Abstract
Transposable elements (TEs) are nucleotide sequences found in most studied genomes. These elements are highly diversified and have a large variation in nucleotide structure and mechanisms of transposition. hobo is a member of class II, belonging to hAT superfamily, described inDrosophila melanogaster, and it presents in its Open Reading Frame, a repetitive region encoding the amino acids threonine-proline-glutamic acid (TPE), which shows variability in the number of repeats in some regions of the world. Due to this variability some evolutionary scenarios of the hobo element are discussed, such as the scenario of the invasion of hobo element in populations ofD. melanogaster. In the present study, we investigated 22 DNA sequences of D. melanogaster and seven sequences ofD. simulans, both from South America, to check the number of repetitions of TPE, in order to clarify the evolutionary scenario of thehobo element in these populations. Our results showed a monomorphism in populations of both species in South America, with only three TPE repeats. Hence, we discuss and propose an evolutionary scenario of the invasion of the hobo element in populations of D. melanogaster and D. simulans.
Keywords: transposable elements, transposons, hAT, genomic evolution, molecular evolution
Transposable Elements (TEs) are sequences of DNA that have the intrinsic ability to move in the host genome (Biémont and Vieira, 2006). This ability gives TEs a wide distribution in the genome of eukaryotes. In humans, 45% of genetic material is composed of TEs, while in Drosophila melanogaster about 20% and 80% of genome in maize (International Human Genome Sequencing Consortium, 2001; Meyers et al., 2001; Biémont and Vieira, 2006). Furthermore, the TEs can be involved in events of Horizontal Transfer (HT), a process by which the genetic material is transferred to another who is not his or her descendant (Capy et al., 1994).
TEs are considered mutagenic agents causing structural and regulatory changes in genes. Also, they promote chromosome rearrangements and epigenetic modifications (Biémont 2010; Hua-Van et al., 2011). TEs show a great diversity in nucleotide sequence and structural organization and are classified based in these characteristics. The first classification was proposed by Finnegan (1989) who divided TEs into two major groups, using the transposition mechanisms and the type of intermediary in the transposition process as criteria. Class I, or retrotransposons, utilize RNA as intermediary and use a mechanism called "copy-and-paste" to replicate. Class II, or transposons, use DNA as intermediary and use a mechanism called "cut-and-paste". Wickeret al. (2007) proposed a new classification system, retaining the two described classes and creating new inclusive categories such as subclass, order, super-family, family and subfamily based on sequence similarities and structural relationships.
The superfamily hAT belongs to class II. Its name is given by the initial of the first elements described and included in this superfamily:hobo from D. melanogaster, Acfrom maize and TAM 3 from Antirrhinum majus (Warren et al., 1994). These elements have characteristics in common, such as the presence of terminal inverted repeats (TIRs), generally 12 base pairs (bp) long and produce a duplication of the target site (TSDs) of eight bp. The hAT superfamily is old and their wide occurrence in higher taxonomic groups (animals, plants and fungi) suggests the results of many HT events (Rubin et al., 2001).
The hobo element described in D. melanogaster by McGinnis et al. (1983) is observed in three different forms in the host genome: i) as a canonical and autonomous element, ii) canonical elements with internal deletion, and iii) the "relic" elements that are degenerated copies (Ortiz and Loreto, 2008). Evidence has shown that the autonomous elements as well as those sequences showing internal deletions are recent in the genomes, but the "relics" are old and probably the result of ancient invasions (Periquet et al., 1994; Simmons et al., 1998; Galindo et al., 2001; Torres et al., 2006).
hobo has in its Open Reading Frame (ORF) a repetitive sequence ACTCCAGAA that encodes the threonine-proline-glutamic acid amino acids (TPE) and presents variability in the number of copies of this repetitions (Calvi et al., 1991; Bazin and Higuet, 1996; Souames et al., 2003a, b). The majority of natural populations of D. melanogaster are monomorphic, with three copies of repetitions, while others are polymorphic. These are found in the center of Europe, in equatorial Africa and western South America, with differences in the number of copies, ranging from three to nine repetitions (Bazin and Higuet, 1996; Bonnivardet al., 2000). A temporal and geographic distribution pattern relative to TPE copy number have been observed in the populations of D. melanogaster . From this pattern, some evolutionary scenarios are proposed. For example, it is suggested that the first invasion of hobo elements in populations of D. melanogaster, throughout the world, was made by elements containing three repeats (Periquet et al., 1989), followed by another invasion ofhobo elements containing five to seven repetitions of TPE (Bonnivard et al., 2000; Souames et al., 2003a, b). The TPE region of hoboelements also appears to be related to the invasive ability of the element. Comparative studies with transgenic strains of D. melanogaster, containing 3 or 5 TPE repeats suggests that the TPE region has a relationship with transposition or activation of hobo elements, such that copies of hoboelements containing three TPE increased the transposition activity in the host genome compared to copies containing five repetitions (Souameset al., 2003a).
The hobo element has an interesting history in relation to this pattern. Research with old strains of D. melanogaster of South America (collected before 1950) showed that the canonical element was absent in these populations, while recent strains present the element. This suggests that the element has been introduced in D. melanogaster by HT (Periquet et al., 1989; Boussy and Daniels, 1991; Pascual and Periquet, 1991). Streck et al. (1986) and Boussy and Daniels (1991) described the hobo element in D. simulans, and the evolutionary history of the element in this species is quite similar to the history in D. melanogaster. However, these authors suggest that D. simulans may have been the donor of the element toD. melanogaster, as the hobo element is similar between the two species, and yet it apparently was present in D. simulans before D. melanogaster.
Thus, using the TPE region of the hobo element as a marker, the objective of this study was to investigate the polymorphism in populations of D. melanogaster in South America, trying to establish a geographic distribution scenario of the element in these populations. We also analyzed the TPE regions in Brazilian populations of D. simulans trying to determine the monomorphic condition of three repetitions of TPE, as supposed by the invasion theory ofhobo elements in D. melanogaster from D. simulans.
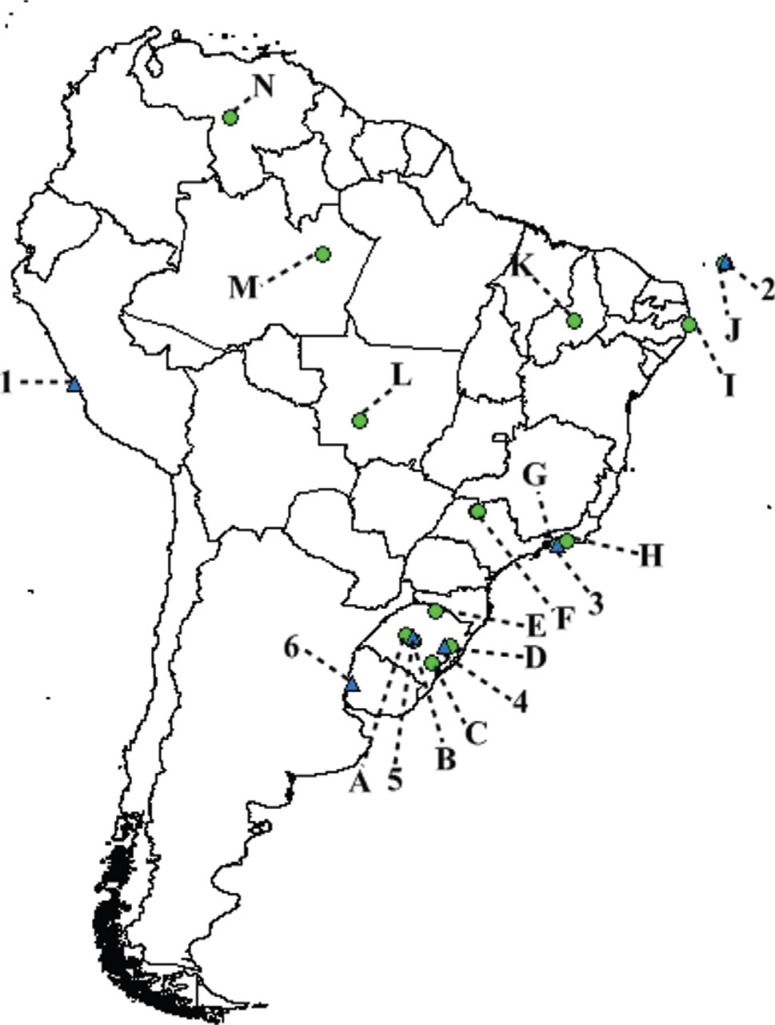
We utilized populations of D. melanogaster and D. simulans from different locations in South America and Europe (see Table 1 and Figure 2). European populations were used as a control since they are known to possess polymorphisms in the TPE region. The polymorphism analyses were made by Sanger DNA sequencing and polyacrylamide gel electrophoresis (PAGE). DNA extraction was performed in approximately 15 individuals of each population studied according to the protocol of Oliveira et al.(2009).
Table 1. Table showing the number of repetitions of TPE in populations of D. melanogaster and D. simulans in South America and the United States, including four European populations.
| Population | Species | Collection site; (State / Province); country | TPE repeats | Method used to obtain the number of repetitions of TPE |
|---|---|---|---|---|
| 1 | D. melanogaster | Rio de Janeiro; (RJ); Brazil | 3 | Sequencing |
| 2 | D. melanogaster | Erechim; (RS); Brazil | 3 | Sequencing and PAGE |
| 3 | D. melanogaster | São José do Rio Preto; (SP); Brazil | 3 | PAGE |
| 4 | D. melanogaster | São José do Rio Preto; (SP); Brazil | 3 | PAGE |
| 5 | D. melanogaster | São José do Rio Preto; (SP); Brazil | 3 | Sequencing and PAGE |
| 6 | D. melanogaster | Santa Maria; (RS); Brazil | 3 | Sequencing |
| 7 | D. melanogaster | Porto Alegre; (RS); Brazil | 3 | Sequencing and PAGE |
| 8 | D. melanogaster | Rio Grande; (RS); Brazil | 3 | Sequencing and PAGE |
| 9 | D. melanogaster | Tangará da Serra; (MT); Brazil | 3 | Sequencing and PAGE |
| 10 | D. melanogaster | Jari; (RS); Brazil | 3 | Sequencing and PAGE |
| 11 | D. melanogaster | Serra da Capivara; (PI); Brazil | 3 | Sequencing |
| 12 | D. melanogaster | Manaus; (AM); Brazil | 3 | Sequencing and PAGE |
| 13 | D. melanogaster | Fernando de Noronha; (PE); Brazil | 3 | Sequencing |
| 14 | D. melanogaster | Fernando de Noronha; (PE); Brazil | 3 | Sequencing |
| 15 | D. melanogaster | Recife; (PE); Brazil | 3 | Sequencing |
| 16 | D. melanogaster | Ilha Governador; (RJ); Brazil | 3 | Sequencing |
| 17 | D. melanogaster | Santa Teresa; (RJ); Brazil | 3 | Sequencing |
| 18 | D. melanogaster | Venezuela | 3 | Sequencing |
| 19 | D. melanogaster | Vienna; Austria | 3 | Sequencing and PAGE |
| 20 | D. melanogaster | Athens; (Attica); Greece | 3 | Sequencing and PAGE |
| 21 | D. melanogaster | Prunay; (Marne); France | 5 | Sequencing and PAGE |
| 22 | D. melanogaster | La Broche; (Upper Normandy); France | 5 | Sequencing and PAGE |
| 23 | D. simulans | Eldorado do Sul; (RS); Brazil | 3 | PAGE |
| 24 | D. simulans | Farrapos; (Río Negro); Uruguay | 3 | PAGE |
| 25 | D. simulans | Fernando de Noronha; (PE); Brazil | 3 | PAGE |
| 26 | D. simulans | Santa Maria; (RS); Brazil | 3 | PAGE |
| 27 | D. simulans | Lima; (Lima); Peru | 3 | PAGE |
| 28 | D. simulans | Havaí; (HI); United States | 3 | PAGE |
| 29 | D. simulans | Rio de Janeiro; (RJ); Brazil | 3 | PAGE |
TPE Repeats: Repeats of TPE found in populations; Method used for obtaining the number of repetitions of TPE: Result obtained by 7% Polyacrylamide Gel Electrophoresis and/or Sanger Sequencing.
Figure 2. Map of South America with the sampling points of the populations ofD. melanogaster (green circles, represented by letters) andD. simulans (blue triangles, represented by numbers), except for the population from Hawaii. (1) Lima, Peru; (2) Fernando de Noronha, Brazil; (3) Rio de Janeiro, Brazil; (4) Eldorado do Sul, Brazil; (5) Santa Maria, Brazil; (6) Farrapos; Uruguay; (A) Jari, Brazil; (B) Santa Maria, Brazil; (C) Rio Grande, Brazil; (D) Porto Alegre, Brazil; (E) Erechim, Brazil; (F) São José do Rio Preto, Brazil; (G) Ilha do Governador, Brazil; (H) Rio de Janeiro, Brazil; (I) Recife, Brazil; (J) Fernando de Noronha, Brazil; (K) Serra da Capivara, Brazil; (L) Tangará da Serra, Brazil; (M) \Manaus, Brazil; (N) Santa Rosa, Venezuela.
The polymerase chain reaction (PCR) was performed under the following conditions: 35 cycles of 15 s at 94°C, 15 s at 53.5°C, and 15 s at 72°C, using the primers TPEf −5'TGCATTTCACAAATTCAAGTCC, TPEr −5'CTTCCCAAAGCCAGTA. These primers were designed from the sequence of the canonical hobo element (GenBank access number X04705), using the Primer3 software. This primer pair amplified the fragment of approximately 240 bp over-passing the TPE region.
The products obtained by the PCR reaction were precipitated in 13% polyethylene glycol (PEG8000) and NaCl (1.6 M) and then sequenced directly using a MegaBACE 500 automated sequencer and the DYEnamic ET® kit (Amersham) according to the manufacturer's protocol.
The sizes of PCR fragments were also estimated by non-denaturing 7% PAGE performed according to the protocol described by Sanguinettiet al. (1994). These gels measured 21x23 cm with a thickness of 2 mm and electrophoresis was done at 50V for 12 h. The DNA bands were detected using silver staining following the protocol described by Bassam and Gresshoff (2007).
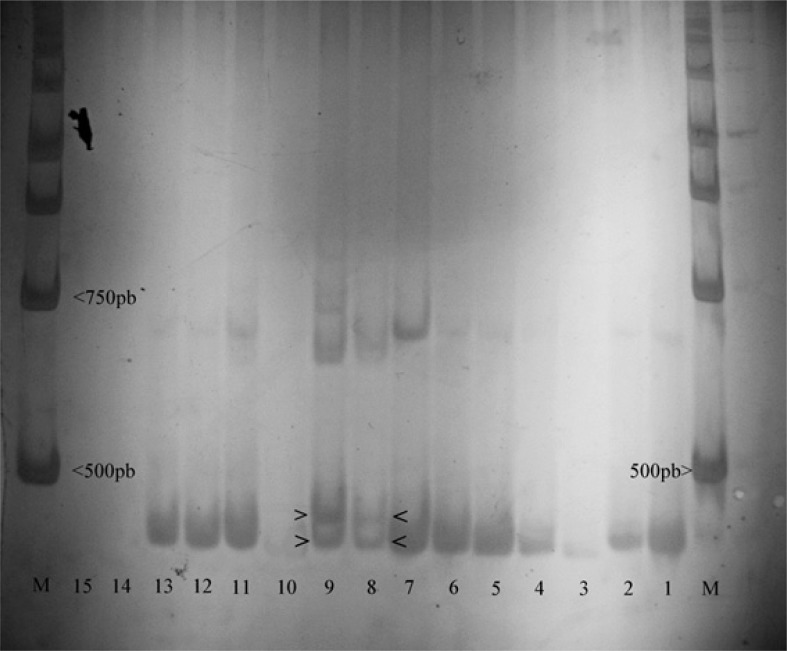
For 22 populations of D. melanogaster we sequenced the PCR product containing the TPE region. This showed that all South American populations had three repeats of TPE, while two European populations, Prunay and La Broche, had five. The other two European populations had three repeats (Table 1). Eleven of the populations that had the TPE region sequenced were also analyzed by PAGE. Two Brazilian populations that were analyzed by PAGE only showed the band characteristic of three TPE repeats (Figure 1). These data suggest that the South American populations of D. melanogaster were monomorphic for three TPE repeats inhobo transposase. In addition, Table 1 shows the origins (collection sites) of the studied populations, the number of TPE repetitions and methods that were used for determining the number of repetitions. The geographic distribution of the South American population sampled is depicted in Figure 2.
Figure 1. Polyacrylamide gel photo showing the differences in migration patterns of bands of European and Brazilian populations of D. melanogaster. (M) markers; (< 500 bp) marker band representing sequence of 500 bp; (< 750pb) marker band representing sequence 750pb; (>) Marking two bands presented by the two European populations, representing regions with polymorphism in the TPE region; (15) Application of negative sample; (14) empty well; (13) Jari, Rio Grande do Sul; (12) Athens, Greece; (11) Vienna, Austria; (10) empty well; (9) Prunay, France; (8) La Broche, France; (7) Erechim, Rio Grande do Sul; (6) Manaus, Amazonas; (5) Porto Alegre, Rio Grande do Sul; (4) Tangara da Serra, Mato Groso; (3) São José do Rio Preto, Sao Paulo, collection point two; (2) São José do Rio Preto, Sao Paulo, collection point 3; (1) São Jose do Rio Preto, Sao Paulo, collection point 4.
Seven populations of D. simulans were analyzed by PAGE electrophoresis and all showed only the band corresponding to three TPE repetitions (Table 1). The populations of D. simulans were similar in their band patterns with Brazilian populations ofD. melanogaster and showed no polymorphism. The geographic distribution of these studied populations can also be found in Figure 2.
As proposed by Periquet et al.(1989), American populations of D. melanogaster did not contain the hobo element before the 1950s, being characterized as strains E (empty). Thereafter, the element was found in American populations and these were characterized as H (hobo) strains. Bonnivard et al. (2000) and Periquet et al. (1989) argued that European populations began to present the hobo element since the 1960s, proposing a direction for the invasion of hobo element. Periquet et al. (1989) suggested that the invasion of the hobo element began in America, and from it, the populations carrying the hobo element spread all over the world. This element contained three TPE repeats and is commonly found around the world. Bonnivard et al. (2000, 2002) added to the evolutionary history of the invasive hobo element of D. melanogaster the hypothesis of a subsequent invasion of the element containing five or seven TPE repeats. They found polymorphic populations in three regions, central Europe, southern Africa and western South America. In North America, Bonnivardet al. (2000) studied 13 populations and found only 3 repeats in nine populations and 3 and 5 repeats in other four populations. From this, the authors suggested that the possible origin of this invasion happened in South America, since in North Africa only populations containing three regions were found. However, North Africa is the geographic region that promotes a connection between the European continent and southern Africa, thus the authors excluded these two regions as possible origins of invasion, suggesting that South America would be the region of origin for the second invasion.
Our results argue against the supposition for the origin of the second invasion, since, like the region intermediate between Europe and southern Africa, our study area, in the East of America also had no polymorphism, hence excluding South America as a possible area of origin for the second invasion.
Thus, of the three regions containing polymorphisms, none is a candidate for the origin of the invasion. From this standoff we suggest that the second invasion of thehobo element may not have occurred in South America or in equatorial Africa, and the polymorphism found in these regions may be due to new mutations occurring in these regions followed by selection or genetic drift.
Souames et al. (2003a) analyzed the mutation rate of TPE repetitions of the hobo element during 20 generations of D. melanogaster and found that the mutation rate in this region was a hundred times higher when compared to other regions of neutral microsatellite genome of D. melanogaster . Thus, we suggest that the polymorphism found in western South America and southern Africa could be the result of mutations that were selected or suffered genetic drift, because none of these places has the potential to be the geographic region of origin of the supposed second invasion.
Unlike these regions, Central Europe has certain peculiarities not shared by South America and Africa. In Europe, the TPE polymorphism frequency is much higher than in any other region and, in addition, presents many kinds of polymorphisms with copies ranging from three repeats and others containing up to ten repetitions (Bonnivard et al., 2002). Thus it is possible that this polymorphism is a local one generated by new mutations.
The monomorphism found in D. melanogaster in the TPE regions was shared by D. simulans . So far, the TPE region of D. simulanshas not been widely studied, but for this current study it is important to discuss the supposed donor of the hobo element in D. melanogaster, the sister species, D. simulans . Boussy and Daniels (1991) indicated a direction of the origin of thehobo element in D. melanogaster, indicating that the donor species of the element would be the sister species D. simulans, and this direction of invasion is supported for two reasons:hobo would be present in D. simulans beforeD. melanogaster and the high similarity of the element between the two species. Periquet et al.(1989) and Bonnivard et al.(2000, 2002) argued that the invasion of the hobo element in populations of D. melanogasterhappened in the 1950s in Central America and the hobo element had three repetitions of TPE, so for D. simulans to be the donor element it would also have to present the hobo element with three repetitions. Our results supported the hypothesis proposed by Boussy and Daniels (1991) of D. simulans as the donor of thehobo element to D. melanogaster because all populations studied had three replications of TPE (Figure 2 and Table 1). In this way we can infer with more clarity that D. simulans was possibly the donor of thehobo element to D. melanogaster.
The TPE region has been used by the scientific community because it contains information about the history of the hobo element and its dynamics. Based on the TPE repeats, the evolutionary scenario of the hobo element was proposed and their history was better understood (Calvi et al., 1991; Bazin and Higuet, 1996; Bonnivard et al., 2000, 2002;). Our results are in accordance with the hypothesis of multiple origins for the second hobo invasion, as suggested by Bonnivard et al.(2000).
Acknowledgments
We thank Dr. Jean David, Dr. Claudia Rohde and Dr. Blanche Bitner-Mathe for sending us fly strains. This study was supported by research grants and fellowships from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and FAPERGS (Fundação de Amparo a pesquisa do Estado do Rio Grande do Sul).
Footnotes
Associate Editor: Louis Bernard Klaczko
References
- Bassam BJ, Gresshoff PM. Silver staining DNA in polyacrylamide gels. Nat Protoc. 2007;2:2649–2654. doi: 10.1038/nprot.2007.330. [DOI] [PubMed] [Google Scholar]
- Bazin C, Higuet D. Lack of correlation between dysgenic traits in thehobo system of hybrid dysgenesis in Drosophila melanogaster . Genet Res. 1996;67:219–226. doi: 10.1017/s001667230003370x. [DOI] [PubMed] [Google Scholar]
- Biémont CA. Brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biémont C, Vieira C. Junk DNA as an evolutionary force. Nature. 2006;443:521–524. doi: 10.1038/443521a. [DOI] [PubMed] [Google Scholar]
- Bonnivard E, Bazin C, Denis B, Higuet D. A scenario for the hobo transposable element invasion, deduced from the structure of natural populations ofDrosophila melanogaster using tandem TPE repeats. Genet Res. 2000;75:13–23. doi: 10.1017/s001667239900395x. [DOI] [PubMed] [Google Scholar]
- Bonnivard E, Bazin C, Higuet D. High polymorphism of TPE repeats within natural populations ofD. melanogaster: a gradient of the 5TPEhobo in Western Europe. Mol Biol Evol. 2002;19:2277–2284. doi: 10.1093/oxfordjournals.molbev.a004051. [DOI] [PubMed] [Google Scholar]
- Boussy IA, Daniels SB. hobo transposable elements in Drosophila melanogaster and D. simulans. Genet Res. 1991;58:27–34. doi: 10.1017/s0016672300029578. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Hong TJ, Findley SD, Gelbart WM. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants:hobo, Activator andTam3 . Cell. 1991;66:465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- Capy P, Anxolabérè D, Langin T. The strange phylogenies of transposable elements: are horizontal transfers the only explanation? Trends Genet. 1994;10:7–12. doi: 10.1016/0168-9525(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Bigot Y, Sánchez MD, Periquet G, Pascual L. Sequences homologus to the hobo transposable element in E strains of Drosophila melanogaster . Mol Biol Evol. 2001;18:1532–1539. doi: 10.1093/oxfordjournals.molbev.a003939. [DOI] [PubMed] [Google Scholar]
- Hua-Van A, Le Rouzic A, Boutin TS, File EJ, Capy P. The struggle for life of the genome's selfish architects. Biol Direct. 2011;6:19–19. doi: 10.1186/1745-6150-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium A physical map of the human genome. Nature. 2001;409:860–921. [Google Scholar]
- McGinnis W, Shermoen AW, Beckendorf S. A transposable element inserted just 5' to aDrosophila glue protein gene alters gene expression and chromatin structure. Cell. 1983;34:75–84. doi: 10.1016/0092-8674(83)90137-x. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Tingey SV, Morgante M. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 2001;11:1660–1676. doi: 10.1101/gr.188201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira LFV, Wallau GL, Loreto LSV. Isolation of high quality DNA: a protocol combining "rennet" and glass milk. Electr J Biotechnol. 2009;12:1–6. [Google Scholar]
- Ortiz MF, Loreto ELS. The hobo-related elements in themelanogaster species group. Genet Res. 2008;90:243–252. doi: 10.1017/S0016672308009312. [DOI] [PubMed] [Google Scholar]
- Pascual L, Periquet G. Distribution of hobo transposable elements in natural populations of Drosophila melanogaster . Mol Biol Evol. 1991;8:282–296. doi: 10.1093/oxfordjournals.molbev.a040649. [DOI] [PubMed] [Google Scholar]
- Periquet G, Hamelin MH, Bigot Y, Lepissier A. Geographical and historical patterns of distribution ofhobo elements in Drosophila melanogaster populations. J Evol Biol. 1989;2:223–229. [Google Scholar]
- Periquet G, Lemeunier F, Bigot Y, Hamelin MH, Bazin C, Ladevèze V, Eeken J, Galindo MI, Pascual L, Boussy I. The evolutionary genetics of the hobotransposable elements in Drosophila melanogastercomplex. Genetica. 1994;93:79–90. doi: 10.1007/BF01435241. [DOI] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA. Structure and evolution of the hAT transposon superfamily. Genetics. 2001;158:949–957. doi: 10.1093/genetics/158.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanguinetti CJ, Dias E, Neto, Simpson AJG. Rapid silver staining and recovery of PCR products separated on polycrylamide gels. Biotechniques. 1994;17:914–921. [PubMed] [Google Scholar]
- Simmons GM, Plummer D, Simon A, Boussy IA, Frantsve J, Itoh M. Horizontal and vertical transmission ofhobo-related sequences between Drosophila melanogaster e Drosophila simulans. In: Syvanen M, Kado CI, editors. Horizontal Gene Transfer. Chapman and Hall; New York: 1998. pp. 285–294. [Google Scholar]
- Souames S, Bazin C, Bonnivard E, Higuet D. Behavior of the hobo transposable element with regard to TPE repeats in transgenic lines of Drosophila melanogaster . Mol Biol Evol. 2003a;20:2055–2066. doi: 10.1093/molbev/msg221. [DOI] [PubMed] [Google Scholar]
- Souames S, Bonnivard E, Bazin C, Higuet D. High mutation rate of TPE repeats: a microsatellite in the putative transposase of the hobo element inDrosophila melanogaster . Mol Biol Evol. 2003b;20:1826–1832. doi: 10.1093/molbev/msg194. [DOI] [PubMed] [Google Scholar]
- Streck RD, MacGaffey JE, Beckendorf SK. The structure of hobo transposable elements and their insertion sites. EMBO J. 1986;5:3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres FP, Fonte LFM, Valente VLS, Loreto ELS. Mobilization of a hobo-related sequence in the genome of Drosophila simulans . Genetica. 2006;126:101–110. doi: 10.1007/s10709-005-1436-1. [DOI] [PubMed] [Google Scholar]
- Warren WD, Atkinson PW, O'Brochta D. The Hermes transposable element from the house fly, Musca domestica, is a short inverted repeat-type element of thehobo, Ac, and Tam3 (hAT) element family. Genet Res. 1994;64:87–97. doi: 10.1017/s0016672300032699. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]