Abstract
The host-encoded Perforin-2 (encoded by the macrophage-expressed gene 1, Mpeg1), which possesses a pore-forming MACPF domain, reduces the viability of bacterial pathogens that reside within membrane-bound compartments. Here, it is shown that Perforin-2 also restricts the proliferation of the intracytosolic pathogen Listeria monocytogenes. Within a few hours of systemic infection, the massive proliferation of L. monocytogenes in Perforin-2−/− mice leads to a rapid appearance of acute disease symptoms. We go on to show in cultured Perforin-2−/− cells that the vacuole-to-cytosol transitioning of L. monocytogenes is greatly accelerated. Unexpectedly, we found that in Perforin-2−/− macrophages, Listeria-containing vacuoles quickly (≤15 min) acidify, and that this was coincident with greater virulence gene expression, likely accounting for the more rapid translocation of L. monocytogenes to its replicative niche in the cytosol. This hypothesis was supported by our finding that a L. monocytogenes strain expressing virulence factors at a constitutively high level replicated equally well in Perforin-2+/+ and Perforin-2−/− macrophages. Our findings suggest that the protective role of Perforin-2 against listeriosis is based on it limiting the intracellular replication of the pathogen. This cellular activity of Perforin-2 may derive from it regulating the acidification of Listeria-containing vacuoles, thereby depriving the pathogen of favorable intracellular conditions that promote its virulence gene activity.
INTRODUCTION
Both extracellular bacteria and virus-infected cells are targeted by innate defense responses that employ pore-forming proteins (1). Extracellular bacteria that become bound with the complement factor C3b and the C5b-8 complex trigger the polymerization of C9, resulting in a doughnut-shaped pore with a diameter of 100 Å that constitutes the membrane attack complex (MAC) (2–4). Similarly, virus-infected cells are recognized and eliminated by natural killer (NK) and cytotoxic T lymphocytes (CTL) that, as part of their respective killing programs, secrete Perforin-1, which forms a cluster of lethal pores in the membrane of the infected cell (5, 6). The complement proteins C6 to C9 and Perforin-1 all possess a membrane-attack-complex-perforin (MACPF) domain, which mediates the homopolymerization process that drives pore formation.
A gene predicted to encode a third MACPF-containing protein, macrophage expressed gene-1 (Mpeg1), recently has been described in a number of invertebrates and zebrafish and plays a role in innate immune responses in these species to bacterial pathogens (7–11). Phylogenic analyses indicate that the MACPF domain of Mpeg1 is the ancestor of the MACPF domains in the complement and Perforin-1 proteins (12). Interestingly, although homologous Mpeg1 genes are found in most metazoan genomes spanning from sponges to humans, Mpeg1, or MACPF-encoding genes more generally, so far have not been identified in nonmetazoan clades of eukaryotes. However, the MACPF domain itself bears a striking structural similarity to the pore-forming domains of cholesterol-dependent cytolysins (CDCs) expressed by Gram-positive bacteria, indicating the evolutionary relatedness of these pore-forming domains (13). Owing to shared evolutionary history and functional similarities, the MACPF and CDC have been proposed to be in the same family of pore-forming proteins (14).
We have recently shown that the mammalian MPEG1 homolog is constitutively expressed in macrophages, whereas in fibroblast- and endothelium-derived cells its expression is induced by interferons as well as infection with intracellular bacterial pathogens (15, 16). In mice and humans the MPEG1 gene product, which is designated Perforin-2, is a 72-kDa protein possessing an MACPF domain in addition to a putative transmembrane and cytosolic domains. In cell-based infection studies, the proliferation of a number of intracellular bacterial pathogens is greatly enhanced when cellular Perforin-2 levels are reduced, suggesting that Perforin-2 targets vacuole- and plasma membrane-bound bacteria (15). Additionally, at least one intracellular pathogen, Chlamydia trachomatis, has evolved to actively block inductive Perforin-2 expression in epithelial cells (16). Thus, it appears that while Perforin-2 targets intracellular bacteria, it also can be targeted itself by pathogens in order to enhance their intracellular replication.
Intracellular bacterial pathogens can take one of two strategies following their internalization into the primary membrane-bound vacuole of eukaryotic cells. Pathogens such as Mycobacterium and Chlamydia remain within this vacuole, from which they are capable of withstanding the various antimicrobial factors that are released into these cellular compartments and/or modify the trafficking of these compartments. In contrast, intracellular pathogens such as Listeria, Shigella, Burkholderia, and Rickettsia rapidly escape from the initial vacuole compartment, which releases the bacteria directly into the cytosol of the infected cell (17). These pathogens have independently evolved to not only gain access to the cytosol but also to harness actin polymerization to move within the cytosol, eventually allowing them to penetrate into adjacent cells.
The best characterized of these cytosolic pathogens, Listeria monocytogenes, secretes both phospholipases and a pore-forming toxin, listerolysin O (LLO), that together rapidly destroy the barrier function of the primary vacuolar membrane. LLO possesses an MACPF/CDC domain (described above) and is required for the intracellular proliferation of L. monocytogenes in both cell- and mouse-based infection models (18). Here, we examined whether host-encoded Perforin-2, which, like LLO, possesses a MACPF/CDC domain, plays a role in the cellular and organismal infection of L. monocytogenes.
MATERIALS AND METHODS
Mice and bacterial strains.
Perforin-2−/− mice (C57BL/6 × 129X1/SvJ) and a derivative mouse embryonic fibroblast (MEF) cell line contain an inactivating insertion in the Mpeg1 locus (19). The green fluorescent protein (GFP)-expressing wild-type Listeria monocytogenes 10403S strain was provided by Daniel Portnoy. The L. monocytogenes 10403S strain NF-L851 possesses an in-frame deletion in actA (20) and is transformed with a plasmid (pAM401) containing the gfp gene driven by the actA promoter. The L. monocytogenes 10403S strain NF-L1177 contains a prfA G145S gene that expresses a constitutively active PrfA transcription factor (21).
Mouse infections.
Mice were treated humanely in strict accordance with federal and state government guidelines and with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (22), and their use was approved for this entire study by the University of Miami institutional animal care and use committee (protocol number 14-134). Mice were infected intravenously with either 10,000 or 150,000 CFU of exponentially growing L. monocytogenes (optical density of ∼0.2) propagated in brain heart infusion media at 37°C, and at either 3, 18, or 72 h postinfection (hpi), infected mice were humanely euthanized. Spleens and livers then were removed and homogenized in sterile water containing 0.05% Triton X-100 by either a tissue disruptor (spleens) or by grinding through a fine wire mesh (livers). The resulting homogenates were diluted further with water to fully lyse individual cells to release intracellular L. monocytogenes, and the resulting dilutions were plated to determine bacterial titers. Serum was collected at 18 hpi, and levels of interleukin-6 (IL-6), monocyte chemoattractant protein 1 (MCP-1), gamma interferon (IFN-γ), and tumor necrosis factor alpha (TNF-α) were determined by a cytometric bead array assay and analyzed by flow cytometry (BD Biosciences).
Cell infections.
The intracellular proliferation of L. monocytogenes was evaluated by CFU assays (23) in which MEFs were seeded in 24-well culture dishes at 3 × 105 cells per well and 36 h later were either left untreated or were treated for 14 h with IFN-γ at 100 U/ml. All cell treatments and/or infections were performed in triplicate. Cells then were infected with exponentially growing L. monocytogenes (prepared as described above) at a multiplicity of infection (MOI) of 5. At 1 hpi, the cell medium was replaced with fresh medium containing 2 μg/ml gentamicin (15750-060; Gibco) to kill extracellular L. monocytogenes. Control experiments established that this concentration of gentamicin kills >99.992% of extracellular L. monocytogenes in less than 5 min. Intracellular (i.e., gentamicin-protected) L. monocytogenes numbers were determined at various time points thereafter by removing medium from wells and adding 1 ml distilled sterile water for 30 s and then plating the resulting dilutions on semisolid LB media to enumerate CFU. Short interfering RNA (siRNA)-mediated knockdown of Perforin-2 expression in MEFs was performed as described previously (15). Briefly, three 19-nucleotide siRNA duplexes complementary to various regions of Mpeg1 or a scrambled siRNA control was transfected into ∼2 × 106 cells using Amaxa Nucleofector (Lonza). Cells then were plated into 24-well cell culture dishes at 3 × 105 cells per well and infected as described above. Peritoneal exudate macrophages (PEMs) were isolated from Perforin-2+/+ and Perforin-2−/− mice as described previously (6) and assessed for viability using Live/Dead fixable near-infrared dead cell stain (Life Technologies) and analyzed by flow cytometry using an LSRII (BD). Live cells were stained with the antibodies allophycocyanin (APC), clone M1/70 (Tonbo Bioscience), and phycoerythrin (PE)-cy7, clone BM8 (eBioscience), to measure the surface expression of the macrophage-specific markers CD11b and F4/80, respectively. Flow cytometry-derived data were analyzed by FlowJo software (TreeStar). PEMs were seeded either at low density on glass coverslips (for microscopy) or onto 24-well tissue culture dishes (5 × 105 cells for CFU assays). Two hours following their seeding, adherent PEMs were infected as described above. Cell experiments using pHrodo (Invitrogen) were performed as described previously (24). Briefly, macrophages were washed with phosphate-buffered saline (PBS) plus 10% fetal bovine serum (FBS) and then labeled with pHrodo at a final concentration of 10 μg/ml. For the simultaneous labeling and infection experiments, bacteria were washed with PBS and reconstituted in PBS plus 10% FBS and then added to the macrophages.
Fluorescent and scanning electron microscopy.
Following infections, macrophage genotypes were blinded to ensure unbiased analyses. Macrophages were fixed with 3.7% paraformaldehyde for 12 min before being washed and mounted onto precleaned glass slides. 4′,6-diamidino-2-phenylindole (DAPI) was included in the ProLong gold mounting medium (Invitrogen). An Olympus fluorescence BX61 microscope was used that was equipped with Nomarski differential interference contrast (DIC) optics, a Uplan S Apo 100× objective (numeric aperture, 1.4), a Roper CoolSnap HQ camera, Sutter Lambda 10-2 excitation and emission filters, and a 175-W Xenon remote source. Intelligent Imaging Innovations Slidebook 4.01 for the Mac was used for image capture. For all cells analyzed, a series of optical Z sections (0.35 μm) were imaged, and prior to analysis, individual stacks were deconvolved using the nearest-neighbor algorithm. Representative projected images were chosen to be included in the figures. ImageJ software was used to quantify the fluoresce signal per infected cell or per bacterium. The fluorescence signal was divided by the area of the cell or bacterium to generate a signal/area ratio that was termed fluorescence intensity in arbitrary units. The percentage of bacteria colocalized with pHrodo* (red-emitting pHrodo) was generated by counting >50 cells in at least three independent experiments, and the number of macrophage-associated bacteria associated with pHrodo* signal was divided by the total number of bacteria. PEMs plated and infected on coverslips were prepared for scanning electron microscopy (SEM) by being fixed with 4% paraformaldehyde and then dehydrated through a graded series of ethanol washes (10 min each of 35%, 70%, 95%, and 100%) and then treated with hexamethyldisilazane (HMDS) for 15 min. Samples were air dried and coated with an ultrathin layer of gold using a Denton Desk V sputter and etch unit and then imaged with a Joel JSM-6010LA analytic scanning electron microscope under high vacuum using a 9-kV accelerating voltage, 40 spot size, and 9-mm working distance.
RESULTS
Perforin-2−/− mice rapidly develop listeriosis following systemic infection.
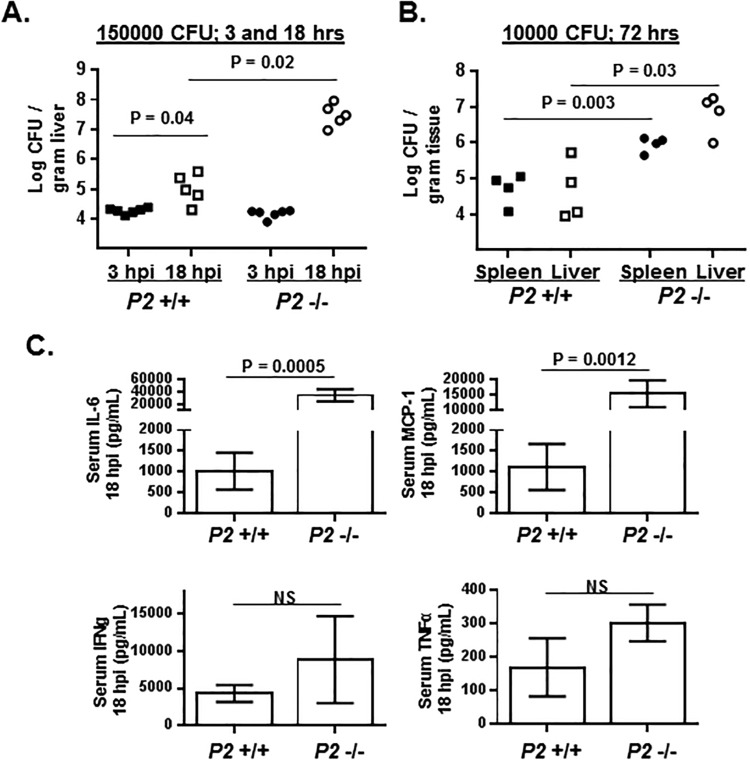
Perforin-2+/+ and Perforin-2−/− mice were infected intravenously with 150,000 CFU L. monocytogenes, and within 12 hpi Perforin-2−/− mice began to present symptoms of murine listeriosis (retarded movements, ruffled coats, and hunched backs) that, by 18 hpi, had developed to such a degree that it was necessary to euthanize the mice. In contrast, similarly infected Perforin-2+/+ mice showed absolutely no visible signs of disease and were indistinguishable in their behavior from uninfected control mice. Bacterial burdens in the livers, where blood-borne L. monocytogenes accumulates and establishes infection, of mice infected for either 3 hpi (prior to any signs of disease) or 18 hpi were analyzed to determine whether enhanced disease in Perforin-2−/− mice was accompanied by greater proliferation of the pathogen. After 3 hpi there was a comparable level of liver colonization of Perforin-2+/+ and Perforin-2−/− mice; however, during the succeeding 15 h of infection the bacterial burdens in the Perforin-2+/+ mice increased 8.5-fold, whereas bacterial burdens in the Perforin-2−/− mice increased by 2,500-fold (Fig. 1A).
FIG 1.
Perforin-2 limits the proliferation of L. monocytogenes in mice. (A) Perforin-2+/+ (P2 +/+) and Perforin-2−/− (P2 −/−) mice were infected intravenously with 1.5 × 105 CFU of L. monocytogenes. After either 3 or 18 h postinfection (hpi), mice were humanely euthanized and the bacterial burdens of the liver were enumerated by a CFU assay. Plotted are the CFU/gram of liver of individual mice (n = 6 per genotype for 3-hpi cohorts; n = 5 per genotype for 18-hpi cohorts). (B) Perforin-2+/+ and Perforin-2−/− mice were infected intravenously with 1 × 104 CFU of L. monocytogenes, and bacterial burdens of the spleen and liver were enumerated by CFU assay at 72 hpi. (C) Serum cytokine levels following 18 h of infection of mice analyzed for panel A. Serum levels of the indicated cytokines were below the level of detection in both uninfected Perforin-2+/+ and Perforin-2−/− mice. (P values were calculated using Student t test of a single representative experiment performed two times.)
In a lower-dose infection (10,000 CFU) of longer duration (72 h), Perforin-2+/+ mice showed no outward signs of disease. In contrast, Perforin-2−/− mice presented the typical symptoms of murine listeriosis noted above. Consistent with these differences in disease presentation, there were ∼20- and ∼100-fold differences in the bacterial burdens in the spleen and liver, respectively, between Perforin-2+/+ and Perforin-2−/− mice (Fig. 1B). Coincident with this accelerated infection, the Perforin-2−/− mice had significantly higher serum levels of the inflammatory cytokines IL-6 and MCP-1/CCL2 than Perforin-2+/+ mice after 18 h of infection (Fig. 1C). The levels of IFN-γ and TNF-α also were elevated (but not significantly so) in Perforin-2−/− mice compared to those in Perforin-2+/+ mice (Fig. 1C). These cytokines were not at detectable levels in the serum of uninfected Perforin-2+/+ or Perforin-2−/− mice. These data suggest that Perforin-2 is involved in limiting L. monocytogenes proliferation during the very earliest stages of systemic infection.
Perforin-2 limits the replication of L. monocytogenes in cells.
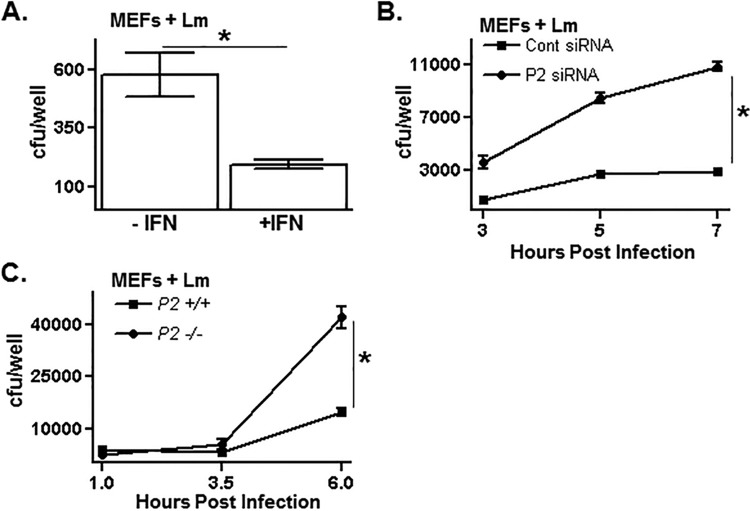
The level of intracellular survival and replication of L. monocytogenes in cultured mammalian cells is determined by both L. monocytogenes- and host-encoded factors. Treating MEFs with IFN-γ significantly limits the intracellular replication of L. monocytogenes as determined by a CFU assay (Fig. 2A). Recently, we showed that Perforin-2 is one of the hundreds of genes whose expression is increased in MEFs and other cells following IFN-γ treatment (15). To test whether Perforin-2 plays a role in limiting L. monocytogenes replication in IFN-γ-treated MEFs, Perforin-2 expression levels were reduced using siRNAs (15). At all time points tested, there was an ∼5-fold greater number of viable L. monocytogenes organisms recovered from Perforin-2 knockdown cells than from control cells (Fig. 2B). This finding suggests that Perforin-2 is a key determinant among the IFN-γ-inducible repertoire of host factors that limit intracellular L. monocytogenes proliferation.
FIG 2.
Perforin-2 limits the replication of L. monocytogenes in cells. (A) Untreated or interferon-treated mouse embryonic fibroblasts (MEFs) were infected with L. monocytogenes for 1 h and then treated with gentamicin to kill extracellular L. monocytogenes. Following an additional 2 h of infection, MEFs were lysed and the number of viable intracellular L. monocytogenes organisms was enumerated by a CFU assay. (B) MEFs were transfected with either a control or Perforin-2-specific siRNA, treated with interferon, subsequently infected with L. monocytogenes for 1 h, and then treated with gentamicin to kill extracellular L. monocytogenes. Infected MEFs were lysed at the indicated time points, and the number of viable intracellular L. monocytogenes organisms was determined by a CFU assay. Cont, control. (C) Perforin-2+/+ and Perforin-2−/− MEFs were infected with L. monocytogenes for 1 h and then treated with gentamicin to kill extracellular L. monocytogenes. The number of viable intracellular L. monocytogenes organisms at the indicated time points was determined by a CFU assay. An asterisk indicates a P value of <0.05 as calculated using Student t test of a single representative experiment performed multiple times.
The role Perforin-2 plays in L. monocytogenes infection was tested further using MEFs and primary macrophages derived from Perforin-2−/− mice described above. At early infection time points, Perforin-2+/+ and Perforin-2−/− MEFs were infected at comparable levels, indicating that the attachment and cell invasion phases of L. monocytogenes infection are not affected by Perforin-2 (Fig. 2C). However, at infections times greater than 3.5 h, there were considerably more viable L. monocytogenes organisms recovered from Perforin-2−/− MEFs than from Perforin-2+/+ MEFs.
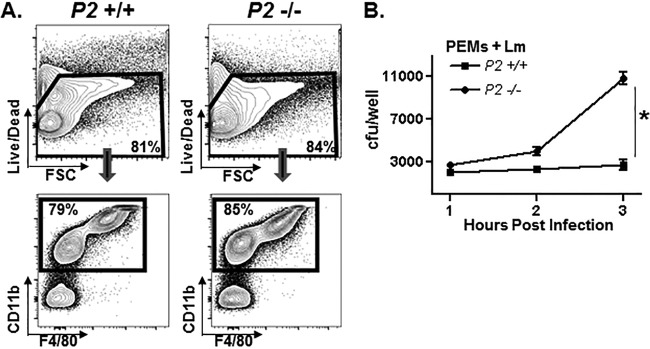
PEMs isolated from Perforin-2+/+ and Perforin-2−/− mice were phenotypically similar in terms of expression levels of macrophage-specific surface markers (Fig. 3A). Similar to MEFs, Perforin-2+/+ and Perforin-2−/− PEMs initially were infected equally well, but starting at around 2 h of infection, the recovery of viable L. monocytogenes from Perforin-2−/− macrophages exceeded that which was recovered from Perforin-2+/+ PEMs (Fig. 3B). Although the infection kinetics differ between MEFs and PEMs in the foregoing assays, it is clear for both cell types that Perforin-2 plays a significant role in restricting the intracellular replication of L. monocytogenes.
FIG 3.
Perforin-2 limits the replication of L. monocytogenes in macrophages. (A) Total peritoneal cells isolated from Perforin-2+/+ and Perforin-2−/− mice were assessed for viability (upper), and the percentage of live macrophages (PEMs) was determined by macrophage-specific surface markers CD11b and F4/80 (lower). FSC, forward scatter. (B) PEMs isolated from Perforin-2+/+ and Perforin-2−/− mice were infected in vitro with L. monocytogenes for 1 h and then treated with gentamicin to kill extracellular L. monocytogenes. The number of viable intracellular L. monocytogenes organisms at the indicated time points was determined in a CFU assay. An asterisk indicates a P value of <0.05 as calculated using Student t test of a single representative experiment performed multiple times.
Perforin-2 limits the translocation of L. monocytogenes into the cytosol.
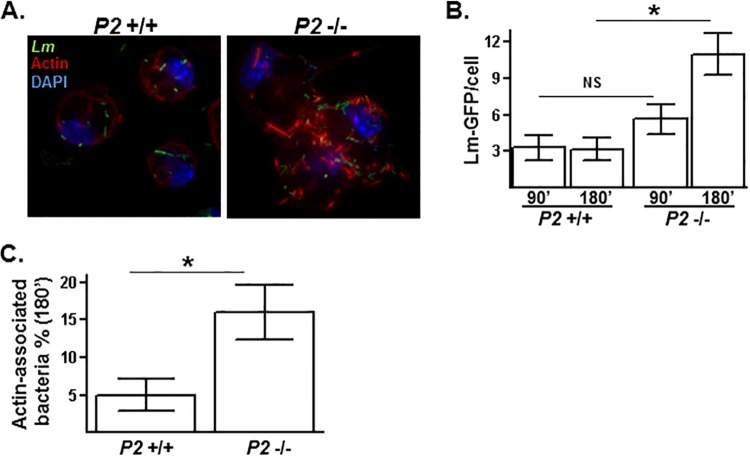
Following the invasion of the cell, L. monocytogenes briefly resides in a membrane-bound phagocytic vacuole before being released into the cytosol. Upon entry into the cytosol, the surface-exposed virulence factor ActA binds monomeric actin as well as the actin polymerization factors onto the exterior of L. monocytogenes (25). Cytosolic L. monocytogenes initially becomes embedded within a cloud of actin, which eventually leads to the emergence of L. monocytogenes possessing actin “tails” due to the preferential polymerization at one pole of the bacterial cell. Therefore, within infected cells, vacuole-bound L. monocytogenes can readily be distinguished from cytosolic-residing L. monocytogenes by staining for actin. In examining individual cells infected with L. monocytogenes, there were substantially fewer actin-associated L. monocytogenes organisms in Perforin-2+/+ macrophages than Perforin-2−/− macrophages following 3 h of infection (Fig. 4A). Additionally, a substantial fraction of cytosolic L. monocytogenes in Perforin-2−/− macrophages (but not Perforin-2+/+ macrophages) had transitioned from being embedded within actin clouds to possessing actin tails, suggesting more rapid kinetics of the L. monocytogenes infection process in the absence of Perforin-2. Shorter infection assays were analyzed to compare the rate of infection kinetics in Perforin-2+/+ and Perforin-2−/− macrophages. Following 90 min of infection, Perforin-2+/+ and Perforin-2−/− macrophages were infected similarly both in terms of the average total number of intracellular L. monocytogenes organisms per cell (Fig. 4B) and the fraction of those L. monocytogenes organisms associated with actin. However, between 90 and 180 min of infection, significant differences arose between the Perforin-2+/+ and Perforin-2−/− macrophages in terms of both the total number of intracellular L. monocytogenes organisms per cell (2-fold greater in Perforin-2−/− than Perforin-2+/+) (Fig. 4B) and the fraction of actin-associated L. monocytogenes (3-fold greater in Perforin-2−/− than Perforin-2+/+ macrophages) (Fig. 4C). These data show that Perforin-2 limits the intracellular replication of L. monocytogenes by inhibiting its translocation into the cytosol.
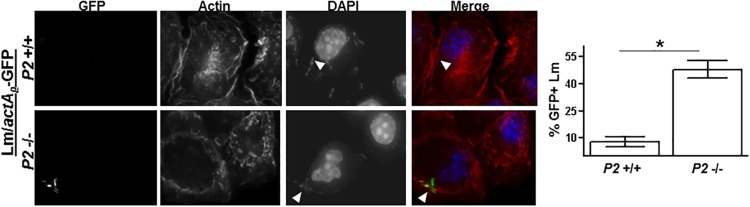
FIG 4.
Perforin-2 inhibits the release of L. monocytogenes into the cytosol. (A) PEMs isolated from Perforin-2+/+ and Perforin-2−/− mice were infected in vitro with L. monocytogenes constitutively expressing GFP (Lm-GFP) for 1 h and then treated with gentamicin to kill extracellular L. monocytogenes. After an additional 2 h of infection, PEMs were stained for actin (red) and nuclei (purple). (B) Perforin-2+/+ and Perforin-2−/− PEMs were infected with L. monocytogenes-GFP for 1 h and then treated with gentamicin to kill extracellular L. monocytogenes. After an additional 30 and 120 min of infection, PEMs were stained for actin. Plotted is the total number intracellular L. monocytogenes-GFP (n > 150) organisms associated with >40 infected PEMs. (C) The percentage of cytosolic L. monocytogenes-GFP organisms was determined by dividing actin-positive L. monocytogenes-GFP organisms by the total number of intracellular L. monocytogenes-GFP organisms. An asterisk indicates a P value of <0.05 as calculated using Student t test of a single representative experiment performed multiple times. NS, not significant.
Perforin-2 limits infection-induced expression of ActA and vacuole acidification.
The various steps of the L. monocytogenes infection cycle (i.e., invasion, cytosolic translocation, and actin-based motility) are driven by virulence factors whose expressions are tightly regulated. The relatively higher rate of cytosolic translocation and cloud-to-tail transitioning observed in Perforin-2−/− macrophages indicates that the expression levels and/or functioning of the virulence factors mediating these intracellular activities differ in L. monocytogenes-infecting Perforin-2+/+ and Perforin-2−/− macrophages. The promoter controlling the transcription of the ActA-encoding locus (actAp) becomes activated following invasion of the cell (26, 27). An L. monocytogenes strain possessing a GFP-encoding reporter gene under the control of actAp (termed the L. monocytogenes-actAp-GFP strain) was used to determine whether there are differences in ActA expression during infection of Perforin-2+/+ and Perforin-2−/− macrophages. L. monocytogenes-actAp-GFP bacteria grown in tissue culture media in the absence of macrophages do not express detectable levels of GFP (not shown). After 150 min of infection of Perforin-2+/+ macrophages, only a small fraction (<10%) of intracellular L. monocytogenes-actAp-GFP bacteria express detectable GFP (Fig. 5). In contrast, in Perforin-2−/− macrophages nearly half (45%) of L. monocytogenes-actAp-GFP bacteria express detectable amounts of GFP. Enhanced actAp activity likely accounts for the more rapid cloud-to-tail transitioning observed in Perforin-2−/− macrophages.
FIG 5.
Perforin-2 inhibits L. monocytogenes virulence gene promoter activity. PEMs were infected with an L. monocytogenes strain possessing an actAp-GFP reporter gene (L. monocytogenes-actAp-GFP; see the text for details). After 1 h of infection, PEMs were treated with gentamicin to kill extracellular L. monocytogenes. After an additional 1.5 h of infection, PEMs were stained for actin and nuclei. Arrows indicate L. monocytogenes-actAp-GFP expressing the GFP reporter protein. Shown to the right is the percentage of L. monocytogenes-actAp-GFP bacteria (n > 50) that were positive for green fluorescence. An asterisk indicates a P value of <0.05 as calculated using Student t test of a single representative experiment performed multiple times.
Enhanced ActA expression levels in Perforin-2−/− macrophages could be driven by more favorable conditions (for the pathogen) during the intravacuolar phase of its infection cycle. The vacuole-to-cytosol transition of L. monocytogenes is mediated in part by secreted virulence factors that become highly active at the reduced luminal pH of the primary vacuole (28, 29). To monitor the pH of vacuoles of Perforin-2+/+ and Perforin-2−/− macrophages, cells were labeled with an ∼10-kDa dextran conjugated with a pH-sensitive rhodamine derivative (pHrodo). The acidification of this reagent leads to a linear increase in its fluorescence, starting at pH 6.8 and reaching a maximum at pH 5. When added to cells pHrodo is internalized by bulk-phase endocytosis, and as it is trafficked from early endosomal to lysosomal compartments in the perinuclear region, it becomes increasingly fluorescent due to the reduction in pH. There were no detectable differences between uninfected Perforin-2+/+ and Perforin-2−/− macrophages following a 45-min labeling period, indicating that neither bulk-phase endocytosis nor endosomal acidification differs between these cells (Fig. 6A). When Perforin-2+/+ macrophages were infected simultaneously with L. monocytogenes and labeled with pHrodo for 15 min, there was not a single observed L. monocytogenes organism in an acidified compartment (i.e., no colocalization of L. monocytogenes and red-emitting pHrodo) (Fig. 6B). In striking contrast, 30% of L. monocytogenes organisms infecting Perforin-2−/− macrophages were contained in highly acidic compartments, as indicated by the colocalization of L. monocytogenes and intense pHrodo fluorescence. This rapid (≤15 min) and extreme acidification in Perforin-2−/− macrophages was highly localized in individual cells such that many cells were observed which contained L. monocytogenes in both nonacidified and acidified compartments. Furthermore, the degree of acidification observed in L. monocytogenes-containing compartments of Perforin-2−/− macrophages was entirely absent from Perforin-2+/+ macrophages, even after prolonged (≥15 min) infection periods. Analyzing these early infection events by scanning electron microscopy (SEM) showed that structurally the L. monocytogenes cellular invasion process appeared similar in Perforin-2+/+ and Perforin-2−/− macrophages; however, it appeared to occur more rapidly in Perforin-2−/− macrophages, as indicated by L. monocytogenes being more highly embedded within the Perforin-2−/− macrophage membrane than the Perforin-2+/+ macrophage membrane following 30 min of infection (Fig. 6C). Together, these data indicate substantial differences in the immediate-early responses of Perforin-2+/+ and Perforin-2−/− macrophages to L. monocytogenes infection and suggest that Perforin-2−/− cells provide a more favorable environment for L. monocytogenes virulence protein expression and/or activity.
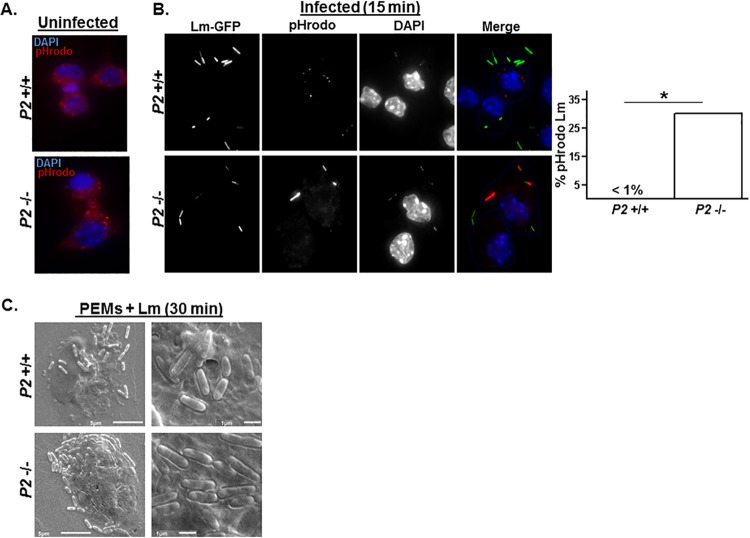
FIG 6.
Perforin-2 inhibits L. monocytogenes vacuole acidification. (A) Uninfected Perforin-2+/+ and Perforin-2−/− PEMs were labeled for 45 min with dextran-conjugated pH-sensitive rhodamine derivative (pHrodo) that emits red fluorescence in increasingly acidic late endosomes/lysosomal perinuclear compartments. (B) Perforin-2+/+ and Perforin-2−/− PEMs were labeled simultaneously with pHrodo and infected with L. monocytogenes-GFP for 15 min and then analyzed by fluorescence microscopy. (Intense pHrodo signal in the Perforin-2−/− image diminishes the GFP-based signal.) Plotted to the right is the percentage of L. monocytogenes-GFP colocalized with acid-positive pHrodo (red) calculated by counting >200 bacteria associated with >75 infected cells in multiple experiments. (C) Perforin-2+/+ and Perforin-2−/− PEMs were infected with L. monocytogenes-GFP for 30 min and then analyzed by scanning electron microscopy (SEM) at a magnification of either ×3,500 (left) or ×10,000 (right).
Hyperexpression of L. monocytogenes virulence factors overcomes Perforin-2-mediated intracellular growth restriction.
If Perforin-2 limits intracellular L. monocytogenes replication by acting to inhibit the induction of virulence gene expression, then it would be expected that L. monocytogenes mutant strains that constitutively express high levels of virulence factors would have similar infection dynamics in Perforin-2+/+ and Perforin-2−/− cells. The major transcriptional regulator of virulence gene expression in L. monocytogenes, PrfA, is itself activated posttranslationally following infection (30). L. monocytogenes strains expressing constitutively active PrfA variants (encoded by prfA* alleles) are hypervirulent in both cell culture and animal infection models (21, 31). Perforin-2+/+ and Perforin-2−/− macrophages were infected with one such prfA* strain that expresses a PrfA containing a G145S mutation. Consistent with earlier published findings, there was much greater replication of the prfA* strain in Perforin-2+/+ macrophages than in the wild-type L. monocytogenes strain (Fig. 7A). There was also a high level of replication of the prfA* strain in Perforin-2−/− macrophages, similar to that observed in Perforin-2+/+ macrophages and in contrast to wild-type L. monocytogenes, which, as shown earlier, has reduced replication in Perforin-2-expressing macrophages (Fig. 7A). Consistent with the replication data, there was a comparable level of cytosolic invasion of the prfA* strain in Perforin-2+/+ and Perforin-2−/− cells, in contrast to wild-type L. monocytogenes which, again as shown earlier, has reduced access to the cytosol in Perforin-2-expressing cells (Fig. 7B). Although it should be noted that the overexpression of virulence genes may mask other differences between Perforin-2+/+ and Perforin-2−/− cells, these data are consistent with a model that Perforin-2 limits L. monocytogenes intracellular replication by inhibiting the expression of its virulence genes.
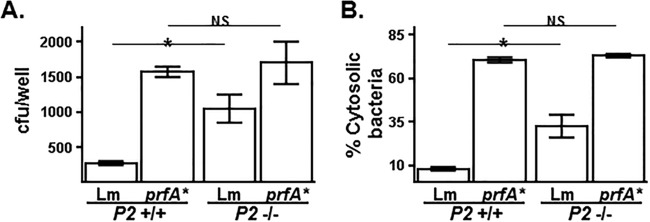
FIG 7.
Hyperexpression of L. monocytogenes virulence genes overcomes the Perforin-2-mediated restriction of L. monocytogenes replication. PEMs isolated from Perforin-2+/+ and Perforin-2−/− mice were infected in vitro with either wild-type L. monocytogenes or an isogenic mutant strain that constitutively hyperexpresses virulence factors (prfA*; see the text) for 1 h and then treated with gentamicin to kill extracellular L. monocytogenes. After an additional 0.5 h of infection, PEMs were analyzed for either viable intracellular bacterial levels by CFU assay (A) or cytosolic bacteria (B) as described in the legend to Fig. 4A. An asterisk indicates a P value of <0.05 as calculated using Student t test of a single representative experiment performed multiple times.
DISCUSSION
Here, we showed that a host cell-encoded MACPF/CDC-containing protein, Perforin-2, acts to restrict both the replication of L. monocytogenes in cells and the systemic proliferation of this bacterial pathogen in the mouse. As mentioned in the introduction, L. monocytogenes employs an MACPF/CDC-containing protein of its own, LLO, to damage the vacuolar membrane to gain access to its replication niche in the cytosol. Thus, in light of the findings presented here, the outcome of the host-L. monocytogenes interaction is largely dependent on the relative activities of these two MACPF/CDC proteins.
We provide evidence that Perforin-2 may be involved in cellular processes that impact the host-pathogen interaction. L. monocytogenes follows a well-characterized modus operandi following its internalization within the primary vacuole of the infected cell. If Perforin-2 only acted to increase the mortality of L. monocytogenes during its intravacuole transitioning to the cytosol, it would be expected that the kinetics of the cellular infection process (in terms of events per unit of time) would be similar between Perforin-2+/+ and Perforin-2−/− cells. However, what was observed was that in Perforin-2−/− cells the pace at which these events occurred, on a per-bacterium basis, was increased. These events included the emergence of L. monocytogenes from the primary vacuole, the activation of the actA promoter, and the level of actin binding and polymerization into tails. A clear indication that there are qualitative differences between Perforin-2+/+ and Perforin-2−/− cells in their interaction with L. monocytogenes revealed itself when the endosomal acidification process was analyzed. After a very brief 15-min infection period, Perforin-2−/− macrophages possessed L. monocytogenes-containing vacuoles that were highly acidic; compartments of comparable acidity were entirely absent from Perforin-2+/+ macrophages. This difference between Perforin-2+/+ and Perforin-2−/− macrophages was observed only in L. monocytogenes-containing compartments, whereas there were no obvious differences in general endosomal trafficking between Perforin-2+/+ and Perforin-2−/− macrophages.
The rapid acidification of L. monocytogenes-containing vacuoles in Perforin-2−/− macrophages almost certainly plays a role in the quickened pace of the infection process, since both the expression and activity of L. monocytogenes virulence factors are stimulated by low pH (28, 29). In addition to the already-mentioned pH-dependent activity of the master transcription factor PrfA that directly regulates virulence gene expression (e.g., the gene promoter for actA), the membrane-disrupting activity of LLO is strongly enhanced when the pH falls below 5.5. This pH dependency differentiates LLO from other MACPF/CDC-containing toxins of Gram-positive pathogens (32–34). Specifically, it has been shown recently that reduced pH promotes the transitioning of LLO from a prepore to pore-forming conformation (35). The hypothesis that Perforin-2 impacts L. monocytogenes infection in cells through its effects on virulence factor activity was supported by the finding that L. monocytogenes strains expressing constitutively high levels of virulence factors had comparable infection dynamics in Perforin-2+/+ and Perforin-2−/− cells.
Precisely how Perforin-2 prevents the rapid acidification of L. monocytogenes-containing vacuoles remains to be determined. The pH of these compartments is controlled by both vacuolar ATPases (V-ATPases) that translocate protons into the luminal space as well as the back-flux (or leakage) of protons back into the cytosol that is mediated by various mechanisms (36). In situ measurements of the phagosomal pH of peritoneal mouse macrophages (the same as the cells used in this study) found that under homeostatic conditions, the back-fluxing of protons reduces the phagosomal pH by a rate of 0.09/min (37). The regulation of V-ATPases and proton back-fluxing is complex and is targeted by a number of intracellular pathogens (36). Perforin-2 may inhibit the rapid reduction of pH of L. monocytogenes-containing vacuoles by negatively regulating V-ATPases and/or by promoting proton back-fluxing. What is clear is that host-encoded Perforin-2 and pathogen-encoded LLO, which both possess evolutionarily related MACPF/CDC domains, compete for dominance during the very initial stages of infection.
ACKNOWLEDGMENTS
We thank George Munson, Emily Clark, and Becky Adkins for their generous assistance.
E.R.P. and K.S. conceived and coordinated the overall study. R.M., W.B., N.S., J.B., M.B., and K.S. designed, performed, and analyzed the experiments. N.S., C.M.B., E.A.D., N.E.F., and W.N.K. provided technical assistance and reagents. K.S. wrote the manuscript, and all authors approved the final version.
This paper is dedicated to the memory of our co-lead author and inspiration leader, Eckhard R. Podack, who died during the preparation of the manuscript.
REFERENCES
- 1.McCormack R, de Armas L, Shiratsuchi M, Podack ER. 2013. Killing machines: three pore-forming proteins of the immune system. Immunol Res 57:268–278. doi: 10.1007/s12026-013-8469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Podack ER, Tschopp JM. 1982. Polymerization of the ninth component of complement (C9): formation of poly(C9) with a tubular ultrastructure resembling the membrane attack complex of complement. Proc Natl Acad Sci U S A 79:574–578. doi: 10.1073/pnas.79.2.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tschopp J, Müller-Eberhard HJ, Podack ER. 1982. Formation of transmembrane tubules by spontaneous polymerization of the hydrophilic complement protein C9. Nature 298:534–538. doi: 10.1038/298534a0. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber RD, Morrison DC, Podack ER, Müller-Eberhard HJ. 1979. Bactericidal activity of the alternative complement pathway generated from 11 isolated plasma proteins. J Exp Med 149:870–882. doi: 10.1084/jem.149.4.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Podack ER, Dennert G. Assembly of two types of tubules with putative cytolytic function by cloned natural killer cells. Nature 302:442–445. [DOI] [PubMed] [Google Scholar]
- 6.Dennert G, Podack ER. 1983. May 1 Cytolysis by H-2-specific T killer cells. Assembly of tubular complexes on target membranes. J Exp Med 157:1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiens M, Korzhev M, Krasko A, Thakur NL, Perović-Ottstadt S, Breter HJ, Ushijima H, Diehl-Seifert B, Müller IM, Müller WE. 2005. July 29 Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway. Induction of a perforin-like molecule. J Biol Chem 280:27949–27959. [DOI] [PubMed] [Google Scholar]
- 8.Mah SA, Moy GW, Swanson WJ, Vacquier VD. 2004. A perforin-like protein from a marine mollusk. Biochem Biophys Res Commun 316:468–475. doi: 10.1016/j.bbrc.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 9.Wang GD, Zhang KF, Zhang ZP, Zou ZH, Jia XW, Wang SH, Lin P, Wang YL. 2008. Molecular cloning and responsive expression of macrophage expressed gene from small abalone Haliotis diversicolor supertexta. Fish Shellfish Immunol 24:346–359. doi: 10.1016/j.fsi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 10.He X, Zhang Y, Yu Z. 2011. An Mpeg (macrophage expressed gene) from the Pacific oyster Crassostrea gigas: molecular characterization and gene expression. Fish Shellfish Immunol 30:870–876. doi: 10.1016/j.fsi.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Benard EL, Racz PI, Rougeot J, Nezhinsky AE, Verbeek FJ, Spaink HP, Meijer AH. 2015. Macrophage-expressed perforins mpeg1 and mpeg1.2 have an anti-bacterial function in zebrafish. J Innate Immun 7:136–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Angelo ME, Dunstone MA, Whisstock JC, Trapani JA, Bird PI. 2012. May 2 Perforin evolved from a gene duplication of MPEG1, followed by a complex pattern of gene gain and loss within Euteleostomi. BMC Evol Biol 12:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosado CJ, Buckle AM, Law RH, Butcher RE, Kan WT, Bird CH, Ung K, Browne KA, Baran K, Bashtannyk-Puhalovich TA, Faux NG, Wong W, Porter CJ, Pike RN, Ellisdon AM, Pearce MC, Bottomley SP, Emsley J, Smith AI, Rossjohn J, Hartland EL, Voskoboinik I, Trapani JA, Bird PI, Dunstone MA, Whisstock JC. 2007. A common fold mediates vertebrate defense and bacterial attack. Science 317:1548–1551. [DOI] [PubMed] [Google Scholar]
- 14.Rosado CJ, Kondos S, Bull TE, Kuiper MJ, Law RH, Buckle AM, Voskoboinik I, Bird PI, Trapani JA, Whisstock JC, Dunstone MA. 2008. The MACPF/CDC family of pore-forming toxins. Cell Microbiol 10:1765–1774. doi: 10.1111/j.1462-5822.2008.01191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormack R, de Armas LR, Shiratsuchi M, Ramos JE, Podack ER. 2013. Inhibition of intracellular bacterial replication in fibroblasts is dependent on the perforin-like protein (perforin-2) encoded by macrophage-expressed gene 1. J Innate Immun 5:185–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields KA, McCormack R, de Armas LR, Podack ER. 2013. Perforin-2 restricts growth of Chlamydia trachomatis in macrophages. Infect Immun 81:3045–3054. doi: 10.1128/IAI.00497-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray K, Marteyn B, Sansonetti PJ, Tang CM. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 18.Portnoy DA, Jacks PS, Hinrichs DJ. 1988. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med 167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormack R, de Armas LR, Shiratsuchi M, Fiorentino D, Olsson M, Lichtenheld M, Morales A, Lyapichev K, Gonzalez L, Strbo N, Sukumar N, Stojadinovic O, Plano G, Tomic-Canic M, Kirsner R, Russell D, Podack ER. 2015. Perforin-2 is essential for intracellular defense of parenchymal cells and phagocytes against pathogenic bacteria. eLife 24:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brundage RA, Smith GA, Camilli A, Theriot JA, Portnoy DA. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc Natl Acad Sci U S A 90:11890–11894. doi: 10.1073/pnas.90.24.11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruno JC Jr, Freitag NE. 2010. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One 5:e15138. doi: 10.1371/journal.pone.0015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 23.Shrestha N, Boucher J, Bahnan W, Clark ES, Rosqvist R, Fields KA, Khan WN, Schesser K. 2013. The host-encoded Heme Regulated Inhibitor (HRI) facilitates virulence-associated activities of bacterial pathogens. PLoS One 8:e68754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahnan W, Boettner DR, Westermark L, Fällman M, Schesser K. 2015. Pathogenic Yersinia promotes its survival by creating an acidic fluid-accessible compartment on the macrophage surface. PLoS One 10:e0133298. doi: 10.1371/journal.pone.0133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocks C, Gouin E, Tabouret M, Berche P, Ohayon H, Cossart P. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521–531. [DOI] [PubMed] [Google Scholar]
- 26.Freitag NE, Jacobs KE. 1999. Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect Immun 67:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moors MA, Levitt B, Youngman P, Portnoy DA. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect Immun 67:131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beauregard KE, Lee KD, Collier RJ, Swanson JA. 1997. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J Exp Med 186:1159–1163. doi: 10.1084/jem.186.7.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glomski IJ, Gedde MM, Tsang AW, Swanson JA, Portnoy DA. 2002. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J Cell Biol 156:1029–1038. doi: 10.1083/jcb.200201081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freitag NE, Rong L, Portnoy DA. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect Immun 61:2537–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miner MD, Port GC, Freitag NE. 2008. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology 154:3579–3589. doi: 10.1099/mic.0.2008/021063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bavdek A, Kostanjšek R, Antonini V, Lakey JH, Dalla Serra M, Gilbert RJ, Anderluh G. 2012. pH dependence of listeriolysin O aggregation and pore-forming ability. FEBS J 279:126–141. doi: 10.1111/j.1742-4658.2011.08405.x. [DOI] [PubMed] [Google Scholar]
- 33.Schuerch DW, Wilson-Kubalek EM, Tweten RK. 2005. Molecular basis of listeriolysin O pH dependence. Proc Natl Acad Sci U S A 102:12537–12542. doi: 10.1073/pnas.0500558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomura T, Kawamura I, Kohda C, Baba H, Ito Y, Kimoto T, Watanabe I, Mitsuyama M. 2007. Irreversible loss of membrane-binding activity of Listeria-derived cytolysins in non-acidic conditions: a distinct difference from allied cytolysins produced by other Gram-positive bacteria. Microbiology 153(Part 7):2250–2258. doi: 10.1099/mic.0.2007/005843-0. [DOI] [PubMed] [Google Scholar]
- 35.Podobnik M, Marchioretto M, Zanetti M, Bavdek A, Kisovec M, Cajnko MM, Lunelli L, Dalla Serra M, Anderluh G. 2015. Plasticity of lysteriolysin O pores and its regulation by pH and unique histidine. Sci Rep 5:9623. doi: 10.1038/srep09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huynh KK, Grinstein S. 2007. Regulation of vacuolar pH and its modulation by some microbial species. Microbiol Mol Biol Rev 71:452–462. doi: 10.1128/MMBR.00003-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukacs GL, Rotstein OD, Grinstein S. 1991. Determinants of the phagosomal pH in macrophages. In situ assessment of vacuolar H(+)-ATPase activity, counterion conductance, and H+ “leak.” J Biol Chem 266:24540–24548. [PubMed] [Google Scholar]