Abstract
Active invasion into nonphagocytic host cells is central to Salmonella enterica pathogenicity and dependent on multiple genes within Salmonella pathogenicity island 1 (SPI-1). Here, we explored the invasion phenotype and the expression of SPI-1 in the typhoidal serovar S. Paratyphi A compared to that of the nontyphoidal serovar S. Typhimurium. We demonstrate that while S. Typhimurium is equally invasive under both aerobic and microaerobic conditions, S. Paratyphi A invades only following growth under microaerobic conditions. Transcriptome sequencing (RNA-Seq), reverse transcription-PCR (RT-PCR), Western blot, and secretome analyses established that S. Paratyphi A expresses much lower levels of SPI-1 genes and secretes lesser amounts of SPI-1 effector proteins than S. Typhimurium, especially under aerobic growth. Bypassing the native SPI-1 regulation by inducible expression of the SPI-1 activator, HilA, considerably elevated SPI-1 gene expression, host cell invasion, disruption of epithelial integrity, and induction of proinflammatory cytokine secretion by S. Paratyphi A but not by S. Typhimurium, suggesting that SPI-1 expression is naturally downregulated in S. Paratyphi A. Using streptomycin-treated mice, we were able to establish substantial intestinal colonization by S. Paratyphi A and showed moderately higher pathology and intestinal inflammation in mice infected with S. Paratyphi A overexpressing hilA. Collectively, our results reveal unexpected differences in SPI-1 expression between S. Paratyphi A and S. Typhimurium, indicate that S. Paratyphi A host cell invasion is suppressed under aerobic conditions, and suggest that lower invasion in aerobic sites and suppressed expression of immunogenic SPI-1 components contributes to the restrained inflammatory infection elicited by S. Paratyphi A.
INTRODUCTION
Salmonella enterica is a highly diverse and ubiquitous pathogen containing more than 2,600 different serovars classified by their antigenic presentation (1). Salmonella serovars differ by their host specificity and by the clinical syndromes they cause, ranging from asymptomatic carriage to invasive systemic disease. While many nontyphoidal Salmonella (NTS) serovars, such as Typhimurium and Enteritidis, are generalist pathogens with broad host specificity, a few S. enterica serovars, including Typhi, Sendai, and Paratyphi A, are highly adapted to humans. These specialist pathogens, collectively referred to as typhoidal Salmonella serovars, are the causative agents of enteric fever, posing an estimated global annual burden of over 27 million cases, resulting in more than 200,000 deaths (2). While NTS cause inflammatory gastroenteritis that is confined to the terminal ileum and colon in immunocompetent patients, typhoidal serovars do not induce a strong inflammatory response during the initial invasion of the intestinal mucosa (3–5). This noninflammatory phase associated with typhoidal infections is thought to facilitate its dissemination to systemic sites (6).
Regardless of the clinical manifestation, both typhoidal and NTS serovars initially adhere to and invade the intestinal epithelium of the small intestine (7), and active invasion into nonphagocytic cells is pivotal for the pathogenicity of all Salmonella serovars. The main mechanism used by Salmonella to enter host cells is the trigger mechanism, which induces cytoskeletal rearrangements known as membrane ruffles (8). This pathway is mediated by the evolutionarily conserved type three secretion system (T3SS) on Salmonella pathogenicity island 1 (SPI-1) and a wide array of translocated effectors. These virulence proteins are directly injected into the host cell cytoplasm and facilitate Salmonella engulfment and transport through the intestinal barrier (9). The expression of the T3SS-1 and its related genes is affected by environmental conditions and is tightly controlled by different regulators, including HilA, which functions as the master regulator of SPI-1.
Following Salmonella invasion into the lamina propria, pathogen-associated molecular patterns (PAMPs) are detected by host pattern recognition receptors. PAMPs such as lipopolysaccharide (LPS), flagellin, and bacterial DNA can trigger Toll-like receptor 4 (TLR4), TLR5, and TLR9, respectively. PAMPs also are recognized by cytosolic NOD-like receptors (NLR), including NLRC4, which is triggered by the T3SS-1 components PrgJ and PrgI and flagellin. TLR and NLR signaling leads to the activation of the innate immune system and inflammatory response by secretion of proinflammatory cytokines and recruitment of neutrophils and macrophages to the site of infection (reviewed in references 10 and 11). In recent years, the SPI-7-encoded Vi capsular polysaccharide and its associated regulator, TviA (which also functions as a negative regulator of flagella and T3SS-1 genes), were shown to play a key role in S. Typhi virulence and in its ability to evade the innate immune response (12–14). Nevertheless, since SPI-7 is absent from S. Paratyphi A, the mechanisms underlying the typhoid-like behavior of S. Paratyphi A are unclear.
In order to better understand the virulence phenotype of S. Paratyphi A, we sought to compare the invasive phenotype and the expression pattern of SPI-1 in S. Paratyphi A to those of S. Typhimurium. Given that the clinical outcome of human infection by these serovars is markedly different, we hypothesized that their host cell invasion is regulated differently. Here, we established that while both serovars are hyperinvasive when grown under microaerobic conditions, S. Paratyphi A is noninvasive when grown aerobically. Furthermore, we found that S. Paratyphi A expresses lower levels of SPI-1 genes and secretes lesser amounts of SPI-1 effectors than S. Typhimurium. The inducible expression of HilA in S. Paratyphi A resulted in a much higher level of SPI-1 gene expression, improved invasion into epithelial cells, stronger disruption of epithelial integrity, and induction of proinflammatory cytokine secretion. Collectively, our results reveal unexpected differences in the expression amplitude of SPI-1 genes and the invasion phenotype between S. Paratyphi A and S. Typhimurium. We propose that the observed differences contribute to the distinct disease manifestations caused by NTS and typhoidal Salmonella.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains utilized in this study are listed in Table S1 in the supplemental material. Bacterial cultures were routinely maintained in Lennox Luria-Bertani (LB; containing 86.2 mM NaCl; BD Difco) medium at 37°C supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, 25 μg/ml chloramphenicol, and 170 mM or 300 mM NaCl where indicated. S. Typhimurium SL1344 and S. Paratyphi A 45157 were grown aerobically to the late logarithmic phase by diluting an overnight culture 1:100 into 250-ml flasks containing 10 ml fresh LB and incubating it at 37°C with shaking (300 rpm) for 2.5 h, reaching an optical density at 600 nm (OD600) of 1.2 to 1.5. To obtain microaerobic stationary-phase cultures, bacteria that were grown for 8 h in LB with aeration were diluted 1:100 into 10 ml fresh LB and incubated in 15-ml tubes (caps were on but not tightly screwed) without shaking for 16 h. To obtain microaerobic late-logarithmic-phase bacteria, overnight cultures were diluted 1:100 and grown for 2 h in LB with aeration. Ten-milliliter aliquots from the aerobically grown cultures were placed in 15-ml tubes that were incubated in a sealed gas jar with a CampyGen 2.5-liter sachet to create microaerobic conditions (6.2% to 13.2% O2) and grown in the jar for an additional 3 h.
Cloning and mutant construction.
All primers used in this study are listed in Table S2 in the supplemental material. The gene hilA from S. Typhimurium SL1344 was cloned under an arabinose-inducible promoter into the low-copy-number vector pBAD18 and introduced into S. Paratyphi A. C-terminally two-hemagglutinin (2HA)-tagged versions of SipB, SopB, SptP, SteA, SopE2, and PrgJ from S. Typhimurium or S. Paratyphi A were constructed within pWSK29 or pACYC184.
Tissue culture.
All cell lines were purchased from the American Type Culture Collection. The Caco-2 cell line was grown in Dulbecco's modified Eagle's medium (DMEM)–F-12 medium supplemented with 20% FBS and 2 mM l-glutamine. HeLa cells were cultured in high-glucose (4.5 g/liter) DMEM supplemented with 10% heat-inactivated FBS, 1 mM pyruvate, and 2 mM l-glutamine. Epithelial cells were seeded at 5 × 104 in a 24-well tissue culture dish 18 to 24 h before bacterial infection. Host cells were infected with Salmonella cultures at a multiplicity of infection (MOI) of ∼1:50. Infection experiments were carried out using the gentamicin protection assay as previously described (15). Salmonella invasion was determined by the number of intracellular Salmonella cells at 2 h postinfection (p.i.) divided by the number of infecting bacteria. Adhesion was determined using cytochalasin D to inhibit actin cytoskeleton rearrangement and inhibit bacterial cell invasion in an actin-dependent manner. Cells were incubated with fresh medium containing 1 μg/ml cytochalasin D 1 h before starting the infection. Bacteria were added and allowed to adhere for 30 min in the presence of 1 μg/ml cytochalasin D. Cells were washed four times with phosphate-buffered saline (PBS) and harvested by the addition of lysis buffer. Salmonella adhesion was determined by the number of adherent Salmonella cells at 30 min p.i. divided by the number of infecting bacteria. All cell lines were cultured at 37°C in a humidified atmosphere with 5% CO2.
TEER and cytokine secretion.
The epithelial integrity of Caco-2 cells seeded (2.5 × 105 cells/cm2) on polycarbonate, 0.4-μm-pore-size tissue culture inserts in 24-well plates (Corning Life Sciences) was verified by measuring the transepithelial electrical resistance (TEER) with an epithelial volt-ohm meter (World Precision Instruments). Medium was changed every other day during the polarization process, and cells were infected after 21 days, when a minimum TEER of ≥1,500 Ω cm−2 was achieved. For invasion assay, filter-grown monolayers (apical volume, 200 μl; basolateral volume, 600 μl) first were equilibrated with DMEM for 2 h under cell culture conditions (37°C, 5% CO2). Subsequently, apical DMEM was removed and replaced with a 100-μl volume of DMEM containing bacteria. After exposure to Salmonella for 2 h, monolayers were washed once with 800 ml of DMEM with 100 μg/ml gentamicin, and incubation was continued for 1 h. Changes in TEER were measured 0, 3, 6, 8, 10, and 12 h p.i. For cytokine release assays, Caco-2 cell supernatants were removed 2 h after the addition of Salmonella and assayed for interleukin-8 (IL-8) and macrophage inflammatory protein 3 alpha (Mip3α) via a sandwich enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (RayBio human IL-8 and Mip3α ELISA kits).
Western blotting.
Bacterial pellets were resuspended in 1× SDS-PAGE sample buffer. Boiled samples were separated on 12% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane (Bio-Rad Laboratories). Blots were probed with anti-2HA antibody (ab18181; Abcam) or anti-DnaK antibody (ab69617; Abcam). Goat anti-mouse antibody conjugated to horseradish peroxidase (ab6721; Abcam) was used as a secondary antibody, followed by detection with enhanced chemiluminescence reagents (Amersham Pharmacia).
Analysis of secreted proteins.
Overnight cultures were diluted 1:100 in LB and grown for about 5.5 h to an OD600 of 2.4 to 2.6. Eight milligrams of human cytochrome C was added to five milliliters of OD-normalized cultures as a spike-in control, followed by centrifugation at 13,000 rpm for 5 min. The obtained supernatant was filtered through a 0.22-mm syringe filter, and prechilled trichloroacetic acid (TCA) was added to a final concentration of 10%. The supernatants were placed on ice overnight and then centrifuged at 13,000 rpm for 45 min at 4°C. The pellets were washed with acetone, allowed to dry, and dissolved in SDS sample buffer. All protein samples were boiled for 5 min, and equal volumes of 25 μl from all samples were separated on 12% polyacrylamide gels using SDS-PAGE. Bands were visualized by staining with 0.05% Coomassie brilliant blue.
Mass spectrometry analysis.
Secreted fractions obtained from S. Typhimurium SL1344 and S. Paratyphi A 45157 cultures grown aerobically in LB for 5.5 h were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a Q Exactive plus mass spectrometer (Thermo). The mass spectrometry data were analyzed and quantified using MaxQuant software 1.5 (www.maxquant.org) for peak picking identification and quantitation using the Andromeda search engine, searching against the S. Paratyphi A or S. Typhimurium sections of the UniProt database as previously described (15). Data transformation (log2) and statistical tests were done using Preseus 1.4.0.20 software.
RT-PCR.
For reverse transcription-PCR (RT-PCR), RNA was extracted from Salmonella cultures grown aerobically to the late logarithmic phase using the Qiagen RNAprotect bacteria reagent and the RNeasy minikit (Qiagen) according to the manufacturer's instructions, including an on-column DNase digest. Purified RNA was treated secondarily with RNase-free DNase I followed by ethanol precipitation, and 200 ng of DNase I-treated RNA was subjected to a first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad Laboratories). Real-time PCRs and data analysis were performed as recently described (15).
Transcriptome analysis.
Total RNA was extracted using the RNeasy minikit (Qiagen) from two independent replicate cultures (at different days) of S. Typhimurium SL1344 and S. Paratyphi A 45157 grown aerobically to the late logarithmic phase (OD600 of 1.2 to 1.5). Total RNA also was extracted from two independent cultures grown to stationary phase in LB supplemented with 0.17 M NaCl under microaerobic conditions as explained above. rRNA was subtracted from 5 μg total RNA using the Ribo-Zero rRNA removal kit (bacteria) (MRZMB126; Illumina, San Diego, CA) by following the manufacturer's recommendations. One hundred nanograms of the subtracted RNA was used as the input to make a sequencing library using the TruSeq RNA sample preparation kit v2 (RS-122-2001; Illumina, San Diego CA) by following the manufacturer's recommendations. The RNA libraries were sequenced on an Illumina HiSeq 2500 in the paired-end mode with a read length of 100 bp at the UCI Genomics High-Throughput Facility (University of California Irvine, Irvine, CA). A total of ∼153 million paired-end reads were obtained across the eight RNA libraries (2 serovars by 2 growth conditions by 2 biological repeats), of which, depending on the samples, 59 to 99.5% were non-rRNA reads. Adaptor removal and quality trimming were performed as previously explained (15). The quality-trimmed reads were aligned to the respective reference genomes using Bowtie2 (ver 2.0.6) (16). Read counts within annotated features were generated using featureCounts (17). Genes differentially expressed between the two serovars were identified using GLM models as implemented in edgeR (18). Trimmed mean of M value (TMM) normalization as implemented in edgeR was performed to eliminate composition biases between libraries and normalized transcriptome sequencing (RNA-Seq) counts.
Virulence experiments in mice.
Six- to seven-week-old female C57/BL6 mice were purchased from Harlan Laboratories and housed at the Sheba Medical Center animal facility under specific-pathogen-free conditions. Experiments in this study were approved and carried out according to the national animal care guidelines and the institutional ethics committee of the Sheba Medical Center (approval no. 601/10). Groups of 5 or 6 mice were given streptomycin (20 mg per mouse) by oral gavage 24 h prior to infection with 1 × 108 to 2 × 108 CFU in 0.2 ml saline. Bacterial cultures were grown aerobically to the late logarithmic phase in LB supplemented with 100 μg/ml ampicillin and 50 mM arabinose. Animals were supplied with food and water ad libitum. At 1 and 3 days p.i., mice were sacrificed and tissue samples from the intestinal tract, spleen, and liver were aseptically removed, homogenized in 0.8 ml saline using a BeadBlaster 24 homogenizer (Benchmark Scientific), and plated on xylose lysine deoxycholate (XLD) agar plates supplemented with 100 μg/ml ampicillin for CFU enumeration. For mice sacrificed at 72 h p.i., 1 mg/ml arabinose and 0.3 mg/ml ampicillin were added to drinking water.
Histological procedures.
Tissues were fixed in 10% neutral buffered formalin and then embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin (H&E). Tissue pathology was scored as previously described (19). Briefly, histopathological changes were quantified by scoring the presence of luminal epithelial cells or neutrophils, the integrity of the epithelial barrier, the level of infiltrating immune cells in the mucosa or submucosa, and the formation of submucosal edema.
Immunofluorescence.
Formalin-fixed tissue sections (5 μm) were deparaffinized and rehydrated. After antigen retrieval in citrate buffer and blocking, slides were incubated with anti-S. Typhimurium antiserum (BD Biosciences), anti-CD68 (Abcam, Cambridge, United Kingdom), and anti-myeloperoxidase (MPO) (Thermo Fisher Scientific, Schwerte, Germany) antibodies, followed by fluorescently labeled secondary antibodies (Invitrogen, Carlsbad, CA). Nuclei were stained using 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Images were obtained and analyzed with an Axio Observer microscope and AxioVision software (Carl Zeiss AG, Oberkochen, Germany).
BioProject accession number.
RNA-Seq data were deposited in the Sequence Read Archive at the NCBI under BioProject number PRJNA304665.
RESULTS
Host cell invasion by S. Paratyphi A is impaired under aerobic conditions.
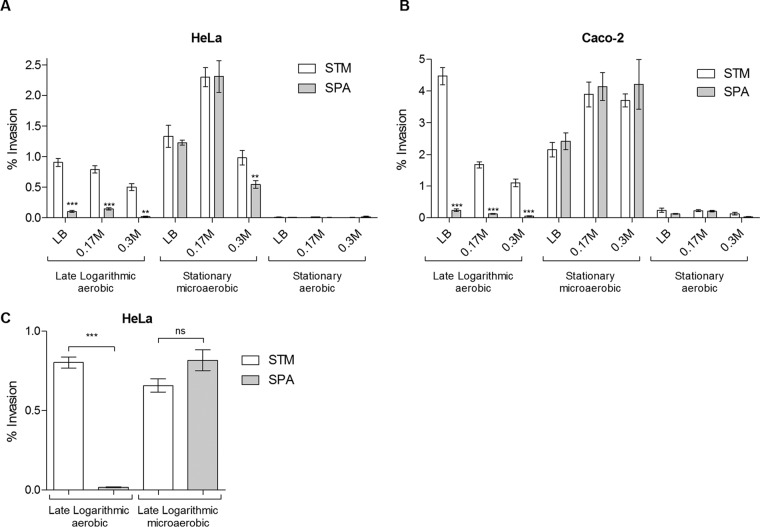
Considering the pivotal role of nonphagocytic host cell invasion in the pathogenicity of all S. enterica serovars (recently reviewed in reference 20), we asked if the typhoidal serovar S. Paratyphi A and the nontyphoidal serovar S. Typhimurium differ in their primary ability to invade epithelial cells. Previously, different growth conditions were shown to affect S. enterica invasion in vitro, including growth phase (21, 22), osmolarity of the medium (23–25), and oxygen concentration (24, 26). Thus, the phenotypes of S. Paratyphi A and S. Typhimurium invasion into HeLa and Caco-2 epithelial cell lines were compared under various growth phase (late logarithmic versus stationary), osmolarity (Lennox LB broth supplemented with 0, 0.17, and 0.3 M NaCl), and oxygen level (aerobic versus microaerobic) conditions. These experiments showed similar results in both cell lines, yielding the following summary: (i) both serovars are noninvasive when grown to the stationary phase under aerobic conditions; (ii) both serovars are hyperinvasive when grown to the stationary phase under microaerobic conditions; and (iii) while S. Typhimurium was highly invasive when grown aerobically to the late logarithmic phase in LB, S. Paratyphi A was noninvasive under these conditions (Fig. 1A and B). Remarkably, noninvasive S. Paratyphi A cultures that were grown to the logarithmic phase under aerobic conditions were able to invade to levels similar to those of S. Typhimurium just by transferring the culture to a microaerobic environment for 3 h prior to cell infection (Fig. 1C). We concluded from these experiments that S. Paratyphi A, in contrast to S. Typhimurium, is not invasive under aerobic growth conditions and that the invasive phenotype is induced under microaerobic environments.
FIG 1.
Effect of environmental conditions on S. Paratyphi A and S. Typhimurium invasion. Salmonella strains were grown at 37°C in Lennox LB supplemented with 0, 0.17, and 0.3 M NaCl. Cultures were grown for 16 h aerobically to an OD600 of 4.5 (stationary aerobic), diluted 1:100, grown for 2.5 h under aerobic conditions to an OD600 of 1.2 to 1.4 (late logarithmic aerobic) or for 16 h statically in capped tubes to an OD600 of 0.6 (stationary microaerobic), and used to infect HeLa cells (A) or Caco-2 cells (B). Data represent the means and standard errors of the means (SEM) from at least three biological replicates under each condition. (C) Overnight S. Typhimurium SL1344 and S. Paratyphi A 45157 cultures were diluted 1:100 into fresh LB broth and grown for 2 h to early logarithmic phase under aerobic conditions. At 2 h postinoculation, these cultures were split in two. One portion of the culture continued growth under aerobic conditions for an additional 1 h (growing for 3 h in total to an OD600 of 1.2), and the other portion was transferred to microaerobic conditions for 3 more hours (growing for 5 h in total to an OD600 of 0.6). All four cultures were used to infect HeLa cells. Data represent the means and SEM from 11 biological replicates. In all experiments, invasion was determined at 2 h postinfection using the gentamicin protection assay and is shown as the percentage of intracellular bacteria (CFU) from the infection inoculum. An unpaired t test with two tails was used to determine the significance of the differences between S. Typhimurium (STM) and S. Paratyphi A (SPA) invasion. **, P < 0.01; ***, P < 0.0001; ns, not significant.
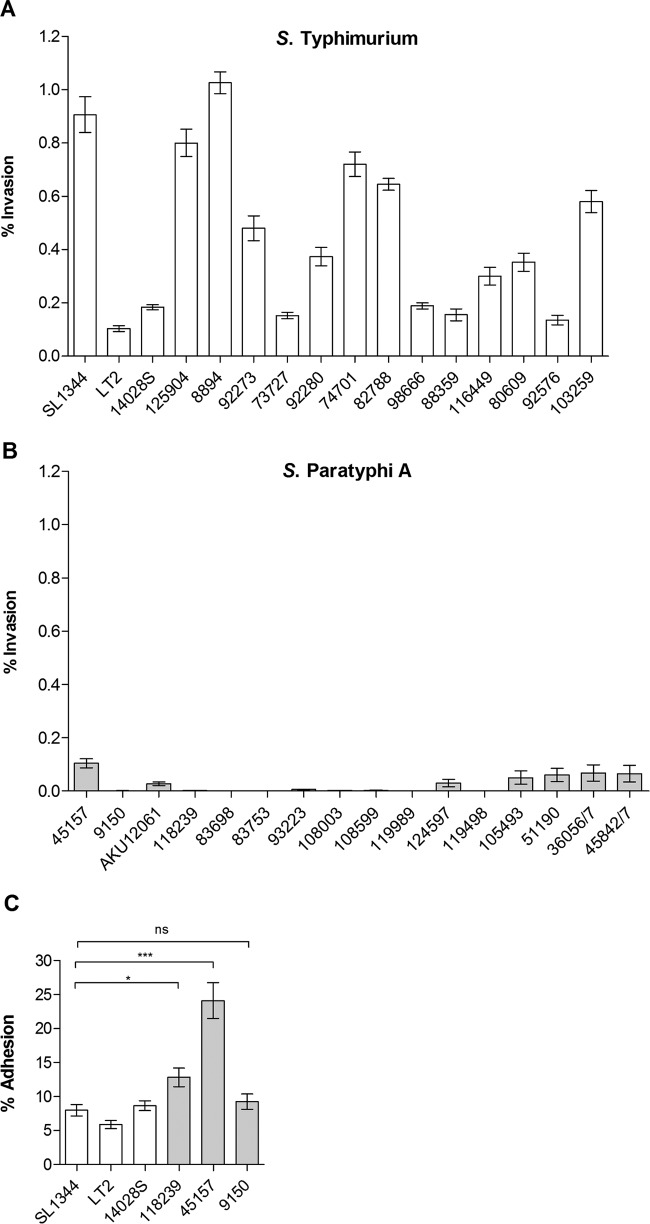
Poor invasion of S. Paratyphi A grown aerobically to the late exponential phase was unexpected, given that these conditions are known to be effective SPI-1-inducing conditions for S. Typhimurium (22, 27). To ensure that the impaired invasion of S. Paratyphi A is not specific to the examined strain (45157) only, we further compared 16 S. Paratyphi A and 16 S. Typhimurium strains for their ability to invade HeLa cells under the above-described conditions. Although some intraserovar variation in the extent of the invasion was observed among S. Typhimurium strains (with exceptionally low invasion by the LT2 strain, which is attenuated due to an altered rpoS allele [28]), serovar Typhimurium presented an average invasion rate of 0.5%. In contrast, a universally impaired invasion of all 16 S. Paratyphi A strains was seen, presenting, on average, a 17-fold lower level of invasion than S. Typhimurium (P < 0.0001) (Fig. 2A and B). We concluded from these experiments that while S. Typhimurium and S. Paratyphi A invade nonphagocytic cells similarly well when grown under microaerobic conditions, when grown aerobically to the late exponential phase, S. Paratyphi A, but not S. Typhimurium, is markedly impaired in host cell invasion.
FIG 2.
Invasion of S. Paratyphi A is impaired at the logarithmic phase under aerobic conditions. Sixteen S. Typhimurium strains (A) and 16 S. Paratyphi A strains (B) were grown in LB aerobically to the late logarithmic phase (OD600 of 1.2 to 1.4). Invasion was determined at 2 h p.i. using the gentamicin protection assay. (C) Adhesion was determined in the presence of cytochalasin D and is shown as the percentage of cell-associated bacteria from the total number of CFU used to infect the cells. Bars represent the mean adhesion and SEM from at least three biological replicates. *, P < 0.05; ***, P < 0.0001; ns, not significant.
To exclude the possibility that the impaired invasion of S. Paratyphi A is due to the lower ability of this pathogen to attach to host cells, for example, due to different composition or different expression levels of fimbrial (21) or nonfimbrial (29) adhesins, we infected HeLa cells in the presence of cytochalasin D, which allows Salmonella adhesion but not invasion into epithelial cells. Different S. Paratyphi A strains displayed similar or even superior adhesion compared to that of S. Typhimurium (Fig. 2C), indicating that the impaired invasion of S. Paratyphi A under these conditions is not the result of impaired attachment to host cells but is due to other mechanisms.
Impaired S. Paratyphi A invasion under aerobic conditions is T3SS-1 dependent.
Having established that S. Paratyphi A presents poor invasion following growth under aerobic conditions, we next asked if T3SS-1-dependent invasion actually occurs under these conditions. To this end, we determined the invasion of two mutant strains harboring an in-frame deletion in structural T3SS-1 genes (invA and invG) relative to the wild-type background. We reasoned that if S. Paratyphi A still can invade in a T3SS-1-dependent manner, an additive effect will be observed and the invasion of these mutants will be lower than the basal invasion of the wild-type strain. Invasion experiments using Caco-2 cells proved that the lack of a functional T3SS-1 (invA or invG strains) does not reduce the already poor invasion of S. Paratyphi A under aerobic conditions (Fig. 3A). In contrast, it was clear that T3SS-1 is functional during S. Typhimurium invasion under all conditions (Fig. 3C and D) and during S. Paratyphi A invasion under microaerobic conditions (Fig. 3B). We concluded that T3SS-1-dependent invasion does not occur during S. Paratyphi A growth under aerobic conditions.
FIG 3.
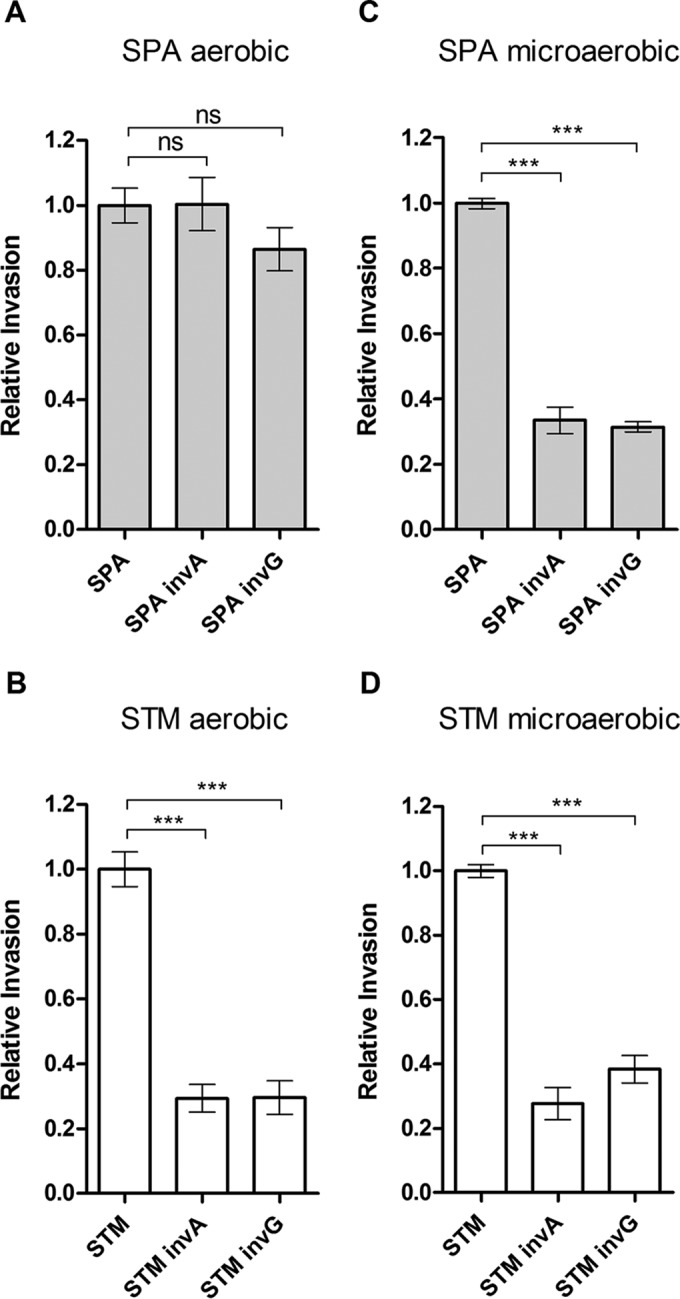
T3SS-1 is not functional in S. Paratyphi A grown to the late logarithmic phase aerobically. S. Typhimurium SL1344 (STM) and S. Paratyphi A 45157 (SPA) and their isogenic invA and invG null mutant strains were grown to the late logarithmic phase aerobically (A and B) or in LB supplemented with 0.17 M NaCl under microaerobic conditions (C and D), as described for Fig. 1, and used to infect Caco-2 cells. Under these conditions, the invasion rates of S. Paratyphi A were 0.06% ± 0.02% and 5.1% ± 1.7% (from the infection inoculum), respectively. Salmonella invasion was determined at 2 h p.i. using the gentamicin protection assay and is shown relative to the invasion of the wild-type background. One-way analysis of variance (ANOVA) with Dunnett's multiple-comparison test was implemented to compare the invasion of the different strains to that of the wild type. ***, P < 0.0001; ns, not significant.
S. Paratyphi A expresses lower levels of SPI-1 genes and secretes less T3SS-1 effectors than S. Typhimurium.
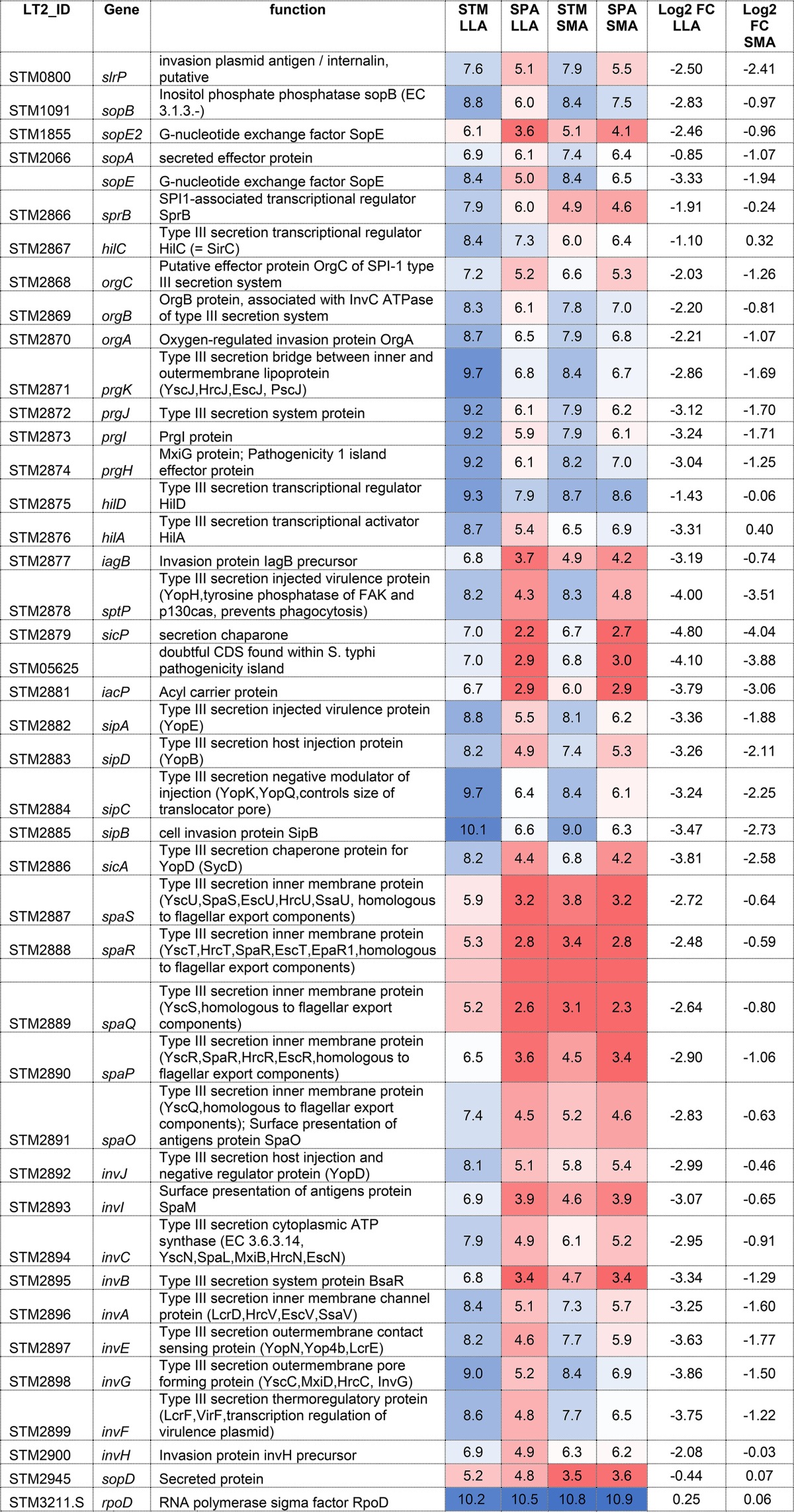
To better understand the impaired invasion of S. Paratyphi A under aerobic conditions, RNA was extracted from S. Typhimurium and S. Paratyphi A cultures grown aerobically to the late logarithmic phase and from Salmonella cultures grown to the stationary phase under microaerobic conditions. Subsequently, Salmonella RNA from both serovars and under both conditions was sequenced to determine differences in the transcriptional landscape. RNA-Seq analysis showed that under the aerobic late-logarithmic-growth conditions, 40 SPI-1 genes and related effectors are expressed at 2- to 45-fold (mean, 6.25-fold) lower levels in S. Paratyphi A than in S. Typhimurium, with the exception of sopD, which was expressed at similar levels in both serovars. At the stationary phase under microaerobic conditions, the difference was smaller and S. Paratyphi A presented, on average, 2-fold lower SPI-1 expression than S. Typhimurium. Housekeeping genes like the RNA polymerase sigma factor, rpoD, were found to be transcribed at very similar levels in both serovars, indicating the reduced expression of SPI-1 genes is specific (Table 1; also see Fig. S1A in the supplemental material).
TABLE 1.
Transcription of the SPI-1 regulon in S. Paratyphi A and S. Typhimuriuma
Gene tags in S. Typhimurium LT2 are shown in the first column. STM and SPA columns represent averaged FPKM (fragments per kilobase per million reads) values on a log2 scale for SPI-1 genes and rpoD as a reference control assayed from two independent biological replicates. RNA was extracted from S. Typhimurium SL1344 (STM) and S. Paratyphi A 45157 (SPA) cultures grown to late logarithmic phase at 37°C under aerobic conditions (LLA) and from cultures grown to the stationary phase under microaerobic conditions (SMA) as explained in Materials and Methods. Log2 FC represents the log2 fold change of the gene expression in SPA compared to that in STM. Transcription data are color-coded such that higher levels of transcription are colored in shades of blue and lower transcription is shaded in red.
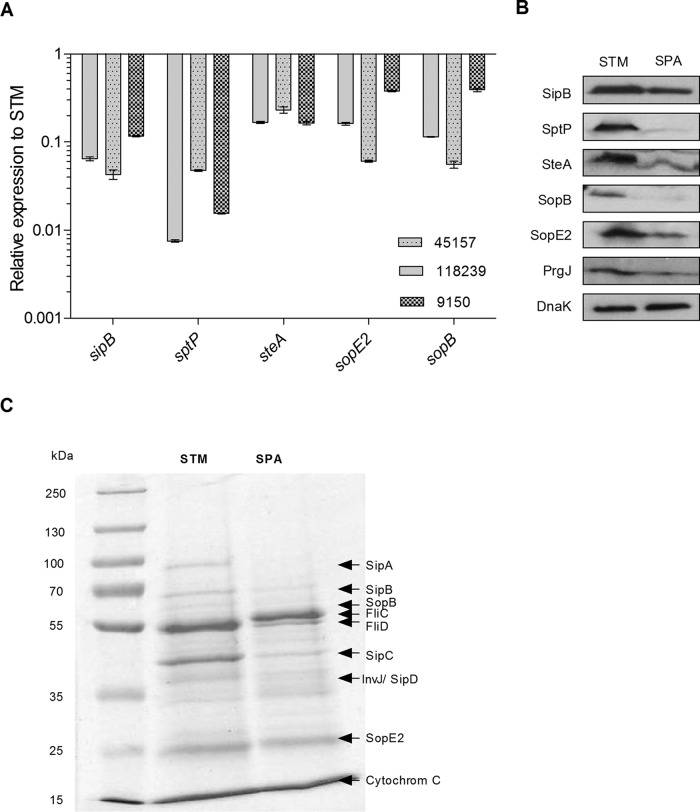
Quantitative reverse transcription-PCR (qRT-PCR) confirmed these results and showed that the T3SS-1 effector genes sptP, steA, sopE2, sipB, and sopB are expressed at 2.5- to 130-fold lower levels in three different strains of S. Paratyphi A than in S. Typhimurium (Fig. 4A). Furthermore, Western blotting using immunostaining against SipB, SptP, SteA, SopB, SopE2 (effectors), and PrgJ (T3SS-1 structural protein) tagged with a hemagglutinin epitope (2HA tag) confirmed the RNA-Seq and qRT-PCR results on the protein level and demonstrated lower cytoplasmic levels of these T3SS-1 proteins in S. Paratyphi A than in S. Typhimurium grown to late logarithmic phase under aerobic conditions (Fig. 4B).
FIG 4.
S. Paratyphi A grown aerobically to the late logarithmic phase expresses lower levels of SPI-1 genes than S. Typhimurium. (A) Total RNA was harvested from S. Typhimurium SL1344 and three S. Paratyphi A strain (45157, 118239, and 9150) cultures grown aerobically to the late logarithmic phase at 37°C, and extracted RNA was subjected to qRT-PCR. The fold change in the abundance of SPI-1 gene transcripts (normalized to rpoD) in S. Paratyphi A strains relative to their expression in S. Typhimurium is shown. The indicated values present the means and the SEM from three independent RT-PCR experiments from two independent RNA extractions. (B) SDS-PAGE Western blot analysis of bacterial cell lysate from S. Typhimurium SL1344 (STM) and S. Paratyphi A 45157 (SPA) strains grown aerobically to the late logarithmic phase. Protein fractions were probed using anti-HA antibody and anti-DnaK antibody as a control. (C) S. Typhimurium SL1344 and S. Paratyphi A 45157 cultures were grown in LB for 5.5 h and normalized to an OD600 of 2.4 to 2.6. Exogenous human cytochrome c (8 mg) was added to the bacterial cultures as a precipitating and loading control. Supernatant (5 ml) cultures were precipitated by TCA, and equal amounts (25 μl) from the precipitated fractions were separated on an SDS-12% polyacrylamide gel and stained with Coomassie G-250.
qRT-PCR also confirmed the different expression patterns between the growth conditions and showed more similar expression of representative SPI-1 genes (especially invA, invF, sopB, and sopE2) between S. Paratyphi A and S. Typhimurium during microaerobic growth to stationary phase than during late logarithmic phase under aerobic conditions (see Fig. S1B in the supplemental material).
To further study differences in the secretion of the T3SS-1 effectors, biochemical and proteomics approaches were taken. The filtered cell-free supernatants from S. Typhimurium and S. Paratyphi A cultures grown aerobically to the late logarithmic phase were precipitated by trichloroacetic acid (TCA), and their entire secretomes were compared using SDS-PAGE. This analysis showed significantly smaller amounts of proteins with the expected molecular weight of the SPI-1 effectors SipA, SipB, SopB SipC, and SopE2 in the secreted fraction of S. Paratyphi A than in that of S. Typhimurium (Fig. 4C).
To analyze differences in these secretomes quantitatively, two independent proteomics experiments using liquid chromatography-tandem mass spectrometry (LC-MS/MS) were conducted using the supernatant from late logarithmic aerobic growth. Mass spectrometry results showed that with the exception of OrgC and SopD, 17 other secreted T3SS-1 proteins either were absent or were present as 1.8- to 752-fold smaller amounts in the supernatants of S. Paratyphi A than S. Typhimurium (Table 2). As expected, LC-MS/MS failed to detect any levels of GtgE, SlrP, AvrA, SteB, and SopA, which are not encoded by or are inactivated in S. Paratyphi A, but the effector SopE also was absent from the S. Paratyphi A supernatant, while it was present in the S. Typhimurium supernatant. Collectively these results indicated that S. Paratyphi A expresses and secretes significantly lower levels of SPI-1 effectors than S. Typhimurium, especially during the late logarithmic phase under aerobic conditions.
TABLE 2.
Differences in the secretion of T3SS-1 substrates between S. Paratyphi A and S. Typhimurium
| Protein name | STM tag | SPA tag | Avg LFQ intensity ina: |
Fold change between SPA and STM | Protein function | |
|---|---|---|---|---|---|---|
| SPA | STM | |||||
| PrgJ | E1WAB7 | Q5PEB5 | 28.553 | 31.137 | 0.167 | T3SS apparatus |
| PrgI | E1WAB8 | Q5PEB4 | 27.609 | 30.192 | 0.167 | T3SS apparatus |
| InvJ | E1WAD6 | Q5PED2 | 29.287 | 31.703 | 0.187 | Surface presentation of antigen protein, associated with type III secretion and virulence |
| OrgC | E1WAB3 | Q5PEA4 | 28.383 | 26.611 | 3.414 | Putative T3SS effector protein |
| SipA | E1WAC6 | Q5PEC3 | 30.566 | 35.755 | 0.027 | Cell invasion protein |
| SipB | E1WAC9 | B5BEU3 | 30.219 | 33.171 | 0.129 | Pathogenicity island 1 T3SS effector protein |
| SipC | E1WAC8 | Q5PEC1 | 32.385 | 34.753 | 0.194 | Cell invasion protein |
| SipD | E1WAC7 | E1WAC7 | 27.860 | 31.020 | 0.112 | Pathogenicity island 1 T3SS apparatus; part of the translocon |
| SopB (SigD) | E1W7C2 | Q5PGB2 | 31.336 | 32.161 | 0.564 | Cell invasion protein |
| SopE2 | E1WG92 | B5BHA1 | 26.088 | 29.083 | 0.125 | T3SS effector protein |
| SopD | E1WAI9 | Q5PEI0 | 28.000 | 27.529 | 1.194 | T3SS effector protein |
| SteA | E1WBR4 | Q5PHW4 | 20.808 | 28.191 | 0.006 | T3SS effector protein |
| SptP | E1WAC3 | B5BET6 | 20.081 | 29.636 | 0.001 | T3SS effector protein |
| GtgE | E1W787 | NDb | 0.000 | 25.543 | Bacteriophage-encoded virulence factor | |
| SlrP | E1W9T7 | ND | 0.000 | 25.640 | T3SS effector protein | |
| AvrA | E1WAB0 | ND | 0.000 | 16.117 | T3SS effector protein | |
| SteB | E1WBW0 | ND | 0.000 | 16.144 | T3SS effector protein | |
| SopA | E1WGZ9 | ND | 0.000 | 30.101 | T3SS effector protein | |
| SopE | E1WJI8 | ND | 0.000 | 33.209 | T3SS effector protein | |
The ratio of normalized (label-free quantification [LFQ]) protein intensities between S. Paratyphi A (SPA) and S. Typhimurium (STM) as determined by LC-MS/MS.
ND, not detected.
hilA induction increases SPI-1 expression and invasion by S. Paratyphi A but not by S. Typhimurium.
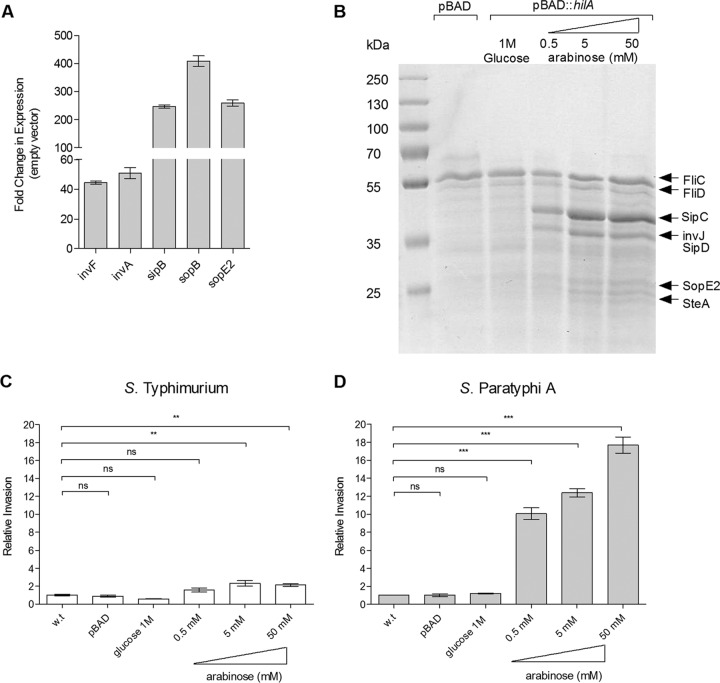
The lower expression and secretion of SPI-1 effectors suggested an explanation for the impaired invasion of S. Paratyphi A at late logarithmic phase under aerobic conditions. Therefore, we predicted that bypassing the native regulation and artificially inducing the expression of SPI-1 genes would increase S. Paratyphi A invasion under these conditions. For this purpose, we cloned the SPI-1 master regulator, hilA, under the control of an arabinose-inducible pBAD promoter and introduced this construct into S. Paratyphi A. The quantification of SPI-1 transcripts by qRT-PCR showed that the induction of hilA in S. Paratyphi A resulted in a 40- to 400-fold upregulation of SPI-1 genes (Fig. 5A). The increased secretion of SPI-1 effectors was further shown in a dose-dependent manner by SDS-PAGE and Coomassie blue staining of the S. Paratyphi A secretome prepared from cultures carrying pBAD::hilA in the presence of increasing arabinose concentrations (Fig. 5B). Furthermore, invasion experiments demonstrated up to an 18-fold increase in the invasion of S. Paratyphi A expressing inducible hilA in an arabinose dose-dependent manner. In contrast, S. Typhimurium expressing the same construct showed only a marginal invasion improvement that reached 2-fold at the maximum compared to the level for the uninduced strain (Fig. 5C). These results indicated that SPI-1 expression and the invasion of S. Paratyphi A can be significantly improved by the induction of hilA and supported the notion that the low expression of SPI-1 genes contributes to the impaired invasion of S. Paratyphi A under aerobic conditions.
FIG 5.
Induced expression of SPI-1 increases S. Paratyphi A invasion. (A) Total RNA was harvested from S. Paratyphi A 45157 carrying pBAD18 or pBAD::hilA grown in LB supplemented with 100 μl/ml ampicillin and 50 mM arabinose to late logarithmic phase aerobically and was subjected to qRT-PCR. The fold change in the abundance of SPI-1 gene transcripts (normalized to rpoD) in S. Paratyphi A harboring pBAD::hilA is shown relative to their expression in S. Paratyphi A carrying pBAD18. The indicated values present the means and the SEM from three independent RT-PCR experiments from two independent RNA extractions. (B) S. Paratyphi A cultures were grown aerobically in LB to logarithmic phase and normalized to an OD600 of 1.2. Secreted fractions from S. Paratyphi A carrying pBAD18 (lane 2) or pBAD::hilA grown in the presence of 1 M glucose (lane 3) or increasing arabinose concentrations (lanes 4 to 6) were separated on SDS-12% polyacrylamide gels and stained with Coomassie G-250. (C) S. Typhimurium SL1344 (wild type) and its isogenic strains carrying pBAD or pBAD::hilA were grown in LB, LB supplemented with 1 M glucose, or LB with increasing concentrations of arabinose under aerobic conditions and used to infect HeLa cells. Invasion is shown relative to that of the wild-type strain (grown in LB supplemented with 50 mM arabinose), presenting a 3.7% ± 0.76% invasion rate (from the infecting inoculum) under these conditions. (D) S. Paratyphi A 45157 and its isogenic strains carrying pBAD or pBAD::hilA were grown and used to infect HeLa cells as described for panel C. Invasion is shown relative to that of the S. Paratyphi A wild-type strain, presenting, on average, 0.05% ± 0.001% invasion. ANOVA with Dunnett's multiple-comparison test was implemented to compare the invasion of the different strains to that of the wild type. The results represent the means and SEM from at least three biological replicates. **, P < 0.01; ***, P < 0.0001; ns, not significant.
hilA induction in S. Paratyphi A but not in S. Typhimurium leads to enhanced disruption of polarized epithelia and cytokine secretion.
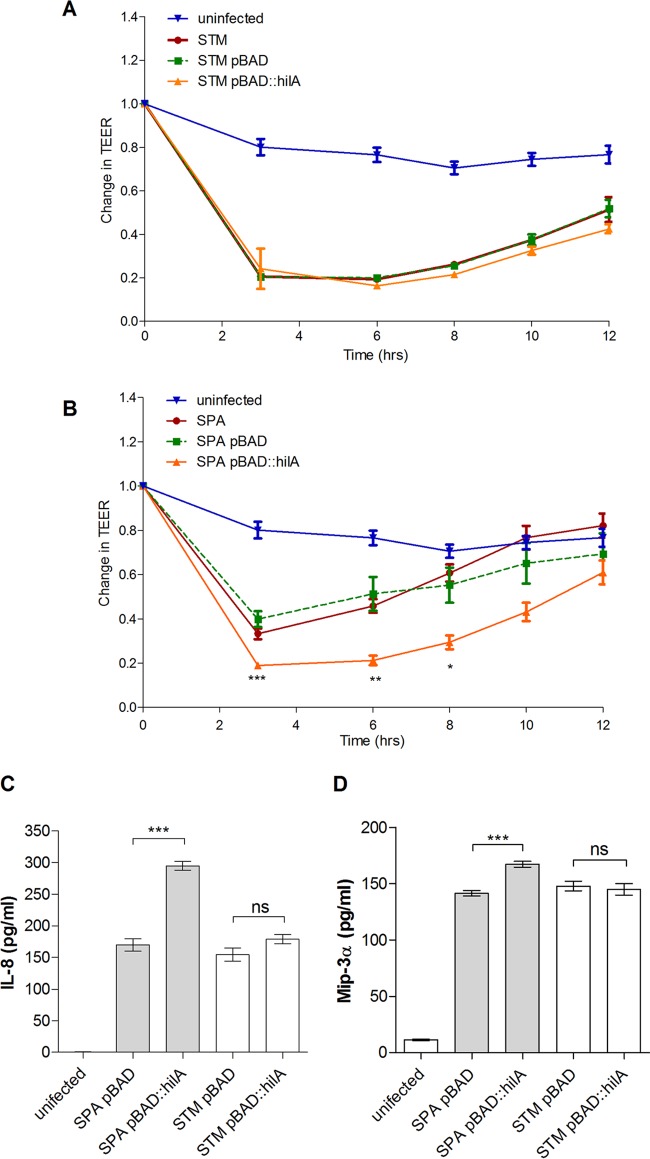
To further investigate how SPI-1 induction affects S. Paratyphi A interactions with host cells, we infected a polarized monolayer of Caco-2 cells and monitored the change in its transepithelial electrical resistance (TEER) over time. In this experimental system, Salmonella invasion disrupts the integrity of the polarized epithelium that is shown as a decrease in TEER. Polarized cells were infected with S. Typhimurium and S. Paratyphi A expressing pBAD::hilA, as well as with the parental strains and with strains carrying the empty vector (pBAD18), all grown aerobically in the presence of 50 mM arabinose. These experiments showed that while hilA induction in S. Typhimurium triggered the same changes in TEER as in the parental strain (Fig. 6A), the overexpression of hilA in S. Paratyphi A augmented TEER decline compared to levels in cells infected with in the parental strain or with the empty vector, indicating the enhanced disruption of the epithelial monolayer (Fig. 6B). It is worth noting that the reduction in TEER in cells infected with S. Paratyphi A expressing pBAD::hilA was similar to or even slightly greater than the levels obtained following S. Typhimurium infections.
FIG 6.
Induced expression of hilA increases epithelium disruption and secretion of proinflammatory cytokines by S. Paratyphi A. The integrity of the epithelial monolayer was determined for polarized Caco-2 cells infected at an MOI of 1:10 at 0, 3, 6, 8, 10, and 12 h p.i. and is shown as the change in transepithelial electrical resistance (TEER) from the time of infection (T0). S. Typhimurium SL1344 (A) and S. Paratyphi A 45157 (B) strains were grown under aerobic conditions to the late logarithmic phase in the presence of 100 μl/ml ampicillin and 50 mM arabinose. Data represent the means and SEM from three to five infections. An unpaired t test with two tails was used to determine the significance of the differences between S. Paratyphi A/pBAD18 and S. Paratyphi A/pBAD::hilA measurements. At 2 h p.i., supernatant was taken from the polarized Caco-2 cells and the concentration of secreted IL-8 (C) and Mip3α (D) was measured by ELISA. Data shown represent the means and standard deviations from three biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.0001; ns, not significant.
In close agreement with these results, polarized Caco-2 cells that were infected with S. Paratyphi A expressing pBAD::hilA were found to secrete about 2-fold more of the proinflammatory cytokine IL-8 than cells infected with S. Paratyphi A carrying pBAD18. In contrast, we did not find any significant additional increase in IL-8 secretion in cells infected with S. Typhimurium carrying pBAD::hilA (Fig. 6C) compared to cells infected with S. Typhimurium carrying pBAD18. Similarly, cells infected with S. Paratyphi A overexpressing hilA also secreted moderately but significantly increased levels (∼18% more) of Mip3α, while the elevated secretion of Mip3α was not observed in cells infected with S. Typhimurium overexpressing hilA (Fig. 6D). Collectively, we concluded from these experiments that the induced expression of hilA in S. Paratyphi A, but not in S. Typhimurium, enhances host cell invasion, the disruption of epithelial integrity, and the secretion of proinflammatory cytokines in vitro.
Overexpression of hilA increases S. Paratyphi A pathogenicity in vivo.
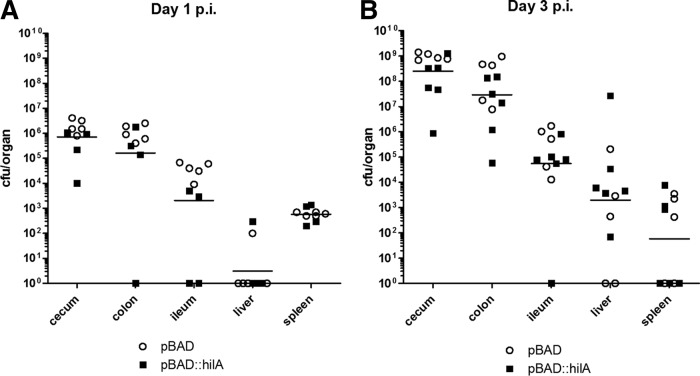
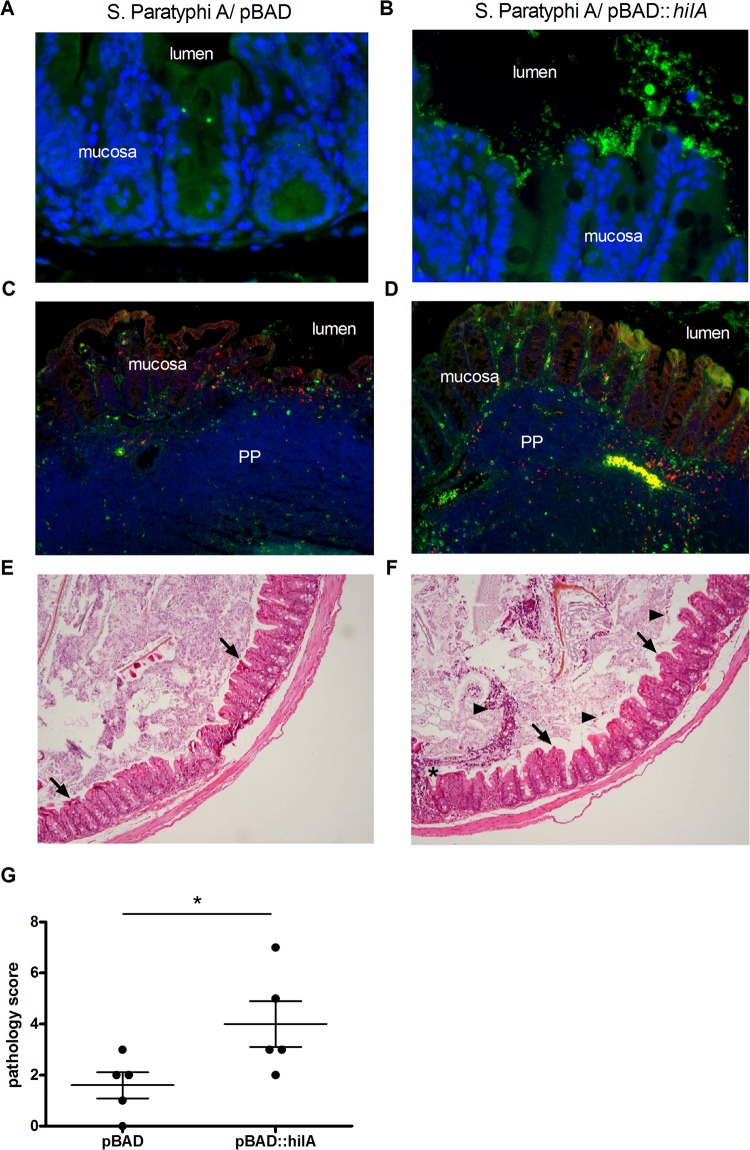
To assess the effect of SPI-1 induction on S. Paratyphi A virulence in vivo, we used streptomycin-pretreated C57BL/6 mice and infected them orally with ∼108 CFU of S. Paratyphi A expressing pBAD::hilA or with a strain carrying the empty pBAD18 vector. Despite the fact that S. Paratyphi A is a human-specific pathogen, using the streptomycin mouse model, a profound colonization of S. Paratyphi A can be obtained (30). Similar bacterial loads were recovered from both the hilA-expressing and control groups. At day one p.i., an average of 1.4 × 106 and 9.5 × 105 CFU were isolated from the cecum and the colon, respectively, and 2.5 × 104 CFU were isolated from the ileum of mice infected with each strain (Fig. 7A). At day three p.i., the mean bacterial load for both strains in the cecum and colon increased to more than 3 × 108 and 5 × 107 in the cecum and colon, respectively, and 1.9 × 105 in the ileum (Fig. 7B), indicating the establishment of substantial intestinal colonization and luminal growth by S. Paratyphi A. Interestingly, although we did not observe higher bacterial burden in mice infected with the inducible hilA strain, the immunofluorescence of cecal sections demonstrated higher numbers of S. Paratyphi A organisms associated with the epithelium (Fig. 8A and B), more bacteria invading the tissue, and significantly increased recruitment of macrophages and neutrophils in the Peyer's patches of mice infected with S. Paratyphi A overexpressing hilA compared to the empty vector (Fig. 8C and D). Additionally, blinded pathology scoring of cecum sections showed significantly higher pathology, with more dead cells in the lumen, more inflammatory infiltrate, and desquamation in the mice infected with hilA-induced S. Paratyphi A (Fig. 8E to G). Overall, the in vivo results demonstrate that the streptomycin-treated mouse model is useful to study S. Paratyphi A intestinal colonization and indicate a moderate yet significantly higher level of histopathological changes in mice infected with S. Paratyphi A overexpressing hilA. These observations are consistent with the in vitro results showing elevated SPI-1 expression in S. Paratyphi A overexpressing hilA and enhanced cell invasion, epithelial integrity disruption, and secretion of proinflammatory cytokines following infection with this strain.
FIG 7.
S. Paratyphi A colonizes the intestine of streptomycin-treated mice. Groups of C57BL/6 mice were treated with streptomycin 1 day preinfection and inoculated orally with 1 × 108 to 2 × 108 CFU of S. Paratyphi A carrying pBAD18 (circles) or pBAD::hilA (closed squares) grown to the late logarithmic phase under aerobic conditions. Mice from day one (A) and day three (B) p.i. were sacrificed, and homogenized tissues were plated on XLD plates supplemented with ampicillin to determine bacterial load per organ. Geometric means from both groups are shown as horizontal lines.
FIG 8.
hilA-overexpressing S. Paratyphi A induces greater pathology and increased host immune response in the mouse model. Cecal sections were taken at day one p.i. from streptomycin-pretreated mice that were infected with S. Paratyphi A carrying pBAD18 (A, C, and E) or pBAD::hilA (B, D, and F). (A and B) Immunofluorescence staining of cecal sections. Nuclei were stained with DAPI (blue), and S. Paratyphi A organisms were stained with anti-Salmonella serum (green). Original magnification, ×400. (C and D) Staining against CD68 (macrophages; red) and MPO (neutrophils; green) was conducted as explained in Materials and Methods, and nuclei were stained with DAPI (blue). The lumen, mucosa, and Peyer's patches (PP) are indicated. Original magnification, ×100. (E and F) Representative micrographs of hematoxylin and eosin staining of cecal sections (original magnification of ×100). Desquamations of the epithelium are indicated by arrows, ulceration is shown by an asterisk, and dead cells in lumen are indicated by arrowheads. (G) Histopathological scoring was assessed by the determination of the infiltration of inflammatory cells together with the evaluation of epithelial damage. *, P < 0.05.
DISCUSSION
Human infection with different S. enterica serovars may result in distinct clinical manifestations. While infection with the majority of the subspecies-1 serovars induces an inflammatory gastroenteritis, three human-restricted serovars, Sendai, Typhi and Paratyphi A, elicit a mild inflammatory response and a systemic disease which is entirely different from the clinical presentation of gastroenteritis (31).
To better understand the mechanisms underlying the distinct pathogenicity of S. Paratyphi A compared to that of S. Typhimurium, we chose to focus on their invasion into nonphagocytic cells and the expression pattern of SPI-1, which is responsible for this virulence-associated phenotype. Interestingly, we found that while S. Typhimurium growing to the late logarithmic phase aerobically in LB (considered SPI-1-inducing conditions) results in highly invasive bacteria, S. Paratyphi A was hardly invasive under these conditions. Remarkably, the stimulation of noninvasive, aerobically grown S. Paratyphi A culture under microaerobic conditions for 3 h fully induced S. Paratyphi A invasive ability, indicating that host cell invasion by S. Paratyphi A is repressed under aerobic conditions.
Independent experimental approaches, including RNA-Seq, RT-PCR, Western blotting, and LC-MS/MS, to the S. Paratyphi A secreted fraction established that the expression of SPI-1 genes and the secretion of SPI-1 effectors are significantly lower in S. Paratyphi A than in S. Typhimurium. Based on these results, we hypothesized that the impaired invasion of S. Paratyphi A at the late logarithmic phase under aerobic conditions is due to the particularly low expression of SPI-1 genes. Consistent with this idea, when we bypassed the natural SPI-1 control and artificially upregulated SPI-1 genes by induced expression of hilA in S. Paratyphi A, we were able to largely increase the expression of SPI-1 effector genes, their secretion into the medium, and the invasion phenotype in an inducer (arabinose) dose-dependent manner. Furthermore, S. Paratyphi A, but not S. Typhimurium, caused an enhanced disruption of epithelial integrity and IL-8 and Mip3α secretion in polarized epithelial cells when overexpressing hilA.
Using streptomycin-pretreated mice, we were able to establish significant intestinal colonization and luminal replication of S. Paratyphi A. These results further emphasize that outcompeting the intestinal microbiota is a prerequisite for efficient Salmonella colonization (32–35). It is worth noting that intestinal colonization was similar between S. Paratyphi A strains expressing inducible hilA and those carrying the empty vector, suggesting that additional HilA-independent factors play an important role in mouse colonization by S. Paratyphi A. Nevertheless, despite similar colonization levels, we found moderately higher pathology and stronger mucosal immune response in mice infected with hilA-induced S. Paratyphi A. Previously, Suar et al. reported similar levels of infection in the same mouse model using different strains of S. Paratyphi A (30). Together, these results demonstrate that the streptomycin-pretreated mice can be used as an efficient, inexpensive model to study S. Paratyphi A colonization in vivo.
Collectively, our results indicate that, under (late-logarithmic) aerobic conditions, S. Paratyphi A expresses much lower levels of SPI-1 genes than S. Typhimurium and does not invade epithelial cells. Hence, under these conditions, S. Paratyphi A is expected to evoke a weaker host immune response than S. Typhimurium due to three reasons: (i) reduced S. Paratyphi A invasion will induce a lower epithelial innate immune response, including lesser secretion of proinflammatory cytokines, such as IL-1β and IL-8 (36); (ii) lower expression of T3SS-1-related PAMPs, in particular, PrgJ and PrgI, recognized by NLRC4, will lead to a lower activation of the inflammasome (37); and (iii) lower expression of SPI-1 effectors known to contribute to intestinal inflammation, including SipA, SopB, SopD, SopE, and SopE2 (38), will moderate the pathogen-induced inflammation in an IL-1β- and NF-κB-dependent manner. Such T3SS-1-mediated inflammation can be even further reduced by the absence of sopA, which is missing from the S. Paratyphi A genome (39).
S. Paratyphi A is likely to encounter relatively oxygenated microenvironments while moving from the microaerobic (or even anaerobic) gastrointestinal tract lumen to invade deeper intestinal tissues, in which their ambient oxygen tension can reach 57.6 ± 2.3 mm Hg or 7.6% ± 0.3% partial O2 pressure (40), or at zones adjacent to the mucosal surface that become oxygenated by O2 diffusion from the capillary network at the villous tips (41, 42). Various oxygen concentrations at different anatomical sites of the intestine were shown to govern the T3SS activity of Shigella flexneri and consequently its cell invasion and virulence (41). Our results, showing poor invasion in S. Paratyphi A but not in S. Typhimurium grown aerobically, suggest that S. Paratyphi A regulates its invasion in response to the oxygen tension as well.
In recent years, two distinct mechanisms were demonstrated to facilitate host immune evasion of a different important typhoidal serovar, namely, S. Typhi. These pathways involve S. Typhi-specific genes contained within SPI-7. The first mechanism requires the expression of the Vi polysaccharide capsule of S. Typhi, which prevents complement activation and neutrophil-mediated clearance of S. Typhi (43). Furthermore, the Vi antigen also was shown to shield S. Typhi LPS and reduces the TLR4-dependent immune response (12, 44). A second mechanism used by S. Typhi to evade the innate immune system is the regulator TviA, which positively regulates the Vi capsule genes but also negatively regulates flagellar and T3SS-1 genes (14). Interestingly, the expression of TviA is repressed at high osmolarity (such as that found in the intestinal lumen) and induced in low-osmolarity environments (e.g., in the intestinal mucosa), allowing S. Typhi to be noncapsulated, flagellated, and invasive at the lumen but capsulated with low expression of flagellar and T3SS-1 genes in the intestinal epithelium (11, 45). Since S. Paratyphi A lacks SPI-7, it is tempting to speculate that S. Paratyphi A has evolved to use ambient oxygen tension as a signal to downregulate the invasion phenotype and suppress the expression of SPI-1-related PAMPs to facilitate bacterial dissemination through innate immune evasion.
The coordination of the SPI-1 regulon is complex and is fine-tuned by many regulators encoded by SPI-1 (HilA, HilD, HilC, and InvF) as well as outside SPI-1 (BarA/SirA, PhoP/Q, and CsrA) (26). Another regulator involved in O2 sensing shown to control SPI-1 genes in S. Typhimurium is FNR (46). In S. flexneri, FNR also was found to mediate its T3SS expression and invasion in response to oxygen levels in the intestine (41). However, since the FNR sequence is identical in S. Typhimurium and S. Paratyphi A and RNA-Seq analysis revealed similar expression levels in both serovars (data not shown), it is likely that the repressed T3SS-1 in S. Paratyphi A is mediated by an as-yet-unknown mechanism(s) other than FNR.
Here, we established that S. Paratyphi A expresses lower levels of SPI-1 genes than S. Typhimurium. Recently, we showed that the flagellum motility regulon also is expressed at lower levels in S. Paratyphi A and that typhoidal serovars generally are less motile than S. Typhimurium (15). Differential expression of these associated regulons seems to be specific, as unrelated housekeeping genes (rpoD, for example) were found to be transcribed at very similar levels. Moreover, we previously demonstrated that S. Paratyphi A, but not S. Typhimurium, invasion into epithelial cells responds to temperature stimuli and is impaired at elevated physiological temperatures equivalent to fever (39 to 42°C). Under these fever-like conditions, the impaired invasion also was shown to be associated with the downregulation of the T3SS-1 genes and classes II and III of the flagellum chemotaxis regulon (47). Collectively, these results demonstrate significantly lower expression of flagellar and T3SS-1 genes, which can serve as PAMPs, in S. Paratyphi A than in S. Typhimurium, and that their invasion and motility programs respond differently to certain environmental or physiological cues. We hypothesize that restrained invasion and controlled PAMP expression prevent strong intestinal inflammation, in contrast to the inflammatory response elicited by NTS, and therefore play a key role in the distinct diseases caused by typhoidal versus NTS serovars in humans.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tamar Ziv from the Smoler Proteomics Center at the Technion-Israel Institute of Technology, Haifa, Israel, for her valuable help with the proteomic analysis. We are grateful for the staff of the Ministry of Health Central Laboratories and specifically to Israel Nissan, Lea Valinsky, and Vered Agmon for sharing Salmonella clinical isolates. We thank WeiPing Chu and Steffen Porwollik for assistance with RNA-Seq. We thank Janin Braun for excellent technical help.
This work was supported by the German-Israeli Foundation for Scientific Research and Development (grant 1096-39.11/2010 to O.G.-M. and G.A.G.), the Israel Science Foundation (grant 999/14 to O.G.-M.), the Deutsche Forschungsgemeinschaft (grant GR2666/5-1) and DZIF to G.A.G., the National Institutes of Health (grants HHSN272200900040C, AI039557, AI052237, AI073971, AI075093, AI077645, AI083646, USDA 2009-03579, and 2011-67017-30127 to M.M.), the Binational Agricultural Research and Development Fund (M.M.), and the Center for Produce Safety (M.M.).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01461-15.
REFERENCES
- 1.Guibourdenche M, Roggentin P, Mikoleit M, Fields PI, Bockemuhl J, Grimont PA, Weill FX. 2010. Supplement 2003-2007 (no. 47) to the White-Kauffmann-Le Minor scheme. Res Microbiol 161:26–29. doi: 10.1016/j.resmic.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull World Health Organ 82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 3.Kraus MD, Amatya B, Kimula Y. 1999. Histopathology of typhoid enteritis: morphologic and immunophenotypic findings. Mod Pathol 12:949–955. [PubMed] [Google Scholar]
- 4.Nguyen QC, Everest P, Tran TK, House D, Murch S, Parry C, Connerton P, Phan VB, To SD, Mastroeni P, White NJ, Tran TH, Vo VH, Dougan G, Farrar JJ, Wain J. 2004. A clinical, microbiological, and pathological study of intestinal perforation associated with typhoid fever. Clin Infect Dis 39:61–67. doi: 10.1086/421555. [DOI] [PubMed] [Google Scholar]
- 5.Sprinz H, Gangarosa EJ, Williams M, Hornick RB, Woodward TE. 1966. Histopathology of the upper small intestines in typhoid fever. Biopsy study of experimental disease in man. Am J Dig Dis 11:615–624. [DOI] [PubMed] [Google Scholar]
- 6.House D, Bishop A, Parry C, Dougan G, Wain J. 2001. Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis 14:573–578. doi: 10.1097/00001432-200110000-00011. [DOI] [PubMed] [Google Scholar]
- 7.Liu SL, Ezaki T, Miura H, Matsui K, Yabuuchi E. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect Immun 56:1967–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou D, Galan J. 2001. Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect 3:1293–1298. doi: 10.1016/S1286-4579(01)01489-7. [DOI] [PubMed] [Google Scholar]
- 9.Agbor TA, McCormick BA. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol 13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong HK, Parry CM, van der Poll T, Wiersinga WJ. 2012. Host-pathogen interaction in invasive Salmonellosis. PLoS Pathog 8:e1002933. doi: 10.1371/journal.ppat.1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keestra-Gounder AM, Tsolis RM, Bäumler AJ. 2015. Now you see me, now you don't: the interaction of Salmonella with innate immune receptors. Nat Rev Microbiol 13:206–216. doi: 10.1038/nrmicro3428. [DOI] [PubMed] [Google Scholar]
- 12.Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Bäumler AJ. 2005. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun 73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winter SE, Winter MG, Poon V, Keestra AM, Sterzenbach T, Faber F, Costa LF, Cassou F, Costa EA, Alves GE, Paixao TA, Santos RL, Bäumler AJ. 2014. Salmonella enterica serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS Pathog 10:e1004207. doi: 10.1371/journal.ppat.1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Russmann H, Bäumler AJ. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol 74:175–193. doi: 10.1111/j.1365-2958.2009.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elhadad D, Desai P, Rahav G, McClelland M, Gal-Mor O. 2015. Flagellin is required for host cell invasion and normal Salmonella pathogenicity island 1 expression by Salmonella enterica serovar Paratyphi A. Infect Immun 83:3355–3368. doi: 10.1128/IAI.00468-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grassl GA, Valdez Y, Bergstrom KS, Vallance BA, Finlay BB. 2008. Chronic enteric salmonella infection in mice leads to severe and persistent intestinal fibrosis. Gastroenterology 134:768–780. doi: 10.1053/j.gastro.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 20.LaRock DL, Chaudhary A, Miller SI. 2015. Salmonellae interactions with host processes. Nat Rev Microbiol 13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop A, House D, Perkins T, Baker S, Kingsley RA, Dougan G. 2008. Interaction of Salmonella enterica serovar Typhi with cultured epithelial cells: roles of surface structures in adhesion and invasion. Microbiology 154:1914–1926. doi: 10.1099/mic.0.2008/016998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steele-Mortimer O. 2008. Infection of epithelial cells with Salmonella enterica. Methods Mol Biol 431:201–211. [DOI] [PubMed] [Google Scholar]
- 23.Arricau N, Hermant D, Waxin H, Ecobichon C, Duffey PS, Popoff MY. 1998. The RcsB-RcsC regulatory system of Salmonella typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol Microbiol 29:835–850. doi: 10.1046/j.1365-2958.1998.00976.x. [DOI] [PubMed] [Google Scholar]
- 24.Bajaj V, Lucas RL, Hwang C, Lee CA. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 25.Tartera C, Metcalf ES. 1993. Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal cells. Infect Immun 61:3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altier C. 2005. Genetic and environmental control of salmonella invasion. J Microbiol 43:85–92. [PubMed] [Google Scholar]
- 27.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilmes-Riesenberg MR, Foster JW, Curtiss R III. 1997. An altered rpoS allele contributes to the avirulence of Salmonella typhimurium LT2. Infect Immun 65:203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerlach RG, Claudio N, Rohde M, Jackel D, Wagner C, Hensel M. 2008. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell Microbiol 10:2364–2376. doi: 10.1111/j.1462-5822.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- 30.Suar M, Jantsch J, Hapfelmeier S, Kremer M, Stallmach T, Barrow PA, Hardt WD. 2006. Virulence of broad- and narrow-host-range Salmonella enterica serovars in the streptomycin-pretreated mouse model. Infect Immun 74:632–644. doi: 10.1128/IAI.74.1.632-644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabbagh SC, Forest CG, Lepage C, Leclerc JM, Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett 305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 32.Endt K, Stecher B, Chaffron S, Slack E, Tchitchek N, Benecke A, Van Maele L, Sirard JC, Mueller AJ, Heikenwalder M, Macpherson AJ, Strugnell R, von Mering C, Hardt WD. 2010. The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog 6:e1001097. doi: 10.1371/journal.ppat.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 5:2177–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Bäumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyland KA, Kohrt L, Vulchanova L, Murtaugh MP. 2006. Mucosal innate immune response to intragastric infection by Salmonella enterica serovar Choleraesuis. Mol Immunol 43:1890–1899. doi: 10.1016/j.molimm.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Sellin ME, Muller AA, Felmy B, Dolowschiak T, Diard M, Tardivel A, Maslowski KM, Hardt WD. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe 16:237–248. doi: 10.1016/j.chom.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 38.McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr Opin Microbiol 12:117–124. doi: 10.1016/j.mib.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, Harkins CR, Wang C, Nguyen C, Berghoff A, Elliott G, Kohlberg S, Strong C, Du F, Carter J, Kremizki C, Layman D, Leonard S, Sun H, Fulton L, Nash W, Miner T, Minx P, Delehaunty K, Fronick C, Magrini V, Nhan M, Warren W, Florea L, Spieth J, Wilson RK. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet 36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- 40.Carreau A, El Hafny-Rahbi B, Matejuk A, Grillon C, Kieda C. 2011. Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia. J Cell Mol Med 15:1239–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marteyn B, West NP, Browning DF, Cole JA, Shaw JG, Palm F, Mounier J, Prevost MC, Sansonetti P, Tang CM. 2010. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465:355–358. doi: 10.1038/nature08970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres Filho IP, Leunig M, Yuan F, Intaglietta M, Jain RK. 1994. Noninvasive measurement of microvascular and interstitial oxygen profiles in a human tumor in SCID mice. Proc Natl Acad Sci U S A 91:2081–2085. doi: 10.1073/pnas.91.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wangdi T, Lee CY, Spees AM, Yu C, Kingsbury DD, Winter SE, Hastey CJ, Wilson RP, Heinrich V, Bäumler AJ. 2014. The Vi capsular polysaccharide enables Salmonella enterica serovar typhi to evade microbe-guided neutrophil chemotaxis. PLoS Pathog 10:e1004306. doi: 10.1371/journal.ppat.1004306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Bäumler AJ. 2008. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol 10:876–890. doi: 10.1111/j.1462-5822.2007.01090.x. [DOI] [PubMed] [Google Scholar]
- 45.Winter SE, Winter MG, Godinez I, Yang HJ, Russmann H, Andrews-Polymenis HL, Bäumler AJ. 2010. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog 6:e1001060. doi: 10.1371/journal.ppat.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, Libby SJ, McClelland M, Hassan HM. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J Bacteriol 189:2262–2273. doi: 10.1128/JB.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elhadad D, McClelland M, Rahav G, Gal-Mor O. 2015. Feverlike temperature is a virulence regulatory cue controlling the motility and host cell entry of typhoidal Salmonella. J Infect Dis 212:147–156. doi: 10.1093/infdis/jiu663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.