Abstract
Cytochrome P450 (CYP) enzymes for which there is no functional information are considered “orphan” CYPs. Previous studies showed that CYP20A1, an orphan, is expressed in human hippocampus and substantia nigra, and in zebrafish (Danio rerio) CYP20A1 maternal transcript occurs in eggs, suggesting involvement in brain and in early development. Moreover, hyperactivity is reported in humans with chromosome 2 microdeletions including CYP20A1. We examined CYP20A1 in zebrafish, including impacts of chemical exposure on expression. Zebrafish CYP20A1 cDNA was cloned, sequenced, and aligned with cloned human CYP20A1 and predicted vertebrate orthologs. CYP20A1s share a highly conserved N-terminal region and unusual sequences in the I-helix and the heme-binding CYP signature motifs. CYP20A1 mRNA expression was observed in adult zebrafish organs including liver, heart, gonads, spleen and brain, as well as eye and optic nerve. Putative binding sites in proximal promoter regions of CYP20A1s, and response of zebrafish CYP20A1 to selected nuclear and xenobiotic receptor agonists, point to up-regulation by agents involved in steroid hormone response, cholesterol and lipid metabolism. There also was a dose-dependent reduction of CYP20A1 expression in embryos exposed to environmentally relevant levels of methylmercury. Morpholino knockdown of CYP20A1 in developing zebrafish resulted in behavioral effects, including hyperactivity and a slowing of the optomotor response in larvae. The results suggest that altered expression of CYP20A1 might be part of a mechanism linking methylmercury exposure to neurobehavioral deficits. The expanded information on CYP20A1 brings us closer to “deorphanization”, that is, identifying CYP20A1 functions and its roles in health and disease.
Keywords: Cytochrome P450 20A1, behavioral disorders, methylmercury, zebrafish
Introduction
In animals, enzymes of the cytochrome P450 (CYP) superfamily (Nelson, 2009) catalyze oxidation reactions as well as reductions and rearrangements with a vast array of endogenous and exogenous compounds, often with high regio- and stereo-selectivity (Lamb and Waterman, 2013). These activities serve a wide range of physiological and toxicological functions. In vertebrate genomes, the number of protein-coding CYP genes range from around 40 to more than 100, in 19 gene families (Nelson et al., 2013). While functions are known for many human CYPs, the physiological substrate(s) and function of a substantial number of vertebrate CYPs remain unknown. In humans, these so-called “orphan P450s” notably include CYP4F22, CYP4V2 and CYP20A1 (Stark and Guengerich, 2007). In the model species zebrafish (Danio rerio), there are few CYPs for which function has been shown. Functions can be inferred for many CYPs in gene families 5-51, based on activities of known mammalian orthologs, yet predicted function has been confirmed in few cases (e.g., the lanosterol 14α-demethylase CYP51) (Morrison et al., 2014). In gene families 1-3, there has been much lineage specific expansion, resulting in co-orthologs, most with unknown function (Kubota et al., 2013). Thus, many zebrafish CYPs still are properly considered orphans, including CYP20A1.
The sole member of the CYP20 family, CYP20A1, is found in a single copy in human, zebrafish and other vertebrate genomes sequenced to date. The catalytic function of CYP20A1 is unknown. Recombinant human CYP20A1 has been tested for activity (Stark et al., 2008), however, no oxidation reaction was detected with several endogenous compounds (steroids, fatty acids, neurotransmitters) or exogenous chemicals (i.e., terfenadine, clotrimazole) that are substrates for some other CYPs. Although limited information is available on this orphan P450, it was found to be relatively highly expressed in human hippocampus and substantia nigra (Stark et al., 2008). These two brain regions are associated with learning and memory, and are involved in neurodegenerative diseases (Wirdefeldt et al., 2011; Zhou et al., 2008). High levels of CYP20A1 transcript occur also in unfertilized eggs (Goldstone et al., 2010) and in notochord (Thisse and Thisse, 2004) of developing zebrafish, and during embryonic development of mouse (Choudhary et al., 2003).
An important observation derives from conditions associated with a microdeletion on chromosome 2 in humans. Patients diagnosed with microdeletions in the 2q33 chromosome region where CYP20A1 is located variously show psychomotor retardation, hyperactivity and bouts of anxiety, among other conditions (Balasubramanian et al., 2011; Tomaszewska et al., 2013). The observations together suggest participation of CYP20A1 early in vertebrate development, and possible involvement in brain functions and behavior. However, these possibilities have not been examined experimentally.
In the present study, we employed zebrafish to examine features of CYP20A1 that could bear on its functions. Zebrafish CYP20A1 was cloned and sequenced, and the inferred primary structure was compared to that of the cloned human enzyme, and to CYP20A1 coding sequences found in vertebrate genomes. We determined the organ distribution of transcript in adult zebrafish, and analyzed the transcriptional responses to agonists for several nuclear receptors and the aryl hydrocarbon receptor (AHR) that are prominent in regulating expression of a number of CYP genes in vertebrates (Honkakoski and Negishi, 2000). The latter analysis was complemented by a search for putative binding sites recurring in proximal promoters of vertebrate CYP20A1 genes. We also conducted studies of CYP20A1 mRNA expression in zebrafish embryos with the potent environmental toxicant methylmercury (MeHg). Finally, early-developing zebrafish were screened for morphological and behavioral effects resulting from knockdown of CYP20A1 expression using morpholine-substituted oligonucleotides. The results together provide new information and perspective on the regulation and function of CYP20A1, including possible involvement in neurobehavioral disorders, and the effects of chemicals linked to such disorders.
Material and Methods
Animals
Experiments were conducted on early-developing and sexually mature zebrafish (Danio rerio) of the Tupfel Long-fin (TL), Tropical 5D and AB strains, and were approved by the Animal Care and Use Committee of the Woods Hole Oceanographic Institution, Oregon State University and Universite Catholique de Louvain, respectively. The associated Animal Welfare Assurance Numbers are respectively A3630-01, A3229-01 and 1458701. Fish were euthanized by immersion in a bath of tricaine methanesulfonate. Fish maintenance and breeding procedures were previously described (Jonsson et al., 2007a; Jonsson et al., 2007b). cDNA Cloning and Sequencing
Primers were designed in the proximal 5’ and 3’ untranslated regions of zebrafish CYP20A1 transcript [GenBank: NM_213332.1]. Primer sequences were 5’-CTGATGGTCATTGTAGACG-3’ (F) and 5’-TCATGGATGTTGGAGTGG-3’ (R). We used the Advantage 2 PCR kit from Clontech (Mountain View, USA) to amplify the coding sequence from TL zebrafish liver cDNA with 1 μM of each primer; the thermal profile was: 94°C for 1 min, [94°C for 30 sec, 58°C for 3 min] for 35 cycles, and 68°C for 7 min. PCR products were purified with the QIAquick Gel Extraction Kit from Qiagen (Valencia, USA) and cloned into pGEM-T Easy from Promega (Madison, USA) as per kit instructions. Mach-1 competent cells from Invitrogen (Carlsbad, USA) were transformed, and, following overnight incubation of selected ampicillin-resistant white colonies in liquid Luria-Bertani medium with antibiotic, plasmids were isolated with the QIAprep Spin Miniprep Kit from Qiagen (Valencia, USA). Three clones were sent to Eurofins MWG Operon (Louisville, USA) for forward and reverse sequencing with T7 primers (1 μg of plasmid DNA), and the full-length sequence was then assembled and aligned with [GenBank: NM_213332.1] using MacVector 12.0.2 (Oxford Molecular Group, Madison, USA). Secondary structure prediction
Amino acid sequences of CYP20A1 proteins were retrieved from the Ensembl database (Flicek et al., 2013) and aligned with MacVector 12.0.2 software (Oxford Molecular Group, USA) (see Supplemental figure S1 for sequence identifiers). Note that the protein sequence inferred from the TL zebrafish transcript we cloned was fully identical to that predicted from the genome [Ensembl: ENSDARP00000003222]. The specific motifs of CYP20A1 secondary structure (i.e. α-helices, β-strands, transmembrane domain, signal peptide) were predicted with two different online softwares, and overlapping predictions were chosen to construct the consensual secondary structure of the orphan P450. The α-helices were predicted with NetSurfP 1.1 (Petersen et al., 2009) and Psipred 3.0 (Bryson et al., 2005) at a cut-off of 80%. Putative β strands were localized with PredictProtein (Rost et al., 2004) and BETApro (Cheng and Baldi, 2005) under default settings. Hypothetical transmembrane domains were identified with PredictProtein (Rost et al., 2004) and TMHMM 2.0 (Krogh et al., 2001) at a cut-off of 80%. The possible presence of a signal peptide in the N-terminal portion of CYP20A1 was investigated with pSORTII (Nakai and Horton, 1999) and SignalP 4.1 (Petersen et al., 2011) under default settings. The locations of the P450 signature motifs (i.e., I helix, K helix and heme-binding region) were analyzed based on the information available in reviews (Sezutsu et al., 2013; Werck-Reichhart and Feyereisen, 2000). The motifs specific to zebrafish and human CYP20A1 protein also were compared to those of all other zebrafish and human protein-coding CYPs available in the Ensembl database (data not shown). A subset of the zebrafish CYPs was used as basis for the comparison of CYP20A1 signature motifs shown in Table 1. The sequence identifiers for the selected non-CYP20 isoforms are shown in Supplemental table S6. CYP20A1 heterologous protein expression
Table 1.
P450 signature motifs in CYP20A1 and other selected CYPs in zebrafish.
| P450 isoform | I helix motif [G/AGXE/DTX] | K helix motif [EXXR] | Heme binding motif [FXXGXR/HXCXG] |
|---|---|---|---|
| CYP20A1 | AGCVIT | ETVR | F-SGSQACPE |
| CYP1A | AGFDTI | EIFR | FGLGKRRCIG |
| CYP2AA2 | AGTDTT | EIQR | FSLGPRACLG |
| CYP3C1 | GGYETT | ESMR | FGLGPRNCIG |
| CYP5A1 | AGYETS | ESLR | FGAGPRSCVG |
| CYP7A1 | ASQGNT | EAMR | FGSGVTKCPG |
| CYP7C1 | ASVGNT | ESLR | FGSGATQCPG |
| CYP8A1 | VTQGNA | ETLR | WGTEDNLCPG |
| CYP8B1 | ASQGNT | ETLR | WGAGTTMCPG |
| CYP8B2 | ASQGNT | ETLR | WGAGTTMCPG |
| CYP8B3 | ASQGNT | ETLR | WGAGTTMCPG |
| CYP11C1 | GGVDTT | ETLR | FGFGSRQCVG |
| CYP39A1 | ASLANA | EAIR | FGGGKNQCPG |
| CYP51 | AGQHTS | ETLR | FGAGRHRCIG |
Underlined residues in some CYP isoforms (including CYP20A1) are those differing from the classical P450 signature motif reported in the heading of each column. See sources in Table S6.
Zebrafish CYP20A1 was heterologously expressed in E. coli, as before (Barnes et al., 1991). In essence, expression was obtained by replacing the putative membrane anchor N-terminal region with a membrane anchor from Bos taurus CYP17A1. Preparation of the samples and spectrophotometric analysis of the reduced, CO-ligated protein was done as before (Morrison et al., 2014).
In silico promoter analysis
The proximal promoter regions of CYP20A1s from the zebrafish, frog, chicken, rat and human were screened to identify putative binding sites for transcription factors (TFs) possibly involved in regulation of the orphan P450. Information on gene sequence was retrieved from the Ensembl database (Flicek et al., 2013). However, in the case of X. tropicalis, we used assembly 7.1 from the Joint Genome Institute available through Xenbase (James-Zorn et al., 2013). A region of 10 kb upstream of the start codon was selected for analysis with MatInspector software (Cartharius et al., 2005). The 10kb proximal promoter regions of vertebrate CYP20A1 genes were searched with MatInspector for putative binding sites having either 100% core sequence identity to known human TF binding sites, or a less stringent 80% core sequence identity. The putative TF binding sites that occurred in common in the different species’ CYP20A1s were classified in two groups, one for nuclear receptor TFs and the other for non-nuclear receptor TFs. Data for the latter group were subjected to an additional analysis. That is, the Uniprot database (Bairoch et al., 2005) was interrogated to identify recurring gene ontology terms in the pool of these recurring non-nuclear receptor TFs. Gene ontology information was retrieved for rat and human TFs whenever possible. The database generated was interrogated to identify the cell types, biological systems and processes over-represented in this pool of non-nuclear receptor TFs.
Organ distribution
Transcript abundance was analyzed in various organs of TL zebrafish by means of quantitative real-time PCR (qPCR), using liver as a reference tissue. The protocol of fish dissection followed that of Gupta & Mullins (Gupta and Mullins, 2010), using MS-222 for euthanasia. For the first set of samples, 12 males and 12 females were dissected to obtain liver, intestine, gonad, heart, kidney, whole brain and eye; organs were pooled from four males and from four females, giving three pools for each sex. Statistical analyses were conducted on tissues of males and females separately. For the second set, 12 males were dissected and tissues pooled to obtain three pools each of liver, spleen, forebrain, midbrain, hindbrain and optic nerve. All samples were flash-frozen in liquid nitrogen upon collection and stored at −70°C until processing. RNA was extracted with the Aurum Total Fatty and Fibrous Tissue kit from Bio-Rad (Hercules, USA) as per kit instructions. Total RNA was quantified by spectrophotometry and purity checked with a Nanodrop ND-1000 (Wilmington, USA) at OD260/280 and OD260/230. Reverse transcription of 1 μg (first set) or 150 ng (second set) of total RNA was done with the iScript cDNA Synthesis kit from Bio-Rad, as per kit instructions. QPCR were run on a MyIQ single-color real-time thermocycler with IQ Sybr green Supermix from Bio-Rad. The E-ΔΔCt method was used for relative quantification (Livak and Schmittgen, 2001), using the geometric mean of aryl hydrocarbon receptor nuclear translocator 2 (arnt2) and elongation factor 1 alpha (ef1α) Ct values for normalization (Kubota et al., 2013; Morrison et al., 2014). Primers were: 5’-TACAGGAGGTGGAAGGAAAGGTG-3’ (F) and 5’-GACGACCACCAAGGGCATAGATAAC-3’ (R) for CYP20A1.
Exposure to nuclear and cytosolic receptor agonists, and methylmercury
Experimental exposures of zebrafish to selected nuclear and xenobiotic receptor agonists were conducted on pools of 15 TL embryos, in 60 ml of 0.3X Danieau's solution. Exposures began at 24 hours post fertilization (hpf), and lasted 24 h (up to 48 hpf) or 48 h (up to 72 hpf). Tested chemicals in solvent carrier (dimethylsulfoxide or ethanol) or solvent carrier alone were added at a final volume of 0.1%. Supplemental table S2 provides a list of the tested chemicals and their known action on nuclear or xenobiotic receptors in mammals and zebrafish, to the best of our knowledge. Ethanol was used as solvent only for exposures to α-tocopherol, arachidonic, docosahexaenoic and eicosapentaenoic acids. Exposures to photosensitive agonists were conducted for 24 h at nominal concentrations of 0.1 – 1 – 10 μM. Other exposures were conducted for 48 h at 0.1 – 1 – 3.2 μM, except that PCB 126 was tested for 48 h at doses (1 – 10 – 32 nM) that elicit CYP1 induction in zebrafish without compromising viability (Jonsson et al., 2007a; Jonsson et al., 2007b). All chemicals were of ≥ 95% purity. Following incubations, TL embryos were quickly washed with 0.3X Danieau's solution, frozen in liquid nitrogen and stored at −70C° until RNA extraction. Sample preparation and qPCR analysis were the same as used in the tissue distribution analysis, except that extraction was performed with Aurum Total RNA Mini kit. One μg of total RNA was used for reverse transcription.
Experimental exposures of zebrafish to methylmercury (MeHg) chloride were conducted on pools of 15 AB embryos, in 60 ml of 1X E3 medium, at the Institute of Life Sciences (Universite Catholique de Louvain). Exposure to 10 and 50 μg l−1 MeHg began at 24 hpf and embryos were sampled at 48 hpf, a protocol that does not result in embryo mortality (Hassan et al. 2012). Solvent carrier was distilled water (0.1 % final volume). Following incubations, AB embryos were washed with E3 medium, frozen in liquid nitrogen and stored at −70°C until RNA extraction. We used the exact same procedure of RNA extraction and cDNA synthesis as for TL embryos above. The qPCR experiments were run on a StepOne Plus real-time thermocycler with SYBR Select Master Mix, both from Applied Biosystems. We confirmed that this change of qPCR protocol did not affect the specificity or efficiency of amplification using the same reference genes and method of relative quantification as in TL exposure studies above. Morpholino-based CYP20A1 knockdown
Fertilized zebrafish eggs were injected at the 2- to 4-cell stage with fluorescein-tagged morpholino oligonucleotides (MO) from Gene Tools (Philomath, USA), complementary to a region encompassing the CYP20A1 start codon (5’-AATCTAGCATTTTTGCTTTTCAGGGA-3’). Morpholino knockdown is transient but effects can persist to 5 or more days post fertilization. Controls for the specificity of gene knockdown consisted of embryos injected with a control MO (5’-CCTCTTACCTCAGTTACAATTTATA-3’), and non-injected embryos. Hence, three groups were defined for phenotyping studies, non-injected, control MO-injected, and CYP20A1 MO-injected fish. Microinjection of 2.1 nl of 0.18 mM MO followed the protocol previously described (Jonsson et al., 2007a). The efficacy of CYP20A1 MO to block translation of the CYP20A1 was verified by inhibition of in vitro protein synthesis with a TnT assay from Promega (the inhibition of in vitro synthesis was about 50%; see Supplemental Figure S2). We used only those embryos showing high-level fluorescence at 24 hpf, indicating abundant MO in the embryo.
For the Optomotor and Movement Tracking behavioral studies with the TL strain (described below), larvae were taken from each separate clutch or batch of TL zebrafish and distributed into three groups (i.e., in non-injected, control MO-injected, and CYP20A1 MO-injected groups), with 30 individuals in each group. Initially, early-developing zebrafish were kept in 0.3X Danieau's solution for three days and 72 hpf embryos were then transferred in system water. Larvae from each group were fed once at 4 days post-fertilization (dpf) with a mixture of spirulina and 30-μm Hathchfry Encapsulon diet from Argent (Redmond, USA). Behavior assays were conducted at 5 dpf. TL fish in the various groups were examined daily by microscopy for any morphological abnormalities
Optomotor assay
Six separate clutches or batches of TL zebrafish larvae were used to study the optomotor response (OMR). With each batch, OMR was assessed on 20 larvae from each of the non-injected, control MO-injected, and CYP20A1 MO-injected groups. The 20 larvae were randomly selected from the 30 larvae in each group. A transparent test chamber (25 cm long), placed on top of a computer monitor, was filled with system water at 26.5 ± 1.5°C. A pattern of black and white stripes, moving with a period of 6.5 cm and a frequency of 0.5 Hz, was displayed on the monitor. Larvae were assayed individually. Since zebrafish respond to low spatial frequencies by low velocities of optomotor response (Maaswinkel and Li, 2003), individuals were tested over 1-min periods and distances travelled were recorded every 15 s. There were no signs of aliasing under these conditions, and the responsiveness and distance travelled by control larvae were not significantly increased when the frequency of wave motion was doubled (data not shown). All trials were conducted between 3:00 and 7:00 PM, and larvae from one group were tested before those from the other two groups. To rule out a possible effect of time on the optomotor responses, three batches were tested, with the non-injected controls tested first and the CYP20A1 MO fish tested last, and three others were conducted in the reverse order. Similar patterns of OMR were observed in all experiments.
Movement tracking and activity quantitation
Behavioral studies were also conducted with TL zebrafish, using an automated video tracking system from ViewPoint Life Sciences (Montreal, Canada). For each experimental group (i.e., non-injected, control MO-injected, and CYP20A1 MO-injected), 8 larvae were randomly selected from among the 10 individuals that were not tested in the OMR assay. These larvae were transferred individually in the wells of a 24-well plate filled with 2 ml of system water (26.5 ± 1.5°C). Motion and activity were recorded in consecutive 10-min videos under white light illumination. The first session (tracking) was started 5 min after larvae were introduced in the plate. The second session (activity) was started 5 min after completion of the first recording. For the tracking experiment, thresholds were: 0.5 cm.s−1 for large movement and 0.1 cm.s−1 for inactivity. For activity analysis, thresholds were 10 pixels per frame for burst and 1 for freeze.
High-throughput Morphological assessment and Photomotor response
As noted above, analysis of the CYP20A1-MO effects on TL strain fish included a daily microscopic monitoring of morphology. We also addressed morphological phenotypes using a high-throughput approach, conducted at 24 hpf (1 day) and 120 hpf (5 days) on non-injected, control MO-injected and CYP20A1 MO-injected Tropical 5D strain zebrafish at the Department of Environmental and Molecular Toxicology, Oregon State University. For this study, at 4 hours post-fertilization (hpf) chorions were removed enzymatically with pronase, and embryos individually transferred to the wells of a 96-well plate with 100 μl of embryo medium at 28°C. Plates were sealed and protected from light for up to 120 hpf (Truong et al., 2014). Tropical 5D zebrafish from each of the three experimental groups also were assessed at 24 hpf for photomotor response according to previously published procedures (Truong et al., 2014).
Statistical analyses
Statistical analyses for results obtained with the TL and AB strains were conducted at the α = 0.05 level of significance with the Statistica 7.1 package from Statsoft (Tulsa, USA). Data were tested for normality by Kolmogorov-Smirnov test, and for homoscedasticity by Levene's test. Since multiple independent experiments were necessary to investigate CYP20A1 expression at all doses of each chemical in exposure studies (except for MeHg), qPCR data were compared by two-tailed Student's t-test with batch-specific solvent controls. The MeHg dose-response data were analyzed by one-way ANOVA followed by post hoc LSD Fisher's test. The non-parametric Kruskal-Wallis ANOVA, followed by Z-adjusted Mann-Whitney U test, was used for statistical analyses of the tissue distribution and OMR studies. Data for movement tracking and activity quantitation of TL larvae were analysed by one-way ANOVA followed by post-hoc LSD Fisher's test. Results were expressed either as mean ± SEM or mean ± SD, and graphs generated with Sigma Plot 8.0 software from Systat (San Jose, USA). Statistical analyses for results with Tropical 5D fish were conducted as before (Truong et al., 2014).
Results
Cloning and sequence analysis of zebrafish CYP20A1
Primers designed to the proximal 5’ and 3’ untranslated regions of zebrafish CYP20A1 transcript were used to amplify a full-length coding sequence from the Tupfel Long-fin (TL) strain. A protocol with a low annealing temperature was selected to ensure that possible strain-specific polymorphism in the untranslated regions did not impede primer binding and PCR efficiency. A bright band of the expected size was obtained, and results of sequencing revealed that the transcript from the TL strain was fully identical to the cDNA sequence of zebrafish CYP20A1 available in various databases (GenBank: NM_213332.1; Ensembl: ENSDART00000019325). The inferred amino acid sequence of our cloned zebrafish CYP20A1 shares sequence identities ranging from 59-66 % with CYP20A1 found in genomes of tetrapods, and from 69-82% with CYP20A1 from genomes of other fishes (Supplemental table S1). Such percentages were to be expected for members of a given vertebrate P450 subfamily.
Sequence structure and P450 signature motifs
The amino acid sequence derived from the cloned zebrafish CYP20A1 and the cloned human CYP20A1 (Stark et al., 2008) are aligned in Fig. 1. Features of protein structure that are alpha-helices (n = 13), beta strands (n = 8) and P450 signature motifs (n = 3; see Table 1 headings) were identified based on alignment with other protein-coding CYPs from human and in silico predictions. As shown in Fig. 1, a conserved N-terminal segment of about 30 residues that comprises a transmembrane domain typical of microsomal P450s is found in human and zebrafish CYP20A1s. A signal peptide -perhaps targeting the protein to mitochondria- also is predicted to occur, just after the transmembrane domain. P450 signature motifs are identified in the I-helix, the K helix and the heme-binding region. However, the amino acid sequences in two of these motifs have features that are unusual compared to most other animal CYPs. Thus, the I helix signature of zebrafish CYP20A1 (aa 276-281, AGCVIT) diverges at the fourth and fifth positions, notably lacking the usually conserved Thr at the fifth position, which is involved in the protonation of oxygen during the P450 catalytic cycle (Sezutsu et al., 2013). The heme-binding region of the zebrafish CYP20A1 (aa 403-411, FSGSQACPE) is one residue shorter than in most P450s, and displays an unusual sequence around the conserved Cys that serves as the apical heme iron ligand. Similar divergence from typical P450 motifs is seen in the I-helix and heme binding peptide of human CYP20A1.
Figure 1. Alignment of zebrafish and human CYP20A1 proteins.
Locations of P450 signature motifs and predicted secondary structure are illustrated. Yellow boxes identify ahelices and blue boxes β-strands. Green line locates the position of the predicted transmembrane region, and dashed purple line that of a putative signaling peptide suggested to target the protein to mitochondria.
The amino acid sequences of the cloned zebrafish and human CYP20A1s also were aligned with the CYP20A1s of selected other vertebrate species, gleaned from genome databases (Supplemental figure S1). This alignment showed the presence of a conserved N-terminal segment and the unusual sequence features of the I-helix and the heme binding motifs in these other species, similar to those in zebrafish and human.
We compared the amino acid sequences of the three signature motifs in zebrafish CYP20A1 to those in other zebrafish CYP families (Table 1). Divergence at the fourth and fifth positions of the I helix motif like that in CYP20A1 appears as well in CYP7A1 (cholesterol 7α-hydroxylase), CYP8A1 (prostacyclin synthase), CYP8B1 (sterol 12α-hydroxylase), and several related zebrafish orphan CYPs (i.e., CYP7C1, CYP8B2 and CYP8B3), as well as CYP39A1 (oxysterol 7α-hydroxylase). Thus, the fourth position of the I-helix motif in all of these P450s has a hydrophobic rather than an acidic residue, and the fifth position does not contain a Thr residue. Instead, an Asp residue is found in all of the above-mentionned CYPs except CYP20A1 (which has an Ile residue). Most of these other CYPs do have a Thr at the 6th position, raising a question about whether that Thr might fulfill a role in proton transfer to the oxygen. However, CYP8A1 and CYP39A1 do not have a Thr at either the 5th or the 6th positions.
Divergence from the classical motif of the heme-binding region was also identified for CYP7A1, CYP8A1, CYP8B1, CYP39A1 (Table 1), and the related CYP7 and CYP8 orphans. However, these sequences were not identical to that seen in CYP20A1, which has a unique heme-binding region. Thus, among zebrafish P450s, CYP20A1 is the only isoform with one residue missing at the beginning of the motif, a glutamine (neutral residue) instead of a basic residue at the center of the motif, and a glutamic acid instead of a glycine (non polar residue) at the terminal position. The same features distinguish the CYP20A1 from other P450s in human.
CYP20A1 heterologous protein expression
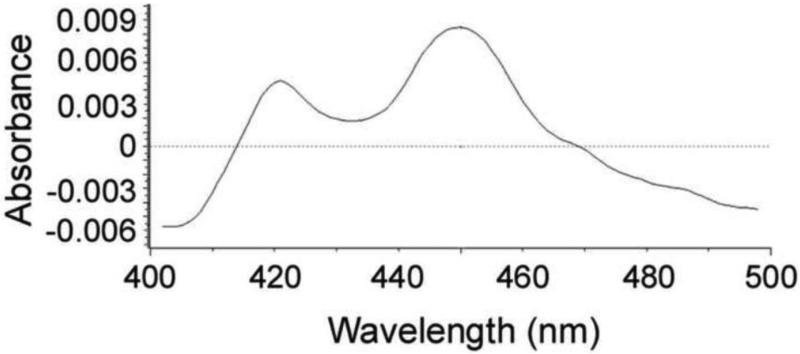
Zebrafish CYP20A1 was heterologously expressed in E. coli to determine whether a protein with a native P450 spectrum could be obtained, given the unusual sequence features. A reduced CO-difference spectrum typical of P450 was obtained (Fig. 2), which indicates expression of native protein. The presence of a peak at 420 nm in the CO-difference spectrum indicates partial loss of the cysteine thiolate linkage to the heme, which is sometimes the result of protein misfolding of non-native (heterologous) proteins in E. coli. However, we also observed that the 450 nm peak was rapidly converted to 420 nm, which suggests that the ferrous – CO complex is unstable, a condition observed with prostacyclin synthase (CYP8A1), an enzyme that acts on substrates that carry their own oxygen (Yeh et al., 2005). We did examine the expressed protein for Type-1 (substrate) binding speotra with cholesterol and arachidonic acid, but these preliminary studies failed to detect any binding to recombinant zebrafish CYP20A1 (authors’ unpublished information).
Figure 2. Reduced, CO-difference spectrum of heterologously expressed zebrafish CYP20A1.
Zebrafish CYP20A1 was expressed in E. coli cells after N-terminal modification. The reduced difference spectrum shows a peak at 450nm, as expected, as well as a peak at 420 nm indicating partial loss of the cysteine thiolate linkage to the heme, possibly due to the non-native protein expression in E coli.
Tissue distribution and developmental expression of CYP20A1
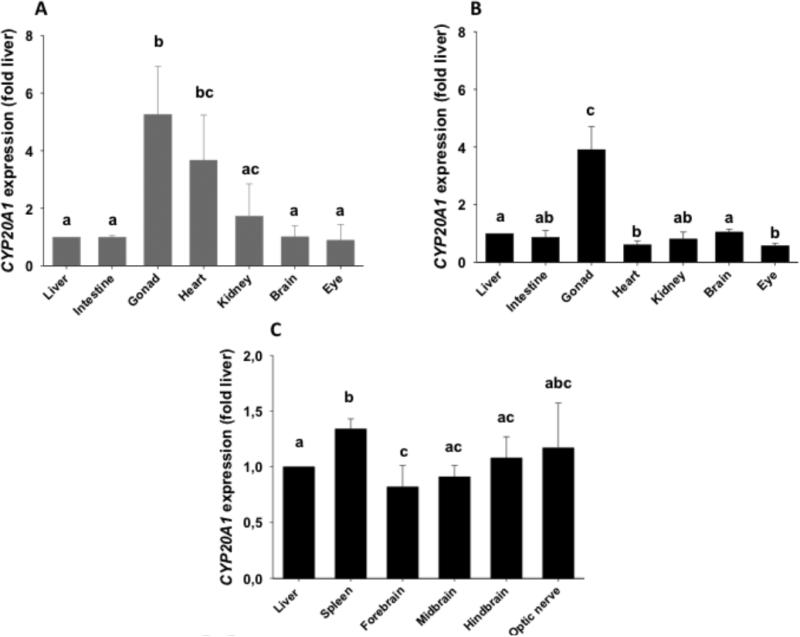
Real-time quantitative PCR (qPCR) analysis was used to determine levels of CYP20A1 transcript in organs from male and female zebrafish. Two sets of samples were analyzed, with tissues pooled from multiple individuals. In the first set, both males and females were examined. In females, there was a significantly higher level of expression in ovary and heart than liver, intestine, kidney, brain and eye (Fig. 3A). In corresponding male samples, transcript levels in testis were higher than in the other tissues examined, and levels of expression were lower in heart and eye than in liver or brain (Fig. 3B). In the second set, we examined organs from male only. Expression levels were compared in different parts of the brain (forebrain, midbrain and hindbrain), optic nerve, spleen and liver. CYP20A1 transcript was slightly more abundant in spleen and optic nerve than in the other tissues (Fig. 3C).
Figure 3. Expression of CYP20A1 transcript in zebrafish tissues.
For both sexes, transcript levels were measured by real-time quantitative PCR (qPCR) using a double reference gene approach (elongation factor 1α and aryl hydrocarbon receptor nuclear translocator 2), and data are expressed relative (i.e., fold change) to transcript content in liver. Pools of organs were prepared and qPCR was run on three pools each for females (Figure 3A) and males (Figure 3B). In the second set, qPCR was run on three pools from males only (Figure 3C). Data are presented as mean ± SD. Different letters indicate significant difference of expression in organs of males and females separately (Kruskal-Wallis ANOVA with post-hoc Z-adjusted Mann-Whitney U test).
Earlier we had reported on the levels of most CYP transcripts during zebrafish development, covering the period from 3 to 48 hours hpf (Goldstone et al., 2010). However, the levels of CYP20A1 were not readily discernible in the figures in that report. Values for CYP20A1 were extracted from the previous microarray data, showing that CYP20A1 transcript was more abundant in embryos during the segmentation period (12-24 hpf) (Supplemental Figure 3).
Reponses to chemical treatment
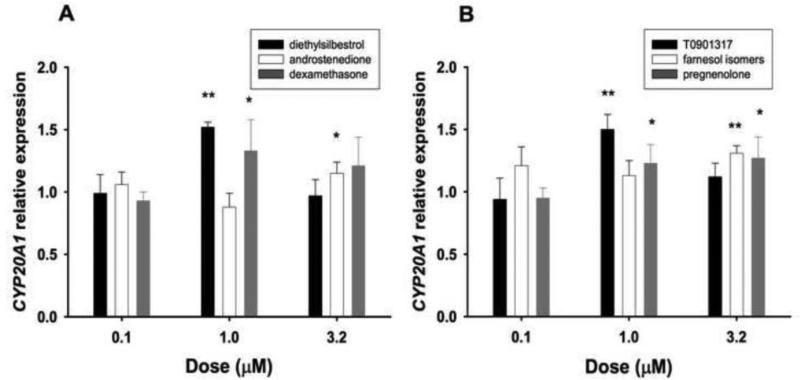
Possible effects on CYP20A1 expression were assessed in zebrafish embryos that had been exposed to agonists for selected nuclear and xenobiotic receptors. Emphasis was put on agonists for receptors involved in steroid function (androgen, estrogen and glucocorticoid receptors - AR, ER, and GR respectively), cholesterol and lipid metabolism (liver X, farnesoid X and peroxisome-proliferator-activated receptors - LXR, FXR, and PPAR respectively), response to xenobiotics (pregnane X receptor, PXR, and the aryl hydrocarbon receptor, AHR), and morphogenesis (retinoic acid receptor RAR) (see Supplemental table S2). Dose-response studies with these chemicals were conducted in zebrafish embryos with exposure beginning at 24 hpf, and sampling at 48 or 72 hpf (i.e., exposure for 24 or 48 h). Significant but modest increases of transcript abundance (about 150% of solvent controls) were observed in embryos exposed for 48 hr to 1 μM of diethylstilbestrol (Fig. 4A), a known ER agonist in zebrafish, or T0901317 (Fig. 4B), a known LXR-FXR agonist in zebrafish. At the highest dose, farnesol isomers (FXR agonists in zebrafish) and pregnenolone (PXR agonist in zebrafish) also led to a mild increase (i.e., about 30%) of CYP20A1 mRNA content in embryos (48 h exposure) (Fig. 4B). A similar increase was detected in embryos exposed to 1 μM dexamethasone (Fig. 4A), a GR agonist in zebrafish. Slight yet significant increases of expression (i.e., 10-20%) were also observed with 10 μM of docosahexaenoic acid (24 h exposure) (Supplemental table S3), a PPAR agonist in zebrafish, 1 μM of pregnenolone and 3.2 μM of the AR agonist androstenedione (48 h exposures) (Fig. 4). Expression of CYP20A1 decreased by about 10-20% in embryos exposed to doses up to 3.2 μM all-trans retinoic acid (24 h exposures) (Supplemental table S3), a RAR agonist in zebrafish. (At 10 μM, all-trans retinoic acid treatment resulted in 98% mortality.)
Figure 4. Expression of CYP20A1 in zebrafish embryos exposed to agonists for nuclear receptors.
(a) Responses to diethylstilbestrol (black), androstenedione (white), and dexamethasone (grey). (b) Responses to T0901317 (black), farnesol isomers (white), and 5-pregnen-3β-ol-20-one (grey). Transcript levels were measured by real-time quantitative PCR (qPCR) using a double reference gene approach (elongation factor 1α and aryl hydrocarbon receptor nuclear translocator 2). Exposures were from 24 to 72 hours post-fertilization on pools of 15 embryos in 60 ml of 0.3X Danieau's solution with 0.1% of dimethylsulfoxide as carrier. Results were expressed as fold change from results obtained with carrier alone. Data are presented as mean ± SD of 4 replicates. Since the various doses of each chemical were tested in separate experiments, analyses were conducted with a 2-tailed Student's t-test. Asterisks indicate levels of statistical significance (* for p ≤ 0.05; ** for p ≤ 0.01).
We also examined response to the non-ortho-substituted (dioxin-like) polychlorinated biphenyl (PCB) 126, an AHR agonist (Supplemental table S2). Exposures were for 48 h, from 24 to 72 hpf. Nanomolar doses of PCB 126, which are highly effective at inducing the target gene CYP1A in zebrafish (Jonsson et al., 2007b), did not elicit any change in CYP20A1 expression (not shown).
To assess further the possible involvement of nuclear receptors and other transcription factors (TFs) in the regulation of CYP20A1 expression, we used an in silico search to predict putative TF binding sites. A 10 kb proximal promoter region of the zebrafish and human CYP20A1 genes was examined, along with the promoters of CYP20A1 predicted in genomes of several other vertebrate species (rat, chicken, frog). We screened these regions for the presence of possible binding sites (i.e., response elements) for nuclear and xenobiotic receptors. When restricting the search to putative binding sites exactly matching the human core sequences, several were found to occur in proximal promoters of all the vertebrate CYP20A1 genes examined. These response elements are for the vertebrate steroidogenic factor 1 (SF1), the estrogen receptors and estrogen-related receptors (ERs and ERRs), the retinoic acid receptor-related orphan receptor alpha (RORα), the peroxisome proliferator-activated receptors (PPARs) and glucocorticoid receptor (GR) (Table 2). When a less stringent cut-off of core sequence similarity was applied (i.e., 80%), response elements were identified also for other nuclear receptors. These include the androgen receptor (AR), the progesterone receptor (PR), the farnesoid X receptor (FXR), the hepatocyte nuclear factor 4 (HNF4) and the retinoid receptors (RAR and retinoid X receptor – RXR) (Supplemental table S4). At 70% core sequence identity, binding sites were identified for the liver X receptor (LXR) but not AHR (data not shown).
Table 2.
Recurring putative binding sites for families of nuclear receptors in proximal promoters of CYP20A1s in vertebrates.
| Matrix family | Danio rerio | Xenopus tropicalis | Gallus gallus | Rattus norvegicus | Homo sapiens |
|---|---|---|---|---|---|
| Steroidogenic Factor (V$SF1F) | 2 | 11 | 9 | 6 | 6 |
| Estrogen Response (V$EREF) | 3 | 5 | 8 | 7 | 2 |
| v-ERB and RAR-related Orphan Receptor Alpha (V$RORA) | 3 | 5 | 5 | 7 | 13 |
| Peroxisome Proliferator-activated Receptor (V$PERO) | 6 | 22 | 14 | 9 | 13 |
| Glucocorticoid Response and Related (V$GREF) | 8 | 12 | 14 | 14 | 7 |
Proximal promoter analyses also identified putative binding sites for numerous TFs that were not nuclear or xenobiotic receptors. Gene ontology terms were compiled from the Uniprot database (Bairoch et al., 2005) for these recurring TFs, and keyword searches conducted to determine the proportion of TFs putatively involved in CYP20A1 regulation that were associated with a particular cell type, biological system or process. At 100% core sequence identity, TFs active in the brain, blood cells (among which 77% were notably involved in immunity; data not shown), skeletal system, eye, sensory system, germ cells and stem cells were over-represented, as were TFs involved in neurogenesis, cell migration, neurulation and anterior/posterior patterning. Wingless (Wnt), Smoothened and Notch were the major developmental signaling pathways represented (Supplemental Table S5). Similar observations (i.e., enrichment of ontology terms) were made at 80% core sequence identity (data not shown).
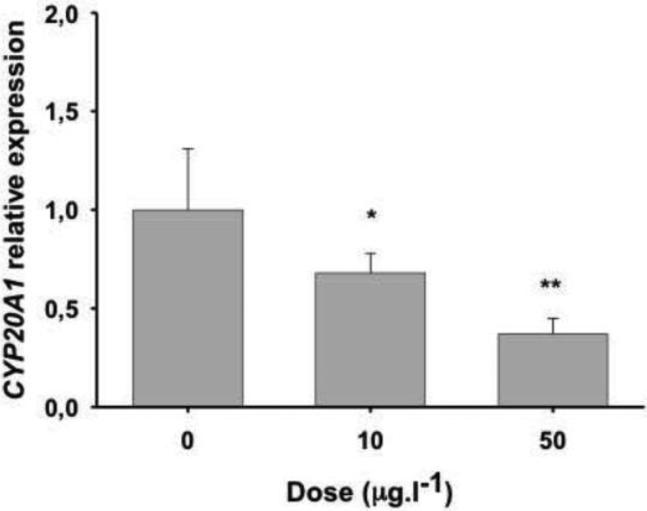
To assess the influence of environmental toxicants on CYP20A1 expression, we examined zebrafish embryos exposed to MeHg chloride (10 and 50 μg.l−1) from 24 and 48 hpf (Hassan et al., 2012). The MeHg elicited dose-dependent reduction in CYP20A1 mRNA levels in zebrafish embryos (Fig. 5). With the higher dose tested, CYP20A1 transcript abundance was less than half the level in solvent controls, and was significantly lower than the levels obtained with a 10 μg.l−1 dose (p = 0.048; not shown).
Figure 5. Expression of CYP20A1 in zebrafish embryos exposed to methylmercury chloride.
Transcript levels were measured by real-time quantitative PCR (qPCR) using a double reference gene approach (elongation factor 1 α and aryl hydrocarbon receptor nuclear translocator 2). Exposures were from 24 to 48 hours post-fertilization on pools of 15 embryos in 60 ml of 1X E3 medium with 0.1% of distilled water as carrier. Results are expressed relative (i.e., fold change) to those obtained with carrier alone. Data are presented as mean ± SD of 4 replicates. Analysis of the difference in CYP20A1 expression from solvent controls was conducted with one-way ANOVA followed by post hoc LSD Fisher's test. Asterisks indicate levels of statistical significance (* for p ≤ 0.05; ** for p ≤ 0.01).
Knockdown of CYP20A1 and phenotypic effects
To gain insight into the function of CYP20A1 protein in zebrafish, we used a morpholino (MO) gene knockdown approach. Morpholinos targeted to the CYP20A1 translation start site, which inhibits CYP20A1 protein synthesis, were injected into 2- to 4-cell stage embryos and morphological and behavioral phenotypes screened at various times. In these studies we used two approaches, with different strains, for assessing morphological and behavioral effects of MO treatment. With TL zebrafish, morphology was assessed daily daily, and no obvious effects were seen in fish that were uninjected, control MO-injected or CYP20A1 MO-injected. Morphology was examined also in a high-throughput analysis using the Tropical 5D wild-type strain. In those latter analyses embryos injected with coded CYP20A1 or control MOs, and non-injected controls, were examined in a blind test at 24 hpf and at five days post-fertilization (dpf). At 24 hpf, somites and notochord appeared normal in both control MO and CYP20A1 MO embryos. Viability was unaffected, however, CYP20A1 MO-injected embryos exhibited a delayed development (data not shown). There were no morphological abnormalities noticed at 5 dpf in either the control MO or the CYP20A1 MO groups, i.e., yolk sac, body axis, eye, snout, jaw, otic vesicle, heart, brain, fins, trunk, swim bladder, notochord, pigmentation and circulation appeared normal.
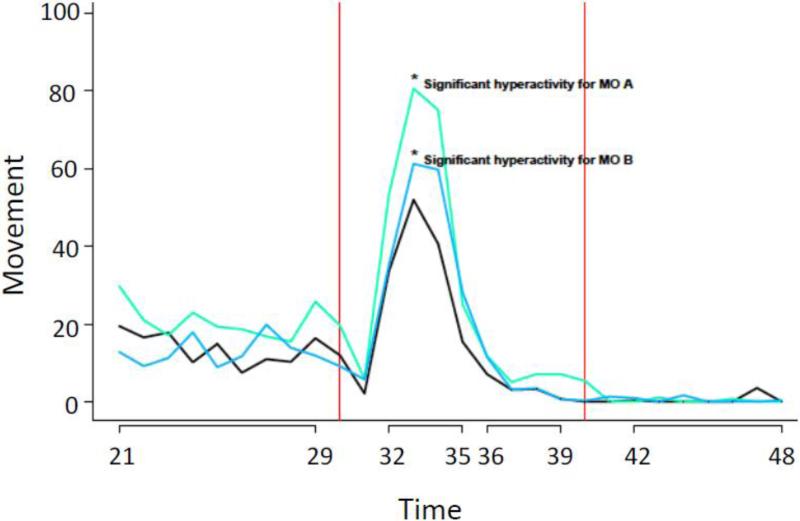
We examined behavioral effects of CYP20A1 knockdown in a blind test at an early stage of development, 24 hpf. For this assay, we employed the Tropical 5D strain injected with coded CYP20A1 MO or control MO as above, and movement (tail flexion) was examined in ovo (Truong et al., 2014). Analysis revealed a greater in ovo activity in the CYP20A1 MO treated embryos, in both the dark-baseline and light-excitatory phases (Figure 6).
Figure 6. In ovo tail flexion movement response in zebrafish embryos treated with CYP20A1 morpholino.
In ovo tail flexion movement was automatically analyzed at 24 hours post-fertilization in zebrafish embryos of the Tropical 5D wild-type strain after injection with CYP20A1 morpholino (MO A) (top line) or control morpholino (MO B). Embryos that were not injected (Uninj) were also used in the blind test. Individuals were placed one per well of a 96-well plate. The red vertical lines indicate the time of a single bright, visible flashlight. The Y-axis values are an index of tail flexion (i.e., pixel changes between successive frames), and the X-axis values are given in seconds. Asterisks and labels indicate the statistically significant hyperactivity associated with CYP20A1 morpholino in both the baseline (dark, un-stimulated) phase and in the first excitatory period (i.e., following first pulse of bright light), and with control morpholino in just the first excitatory period.
For later stage behavioral studies, TL strain zebrafish (the strain from which CYP20A1 had been cloned) were injected with control or CYP20A1 MOs at the 2- to 4-cell stage and at five dpf were screened for an optomotor response, and for locomotory activity with an automated tracking system. Non-injected TL zebrafish embryos were similarly tested.
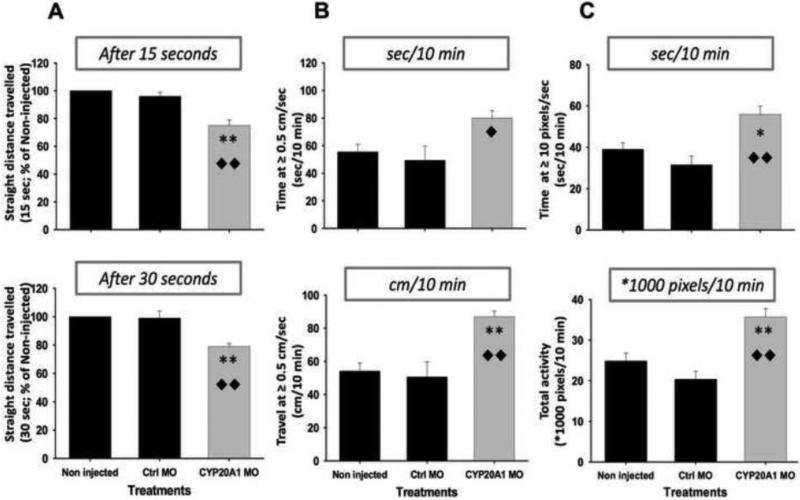
We first tested for the optomotor response (OMR), a test for visual responsiveness in which fish larvae follow waves of vertical black and white stripes displayed on the fish tank bottom. Larvae from the three experimental groups were tested individually over 1-min periods, and the distance they travelled was recorded every 15 s. On average, larvae in the CYP20A1 MO group travelled about 20% less distance than those injected with control MO, or not injected (Fig. 7A). A substantial number of larvae in the CYP20A1 MO group did not initiate an oriented movement after 15 or 30 s of stimulation, indicating latency or reduced responsiveness of these fish to the visual stimulus. The CYP20A1 MO treatment did not appear to affect the locomotory capacity of larvae that did respond to the stimulus, based on distances traveled per unit time once the response began.
Figure 7. Optomotor and locomotory effects of CYP20A1 knockdown in zebrafish larvae.
TL embryos were injected at the two-to-four cell stage with a morpholino (MO) specific for CYP20A1 or a control morpholino (Ctrl MO). Batch-specific embryos that were not injected were also used (Non injected). Assays were conducted on 5 day-old fish. In the optomotor response study (OMR; Figure 7A), a sine wave of black and white stripes (6.5 cm periodicity; 0.5 Hz) was tested (20 larvae per replicate per group; tested individually) and the sum of the straight distances travelled was used to compare OMR between groups every 15 s, up to 1 min. Upper and lower panels show results from the first time bins as percentages of the straight distance travelled relative to non-injected larvae. The automated ViewPoint Zebrabox® was used to track locomotion (Figure 7B) and activity (Figure 7C) of the larvae in 24-well plates under constant illumination. 8 larvae from each group were simultaneously tested in consecutive 10-minute movie sessions (one for movement tracking and one for activity quantitation). For movement tracking, thresholds were: 0.5 cm.s−1 for large and 0.1 for no movement. Upper and lower panels of Figure 7B respectively depict the time and distance travelled by larvae at high speed (i.e., ≥ 0.5 cm.s−1). For activity quantitation, thresholds were: 10 pixels per frame for burst and one for freeze. Upper and lower panels of Figure 7C respectively depict the time spent by larvae in a hyperactive state (i.e., ≥ 10 pixels per frame) and their total level of activity (i.e., area under curve). Data are presented as mean ± SEM of six replicates for OMR and three replicates for tracking and activity, and were analyzed with Kruskal-Wallis ANOVA followed by Z-adjusted Mann-Whitney U test (OMR) or one-way ANOVA followed by post hoc LSD Fisher's test (tracking-and-activity). Differences of CYP20A1 MO groups from non-injected controls are indicated by asterisks, (* p ≤ 0.05; ** p ≤ 0.01) and from Ctrl MO by diamonds (◆ p ≤ 0.05; ◆◆ p ≤ 0.01).
We then investigated the spontaneous locomotory behavior of CYP20A1 MO-injected TL larvae, using an automated tracking system (Viewpoint Zebrabox). In three independent experiments, eight larvae from each group (CYP20A1 MO-injected, control MO-injected and, non-injected) were tested simultaneously in 24-well plates, under constant illumination. Two 10-min movies were recorded, one for tracking movement (locomotion or directed motion) and the other for “activity quantitation”, quantitation of general activity. In the directed motion assay,CYP20A1 MO-injected larvae spent significantly more time at high speed (≥ 0.5 cm.s−1) than either non-injected or control MO-injected larvae, and also travelled more distance at high speed (Fig. 7B). Hyperactivity or bursts of activity, evident as significantly longer periods of time they spent at a high activity state, were typical of these larvae. This was reflected also in the higher level of total activity, expressed as integrated values (Fig. 7C).
Discussion
In these studies, we cloned and sequenced the cDNA of CYP20A1 of the zebrafish TL strain and found it identical to the zebrafish CYP20A1 coding sequence available in databases, and with a high % identity to the cloned human CYP20A1. Unusual features of the CYP20A1 sequence have been suggested before, based largely on inferences from genome sequences (Sezutsu et al., 2013). The comparison of cloned zebrafish and human CYP20A1s establishes that these unusual features are present in the expressed proteins from both species. Zebrafish and human CYP20A1s have a highly conserved N-terminal motif that not only contains the transmembrane domain typical of microsomal P450s, but also have an adjacent motif that could possibly act as a signal peptide for mitochondrial targeting. This suggests that CYP20A1s might have a bimodal distribution in the ER and mitochondria, similarly to some other vertebrate P450s (Avadhani et al., 2011). Further work is needed to determine whether there is dual subcellular localization of CYP20A1s.
The zebrafish CYP20A1 protein and the human ortholog contain a typical EXXR motif in the K-helix, thought to be required for proper folding and heme inclusion (Sezutsu et al., 2013). However, the I-helix and heme-binding regions were found to diverge significantly from the classical P450 signature motifs. The absence of one residue at the beginning of the heme-binding motif, and the Glu instead of a conserved Gly at the terminal portion, are unique to CYP20A1 when compared to other vertebrate P450s. Replacement of a non polar (Gly) by a negatively charged (Glu) residue at the end of the heme-binding motif is intriguing considering that the site where this change occurs is thought to be in close vicinity to the catalytic Cys in the tertiary structure (Otyepka et al., 2007). The presence of a Gln residue at a central site of the heme-binding motif, where usually an Arg or His residue is found, is also intriguing. The signature Arg or His residue is responsible for the hydrogen bonding with one heme propionate (Otyepka et al., 2007). It has been suggested that these hydrogen bonds not only can influence the stability of hemeproteins, but also their reactivity and ligand selection (Ramos-Santana and Lopez-Garriga, 2012). Note that Gln is also able to generate a hydrogen bond with the heme propionate. These features imply there could be unusual catalytic properties of CYP20A1s, as previously suggested (Sezutsu et al., 2013). In preliminary studies we failed to detect any binding of cholesterol or arachidonic acid to recombinant zebrafish CYP20A1. Likewise, Stark et al. (Stark et al., 2007) did not detect any activity of human CYP20A1 with cholesterol or arachidonic acid or any of 9 other possible substrates, including neurotransmitters. However, in a proteomic mapping of cholesterol-interacting proteins CYP20A1 was identified as possibly interacting with trans-sterol (Hulce et al., 2013). The success at expressing recombinant zebrafish CYP20A1 will facilitate the search for substrates, and could provide for engineering to test possible functions.
The divergence observed in the I-helix, especially the absence of the conserved threonine at the 5th position of the motif, is similar to what is observed in zebrafish and human CYP7s and CYP8s. These sequences differ from zebrafish CYP20A1 in having a Ser residue at the second position of the motif in most cases (although a Ser is present in human CYP20A1), a Gly residue at the fourth and an Asn residue at the fifth position. While the functional significance of these I-helix features of CYP20A1 is unknown, it is notable that the presence of an Ile residue rather than Thr at the 5th position of the I-helix motif also occurs in human CYP5A1 (thromboxane synthase). Reverse-engineering experiments with human CYP5A1 have demonstrated a specific role for the Ile residue in keeping rates of product formation by the enzyme at low levels (Meling et al., 2015). The tight control of catalysis by CYP5A1 suggests that the Ile in CYP20A1 could perhaps act similarly to limit product formation by this enzyme. Thus, too much or too little CYP20A1 activity could be detrimental.
Tissue distribution in reproductive, immune and nervous systems
The tissue distribution of CYP20A1 transcript in zebrafish was somewhat different from that observed with rodents. Thus, in male mouse, abundant CYP20A1 expression was found in spleen, followed by skeletal muscle, brain, heart, lung, kidney, testis and liver (Choudhary et al., 2003). In male rat, CYP20A1 was most strongly expressed in liver, followed by lung, spleen, kidney, heart, brain and testis (Takiguchi et al., 2010). Regardless of these differences, studies with zebrafish, human (Stark et al., 2008) and rodents are consistent in pointing to relatively high-level expression for CYP20A1 in reproductive, immune and nervous tissues. These patterns are consistent with the putative TF binding sites in CYP20A1 proximal promoters.
The tissue distribution in zebrafish brain and eye also suggests a role for CYP20A1 in development and maintenance and/or function of the nervous system, supporting the inference from high level expression in human hippocampus, substantia nigra and retinal pigment epithelium (Stark et al., 2008; Strunnikova et al., 2010). It also has been noted that traumatic injury to both hippocampus and retinal pigment epithelium resulted in significant up-regulation of CYP20A1 expression in these rodent tissues (Birnie et al., 2013; Hadziahmetovic et al., 2012). Our observation that the optomotor response was slowed in the MO knockdown larvae is consistent with some role in visual processes. Refining the localization of CYP20A1 expression in zebrafish brain is an objective for further studies.
The data on CYP20A1 expression in the literature also reveal an enrichment of the transcript in human neural progenitor cells derived from embryonic stem cells (Chan et al., 2015) and expression, along with a limited set of other CYP genes, in human cord blood hematopoietic and early progenitor cells (Xu et al., 2014) These observations support a role for CYP20A1 at early stages of cell differentiation in some tissues.
Response to agonists for nuclear and xenobiotic receptors
At micromolar doses, agonists for some nuclear receptors modestly, but significantly, increased CYP20A1 transcript levels in zebrafish embryos. The responses conform to predictions based on the presence of cis-response elements in the proximal promoter region of CYP20A1s. In silico and in vivo studies point to regulation of CYP20A1 expression by nuclear receptors involved in steroid response, cholesterol and lipid metabolism, and down-regulation by the morphogen all-trans retinoic acid.
Among NR agonists, the highest increases of CYP20A1 expression were seen with diethylstilbestrol (DES) and T0901317. In zebrafish, DES is a potent ER agonist (Gorelick and Halpern, 2011). In mammals, it also acts as an inverse agonist for ERRs (Coward et al., 2001). To our knowledge, there are no reports of antagonism of zebrafish ERRs by DES. However, putative binding sites were specifically predicted for ERRα in proximal promoters of the vertebrate CYP20A1s at a cut-off of 80% of the human core sequence identity. ERs exert functions in reproduction and neuronal protection (Dahlman-Wright et al., 2006), while ERRs control the diurnal expression of genes involved in fatty acid, cholesterol and bile acid metabolism (Giguere et al., 2011).
An increase of CYP20A1 expression in zebrafish embryos also was observed with T0901317 exposure. T0901317 is a benzenesulfonamide that is a potent agonist for LXR in mammals, and also is an agonist for PXR and FXR (Houck et al., 2004; Mitro et al., 2007) and an inverse agonist for RORα, and RORγ (Kumar et al., 2010). T0901317 appears to be an agonist for zebrafish LXR and FXR, but not PXR (Krasowski et al., 2011; Reschly et al., 2008). In silico studies predicts the presence of binding sites for FXR and RORα in promoters of vertebrate CYP20A1. A less stringent cut-off also identified binding sites for LXR (i.e., 70% similarity with human set of LXR binding sites). These observations suggest that a diversity of nuclear receptors could have been involved in the T0901317-mediated increase of CYP20A1 expression. The LXRs and FXR are involved in sterol and bile acid homeostasis (Kalaany and Mangelsdorf, 2006), while RORs, which bind cholesterol, exert functions in brain and eye development, and immunity (Jetten, 2009). In contrast, there was no effect on CYP20A1 expression by nanomolar doses of PCB 126, or 2,3,7,8-tetrachlorodibenzodioxin (data not shown), pointing to a lack of involvement of the AHR. Immunoassay with antibodies to zebrafish CYP20A1 would help to define localization of protein and responses to chemical treatment.
CYP20A1 knockdown effects
The potential involvement of CYP20A1 in neurobehavioral disorders is indicated not only by the expression in zebrafish, rodent and human brain, but also by the effects of CYP20A1 knockdown investigated here. Zebrafish embryos in which CYP20A1 translation was partly inhibited showed developmental delay at 24 hpf, and behavioral effects were evident at 1 and 5 dpf. The latency in initiating an appropriate swimming response due to CYP20A1 knockdown in the OMR assay can be described as a defective visuo-motor integration or coordination, or an increased anxiety (Kalueff et al., 2014), rather than an impaired locomotory capacity. Larvae of the CYP20A1 MO group also exhibited a substantial increase of activity and swimming speed compared to the two control groups in the automated tracking-and-activity experiments. The possibility that the confined settings of the multiwell plates elicited these phenotypes in the CYP20A1 knockdown fish cannot be ruled out; thus, these bursts of activity and higher swimming speeds could have been driven by anxiety (Maximino et al., 2010).
Intriguingly, these phenotypic effects of CYP20A1 knockdown in zebrafish are strongly reminiscent of those seen in human patients diagnosed with microdeletions in the 2q33 chromosome region where CYP20A1 is located. Patients with this rare syndrome variously show psychomotor retardation, hyperactivity and bouts of anxiety, and a delayed visuomotor coordination was specifically reported once (Balasubramanian et al., 2011; Tomaszewska et al., 2013). Analyzing the specific regions deleted, which are displayed in Balasubramian et al. (Balasubramanian et al., 2011), reveals that the majority of patients (5-6 of 7) with deletions that included CYP20A1 presented with hyperactivity, while this phenotype was rarely observed in patients with deletions that excluded CYP20A1 (1-3 of 13). The present results with knockdown of CYP20A1 in zebrafish, and the 2q33 microdeletion patients, suggests that this orphan P450 could well be involved in neurobehavioral hyperactivity.
Response to methylmercury
The possible effects of a neurotoxicant on CYP20A1 expression were assessed by exposing zebrafish embryos to MeHg, among the chemicals suggested to contribute to hyperactivity disorder (Grandjean and Landrigan, 2014). These experiments revealed a dose-dependent reduction of CYP20A1 transcript abundance following 24 h exposure of zebrafish, beginning at 24 hpf. The higher dose tested resulted in the strongest decline of CYP20A1 expression. Notably, the MeHg concentration threshold in human cord blood that is associated with the detection of behaviors typical of attention deficit hyperactivity disorder (ADHD) during childhood is very close to the lowest dose we tested (Boucher et al., 2012). Providing that this effect on CYP20A1 expression also occurs in human and translates into changes at the protein level, it is tempting to envisage a link between prenatal exposure to MeHg, CYP20A1 repression (with detrimental developmental consequences) and the onset of ADHD. We are investigating further the mechanism of the MeHg effect on CYP20A1, and how CYP20A1 may function in neurological processes. Such studies will include direct comparison of behavioral change and CYP20A1 expression following MeHg exposures, as well as CYP20A1 responses to other neurotoxicants.
Conclusion
In this study, we cloned and examined properties of the zebrafish orphan P450 CYP20A1. Peculiar structural features of zebrafish, human and other vertebrate CYP20A1s are clues to an unusual catalytic function. The expression patterns of CYP20A1 transcript, and putative transcription factor binding sites suggest potential involvement in reproductive, immune and neuronal functions, as well as steroid response, lipid and cholesterol metabolism. A possible involvement in neuronal processes is strongly indicated by the behavioral effects occurring in zebrafish larvae treated with a morpholino against CYP20A1. The effects of knockdown - especially hyperactivity - are reminiscent of those in humans who have a microdeletion that includes loss of CYP20A1 gene. The MeHg-mediated reduction of CYP20A1 transcript levels in zebrafish embryos points to the possibility that disruption of CYP20A1 expression may be involved in some neurobehavioral disorders. Studies of CYP20A1 expression with additional toxicants are underway. Future studies will investigate whether over-expression can reverse behavioral deficits seen with MO treatments or MeHg, or whether overexpression also could be detrimental, which could occur if tight regulation of the enzyme might be necessary. The results presented here together shed new light on the possible biological and toxicological significance of the orphan CYP20A1 that has thus far resisted “deorphanization”.
Supplementary Material
Highlights.
The “orphan” CYP20A1 was cloned from zebrafish and its sequence analyzed.
Knockdown of CYP20A1 reduced an optomotor response and elicited bursts of activity.
Effects of knockdown resemble some features of a microdeletion of CYP20A1 in human.
Expression of CYP20A1 was downregulated by the neurotoxicant methylmercury.
CYP20A1 may be involved in neurobehavioral processes and effects of some chemicals. .
Acknowledgements
This work was supported in part by NIH Superfund Research Program at Boston University, grant P42ES007381 from the National Institute of Environmental Health Sciences (NIEHS) (JJS, JVG, AK and BL), by a Belgian-American Educational Foundation (BAEF) Postdoctoral Fellowship (BL), the Japan Society for the Promotion of Science Postdoctoral Fellowship and Postdoctoral Fellowship for Research Abroad to AK (nos. 4313 and 820, respectively), and NIEHS grant P30ES000210. Additional support (JJS) was provided by NIH grant P01ES021923 and National Science Foundation Grant OCE-1314642 through the Woods Hole Center for Oceans and Human Health, and from the Aquastress project funded by the BELSPO; Belgian Federal Science Policy Office, P7/31. We gratefully acknowledge the assistance of Matthew Takata for preparation of RNA from tissue samples, Sibel Karchner and Diana Franks for help with TnT assay, Rachel Harbeitner for assistance with qPCR, and Victoria Starczak for help with statistics.
Abbreviations
- AHR
aryl hydrocarbon receptor
- AR
androgen receptor
- arnt2
aryl hydrocarbon receptor nuclear translocator 2
- CYP
cytochrome P450
- DES
diethylstilbestrol
- dpf
days post-fertilization
- ef1α
elongation factor 1 alpha
- ER
estrogen receptor
- ERR
estrogen-related receptor
- FXR
farnesoid X receptor
- GR
glucocorticoid receptor
- HNF4
hepatocyte nuclear factor 4
- hpf
hours post-fertilization
- LXR
liver X receptor
- MeHg
methylmercury
- MO
morpholino
- OMR
optomotor response
- PCB
polychlorinated biphenyl
- PPAR
peroxisomeproliferator-activated receptor
- PR
progesterone receptor
- PXR
pregnane X receptor
- qPCR
real-time quantitative PCR
- RAR
retinoic acid receptor
- ROR
RAR-related orphan receptor
- RXR
retinoid X receptor
- SF1
vertebrate steroidogenic factor
- TF
transcription factor
- TL
Tupfel Long-fin zebrafish strain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avadhani NG, Sangar MC, Bansal S, Bajpai P. Bimodal targeting of cytochrome P450s to endoplasmic reticulum and mitochondria: the concept of chimeric signals. Febs J. 2011;278:4218–4229. doi: 10.1111/j.1742-4658.2011.08356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS. The Universal Protein Resource (UniProt). Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian M, Smith K, Basel-Vanagaite L, Feingold MF, Brock P, Gowans GC, Vasudevan PC, Cresswell L, Taylor EJ, Harris CJ, Friedman N, Moran R, Feret H, Zackai EH, Theisen A, Rosenfeld JA, Parker MJ. Case series: 2q33.1 microdeletion syndrome--further delineation of the phenotype. J Med Genet. 2011;48:290–298. doi: 10.1136/jmg.2010.084491. [DOI] [PubMed] [Google Scholar]
- Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 alpha-hydroxylase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnie M, Morrison R, Camara R, Strauss KI. Temporal changes of cytochrome P450 (Cyp) and eicosanoid-related gene expression in the rat brain after traumatic brain injury. BMC Genomics. 2013;14:303. doi: 10.1186/1471-2164-14-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher O, Jacobson SW, Plusquellec P, Dewailly E, Ayotte P, Forget-Dubois N, Jacobson JL, Muckle G. Prenatal methylmercury, postnatal lead exposure, and evidence of attention deficit/hyperactivity disorder among Inuit children in Arctic Quebec. Environmental health perspectives. 2012;120:1456–1461. doi: 10.1289/ehp.1204976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chan SF, Huang X, McKercher SR, Zaidi R, Okamoto SI, Nakanishi N, Lipton SA. Transcriptional profiling of MEF-2 regulated genes in human neural progenitor cells derived from embryonic stem cells. Genomics Data. 2015;3:24–27. doi: 10.1016/j.gdata.2014.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Baldi P. Three-stage prediction of protein beta-sheets by neural networks, alignments and graph algorithms. Bioinformatics. 2005;21(Suppl 1):i75–84. doi: 10.1093/bioinformatics/bti1004. [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Schenkman JB, Sarfarazi M, Stoilov I. Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Archives of biochemistry and biophysics. 2003;414:91–100. doi: 10.1016/s0003-9861(03)00174-7. [DOI] [PubMed] [Google Scholar]
- Coward P, Lee D, Hull MV, Lehmann JM. 4-Hydroxytamoxifen binds to and deactivates the estrogen-related receptor gamma. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8880–8884. doi: 10.1073/pnas.151244398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman-Wright K, Cavailles V, Fuqua SA, Jordan VC, Katzenellenbogen JA, Korach KS, Maggi A, Muramatsu M, Parker MG, Gustafsson JA. International Union of Pharmacology. LXIV. Estrogen receptors. Pharmacol Rev. 2006;58:773–781. doi: 10.1124/pr.58.4.8. [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V, Dufour CR, Eichner LJ, Deblois G, Cermakian N. Estrogen-related receptor alpha, the molecular clock, and transcriptional control of metabolic outputs. Cold Spring Harb Symp Quant Biol. 2011;76:57–61. doi: 10.1101/sqb.2011.76.011031. [DOI] [PubMed] [Google Scholar]
- Goldstone JV, McArthur AG, Kubota A, Zanette J, Parente T, Jonsson ME, Nelson DR, Stegeman JJ. Identification and developmental expression of the full complement of Cytochrome P450 genes in Zebrafish. BMC Genomics. 2010;11:643. doi: 10.1186/1471-2164-11-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick DA, Halpern ME. Visualization of estrogen receptor transcriptional activation in zebrafish. Endocrinology. 2011;152:2690–2703. doi: 10.1210/en.2010-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean P, Landrigan PJ. Neurobehavioural effects of developmental toxicity. Lancet Neurol. 2014;13:330–338. doi: 10.1016/S1474-4422(13)70278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta T, Mullins MC. Dissection of organs from the adult zebrafish. J Vis Exp. 2010 doi: 10.3791/1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadziahmetovic M, Kumar U, Song Y, Grieco S, Song D, Li Y, Tobias JW, Dunaief JL. Microarray analysis of murine retinal light damage reveals changes in iron regulatory, complement, and antioxidant genes in the neurosensory retina and isolated RPE. Invest Ophthalmol Vis Sci. 2012;53:5231–5241. doi: 10.1167/iovs.12-10204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan SA, Moussa EA, Abbott LC. The effect of methylmercury exposure on early central nervous system development in the zebrafish (Danio rerio) embryo. J Appl Toxicol. 2012;32:707–713. doi: 10.1002/jat.1675. [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. Regulation of cytochrome P450 (CYP) genes by nuclear receptors. The Biochemical journal. 2000;347:321–337. doi: 10.1042/0264-6021:3470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck KA, Borchert KM, Hepler CD, Thomas JS, Bramlett KS, Michael LF, Burris TP. T0901317 is a dual LXR/FXR agonist. Mol Genet Metab. 2004;83:184–187. doi: 10.1016/j.ymgme.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Hulce JJ, Cognetta AB, Niphakis MJ, Tully SE, Cravatt BF. Proteome-wide mapping of cholesterol-interacting proteins in mammalian cells. Nat Methods. 2013;10:259–264. doi: 10.1038/nmeth.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Zorn C, Ponferrada VG, Jarabek CJ, Burns KA, Segerdell EJ, Lee J, Snyder K, Bhattacharyya B, Karpinka JB, Fortriede J, Bowes JB, Zorn AM, Vize PD. Xenbase: expansion and updates of the Xenopus model organism database. Nucleic Acids Res. 2013;41:D865–870. doi: 10.1093/nar/gks1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson ME, Jenny MJ, Woodin BR, Hahn ME, Stegeman JJ. Role of AHR2 in the expression of novel cytochrome P450 1 family genes, cell cycle genes, and morphological defects in developing zebra fish exposed to 3,3',4,4',5-pentachlorobiphenyl or 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicological sciences : an official journal of the Society of Toxicology. 2007a;100:180–193. doi: 10.1093/toxsci/kfm207. [DOI] [PubMed] [Google Scholar]
- Jonsson ME, Orrego R, Woodin BR, Goldstone JV, Stegeman JJ. Basal and 3,3',4,4',5-pentachlorobiphenyl-induced expression of cytochrome P450 1A, 1B and 1C genes in zebrafish. Toxicology and applied pharmacology. 2007b;221:29–41. doi: 10.1016/j.taap.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kalueff AV, Stewart AM, Gerlai R. Zebrafish as an emerging model for studying complex brain disorders. Trends in pharmacological sciences. 2014;35:63–75. doi: 10.1016/j.tips.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasowski MD, Ai N, Hagey LR, Kollitz EM, Kullman SW, Reschly EJ, Ekins S. The evolution of farnesoid X, vitamin D, and pregnane X receptors: insights from the green-spotted pufferfish (Tetraodon nigriviridis) and other non-mammalian species. BMC Biochem. 2011;12:5. doi: 10.1186/1471-2091-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kubota A, Bainy AC, Woodin BR, Goldstone JV, Stegeman JJ. The cytochrome P450 2AA gene cluster in zebrafish (Danio rerio): expression of CYP2AA1 and CYP2AA2 and response to phenobarbital-type inducers. Toxicology and applied pharmacology. 2013;272:172–179. doi: 10.1016/j.taap.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, Garcia-Ordonez RD, Burris TP, Griffin PR. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethy l]phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Molecular pharmacology. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb DC, Waterman MR. Unusual properties of the cytochrome P450 superfamily. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120434. doi: 10.1098/rstb.2012.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maaswinkel H, Li L. Spatio-temporal frequency characteristics of the optomotor response in zebrafish. Vision Res. 2003;43:21–30. doi: 10.1016/s0042-6989(02)00395-4. [DOI] [PubMed] [Google Scholar]
- Maximino C, de Brito TM, da Silva Batista AW, Herculano AM, Morato S, Gouveia A., Jr. Measuring anxiety in zebrafish: a critical review. Behav Brain Res. 2010;214:157–171. doi: 10.1016/j.bbr.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Meling DD, Zelasko S, Kambalyal A, Roy J, Das A. Functional role of the conserved i-helix residue I346 in CYP5A1-Nanodiscs. Biophys Chem. 2015;200-201:34–40. doi: 10.1016/j.bpc.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Mitro N, Vargas L, Romeo R, Koder A, Saez E. T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS letters. 2007;581:1721–1726. doi: 10.1016/j.febslet.2007.03.047. [DOI] [PubMed] [Google Scholar]
- Morrison AM, Goldstone JV, Lamb DC, Kubota A, Lemaire B, Stegeman JJ. Identification, modeling and ligand affinity of early deuterostome CYP51s, and functional characterization of recombinant zebrafish sterol 14alpha-demethylase. Biochimica et biophysica acta. 2014;1840:1825–1836. doi: 10.1016/j.bbagen.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Nelson DR. The cytochrome p450 homepage. Hum Genomics. 2009;4:59–65. doi: 10.1186/1479-7364-4-1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Goldstone JV, Stegeman JJ. The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120474. doi: 10.1098/rstb.2012.0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otyepka M, Skopalik J, Anzenbacherova E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Biochimica et biophysica acta. 2007;1770:376–389. doi: 10.1016/j.bbagen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Ramos-Santana BJ, Lopez-Garriga J. Tyrosine B10 triggers a heme propionate hydrogen bonding network loop with glutamine E7 moiety. Biochemical and biophysical research communications. 2012;424:771–776. doi: 10.1016/j.bbrc.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschly EJ, Ai N, Welsh WJ, Ekins S, Hagey LR, Krasowski MD. Ligand specificity and evolution of liver X receptors. J Steroid Biochem Mol Biol. 2008;110:83–94. doi: 10.1016/j.jsbmb.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezutsu H, Le Goff G, Feyereisen R. Origins of P450 diversity. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120428. doi: 10.1098/rstb.2012.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Guengerich FP. Characterization of orphan human cytochromes P450. Drug Metab Rev. 2007;39:627–637. doi: 10.1080/03602530701467708. [DOI] [PubMed] [Google Scholar]
- Stark K, Wu ZL, Bartleson CJ, Guengerich FP. mRNA distribution and heterologous expression of orphan cytochrome P450 20A1. Drug Metab Dispos. 2008;36:1930–1937. doi: 10.1124/dmd.108.022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, Sergeev Y, Chen W, Edwards AO, Stambolian D, Abecasis G, Swaroop A, Munson PJ, Miller SS. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Human molecular genetics. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi M, Darwish WS, Ikenaka Y, Ohno M, Ishizuka M. Metabolic activation of heterocyclic amines and expression of CYP1A1 in the tongue. Toxicological sciences : an official journal of the Society of Toxicology. 2010;116:79–91. doi: 10.1093/toxsci/kfq087. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast release clones: a high throughput expression analysis. ZFIN direct data submision. 2004 ( http://zfin.org)
- Tomaszewska A, Podbiol-Palenta A, Boter M, Geisler G, Wawrzkiewicz-Witkowska A, Galjaard RJ, Zajaczek S, Srebniak MI. Deletion of 14.7 Mb 2q32.3q33.3 with a marfanoid phenotype and hypothyroidism. Am J Med Genet A. 2013;161:2347–2351. doi: 10.1002/ajmg.a.36076. [DOI] [PubMed] [Google Scholar]
- Truong L, Reif DM, St Mary L, Geier MC, Truong HD, Tanguay RL. Multidimensional in vivo hazard assessment using zebrafish. Toxicological sciences : an official journal of the Society of Toxicology. 2014;137:212–233. doi: 10.1093/toxsci/kft235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1:REVIEWS3003. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirdefeldt K, Adami HO, Cole P, Trichopoulos D, Mandel J. Epidemiology and etiology of Parkinson's disease: a review of the evidence. Eur J Epidemiol. 2011;26(Suppl 1):S1–58. doi: 10.1007/s10654-011-9581-6. [DOI] [PubMed] [Google Scholar]
- Xu S, Ren Z, Wang Y, Ding X, Jiang Y. Preferential expression of cytochrome CYP CYP2R1 but not CYP1B1 in human cord blood hematopoietic stem and progenitor cells. Acta Pharm Sin B. 2014;4:464–469. doi: 10.1016/j.apsb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh HC, Hsu PY, Wang JS, Tsai AL, Wang LH. Characterization of heme environment and mechanism of peroxide bond cleavage in human prostacyclin synthase. Biochimica et biophysica acta. 2005;1738:121–132. doi: 10.1016/j.bbalip.2005.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Dougherty JH, Jr., Hubner KF, Bai B, Cannon RL, Hutson RK. Abnormal connectivity in the posterior cingulate and hippocampus in early Alzheimer's disease and mild cognitive impairment. Alzheimers Dement. 2008;4:265–270. doi: 10.1016/j.jalz.2008.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.