
| FIB only | 2 (100) | 2 | 0.12 | |||
| FIB total | 25 (16) | 43 (28) | 47 (30) | 40 (25) | 155 | <0.001 |
| FIB-FII | 8 (22) | 25 (69) | 3 (8) | 36 | <0.001 | |
| FIB-FIC-FII | 2 (12) | 15 (88) | 17 | <0.001 | ||
| FIC total | 2 (12) | 15 (88) | 17 | <0.001 | ||
| FII only | 4 (67) | 1 (17) | 1 (17) | 6 | 0.12 | |
| FII total | 25 (13) | 45 (23) | 49 (25) | 80 (40) | 199 | 0.02 |
| H | 5 (100) | 5 | 0.001 | |||
| I | 4 (25) | 6 (38) | 3 (19) | 3 (19) | 16 | 0.09 |
| N | 2 (15) | 7 (54) | 4 (31) | 13 | 0.15 | |
| P | 2 (100) | 2 | 0.12 | |||
| Q | 1 (11) | 7 (78) | 1 (11) | 9 | 0.005 | |
| R | 1 (100) | 1 | 0.36 | |||
| X-like | 7 (58) | 2 (17) | 3 (25) | 12 | 0.06 | |
| Y | 1 (8) | 1 (8) | 2 (17) | 8 (67) | 12 | 0.25 |
Boldface indicates statistically significant differences (P < 0.05).
A specific FII variant (GenBank nucleotide sequence accession no. AY458016; pC15-1a; consistent with pMLST allele 2) was significantly associated with clade C2 (48/82 C2 isolates versus 18/153 non-C2 isolates, P < 0.001, Fisher exact test). Within clade C2, a further 23 isolates had eight different FII_AY458016-like variants containing up to 12 SNVs among them; almost all of these variants were in isolates with FIA-FIB-FII replicon combinations (see Fig. S3 in the supplemental material). Of the 11 clade C2 isolates without FII_AY458016-like replicon variants, four contained a plasmid with a different FII replicon (GenBank nucleotide sequence accession no. AJ851089; pRSB107, 35 SNVs different from FII_AY458016; consistent with pMLST allele 1), five had chromosomally integrated blaCTX-M-15 (of which four also contained an FII_ AJ851089-like plasmid), one was blaCTX-M negative, and one contained deletions in blaCTX-M-15. There were only nine clade C2 isolates with FII_AY458016-like replicons but no blaCTX-M-15. The different FII replicon associated with blaCTX-M-15 in clade C2, FII_AJ851089, was also clade associated, being found predominantly in clades A (13/25 isolates, 52%) and C1 (41/57, 72%) rather than B (12/51, 24%) and C2 (8/82, 10%) (P < 0.0001, Fisher exact test) (see Fig. S3). Overall, this strongly suggests the ancestral acquisition of the FII_AY458016 replicon within clade C2, its association with blaCTX-M-15 and the expansion of the clade, its evolution in the presence of FIA-FIB replicons, and its sporadic loss.
Plasmid transformants demonstrate similarities and differences in blaCTX-M-15 plasmids from ST131 clades and other sequence types.
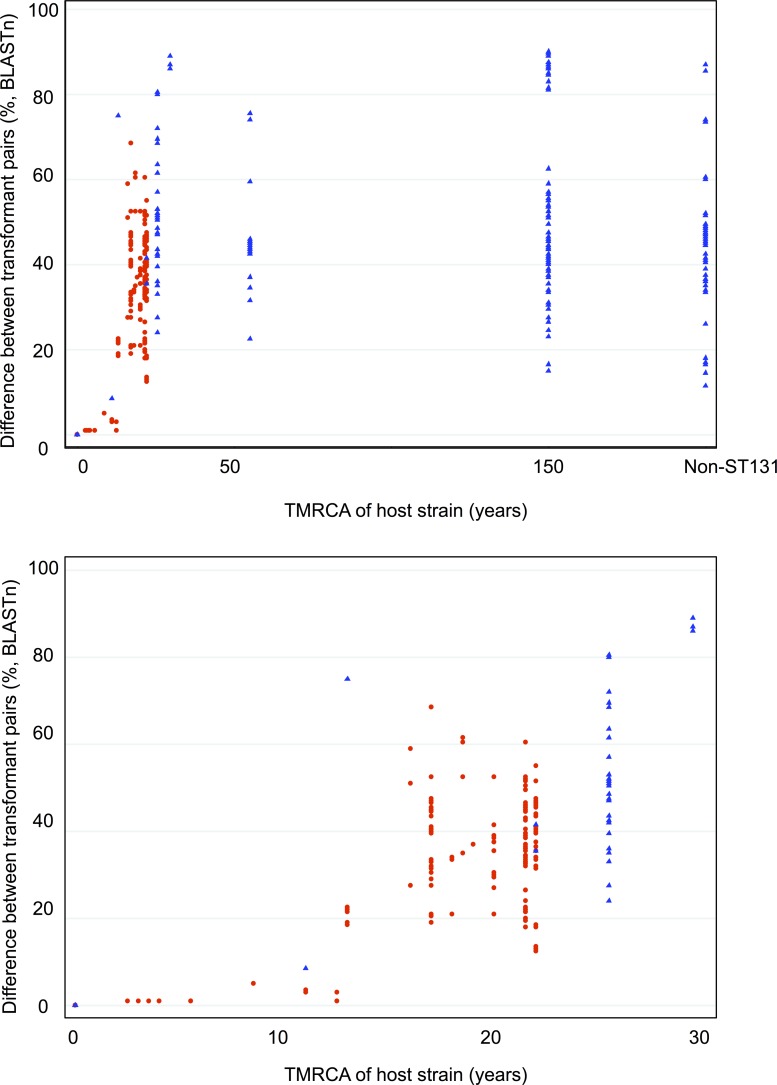
Sequence data were generated for 30 transformed blaCTX-M plasmids (denoted as <host isolate name> _T; relevant source strains are labeled “T” in Fig. 1): four from clade A, containing blaCTX-M-15 (n = 1), blaCTX-M-14 (n = 2), and blaCTX-M-27 (n = 1); one from clade B, containing blaCTX-M-55; three from clade C1, containing blaCTX-M-14 (n = 2) and blaCTX-M-24 (n = 1); 20 from clade C2, containing blaCTX-M-15; and two blaCTX-M-15 plasmids from non-ST131 isolates (see Table S2 in the supplemental material). The mean percent pairwise differences in plasmid sequence between all plasmid pairs were compared with the divergence times of the corresponding host strains. This demonstrated that all transformed blaCTX-M plasmids shared at least 10% homology but could be genetically divergent (Fig. 4), plasmids found in different STs could be very similar (up to ~90% sequence homology), and plasmid genetic similarity correlated with host strain divergence time for recently diverged host strains (up to ~30 years) but was much more variable for more remotely diverged host strains.
FIG 4 .
Mean pairwise percent difference between all transformed plasmid sequence pairs plotted against time to most recent common ancestor (TMRCA) for the two strains hosting the respective transformant plasmids. Red circles indicate pairs where both strains are in C2; blue triangles indicate pairs where one or both strains are outside C2. The lower panel represents the same data but limited to strains with a TMRCA of less than 30 years.
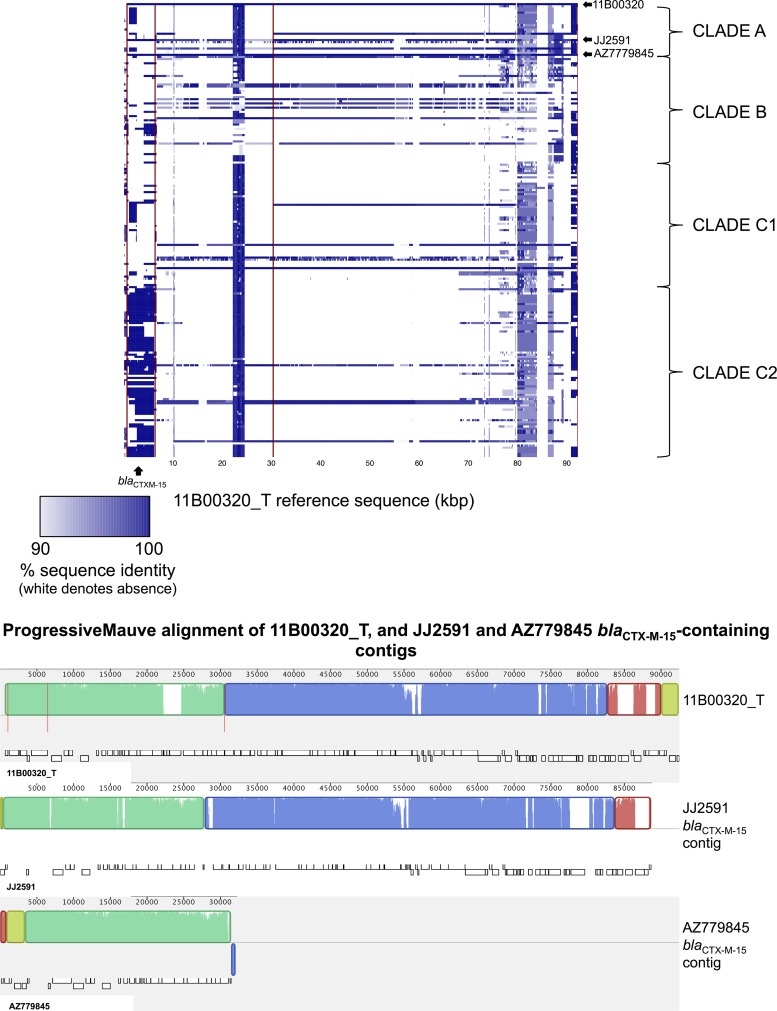
Most transformed blaCTX-M plasmids were IncF, except in two cases (11B00320_T and la_7619_T). BLASTn-based comparisons revealed that the clade A blaCTX-M-15 IncI plasmid (11B00320_T; isolated in Mae Sot, Thailand-Myanmar border) was circulating in a limited fashion (Fig. 5) but with substantial sequence homology to blaCTX-M-15-containing contigs from the two other clade A blaCTX-M-15-positive isolates (JJ2591, Minneapolis, MN, USA, and AZ779845, Spain). Although we did not have transformants or specific plasmid sequences for these, the blaCTX-M-15-containing contig assembled for JJ2591 was 88,693 bp long and very similar to the 11B00320_T assembly, whereas the AZ779845 blaCTX-M-15-containing contig was 32,228 bp long and likewise highly similar in structure (Fig. 5). These data suggest that an IncI-CTX-M-15 plasmid is responsible for sporadic, horizontal introductions of blaCTX-M-15 into ST131 with a wide geographic distribution.
FIG 5 .
(Top) BLASTn-based comparison across the ST131 data set, using the blaCTX-M-15-containing 11B00320_T as a reference. Color represents degree of presence/absence of identity to the 11B00320_T sequence on an isolate-by-isolate basis per row. Rows/isolates are arranged as in the Fig. 1 phylogeny. (Bottom) ProgressiveMauve alignment of 11B00320_T and the CTX-M-15-containing contigs for two other isolates in clade A. Alignments with substantial homology are represented as similarly colored blocks (“locally collinear blocks”); white regions within these blocks represent low homology. Vertical red lines represent contig breaks.
Nineteen of 20 transformed blaCTX-M-15 plasmids from clade C2 contained an FII_ AY458016-like replicon, supporting the association of IncFII_AY458016 with blaCTX-M-15. Sequence comparisons among 17 (of 20 total) plasmids from clade C2 that contained IncFII_AY458016 identified a significant degree of homology (Fig. 6) (excluding 8A16G_T, 11B01979_T, and 19B19L_T; see Materials and Methods). However, only eight coding sequences were shared with 100% nucleotide similarity, including blaCTX-M-15, blaOXA-1, aac(6')-Ib-cr, a glucose-1-phosphatase-like-enzyme, a CAAX amino-terminal protease self-immunity protein, a hypothetical phage protein, and a pemI/pemK plasmid addiction system. This lack of gene conservation suggests that significant genetic exchange and rearrangement occur among these plasmids as they evolve within the subclade.
FIG 6 .
ProgressiveMauve alignment of assembled contigs, ordered with respect to pP46212, for 17 blaCTX-M-15 FII plasmids derived from sequenced transformants, all in clade C2/H30Rx. Plasmids are ordered with respect to the position of their host strains in the main phylogeny (except pP46212 [Fig. 1]). Alignments with substantial homology are represented as colored blocks (“locally collinear blocks”) and are linked with colored lines; white regions within these blocks represent low homology.
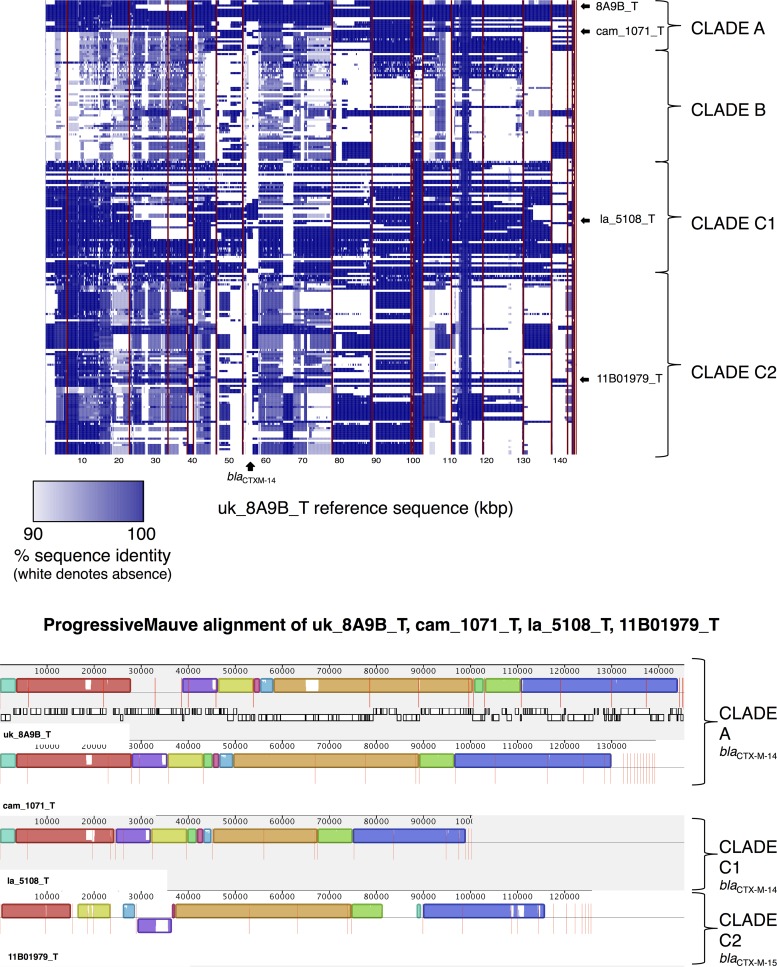
Genetic comparisons among the transformed blaCTX-M-14/14-like plasmids revealed that three shared strikingly similar genetic structures, two of which (uk_8A9B_T, Oxford, United Kingdom, and cam_1071_T, Siem Reap, Cambodia) were identified in clade A, in host strains with a TMRCA within the last 15 years, and one in clade C1 (la_5108_T, Vientiane, Laos) (Fig. 7). BLASTn-based comparisons across all 215 ST131 sequences demonstrated that many isolates in clades A (predominantly subcluster [i]) and C1 apparently contained stretches of highly similar genetic structures, as did small numbers of isolates in clade C2 (Fig. 7). 11B01979_T (Mae Sot, Thailand-Myanmar border), a transformed blaCTX-M-15 plasmid in clade C2, also showed significant homology to uk_8A9B_T, cam_1071_T, and la_5108_T (Fig. 7), suggesting that both blaCTX-M-14 and blaCTX-M-15 variants can be accommodated on the same plasmid background.
FIG 7 .
(Top) BLASTn-based comparisons across the ST131 data set, using the blaCTX-M-14-containing uk_8A9B_T as a reference. Color represents degree of presence/absence of identity to the uk_8A9B_T sequence on an isolate-by-isolate basis per row. Rows/isolates are arranged as in the Fig. 1 phylogeny. (Bottom) ProgressiveMauve alignment of uk_8A9B_T, cam1071_T, la_5108_T, and 11B01979_T, with the last three ordered using uk_8A9B_T as a reference and contig boundaries represented as vertical red lines. Alignments with substantial homology are represented as colored blocks (“locally collinear blocks”); white regions within these blocks represent low homology. Vertical red lines represent contig breaks.
The isolates containing blaCTX-M-55 and blaCTX-M-24 (one-SNV derivatives of blaCTX-M-15 and blaCTX-M-14, respectively) apparently resulted from discrete plasmid acquisition and/or blaCTX-M transposition events within ST131 (see Fig. S4 in the supplemental material). These were not therefore shown to represent blaCTX-M evolution within established blaCTX-M-15 or blaCTX-M-14 plasmid backgrounds.
DISCUSSION
Our WGS analysis of the largest (n = 215) and most diverse collection of ST131 isolates to date establishes that the global emergence of drug-resistant clades (C1/H30, C2/H30Rx) occurred approximately 25 years ago, most likely in a North American context and consistent with strong selection pressure exerted by the widespread introduction and use of fluoroquinolones and extended-spectrum cephalosporins. Interestingly, this appears to be at odds with the previous observation that ESBLs predominating in North America in the 1990s and early 2000s were mostly blaTEM or blaSHV variants (23); however, the studies summarized in this review were mostly undertaken in nosocomial and/or critical care settings, investigated non-E. coli Enterobacteriaceae, used phenotypic screening methods that may have missed the emergence of blaCTX-M in E. coli (e.g., higher extended-spectrum cephalosporin breakpoints, focused on beta-lactam/beta-lactamase inhibitor-resistant isolates), or did not test specifically for blaCTX-M variants.
Although members of each ST131 clade have dispersed globally, our data indicate that within specific geographic regions, smaller clonal ST131 outbreaks occur at all genetic levels (gene, flanking context, plasmid, and host strain), supporting the hypothesis that both horizontal gene transfer and clonal expansion have contributed to the global dissemination of this sequence type. The estimated molecular evolutionary rate of ST131 (1.00 mutation per genome per year) is similar to previous estimates from ST131 (24) and the species overall (25), strongly suggesting that ST131’s epidemiological success is not due to a higher-than-average mutation rate.
Our study shows that the apparent persistence of particular blaCTX-M variants within specific ST131 clades is due to diverse mechanisms. These include (i) acquisition of a blaCTX-M-containing plasmid by a specific host strain subcluster, followed by evolution and spread across geographic regions (e.g., clade A blaCTX-M-14 subcluster [i] [Fig. 6; see also Fig. S2 in the supplemental material]); (ii) multiple discrete acquisition events involving blaCTX-M-containing plasmids (e.g., blaCTX-M-55 and blaCTX-M-24 [see Fig. S4]; different blaCTX-M-14 clusters); (iii) horizontal transfer of common plasmid structures across clades (e.g., the IncI blaCTX-M-15 plasmid [Fig. 5]); and (iv) chromosomal integration of blaCTX-M and evolution by descent (e.g., blaCTX-M-15 [Fig. 2]; blaCTX-M-14). Despite this high degree of genetic plasticity, we also found clear structuring of blaCTX-M variants and plasmid content, with the near-complete absence of blaCTX-M in clade B and associations of blaCTX-M-14/14-like variants with clade A and clade C1/H30R, of blaCTX-M-15 with clade C2/H30Rx, and of specific combinations of IncF replicons with certain clades. This supports the hypothesis that some plasmid replicons are acquired and persist stably within clades. Although the evolutionary dynamics of plasmid-host combinations remain to be clearly elucidated, coevolution of host and plasmid in the case of C2/H30Rx appears to have ameliorated costs to the host and facilitated persistence of the replicon (26, 27), with ongoing conjugative exchange of genetic material. The relative contribution of changing environmental influences on this coevolution is unclear; it may also be affected by a host-plasmid “arms race” in a microevolutionary version of the “Red Queen Hypothesis” (antagonistic coevolution) (28, 29).
The almost ubiquitous presence of blaCTX-M-15 in clade C2/H30Rx is most striking and is strongly associated with the presence of an IncFII_AY458016-like replicon. Previous smaller studies have found that blaCTX-M-15 is frequently part of a 2,971-bp ISEcp1-blaCTX-M-15-ORF477 transposition unit, with blaCTX-M-15 located 48 bp downstream of the ISEcp1 IRR-R, and that this is commonly nested within a Tn2 element (30). One hypothesis is that an IncFII_AY458016 ancestral plasmid was acquired by a fluoroquinolone-resistant C1 host strain approximately 25 years ago and subsequently incorporated one of these blaCTX-M-15 transposition units. In response to the widespread clinical use of extended-spectrum cephalosporins and fluoroquinolones, the C2/H30Rx clade has expanded, and within it, blaCTX-M-15 has been mobilized through further transposition events (e.g., to the chromosome) and rearrangement/recombination among IncFII-like plasmids, much of this associated with IS26 (31) (Fig. 3). The persistence of the IncFII_AY458016-like replicon in C2 may be attributable, at least in part, to its association with a plasmid addiction system (pemI/pemK) (32), whereas its ongoing evolution is potentially linked to the concomitant presence of FIA/FIB replicons on blaCTX-M-15 plasmids (see Fig. S3 in the supplemental material) (33). Alternative hypotheses could be envisioned, e.g., multiple, clade C2-restricted acquisitions of different blaCTX-M-15-containing FII_AY458016-like plasmids or recurrent ISEcp1-blaCTX-M-15-ORF477 unit acquisitions. These seem less likely, however, because (i) there are no geographic or major genotypic distinctions between clades C1 and C2 to explain why this would occur, (ii) there is a degree of homology in the flanking contexts around the gene throughout the clade, and (iii) flanking context/transformed plasmid structures also appear to be consistent within C2 subclusters.
Our novel comparison of transformed, sequenced plasmids demonstrates that a substantial degree of similarity can exist among blaCTX-M plasmids found in different clades and STs. This indicates that between-clade/ST transfer of these resistance plasmids occurs and that care is needed when inferring plasmid evolution by descent (Fig. 4). The observed plasmid similarity across geography in the context of host strain phylogenetic clustering and homology in regions flanking blaCTX-M (as demonstrated here) is much more likely to represent ancestral plasmid acquisition and subsequent evolution by descent rather than multiple acquisition events but still needs to be interpreted with caution, as it may, for example, represent exposure to a common, global, plasmid reservoir.
The study has several limitations. First is the inability with short-read sequencing and limited transformant sequencing to assess fully the flanking regions and plasmid structures across the entire data set. In particular, the BLASTn-based heat maps across the wider data set represent not genetic contiguity of plasmid structures within isolates as such but instead overall plasmid sequence presence/absence. Second, results from de novo assemblies of these short-read data also must be interpreted cautiously, as these assembly methods are known to increase the number of SNVs compared with mapping-based approaches and may result in misinterpretations of genetic structures, particularly repetitive regions (34). Third, again relating to the limitations of short-read data, the transformant plasmid sequences comprise multiple contigs, precluding certainty as to the plasmids’ exact structure. More extensive future use of long-read sequencing (e.g., PacBio) could help resolve this. Fourth, many of our H30Rx/C2 clade transformed CTX-M plasmids were from a single United Kingdom center; however, the genetic flanking contexts identified here have also been found in plasmid sequences from other national and international locations (30, 35, 37), suggesting that these are dispersed more widely and that our results are likely generalizable.
In summary, our analysis strongly suggests that the emergence of the C2/H30Rx clade within ST131 has been driven by the acquisition of a specific FII plasmid, which has subsequently undergone major genetic restructuring within its globally dispersing bacterial host. The initial acquisition event occurred approximately 25 years ago, possibly associated with the widespread clinical introduction of extended-spectrum cephalosporins and fluoroquinolones, which would have exerted significant selection pressure for persistence of chromosomal fluoroquinolone mutations and presence of blaCTX-M. Sporadic gain/loss events involving other, non-FII blaCTX-M-15 plasmids have also occurred but have not dominated. Similar processes may be driving the more recent emergence of sublineages of ST131 with blaCTX-M-14 and blaCTX-M-27, as described in Japan (36), although for blaCTX-M-14, these appear to have occurred on at least two occasions (clades A and C1/H30R [Fig. 1]). This study highlights the global imperative to reduce antimicrobial selection pressures; the capacity of these resistance plasmids for genetic reassortment; the important role of certain insertion sequences, such as IS26, in facilitating horizontal mobility of resistance determinants; and the possibility of targeting specific replicons in an attempt to limit the spread of important resistance gene mechanisms.
MATERIALS AND METHODS
Sample collection, sequencing, and sequence read processing.
Isolates were obtained from wider collections held in several centers: the Shoklo Malaria Research Unit, Mae Sot, Thailand; the Lao-Oxford-Mahosot Hospital Wellcome Trust Research Unit, Vientiane, People’s Democratic Republic of Laos; the Cambodia-Oxford Medical Research Unit, Angkor Hospital for Children, Siem Reap, Cambodia; and the Microbiology Laboratory, Oxford University Hospitals NHS Trust, Oxford, United Kingdom. No two isolates were taken from the same individual. In addition, seven isolates collected from clinical samples across Canada between 2006 and 2008 and one isolate recovered from poultry in 2006 were included. DNA was extracted as previously described (38). Sequence data for the eight AstraZeneca strains had been generated from a series of isolates collected by International Health Management Associates, Inc., as part of a global resistance survey; the data for the Price strains were as previously described (13). Sequencing was performed using either the Illumina HiSeq or the MiSeq sequencer (100- or 151-bp paired-end reads [details for non-Price strains are in Table S1 in the supplemental material]). Sequence type was confirmed using BLASTn-based (39) in silico multilocus sequence typing (MLST) of de novo-assembled WGS data (40).
Properly paired sequence reads were mapped using Stampy v1.0.17 (without Burrows-Wheeler Aligner premapping, using an expected substitution rate of 0.01) to a fully sequenced E. coli ST131 reference (E. coli O150:H5 SE15; RefSeq NC_013654), in order to limit bias introduced by mapping to a more divergent reference. Repetitive regions (166,828 bases, 3.5%) of the reference were identified using self-self BLASTn analysis with default settings; these regions were then masked prior to mapping and base calling. Single-nucleotide variants (SNVs) were determined across all mapped nonrepetitive sites using SAMtools (version 0.1.18) mpileup. mpileup was run twice to separate high-quality base calls from low-quality base calls: first, with options “-E -M0 -Q25 -q30 -m2 -D -S” and otherwise default settings, and second, with options “-B -M0 -Q0 -q0 -m2 -D -S” and otherwise default settings. Vcf files of annotated variant sites were created using GATK (v1.4.21). Base calls derived from these two Vcf files were then retained only if (i) the proportion of high-quality bases supporting the call was ≥90%, and ≥5 high-quality bases were required as a minimum; (ii) the root of the mean square mapping quality of reads covering a putative variable site was ≥30; (iii) the Phred scaled quality supporting a base call was ≥25; and (iv) reads spanning the putative variable site were made up of ≥35% high-quality bases. Core variable sites (base called in all sequences, excluding “N” or “-” calls) derived from mapping to the SE15 reference were “padded” with invariant sites in a proportion consistent with the GC content and length of the reference genome (4.72 Mb, 51% average GC content), to generate a modified alignment of input sequences for our phylogenetic analyses (see below).
De novo assemblies were generated using Velvet with the VelvetOptimiser wrapper (n = 211) (41) (http://bioinformatics.net.au/software.velvetoptimiser.shtml) or A5-MiSeq (42). The latter was used in cases where the number of assembled bases was below the expected assembly size of 4 to 5.5 Mb (n = 4 [strains la_12107_3, can_70883, can_1731_01, and can_1070] in which the median optimized assembly size with Velvet was 16,004 bases and the median number of contigs was only six). Using A5-MiSeq, assemblies for these four strains were generated with an appropriate median size of 5,143,908 bp and 269 contigs.
Identification/characterization of blaCTX-M and genetic context, gyrA mutations, and fimH typing.
BLASTn analysis of de novo assemblies was used to identify: (i) blaCTX-M presence and variants (in-house reference gene database) (38); (ii) genetic context for blaCTX-M, by extracting and annotating contigs containing blaCTX-M variants using PROKKA and ISFinder (manual annotation) (43, 44); (iii) chromosomal gyrA mutations in the quinolone resistance-determining region known to be responsible for conferring most resistance to fluoroquinolones; (iv) fimH presence and variant (45); and (v) Inc type using the downloaded PlasmidFinder (46) and pMLST databases (available at http://pubmlst.org/plasmid/) (47). Genetic contexts for blaCTX-M were classified as chromosomal if annotations for regions flanking blaCTX-M were found to be consistently chromosomal in other E. coli strains in GenBank and plasmid if these were associated specifically with plasmids (e.g., tra genes); otherwise, they were classified as unknown. IncFII_AY458016-like sequences were extracted, aligned, and visually inspected to confirm variant types using Geneious (version 7.1.9; Biomatters Ltd., Auckland, New Zealand) (48).
ST131 chromosomal phylogenetic comparisons using ClonalFrame, BEAST, and BASTA.
ExPEC bacteria are recombinogenic and contain recombination hot spots with higher-than-average recombination rates (49). Recombination can obscure the clonal phylogenetic signal, and we therefore initially analyzed the alignment of sequences with ClonalFrame (50) to identify recombinant regions. Three separate runs were performed on the alignment of ST131 sequences, with the following settings: 2,000 burn-in iterations, 2,000 Monte Carlo Markov chain (MCMC) iterations following the burn-in period, and 2 iterations between recording parameter values in the posterior sample (the thinning interval). Convergence of the runs was assessed by comparing the similarities of the run outputs. SNVs within regions identified as recombinant from the consensus ClonalFrame output were ignored in the subsequent BEAST/BASTA analyses.
Using the modified alignment of ST131 sequences generated following the ClonalFrame analysis, mutation rate estimates across ST131 and a time-scaled phylogeny were calculated in BEAST (51). The model parameters were (i) a generalized time-reversible nucleotide substitution model; (ii) four relative rates of mutation across sites, allowing for all sites to be subject to mutation (i.e., the proportion of invariant sites fixed at 0%); (iii) a strict molecular clock estimating a uniform evolutionary rate across all branches of the tree; and (iv) a constant population size. Triplicate runs with 30 million iterations were performed, with 10% discounted as burn-in. Run convergence and mixing were assessed by inspecting the run log files in Tracer v1.5 (http://beast.bio.ed.ac.uk); adequate convergence of run statistics and mixing for each run and effective sample sizes (ESSs) for all parameters greater than 200 were required for an analysis to be considered adequate, in line with recommendations in the BEAST tutorials on the developers’ website (http://beast.bio.ed.ac.uk). We explored the application of several other models in BEAST incorporating the relaxed clock and variable population growth (exponential, logistic, and Bayesian skyride), but these either failed to converge, showed poor mixing, or had effective sample size (ESS) estimates of <200 and were therefore not considered robust.
We used the phylogeographic method BASTA (20) in the Bayesian phylogenetic package BEAST 2.2.1 (52) to infer patterns and rates of migration between geographical regions from the genome alignment, collection dates, and sampling locations. Initially, we grouped samples into three discrete locations, North America, Southeast Asia, and Europe, and disregarded samples from South America and Australasia because of the small sample numbers. Due to the nonrandom sampling scheme, we estimated only a single effective population size, equal for all locations, and a symmetric migration rate matrix. The analysis was run for 108 MCMC steps. We subsequently reran the analysis including a fourth, unsampled deme, using the same model parameters, to determine whether this altered the outcome.
Plasmid transformations, sequencing, and analyses.
Plasmid transformants were generated from 30 strains chosen on the basis of tree topology and association with CTX-M variants, aiming to transform at least one plasmid from each of the major CTX-M variant clusters. Two blaCTX-M-containing plasmids from non-ST131 E. coli (one ST617/blaCTX-M-15 and one ST405/blaCTX-M-55) were also transformed and sequenced as an external comparison.
Plasmid DNA was extracted from subcultures of frozen stock grown overnight on blood agar, followed by selective culture of a single colony in lysogeny broth (BD/Difco LB broth; Miller [Luria-Bertani]; catalog no. 244620) with ceftriaxone at 1 µg/ml. DNA extraction was performed using the Qiagen plasmid minikit (Qiagen, Venlo, Netherlands), in accordance with the manufacturer’s instructions, with the addition of Glycoblue coprecipitate (Life Technologies, Carlsbad, CA, USA) to the DNA eluates prior to isopropanol precipitation to enable better visualization of the DNA pellet. Plasmid DNA was redissolved in distilled water and then typically electroporated on the same day or stored in the refrigerator prior to electroporation within 24 h.
Commercially prepared DH10B E. coli (ElectroMAX DH10B cells; Invitrogen/Life Technologies, Carlsbad, CA) was used as the recipient cell strain for plasmid electroporation, because of its high transformation efficiency and the fact that the strain has been fully sequenced (NCBI RefSeq NC_010473.1) (53). Electroporation was performed with a MicroPulser electroporator (Ec2 settings). Transformant cell suspensions were cultured on selective agar (Luria-Bertani agar plus ceftriaxone [1 µg/ml]), with appropriate controls.
Sequencing was performed on the Illumina HiSeq or MiSeq sequencer, generating 150- or 300-base paired-end reads (see Table S2 in the supplemental material). Sequencing reads from the isolate from which the transformed plasmid had been obtained were mapped back to the transformed plasmid assembly in order to ascertain the reliability of the assembly in each case. Reads were assembled using A5/A5-MiSeq (42), and assembled contigs were annotated with PROKKA (43). The median plasmid assembly size was 122,786 (range, 72,449 to 171,919), with a median of 22 contigs in each assembly (range, 1 to 33). Using longer reads (300 bp; MiSeq platform) resulted in a significantly smaller number of contigs per assembly (median, 17 versus 25; rank sum P = 0.003). Mapping was used to assess the reliability of our plasmid constructs and reflected the content present in each transformed and assembled resistance plasmid, with the exception of 8A16G_T.
A single strain (P46212) from the data set was also sequenced using long-read technology (PacBio); the CTX-M-15 plasmid (pP46212) from this strain was assembled into a single, circularized contig as described elsewhere (A. E. Sheppard, N. Stoesser, D. J. Wilson, R. Sebra, A. Kasarskis, L. W. Anson, A. Giess, L. J. Pankhurst, A. Vaughan, C. J. Grim, H. L. Cox, A. J. Yeh, Modernising Medical Microbiology Informatics Group, C. D. Sifri, A. S. Walker, T. E. A. Peto, D. W. Crook, and A. J. Mathers, submitted for publication).
Plasmid content across the data set was investigated in a number of ways. First, the transformed plasmid sequences that we generated were used as references against which BLASTn-based comparisons for degree of presence/absence were made for the whole data set. We used default BLASTn settings to compare the de novo assembly for each ST131 isolate with each respective, concatenated plasmid reference sequence. Sequence identity of BLASTn hits across the plasmid reference was plotted using the heatmap.2 package in R, with a minimum threshold of 90% identity for plotting. For simplicity, values were averaged over 100-bp bins (script available at https://github.com/aesheppard/plasmid_comp).
Second, comparisons between each pair of transformed plasmid assemblies were undertaken, again using BLASTn with default settings. For the query sequence in each comparison, the percentage of sites contained in hits (counting overlapping hits only once) was identified from parsed blast output. For each pair, two percentage of homology statistics were generated, taking each member of the pair as a reference in turn, to account for differences in length (script available at https://github.com/aesheppard/plasmid_comp). The mean percent divergence for each plasmid sequence pair was then plotted against the time to most recent common ancestor (TMRCA) of the two host strains containing those transformed plasmid sequences (derived from the time-scaled tree) in Stata (SE) (StataCorp, Texas, USA; version 11.2).
Third, for visualization, plasmid sequences were compared using ProgressiveMauve (54), with assembled contigs reordered with respect to the pP46212 PacBio-generated CTX-M-15 plasmid reference, using the “Move contigs” tool. For this, three transformed plasmid sequences were excluded: 8A16G_T because of issues surrounding the assembly, 11B01979_T because it was virtually identical to transformed blaCTX-M-14 plasmid sequences in clade A, and 19B19L_T because it lacked an FII replicon. Finally, annotated, transformed plasmid sequences were clustered using CD-Hit (55) [-c 1.0 -n 5 -d 0 -g 1], to identify whether any coding sequences were shared and whether there might be any biological significance associated with these on the basis of their annotations.
Sequencing data resources.
The positions for called, variable sites across the data set (with respect to the reference SE15 E. coli genome) are listed in Text S1 in the supplemental material, and the positions in recombinant regions (and therefore not included in the phylogenetic analyses) are listed in Text S2. Contigs for the de novo assemblies for the transformed plasmid sequences are in Text S3, and those for all the new isolates are freely downloadable at http://modmedmicro.nsms.ox.ac.uk/stoesser-n-et-al/.
Accession numbers.
Sequencing data for the new isolates sequenced for this study have been deposited in the NCBI Short Read Archive (BioProject number PRJNA297860, 108 ST131 sequences and 30 blaCTX-M plasmid transformants [see Tables S1 and S2 in the supplemental material]). The uk_P46212 sequence assembled using PacBio is available from GenBank (accession numbers CP013658 [chromosome] and CP013657 [CTX-M-15 plasmid]).
SUPPLEMENTAL MATERIAL
Genetic contexts of clade A-associated blaCTX-M-14/14-like variants. Many contexts are limited by the extent of the assembled region around the blaCTX-M-14/14-like gene (marked with “X”). For all aligned, similarly colored regions, sequence homology is preserved; curly brackets cluster those isolates with identical flanking sequences. Flanking contexts not shown for isolates with known chromosomal integration or for blaCTX-M-negative/non-blaCTX-M-14/14-like isolates in the subclusters. Coloring of isolate names reflects geographic locations (blue, North America; red, Europe; green, Southeast Asia; yellow, Australasia). Download
Genetic contexts of clade C1-associated blaCTX-M-14/14-like variants. Many contexts are limited by the extent of the assembled region around the blaCTX-M-15 gene (marked with “X”). For all aligned, similarly colored regions, sequence homology is preserved; curly brackets cluster those isolates with identical flanking sequences. Flanking contexts not shown for isolates with known chromosomal integration or for blaCTX-M-negative/non-blaCTX-M-14/14-like isolates in the subcluster (blue, North America; red, Europe; green, Southeast Asia; yellow, Australasia). Download
Inc types identified in whole-isolate sequencing data, plotted with respect to ST131 host strain phylogeny. Blast match (%) denotes a composite score of percent matched length and percent homology to reference Inc sequence, with highest percent score/contig hit represented. Matches of <80% were excluded. Reference Inc sequences were downloaded from the PlasmidFinder database; those that were present (Blast match of ≥80%) in at least one isolate are represented on the x axis. Download
BLASTn-based comparisons across the ST131 data set, using la_12107-3_T and la_5220-3_T as references. Color represents degree of presence/absence of corresponding reference sequence on an isolate-by-isolate basis per row. Rows/isolates arranged as in the Fig. 1 phylogeny. Download
List of positions for called, variable sites across the data set, with respect to the reference SE15 E. coli genome. Download
List of positions identified as being in recombinant regions through the ClonalFrame analysis. Download
Fasta file of contigs for the de novo assemblies for the transformed plasmid sequences. Download
Details of newly sequenced ST131 strains included in the analysis.
Details of transformed blaCTX-M plasmid sequences.
ACKNOWLEDGMENTS
We are grateful to the patients and staff at the health care, microbiology laboratory, and research units contributing isolates to this study, including Nicholas Day of the Mahidol-Oxford Tropical Medicine Research Unit, Bangkok, Thailand; Paul Newton and David Dance of the Lao-Oxford-Mahosot Hospital-Wellcome Trust Research Unit, Vientiane, Laos; the Ped Study Team and Microbiology Laboratory at Patan Hospital, Kathmandu, Nepal; and Francois Nosten of the Shoklo Malaria Research Unit, Mae Sot, Thailand. We thank Peter Donnelly and the staff at the Sequencing Center, Wellcome Trust Center for Human Genetics, Oxford, United Kingdom, for their sequencing work and Laura Matseje of the Public Health Agency of Canada for sharing her laboratory protocol for plasmid transformation. We are grateful to Johann Pitout, Nicholas Day, Amy Mathers, and Chris Parry for their critical review of the draft manuscript.
This material is based in part upon work supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant no. 1 I01 CX000192 01 (J.R.J.), and NIH R01 AI106007 (E.S.). A.R.M. is supported through funding from the Canadian Institutes of Health Research (MOP-114879). N.S. was funded through a Wellcome Trust Clinical Research Fellowship during this study (099423/Z/12/Z).
J.R.J. has received grants and/or consultancies from Actavis, ICET, Janssen/Crucell, Merck, Syntiron, and Tetraphase. J.R.J., L.B.P., and E.S. have submitted patent applications pertaining to tests for specific E. coli strains. The other authors have no specific conflicts of interest to declare.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW, Modernizing Medical Microbiology Informatics Group (MMMIG). 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7(2):e02162-15. doi:10.1128/mBio.02162-15.
REFERENCES
- 1.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 3.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev 35:736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, Junka AF, Maczynska B, Denamur E. 2014. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol 52:1358–1365. doi: 10.1128/JCM.03502-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 7.Mathers AJ, Peirano G, Pitout JD. 2015. Escherichia coli ST131: the quintessential example of an international multiresistant high-risk clone. Adv Appl Microbiol 90:109–154. doi: 10.1016/bs.aambs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisse S, Diancourt L, Laouénan C, Vigan M, Caro V, Arlet G, Drieux L, Leflon-Guibout V, Mentré F, Jarlier V, Nicolas-Chanoine MH, Coli β Study Group . 2012. Phylogenetic distribution of CTX-M- and non-extended-spectrum-beta-lactamase-producing Escherichia coli isolates: group B2 isolates, except clone ST131, rarely produce CTX-M enzymes. J Clin Microbiol 50:2974–2981. doi: 10.1128/JCM.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantón R, González-Alba JM, Galán JC. 2012. CTX-M enzymes: origin and diffusion. Front Microbiol 3:110. doi: 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naseer U, Sundsfjord A. 2011. The CTX-M conundrum: dissemination of plasmids and Escherichia coli clones. Microb Drug Resist 17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 12.Novais A, Pires J, Ferreira H, Costa L, Montenegro C, Vuotto C, Donelli G, Coque TM, Peixe L. 2012. Characterization of globally spread Escherichia coli ST131 isolates (1991 to 2010). Antimicrob Agents Chemother 56:3973–3976. doi: 10.1128/AAC.00475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price LB, Johnson JR, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Billig M, Chattopadhyay S, Stegger M, Andersen PS, Pearson T, Riddell K, Rogers P, Scholes D, Kahl B, Keim P, Sokurenko EV. 2013. The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. mBio 4:e00377-00313. doi: 10.1128/mBio.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Bae IK, Jeong SH, Chang CL, Lee CH, Lee K. 2011. Characterization of IncF plasmids carrying the blaCTX-M-14 gene in clinical isolates of Escherichia coli from Korea. J Antimicrob Chemother 66:1263–1268. doi: 10.1093/jac/dkr106. [DOI] [PubMed] [Google Scholar]
- 15.Rodríguez I, Thomas K, Van Essen A, Schink AK, Day M, Chattaway M, Wu G, Mevius D, Helmuth R, Guerra B, SAFEFOODERA-ESBL Consortium . 2014. Consortium S-E: chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int J Antimicrob Agents 43:553–557. doi: 10.1016/j.ijantimicag.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 16.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jean SS, Hsueh PR. 2011. High burden of antimicrobial resistance in Asia. Int J Antimicrob Agents 37:291–295. doi: 10.1016/j.ijantimicag.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Toh H, Oshima K, Toyoda A, Ogura Y, Ooka T, Sasamoto H, Park SH, Iyoda S, Kurokawa K, Morita H, Itoh K, Taylor TD, Hayashi T, Hattori M. 2010. Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J Bacteriol 192:1165–1166. doi: 10.1128/JB.01543-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Maio N, Wu CH, O’Reilly KM, Wilson D. 2015. New routes to phylogeography: a Bayesian structured coalescent approximation. PLoS Genet 11:e1005421. doi: 10.1371/journal.pgen.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz J. 2003. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 51:1109–1117. doi: 10.1093/jac/dkg222. [DOI] [PubMed] [Google Scholar]
- 22.Poirel L, Lartigue MF, Decousser JW, Nordmann P. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother 49:447–450. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bush K. 2008. Extended-spectrum beta-lactamases in North America, 1987–2006. Clin Microbiol Infect 14(Suppl 1):134–143. doi: 10.1111/j.1469-0691.2007.01848.x. [DOI] [PubMed] [Google Scholar]
- 24.Reeves PR, Liu B, Zhou Z, Li D, Guo D, Ren Y, Clabots C, Lan R, Johnson JR, Wang L. 2011. Rates of mutation and host transmission for an Escherichia coli clone over 3 years. PLoS One 6:e26907. doi: 10.1371/journal.pone.0026907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochman H. 2003. Neutral mutations and neutral substitutions in bacterial genomes. Mol Biol Evol 20:2091–2096. doi: 10.1093/molbev/msg229. [DOI] [PubMed] [Google Scholar]
- 26.Bahl MI, Hansen LH, Sørensen SJ. 2009. Persistence mechanisms of conjugative plasmids. Methods Mol Biol 532:73–102. doi: 10.1007/978-1-60327-853-9_5. [DOI] [PubMed] [Google Scholar]
- 27.Harrison E, Guymer D, Spiers AJ, Paterson S, Brockhurst MA. 2015. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr Biol 25:2034–2039. doi: 10.1016/j.cub.2015.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Harrison E, Brockhurst MA. 2012. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol 20:262–267. doi: 10.1016/j.tim.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GD. 2014. Running with the Red Queen: the role of biotic conflicts in evolution. Proc Biol Sci 281:20141382. doi: 10.1098/rspb.2014.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partridge SR, Zong Z, Iredell JR. 2011. Recombination in IS26 and Tn2 in the evolution of multiresistance regions carrying blaCTX-M-15 on conjugative IncF plasmids from Escherichia coli. Antimicrob Agents Chemother 55:4971–4978. doi: 10.1128/AAC.00025-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carattoli A. 2013. Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Sýkora P. 1992. Macroevolution of plasmids: a model for plasmid speciation. J Theor Biol 159:53–65. doi: 10.1016/S0022-5193(05)80767-2. [DOI] [PubMed] [Google Scholar]
- 34.Stoesser N. 2014. Applications of whole genome sequencing to understanding the mechanisms, evolution and transmission of antibiotic resistance in Escherichia coli and Klebsiella pneumoniae. Ph.D. thesis University of Oxford, Oxford, United Kingdom. [Google Scholar]
- 35.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum beta-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S, Kyoto-Shiga Clinical Microbiology Study Group . 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. doi: 10.1093/jac/dkv017. [DOI] [PubMed] [Google Scholar]
- 37.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. 2009. Complete nucleotide sequences of plasmids pEK204, pEK499, and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother 53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoesser N, Batty EM, Eyre DW, Morgan M, Wyllie DH, Del Ojo Elias C, Johnson JR, Walker AS, Peto TE, Crook DW. 2013. Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J Antimicrob Chemother 68:2234–2244. doi: 10.1093/jac/dkt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 43.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 44.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weissman SJ, Johnson JR, Tchesnokova V, Billig M, Dykhuizen D, Riddell K, Rogers P, Qin X, Butler-Wu S, Cookson BT, Fang FC, Scholes D, Chattopadhyay S, Sokurenko E. 2012. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 78:1353–1360. doi: 10.1128/AEM.06663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton A, Markowitz S, Duran C, Thierer T, Ashton B, Metjies P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Didelot X, Méric G, Falush D, Darling AE. 2012. Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. BMC Genomics 13:256. doi: 10.1186/1471-2164-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. Beast 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Durfee T, Nelson R, Baldwin S, Plunkett G III, Burland V, Mau B, Petrosino JF, Qin X, Muzny DM, Ayele M, Gibbs RA, Csörgo B, Pósfai G, Weinstock GM, Blattner FR. 2008. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol 190:2597–2606. doi: 10.1128/JB.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genetic contexts of clade A-associated blaCTX-M-14/14-like variants. Many contexts are limited by the extent of the assembled region around the blaCTX-M-14/14-like gene (marked with “X”). For all aligned, similarly colored regions, sequence homology is preserved; curly brackets cluster those isolates with identical flanking sequences. Flanking contexts not shown for isolates with known chromosomal integration or for blaCTX-M-negative/non-blaCTX-M-14/14-like isolates in the subclusters. Coloring of isolate names reflects geographic locations (blue, North America; red, Europe; green, Southeast Asia; yellow, Australasia). Download
Genetic contexts of clade C1-associated blaCTX-M-14/14-like variants. Many contexts are limited by the extent of the assembled region around the blaCTX-M-15 gene (marked with “X”). For all aligned, similarly colored regions, sequence homology is preserved; curly brackets cluster those isolates with identical flanking sequences. Flanking contexts not shown for isolates with known chromosomal integration or for blaCTX-M-negative/non-blaCTX-M-14/14-like isolates in the subcluster (blue, North America; red, Europe; green, Southeast Asia; yellow, Australasia). Download
Inc types identified in whole-isolate sequencing data, plotted with respect to ST131 host strain phylogeny. Blast match (%) denotes a composite score of percent matched length and percent homology to reference Inc sequence, with highest percent score/contig hit represented. Matches of <80% were excluded. Reference Inc sequences were downloaded from the PlasmidFinder database; those that were present (Blast match of ≥80%) in at least one isolate are represented on the x axis. Download
BLASTn-based comparisons across the ST131 data set, using la_12107-3_T and la_5220-3_T as references. Color represents degree of presence/absence of corresponding reference sequence on an isolate-by-isolate basis per row. Rows/isolates arranged as in the Fig. 1 phylogeny. Download
List of positions for called, variable sites across the data set, with respect to the reference SE15 E. coli genome. Download
List of positions identified as being in recombinant regions through the ClonalFrame analysis. Download
Fasta file of contigs for the de novo assemblies for the transformed plasmid sequences. Download
Details of newly sequenced ST131 strains included in the analysis.
Details of transformed blaCTX-M plasmid sequences.