Abstract
Single-cell DNA sequencing methods are challenged by poor physical coverage, high technical error rates and low throughput. To address these issues, we developed a single-cell DNA sequencing protocol that combines flow-sorting of single nuclei, time-limited multiple-displacement amplification (MDA), low-input library preparation, DNA barcoding, targeted capture and next-generation sequencing (NGS). This approach represents a major improvement over our previous single nucleus sequencing (SNS) Nature Protocols paper in terms of generating higher-coverage data (>90%), thereby enabling the detection of genome-wide variants in single mammalian cells at base-pair resolution. Furthermore, by pooling 48–96 single-cell libraries together for targeted capture, this approach can be used to sequence many single-cell libraries in parallel in a single reaction. This protocol greatly reduces the cost of single-cell DNA sequencing, and it can be completed in 5–6 d by advanced users. This single-cell DNA sequencing protocol has broad applications for studying rare cells and complex populations in diverse fields of biological research and medicine.
INTRODUCTION
The development of NGS methods has greatly improved our understanding of genomics in many fields of biology and medicine1. As the cost of NGS continues to decrease, it is now feasible to perform large-scale sequencing studies, such as The Cancer Genome Atlas (TCGA) and the 1000 Genomes Project, that aim to discover novel mutations and variants in large patient cohorts2–5. Although NGS can identify genomic differences between individual patients, the genomic differences within individual samples is often missed6. The central issue is that NGS methods require micrograms of DNA as input material, and these bulk samples consist of millions of cells. Consequently, the data generated from NGS methods often reflect an average signal from a complex population, and therefore they cannot accurately resolve population substructure. This problem is particularly acute in heterogeneous populations of cells, such as tumors in which genomic intratumor heterogeneity is common7–9. To address this problem, single-cell DNA sequencing methods have been developed, and they have shown great utility in resolving intratumor heterogeneity and reconstructing clonal evolution during tumor growth10–15. For a detailed review of single-cell sequencing (SCS) applications in cancer and other fields, please refer to review articles (refs. 16–20).
Development of the protocol
SCS protocols can be broken down into three stages: (i) single-cell isolation, (ii) genome or transcriptome amplification and (iii) NGS.
A number of methods have been developed to accomplish all of these stages, and the most appropriate approach will depend largely on the sample type. Although most studies sequence DNA obtained from whole cells, this protocol involves sequencing DNA from single nuclei. This approach has the major advantage of allowing single-cell information to be obtained from archival frozen tissues, in which the cell surface structure is ruptured during the freeze-thaw cycle but the nuclear membrane remains intact. Sequencing from nuclei is also advantageous for fresh tissues for which the physical separation of single cells is difficult, such as brain tissues.
The first published single-cell DNA sequencing method, SNS, combined flow-sorting of nuclei, degenerate-oligonucleotide-primer PCR (DOP-PCR) and sparse NGS to measure copy number profiles of individual cells at high (54 kb) resolution21. A detailed protocol for SNS was previously published in Nature Protocols22. Although it is effective for measuring copy number in large intervals, DOP-PCR is inherently limited to the generation of sparse coverage data (~10% of the genome), which does not provide sufficient data to measure genome-wide mutations that differ between single cells at base-pair resolution.
To improve coverage performance, we developed a method called Nuc-seq, to perform whole-genome SCS23. With this method, we have shown that it is possible to detect single-nucleotide variants (SNVs) and small insertions/deletions (indels) in individual mammalian cells at genomic resolution. The Nuc-seq method combines flow-sorting of single cells, time-limited MDA using Φ29, library construction using Tn5 transposases, limited PCR amplification and Illumina sequencing to generate high-coverage (>90%) data sets from single-cell genomes. In this approach, we flow-sort single nuclei in the G2/M phase of the cell cycle, which doubles the starting amount of input DNA (four copies of the genome), thereby increasing the probability of amplifying both variant alleles during whole-genome amplification (WGA). Our data showed that sequencing G2/M cells increases the detection efficiency for SNVs by 5.66% and reduces the allelic dropout rate (ADR) by 9.30%, which are the major sources of technical error in SCS methods. This method also greatly improved the physical coverage performance to more than 90% for SCS, which is due to the use of a Φ29 bacteriophage polymerase for WGA by MDA.
We further improved our SCS method by developing single-nucleus exome sequencing (SNES) to eliminate the use of transposase and its associated integration bias24. By capturing the exonic regions of the genome, SNES enabled more single cells to be multiplexed in parallel in a single sequencing lane of the HiSeq 2000 instrument (Illumina), thereby reducing cost and time for processing samples. In this previous study, we showed that SNES can achieve 95.94% of coverage breadth of the exome in single cells, low ADRs of 21.52% and high detection efficiencies of 92.37% for SNVs in an isogenic population. Moreover, we show that SNES can be applied to sequence either G1/0 or G2/M cells, thereby enabling its application to sequencing normal cells with low proliferation rates. However, it is important to consider that biasing toward G2/M cells may miss profiling cells with low proliferation rates.
In the current protocol, we have further refined SNES by using DNA barcoding to multiplex 48–96 single cells into single sequencing reactions to further increase throughput and to reduce costs of SCS. This is achieved by performing targeted capture on a panel of 200 cancer-associated genes, resulting in high coverage depth (average 255×) and reducing the cost of sequencing. This protocol can readily be adapted to any targeted capture panel to genotype hundreds of single cells in parallel. The protocol can be applied for the analysis of G2/M cells or cells at any stage of the cell cycle.
In summary, the high-coverage SNS approach using MDA described in this protocol was optimized to increase physical coverage, and it is distinct from the previous SNS protocol that uses DOP-PCR for WGA22. MDA results in high coverage breadth, but at the cost of nonuniform amplification, and thus it is recommended for the detection of point mutations and indels at base-pair resolution. In contrast, the DOP-PCR amplifies the genome more uniformly but at sparse (~10%) targeted regions during PCR.
Therefore, we recommend using the protocol reported here to detect point mutations and indels, while using the SNS protocol22 for detecting copy number profiles by read counting from sparse coverage depth.
Overview of the experimental procedure
Nuclear suspensions are prepared from fresh or frozen tissue and stained with NST-DAPI buffer for flow-sorting. Direct comparisons of DAPI-stained nuclei versus unstained nuclei show no differences in the technical error rates. Nuclei are gated by total DNA content into G1/0 or G2/M populations and deposited singly into a 96-well plate containing lysis buffer. Genomic DNA from individual nuclei is amplified by MDA using the Φ29 polymerase and modified random hexamers (see Reagents). Quality control (QC) for WGA efficiency is performed by qPCR or PCR using a panel of 22 chromosome-specific primers, and only single cells with >20 amplicons are selected for library preparation and NGS. Illumina sequencing libraries are constructed by TA cloning using the New England BioLabs (NEB) library preparation reagents. The protocol describes in detail the DNA library preparation, barcoding and the purification and quantification steps. During the ligation step, we add unique 8-bp barcodes in order to multiplex libraries together before targeted capture. The process for generating barcoded adapters is described in Box 1. For experiments in which commercial kits are used, we follow the manufacturers’ protocols with minor modifications. The bar-coded libraries are then pooled for targeted capture of genomic regions. Targeted capture can be performed using a variety of platforms from different manufacturers. We provide a detailed protocol for targeted capture of a 1-Mb cancer gene panel that was synthesized by Roche NimbleGen. The captured libraries then undergo a final PCR enrichment step to amplify sufficient DNA for NGS on the Illumina platform. An overview of the protocol is shown in Figure 1.
Figure 1.
Protocol overview. Step 1: tissue or cells are lysed and nuclear suspensions are prepared and stained with DAPI. Steps 2–9: nuclei are flow-sorted using FACS by gating the G1/0 or G2/M distributions based on total DNA content, and single nuclei are deposited into individual wells in a 96-well plate. Steps 10 and 11: the nuclear membrane is lysed.
Steps 12 and 13: WGA using MDA is performed using the Φ29 polymerase. Steps 14–16: for quality control of WGA efficiency, each WGA reaction is screened with a 22-amplicon qPCR panel using chromosome-specific primer pairs. Steps 17–61: barcoded libraries are prepared from each single-cell WGA reaction. Steps 62–103: barcoded libraries (12–96) are pooled together into a single reaction followed by targeted capture. Step 104: the pooled libraries are then used for NGS on the Illumina platform.
Overview of the data analysis procedure
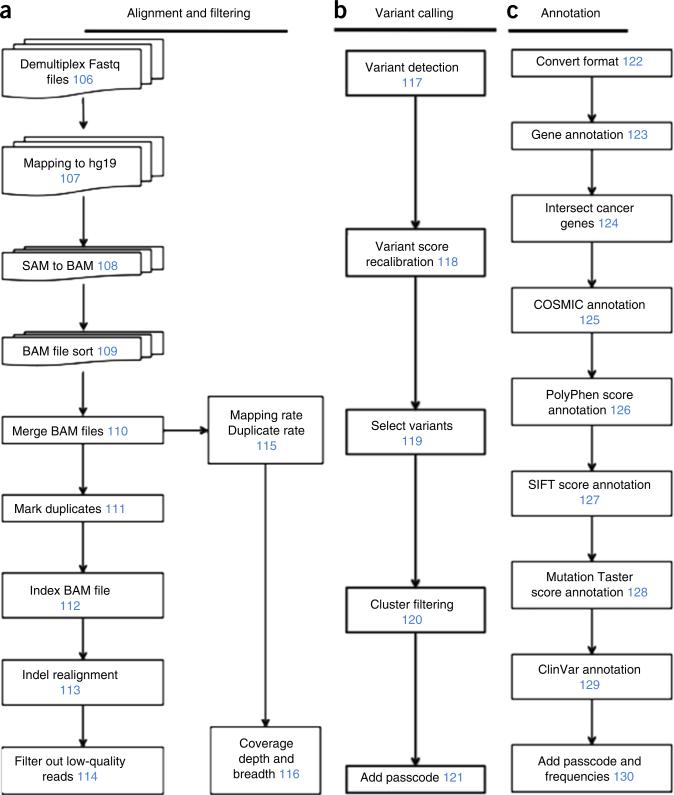
The FASTQ file containing all of the NGS data is demultiplexed into individual FASTQ files using our in-house software (deplexer.pl). Individual FASTQ files are aligned to the human genome reference assembly GRCh37 (HG19) using Bowtie 2, and they are converted to BAM files using SAMtools25,26. BAM files are then processed by Picard to remove PCR duplicates. Re-alignment is performed around indel regions using the Genome Analysis Toolkit (GATK)27. Sequencing reads with mapping quality lower than 40 are removed. To calculate coverage metrics, we use an in-house Perl script (cal_coverage_metrics.pl; Supplementary Software), which uses BEDTools to get coverage depth at each site and to calculate overall coverage depth and coverage breadth28. After running GATK to perform variant calling, we generate a multi-cell VCF file. Subsequently, GATK is run to perform variant quality score recalibration on the VCF file. We then filter mutations that are only detected in one single cell. We filter mutations in clustered regions, in which multiple mutations are detected within a 10-bp window. Finally, we annotate the variants using ANNOVAR to classify them as synonymous, nonsynonymous, intergenic, splice-site variants, frameshift or nonframeshift, and to estimate the damaging effect on protein function using SIFT and PolyPhen scores29–31. The outline of the data processing pipeline, variant detection and annotation steps are shown in Figure 2. Software and scripts we developed and used for data analysis are provided as Supplementary Software.
Figure 2.
Data processing pipeline. (a) Alignment of sequencing reads to the reference genome and filtering by quality metrics. (b) Detection of DNA variants and filtering of technical artifacts. (c) Annotation of variants using integrated databases and protein damage–prediction algorithms. Numbers in blue indicate Step numbers in the PROCEDURE.
Applications of the protocol
This protocol has applications in many broad fields of biology and medicine for investigating genomic diversity in complex populations, and profiling rare cells. Fields with major applications include neurobiology, immunology, microbiology, development and cancer research. Clinical applications include prenatal genetic diagnostics, cancer diagnostics and noninvasive monitoring7–9. In cancer research, these protocols have a number of applications for studying transformation, invasion, metastasis and chemotherapy resistance evolution16,32. Several studies have already shown the value of applying SCS methods to the study of circulating tumor cells in the blood in order to understand metastasis and clinical applications for noninvasive monitoring6. However, most of the aforementioned applications are speculative, and they have not yet been investigated. In our own work, we have applied these tools to the study of intratumor heterogeneity and mutational evolution in primary breast cancer patients and genomic mosaicism in normal fibroblast cell lines23.
Alternative methods for single-cell DNA sequencing
Several other approaches have been developed for DNA SCS. The Beijing Genome Institute (BGI) developed an MDA-based approach, which showed high ADRs of 46% for SCS of exomes14. Another method called multiple annealing and looping-based amplification cycles (MALBAC) was developed that circularizes DNA molecules by combining an MDA and DOP-PCR approach33. MALBAC performs WGA using both a Bst polymerase with no proofreading activity and a PyroPhage polymerase with proofreading activity. Consequently, the false-positive error rate is substantially increased compared with WGA protocols that use only Φ29 for MDA. Our own group has developed protocols for single-cell copy number profiling (SNS) that combine flow-sorting of nuclei, DOP-PCR and NGS21,22. In general, the data from these methods suggest that DOP-PCR– and MALBAC-based approaches provide better data for detecting copy number profiles, whereas the MDA-based approaches are better for detecting DNA mutations at base-pair resolution (point mutations and indels)16,17.
Limitations of the protocol
This protocol achieves optimal performance when G2/M single cells are isolated. However, in noncycling populations (e.g., neurons), it may not be possible to isolate G2/M cells. Alternatively, G1/0 cells can be used with this protocol. However, this may lead to a small increase (~10%) in the ADR, as previously shown in normal fibroblast cell cultures24.
Because of the use of Φ29 polymerase to perform WGA of single cells, different regions with varying amounts of GC content and repetitive sequence may amplify unevenly. This results in nonuniform coverage and makes it difficult to measure copy number from sequence read depth.
False-positive errors that arise during WGA may be mistaken for real biological variants during data processing and analysis. However, their distribution is largely random from cell to cell; therefore, they can be mitigated by considering only SNVs that are detected in two or more single cells at the same nucleotide site.
All SCS methods (including this protocol) generate false-positive and false-negative technical errors. Once mutations are discovered, it is important to distinguish between biological heterogeneity and technical variability by performing targeted validation experiments (e.g., deep sequencing or droplet digital PCR34).
Experimental design
Sample requirements and preparation
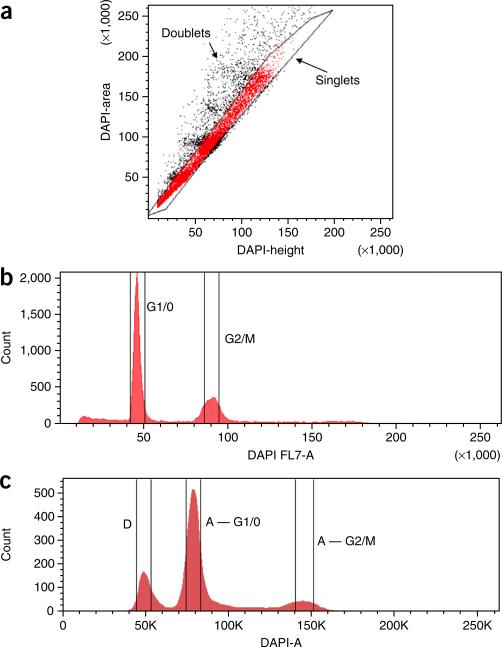
Our protocol is optimized for the isolation of single nuclei by flow-sorting nuclear suspensions prepared from human tumor tissues. For these experiments, we recommend using frozen or fresh tissue to prepare nuclear suspensions. However, it is worth noting that this protocol can also be applied broadly to any tissue type. When cell suspensions have already been prepared (e.g., cell culture or blood samples), users can alternatively flow-sort whole cells without first preparing nuclear suspensions. In such cases, staining with DAPI is not necessary for gating cells by ploidy. Users should be aware that the quality of tissue is important for the success of these experiments. Tissues that have undergone multiple freeze-thaw cycles can suffer DNA degradation and generate poor-quality sequencing data. Although fresh or snap-frozen tissues are preferred for this protocol, fixed tissues in ethanol or methanol may also be used. However, we do not recommend using formalin-fixed samples for these experiments, which often have double-stranded breaks and single-stranded nicks in the DNA. It is also important to avoid aggregate cells during flow-sorting by plotting DAPI area and DAPI height to exclude ‘doublets’ or multiple-nuclei clusters (Fig. 3).
Figure 3.
Flow-sorting and gating for single nucleus isolation. (a) Nuclei are flow-sorted and gated by DAPI area and DAPI height to avoid collecting multiple nuclei that are stuck together. (b,c) DAPI area versus nuclei count is plotted showing gates for G1/0 or G2/M distributions from (b) a normal fibroblast cell line and (c) a breast tumor sample with an aneuploid subpopulation. K indicates thousands.
Sequencing and QC
When sequencing single tumor cells, we recommend sequencing the bulk tumor sample and normal tissue in parallel. The matched normal tissue is important for identifying germline mutations that can be filtered from the single tumor cells in order to distinguish somatic mutations. The bulk tumor cell provides a comparative data set in which the frequencies of somatic mutations are expected to correlate with the number of single cells that carry the specific mutations. When preparing the bulk tissue samples, at least 100 ng of DNA is required to generate a library without WGA, which may introduce technical errors in the final data set. In the ANTICIPATED RESULTS section, we demonstrate results from an isogenic cell line, in which we can assume that the variants present in the bulk sequencing are also present in all of the single cells. This allows accurate calculations of the ADR and detection efficiency in order to understand the technical error rates of the approach before data analysis.
In our previous study, we determined that 3 h of WGA using Φ29 is the optimal time frame for producing sufficient DNA from single cells for preparing NGS libraries24. However, users can optimize the amplification time to obtain more or less DNA for different downstream applications.
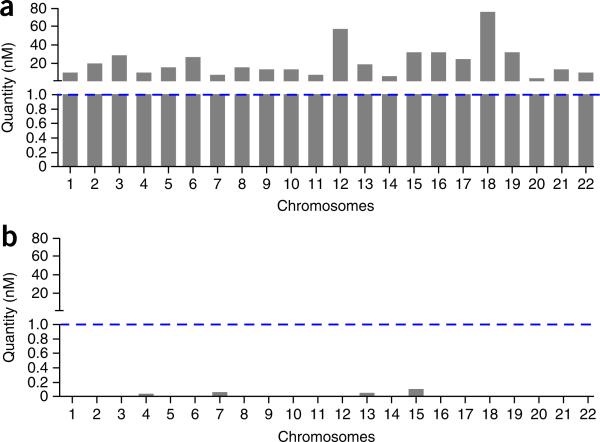
To avoid sequencing WGA reactions that consist of bacterial DNA, or WGA reactions with poor uniformity of coverage, it is important to perform a QC step in the protocol, in which 22 chromosome-specific primers are used to amplify DNA from each single-cell WGA reaction (Fig. 4). Alternatively, users may choose a smaller subset of these primer pairs (e.g., n = 8 or n = 12) or use a 384-well plate to increase throughput in larger sample sets. Single cells with poor WGA performance (<20 amplicons) should be excluded from subsequent library construction and NGS analysis. For standards used in the qPCR QC step, we use genomic DNA that has been previously analyzed using a fluorimeter (e.g., Qubit, Life Technologies).
Figure 4.
WGA quality control using qPCR panels. For each single cell, a series of qPCRs is performed using 22 primer pairs that target each chromosome independently. A concentration of 1 nM is used as the threshold for a positive amplification reaction indicated by a blue dotted line. (a) Bar plot showing a single nucleus with positive amplification of all 22 chromosomes. (b) Bar plot showing a single nucleus that has no amplification of any chromosomes after WGA. This WGA reaction will not be used for subsequent library construction and sequencing.
Sonication
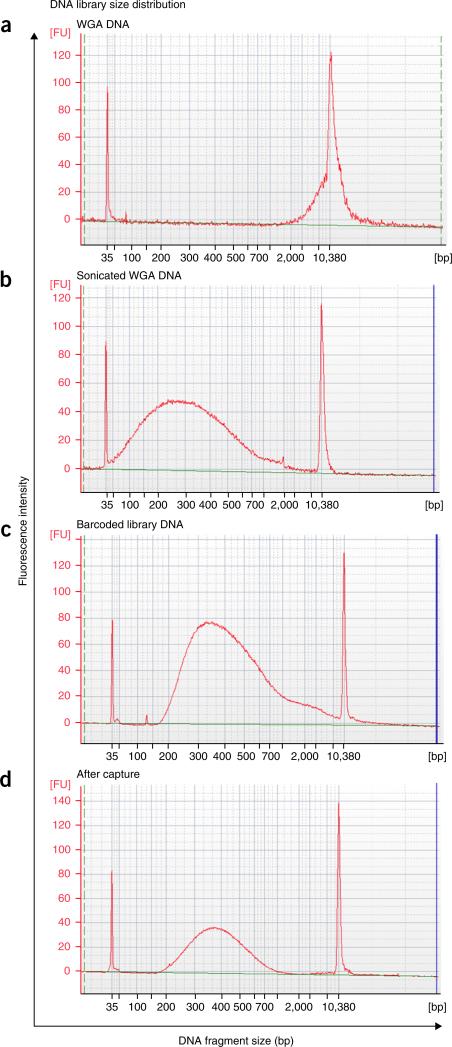
To construct sequencing libraries, genomic DNA must first be sheared into suitably sized fragments; we achieve this using an acoustic sonicator. We have determined that 250 bp is the optimal fragment size for hybridization to 100-mer probes during targeted capture (Fig. 5). However, users can choose to increase or decrease the fragment size for specific experimental needs. Table 1 lists the parameters for obtaining different fragment sizes from Φ29-amplified DNA using the Covaris Sonicator S220 (see also the manufacturer's user guide). Alternatives to acoustic sonication for fragmenting DNA include the use of DNA fragmentase enzymes or nebulization.
Figure 5.
DNA library insert size distributions at different steps during the protocol. Bioanalyzer microchip gel electrophoresis plots of DNA size distributions. (a) DNA size distribution of a single nucleus after WGA by MDA. (b) DNA size distribution after acoustic sonication of WGA DNA to a mean size of 250 bp. (c) Library insert size distribution of a single nucleus after the addition of barcoded sequencing adapters and PCR enrichment. (d) Library size distribution after targeted capture and final PCR enrichment.
TABLE 1.
Sonication parameters.
| DNA fragment size | 200 | 250 (Preferred) | 300 | 400 | 500 |
|---|---|---|---|---|---|
| Peak incident power (W) | 175 | 157 | 140 | 140 | 105 |
| Duty factor (%) | 10 | 10 | 10 | 10 | 5 |
| Cycles per burst | 200 | 200 | 200 | 200 | 200 |
| Treatment time (s) | 180 | 130 | 80 | 55 | 80 |
Targeted capture and sequencing output
To multiplex 48–96 single cells into one sequencing lane, libraries must be barcoded with an 8-bp sequence, pooled together and captured in a single targeted capture reaction. When only a few samples are multiplexed (such as in exome capture), the selection of barcodes must be pooled in specific combinations to ensure index diversity during NGS on the Illumina platform. In the final combination of the barcode pooling, each nucleotide position must contain at least one A or C and one G or T (Supplementary Table 1).
In our protocol, we implemented a panel of 200 cancer-associated genes that was previously developed by Chen et al.35. We use capture probes synthesized by Roche NimbleGen; however, users can choose a different gene-capture panel according to their experimental requirements. Currently, our capture region is 1 Mb. Users can design a larger capture region at the cost of decreasing coverage depth. To address this issue, users can multiplex fewer cells to maintain the optimal coverage depth for detecting variants. The data shown in the ANTICIPATED RESULTS section are generated from an Illumina HiSeq 2000 sequencing machine. If the user performs NGS using an updated machine (e.g., Illumina HiSeq 3000), the sequencing output will be higher, allowing for a larger capture region while maintaining the same cell number.
Sequencing metrics evaluation
After sequencing, basic sequencing metrics should be calculated and the quality of the single-cell data should be assessed, including the number of reads per cell, PCR duplicates, coverage uniformity and coverage depth. In addition, the user may want to determine the technical error rates of their approach using an isogenic cell line. Below we describe how to calculate technical error rates (the ADR and the false-positive rate (FPR)) and detection efficiencies. In these calculations, both sites (reference and single cell) require a minimum of 6× coverage depth to call variants.
The ADR is defined as the mean fraction of homozygous sites in the single-cell samples (Homs) in which the matched population reference sample is heterozygous (Hetp) at the same nucleotide site.
The FPR is defined as the number of heterozygous sites in the single-cell samples (Hets) divided by the number of sites in the population reference sample that are homozygous (Homp) for the reference allele at the same nucleotide site.
The detection efficiencies can be calculated from the VCF4 variant files after the filtering steps are performed. The filtered VCF4 file must first be partitioned into a separate file for SNVs. For each line in the VCF file, we then add a binary string indicating the absence or presence of each variant in the single-cell samples or the reference population sample. For each variant site in the population sample, we identify variant sites in the single-cell samples in which sufficient coverage depth (≥6×) is present. From the binary string, we determine whether the variant is present or absent in each single cell relative to the population reference sample. We define each variant as being detected if the reference allele is AB and the single-cell data are either AB or BB. The mean detection efficiencies for indels and SNVs are then computed across all of the single-cell samples.
Data analysis
The bioinformatics pipeline is designed and optimized for single-cell DNA sequencing data. Because SCS methods often have errors that are higher than conventional sequencing techniques, we have incorporated several optimization and filtering steps in the variant calling pipeline. For samples mapped with Bowtie 2, we typically find that >70% of reads from SCS experiments have mapping quality scores ≥40. To prevent detecting errors generated from sequencing, we filter out reads with poor mapping quality that are lower than 40; this cutoff can be adjusted at Step 114 according to specific experimental preferences.
In this protocol, we describe the steps to calculate coverage metrics. Step 116 uses an in-house Perl script (cal_coverage_metrics.pl) to calculate coverage breadth and coverage depth using SAMtools and BEDTools. This protocol uses a BED file for the gene-capture panel of 200 cancer-associated genes (IPCT; Supplementary Data). Users will need to replace the BED file with their own targeted cancer gene panel or exome capture region file, depending on the targeted capture platform that was used.
A common technical artifact in SCS data is a genomic region with clusters of false-positive (FP) mutations due to poorly mapped regions. To remove these FP artifacts, we filter out ‘clustered regions’ from the VCF files in which more than one SNV or indel is detected within a 10-bp window using a custom Perl script (filter_clustered_mutations.pl) at Step 120. Users can decide to adjust the 10-bp window size for cancers in which very high mutation frequencies are expected, or they can skip this step altogether if too many variants are being filtered.
MATERIALS
REAGENTS
Cell lines ! CAUTION Cell lines should be regularly checked for authenticity and the absence of Mycoplasma.
Fresh or frozen human tissue ! CAUTION Appropriate approvals are required from users’ institutions.
Φ29 Polymerase (10 units per 1 μl; NEB, cat. no. M0269L; Polymerase buffer is included)
dNTP set, 100 mM (NEB, cat. no. N0446S)
PBS 1×
Qubit dsDNA high-sensitivity assay kit (Invitrogen, cat. no. Q32854) ! CAUTION The dye is light sensitive. Keep away from light.
NEBNext end repair module (NEB, cat. no. E6050L)
NEBNext dA-tailing module (NEB, cat. no. E6053L)
NEBNext quick ligation module (NEB, cat. no. E6056L)
NEBNext high-fidelity 2× PCR master mix (NEB, cat. no. M0541L)
SYBR Fast qPCR master mix (KAPA, cat. no. KK4835)
SeqCap EZ Choice library (Roche NimbleGen)
SeqCap hybridization and wash kit (Roche NimbleGen, cat. no. 05634253001)
SeqCap EZ pure capture bead kit (Roche NimbleGen, cat. no. 06 977 952 001)
HiFi HotStart mix, 2× (KAPA, cat. no. KK2602)
DTT (Fisher Scientific, cat. no. BP172-5) ! CAUTION Direct contact can cause irritation.
Tris-HCl, 1 M (Life Technologies, 15567-027)
KOH (Fisher Scientific, cat. no. P251-500) ! CAUTION It may cause burn by exposure. Make a 1 M stock, divide it into aliquots and store them at −20 °C.
DTT (Fisher Scientific, cat. no. BP172-5) ! CAUTION It may cause irritation to skin. Make a 1 M stock, filter-sterilize the solution, divide it into aliquots and store them at −20 °C.
KCl (VWR International, cat. no. 101189-496)
HCl (EWD, cat. no. HX0603-4) ! CAUTION It causes severe skin burns and eye damage.
NaCl (Fisher Scientific, cat. no. BP358-1)
Tris base (Fisher Scientific, BP154-1)
CaCl2 (Sigma-Aldrich, cat. no. 21115-1ML)
MgCl2 (Sigma-Aldrich, cat. no. 63069-100ML)
BSA (Sigma-Aldrich, cat. no. A2058-100G)
Nonidet P-40 (US Biological, cat. no. N3500)
DAPI (Invitrogen, cat. no. D1306) ! CAUTION Keep it away from light.
Tween 20 (Sigma-Aldrich, cat. no. 11332465001)
Ethanol (Fisher Scientific, cat. no. BP2818500)
PBS (Sigma-Aldrich, cat. no. 79378)
NNNN*N*N random hexamer; order NNNN*N*N random hexamer (Integrated DNA Technologies). Asterisks represent a phosphorothioate linkage. Make a 1 mM working stock in water or elution buffer and store it at −20 °C
Ethanol; make 80% (vol/vol) ethanol with water. Make it freshly each time ! CAUTION Ethanol is flammable. Keep it away from open flame.
Annealed barcoded adapters, 10 μM (Box 1). Store them at −20 °C until use
P5 adapter; order oligos from Integrated DNA Technologies. Dilute the oligos to 100 μM in sterile water. Store them at −20 °C until use. 5-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T-3 (* = phosphorothioate)
Barcoded P7 adapter; order oligonucleotides from Integrated DNA Technologies. Dilute the oligos to 100 μM in sterile water. Store them at −20 °C until use (see sequences in Supplementary Table 2)
qPCR primers; order qPCR primer sequences (see sequence in Supplementary Table 3). Dilute and mix forward and reverse primers for each chromosome into individual tubes at a concentration of 5 nM. Store them at −20 °C until use
Adapter PCR primers; order sequences from Integrated DNA Technologies. Dilute the oligos to 5 μM in sterile water. Store them at −20 °C until use. Primer_F: 5-AATGATACGGCGACCACCGAGATCTACAC-3; Primer_R: 5-CAAGCAGAAGACGGCATACGAGAT-3
Bioinformatics software
Bowtie 2: http://sourceforge.net/projects/bowtie-bio/files/bowtie2/2.2.4/
SAMtools: http://sourceforge.net/projects/samtools/files/samtools/
Picard: https://github.com/broadinstitute/picard/releases/tag/1.128
GATK: https://www.broadinstitute.org/gatk/download/auth?package=GATK requires free registration
ANNOVAR: http://www.openbioinformatics.org/annovar/annovar_download.html
Integrated Genome Viewer: http://www.broadinstitute.org/software/igv/download
Custom software and scripts developed in our laboratory (Supplementary Software)
EQUIPMENT
AirClean 600 PCR workstation (AirClean, cat. no. AC632LFUVC) ! CAUTION UV is harmful to uncovered eyes and skin.
Bio-Rad T100 thermal cycler (Bio-Rad, cat. no. 186-1096)
PCR strips, 0.2 ml: Thermo Scientific 0.2-ml Thermo-Strip (Thermo Scientific, cat. no. AB-1182)
Pulse-spinning: Denville mini-mouse II microcentrifuge (Denville, cat. no. C0801-N)
Tube, 1.5 ml: Sigma-Aldrich Eppendorf Safe-Lock microcentrifuge tubes (Sigma-Aldrich, cat. no. T9661)
Nylon mesh filter, 36 μm (Small Parts, cat. no. CMN-0035-D)
Polystyrene round-bottom tube, 5 ml (BD Falcon, cat. no. 352058)
Feather disposable scalpel no. 11 (Graham-Field, cat. no. 2975#11)
Cell culture dish, 60 × 15mm (Corning, cat. no. 430196)
FACSAria II (BD Biosciences)
SPHERO Supra rainbow midrange fluorescent particles (Spherotech, cat. no. 556298)
PCR plate, 96 well: Thermo-tube plate (Thermo Scientific, cat. no. AB-0731)
Adhesive sealing sheets (Thermo Scientific, cat. no. AB-0558)
Cold block: Eppendorf PCR cooler iceless cold storage system for a 96-well plate and PCR tubes (Sigma-Aldrich, cat. no. Z606634-1EA) ▲ CRITICAL Place the cold block at −20 °C 1 h before use.
accuSpin micro 17 microcentrifuge (Fisher Scientific, cat. no. 13-100-675)
Zymo DNA Clean & Concentrator-5 columns kit (Zymo, cat. no. D4004)
Qubit 2.0 fluorometer (Invitrogen, cat. no. Q32866)
Covaris microTUBE AFA fiber pre-slit snap-cap (Covaris, cat. no. 520045)
Covaris S220 sonicator (Covaris)
PRISM real-time machine (ABI 7900HT)
MicroAmp optical 96-well reaction plate (ABI B8010560)
ThermalSeal RT2 film for qPCR (Phenix LMT-RT)
Digital dry block heater (VWR International, cat. no. 12621-084)
Precision digital circulating water bath (Thermo Scientific, cat. no. 2864)
Polypropylene tubes, 0.5 ml (USA Scientific, cat. no. 1405-8100)
Magnetic separation rack, six tube (NEB, cat. no. S1506S)
Magnetic plate, 96 well (Alpaqua, cat. no. A01322)
Digital SpeedVac modular concentrator (Thermo Scientific, cat. no. SPD111V-115)
REAGENT SETUP
NST-DAPI
Mix 800 ml of NST solution (146 mM NaCl, 10 mM Tris base (pH 7.8), 1 mM CaCl2, 21 mM MgCl2, 0.05% (wt/vol) BSA and 0.2% (vol/vol) Nonidet P-40) with 200 ml of DAPI solution (106 mM MgCl2 and 10 mg of DAPI). Filter-sterilize the solution and store it at 4 °C in the dark for up to 1 year.
Lysis buffer
Lysis buffer is 200 mM KOH and 50 mM DTT. To make 150 μl, mix 30 μl of 1 M KOH, 7.5 μl of 1 M filter-sterilized DTT and 112.5 μl of sterile water together. This volume is sufficient for one 96-well plate. ▲ CRITICAL The lysis buffer must be freshly made every time, and it should be placed on ice until use.
Neutralization buffer
Neutralization buffer is 900 mM Tris-HCl (pH 8.3), 300 mM KCl and 200 mM HCl. Autoclave the neutralization buffer. Each 96-well plate requires 144 μl of neutralization buffer. Divide the solution into aliquots and store them at −20 °C for up to 1 year.
Amplification buffer
For each single-cell amplification reaction, make a 45-μl amplification buffer by adding 5 μl of 10× Φ29 buffer, 2.5 μl of 1 mM random hexamers, 0.5 μl of 100 mM dNTP (mix equal amount of dATP, dCTP, dGTP and dTTP together), 36 μl of sterile water and 1 μl of Φ29 buffer polymerase. Freshly prepare the solution each time.
Elution buffer
Elution buffer is 10 mM Tris-Cl at pH 8.5.
Autoclave and store the buffer at room temperature (RT, 23 °C) for up to 1 year.
Tween 20, 0.05% (vol/vol)
Make 0.05% (vol/vol) Tween 20 in ddH2O.
Store the solution at RT for up to 1 month.
EQUIPMENT SETUP
Covaris S220 sonicator for 250 bp
Settings are as follows: peak power 157, duty factor 10%, cycles/burst 200, time 130 s and temperature 4–7 °C (Table 1). ▲ CRITICAL Water should be degassed.
PROCEDURE
Preparation of nuclear suspensions ● TIMING 30 min
-
1|
To prepare the sample for flow-sorting, follow either option A (if you are starting with cultured cells) or option B (if you are starting with fresh or frozen tissue).
-
(A)Cultured cells
-
(i)Trypsinize the cells (2–10 × 105) from the Petri dish using 1 ml of trypsin at 37 °C for 5 min. ! CAUTION The cell lines used in your research should be regularly checked to ensure that they are authentic and not infected with Mycoplasma.
-
(ii)Collect the trypsinized cells and centrifuge the cells at 130g for 5 min at RT.
-
(iii)Aspirate the supernatant and resuspend the cells in 1× PBS to wash them.
-
(iv)Centrifuge the cells at 130g for 5 min at RT and aspirate PBS.
-
(v)Lyse the cells by resuspending them in 1 ml of NST-DAPI buffer (mix by pipetting up and down five times) and incubate them at RT for a minimum of 5 min.
-
(vi)Filter the cells through a nylon-mesh filter into a 5-ml Falcon polystyrene round-bottom tube. Place the tube on ice until flow-sorting at Step 5.
-
(i)
-
(B)Frozen or fresh tissue
-
(i)Cut frozen or fresh tissue and mince it with a scalpel in a 60-cm2 Petri dish with 1 ml of NST-DAPI buffer for 10–15 min until tissue chunks are no longer visible.
-
(ii)Filter the mixture through a nylon-mesh filter into a 5-ml Falcon polystyrene round-bottom tube. Place the tube on ice until flow-sorting at Step 5.
- ▲ CRITICAL STEP Filters may clog during the process, in which case they will need to be replaced with a new filter.
-
(i)
Flow-sorting and genome amplification of single nuclei ● TIMING 5 h
-
2|
Add 150 μl of lysis buffer and 225 μl of sterile PBS to a 1.5-ml tube and vortex to mix the contents. Add 3.5 μl per well of this mixture into a 96-well plate. Cover the plate with adhesive sealing sheet, and place it on a cold block until flow-sorting at Step 8. This mixture provides additional volume to account for pipetting error.
-
3|
Flow-sort 100 beads (fluorescent particles, Spherotech) onto a sealed empty 96-well plate based on forward and side scatter to calibrate the X and Y coordinates for deposits.
▲ CRITICAL STEP This step is important to ensure that cells are accurately deposited into each well.
-
4|
Remove the beads from the cytometer and replace them with a 5-ml Falcon tube of water. Flow the water through the machine for 1 min to wash.
-
5|
Replace water with a 5-ml Falcon tube of nuclei sample (from Step 1). Once the flow-sorting of nuclei has started, set up a plot for DAPI area and DAPI height to gate singlet populations and to exclude doublet nuclei according to Figure 3a.
-
6|
Set up a plot for cell counts and DAPI area. Set gates for isolating single cells from G1/0 or G2/M ploidy distributions according to Figure 3b.
-
7|
(Optional) Flow-sort single nuclei onto a glass slide and mark the underside of the slide. Check the slide under a fluorescent microscope to determine whether single nuclei are being deposited by the flow sorter.
-
8|
Centrifuge the plates from Step 2 (130g, 1 min, RT) to ensure that the lysis buffer solution is in the bottom of the wells. Flow-sort single nuclei into individual wells of this plate (one nucleus per well) using gates demonstrated in Figure 3.
-
9|
Immediately after flow-sorting, seal the plate with adhesive sealing sheet and centrifuge it at 130g for 1 min at RT.
-
10|
Incubate the 96-well plate in a thermocycler at 95 °C for 10 min.
-
11|
Place the plate on a cold block (−20 °C) and pipette 1.5 μl of neutralization buffer into each well. Vortex gently and spin at 130g for 30 s at RT.
-
12|
Pipette 45 μl of amplification buffer (see Reagent Setup) into each well. Vortex gently and spin at 130g for 30 s at RT.
-
13|
Place the plate in a thermocycler at 30 °C for 3 h, followed by 65 °C for 3 min. Hold it at 4 °C. ■ PAUSE POINT Store the plate at −20 °C until use for up to 1 year.
QC for WGA efficiency ● TIMING 1.5 h for four cells
-
14|For each single cell from Step 13, set up 22 individual qPCRs as tabulated below, using the qPCR primer pairs listed in Supplementary Table 3. Set up a qPCR plate for each primer pair and include on each plate a duplicate set of four genomic standards (previously measured by Qubit).
Component Amount per reaction
Final Sample (μl) Standard (μl) Genomic DNA from Step 13 8 - 5 ng Genomic standards (20 or 2 nM, or 0.2 or 0.02 nM) - 1 Water - 7 Forward, 5 nM and reverse primer mix 2 2 0.5 nM KAPA SYBR Fast qPCR master mix, 2× 10 10 Final volume 20 20 -
15|Perform qPCR using the following conditions.
Cycle number Denature Anneal/extension 1 95 °C, 3 min 2-46 95 °C, 20 s 60 °C, 30 s -
16|
Create a standard curve that generates the highest R2 value from the standards (20, 2, 0.2 and 0.02 pM, respectively) performed in duplicate. If the sample is higher than 1 nM, it is considered a positive reaction. Only cells that test positive with 20 or more of the qPCR primer pairs should be retained for use in Step 17.
? TROUBLESHOOTING
Column purification of DNA ● TIMING 1 h
-
17|
Mix amplified DNA from Step 13 with 500 μl of DNA binding buffer (from Zymo kit) in a 1.5-ml Eppendorf tube.
-
18|
Transfer the mixture to a Zymo-spin column. Centrifuge for 1 min at RT at maximum speed (13,000g). Discard the flow-through
-
19|
Add 200 μl of wash buffer to the column. Centrifuge the mixture at maximum speed for 1 min at RT.
-
20|
Repeat the wash (Step 19) one more time for a total of two washes.
-
21|
Discard the collection tube and transfer the column to a clean 1.5-ml tube.
-
22|
Add 30 μl of elution buffer directly to the column membrane. Incubate the mixture at RT for 5 min.
-
23|
Centrifuge the mixture at maximum speed for 2 min at RT.
-
24|
Discard the column and close the tube lid. Quantify purified DNA using Qubit fluorometer according to the manufacturer's instructions.
■ PAUSE POINT Store the samples at −20 °C until use for up to 1 year.
Barcoded library construction: acoustic sonication of WGA DNA ● TIMING 3 min per sample
-
25|
Pipette 500 ng of purified DNA from Step 24 into a Covaris glass tube, and add water to a final volume of 87 μl. Resuspend the DNA thoroughly.
-
26|
Sonicate DNA to 250 bp using the Covaris S220 sonicator; the parameters that we use are outlined in Equipment Setup. See Table 1 for parameters for other fragment sizes.
-
27|
Transfer 85 μl of each sample to a 0.2-ml PCR tube strip.
▲ CRITICAL STEP If there are many samples, users can elect to use a 96-well PCR plate and perform library construction in a plate, instead of in 0.2-ml PCR tube strips.
Barcoded library construction: end repair ● TIMING 45 min
-
28|
To each sample from Step 27, add 10 μl of end repair buffer and 5 μl of end repair enzyme (both are from the NEBNext end repair module). The final volume is 100 μl.
-
29|
Incubate the mixture at 20 °C for 30 min. Hold it at 4 °C for no longer than 1 h until column purification.
Barcoded library construction: column purification of DNA ● TIMING 30 min
-
30|
Add 500 μl of DNA binding buffer to each 100-μl sample from Step 29. The total volume is now 600 μl. Pipette the mixture up and down to mix.
-
31|
Perform column purification as described in Steps 18–21.
-
32|
Add 22 μl of elution buffer directly to the column membrane. Incubate the mixture at RT for 5 min.
-
33|
Centrifuge the mixture at maximum speed (13,000g) for 2 min at RT.
-
34|
Transfer ~21 μl of eluted DNA to a 0.2-ml PCR tube strip.
Barcoded library construction: 3′ adenylation ● TIMING 45 min
-
35|
Place the PCR strips with 21 μl of eluted DNA (from Step 34) on a cold block. Add 2.5 μl of dA-tailing buffer (10×; from the NEBNext dA-tailing module) to the eluted DNA.
-
36|
Add 1.5 μl of Klenow (exo–) enzyme (from NEBNext dA-tailing module) to eluted DNA. The final volume should be 25 μl.
-
37|
Incubate the mixture at 37 °C for 30 min. Hold it at 4 °C for no longer than 1 h until adapter ligation.
Barcoded library construction: adapter ligation ● TIMING 20 min
-
38|Perform adapter ligation by adding the components below.
Component Amount per reaction (μl) Final DNA sample from Step 37 25 Annealed 10 μM barcoded adapters 2 0.4 μM Sterile water 8 Quick T4 DNA ligase (from NEBNext quick ligation module) 5 Quick ligation reaction buffer, 5× (from NEBNext quick ligation module) 10 1× Final volume 50 -
39|
Incubate the mixture at 20 °C for 15 min. Hold it at 4 °C for no longer than 1 h until AMPure bead purification step.
Barcoded library construction: AMPure bead purification ● TIMING 1 h
-
40|
Equilibrate AMPure XP beads to RT for 30 min before use, and then vortex to mix them.
-
41|
Add 50 μl of AMPure beads to the sample from Step 39. Pipette to mix the contents. The total volume should now be 100 μl.
-
42|
Incubate the mixture at RT for 10 min. Pulse-spin it using a microcentrifuge.
-
43|
Place the PCR strip on 96-well magnetic plate for 5 min, and then discard the supernatant by pipetting.
-
44|
Add 200 μl of 80% (vol/vol) ethanol and incubate the mixture at RT for 30 s. Discard the supernatant by pipetting. Repeat this step one more time for a total of two washes.
-
45|
Use a P20 pipette to pipette out and discard the residual supernatant. Air-dry for 10 min on a magnetic plate.
-
46|
Remove the PCR strip from the magnetic plate and add 22 μl of elution buffer; pipette up and down ten times to mix.
-
47|
Incubate the mixture at RT for 5 min.
-
48|
Place it on the magnetic plate for 5 min and then transfer 20 μl of the supernatant to a new 1.5-ml tube. ■ PAUSE POINT Store it at −20 °C until use for no longer than 1 week.
Barcoded library construction library amplification ● TIMING 1 h
-
49|Perform library amplification by adding the components below.
Component Amount per reaction (μl) Final Purified sample from Step 48 20 NEBNext high-fidelity 2× PCR master mix 25 1× Adapter PCR primers (10 μM) 5 1 μM Final volume 50 -
50|
The final volume should now be 50 μl. Gently vortex and pulse-spin the mixture.
-
51|Perform PCR using the following conditions.
Cycle number Denature Anneal Extend Hold 1 98 °C, 30 s 2-9 98 °C, 10 s 65 °C, 30 s 72 °C, 30 s 10 72 °C, 5 min 11 4 °C ▲ CRITICAL STEP During the PCR, equilibrate AMPure XP beads to RT in preparation for Step 52.
■ PAUSE POINT Store at −20 °C until use for no longer than 1 week.
Barcoded library construction: AMPure bead purification ● TIMING 1 h
-
52|
Vortex AMPure beads to mix. Add 50 μl of AMPure beads to the sample from Step 51, and purify as described in Steps 41–45.
-
53|
Remove the tube from the magnetic plate and add 32 μl of elution buffer; pipette the mixture up and down ten times to mix.
-
54|
Incubate the tube at RT for 5 min.
-
55|
Place it on the magnetic plate for 5 min, and then transfer 30 μl of the supernatant to a new tube.
qPCR quantification ● TIMING 2 h
-
56|
Thaw the reagents from the KAPA library quantification kit. Transfer 1 ml of primer premix to the SYBR Green solution. Vortex to mix.
-
57|
For each library, dilute 1 μl of library in 999 μl of 0.05% (vol/vol) Tween 20, creating a 1:1,000 dilution.
-
58|Add the following components onto a MicroAmp optical reaction 96-well plate and seal it using ThermalSeal RT2 film. Gently vortex and pulse-spin the plate. Note that standards should be included in duplicate.
Component Amount per reaction
Sample (μl) Standard (μl) Negative control (μl) Diluted DNA library from Step 55 4 - - Standards (20, 2, 0.2 and 0.02 nM) - 4 - Water 4 4 8 SYBR Green solution with primer premix (from KAPA SYBR Fast qPCR master mix) 12 12 12 Final volume 20 20 20 -
59|Perform qPCR using the following conditions.
Cycle number Denature Anneal/extension 1 95 °C, 5 min 2-36 98 °C, 30 s 60 °C, 45 s -
60|
Create a standard curve that generates the highest R2 value from standards 1–4 (20, 2, 0.2 and 0.02 pM, respectively) performed in duplicate.
-
61|
Use the average of the duplicate sample data points to determine the concentration of the sample within the range of the standards.
? TROUBLESHOOTING
■ PAUSE POINT Store the sample at −20 °C until use, for no longer than 1 month.
Targeted DNA capture: hybridization ● TIMING 3 d
-
62|Add the following components to a new 1.5-ml tube: if 48 samples are pooled, add 50 ng per sample to the tube; if 96 samples are pooled, add 25 ng per sample to the tube. For pooled barcoded P7 adapter, pool only the barcoded P7 adapters that are used in the sample. For example, if only 40 libraries are made using barcoded adapters 1–40, pool only the individual barcoded P7 adapters 1–40, omitting adapters 41–48.
Component Amount per reaction (μl) Final DNA library from Step 55 Variable 1 mg/ml COT human DNA (from Roche NimbleGen kit) 5 5 μg P5 adapter (100 μM) 10 Pooled barcoded P7 adapter (100 μM) 10 Final volume Variable -
63|
Gently vortex and pulse-spin the mixture. Open the tube lid and dry with a SpeedVac on medium-high heat until all of the liquid evaporates.
-
64|
Add 7.5 μl of 2× hybridization buffer and 3 μl of hybridization component A. Pipette up and down to mix.
-
65|
Gently vortex and pulse-spin the mixture. Put the tube on a heat block for 10 min at 95 °C.
-
66|
Pulse-spin and transfer 10.5 μl of the sample to a 0.2-ml PCR tube.
-
67|
Add 4.5 μl of Roche NimbleGen targeted capture to the tube. Flick the tube to mix and pulse-spin it.
-
68|
Incubate the sample in a PCR block at 47 °C for 64–72 h. Set the lid to 57 °C.
Targeted DNA capture: wash ● TIMING 2 h
-
69|
Turn on the water bath to 47 °C.
-
70|
Equilibrate the SeqCapEZ hybridization and wash kit and pure capture bead kit to RT for at least 30 min before use.
-
71|Dilute the following wash buffers.
Buffer Buffer volume (μl) Water volume (ml) Stringent wash, 10× 40 360 Wash buffer 1, 10× 30 270 Wash buffer 2, 10× 20 180 Wash buffer 3, 10× 20 180 Bead wash buffer, 2.5× 200 300 -
72|
Put 400 μl of diluted 10× stringent wash and 100 μl of diluted wash buffer 1 in a preheated 47 °C water bath.
-
73|
Vortex and place 100 μl of SeqCap EZ capture beads in a new 1.5-ml tube.
-
74|
Place it on a six-tube magnetic stand. Remove the supernatant.
-
75|
Add 200 μl of diluted bead wash buffer and vortex it for 10 s. Place the tube on a magnet and remove the supernatant.
-
76|
Repeat Step 75 once more.
-
77|
Repeat Step 75 with 100 μl of bead wash buffer.
-
78|
Keep the beads in a 1.5-ml tube and quickly transfer the hybridization reaction from the PCR block (Step 68) to the beads.
-
79|
Pipette to mix and leave the tube in a 47 °C water bath for 45 min. Vortex the mixture every 15 min.
-
80|
Add 100 μl of heated diluted wash buffer 1, and then pipette to mix.
▲ CRITICAL STEP Steps 81–84 should be performed quickly using the water bath. Place the magnet next to the water bath at RT.
-
81|
Place the tube on the magnet for 5 s. Remove the supernatant.
-
82|
Add 200 μl of heated diluted stringent wash buffer. Pipette to mix, and incubate the mixture in the water bath for 5 min.
-
83|
Place it on the magnet for 5 s. Remove the supernatant.
-
84|
Repeat Steps 82 and 83.
-
85|
Add 200 μl of diluted RT wash buffer 1 and vortex it for 2 min. Pulse-spin the mixture briefly.
-
86|
Place it on the magnet and remove the supernatant.
-
87|
Add 200 μl of diluted RT wash buffer 2 and vortex it for 1 min. Pulse-spin the mixture briefly.
-
88|
Place the tube on the magnet and remove the supernatant.
-
89|
Add 200 μl of diluted RT wash buffer 3 and vortex the mixture for 30 s. Pulse-spin briefly.
-
90|
Place the tube on the magnet and remove the supernatant.
-
91|
Add 50 μl of water to the resuspend beads.
▲ CRITICAL STEP Users do not need to elute samples from beads before PCR (Step 92).
Targeted DNA capture: PCR ● TIMING 2 h
-
92|Add the following components to perform PCR for captured libraries.
Component Amount per reaction (μl) Final Captured sample with beads from Step 91 50 Adapter PCR primers, 20 μM 6 0.6 μM Sterile water 44 KAPA 2× HiFi HotStart mix 100 1× Final volume 200 ▲ CRITICAL STEP Adapter PCR primer concentration is different from the adapter PCR primer concentration used during library construction at Step 49.
-
93|
Split the mixture into two reactions in 0.2-ml PCR tubes with 100 μl each.
-
94|Perform PCR on the reactions from Step 93 using the following program.
Cycle number Denature Anneal Extend Hold 1 98 °C, 45 s 2-13 98 °C, 15 s 60 °C, 30 s 72 °C, 30 s 14 72 °C, 1 min 15 4 °C ▲ CRITICAL STEP During PCR, equilibrate AMPure beads to RT in preparation for Step 95.
Targeted DNA capture: AMPure bead purification ● TIMING 1 h
-
95|
Combine two PCRs from Step 94 into a 1.5-ml tube.
-
96|
Add 360 μl of AMPure beads to the tube and incubate the mixture for 15 min at RT.
-
97|
Place it on a six-tube magnet stand for 3 min and remove the supernatant.
-
98|
Add 600 μl of 80% (vol/vol) EtOH to wash the beads. Incubate the beads for 30 s. Remove and discard the supernatant. Repeat this wash step once more.
-
99|
Use a P20 pipette to remove the remaining supernatant, and allow the beads to air-dry on the magnet for 10 min
-
100|
Resuspend the beads in 32 μl of water. Incubate the mixture at RT for 5 min.
-
101|
Place the tube on the magnet for 5 min and transfer 30 μl of the supernatant to a new 1.5-ml tube.
■ PAUSE POINT Store it at −20 °C until use for no longer than 1 year.
qPCR quantification ● TIMING 2 h
-
102|
Perform library quantification as described in Steps 57–60.
-
103|
Use the average of the duplicate sample data points to determine the concentration of the sample within the range of the standards.
NGS ● TIMING 1 week for HiSeq 3000 or 2 weeks for HiSeq 2000
-
104|
Sequence captured libraries on the Illumina HiSeq platform using 100 paired-end cycles or desired read length.
Bioinformatics processing: alignment and generation of BAM files ● TIMING 4 h with a 3.7-GHz CPU, 8 GB RAM
-
105|To demultiplex the FASTQ file containing all of the sequence reads into individual files for each barcode, first create a text file called barcodes.txt with all the barcodes that were used in the experiments (one barcode per line). An example for 6 barcodes is as follows:
GCCAAGAC GATGAATC GAGTTAGC GACAGTGC GAACAGGC CTAAGGTC
-
106|Transfer the FASTQ file into the same directory as the barcode identifier text file. Run the demultiplexer on the FASTQ file using the deplexer.pl script (Supplementary Software).
$ perl /file_path/deplexer.pl barcodes.txt
-
107|Use Bowtie 2 to align the individual FASTQ files of each cell or sample to the human genome. For each .fastq file, ‘SN’ is the name of the sample; ‘GCTACGC’ is the barcode; ‘L006’ is the lane number of HiSeq 2000 platform; ‘R1’ and ‘R2’ means read 1 or read 2, respectively; and ‘001’ means fastq file 001 for the sample.
$ /file_path/bowtie2 -x /file_path/hg19 -1 /file_path/SN_GCTACGC_L006_R1_001.fastq.gz -2 /file_path/SN_GCTACGC_L006_R2_001.fastq.gz -S /file_path/SN_GCTACGC_L006_R1_001.sam --local --rg-id SN --rg SM:SN --rg PU:SN --rg PL:ILLUMINA --rg LB:SN -p 2
-
108|Convert all SAM files generated at Step 107 to compressed binary BAM files.
$ /file_path/samtools view -bS /file_path/SN_GCTACGC_L006_R1_001.sam -o /file_path/SN_GCTACGC_L006_R1_001.bam
-
109|Sort all the BAM files obtained from Step 108 according to genomic coordinates.
$ /file_path/samtools sort -m 7600000000 /file_path/SN_GCTACGC_L006_R1_001.bam /file_path/SN_GCTACGC_L006_R1_001.sorted
-
110|For samples with multiple BAM files only: merge all BAM files from one cell into a merged BAM file. For samples with multiple sequence libraries, the BAM files of these samples need to be merged with Picard tools because SAMtools does not integrate the RG tag.
$ /file_path/samtools merge /file_path/SN.sorted.bam /file_path/SN_GCTACGC_L006_R1_001.bam /file_path/SN_GCTACGC_L006_R1_002.bam
...
For a sample with multiple sequence libraries, use the following:
$ java -Xmx8g /file_path/MergeSamFiles.jar I=/file_path/ SN_GCTACGC_L006_R1_001.bam I=/file_path/SN_GCTACGC_L006_R1_002.bam O=/file_path/SN.sorted.bam TMP_DIR=/file_path/tmp VALIDATION_STRINGENCY=SILENT AS=true SO=coordinate
-
111|Mark duplicate reads using the Picard tool:
$ java -Xmx8g -jar /file_path/MarkDuplicates.jar I=/file_path/SN.sorted.bam O=/file_path/SN.marked.sorted.bam M=/file_path/SN.marked.sorted.bam.log TMP_DIR=/file_path/tmp VALIDATION_STRINGENCY=SILENT AS=true REMOVE_DUPLICATES=false VERBOSITY=INFO MAX_RECORDS_IN_RAM=10000000
-
112|Index the marked BAM file with SAMtools:
$ samtools index SN.marked.sorted.bam
-
113|
Run indel re-alignment with GATK. Genomic regions with indels have many false-positive errors because of poor alignment with Bowtie. Therefore, a more accurate alignment algorithm is used to re-align sequencing reads in genomic regions that contain potential indels with GATK.
First generate the file of intervals in which potential indels occur:
$ java -Djava.io.tmpdir=/file_path/tmp -Xmx12g -jar /file_path/GenomeAnalysisTK.jar -T RealignerTargetCreator -I /file_path/SN.marked.sorted.bam -o /file_path/SN.marked.sorted.bam.intervals –log /file_path/SN.marked.sorted.bam.intervals.log -R hg19_all.fa -S SILENT -nt 2 -dt BY_SAMPLE -dcov 2500 -l INFO –filterMBQ
Re-align regions within the intervals:
$ java -Djava.io.tmpdir=/file_path/tmp -Xmx12g -jar /file_path/GenomeAnalysisTK.jar -T IndelRealigner -I /file_path/SN.marked.sorted.bam -o /file_path/SN.marked.sorted.re_aln.bam -log /file_path/SN.marked.sorted.re_aln.bam.log -targetIntervals /file_path/SN.marked.sorted.bam.intervals -R /file_path/hg19_all.fa -S SILENT -dt BY_SAMPLE -dcov 2500 -l INFO –filterMBQ
-
114|Filter reads in BAM files with poor mapping quality (MQ < 40):
$ /file_path/samtools view -q 40 -bh /file_path/SN.marked.sorted.re_aln.bam > /file_path/SN.marked.sorted.re_aln.q40.bam
▲ CRITICAL STEP The output BAM file generated at this step is the final BAM file for each cell. All intermediate SAM files and BAM files can be deleted to free up storage space.
Bioinformatics processing: calculating basic data quality metrics ● TIMING 30 min
-
115|Calculate the total number of reads, mapping rates and duplicate rates using SAMtools:
$ /file_path/samtools flagstat /file_path/SN.marked.sorted.re_aln.bam >& /file_path/SN.marked.sorted.re_aln.bam.flag
-
116|Calculate the coverage breadth and depth using SAMtools, BEDTools and the provided Perl script (Supplementary Software). Users need to replace the bed file used in the following example with their own exome capture or target capture bed file.
$ /file_path/samtools view -uF 0x400 -q 1 /file_path/SN.marked.sorted.re_aln.bam | /file_path/BEDTools-Version-2.14.3/bin/coverageBed -abam - -b /file_path/ TargetedRegions_hg19_clean.bed -d >& /file_path/SN.marked.sorted.re_aln.bam.coverage $ perl cal_coverage_metrics.pl SN.marked.sorted.re_aln.bam.coverage SN.marked.sorted.re_aln.bam.coverage.cal
? TROUBLESHOOTING
Bioinformatics processing: variant detection ● TIMING 30 min
-
117|Run GATK to create VCF4 files from the BAM files from Step 114.
$ java -Djava.awt.headless=true -Djava.io.tmpdir=/file_path/tmp -Xmx48g -jar /file_path/GenomeAnalysisTK.jar -T UnifiedGenotyper -glm BOTH --dbsnp /file_path/dbsnp_137.hg19.excluding_sites_after_129.vcf -R /file_path/hg19_all.fa -I /file_path/SN1.marked.sorted.re_aln.q40.bam –I /file_path/SN2.marked.sorted.re_aln.q40.bam -S SILENT -nt 10 -dt BY_SAMPLE -dcov 2500 -l INFO -mbq 20 -rf BadCigar -o /file_path/SN_83.vcf -log /file_path/SN_83.vcf.log -L chr12:1-25000000
All VCF files for each sample are then pooled together into a multisample VCF4 file with the following command:
$ java -Djava.awt.headless=true -Djava.io.tmpdir=/file_path/tmp -Xmx8g -jar /file_path/GenomeAnalysisTK.jar -T CombineVariants -o /file_path/SN.vcf -R file_path/hg19_all.fa –V /file_path/SN_0.vcf –V /file_path/SN_1.vcf ... -V /file_path/SN_204.vcf
-
118|Run GATK to evaluate the quality score of each variant. The following command lists all databases used for this step.
$ java -Xmx32g -jar /file_path/GenomeAnalysisTK.jar -T VariantRecalibrator -input /file_path/SN.vcf -tranchesFile /file_path/SN.vcf.tranchesFile -recalFile /file_path/SN.vcf.recal -rscriptFile /file_path/SN.vcf.R -mode BOTH -R /file_path/hg19_all.fa - resource:hapmap,VCF,known=false,training=true,truth=true,prior=15.0 /file_path/hapmap_3.3.hg19.sites.vcf - resource:omni,VCF,known=false,training=true,truth=false,prior=12.0 /file_path/1000G_omni2.5.hg19.sites.vcf - resource:dbsnp,VCF,known=true,training=false,truth=false,prior=8.0 /file_path/dbsnp_137.hg19.excluding_sites_after_129.vcf -resource:mills,VCF,known=true,training=true,truth=true,prior=12.0 /file_path/Mills_and_1000G_gold_standard.indels.hg19.vcf -an QD -an HaplotypeScore -an MQRankSum -an ReadPosRankSum -an MQ -an DP -an FS -U ALLOW_SEQ_DICT_INCOMPATIBILITY $ java -Xmx32g -jar /file_path/GenomeAnalysisTK.jar -T ApplyRecalibration- input /file_path/SN.vcf -recalFile /file_path/SN.vcf.recal -tranchesFile /file_path/SN.vcf.tranchesFile -o /file_path/SN.filtered.vcf -mode BOTH -R /file_path/hg19_all.fa
-
119|Select and save SNVs and indels into separate VCF4 files
$ java -jar /file_path/GenomeAnalysisTK.jar -T SelectVariants -V /file_path/SN.filtered.vcf -o /file_path/SN.filtered.SNP.vcf -selectType SNP –R file_path/hg19_all.fa $ java -jar /file_path/GenomeAnalysisTK.jar -T SelectVariants -V /file_path/SN.filtered.vcf -o /file_path/SN.filtered.INDEL.vcf -selectType INDEL –R hg19_all.fa
-
120|Use filter_clustered_mutations.pl (Supplementary Software) to filter out mutations that occurred more than once within a 10-bp window.
$ /file_path/perl /file_path/filter_clustered_mutations.pl /file_path/SN.filtered.INDEL.vcf /file_path/SN.filtered.SNP.vcf
▲ CRITICAL STEP This window size can be adjusted for cancers in which very high mutation frequencies are expected, or skipped altogether if too many variants are being filtered at this step.
-
121|Filter out detected mutations on the basis of allelic frequency, and add the passcode string to the end of the VCF4 lines.
$ /file path/perl /file_path/add_passcode_2_filtered_vcf.10andVaried_6and2forD.IPCT.pl /file_path/SN.filtered.cluster_filetered.SNP.vcf /file_path/SN.filtered.cluster_filetered.passcode.SNP.vcf
Use the ‘grep’ command and Perl one-liners to extract interested mutations. For example, assuming that the last sample is the normal sample, the command to grab all somatic mutations that are detected in more than one cell is as follows:
$ grep “<E.....0>” file_path/SN.filtered.cluster_filetered.passcode.SNP.vcf |
perl -e
‘chomp; @b = split(/ \ t/ ); @a = split(//, $b[−1]); $num_cell=0;$size=@a;for($i=2; $i<$size;$i++){if($a[$i]
eq “1” || $a[$i] eq “2” ){$num_cell++;}} if($num_cell > 1){print STDERR
“$_\n”;’
Bioinformatics processing: variant annotation ● TIMING 30 min
-
122|Our annotation pipeline uses ANNOVAR, BEDTools and several Perl scripts to integrate data from several biological databases (Fig. 2). To read a .vcf file as input and to run the analysis described in Steps 122–129 automatically, run the following command line:
$ /file_path/run_annovar.pl /file_path/SN.filtered.cluster_filtered.passcode.SNP.vcf
Alternatively, run commands separately for each database, as described in Steps 122–129. First, convert variant VCF4 file to ANNOVAR format:
$ /file_path/annovar_hg19/convert2annovar.pl --format vcf4old /file_path/SN.filtered.cluster_filetered.passcode.SNP.vcf -outfile /file_path/tmp.avinput
-
123|Annotate with human genes.
$ /file_path/annovar_hg19/annotate_variation.pl -geneanno -buildver hg19 /file_path/tmp.avinput /file_path/annovar_hg19/humandb/
-
124|
Determine whether a mutation intersects with a known cancer gene. We use the Cancer Gene Census gene lists and BEDTools to determine whether the mutation occurs in a known cancer gene.
Create a mutation bed file for BEDTools.
$ cut -f1-3,5 /file_path/tmp.avinput >& /file_path/tmp.avinput.bed
Intersect the mutations with cancer gene coordinates
$ /file_path/BEDTools-Version-2.14.3/bin/intersectBED -a file_path/tmp.avinput.bed -b /file_path/CancerGenes/CancerGenes.bed -wa -wb >& file_path/tmp.avinput.gene6
-
125|Annotate with COSMIC database. We downloaded the latest version of the COSMIC database with ANNOVAR.
$ /file_path/annovar_hg19/annotate_variation.pl -filter -dbtype cosmic70 -buildver hg19 /file_path/tmp.avinput /file_path/annovar_hg19/humandb/
-
126|Annotate with PolyPhen score database.
$ /file_path/annovar_hg19/annotate_variation.pl -filter -dbtype ljb26_pp2hdiv -buildver hg19 /file_path/tmp.avinput /file_path/annovar_hg19/humandb/
-
127|Annotate with SIFT score database.
$ /file_path/annovar_hg19/annotate_variation.pl -filter -dbtype ljb26_sift -buildver hg19 /file_path/tmp.avinput /file_path/annovar_hg19/humandb/
-
128|Annotate with Mutation Taster database.
$ /file_path/annovar_hg19/annotate_variation.pl -filter -dbtype ljb26_mt buildver hg19 /file_path/tmp.avinput /file_path/annovar_hg19/humandb/
-
129|Annotate with ClinVar database.
$ /file_path/annovar_hg19/annotate_variation.pl -filter -dbtype clinvar_20140929 -buildver hg19 /file_path/tmp.avinput /file_path/annovar_hg19/humandb/
■ CRITICAL STEP All the annotation results from Steps 122–129 are combined into one text file using the run_anovar.pl script. For SNPs, the script then extracts the results for synonymous, nonsynonymous, intronic and non_coding RNA mutations and saves them into separate files. For indels, the script extracts frameshift and nonframeshift mutations and saves them to a separate file.
-
130|
Add passcode and allele frequencies to the final annotation file. We developed a Perl script (Supplementary Software) that finds passcodes for each annotated mutation from the input VCF4 file, and calculates allele frequencies for all samples; then, it adds this information to the first two columns of the annotation results file. The number that is input at the end of the following command indicates the number of the columns where chromosome information for each mutation can be found.
For nonsynonymous SNPs:
$ /file_path/combine_passcode.pl /file_path/SN.filtered.cluster_filetered.passcode.SNP.vcf.nonsynonymous /file_path/SN.filtered.cluster_filetered.passcode.SNP.vcf 10
For frameshift indels:
$ /file_path/combine_passcode_INDEL.pl /file_path/SN.filtered.cluster_filetered.passcode.INDEL.vcf.frame /file_path/SN.filtered.cluster_filetered.passcode.INDEL.vcf 10
The format of the allele frequency information is similar to the passcode format. For example, ‘<0.73,0.24,0.19,0.53, 0.67,0.53>’ means that there are six samples and their frequencies are 0.73, 0.24 and so on.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 16 | Low WGA concentration | Reagents and lysis buffers have undergone too many freeze-thaw cycles, or random hexamer oligonucleotides have degraded | Make aliquots of random hexamer oligos to avoid multiple freeze-thaw cycles. Ensure that lysis buffer is freshly made each time. Ensure that DTT has not been frozen and thawed multiple times |
| Thermocycler malfunctioning, leading to inaccurate incubation temperature | Check and calibrate the temperature of the thermocycler | ||
| No positive PCR amplicons are evident at the WGA quality control step | Bacterial contamination in samples or WGA reagents | Ensure that all reagents are sterilized. Perform all work under a pre-PCR workstation. Bleach all surfaces and use gloves. UV-irradiate equipment and reagents for 1 h | |
| No nuclei were deposited into the wells of the 96-well plate | Check to see whether the flow-sorting machine is depositing single nuclei accurately. See Step 7. Check the droplet under a fluorescence microscope for single DAPI-stained nuclei after each deposit | ||
| Nuclei are not deposited at the bottom of the well and do not lyse | It is possible that the nucleus is not deposited at the exact center of the well, but instead on the wall of the well. To avoid this problem, it is crucial that the 96-well plate be centrifuged after flow-sorting, to ensure that the nucleus submerges fully into the lysis buffer | ||
| 61 | Low library concentration after library preparation | Insufficient washing with AMPure beads | Be sure to incubate for at least 10 min at RT after adding the library sample to the AMPure beads. This allows sufficient time for DNA to bind to the beads. Also ensure that the beads are completely clustered at the magnetic side of the tube before pipetting out the supernatant |
| 116 | Reads mapped outside of the targeted region | The library insert size is too large for the targeted capture probes (100 bp) to hybridize efficiently | Ensure that the DNA is fragmented to a mean size of 250 bp. Before library construction, run DNA fragments on the Bioanalyzer system to determine fragment size distributions |
● TIMING
Steps 1–13, preparation, flow-sorting and WGA of single nuclei: 5.5 h
Steps 14–16, QC for WGA efficiency: 1.5 h for four cells
Steps 17–24, column purification and quantification of DNA: 1 h
Steps 25–61, barcoded library construction and qPCR quantification: 1–2 d
Steps 62–103, targeted DNA capture and quantification: ~3 d
Step 104, NGS: 1–2 weeks
Steps 105–130, bioinformatics processing: 4 h per cell using a 3.7-GHz CPU and 8 GB RAM
ANTICIPATED RESULTS
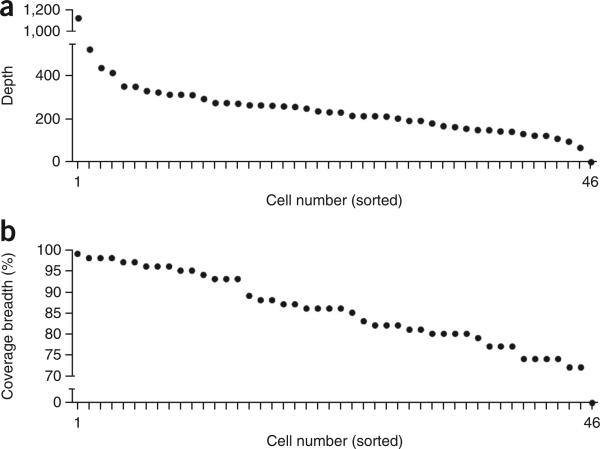
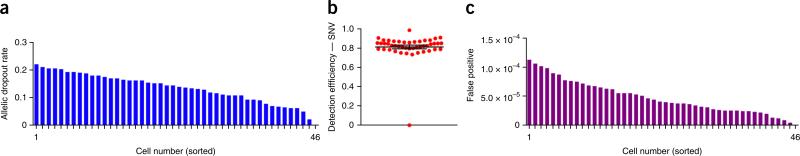
This protocol is expected to generate high-coverage (>90%) single-cell data at targeted regions of the genome that can be used to detect point mutations and indels at base-pair resolution. To establish technical error rates and metrics for this protocol, we applied our method to an isogenic breast cancer cell line (MDA-MB-231) to sequence 46 single cells and two matched bulk populations. We constructed 48 barcoded libraries and pooled together 46 single-cell libraries and two matched population samples into a single reaction for targeted capture using the 1-Mb IPCT panel of 200 cancer-associated genes (Supplementary Table 4). The pooled libraries were sequenced on a single lane on a HiSeq 2000 system (Illumina) at 100-bp paired-end cycles. Our samples showed an average coverage depth of 255× (s.e.m. = 23.54) and coverage breadth of 85% (s.e.m. = 0.02%; Fig. 6). Uneven pooling can lead to occasional samples with low coverage, which should be removed from downstream analysis (e.g., cell number 46; Fig. 6a,b). The average on-target performance for this SCS data set in the capture regions was determined to be 65.03%. Using the variants detected in the isogenic population samples, we calculated the technical error rates for each single cell at sites at which both samples had sufficient coverage depth. The mean ADR for the single-cell data was 13.68% (s.e.m. = 0.79%; Fig. 7a). Next, we calculated the detection efficiency of SNVs in regions in which sufficient coverage (>10×) was found in both the population and single-cell data sets. Our analysis identified an SNV detection efficiency of 82.80% (s.e.m. = 1.9; Fig. 7b). We calculated the mean false positive error rate to be 4.98e−5 (s.e.m. = 4.175e−6; Fig. 7c). This error rate is drastically reduced (squared) by calling mutations concurrently in two or more single cells. Collectively, these data show that high-coverage breadth data and high detection efficiencies can be obtained using the protocol to perform highly multiplexed single-cell targeted DNA sequencing.
Figure 6.
Coverage depth and breadth for 46 multiplexed single cells. (a,b) Coverage depth (a) and coverage breadth (b) were calculated for each single nucleus from a sequencing experiment in which 46 barcoded cells were pooled and sequenced on a single lane for NGS. Single cells were isolated from the MDA-MB231 breast cancer cell line.
Figure 7.
Technical error rate metrics for 46 multiplexed single cells. Technical error rates were calculated for 46 single nuclei sequenced from the MDA-MB-231 breast cancer cell line. (a–c) The allelic dropout rates (a), the SNV detection efficiency for single cells compared with the bulk population (b) and false-positive error rates for each single cell (c).
Supplementary Material
Box 1 | Generation of barcoded sequencing adapters.
Illumina sequencing adapters are designed with 8-bp barcodes to allow multiplexing of single-cell libraries. The adapters consist of two semicomplimentary sequences labeled as P5 and P7. The P7 adapter contains a unique 8-bp barcode, whereas the P5 adapter contains a universal adapter sequence. The single-stranded oligonucleotides must be hybridized together to form a double-stranded sequence that can be used for adapter ligation to the single-cell DNA.
To create barcoded adapters, users must first order the following sequences.
P5 adapter: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC*T
Barcoded P7 adapter: #GATCGGAAGAGCACACGTCTGAACTCCAGTCACGCCAAGACATCTCGTATGCCGTCTTCTGCTTG
The P5 sequence contains a 3′ phosphorothioate linkage between the last two nucleotides (indicated by *). P7 sequence contains a 5′ phosphate group (indicated by #) and an 8-bp barcode sequence (underscored). In order to multiplex 48–96 samples, individual P7 adapters with unique barcodes need to be synthesized (Supplementary Table 2). The user will need to prepare a series of double-stranded adapters by hybridizing unique P7 and universal P5 adapters for each of the 48–96 barcodes.
To generate working barcoded adapters for library construction, P5 adapter and barcoded P7 adapter will need to be annealed together. First prepare a 10 nM mixture of both the P5 and P7 adapters in Tris-HCl. Place the mixture in thermocycler at 95 °C for 5 min, followed by a 1 °C decrease every 2 min, until the final temperature ramps down to 25 °C. Annealed barcoded adapters can be aliquotted in multiple tubes for storage at −20 °C for up to 1 year.
ACKNOWLEDGMENTS
N.E.N. is a Nadia's Gift Foundation Damon Runyon-Rachleff Innovator (DRR-25-13), and also is a T.C. Hsu Endowed Scholar. This work was supported by a gift from the Eric & Liz Lefkofsky Family Foundation.
The study was supported by grants to N.E.N. from the National Cancer Institute (NCI; no. 1RO1CA169244-01), the National Institutes of Health (NIH; no. R21CA174397-01) and an Agilent University Relations Grant. This work was supported by the MD Anderson Cancer Moonshot Knowledge Gap Award, Center for Genetics & Genomics and Center for Epigenetics. M.L.L. is supported by a Research Training Award from the Cancer Prevention and Research Institute of Texas (CPRIT RP140106), and is also supported by the American Legion Auxiliary (ALA) and Hearst Foundations. This work was also supported by the MD Anderson Sequencing Core Facility Grant (no. CA016672) and the Flow Cytometry Facility grant from NIH (no. CA016672). C.K. is supported by the NIH National Center for Advancing Translational Sciences (TL1TR000369 and UL1TR000371) and the ALA. This work was supported by a CPRIT research training award to J.J. (RP101502). We thank F. Meric-Bernstam and K. Eterovic for their support with the cancer gene targeted capture panels. We also thank L. Ramagli, K. Khanna, E. Thompson and H. Tang at the MD Anderson Sequencing Core Facility for supporting the sequencing experiments. We are also grateful to W. Schober and N. Patel at the MD Anderson Flow Core Facility for their support.
Footnotes
AUTHOR CONTRIBUTIONS M.L.L. performed experiments, performed data analysis, prepared figures and wrote the manuscript. Y.W. and N.E.N. performed data analysis and wrote the manuscript. C.K., J.J and E.S. performed experiments. R.G. wrote the software. E.S. performed experiments.
Accession codes. The data from this project has been deposited in the Sequence Read Archive (SRP058890).
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu W, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Navin N, Hicks J. Future medical applications of single-cell sequencing in cancer. Genome Med. 2011;3:31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marusyk A, Almendro V, Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 10.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc. Natl. Acad. Sci. USA. 2014;111:17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu C, et al. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 2014;24:701–712. doi: 10.1038/cr.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni X, et al. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc. Natl. Acad. Sci. USA. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, et al. Single-cell sequencing analysis characterizes common and cell-lineage-specific mutations in a muscle-invasive bladder cancer. Gigascience. 2012;1:12. doi: 10.1186/2047-217X-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Navin NE. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15:452. doi: 10.1186/s13059-014-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Loo P, Voet T. Single cell analysis of cancer genomes. Curr. Opin. Genet. Dev. 2014;24:82–91. doi: 10.1016/j.gde.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Saliba AE, Westermann AJ, Gorski SA, Vogel J. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42:8845–8860. doi: 10.1093/nar/gku555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandberg R. Entering the era of single-cell transcriptomics in biology and medicine. Nat. Methods. 2014;11:22–24. doi: 10.1038/nmeth.2764. [DOI] [PubMed] [Google Scholar]
- 21.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baslan T, et al. Genome-wide copy number analysis of single cells. Nat. Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung ML, Wang Y, Waters J, Navin NE. SNES: single-nucleus exome sequencing. Genome Biol. 2015;16:55. doi: 10.1186/s13059-015-0616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navin NE. Delineating cancer evolution with single-cell sequencing. Sci. Transl. Med. 2015;7:296fs229. doi: 10.1126/scitranslmed.aac8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen K, et al. Clinical actionability enhanced through deep targeted sequencing of solid tumors. Clin. Chem. 2015;61:544–553. doi: 10.1373/clinchem.2014.231100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.