Abstract
Entamoeba histolytica is the protozoan parasite that causes invasive amebiasis, which is endemic to many developing countries and characterized by dysentery and liver abscesses. The virulence of E. histolytica correlates with the degree of host cell engulfment, or phagocytosis, and E. histolytica phagocytosis alters amebic gene expression in a feed-forward manner that results in an increased phagocytic ability. Here, we used a streamlined RNA interference screen to silence the expression of 15 genes whose expression was upregulated in phagocytic E. histolytica trophozoites to determine whether these genes actually function in the phagocytic process. When five of these genes were silenced, amebic strains with significant decreases in the ability to phagocytose apoptotic host cells were produced. Phagocytosis of live host cells, however, was largely unchanged, and the defects were surprisingly specific for phagocytosis. Two of the five encoded proteins, which we named E. histolytica ILWEQ (EhILWEQ) and E. histolytica BAR (EhBAR), were chosen for localization via SNAP tag labeling and localized to the site of partially formed phagosomes. Therefore, both EhILWEQ and EhBAR appear to contribute to E. histolytica virulence through their function in phagocytosis, and the large proportion (5/15 [33%]) of gene-silenced strains with a reduced ability to phagocytose host cells validates the previously published microarray data set demonstrating feed-forward control of E. histolytica phagocytosis. Finally, although only limited conclusions can be drawn from studies using the virulence-deficient G3 Entamoeba strain, the relative specificity of the defects induced for phagocytosis of apoptotic cells but not healthy cells suggests that cell killing may play a rate-limiting role in the process of Entamoeba histolytica host cell engulfment.
INTRODUCTION
Entamoeba histolytica infection, which causes dysentery and amebic liver abscesses, is an enormous public health problem in developing countries (1, 2). In areas where the infection is highly endemic, such as the Mirpur region of Dhaka, Bangladesh, over 50% of children may become infected by 5 years of age (3), yet only about 10% of all infections become symptomatic (4, 5). Isolated strains of E. histolytica have been shown to have different levels of virulence both in vivo and in vitro (6–8), which may in part determine whether or not a particular infection becomes invasive.
Multiple mechanisms contribute to the ability of E. histolytica to destroy the host intestinal mucosa and cause disease (9). Following excystation within the small intestinal lumen, trophozoites attach to host mucous and epithelial cells in the colon, largely through the multisubunit amebic GalNAc lectin (10, 11). Trophozoites also secrete multiple cysteine proteases, which degrade mucin and the extracellular matrix (12–14), and they kill resident host cells through a contact-dependent process that remains poorly understood (15–17). Host cells appear to suffer from a disruption of the cell membrane with rapid changes in intracellular calcium levels (18). This leads to changes in the host cell that closely resemble apoptosis with membrane blebbing, DNA digestion, and activation of caspase 3 (17, 19, 20). Finally, E. histolytica trophozoites phagocytose red blood cells and nucleated host cells, and the ability to phagocytose host cells is strongly associated with virulence (21–23).
Two forms of amebic phagocytosis have been described: a fast variant in which entire cells are taken up intact and a slow variant in which target cells are progressively deformed and torn apart, followed by ingestion of cell fragments (24, 25). In vitro, most intact engulfed cells are apoptotic (26), suggesting that host cell apoptosis facilitates what was previously termed fast amebic phagocytosis. Similarly, intact ingested apoptotic cells and erythrocytes are readily observed in vivo (26–28). This process is partially mediated by exposure of phosphatidylserine on the surface of dying nucleated cells and erythrocytes (26, 29). It is also stimulated by host C1q, which binds to the surface of apoptotic cells and to amebic cell surface calreticulin (30, 31). On the other hand, it is now clear that slow amebic phagocytosis corresponds to the phenomenon recently termed trogocytosis, a process that contributes to cell killing, during which E. histolytica bites off and engulfs pieces of viable cells (32).
By modeling of phagocytosis of apoptotic cells through ingestion of C1q-coated particles, our laboratory previously showed that E. histolytica trophozoites have a program of gene expression changes that is initiated through the act of phagocytosis and that actually increases their phagocytic ability (33). Stimulation with host cells also enhances trogocytosis (32), likely through the same changes in gene expression. Microarray analysis comparing gene expression in phagocytic and nonphagocytic populations of trophozoites identified 121 genes that were upregulated in the phagocytic population (33). Gene clustering based on functional annotation identified strong clusters of genes encoding proteins involved in actin binding and cytoskeletal organization. Here, we use small interfering RNA-based gene silencing of E. histolytica trophozoites to confirm that many of the genes discovered to be upregulated in our microarray analysis have a direct impact on the phagocytic ability. Silencing of 5 of 15 genes significantly decreased the ability of E. histolytica to phagocytose apoptotic host cells. Two of these five genes were labeled with a SNAP tag to allow localization. Both proteins localized to the site of host cell phagocytosis and also to extending pseudopodia.
MATERIALS AND METHODS
Gene silencing.
Selected E. histolytica genes were cloned into the pCR8/GW/TOPO entry vector (Invitrogen) and then shuttled into a custom Gateway destination vector version of the silencing plasmid, psAP-2 (34). This vector was created by inserting a Gateway destination cassette into the existing multiple-restriction site of psAP-2 (Invitrogen) (shown in Fig. S1 in the supplemental material). All primers used for the cloning of vector constructs can be found in Supplemental Information S1 in the supplemental material. G3 strain trophozoites were harvested during mid-log growth and transfected with a silencing plasmid using the Attractene transfection reagent (Qiagen) and M199S medium (M199 medium supplemented with 25 mM HEPES, 5.7 mM cysteine, and 0.5% bovine serum albumin) according to a protocol adapted from that of Petri and colleagues (35). Transfected amoebae were then selected with G418 (Sigma) over a 3-week period, starting with 1.5 μg/ml and ending with 24 μg/ml. Silencing was assessed in these strains using quantitative reverse transcription-PCR (qRT-PCR). All primers for qRT-PCR can be found in Supplemental Information S1 in the supplemental material. Once gene silencing was confirmed, the G418 selection was removed, and then silencing was again confirmed and quantified prior to phenotypic assays.
Cell monolayer adherence assay.
CHO cells were grown to confluence over a 48-h period in 24-well plates. Cell monolayers were washed three times with phosphate-buffered saline (PBS); fixed with 4% paraformaldehyde, which was quenched with 250 mM glycine; and then washed twice with PBS. Amoebae were incubated on ice for 15 min and resuspended at 2.5 × 105 cells per ml in M199S medium, 1 ml of which was applied to the cell monolayer in each well. The plates were then incubated for 30 min at 37°C. Nonadherent amoebae were washed away with 1 ml of cold PBS (twice), and then adherent amoebae were fixed to the cell monolayer with 4% paraformaldehyde. This reaction was quenched with 250 mM glycine, and each well was washed twice with PBS. Amoebae were then labeled using an anti-serine-rich E. histolytica protein (anti-SREHP; clone 10D11) mouse monoclonal antibody (originally described by Teixeira and Huston [36]) and Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes). The plates were imaged using a Nikon eTi-2000 microscope with a programmable, motorized stage. Twenty images per well were taken at a magnification of ×200, and the motorized stage was programmed to randomly chose images from within the central 60% of each well. Adherent cells were then quantified by batch processing using NIH ImageJ software as follows: the images were converted to 8 bits, thresholded, and binarized, and a watershedding algorithm was used to separate individual objects. Automated counts were then obtained by using size exclusion (see Fig. S2 in the supplemental material).
Cell suspension adherence assay.
Adherence to Jurkat T lymphocytes was measured using a modified version of a previously described assay (10). G3 trophozoites in which genes were silenced were incubated on ice for 15 min and then washed twice with M199S medium and resuspended at 105 amoebae per ml in M199S medium. Jurkat T lymphocytes were grown to confluence in T-25 flasks (Corning) and then washed twice with M199S medium and resuspended at 2 × 106 cells per ml in M199S medium. Amoebae (104) were added to 2 × 105 Jurkat T lymphocytes in sterile borosilicate glass culture tubes (Fisher). Each sample was centrifuged at 900 rpm for 5 min at 4°C and then incubated on ice for 60 min to allow adherence to target cells. Following another centrifugation at 900 rpm for 5 min at 4°C, samples were vortexed briefly and the cells were quantified using a hemocytometer. Trophozoites with three or more adherent Jurkat T lymphocytes were counted as adherent. Results represent combined data from six biological replicates performed.
Cytotoxicity assay.
2′,7′-Bis-(2-carboxyethyl)-5 (and -6)-carboxyfluorescein (BCECF) release from Jurkat T lymphocytes was used to measure E. histolytica cytotoxicity using a protocol derived from that of Berninghausen and Leippe (37). Jurkat T lymphocytes were labeled using 10 μM BCECF acetoxymethyl ester (Calbiochem) in PBS for 20 min at 37°C. The cells were then washed three times with PBS and resuspended at 3 × 106 cells per ml in PBS. Amoebae were incubated on ice for 15 min, washed once with PBS, and then resuspended at 3 × 105 cells per ml in PBS. Next, 3 × 105 Jurkat T lymphocytes were moved into a polypropylene tube (12 by 75 mm) with 3 × 104 amoebae. Samples were centrifuged for 3 min at 150 × g and then incubated for 30 min at 37°C. The supernatant was collected following centrifugation at 400 × g for 10 min, and fluorescence was measured using a BioTek PowerWave XS plate reader. Background fluorescence was determined using a lymphocyte-only control, complete lysis was determined using cells lysed with sodium dodecyl sulfate (SDS), and percent specific lysis (cytotoxicity) was calculated according to the following formula: percent cytotoxicity = [(fluorescenceexperimental − fluorescencespontaneous)/(fluorescencedetergent lysed − fluorescencespontaneous)] × 100, where fluorescenceexperimental is the experimental fluorescence, fluorescencespontaneous is the spontaneous fluorescence, and fluorescencedetergent lysed is the fluorescence of detergent-lysed cells. Results represent combined data from six biological replicates.
Apoptotic phagocytosis assay.
Phagocytosis assays were performed using flow cytometry as previously described (26). Jurkat T lymphocytes were first exposed to UV light (using a Fotodyne UV box) for 15 min to induce apoptosis. Immediately following UV exposure, cells were fluorescently labeled by resuspension in 2 ml of 28 μM carboxyfluorescein succinimidyl ester (Sigma) in PBS. Labeling proceeded for 15 min at 37°C, at which time 2 ml of fetal bovine serum was added to quench the labeling reaction. Jurkat cells were resuspended in 35 ml of RPMI medium and incubated at 37°C for 4 h to allow apoptotic maturation. After incubation, Jurkat T lymphocytes were resuspended in cold M199S medium at a cell density of 4 × 106 per ml. Amoebae were placed on ice for 15 min, washed with PBS, and then resuspended in cold M199S medium at a cell density of 2 × 106 cells per ml. Next, 4 × 105 Jurkat T lymphocytes were moved into a polypropylene tube (12 by 75 mm) with 2 × 105 amoebae. Samples were centrifuged for 5 min at 400 × g and then incubated at 37°C for 10 min. Samples were washed twice with 1 ml of PBS–d-Gal and agitated to remove adherent cells. Amoebae were then fixed in 4% paraformaldehyde and resuspended in PBS for analysis by flow cytometry. All flow cytometry was done using a Beckman Coulter EPICS XL-MCL flow cytometer. Amoebae and Jurkat T lymphocytes were differentiated using side and forward scatter measurements. Phagocytic amoebae were defined as those with fluorescence over the background fluorescence, determined using a negative control. Figure S3 in the supplemental material shows the flow cytometry gating scheme used for a typical phagocytosis experiment. The phagocytic index was calculated by multiplying the percentage of phagocytic amoebae by the mean fluorescence for those determined to be phagocytic (38). Results represent combined data from six biological replicates.
Live phagocytosis assay.
Live phagocytosis assays were performed in the same manner as the apoptotic phagocytosis assay, but without UV exposure and incubation of amoebae and Jurkat T lymphocytes for 25 min at 37°C. Results represent combined data from six biological replicates.
SNAP vector construction.
E. histolytica ILWEQ (EhILWEQ; EHI_003930) and E. histolytica BAR (EhBAR; EHI_105240) were cloned (see Supplemental Information S1 in the supplemental material for PCR primers) into the E. histolytica SNAP (EhSNAP) vector (shown in Fig. S1 in the supplemental material) for expression as fusion proteins with an N-terminal SNAP tag. This plasmid uses a previously validated SNAP tag gene sequence with codon usage optimized for E. histolytica expression (39). HM1 strain trophozoites were harvested during mid-log growth and transfected using the Attractene transfection reagent (Qiagen) as described above. Transfected amoebae were then selected with hygromycin (Sigma) over a 3-week period, starting with 1.5 μg/ml and ending with 15 μg/ml.
Fluorescent microscopy.
E. histolytica trophozoites were grown in 15-ml glass tubes with trypsin yeast extract-iron-sulfur-33 (TYI-S-33) medium and induced with 1 mg/ml tetracycline (Sigma) for 24 h prior to addition of SNAP substrate. Six hours prior to use, 100,000 trophozoites were moved into a 2-ml glass tube and SNAP-Cell TMR-Star substrate (New England BioLabs) was added directly to the medium at 500 nM. Thirty minutes prior to use, this medium was replaced with fresh TYI-S-33 medium. Amoebae were then resuspended in M199S medium and applied to a 35-mm glass-bottom dish (Mattek) containing a confluent monolayer of human ileocecal carcinoma cells (HCT-8 cells; courtesy of ATCC) stained with Hoechst 33342 (Sigma). The amoebae and the cell monolayer were incubated for 1 h at 37°C and then fixed with 4% paraformaldehyde. Cell monolayers were imaged using a Nikon eTi-2000 microscope with a 60× (numerical aperture, 1.4) oil immersion objective. Images were taken using a z-spacing of 0.500 μm and then deconvolved using Autoquant X software (MediaCybernetics).
Bioinformatics, statistical analyses, and figure preparation.
Conserved protein domains were identified using the NCBI conserved protein domains database unless otherwise noted, and all E values of ≤10−3 were considered significant. In addition to searching for conserved domains, the Entamoeba protein sequences and TBLASTN were used to search the NCBI nonredundant translated nucleotide database to assess the degree of conservation of the entire protein (E values of ≤10−5 were considered significant). All statistical analyses were done using GraphPad Prism (version 6) software (the specific statistical tests used are indicated in the appropriate figure legends). All figures were prepared using the Adobe Illustrator (version 10.0.3) program.
RESULTS
Large-scale silencing of E. histolytica genes found to be upregulated in phagocytic trophozoites.
Nineteen genes that were previously found to be overexpressed in phagocytic trophozoites were chosen for silencing to determine which ones, if any, had a measurable impact on phagocytosis. Using a custom Gateway vector system based on the psAP-2 vector (34), silencing vectors for all 19 genes were cloned and used to transfect G3 trophozoites. AmoebaDB gene identifiers and the outcomes of gene silencing and preliminary phagocytosis assays for these 19 genes are summarized in Table S1 in the supplemental material. Three transfectants were repeatedly not viable, and one gene was not sufficiently silenced to enable further characterization on the basis of the qRT-PCR results. Of the strains transfected with a plasmid designed to silence 1 of the remaining 15 genes, 6 were identified to have altered phenotypes, on the basis of the preliminary phagocytosis and adherence phenotypic screens. Table 1 shows the decrease in mRNA expression for each of these six strains compared to the level of mRNA expression for empty vector-transfected amoebae. The reduction in gene expression determined using the Gateway silencing vector ranged from 4- to 20-fold. While this degree of gene silencing is adequate for phenotypic characterization, it should be noted that silencing with this plasmid was less robust than it was in prior studies and our own experience using the psAP-2 Gunma vector (34, 40, 41). This may be due to the addition of att recombination sites (each approximately 124 nucleotides in length), which are necessary for Gateway recombination, on either side of the target sequence (42).
TABLE 1.
Reduction in gene expression measured by qRT-PCR
| AmoebaDB gene identifier | AmoebaDB product name | ΔΔCTa |
Mean fold change in expressionc | ||
|---|---|---|---|---|---|
| Biological replicatesb | Mean ΔΔCT | SD ΔΔCT | |||
| EHI_003930 | I/LWEQ domain protein | 4.396, 4.288 | 4.342 | 0.076 | −20.28 |
| EHI_007640 | Hypothetical protein | 4.349, 4.099 | 4.224 | 0.177 | −18.68 |
| EHI_098510 | Hypothetical protein | 2.575, 3.510 | 3.042 | 0.662 | −8.24 |
| EHI_105240 | BAR/SH3 domain-containing protein | 2.540, 1.603 | 2.071 | 0.663 | −4.20 |
| EHI_150430 | Villidin, putative | 3.107, 3.113 | 3.110 | 0.004 | −8.64 |
| EHI_189500 | Calponin homology domain protein, putative | 4.854, 3.536 | 4.195 | 0.932 | −18.32 |
CT, threshold cycle.
Results are from two biological replicates of E. histolytica G3 trophozoites in which the indicated gene was silenced. The qRT-PCR assays were performed on separate days.
Mean fold change in expression for gene-silenced E. histolytica G3 trophozoites compared to that for empty vector-transfected control cells.
Phagocytosis of live and apoptotic host cells.
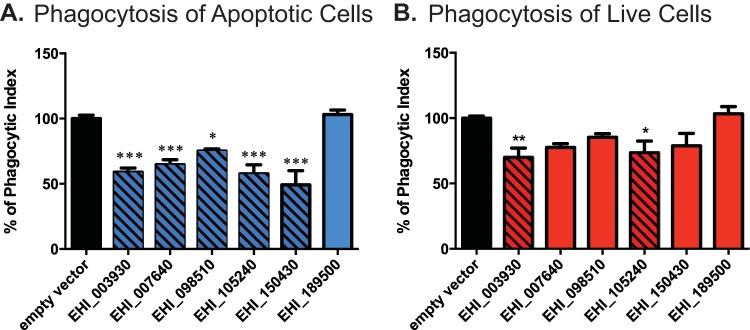
Entamoeba histolytica phagocytoses apoptotic cells more rapidly than healthy cells (26). Thus, to facilitate the comparison of results using apoptotic versus healthy target cells, assay conditions were established to achieve a comparable phagocytic index for engulfment of each during the allotted incubation time. When assayed for the ability to phagocytose apoptotic Jurkat T lymphocytes, five of the six strains showed a clear and statistically significant decrease (Fig. 1A). The reduction in the uptake of apoptotic lymphocytes ranged from roughly 25% (in the strain in which EHI_098510 was silenced) to 50% (in the strain in which EHI_150430 was silenced) in E. histolytica gene-silenced trophozoites, as determined by measurement of the uptake of fluorescently labeled host cells during 10 min of incubation. In contrast, the same gene-silenced strains showed less of a decrease in uptake when assayed for phagocytosis of live Jurkat T lymphocytes (Fig. 1B). Although trophozoites were allowed to phagocytose live lymphocytes for 25 min, which is longer than the amount of time that they were allowed to phagocytose apoptotic lymphocytes, the phagocytic index for the live lymphocytes was decreased with statistical significance for only two strains in which genes were silenced by RNA interference (the strains in which EHI_003930 and EHI_105240 were silenced).
FIG 1.
Phagocytosis of live and apoptotic lymphocytes. (A and B) Phagocytosis of apoptotic (A) and live (B) Jurkat T lymphocytes by G3 trophozoite strains in which the indicated genes were silenced. Jurkat T lymphocytes were labeled with carboxyfluorescein succinimidyl ester prior to incubation with trophozoites; phagocytosis was subsequently quantified using flow cytometry. Results are shown as the phagocytic index calculated as a percentage of that for the control (black bars), as measured via flow cytometry. Significance is based on one-way analysis of variance with Dunnett's multiple-comparison test. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Adherence and cytotoxicity assays.
The method by which host cells are killed and phagocytosed by E. histolytica may follow a sequential process of adherence, cell killing, and phagocytosis (reviewed in reference 9) (15, 26, 29, 43). In order to characterize our silenced E. histolytica strains more fully, therefore, we also tested for defects in adherence and cytotoxicity using previously established methods. Strains of trophozoites in which genes were silenced were assayed for the ability to adhere to a monolayer of cells (Fig. 2A), for the ability to adhere to host cells in suspension (Fig. 2B), and for a cytotoxic effect on labeled Jurkat T lymphocytes (see Fig. S4 in the supplemental material). Two strains of trophozoites, amoebae in which either EHI_003930 or EHI_189500 was silenced, showed an increase in monolayer adherence compared to that for trophozoites transfected with the empty vector. The former strain also showed increased adherence to host cells in suspension. Regarding cytotoxicity, it should be noted that the G3 E. histolytica strain used to facilitate silencing of the expression of numerous genes does not express the genes for amoebapores A and B, which are believed to be involved in host cell killing (44). Despite this limitation, approximately 17% of cells were killed by G3 strain trophozoites transfected with the empty vector control, as defined by the detection of cell membrane disruption by vital dye release. None of the tested strains in which genes were silenced had a further decrease in cytotoxic ability.
FIG 2.
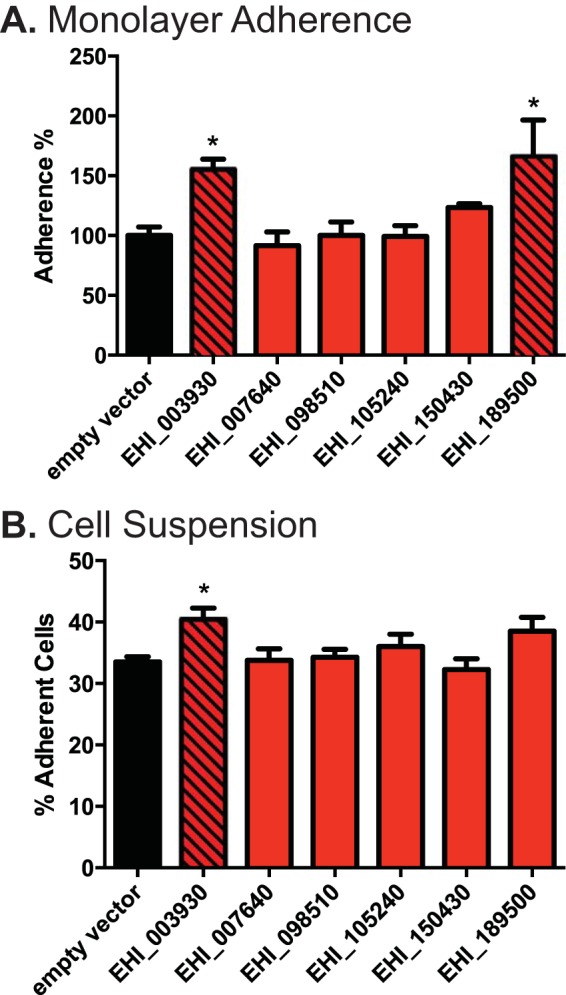
Host cell adherence and cytotoxicity. (A) Measured adherence of gene-silenced trophozoite strains to a cell monolayer. The adherence of each strain is calculated as a percentage of that for the empty vector-transfected control (black bar). (B) Measured adherence of gene-silenced trophozoite strains to Jurkat T lymphocytes in suspension, reported as the percentage of amoebae with three or more adherent host cells. Significance for all graphs was based on one-way analysis of variance with Dunnett's multiple-comparison test. *, P < 0.05.
Fluorescent microscopy.
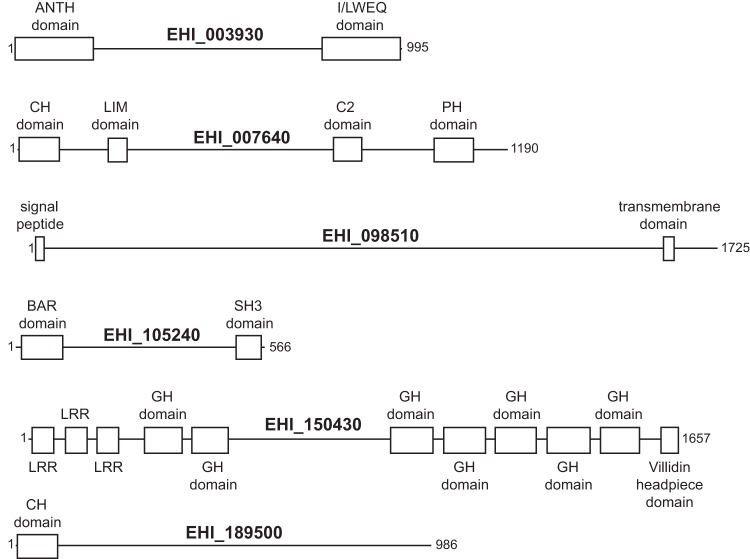
We selected EHI_003930 and EHI_105240 for the localization of their products, because silencing of these two E. histolytica genes produced the largest reductions in phagocytosis and sequence analysis of each indicated the presence of well-conserved protein domains with plausible roles in adherence and/or the control of membrane dynamics. We assigned names on the basis of their predicted domains (Fig. 3; see also Table S3 in the supplemental material for domain E values) and fluorescently labeled them to determine the cellular location of their protein products. EhILWEQ (EHI_003930) and EhBAR (EHI_105240) were both cloned into an E. histolytica-specific SNAP tag expression vector, which expresses each as a fusion with a codon-optimized SNAP protein (39). Trophozoites were then fluorescently labeled using a cell-permeant O6-benzylguanine-derivative SNAP protein substrate, which irreversibly binds to the SNAP protein. To model parasite infection, labeled trophozoites were applied to a confluent monolayer of human ileocecal carcinoma cells. Trophozoites proceeded to attack the cell monolayer and phagocytose the host cells. In contrast to a recent report that E. histolytica rejects cell corpses that it has killed (32), we observed both slow and fast phagocytosis under these conditions, with engulfment of cell pieces (i.e., trogocytosis) and frequent engulfment of apparently intact cells (see Movie S1 in the supplemental material).
FIG 3.
Mapping of predicted domains. All domains were predicted by use of the NCBI conserved domain database, unless noted differently below. Specific E values are given in Table S3 in the supplemental material. CH domain, calponin homology domain; GH domain, gelsolin homology domain; LRR, leucine-rich repeat domain; PH domain, pleckstrin homology domain. The signal peptide was predicted using the SignalP program, and the transmembrane domain was predicted using the TMHMM program (66, 71). The calponin homology domain on EHI_189500 was predicted through the use of AmoebaDB only.
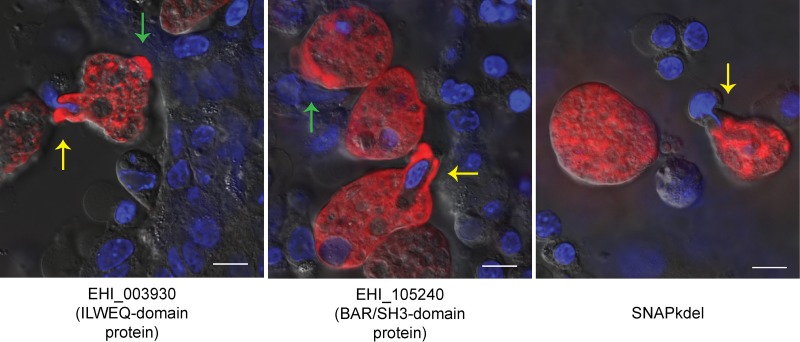
Both EhILWEQ and EhBAR localized to the majority of partially formed phagosomes (Fig. 4, yellow arrows) (6/6 and 10/12 observed events, respectively) but were typically not enriched in areas surrounding fully formed phagosomes (1/13 and 1/36 observed events, respectively). Both proteins were also present within extending pseudopodia (Fig. 4, green arrows). In contrast, trophozoites expressing the SNAP protein with an endoplasmic reticulum (ER) localization sequence (SNAPkdel) showed little to no staining at either of these locations.
FIG 4.
EhBAR (EHI_105240) and EhILWEQ (EHI_003930) localization via SNAP tag labeling. Entamoeba trophozoites expressing EhBAR- and EhILWEQ-SNAP fusion proteins were labeled and fixed following a 60-min incubation on an HCT-8 host cell monolayer. EhBAR and EhILWEQ show enrichment at the site of partially formed phagosomes (yellow arrows), whereas SNAP with an ER localization tag does not. EhBAR and EhILWEQ also show enrichment in extending pseudopodia (green arrows). HCT-8 cell nuclei were stained using cell-permeant Hoechst 33342 (blue), and SNAP fusion proteins were labeled using a cell-permeant TMR-Star substrate (red). Bars = 10 μm.
DISCUSSION
In this study, we used a moderate-throughput gene silencing method in combination with a battery of phenotypic assays to identify E. histolytica genes encoding proteins that function in amebic phagocytosis. A specific role for EhBAR and EhILWEQ during early stages of phagocytosis is implied by the localization of each to the early phagosome but not fully formed phagosomes, as well as by significant reductions in amebic phagocytosis following gene silencing. On the other hand, silencing of expression of either EhBAR or EhILWEQ did not affect amebic cytotoxicity. These data are significant because (i) consistent with the rapid engulfment of apoptotic cells and the engulfment of apparently whole cells seen in vivo (26–28), they provide further evidence that amebic cell killing and phagocytosis occur largely as a sequence of events, and (ii) they further validate previously published microarray data demonstrating the upregulated gene expression following phagocytosis that correlates with an increased phagocytic ability (33).
The genes selected for silencing were not randomly chosen from among those previously shown to be expressed at higher levels in phagocytic cells but, rather, were handpicked on the basis of functional annotation via the use of the NIH DAVID bioinformatics resource and the prediction of homologous domains by the use of Pfam (45, 46). Three of the genes chosen for silencing produced nonviable E. histolytica strains following two attempts at transfection, and it should be noted that these genes might be essential for E. histolytica survival. The six genes that resulted in viable strains after they were silenced and whose silencing showed a significant effect on the phenotype during preliminary screens are described below (Fig. 3 and Table 2).
TABLE 2.
Summary of phenotypes resulting from silencing of expression of each E. histolytica gene
| AmoebaDB identifier | Phenotypea |
||||
|---|---|---|---|---|---|
| Phagocytosis |
Adherence |
Cytotoxicity | |||
| Apoptotic cells | Live cells | Cell monolayer | Cell suspension | ||
| EHI_003930 | ▼ | ▼ | ▲ | — | — |
| EHI_007640 | ▼ | — | — | — | — |
| EHI_098510 | ▼ | — | — | — | — |
| EHI_105240 | ▼ | ▼ | — | — | — |
| EHI_150430 | ▼ | — | — | — | — |
| EHI_189500 | — | — | ▲ | — | — |
▼, downregulation; ▲, upregulation; —, no change in phenotype.
EhBAR (EHI_105240) is predicted to have both an N-terminal Bin-amphiphysin-Rvs (BAR) domain and a C-terminal src homology 3 (SH3) domain. BAR-domain proteins are highly conserved among eukaryotes (Pfam family BAR, Pfam accession no. PF03114]). Proteins with N-terminal amphipathic helical BAR (N-BAR) domains are thought to function as dimers, binding together to both drive and sense membrane curvature. N-BAR-containing proteins, such as the endophilins and the amphiphysins, function in endocytosis (47, 48). The human N-BAR-containing Bin2 protein was recently shown to affect motility, adhesion, and phagocytosis in leukocytes (49). SH3 domains have been shown to bind proline residues and are commonly found in proteins involved in tyrosine kinase signaling (50). E. histolytica contains 55 putative tyrosine kinases, 44 of which are putative receptor tyrosine kinases (51). In our earlier microarray analysis comparing gene regulation between phagocytic and nonphagocytic populations of E. histolytica trophozoites, the SH3 domain was found to be enriched in genes upregulated in phagocytic trophozoites (33).
EhILWEQ (EHI_003930) is predicted to have both I/LWEQ and ANTH domains. On the basis of a TBLASTN search of the translated NCBI nonredundant nucleotide database, proteins with high degrees of homology and the same domain structure are present in other Entamoeba and Dictyostelium species and numerous fungi. The I/LWEQ domain is conserved in proteins from single-celled eukaryotes to mammals and is capable of binding F actin in vivo. It is commonly found in the talin family of eukaryotic proteins that function in cell adhesion (52). The ANTH domain is known to bind the cell membrane and is commonly found in proteins that function in clathrin-mediated endocytosis (53). Although microscopic evidence shows that EhILWEQ localizes to the site of phagocytosis and shows no pattern of vesicular labeling (Fig. 4), we cannot rule out the possibility that EhILWEQ functions in trafficking of proteins within trophozoites.
EHI_150430 is a recently classified homolog of the Dictyostelium discoideum flightless-like 1 protein (FLIL1) (54). The putative protein contains seven gelsolin domains, a leucine-rich repeat domain, and a villidin headpiece domain. While the overall protein structure and arrangement of the domains is variable, villidin proteins are ubiquitous among eukaryotes ranging from single-celled organisms to humans. Gelsolin homology domains are found exclusively in actin binding proteins, so it appears that EHI_150430 also plays a role in cytoskeleton organization.
Our gene silencing screen also identified two hypothetical proteins, EHI_007640 and EHI_098510, that are potentially involved in phagocytosis. The former, EHI_007640, is highly conserved and has homologous C2, pleckstrin, LIM, and calponin domains. C2 domains function in targeting proteins to the cell membrane and bind intracellular calcium, a second messenger shown to be critical in Entamoeba virulence and phagocytosis (55–58). The pleckstrin homology domain is understood to function in phosphoinositide 3-kinase (PI3 kinase) signaling through binding of phosphatidylinositol lipids (59). Ghosh and Samuelson showed that treatment of E. histolytica with wortmannin, a fungal inhibitor of PI3 kinase, inhibited the phagocytosis of bacteria, red blood cells, and mucin-coated beads, strongly implicating PI3 kinase signaling in E. histolytica phagocytosis (60). In addition, Nakada-Tsukui et al. (61) and Byekova et al. (62) have demonstrated that phosphatidylinositol-3,4,5-trisphosphate, a PI3 kinase product, is present in phagosomal cups. LIM domains, which are ubiquitous in eukaryotes, consist of two zinc finger motifs separated by a short linker (63). The E. histolytica protein LimA (EhLimA) has been shown to localize to the plasma membrane and bind actin via its LIM domain (64). The calponin homology domain is also reported to bind F actin and microtubules (65). The other hypothetical protein, EHI_098510, appears to be specific to Entamoeba species and has no significant homologous domains. However, it is the only gene with a predicted signal peptide and a transmembrane domain that was silenced (predicted via the SignalP and TMHMM programs, respectively) (66). EHI_098510 was identified within E. histolytica phagosomes in two prior proteomics studies (67, 68).
EHI_189500 encodes a calponin homology domain containing a protein that is specific to Entamoeba species. This protein has a domain with weak homology to the aforementioned calponin domain. Silencing of EHI_189500 resulted in a strain of trophozoites with an increase in host cell adhesion but no detectable change in host cell engulfment.
One caveat of the microscopy studies demonstrating the localization of EhBAR and EhILWEQ to early phagosomes is that each protein was localized by expressing a SNAP-tagged protein using an episomal system. It is therefore conceivable that either the SNAP tag or protein overexpression caused aberrant protein localization. When these data are considered in the context of the effect of gene silencing on the phenotype (i.e., reduced phagocytosis), however, the localization of each protein to the phagosome is not surprising.
Although none of the silenced strains chosen for phenotypic characterization displayed a significant change in cytotoxicity, these results must also be interpreted with caution. The G3 strain of trophozoites used for silencing was originally described by Bracha et al. to be a unique strain of E. histolytica HM-1:IMSS capable of gene silencing through a then-unknown mechanism (34, 69). Zhang et al. then demonstrated that G3 trophozoites silence gene expression through both chromatin remodeling and activation of an RNA-induced transcriptional silencing complex (70). This method has proven invaluable for E. histolytica research, yet the G3 strain differs from the original HM-1:IMSS strain in that amoebapore proteins, small amphipathic proteins that form holes in target host cell membranes, are permanently silenced in the G3 strain. Specifically, amoebapores A and B are silenced, but amoebapore C is not. As a result, the G3 strain has decreased cytotoxicity compared to that of the HM-1:IMSS strain of E. histolytica, and therefore, any decrease in cytotoxicity would be in addition to the previous reduction (34). Nevertheless, G3 strain trophozoites retain some cytotoxic ability. In the current study, incubation of control G3 strain trophozoites with vital dye-labeled Jurkat T lymphocytes resulted in the specific release of approximately 17% of the total fluorescent signal after 30 min (see Fig. S4 in the supplemental material). This dye release reflects the cell membrane disruption that presumably occurs via an amoebapore-independent mechanism and may be due to the process of trogocytosis, during which host cell pieces are torn off and engulfed (32). Given that each system has limitations, the G3 strain proved invaluable when it was coupled with the custom Gateway silencing vector, which allowed us to silence many candidate genes over a short period of time.
Interestingly, we did not see the same decrease in phagocytosis rates when the uptake of live cells was compared to that of apoptotic host cells. If cell killing is to be considered a sequential process, then an assay of live cell phagocytosis measures the rate of cell adherence, cell killing, and initiation of phagocytosis and then the rate of cell engulfment via a common final pathway. In our in vitro assay, to induce contact, Jurkat T lymphocytes were centrifuged with E. histolytica, which facilitates adherence by putting cells in close proximity. Therefore, our in vitro assay of live cell phagocytosis measures the rate of cell killing, initiation of phagocytosis, and engulfment. In contrast, our assay of apoptotic phagocytosis measures only the rate of initiation of phagocytosis and engulfment. If the rate of cell killing is low compared to the rate of phagocytosis, cell killing may act as the rate-limiting step in the process of host cell killing and phagocytosis. Under these conditions, trogocytosis, which was previously described to be slow E. histolytica phagocytosis but which is essentially a mechanism of cell killing, likely predominates. On the other hand, previous work has shown that phosphatidylserine exposure on the outer cell membrane (a signal of host cell death) stimulates the phagocytosis of Jurkat T lymphocytes and host erythrocytes, and under these conditions, largely intact cells are rapidly engulfed (26, 29). Had initiation of phagocytosis been the rate-limiting step, one would expect that phosphatidylserine exposure would not affect the rate of lymphocyte or erythrocyte uptake. In the context of this previous work, we believe that our results concerning live and apoptotic host cell phagocytosis support cell killing as the rate-limiting step in the process of E. histolytica cell killing and phagocytosis (26, 29).
Finally, our results highlight the redundancy in the E. histolytica phagocytic machinery, since not one of the five strains in which genes were silenced and which showed defects in phagocytosis showed a complete lack of phagocytosis. Trophozoites in which EHI_150430 was silenced had the greatest defect at a 50% phagocytic index compared to that for the empty vector-transfected control. Despite this redundancy, said pathways appear to share a program of feed-forward regulation in response to phagocytosis, in which phagocytosis stimulates the expression of genes that enhance the phagocytic ability (33).
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of our laboratory, in particular, Jose Teixeira and Kovi Bessoff, for their helpful discussions. We also thank the University of Vermont DNA Facility for technical assistance.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01325-15.
REFERENCES
- 1.WHO. 1997. Amoebiasis. Wkly Epidemiol Rec 72:97–100. [PubMed] [Google Scholar]
- 2.Haque R, Huston CD, Hughes M, Houpt E, Petri WA. 2003. Amebiasis. N Engl J Med 348:1565–1573. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 3.Haque R, Ali IM, Petri WA Jr. 1999. Prevalence and immune response to Entamoeba histolytica infection in preschool children in Bangladesh. Am J Trop Med Hyg 60:1031–1034. [DOI] [PubMed] [Google Scholar]
- 4.Blessmann J, Ali IKM, Ton Nu PA, Dinh BT, Ngo Viet TQ, Le Van A, Clark CG, Tannich E. 2003. Longitudinal study of intestinal Entamoeba histolytica infections in asymptomatic adult carriers. J Clin Microbiol 41:4745–4750. doi: 10.1128/JCM.41.10.4745-4750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramos F, Moran P, Gonzalez E, Garcia G, Ramiro M, Gomez A, De Leon MDCG, Melendro EI, Valadez A, Ximenez C. 2005. High prevalence rate of Entamoeba histolytica asymptomatic infection in a rural Mexican community. Am J Trop Med Hyg 73:87–91. [PubMed] [Google Scholar]
- 6.Ankri S, Padilla-Vaca F, Stolarsky T, Koole L, Katz U, Mirelman D. 1999. Antisense inhibition of expression of the light subunit (35 kDa) of the Gal/GalNac lectin complex inhibits Entamoeba histolytica virulence. Mol Microbiol 33:327–337. doi: 10.1046/j.1365-2958.1999.01476.x. [DOI] [PubMed] [Google Scholar]
- 7.Dvorak JA, Kobayashi S, Nozaki T, Takeuchi T, Matsubara C. 2003. Induction of permeability changes and death of vertebrate cells is modulated by the virulence of Entamoeba spp. isolates. Parasitol Int 52:169–173. doi: 10.1016/S1383-5769(02)00090-9. [DOI] [PubMed] [Google Scholar]
- 8.Davis PH, Zhang X, Guo J, Townsend RR, Stanley SL Jr. 2006. Comparative proteomic analysis of two Entamoeba histolytica strains with different virulence phenotypes identifies peroxiredoxin as an important component of amoebic virulence. Mol Microbiol 61:1523–1532. doi: 10.1111/j.1365-2958.2006.05344.x. [DOI] [PubMed] [Google Scholar]
- 9.Sateriale A, Huston CD. 2011. A sequential model of host cell killing and phagocytosis by Entamoeba histolytica. J Parasitol Res 2011:926706. doi: 10.1155/2011/926706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravdin JI, Guerrant RL. 1981. Role of adherence in cytopathic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J Clin Invest 68:1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petri WA Jr, Chapman MD, Snodgrass TL, Mann BJ, Broman J, Ravdin JI. 1989. Subunit structure of the galactose and N-acetyl-d-galactosamine-inhibitable adherence lectin of Entamoeba histolytica. J Biol Chem 264:3007–3012. [PubMed] [Google Scholar]
- 12.Bruchhaus I, Jacobs T, Leippe M, Tannich E. 1996. Entamoeba histolytica and Entamoeba dispar: differences in numbers and expression of cysteine proteinase genes. Mol Microbiol 22:255–263. doi: 10.1046/j.1365-2958.1996.00111.x. [DOI] [PubMed] [Google Scholar]
- 13.Lidell ME, Moncada DM, Chadee K, Hansson GC. 2006. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci U S A 103:9298–9303. doi: 10.1073/pnas.0600623103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruchhaus I, Loftus BJ, Hall N, Tannich E. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot Cell 2:501–509. doi: 10.1128/EC.2.3.501-509.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ravdin JI, Croft BY, Guerrant RL. 1980. Cytopathogenic mechanisms of Entamoeba histolytica. J Exp Med 152:377–390. doi: 10.1084/jem.152.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petri WA Jr, Ravdin JI. 1987. Cytopathogenicity of Entamoeba histolytica: the role of amebic adherence and contact-dependent cytolysis in pathogenesis. Eur J Epidemiol 3:123–136. [DOI] [PubMed] [Google Scholar]
- 17.Huston CD, Houpt ER, Mann BJ, Hahn CS, Petri WA. 2000. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell Microbiol 2:617–625. doi: 10.1046/j.1462-5822.2000.00085.x. [DOI] [PubMed] [Google Scholar]
- 18.Ravdin JI, Moreau F, Sullivan JA, Petri WA Jr, Mandell GL. 1988. Relationship of free intracellular calcium to the cytolytic activity of Entamoeba histolytica. Infect Immun 56:1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragland BD, Ashley LS, Vaux DL, Petri WA Jr. 1994. Entamoeba histolytica: target cells killed by trophozoites undergo DNA fragmentation which is not blocked by Bcl-2. Exp Parasitol 79:460–467. doi: 10.1006/expr.1994.1107. [DOI] [PubMed] [Google Scholar]
- 20.Seydel KB, Stanley SL Jr. 1998. Entamoeba histolytica induces host cell death in amebic liver abscess by a non-Fas-dependent, non-tumor necrosis factor alpha-dependent pathway of apoptosis. Infect Immun 66:2980–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trissl D, Martinez-Palomo A, de la Torre M, de la Hoz R, de Suarez EP. 1978. Surface properties of Entamoeba: increased rates of human erythrocyte phagocytosis in pathogenic strains. J Exp Med 148:1137–1145. doi: 10.1084/jem.148.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orozco E, Guarneros G, Martinez-Palomo A. 1983. Entamoeba histolytica: phagocytosis as a virulence factor. J Exp Med 158:1511. doi: 10.1084/jem.158.5.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez MA, Orozco E. 1986. Isolation and characterization of phagocytosis- and virulence-deficient mutants of Entamoeba histolytica. J Infect Dis 154:27–32. doi: 10.1093/infdis/154.1.27. [DOI] [PubMed] [Google Scholar]
- 24.Lejeune A, Gicquaud C. 1987. Evidence for two mechanisms of human erythrocyte endocytosis by Entamoeba histolytica-like amoebae (Laredo strain). Biol Cell 59:239–245. doi: 10.1111/j.1768-322X.1987.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 25.Lejeune A, Gicquaud C. 1992. Target cell deformability determines the type of phagocytic mechanism used by Entamoeba histolytica-like, Laredo strain. Biol Cell 74:211–216. doi: 10.1016/0248-4900(92)90027-X. [DOI] [PubMed] [Google Scholar]
- 26.Huston CD, Boettner DR, Miller-Sims V, Petri WA. 2003. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect Immun 71:964–972. doi: 10.1128/IAI.71.2.964-972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin JL. 1972. Human amebic dysentery: electron microscopy of Entamoeba histolytica contacting, ingesting, and digesting inflammatory cells. Am J Trop Med Hyg 21:895–906. [PubMed] [Google Scholar]
- 28.Gonzalez-Ruiz A, Haque R, Aguirre A, Castanon G, Hall A, Guhl F, Ruiz-Palacios G, Miles MA, Warhurst DC. 1994. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J Clin Pathol 47:236–239. doi: 10.1136/jcp.47.3.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boettner DR, Huston CD, Sullivan JA, Petri WA. 2005. Entamoeba histolytica and Entamoeba dispar utilize externalized phosphatidylserine for recognition and phagocytosis of erythrocytes. Infect Immun 73:3422–3430. doi: 10.1128/IAI.73.6.3422-3430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira JE, Heron BT, Huston CD. 2008. C1q- and collectin-dependent phagocytosis of apoptotic host cells by the intestinal protozoan Entamoeba histolytica. J Infect Dis 198:1062–1070. doi: 10.1086/591628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaithilingam A, Teixeira JE, Miller PJ, Heron BT, Huston CD. 2012. Entamoeba histolytica cell surface calreticulin binds human C1q and functions in amebic phagocytosis of host cells. Infect Immun 80:2008–2018. doi: 10.1128/IAI.06287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ralston KS, Solga MD, Mackey-Lawrence NM, Somlata Bhattacharya A, Petri WA Jr. 2014. Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature 508:526–530. doi: 10.1038/nature13242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sateriale A, Vaithilingam A, Donnelly L, Miller P, Huston CD. 2012. Feed-forward regulation of phagocytosis by Entamoeba histolytica. Infect Immun 80:4456–4462. doi: 10.1128/IAI.00671-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bracha R, Nuchamowitz Y, Mirelman D. 2003. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot Cell 2:295–305. doi: 10.1128/EC.2.2.295-305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buss SN, Hamano S, Vidrich A, Evans C, Zhang Y, Crasta OR, Sobral BW, Gilchrist CA, Petri WA Jr. 2010. Members of the Entamoeba histolytica transmembrane kinase family play non-redundant roles in growth and phagocytosis. Int J Parasitol 40:833–843. doi: 10.1016/j.ijpara.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixeira JE, Huston CD. 2008. Participation of the serine-rich Entamoeba histolytica protein in amebic phagocytosis of apoptotic host cells. Infect Immun 76:959–966. doi: 10.1128/IAI.01455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berninghausen O, Leippe M. 1997. Necrosis versus apoptosis as the mechanism of target cell death induced by Entamoeba histolytica. Infect Immun 65:3615–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fadok VA, Voelker DR, Campbell PA, Cohen JJ, Bratton DL, Henson PM. 1992. Exposure of phosphatidylserine on the surface of apoptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148:2207–2216. [PubMed] [Google Scholar]
- 39.Sateriale A, Roy NH, Huston CD. 2013. SNAP-tag technology optimized for use in Entamoeba histolytica. PLoS One 8:e83997. doi: 10.1371/journal.pone.0083997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Penuliar GM, Furukawa A, Sato D, Nozaki T. 2011. Mechanism of trifluoromethionine resistance in Entamoeba histolytica. J Antimicrob Chemother 66:2045–2052. doi: 10.1093/jac/dkr238. [DOI] [PubMed] [Google Scholar]
- 41.Teixeira JE, Sateriale A, Bessoff KE, Huston CD. 2012. Control of Entamoeba histolytica adherence involves metallosurface protease 1, an M8 family surface metalloprotease with homology to leishmanolysin. Infect Immun 80:2165–2176. doi: 10.1128/IAI.06389-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim S, Landy A. 1992. Lambda Int protein bridges between higher order complexes at two distant chromosomal loci attL and attR. Science 256:198–203. doi: 10.1126/science.1533056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravdin JI, Guerrant RL. 1982. Separation of adherence, cytolytic, and phagocytic events in the cytopathogenic mechanisms of Entamoeba histolytica. Arch Invest Med (Mex) 13:123–128. [PubMed] [Google Scholar]
- 44.Leippe M. 1997. Amoebapores. Parasitol Today 13:178–183. doi: 10.1016/S0169-4758(97)01038-7. [DOI] [PubMed] [Google Scholar]
- 45.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res 40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 48.Ringstad N, Gad H, Low P, Di Paolo G, Brodin L, Shupliakov O, De Camilli P. 1999. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron 24:143–154. doi: 10.1016/S0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez-Barrena MJ, Vallis Y, Clatworthy MR, Doherty GJ, Veprintsev DB, Evans PR, McMahon HT. 2012. Bin2 is a membrane sculpting N-BAR protein that influences leucocyte podosomes, motility and phagocytosis. PLoS One 7:e52401. doi: 10.1371/journal.pone.0052401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koch CA, Anderson D, Moran MF, Ellis C, Pawson T. 1991. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science 252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 51.Anamika K, Bhattacharya A, Srinivasan N. 2008. Analysis of the protein kinome of Entamoeba histolytica. Proteins 71:995–1006. doi: 10.1002/prot.21790. [DOI] [PubMed] [Google Scholar]
- 52.McCann RO, Craig SW. 1997. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci U S A 94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. 2004. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci 117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 54.Ghoshdastider U, Popp D, Burtnick LD, Robinson RC. 2013. The expanding superfamily of gelsolin homology domain proteins. Cytoskeleton (Hoboken) 70:775–795. doi: 10.1002/cm.21149. [DOI] [PubMed] [Google Scholar]
- 55.Zhang D, Aravind L. 2010. Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes. Gene 469:18–30. doi: 10.1016/j.gene.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Christy NC, Petri WA. 2011. Mechanisms of adherence, cytotoxicity and phagocytosis modulate the pathogenesis of Entamoeba histolytica. Future Microbiol 6:1501–1519. doi: 10.2217/fmb.11.120. [DOI] [PubMed] [Google Scholar]
- 57.Gilchrist CA, Moore ES, Zhang Y, Bousquet CB, Lannigan JA, Mann BJ, Petri WA. 2010. Regulation of virulence of Entamoeba histolytica by the URE3-BP transcription factor. mBio 1:e00057-10. doi: 10.1128/mBio.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ravdin JI, Murphy CF, Guerrant RL, Long-Krug SA. 1985. Effect of antagonists of calcium and phospholipase A on the cytopathogenicity of Entamoeba histolytica. J Infect Dis 152:542–549. doi: 10.1093/infdis/152.3.542. [DOI] [PubMed] [Google Scholar]
- 59.Lemmon MA. 2008. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 60.Ghosh SK, Samuelson J. 1997. Involvement of p21racA, phosphoinositide 3-kinase, and vacuolar ATPase in phagocytosis of bacteria and erythrocytes by Entamoeba histolytica: suggestive evidence for coincidental evolution of amebic invasiveness. Infect Immun 65:4243–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakada-Tsukui K, Okada H, Mitra BN, Nozaki T. 2009. Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica. Cell Microbiol 11:1471–1491. doi: 10.1111/j.1462-5822.2009.01341.x. [DOI] [PubMed] [Google Scholar]
- 62.Byekova YA, Powell RR, Welter BH, Temesvari LA. 2010. Localization of phosphatidylinositol (3,4,5)-trisphosphate to phagosomes in Entamoeba histolytica achieved using glutathione S-transferase- and green fluorescent protein-tagged lipid biosensors. Infect Immun 78:125–137. doi: 10.1128/IAI.00719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kadrmas JL, Beckerle MC. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 64.Wender N, Villalobo E, Mirelman D. 2007. EhLimA, a novel LIM protein, localizes to the plasma membrane in Entamoeba histolytica. Eukaryot Cell 6:1646–1655. doi: 10.1128/EC.00177-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sjoblom B, Ylanne J, Djinovic-Carugo K. 2008. Novel structural insights into F-actin-binding and novel functions of calponin homology domains. Curr Opin Struct Biol 18:702–708. doi: 10.1016/j.sbi.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 67.Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, Sherman NE, Petri WA. 2008. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog 4:e8. doi: 10.1371/journal.ppat.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marion S, Laurent C, Guillen N. 2005. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cell Microbiol 7:1504–1518. doi: 10.1111/j.1462-5822.2005.00573.x. [DOI] [PubMed] [Google Scholar]
- 69.Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. 2006. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog 2:e48. doi: 10.1371/journal.ppat.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang H, Alramini H, Tran V, Singh U. 2011. Nucleus-localized antisense small RNAs with 5′-polyphosphate termini regulate long term transcriptional gene silencing in Entamoeba histolytica G3 strain. J Biol Chem 286:44467–44479. doi: 10.1074/jbc.M111.278184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175–182. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.