Abstract
Non-indigenous plants exhibit different attributes that make them aggressive competitors with indigenous plants and serious threats to biodiversity.Senecio madagascariensis (fireweed, Asteraceae), a native from southern Africa, is a strong competitor in agricultural activities and has toxic alkaloids that may result in high cattle mortality. In Brazil, this weed was collected for the first time in 1995 and has since spread quickly throughout the Pampas region. To better understand the invasion of the fireweed in South America, we used a genetic characterization with internal transcribed spacer (ITS) and microsatellite markers. Based on the ITS data, the southern Brazil populations of S. madagascariensis shared genetic homology with samples taken from the Hawaiian Islands and South Africa. Microsatellite analysis showed the genetic diversity split in two clusters, perhaps intimating the independent introduction of each species into South America. Although fireweed was introduced recently in southern Brazil, the considerable levels of genetic diversity, gene flow, and inbreeding may indicate success in the species establishment in this environment.
Keywords: Microsatellites, ITS, Pampas, weed, spread
Introduction
Invasive plants have different attributes and mechanisms that allow their dispersal and establishment in new locations (López-García and Maillet, 2005; Murray and Phillips, 2010). They may be highly aggressive competitors for light and soil nutrients with indigenous plants (Le Roux et al., 2006). Furthermore, the absence of herbivores and pathogens may lead to the unbridled growth of invasive plants (Keane and Crawley, 2002; Allendorf and Lundquist, 2003). These plants produce a direct impact on the natural ecosystems and are serious threats to global biodiversity (Sala et al., 2000; Allendorf and Lundquist, 2003; Strauss et al., 2006). Studying the early stages of an invasion may reveal additional data on the processes of microevolution (Mooney and Cleland, 2001;Sakai et al., 2001;Lee, 2002; Barrett et al., 2008; Rosenthal et al., 2008).
Blackburn et al. (2011)proposed four main stages in successful invasions: first, a species disperses to a new geographic region (transport and introduction); third, it establishes populations in the new environment (establishment); and finally, it reproduces successfully (spread). To progress through each of these phases, invasive species must overcome barriers that are directly linked to genetic processes, such as the founder effect, genetic drift, gene flow and natural selection. Small founder populations have a limited genetic pool due to the founder effect and genetic drift, which may hinder their establishment in new environments (Sakai et al., 2001). Historically, phenotypic plasticity has been seen as a potentially important mechanism for the success of colonization in environmentally diverse areas and may play a role in invasion (Chun et al., 2009). Adaptive plasticity may contribute to invasive ability by allowing the acclimation of invasive pre-adapted genotypes to diverse environments, as well as buffering against existing genetic variation from selection, thereby reducing the necessity for local adaptation (Sultan, 2003; Thébault et al., 2011). Therefore, adaptive plasticity may lead to phenotypic homeostasis (tolerance) in fitness, which can be important to successful invasion (Rejmánek, 2000; Alpert and Simms, 2002). Another factor that may increase the likelihood of success for an invasive species is the decrease in the founder effect through multiple sources of introduction, as observed in several studies (Novak, 2007; Chun et al., 2009; Ghabooliet al., 2011). High genetic diversity can be beneficial on both ecological and evolutionary timescales: in the short term, high diversity has been shown to improve colonization success (Crawford and Whitney, 2010; Bocket al., 2015). Newly introduced populations often experience a genetic bottleneck, which can have potentially dramatic consequences for their evolution and survival in the new site. However, multiple introductions often restore the lost diversity, frequently resulting in higher levels of variability than in the native range because of admixture of genetically different sources of variation (Bock et al., 2015)
Trade and travel are primarily responsible for the introduction of alien organisms across the globe (Keller et al., 2011). Unfortunately, terrestrial species are inadvertently transported in shipping containers (Lounibos, 2002), facilitating the rapid spread of invasive organisms (Kaluza et al., 2010). Often, weeds (frequently associated with seeds) are recovered as unknown contaminants in baggage. Although biological invasions are serious threats to biodiversity, they are opportunities to understand fundamental ecological and evolutionary processes (Sax et al., 2007).
Senecio madagascariensis Poir., widely known as fireweed, is a member of the Asteraceae family, native to southern Africa (Le Roux et al., 2006). It is characterized as an annual herb, sometimes behaving as a perennial, under favorable conditions. Plants of this species when isolated appear to be fully self-compatible (Le Roux et al., 2010). They produce large numbers of seeds that disperse long distances by wind and are strong competitors for agricultural resources, while the toxic alkaloids they contain may result in death when ingested by animals (Le Rouxet al., 2006). Fireweed was accidentally introduced to Australia c.1918 (Dormontt et al., 2014) and to the Hawaiian Islands in the early 1980s (Le Roux et al., 2006). In these locations, this weed competed strongly with existing pasture flora, ultimately leading to the deterioration of pastures (Le Rouxet al., 2006). In Australia, Sindel et al. (1998) reported that the species has a high degree of competitiveness as a weed, causing great economic losses due to the mortality of cattle.
In Argentina, the first specimen of S. madagascariensis was collected in 1940; the current populations are entirely diploid (López et al., 2008). In Brazil, fireweed was first identified in 1995 (Matzenbacher and Schneider, 2008), and in recent years, it has spread quickly throughout the Pampas in southern areas (see Figure S1 (1.4MB, pdf) ). In other parts of Brazil, the weed does not appear to grow below the southern latitude of 29°S. More recently, there is evidence that fireweed, together with otherSenecio species, has caused great economic losses to livestock in southern Brazil, with 45,500 animals lost annually (Karam et al., 2011). Cattle production has existed in the Pampas region since the 17th century and is the most important environmental change to have occurred in recent decades due to agricultural expansion in South America (Overbecket al., 2007). The effective management of invasive plant species may be facilitated by further research into the biological processes that were challenging to clarify or quantify under field conditions prior to recent advances in molecular genetics. To contribute to a better understanding of the invasion of S. madagascariensis in southern Brazil, we aimed a) to clarify the invasion history including the origin of introduced genotypes and the possibility of multiple introduction events; b) to determine the genetic diversity and the structure of the invasive populations; and c) to examine the gene flow among populations.
Materials and Methods
Sample collection and DNA extraction
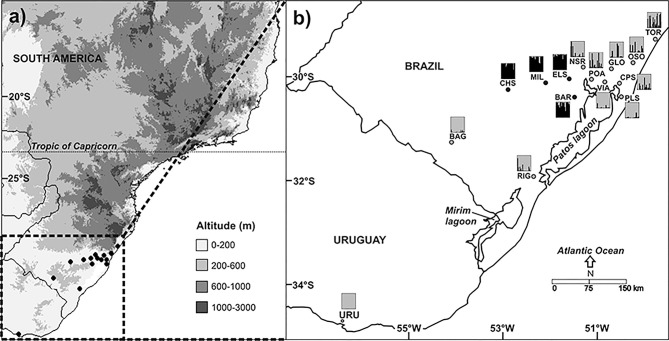
A total of 307 S. madagascariensis individuals were sampled from 15 sites (hereafter referred to as populations) covering the entire geographical range of the distribution for this species in Brazil and Uruguay (Table 1; Figure 1). We collected 20–30 individuals per site, where practical, and the minimum distance between two collection points was 25 km. All populations were found at elevations below 200 m in the Pampas region, preferentially in lowlands close to sea level. The TOR population (~29°S) was the northernmost S. madagascariensis in Brazil. Vouchers were deposited in the ICN Herbarium (at the Universidade Federal de Rio Grande do Sul, Porto Alegre, RS, Brazil), and young leaves were carefully collected in the field for genetic analysis. After drying on silica gel, the leaves were frozen in liquid nitrogen and ground into a fine powder. Subsequently, DNA was extracted from the powdered leaves as described by Roy et al. (1992).
Table 1. Genetic diversity and collection sites information for the Senecio madagascariensis populations sampled.
| Local information | ITS | Microsatellite diversity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | n 1 | n 2 | Collection site | Geographical coordinates | Voucher* | Sequence types# | A | AR | Ho | He | Fis | PA |
| URU | 04 | 04 | URU, Ciudad Del Plata | 34°45' 53″S/56°24' 25'W | 166599 | S3;S4 | 3.00 | 2.71 | 0.412 | 0.577 | 0.316 | 2 |
| POA | 10 | 21 | BRA, Porto Alegre | 30°04' 01″S/51°07' 14″W | 166600 | S1–2; S6; S11–12 | 7.50 | 3.67 | 0.454 | 0.763 | 0.411 | 2 |
| NSR | 11 | 22 | BRA, Nova Santa Rita | 29°50' 45″S/51°16' 38″W | 166601 | S1–2; S6; S13; S17 | 8.13 | 3.48 | 0.377 | 0.731 | 0.490 | 7 |
| RIG | 18 | 24 | BRA, Rio Grande, Povo Novo | 31°56' 31″S/52°18' 39″W | 166604 | S1–2 | 5.88 | 2.94 | 0.352 | 0.652 | 0.465 | 3 |
| VIA | 13 | 21 | BRA, Viamão | 30°08' 15″S/50°52' 05″W | 166605 | S1–2 | 6.75 | 3.28 | 0.452 | 0.698 | 0.358 | 3 |
| CPS | 13 | 22 | BRA, Capivari do Sul | 30°08' 51'S/50°30' 31″W | 166606 | S1–2; S18 | 8.13 | 3.62 | 0.467 | 0.738 | 0.374 | 4 |
| GLO | 10 | 14 | BRA, Glorinha | 29°53' 25″S/50°42' 08″W | 166607 | S1–2 | 6.38 | 3.59 | 0.402 | 0.759 | 0.479 | 0 |
| OSO | 06 | 17 | BRA, Osório | 29°45' 05″S/50°12' 47″W | 166608 | S1–2; S19 | 6.13 | 3.31 | 0.477 | 0.710 | 0.337 | 1 |
| TOR | 08 | 26 | BRA, Torres | 29°18' 17″S/49°46' 21″W | 169679 | S1–2; S10 | 8.00 | 3.58 | 0.548 | 0.756 | 0.279 | 2 |
| PLS | 08 | 12 | BRA, Palmares do Sul | 30°25' 37″S/50°30' 04″W | 169680 | S1–2; S20 | 4.00 | 2.73 | 0.584 | 0.626 | 0.071 | 1 |
| BAG | 07 | 33 | BRA, Bagé | 31°17' 57″S/54°04' 46″W | 170944 | S1;S14 | 8.25 | 3.45 | 0.534 | 0.711 | 0.252 | 5 |
| ELS | 10 | 22 | BRA, Eldorado do Sul | 30°03'58″S/51°33' 27″W | 166602 | S1–2; S6; S7 | 7.50 | 3.34 | 0.597 | 0.722 | 0.177 | 6 |
| BAR | 11 | 21 | BRA, Barra do Ribeiro | 30°25' 22″S/51°28' 03″W | 166603 | S1–2; S9 | 8.13 | 3.67 | 0.451 | 0.764 | 0.417 | 2 |
| MIL | 04 | 18 | BRA, Minas do Leão | 30°08' 59″S/52°04' 18″W | 170943 | S1–2; S8 | 8.50 | 3.85 | 0.551 | 0.771 | 0.292 | 7 |
| CHS | 06 | 30 | BRA, Cachoeira do Sul | 30°16' 30″S/52°56' 13″W | 166946 | S1-2; S16 | 9.13 | 3.61 | 0.529 | 0.747 | 0.295 | 10 |
| Total | 139 | 307 | Average | 7.03 | 3.39 | 0.479 | 0.715 | 0.334 | ||||
Sample size (n 1 = ITS data;n 2 = SSR data); number of alleles per population (A); allelic richness (AR); average observed heterozygosity across loci (He); expected heterozygosity (Ho) and fixation index (FIS); private alleles (PA).
ICN Herbarium, Department of Botany, Biosciences Institute, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil.
Sequence types of samples outside of South América: South África S1; Hawaii S2; Swaziland S21; Madagascar S22-26.
Figure 1. Collection sites. a) Map of South America highlighting the Brazilian state of Rio Grande do Sul and Uruguay. b) Colored dots indicate all collected populations of Senecio madagascariensis Poir. included in this study. The different shapes correspond to the clusters by STRUCTURE based on SSR data (rectangles; bars represent the membership coefficients (Q); K = 2) and NJ dendrogram/network (circles).
Molecular population genetic analysis of ITS
For these analyses, we used 139 samples belonging to 15 populations (Table 1) and 24 GenBank sequences from the following regions: Australia (1); Argentina (1); Madagascar (8); South Africa (7); Swaziland (2); and the Hawaiian Islands (5), respectively (GenBank accession numbers JN789815, U93198, DQ322598-DQ322619). The internal transcribed spacers (ITS 1 and 2) were amplified using primers and reaction conditions described by Desfeux and Lejeune (1996). We used 10% dimethyl sulfoxide (DMSO) to exclude the presence of unstable templates (Buckler et al., 1997; Fuertes-Aguilar and Nieto-Feliner, 2003). All PCR products were purified using a 20% polyethyleneglycol (PEG) precipitation method (Dunn and Blattner, 1987). Sequencing was performed in a MegaBACE 1000 automatic machine using the ET Terminator Kit (GE Healthcare) following the manufacturer's protocol. The sequences were aligned manually using the GeneDoc program (Nicholas et al., 1997) and deposited in GenBank (accession numbers: JQ653968-JQ654097). Following Mäder et al. (2010), the heterozygous ITS sites were not included in the analysis.
We used DnaSP 5 (Librado and Rozas, 2009) to determine the sequence types. Basic sequence statistics, such as haplotype (h) and nucleotide diversity (π) (Nei, 1987), and the analysis of molecular variance (AMOVA;Excoffier et al., 1992) among collection sites using FST(pairwise differences) were obtained in Arlequin 3.5.1.2 (Excoffier and Lischer, 2010). We estimated the evolutionary relationships among the sequences by the Median-Joining method (ε = 0; Bandelt et al., 1999) as implemented in Network 4.6 software. Sequences of S. inaequidens from GenBank were used as the outgroup (DQ322620 and DQ322621).
Molecular population genetic analysis of microsatellite markers
We used all 307 sampled individuals (Table 2) to assess the genetic variability at eight nuclear microsatellite loci (SSR). Details of PCR protocols, isolation, characterization, and internal repeat structures of the S. madagascariensis microsatellite loci used in this study can be found in the study by Le Roux and Wieczorek (2007). In the present work, we analyzed only the eight loci also used by Le Roux et al. (2010). PCR amplifications were performed in 15-μl reaction volumes containing 2.5–5 ng of genomic DNA. Microsatellite genotyping was performed using a MegaBACE 1000 automated sequencer and the ET550-R Size Standards (GE Healthcare). Allele sizes were scored using Genetic Profiler 2.2 software (Amersham Biosciences). All microsatellite loci were screened for null alleles and large allele dropouts using MICRO-CHECKER 2.2.3 (Van Oosterhout et al., 2004).
Table 2. Diversity and neutrality indices in Senecio madagascariensis based on ITS data.
| Parameter | South America | Complete data set* |
|---|---|---|
| Sample size | 139 | 161 |
| No. sequence types | 20 | 26 |
| π (SD) | 0.001 (0.001) | 0.002 (0.002) |
| Genetic diversity (SD) | 0.624 0.02 | 0.763 (0.017) |
| FST | 0.146 (P <0.001) | 0.312 (P <0.001) |
South America samples plus sequences from South Africa (7); Swaziland (2);
Madagascar (8) and Hawaiian Islands (5).
π = Nucleotide diversity.
SD= Standard deviation.
Genetic diversity was estimated for each population in terms of the number of alleles (A) and allelic richness (AR); expected (HE) and observed (Ho) heterozygosity, deviations of genotype frequencies from those expected under Hardy-Weinberg equilibrium (HWE); and the inbreeding coefficient (FIS) using Arlequin and FSTAT 2.9.3 (Goudet, 2001). Additionally, we obtained AMOVA data from the populations usingFST (pairwise differences) in Arlequin.
The ISOLDE program (part of the Genepop 4.1 package) was used to test the relationships between geographic and genetic (FST) distances among populations, with the statistical significance assessed using a Mantel test with 10,000 permutations. To test the structure and to assess the scale of geographic differentiation among populations, we used STRUCTURE 2.3.3 (Falush et al., 2007) with a burn-in of 250,000, a run length of 1,000,000 Markov Chain Monte Carlo (MCMC), and a model allowing for admixture and correlated allele frequencies. We did not use a prioriinformation on population origin. Five independent runs were performed by setting the number of populations (K) from 1 to 15, and an average likelihood value, L(K), was calculated for each K across all runs. Additionally, we calculated ΔK (Evanno et al., 2005) by taking into account the shape of the log-likelihood curve with increasing K and variance among estimates among multiple runs. The runs of individual ancestry coefficients were calculated by the average pairwise similarity of individual assignments across runs using CLUMPP 1.1.2 (Jakobsson and Rosenberg, 2007) and plotted using DISTRUCT 1.1 (Rosenberg, 2004). An unrooted Neighbor-Joining (NJ) dendrogram was constructed from the genetic distance (proportion of shared alleles) calculated with the Microsat program using MEGA 5.0 (Tamura et al., 2011). To verify the existence of migrants between populations, we ran 10,000 MCMC simulations per population in GeneClass2 (Piry et al., 2004) using the Lh/Lmax likelihood computation, where Lh is the likelihood of an individual being assigned to the population from which it was sampled and Lmax is the maximum likelihood for all populations considered. An individual was considered a migrant if the Lh/Lmax P value was below 0.01. Statistical comparisons among populations were performed using the paired Student's t-test, with significance being designated for P <0.05.
Results
Distribution
During the fieldwork, it was possible to observe the rapid spread of S. madagascariensis, particularly in the Rio Grande do Sul Brazilian state. Population growth was observed in different locations, where this fireweed was not registered previously by Matzenbacher and Schneider (2008) (OSO and TOR). In other localities (BAG and CHS), populations were noticeably more prevalent than the local vegetation, forming dense populations in the Pampas region (unpublished data based on fieldwork performed in the last six years, 2009–2014) during the flowering period of fireweed (see Figure S1 (1.4MB, pdf) ).
ITS data
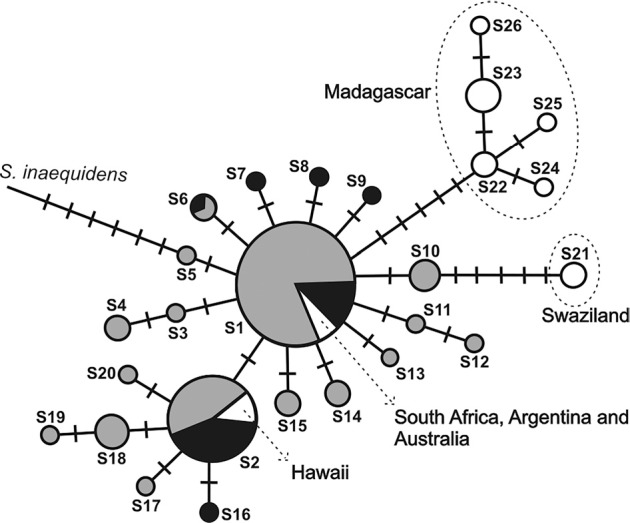
The internal transcribed spacer ITS length varied between 665 and 666 base pairs (bp) in all analyzed samples, with 31 polymorphic sites in the alignment and one insertion/deletion event and 30 substitutions (11 transitions and 19 transversions). Among the 26 sequence types inferred by DNAsp, nucleotide diversity (π) was 0.0023 ± 0.002, while genetic diversity was 0.763 ± 0.017 (Table 2). Of the 26 types of sequences, 20 were sampled from the South American populations; therefore, the value of genetic diversity was similar to the total sample (0.624 ± 0.02) (Table 2). A moderate degree of genetic structure was observed (FST = 0.312;P <0.001) in the global dataset when the populations were clustered into five geographical regions: South America, South Africa, Madagascar, Swaziland, and the Hawaiian Islands. We analyzed the 15 populations from South America independently from the other global populations (Table 1). FSTdecreased to 0.146 (P <0.001), showing a reduction in the genetic structure. Median-joining network analysis verified the absence of a geographic structure in South American populations (Table 1; Figure 2). Among the South American populations, URU was the only population that did not show the central sequence type (S1), while S2 was observed in all populations except URU and BAG. Only RIG, VIA and GLO did not present exclusive sequence types. All South American populations had at least two sequence types. A prominent network feature was the relationship of the South American populations with the sequences from the Hawaiian Islands and South Africa (S1 and S2). The sequences of plants from Madagascar remained in an isolated cluster (sequence types S22-S26), and the Swaziland samples presented only one unique sequence type (S21) (Figure 2).
Figure 2. The evolutionary relationships of Senecio madagascariensis Poir. based on ITS sequence types obtained by median-joining network approach. Gray scale shading identify the geographic origin. Circle sizes are proportional to haplotype frequency. Crossed lines represent substitutions inferred in the branches.
Microsatellite marker
The eight microsatellite loci tested were polymorphic across all 307 individuals from the 15 populations. Across all individuals, the number of alleles per locus ranged from 10 (Se-194) to 44 (Se-136); within populations, the number of alleles ranged from 2 to 17 (mean = 7.03) and allelic richness ranged from 2.71 to 3.75 (mean = 3.39) (Table 1). Expected heterozygosity (HE) ranged from 0.577 to 0.771 (mean = 0.715), observed heterozygosity (HO) from 0.352 to 0.597 (mean = 0.456) and FIS ranged from 0.071 to 0.490 (mean = 0.334) across all loci (Table 1). Except for the Uruguayan population (URU), all populations showed a deficit of heterozygotes under Hardy-Weinberg Equilibrium (α = 0.05), and few null alleles were detected at very low frequencies.
Diversity within population explained ca. 90% of the genetic diversity based on microsatellite (FST = 0.103), similar to the results from the sequence data (ITS). Across fireweed populations, the Mantel test showed a moderately significant positive correlation between genetic and geographic distances (r 2 = 0.37; P<0.001) (see, Figure S2 (546.7KB, pdf) ). The results of Bayesian cluster analyses revealed the presence of structured genetic diversity among groups. Following the method of Evanno et al. (2005), the model-based clustering method implemented in STRUCTURE found two distinct major genetic clusters (K) (Figure 1). A cluster including four populations (ELS, BAR, MIL and CHS) that presented more continental geographic distribution (latitude approximately 30° S) was named "Central Group". The second cluster was composed of the remaining populations, found in the Coastal Plain region (except for BAG), and was named "Coastal Group". We detected some admixture between the two clusters (e.g., POA, NSR, GLO, TOR; Figure 1). The unrooted NJ dendrogram (Figure 3) distinguished two clades of samples, corresponding to the same groups obtained by STRUCTURE, and similarly showed admixture between Central and Coastal groups. Some samples were found in the clade of the opposite group (Figure 3). More precisely, 30 individuals of the Central Group were found in the Coastal clade and 10 individuals of the Coastal Group positioned in the Central clade. Furthermore, AMOVA showed that 38.7% of the variation found refers to the difference between Central and Coastal Groups (FST = 0.387). On average, populations from the Central Group differed significantly from Coastal Group populations in terms of heterozygosity (Ho), inbreeding (FIS) and levels of differentiation among populations (FST). However, there were no significant differences in observed allelic richness (AR) or unbiased gene diversity (Hs) between the two groups (Student's pairedt-test) (Table 3).
Figure 3. The unrooted Neighbor-Joining dendrogram based on the proportion of shared allele distances obtained by SSR genotypes. The branch colors indicate the two groups inferred by the STRUCTURE software. Black: Central group. Gray: Coastal group. *First-generation migrants among groups by estimates of dispersal in GeneClass2.
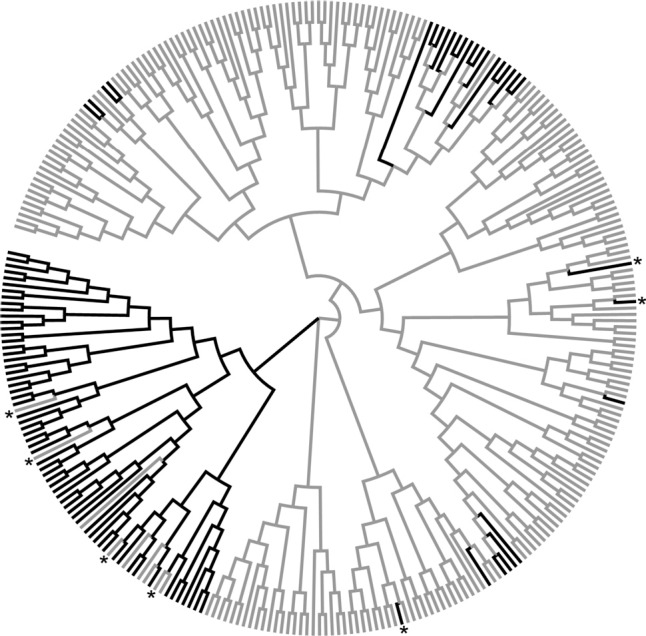
Table 3. Statistical comparison of allelic richness (AR), unbiased gene diversity (Hs), observed heterozygosity (Ho), inbreeding coefficient (FIS) and levels of differentiation among populations (FST) for the two genetic groups of Senecio madagascariensis .
| n | AR | Hs | Ho* | FIS * | FST * | |
|---|---|---|---|---|---|---|
| Coastal group | 216 | 3.305 | 0.739 | 0.460 | 0.348 | 0.073 |
| Central group | 91 | 3.616 | 0.794 | 0.532 | 0.295 | 0.103 |
P <0.05.
n = sample size.
Contemporary estimates of dispersal calculated in GeneClass2 identified 38 of 307 individuals as potential 'first-generation migrants' (P = 0.01). However, given the low genetic structure observed between pairwise populations, the results based on the identification of immigrants should be interpreted with caution because a decrease in genetic differentiation and limited number of loci reduces the ability to correctly identify immigrants. Therefore, we focused on overall patterns of potential migration. Not surprisingly, many of the migrants between Central and Coastal groups correspond with those individuals that show a distinct population pattern in the dendrogram (indicated by asterisks in Figure 3) and STRUCTURE analysis.
Discussion
Origin and preadaptation
Based on the ITS data, the South American populations of fireweed were more closely related to the samples from the Hawaiian Islands and South Africa (sharing the more frequent sequence types, S1, S2), while samples from Madagascar and Swaziland were unique and distant from the others (Figure 2). This suggests that the South American populations most likely originated in South Africa, or alternatively, originated in another country where fireweed is alien (e.g., Australia or the Hawaiian Islands), constituting a secondary invasion. Several works have attributed the arrival of invasive plants via cargo ships from South Africa (McCullough et al., 2006; Lachmuth et al., 2010; Keller et al., 2011). The first hypothesis is the most probable given the large number of trade routes between sub-tropical South America and southern Africa, while the same is not true of Australia or the Hawaiian Islands (Kaluza et al., 2010). Furthermore, S. madagascariensis was only just recently introduced into South America, so there are no dense populations in Australia or on the Hawaiian Islands (Le Rouxet al., 2006), suggesting the difficulty in its migration to a new continent. The populations of fireweed in subtropical South America are located in areas with similar latitude (usually 29–32°S) and altitude (<200 m, predominantly at sea level, Figure 1). Their indigenous populations in South Africa include the harbor area of the KwaZulu-Natal region. This is the indicated source of the invasion into the Hawaiian Islands according to Le Roux et al. (2006). This putative origin may have facilitated the adaptation of these aliens in Brazil, which may be pre-adapted to environments with similar climatic conditions, a great advantage in terms of general survival according to Thébault et al. (2011).
Genetic variability and structure
The ITS data showed many sequences in South American populations of fireweed. Over 75% of the sampled sequence types (20 of 26) were found in South America. However, the most thorough comparison is the quantification of the genetic variability of indigenous S. madagascariensis populations. Although the ITS sequences of S. madagascariensis have exhibited low variability when compared with other groups of plants (e.g., Nettel and Dodd, 2007; Yamaji et al., 2007; Mäder et al., 2010), the difficulty in finding informative molecular markers inSenecio has been previously observed (e.g., Comes and Abbott, 2001; Pelser et al., 2007), as well as in other plants outside their natural habitat (Young and Murray, 2000; Yu et al., 2014). Similarly, several studies, such as those conducted by Lavergne and Molofsky (2007) and by Ghabooli et al. (2011) found lowFST values in invasive populations. The potential high number of private sequence types observed in invasive populations is probably due to the small number of individuals from their region of origin included in this study. Nevertheless, some private sequence types might have originated in loco, after the invasion, since a majority of haplotypes differs from each other by a single mutational step. Unfortunately, a more accurate conclusion requires large-scale sequencing in the region to which fireweed is indigenous (southern Africa), and this information is not available. Therefore, due to insufficient variability, the ITS analysis did not allow us to explore the network relationships among South American populations. Instead, the relationships were analyzed on a global scale, allowing the differentiation of the indigenous populations in southern Africa (South Africa, Swaziland and Madagascar) (Figure 2).
Microsatellite analysis revealed levels of genetic diversity similar (number of alleles, allelic richness and heterozygosity observed) to those obtained from the Hawaiian populations (Le Roux et al., 2010). However, these values were lower than those observed in populations from Australia and South Africa (Dormontt et al., 2014). Nevertheless, as observed by Dormontt et al. (2014), the reduction in the genetic diversity of fireweed does not appear to have hindered its spread, perhaps due to high levels of phenotypic plasticity, changes in gene expression or the 'adequacy' of diversity for populations in adapting to the new conditions. The two clusters observed in the STRUCTURE analysis (Figure 1) and the NJ dendrogram (Figure 3), beyond the distinctive characteristics listed in Table 3, suggest at least two independent introduction events. These results suggest that invasive populations in South America have a genetic diversity that may be related to multiple introduction events, and this may be related to shipping traffic from southern Africa, as previously mentioned. Several studies have shown, using historical and molecular data, that many invasive weeds have significant genetic diversity as a result of multiple introductions; for example: Alliaria petiolata (Bieb.) Cavara and Grande analyzed by Durka et al. (2005) and Phalaris arundinacea L. in Lavergne and Molofsky (2007). Alternatively, we may not reject that the two observed groups are related to a process of genetic drift following the introduction of fireweed into the Pampas. The Mantel test indicates a positive correlation between genetic and geographic diversity; however, due to the recent colonization of fireweed in subtropical South America, it is unlikely that this is related to patterns of structuring by some type of barrier to gene flow, but instead is caused by the founder effect of each population.
Spread and gene flow
Although fireweed was introduced recently into southern Brazil, the field observations in recent years suggest that this weed has already reached the current spreading stage. This rapid success may be related to high production, seed dispersal and preadaptation rather than to early aliens (Le Roux et al., 2010;Thébault et al., 2011). In addition, we found a proportionally higher number of migrants than reported by Le Roux et al. (2010) in the Hawaiian Islands. This difference can be explained by the greater ability of pollen and seeds to disperse in an environment with continuous lowlands, as in the Pampas region (Chaneton and Lavado, 1996). Another factor that may be contributing to the rapid dissemination of S. madagascariensis in the Pampas region is the considerable level of inbreeding observed here. This reproductive feature facilitates rapid population increase and spread. Additionally, Le Rouxet al. (2010) showed that moderate levels of inbreeding are commonly observed in other alien species (e.g.,Young and Murray, 2000; Yu et al., 2014). Inbreeding may be an indirect consequence of inter-breeding between close relatives due to the founder effect and/or excessive self-fertilization (Le Roux et al., 2010). The moderate levels of inbreeding produced heterozygosity values lower than those expected and may provide some evidence of self-fertilization of S. madagascariensis in the Pampas region. In contrast, the gene flow among populations can influence a rapidly increasing population in preventing genetic drift and inbreeding depression (Le Rouxet al., 2010).
Impacts in the Pampas region
Senecio madagascariensis has been very effective in its dispersal and establishment in new locations in the Pampas region. The considerable levels of genetic diversity, gene flow and inbreeding indicate that this species has great potential for successful establishment in new environments in southern Brazil. Fireweed is quickly expanding geographically and in population density. Additionally, fireweed is an increasingly significant threat to indigenous species and livestock production in the Pampas region. Researchers aim to control this and other Senecio species in the Pampas region (Karam et al., 2011, 2013) due to the severe danger that these plants represent for cattle. Our results may underpin future studies about this weed in subtropical South America and also in other locations around the world.
Thus, for a better understanding of the invasion of Senecio madagascariensis into the Pampas region it is still important to investigate more molecular markers and other populations from Uruguay and Argentina. In this regard, the EST collection performed by Prentis et al. (2010) provides many initial candidate-gene markers that can help to answer a number of leading hypotheses in invasion biology.
Acknowledgments
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), the Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS), and the Programa de Pós–Graduação em Genética e Biologia Molecular – Universidade Federal do Rio Grande do Sul (PPGBMUFRGS). We thank Ana L. A. Segatto and Nelson J. R. Fagundes for help in the analyses, Priscilla M. Zamberlan for sequencing facilities, and Fernando C. Karam for personal communications.
Supplementary Material.
The following online material is available for this article:
Figure S1 (1.4MB, pdf) - Senecio madagascariensis in Bagé.
Figure S2 (546.7KB, pdf) - Relationship between geographical (km) and genetic (FST) distances.
This material is available as part of the online article from http://www.scielo.br/gmb
Footnotes
Associate Editor: Marcio de Castro Silva-Filho
References
- Allendorf FW, Lundquist LL. Introduction: population biology, evolution, and control of invasive species. Conserv Biol. 2003;17:24–30. [Google Scholar]
- Alpert P, Simms EL. The relative advantages of plasticity and fixity in different environments: When is it good for a plant to adjust? Evol Ecol. 2002;16:285–297. [Google Scholar]
- Bandelt H–J, Forster P, Röhl A. Median–joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barrett SCH, Colautti RI, Eckert CG. Reproductive systems and evolution during biological invasion. Mol Ecol. 2008;17:373–383. doi: 10.1111/j.1365-294X.2007.03503.x. [DOI] [PubMed] [Google Scholar]
- Blackburn TM, Pyek P, Bacher S, Carlton JT, Duncan RP, Jaroík V, Wilson JRU, Richardson DM. A proposed unified framework for biological invasions. TrendsEcol Evol. 2011;26:333–339. doi: 10.1016/j.tree.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Bock DG, Caseys C, Cousens RD, Hahn MA, Heredia SM, Hübner S, Turner KG, Whitney KD, Rieseberg LH. What we still don't know about invasion genetics. Mol Ecol. 2015;24:2277–2297. doi: 10.1111/mec.13032. [DOI] [PubMed] [Google Scholar]
- Buckler ES, Ippolito A, Holtsford TP. The evolution of ribosomal DNA: divergent paralogues and phylogenetic implications. Genetics. 1997;145:821–832. doi: 10.1093/genetics/145.3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton EJ, Lavado RS. Soil nutrients and salinity after long–term grazing exclusion in a flooding Pampa grassland. J Range Manage. 1996;49:182–187. [Google Scholar]
- Chun YJ, Nason JD, Moloney KA. Comparison of quantitative and molecular genetic variation of native vs. invasive populations of purple loosestrife (Lythrum salicaria L., Lythraceae) Mol Ecol. 2009;18:3020–3035. doi: 10.1111/j.1365-294X.2009.04254.x. [DOI] [PubMed] [Google Scholar]
- Comes HP, Abbott RJ. Molecular phylogeography, reticulation, and lineage sorting in Mediterranean Senecio sect. Senecio(Asteraceae) Evolution. 2001;55:1943–1962. [PubMed] [Google Scholar]
- Crawford KM, Whitney KD. Population genetic diversity influences colonization success. Mol Ecol. 2010;19:1253–1263. doi: 10.1111/j.1365-294X.2010.04550.x. [DOI] [PubMed] [Google Scholar]
- Desfeux C, Lejeune B. Systematics of Euromediterranean Silene(Caryophyllaceae): evidence from a phylogenetic analysis using ITS sequence. C R Acad Sci III. 1996;319:351–358. [PubMed] [Google Scholar]
- Dormontt EE, Gardner MG, Breed MF, Rodger JG, Prentis PJ, Lowe AJ. Genetic bottlenecks in time and space: reconstructing invasions from contemporary and historical collections. PLoS One. 2014;9: doi: 10.1371/journal.pone.0106874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn IS, Blattner FR. Sharons 36 to 40: multi–enzyme, high capacity, recombination deficient replacement vectors with polylinkers and polystuffers. Nucleic Acids Res. 1987;15:2677–2698. doi: 10.1093/nar/15.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durka W, Bossdorf O, Prati D, Auge H. Molecular evidence for multiple introductions of garlic mustard (Alliaria petiolata, Brassicaceae) to North America. Mol Ecol. 2005;14:1697–1706. doi: 10.1111/j.1365-294X.2005.02521.x. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals sing the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population STRUCTURE using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes–Aguilar J, Nieto–Feliner G. Additive polymorphisms and reticulation in an ITS phylogeny of thrifts (Armeria, Plumbaginaceae) Mol Phylogenet Evol. 2003;28:430–447. doi: 10.1016/s1055-7903(02)00301-9. [DOI] [PubMed] [Google Scholar]
- Ghabooli S, Shiganova TA, Zhan A, Cristescu ME, Eghtesadi–Araghi P, MacIsaac HJ. Multiple introductions and invasion pathways for the invasive ctenophore Mnemiopsis leidyi in Eurasia. Biol Invasions. 2011;13:679–690. [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kaluza P, Kölzsch A, Gastner MT, Blasius B. The complex network of global cargo ship movements. J R Soc Interface. 2010;7:1093–1103. doi: 10.1098/rsif.2009.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam FC, Schild AL, Mello JRB. Poisoning by Senecio spp. in cattle in southern Brazil: Favorable conditions and control measures. Pesq Vet Bras. 2011;31:603–609. [Google Scholar]
- Karam FC, Moraes JCF, Schild AL. Sheep grazing for control of Senecio spp. According to infestation level and phenology. Pesq Vet Bras. 2013;33:1109–1115. [Google Scholar]
- Keane RM, Crawley MJ. Exotic plant invasions and the enemy release hypothesis. Trends Ecol Evol. 2002;17:164–170. [Google Scholar]
- Keller RP, Drake JM, Drew MB, Lodge DM. Linking nvironmental conditions and ship movements to estimate invasive species transport across the global shipping network. Divers Distrib. 2011;17:93–102. [Google Scholar]
- Lachmuth S, Durka W, Schurr FM. The making of a rapid plant invader: genetic diversity and differentiation in the native and invaded range of Senecio inaequidens . Mol Ecol. 2010;19:3952–3967. doi: 10.1111/j.1365-294X.2010.04797.x. [DOI] [PubMed] [Google Scholar]
- Lafuma L, Maurice S. Increase in mate availability without loss of self–incompatibility in the invasive species Senecio inaequidens (Asteraceae) Oikos. 2007;116:201–208. [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic diversity and evolutionary potential drive the success of an invasive grass. Proc Natl Acad Sci USA. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux JJ, Wieczorek AM. Isolation and characterization of polymorphic microsatellite markers from fireweed, Senecio madagascariensis Poir. (Asteraceae) Mol Ecol Notes. 2007;7:327–329. [Google Scholar]
- Le Roux JJ, Wieczorek AM, Ramadan MM, Tran CT. Resolving the native provenance of invasive fireweed (Senecio madagascariensis Poir.) in the Hawaiian Islands as inferred from phylogenetic analysis. Divers Distrib. 2006;12:694–702. [Google Scholar]
- Le Roux JJ, Wieczorek AM, Tran CT, Vorsino A. Disentangling the dynamics of invasive fireweed (Senecio madagascariensis Poir. species complex) in the Hawaiian Islands. Biol Invasions. 2010;12:2251–2264. [Google Scholar]
- Lee CE. Evolutionary genetics of invasive species. Trends Ecol Evol. 2002;17:386–391. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- López MG, Wulff AF, Poggio L, Xifreda CC. South African fireweed Senecio madagascariensis(Asteraceae) in Argentina: relevance of chromosome studies to its systematic. Bot J Linn Soc. 2008;158:613–620. [Google Scholar]
- López–García MC, Maillet J. Biological characteristics of an invasive South African species. Biol Invasions. 2005;7:181–194. [Google Scholar]
- Lounibos LP. Invasions by insect vectors of human disease. Annu Rev Entomol. 2002;47:233–266. doi: 10.1146/annurev.ento.47.091201.145206. [DOI] [PubMed] [Google Scholar]
- Mäder G, Zamberlan PM, Fagundes NJR, Magnus T, Salzano FM, Bonatto SL, Freitas LB. The use and limits of ITS data in the analysis of intraspecific variation in Passiflora L. (Passifloraceae) Genet Mol Biol. 2010;33:99–108. doi: 10.1590/S1415-47572009005000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzenbacher NI, Schneider AA. Nota sobre a presença de uma espécie adventícia deSenecio (Asteraceae) no Rio Grande do Sul, Brasil. Rev Bras Bioci. 2008;6:111–115. [Google Scholar]
- McCullough DG, Work TT, Cavey JF, Liebhold AM, Marshall D. Interceptions of nonindigenous plant pests at US ports of entry and border crossings over a 17–year period. Biol Invasions. 2006;8:611–630. [Google Scholar]
- Mooney HA, Cleland EE. The evolutionary impact of invasive species. Proc Natl Acad Sci USA. 2001;98:5446–5451. doi: 10.1073/pnas.091093398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray BR, Phillips ML. Investment in seed dispersal structures is linked to invasiveness in exotic plant species of south–eastern Australia. Biol Invasions. 2010;12:2265–2275. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. 512 [Google Scholar]
- Nettel A, Dodd RS. Drifting propagules and receding swamps: genetic footprints of mangrove recolonization and dispersal along tropical coasts. Evolution. 2007;61:958–71. doi: 10.1111/j.1558-5646.2007.00070.x. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW. GeneDoc: Analysis and Visualization of Genetic Variation. EMBNET.news. 1997;4:14–14. [Google Scholar]
- Novak SJ. The role of evolution in the invasion process. Proc Natl Acad Sci USA. 2007;104:3671–3672. doi: 10.1073/pnas.0700224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeck GE, Müller SC, Fidelis A, Pfadenhauer J, Pillar VD, Blanco CC, Boldrini II, Both R, Forneck ED. Brazil's neglected biome: the South Brazilian Campos. Perspect Plant Ecol Evol Syst. 2007;9:101–116. [Google Scholar]
- Pelser PB, Nordenstam B, Kadereit JW, Watson LE. An ITS phylogeny of Tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon. 2007;56:1077–1104. [Google Scholar]
- Piry S, Alapetite A, Cornuet J–M, Paetkau D, Baudouin L, Estoup A. GeneClass2: a software for genetic assignment and first–generation migrant detection. J Hered. 2004;95:536–539. doi: 10.1093/jhered/esh074. [DOI] [PubMed] [Google Scholar]
- Prentis PJ, Woolfit M, Thomas-Hall SR, Ortiz-Barrientos D, Pavasovic A, Lowe AJ, Schenk PM. Massively parallel sequencing and analysis of expressed sequence tags in a successful invasive plant. Ann Bot. 2010;106:1009–1017. doi: 10.1093/aob/mcq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rejmánek M. Invasive plants: approaches and predictions. Austral Ecol. 2000;25:497–506. [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- Rosenthal DM, Ramakrishnan AP, Cruzan MB. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol Ecol. 2008;17:4657–4669. doi: 10.1111/j.1365-294X.2008.03844.x. [DOI] [PubMed] [Google Scholar]
- Roy A, Frascaria N, MacKay J, Bousquet J. Segregating random amplified polymorphic DNAs (RAPDs) inBetula alleghaniensis . Theor Appl Genet. 1992;85:173–180. doi: 10.1007/BF00222856. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Weller SG, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, et al. The population biology of invasive species. Annu Rev Ecol Syst. 2001;32:305–332. [Google Scholar]
- Sala OE, Chapin FS, Armesto JJ, Berlow E, Bloomfield J, Dirzo R, Huber–Sanwald E, Huenneke LF, Jackson RB, Kinzig A, et al. Global biodiversity scenarios for the year 2100. Science. 2000;287:1770–1774. doi: 10.1126/science.287.5459.1770. [DOI] [PubMed] [Google Scholar]
- Sax DF, Stachowicz JJ, Brown JH, Bruno JF, Dawson MN, Gaines SD, Grosberg RK, Hastings A, Holt RD, Mayfield MM, et al. Ecological and evolutionary insights from species invasions. Trends Ecol Evol. 2007;22:465–471. doi: 10.1016/j.tree.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Sindel BM, Radford IJ, Holtkamp RH, Michael PW. The biology of Australian weeds. 33. Senecio madagascariensis Poir. Plant Prot Q. 1998;13:2–15. [Google Scholar]
- Strauss SY, Lau JA, Carroll SP. Evolutionary responses of natives to introduced species: what do introductions tell us about natural communities? Ecol Lett. 2006;9:354–371. doi: 10.1111/j.1461-0248.2005.00874.x. [DOI] [PubMed] [Google Scholar]
- Sultan SE. Phenotypic plasticity in plants: a case study in ecological development. Evol Dev. 2003;5:25–33. doi: 10.1046/j.1525-142x.2003.03005.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thébault A, Gillet F, Müller-Schärer H, Buttler A. Polyploidy and invasion success: trait trade-offs in native and introduced cytotypes of two Asteraceae species. Plant Ecol. 2011;212:315–325. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO–CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Yamaji H, Fukuda T, Yokoyama J, Pak J–H, Zhou C–Z, Yang C–S, Kondo K, Morota T, Takeda S, Sasaki H, Maki M. Reticulate evolution and phylogeography inAsarum sect. Asiasarum (Aristolochiaceae) documented in internal transcribed spacer sequences (ITS) of nuclear ribosomal DNA. Mol Phylogenet Evol. 2007;44:863–884. doi: 10.1016/j.ympev.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Young AG, Murray BG. Genetic bottlenecks and dysgenic gene flow into re-established populations of the grassland daisy, Rutidosis leptorrhynhoides . Aust J Bot. 2000;43:409–416. [Google Scholar]
- Yu X, He T, Zhao J, Li Q. Invasion genetics of Chromolaena odorata(Asteraceae): extremely low diversity across Asia. Biol Invasions. 2014;16:2351–2366. [Google Scholar]
Internet Resources
- Genepop 4.1 software. [November 12, 2014]. http://kimura.univ-montp2.fr/~rousset/Genepop.htm.
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001. [November 12, 2014]. http://www2.unil.ch/popgen/softwares/fstat.htm.
- Network 4.6 software. [November 12, 2014]. http://www.flux-us-engineering.com.