Abstract
Development of acetic acid-resistant Saccharomyces cerevisiae is important for economically viable production of biofuels from lignocellulosic biomass, but the goal remains a critical challenge due to limited information on effective genetic perturbation targets for improving acetic acid resistance in the yeast. This study employed a genomic-library-based inverse metabolic engineering approach to successfully identify a novel gene target, WHI2 (encoding a cytoplasmatic globular scaffold protein), which elicited improved acetic acid resistance in S. cerevisiae. Overexpression of WHI2 significantly improved glucose and/or xylose fermentation under acetic acid stress in engineered yeast. The WHI2-overexpressing strain had 5-times-higher specific ethanol productivity than the control in glucose fermentation with acetic acid. Analysis of the expression of WHI2 gene products (including protein and transcript) determined that acetic acid induced endogenous expression of Whi2 in S. cerevisiae. Meanwhile, the whi2Δ mutant strain had substantially higher susceptibility to acetic acid than the wild type, suggesting the important role of Whi2 in the acetic acid response in S. cerevisiae. Additionally, overexpression of WHI2 and of a cognate phosphatase gene, PSR1, had a synergistic effect in improving acetic acid resistance, suggesting that Whi2 might function in combination with Psr1 to elicit the acetic acid resistance mechanism. These results improve our understanding of the yeast response to acetic acid stress and provide a new strategy to breed acetic acid-resistant yeast strains for renewable biofuel production.
INTRODUCTION
Lignocellulosic biomass from nonfood stocks such as agricultural and forestry residues has been identified as the prime source for production of renewable biofuels to substitute for conventional fossil fuels in the face of growing demand for energy and rising concerns about greenhouse gas emissions (1–4). Bioconversion of plant cell wall materials by microbial fermentation is typically preceded by harsh (physico)chemical hydrolysis designed to release sugars; this hydrolysis treatment also generates by-products that are toxic to fermenting microorganisms (5, 6). Since hemicellulose and lignin in the plant cell wall are ubiquitously acetylated (7, 8), the typical acidic pretreatment of lignocellulosic biomass generates substantial amounts of acetic acid (with concentrations ranging from 1 g/liter to 15 g/liter) in the resulting hydrolysates (9, 10). Acetic acid severely inhibits cell growth and fermentation activity in Saccharomyces cerevisiae (5, 6, 11–13), the predominant microorganism used in industrial fermentation (14, 15). Therefore, improvement in S. cerevisiae resistance to acetic acid is highly desired and critical for achieving efficient and economically viable bioconversion of cellulosic sugars to biofuels.
The toxic effects of acetic acid in S. cerevisiae have been intensively characterized, and toxicity mechanisms have been proposed (10, 11, 16–20). When the external pH is lower than the pKa of acetic acid (4.7), the undissociated form of acetic acid prevails and can enter the cells simply by diffusion through plasma membranes. The dissociation of acetic acid at neutral cytosolic pH can lead to intracellular acidification (6). As a result, the intracellular pH needs to be recovered by pumping out protons at the expense of ATP hydrolysis, which may induce cell growth arrest and reduced fermentation performance (21). In addition, intracellular anion accumulation can reach high levels and decrease the activity of some key enzymes for glycolysis (16).
However, the genetic basis of the yeast stress response to acetic acid remains unclear, making it difficult to improve the acetic acid resistance in S. cerevisiae. It is known that the yeast response to acetic acid stress involves genome-wide transcriptional changes (22–25). For example, upregulation of various genes involved in glycolysis, the Krebs cycle, and ATP synthesis was identified in yeast cells cultivated in the presence of acetic acid, indicating substantial alterations in carbohydrate and energy metabolism in acetic acid-stressed cells (26). Genome-scale transcriptional analyses suggested that the transcriptional activators Haa1 and Msn2 might be involved in regulating yeast adaptation to acetic acid (21, 26, 27). A large-scale chemical genomics study identified 648 genes whose deletion increased the susceptibility of yeast to acetic acid, and these gene determinants were found to be involved in carbohydrate metabolism, cell wall assembly, amino acid metabolism, internal pH homeostasis, biogenesis of mitochondria, and signaling and uptake of various nutrients (26). Recent studies reported cell-to-cell heterogeneity in acetic acid tolerance (28, 29). It was found that only a fraction of the cells within an isogenic S. cerevisiae population resumed growth under acetic acid stress (28) and that variations in the cytosolic pH of individual cells might contribute to the differences between cells (29). The findings suggested that genes related to cell-to-cell heterogeneity might be a potential pool for the search of genetic targets for improving acetic acid resistance. These prior results illustrate that the acetic acid stress response mechanism in S. cerevisiae is rather complex and involves coordinated regulations of multiple genes. Due to the currently incomplete understanding of relevant genetic and biomolecular networks for the acetic acid stress response, it is challenging to develop acetic acid-resistant yeast strains through knowledge-based rational metabolic engineering (30). In particular, information regarding the genetic targets and perturbation strategy for effectively engineering acetic acid-resistant yeast strains is needed.
This study employed a genomic-library-based inverse metabolic engineering approach to develop S. cerevisiae strains for improved fermentation of cellulosic sugars under acetic acid stress and identify genetic perturbation targets for enhancing the acetic acid resistance in the yeast. Specifically, we first introduced a genome-wide library into a parent S. cerevisiae strain containing an optimized xylose fermentation pathway, so that we could evaluate the transformants in the fermentation of glucose and/or xylose (the two most abundant sugars from lignocellulosic biomass) under acetic acid stress. We then characterized the screened transformants to identify the target of gene perturbation. Here we report on a novel gene target, WHI2. Overexpression of WHI2 substantially enhanced the acetic acid resistance of S. cerevisiae. We show that acetic acid-induced endogenous expression of Whi2 and that deletion of the WHI2 gene resulted in hypersensitivity to acetic acid. We further determined the function of Whi2 and its binding partner Psr1 in eliciting the acetic acid resistance mechanisms in S. cerevisiae. Last, we characterized improved performance of an engineered strain for glucose and/or xylose fermentation in the presence of toxic levels of acetic acid under industrially relevant conditions. The results improve our understanding of the stress response to acetic acid in S. cerevisiae and provide a new strategy for breeding acetic acid-resistant yeast strains for renewable biofuel production from lignocellulosic biomass.
MATERIALS AND METHODS
Strains and plasmids.
All of the strains and plasmids used in this study are summarized in Table 1. The recombinant xylose-fermenting S. cerevisiae strain SR8 (31) was used in this study for glucose and xylose fermentation. The strain was constructed previously (31) through heterologous expression of XYL1 (coding for xylose reductase [XR]), XYL2 (coding for xylitol dehydrogenase [XDH]), and XYL3 (coding for xylulokinase [XK]) from Scheffersomyces stipitis in S. cerevisiae D452-2 (MATα leu2 ura3 his3 can1) and optimization of the expression levels of XR, XDH, and XK, laboratory evolution on xylose, and deletion of ALD6, coding for acetaldehyde dehydrogenase. The auxotrophic marker genes in the strains SR8-trp and SR8-4 (Table 1) were recovered by the CRISPR-cas9 method (32, 33). These strains were kindly provided by Yong-Su Jin's lab. The Escherichia coli TOP10 strain was used for gene cloning and manipulation. The whi2 knockout mutant derived from the laboratory strain BY4741 (GE Healthcare Dharmacon) was used for evaluating the susceptibility of the null mutant versus that of the wild type. The msn2 knockout mutant derived from the strain SR8-trp was used for testing the hypothesis regarding the interaction of Whi2 and Msn2 in eliciting acetic acid resistance.
TABLE 1.
Plasmids and strains
| Plasmid or strain | Description | Reference or source |
|---|---|---|
| Plasmids | ||
| pRS424 | TRP1, a multicopy episomal plasmid | 65 |
| pRS425 | LEU2, a multicopy episomal plasmid | 65 |
| pRS424GPD | pRS424 with the GPD promoter | 65 |
| pRS425GPD | pRS425 with the GPD promoter | 65 |
| pRS424GPD-WHI2 | WHI2 expressed in pRS424GPD | This study |
| pRS425GPD-WHI2 | WHI2 expressed in pRS425GPD | This study |
| pRS424GPD-HAA1 | HAA1 expressed in pRS424GPD | This study |
| pRS424GPD-MSN2 | MSN2 expressed in pRS424GPD | This study |
| pRS424GPD-PSR1 | PSR1 expressed in pRS424GPD | This study |
| pRS425GPD-PSR1 | PSR1 expressed in pRS425GPD | This study |
| Strains | ||
| D452-2 | MATα leu2 his3 ura3 can1 | 66 |
| SR8 | D452-2 expressing XYL1, XYL2, and XKS1 through integration, evolutionary engineering in xylose-containing media, and ALD6 deletion | 31 |
| SR8-trp | SR8 with trp1 as the auxotrophic marker | Developed in Yong-Su Jin's lab |
| S-msn2Δ | msn2 knockout mutant of SR8-trp | This study |
| SR8-4 | SR8 with trp1, leu2, his3, and ura3 as auxotrophic markers | Developed in Yong-Su Jin's lab |
| S-C1 | SR8-trp harboring pRS424GPD | This study |
| S-WHI2 | SR8-trp harboring pRS424GPD-WHI2 | This study |
| S-HAA1 | SR8-trp harboring pRS424GPD-HAA1 | This study |
| S-MSN2 | SR8-trp harboring pRS424GPD-MSN2 | This study |
| S-msn2Δ-C1 | S-msn2Δ harboring pRS424GPD | This study |
| S-msn2Δ-WHI2 | S-msn2Δ harboring pRS424GPD-WHI2 | This study |
| S-C2 | SR8-4 harboring pRS424GPD and pRS425GPD, as a control | This study |
| S-WHI2-c | SR8-4 harboring pRS424GPD-WHI2 and pRS425GPD | This study |
| S-PSR1-c | SR8-4 harboring pRS424GPD-PSR1 and pRS425GPD | This study |
| S-WHI2-PSR1 | SR8-4 harboring pRS424GPD-WHI2 and pRS425GPD-PSR1 | This study |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | |
| BY2-WHI2 | BY4742 harboring pRS425GPD-WHI2 | This study |
| BY2-C1 | BY4742 harboring pRS425GPD | This study |
Enzymes, primers, and chemicals.
Restriction enzymes, DNA-modifying enzymes, and other molecular reagents were obtained from New England BioLabs (Beverly, MA). The reaction conditions were set up following the manufacturer's instructions. All general chemicals and medium components were purchased from Sigma-Aldrich (St. Louis, MO) or Fisher Scientific (Pittsburgh, PA). Primers for both PCR and sequencing were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Table S1 in the supplemental material.
Media and culture conditions.
Yeast strains were routinely cultivated at 30°C in YP medium (10 g/liter of yeast extract and 20 g/liter of peptone) or synthetic complete (SC) medium (6.7 g/liter of yeast nitrogen base and appropriate amino acids or nucleotides) containing 20 g/liter of d-glucose (SCD). SC medium containing 20 g/liter agar and glucose without tryptophan and/or leucine amendment was used to select transformants using TRP1 and/or LEU2 as auxotrophic markers. The E. coli strains were grown in Luria-Bertani medium at 37°C, and 100 μg/ml of ampicillin was added to the medium when required.
Construction of S. cerevisiae genomic library and yeast transformation.
The genomic DNA of the S. cerevisiae S288C strain was used to construct the genomic library as previously described (34). Briefly, genomic DNA fragments (2 to 5 kb) were generated by sonication and ligated into a multicopy plasmid, pRS424 (a yeast episomal plasmid), with TRP1 as an auxotrophic selection marker (35, 36). Plasmid extraction was performed using the QIAprep spin miniprep kit (Germantown, MD). The genomic library was transformed to the S. cerevisiae strain SR8-trp using lithium acetate-polyethylene glycol (LiAc-PEG) methods (37).
Selection of transformants with acetic acid resistance.
Yeast genomic library transformants were inoculated on SC medium agar plates (15-mm diameter), which contained glucose (20 g/liter) or xylose (20 g/liter) and different concentrations of acetic acid (2.0 g/liter, 2.5 g/liter, 3.0 g/liter, and 3.5 g/liter). The pH of the agar plates was adjusted to be 4.0 so that acetic acid was predominantly undissociated. The strain SR8-trp harboring the pRS424GPD plasmid (i.e., the strain S-C1 in Table 1) was used as the control under all conditions. Transformants were isolated from the plates with acetic acid concentrations at which the corresponding plates inoculated with the control strain had no colony grown. Then, the cell growth and sugar consumption rates of the isolated transformants were evaluated in liquid SC medium containing a toxic level of acetic acid and 20 g/liter glucose or 20 g/liter xylose. The initial cell biomass was adjusted to an optical density at 600 nm (OD600) of 0.05. Plasmids from the selected transformants were isolated by a Zyppy plasmid miniprep kit (Zymo Research) and amplified by E. coli transformation. The isolated plasmids were retransformed into the strain SR8-trp to verify the effects of the plasmids on acetic acid resistance.
Insert identification and plasmid and strain construction.
The verified plasmids showing effects in improving acetic acid resistance were sequenced using T3 and T7 primers (see Table S1 in the supplemental material) to determine the nucleotide sequences of the inserted genomic DNA fragments. Sequences were compared to the S. cerevisiae genome sequence to identify the insert sizes and open reading frames (ORFs). To construct overexpression plasmids, the complete open reading frames of the target genes (WHI2, PSR1, HAA1, or MSN2) were amplified by PCR with the primers listed in Table S1. The PCR products were subsequently digested and ligated to appropriate multiple cloning sites of the plasmid pRS424GPD or pSR425GPD. To construct the msn2 mutant strain from SR8-trp, the kanMX marker gene flanked by about 200 bp homologous to upstream and downstream regions of the MSN2 gene was PCR amplified from the genomic DNA of an msn2 knockout strain derived from the laboratory strain BY4741 (GE Healthcare Dharmacon) with primers msn2-F and msn2-R, listed in Table S1. Transformation of the PCR product into SR8-trp was performed using an EZ yeast transformation kit (BIO 101). Positive transformants were selected on YP medium containing 20 g/liter glucose and 300 μg/ml Geneticin (G418). A diagnostic PCR was performed to confirm successful deletion, yielding the strain S-msn2Δ. The overexpression plasmids were transformed into the strain SR8-trp, SR8-4, or S-msn2Δ using an EZ yeast transformation kit.
Protein expression experiments and data analysis.
A yeast green fluorescent protein (GFP) clone collection of S. cerevisiae BY4742 strains (no. 95702; Life Technologies), developed by oligonucleotide-directed homologous recombination to tag each ORF with Aequorea victoria GFP (S65T) in its chromosomal location at the 3′ end (38), was used in protein expression analysis in response to acetic acid stress. Precultured yeast cells with the GFP fusion protein Whi2 were grown in SCD medium to early exponential phase and then were inoculated into 200 μl SCD medium (OD600 = 0.2) in clear-bottom black 96-well plates (Costar, Bethesda, MD) containing different concentrations of acetic acid (0 g/liter, 2 g/liter, and 3 g/liter [pH 4.0]). The plates were then incubated in a microplate reader (Synergy HT multimode; Biotech, Winooski, VT) at 30°C with fast shaking, and the OD600 and GFP signal (fluorescence readings were excitation at 485 nm and emission at 528 nm) were simultaneously measured every 1 h for 6 h. All experiments were performed in triplicate.
The protein expression levels based on GFP signals were calculated using the method previously described (39, 40). The raw data from the OD600 and GFP measurements were corrected by considering the background signals in control with medium only, and the protein expression per cell unit for each measurement was calculated by normalizing the GFP data to the cell number (OD600), with P equal to the GFP measurement over the OD600. The expression of the PGK1 gene product was used as the reference, and the P values for each protein of interest were normalized based on the mean P level of PGK1 corresponding to the same condition on the same plate. The relative expression (RE) for the protein of interest at each time point under each acetic acid exposure condition versus the control condition without acetic acid was calculated as RE = Paa/Pc, where Paa refers to the normalized P in the experimental condition with acetic acid and Pc represents the corrected and normalized P in the control condition without acetic acid.
Reverse transcription-qPCR.
The wild-type strain in early exponential phase was grown in SC medium containing glucose (20 g/liter) and acetic acid (2 g/liter) or in medium without acetic acid (control condition). After 6 h of incubation, cells from triplicate experiments were harvested by centrifugation for 30 s at 1,000 rpm under 4°C, and RNA was immediately extracted by using a PureLink RNA minikit (Life Technologies, NY) according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using an iScript cDNA synthesis kit (Bio-Rad Laboratories, CA). The cDNA was then used for quantitative PCR (qPCR) analysis using the primers in Table S1 in the supplemental material on a CFX Connect real-time system (Bio-Rad Laboratories) and by using iQ SYBR green supermix (Bio-Rad Laboratories). To confirm the amplification specificity, the PCR products were subjected to melting curve analysis and gel electrophoresis. All of the measurements were performed in triplicate for each biological triplicate (n = 3 × 3). Gene expression was calculated by the quantification method (41) as (1 + E)Cq/(1 + Eref)Cq,ref, where E refers to application efficiency and Cq refers to the quantification cycle value.
Fermentation experiments.
Batch fermentation experiments under oxygen-limited conditions were performed in 125-ml Erlenmeyer flasks containing 20 ml medium, and anaerobic batch fermentation experiments were performed in 160-ml serum bottles that contained 20 ml medium and were sealed with butyl rubber stoppers, at 30°C and 100 rpm. The anaerobic fermentation media were prepared by flushing with nitrogen which had passed through a heated, reduced-copper column to remove trace oxygen. Precultured yeast cells in SCD medium were centrifuged and washed with sterilized water and then inoculated into fermentation media containing glucose/xylose and acetic acid. The initial cell densities were adjusted to an OD600 of 1. The initial pH of the medium was adjusted to 4.0. For the anaerobic fermentation experiments, ergosterol and Tween 80 were added to final concentrations of 0.01 g/liter and 0.42 g/liter, respectively (42). Culture samples were taken from fermentation experiments to measure the OD600 values and concentrations of metabolites. For anaerobic fermentation experiments, samples were collected by sterile syringes and 26-gauge BD needles. Yeast cell dry weight was determined using a microwave method as described previously (43). All of the fermentation experiments were performed in duplicate.
As for fermentation with cellulosic hydrolysates, the corn stover hydrolysate was prepared by the National Renewable Energy Laboratory (NREL) (http://www.nrel.gov/biomass/pdfs/47764.pdf) through dilute acid pretreatment. The hydrolysate liquid fraction contained 10.9 g/liter acetic acid, 115 g/liter xylose, and 17 g/liter glucose. The hydrolysate was mixed with YP medium at a 50% (vol/vol) ratio to result in a hydrolysate mixture containing around 3.5 g/liter acetic acid, 40 g/liter xylose, and an adjusted glucose concentration of 20 g/liter. The pH was adjusted to 4.0. The fermentation experiments were set up in flasks under oxygen-limited conditions as described above.
Analytical methods.
Cell growth was monitored by measuring the OD600 using a UV-visible spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). Glucose, xylose, xylitol, glycerol, acetic acid, and ethanol were quantified by a high-performance liquid chromatograph (Agilent Technologies 1200 series) equipped with a refractive index detector and a Rezex ROA-organic acid H+ (8%) column (Phenomenex Inc., CA). The column was eluted with 0.005 N H2SO4 as the mobile phase under the flow rate of 0.6 ml/min at 50°C.
RESULTS
Screening of transformants exhibiting improved acetic acid resistance.
With the goal of obtaining transformants that grew faster with acetic acid stress under both glucose- and xylose-consuming conditions, a method that combined selection on agar plates and screening in liquid medium containing toxic levels of acetic acid was developed. The plates inoculated with the genomic-library transformants had colonies grown on glucose- or xylose-containing agar plates with up to 3 g/liter acetic acid (pH 4.0) within 3 to 5 days. In contrast, the plates inoculated with the control strain S-C1 (Table 1) did not have any colony grown at acetic acid concentrations of >2 g/liter through 2 weeks of incubation. Then 130 fast-growing transformants were isolated from the plates containing 2.5 g/liter or 3 g/liter acetic acid (65 from the glucose-containing plates and 65 from the xylose-containing plates).
We further screened the isolated transformants in liquid SC medium to identify strains with superior resistance to acetic acid under both glucose-consuming and xylose-consuming conditions. Among the 65 transformants isolated from xylose-containing plates, 10 transformants (numbers 7, 32, 34, 35, 36, 38, 41, 49, 52, and 59) grew much faster than the control strain S-C1 in liquid medium containing glucose plus acetic acid (see Fig. S1A in the supplemental material), and 5 out of these 10 (numbers 34, 35, 38, 52, and 59) also exhibited significant improvements in liquid medium containing xylose plus acetic acid (Fig. S1B). As for the 65 transformants isolated from glucose-containing plates, while some transformants showed improved cell growth in medium containing glucose plus acetic acid, no significant improvements were observed when these transformants were grown in medium containing xylose plus acetic acid (data not shown). A possible reason is that the yeast was more sensitive to acetic acid when grown on xylose than on glucose; a similar observation was also reported in a previous study (44).
Plasmids from each of the five selected transformants were isolated and retransformed into the parental strains, respectively, to confirm the effects. Four plasmids (numbers 34, 38, 52, and 59) showed positive effects in the retransformed strains comparable to those in the original transformants in terms of cell growth and sugar consumption under acetic acid stress. The number 35 plasmid did not show improved acetic acid resistance in the retransformed strain, indicating that the increased resistance in the original transformant might be due to genome mutations. Sequence analysis of the four confirmed plasmids revealed that three (numbers 34, 38, and 59) had genome coordinates similar to those of the identified targets (chromosome XV [chrXV], nucleotide [nt] positions 409259 to 412369 for numbers 34 and 38; chrXV, nt positions 410675 to 412722 for number 59), while plasmid number 52 did not harbor any intact open reading frame (chrIX, nt positions 4597 to 5728).
All of the three inserts harbored a complete sequence of the gene WHI2 (chrXV, nt positions 410870 to 412330). The WHI2 gene product in S. cerevisiae is a 55-kDa cytoplasmatic globular scaffold protein (38). Whi2 is involved in coordinating cell growth and proliferation and plays an important role in nutrient-dependent cell cycle arrest (45–48). The protein has also been reported to be required for activation of the general stress response in the yeast (49–51).
Enhanced acetic acid resistance by overexpression of the WHI2 gene.
The effect of overexpression of WHI2 on acetic acid resistance in S. cerevisiae has not been reported before, and the role of Whi2 in the acetic acid stress response is unclear. To determine the effect of WHI2 as a gene perturbation target for improving acetic acid resistance, we introduced a multicopy plasmid overexpressing WHI2 under the control of the constitutive GPD (TDH3) promoter and CYC1 terminator (Table 1) into S. cerevisiae strain SR8-trp or BY4742 (i.e., WHI2 overexpression in different strain backgrounds), yielding strains S-WHI2 and BY2-WHI2 (Table 1), and the corresponding control strains S-C1 and BY2-C1 harbored the plasmid without the WHI2 insert. The strain S-WHI2 had noticeably higher cell growth under acetic acid stress than the control strain S-C1, according to the results of yeast spotting assays on SC agar plates containing glucose (20 g/liter) and different concentrations of acetic acid (Fig. 1A). When grown on plates with xylose as the substrate, the strain S-WHI2 also had substantially higher acetic acid resistance than the control strain (Fig. 1B). Overexpression of WHI2 in another strain background (BY-WHI2) also showed improvement (see Fig. S2 in the supplemental material).
FIG 1.
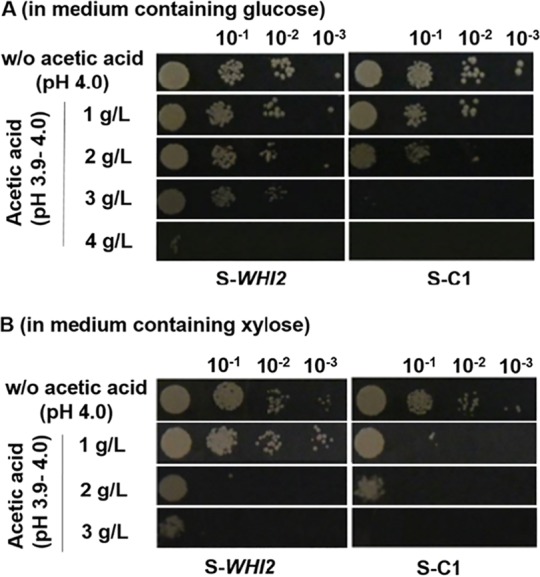
Increased cell growth conferred by overexpression of WHI2. Cells were grown on minimal medium agar plates containing glucose (20 g/liter) (A) or xylose (20 g/liter) (B), amended with various concentrations of acetic acid or without (w/o) acetic acid. Cells of the strain S-WHI2 or the control strain S-C1 were spotted in serial dilutions (diluted by a factor of 10).
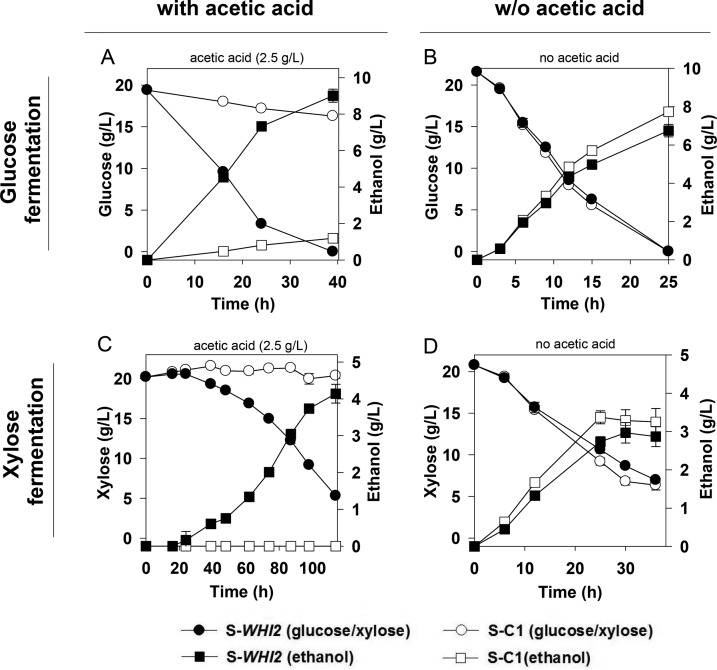
Batch experiments were conducted to characterize the glucose/xylose fermentation performances of the strain S-WHI2 versus those of the control strain S-C1 under acetic acid stress. Figure 2 shows the ethanol production and sugar consumption profiles of the two strains. In glucose fermentation under oxygen-limited conditions, glucose (20 g/liter) was completely consumed within 39 h by S-WHI2 with the presence of 2.5 g/liter acetic acid, while the control strain S-C1 consumed only 3.2 g/liter glucose (Fig. 2A). The specific sugar consumption rate and specific ethanol productivity of the strain S-WHI2 were both >5 times higher than those of the control strain (see Table S2 in the supplemental material). The pH of the fermentation medium was 4.0 initially and decreased to 3.3 at the end of S-WHI2 fermentation; a toxic level of the protonated form of acetic acid prevailed during the fermentation process. It was noted that the fermentation profiles of S-WHI2 and S-C1 had no significant differences under the condition without acetic acid (with a pH of <4.0 during fermentation) (Fig. 2B), suggesting that the improvement brought by WHI2 overexpression is associated with the cellular response to acetic acid stress. It was noted that the ethanol yields in glucose fermentation with acetic acid were higher than the yields without acetic acid for both S-WHI2 and S-C1 (Table S2), probably due to the stimulating effects on glucose fermentation by low concentrations of weak acids as reported in previous studies (52, 53). A similar level of improvement in the fermentation by the strain S-WHI2 versus that of S-C1 was also observed under strict anoxic conditions (Table S2).
FIG 2.
Improved fermentation by the strain S-WHI2 compared to that of the control strain S-C1 in SC medium containing glucose (20 g/liter) (A and B) or xylose (20 g/liter) (C and D) under oxygen-limited conditions with or without acetic acid. Sugar consumption and ethanol production are shown. The results are the means from duplicate experiments; error bars indicating standard deviations are not visible when smaller than the symbol size.
Additionally, xylose fermentation under acetic acid stress also showed significant improvement by overexpression of WHI2. The strain S-WHI2 consumed xylose with a specific rate of 0.245 ± 0.004 g/g cells (dry weight)/h under the oxygen-limited condition, while the control strain did not consume xylose in the experimental time frame (Fig. 2C). In the absence of acetic acid, the two strains had similar xylose fermentation profiles (Fig. 2D), suggesting that WHI2 overexpression did not have a significant impact on the xylose fermentation pathway itself. Xylose fermentation under anaerobic conditions proceeded slowly due to the intrinsic limitation of the xylose-assimilating pathway of the fungi. No cell growth was observed with 2.5 g/liter acetic acid, and thus the lower level of acetic acid (1.5 g/liter) was used in the experiment. There were significant improvements (t test, P < 0.05) in the sugar consumption rate (0.23 ± 0.03 g sugar/g cells ]dry weight]/h) and ethanol productivity (0.074 ± 0.007 g ethanol/g cells [dry weight]/h) in S-WHI2 compared to that in S-C1 (0.195 ± 0.01 g sugar/g cells [dry weight]/h and 0.066 ± 0.001 g ethanol/g cells [dry weight]/h) (see Table S2 in the supplemental material). Overall, the data showed that overexpression of WHI2 might improve the yeast growth and ethanol fermentation from glucose or xylose under acetic acid stress.
The acetic acid concentration in glucose fermentation did not change significantly, most likely due to carbon catabolite repression by glucose, while the acetic acid concentration in xylose fermentation started to decrease after 40 h and was reduced by ∼1 g/liter at the end of the fermentation experiment. Under anoxic conditions, no acetic acid consumption occurred in glucose or xylose fermentation, as S. cerevisiae does not metabolize acetic acid without oxygen (43). Noticeably, when no acetic acid consumption occurred, there was still significant improvement in fermentation by strain S-WHI2 versus that by the control strain S-C1 (Fig. 2; see also Table S2 in the supplemental material), suggesting that the positive impact of WHI2 overexpression was not due to acetic acid consumption.
Acetic acid-induced expression of Whi2.
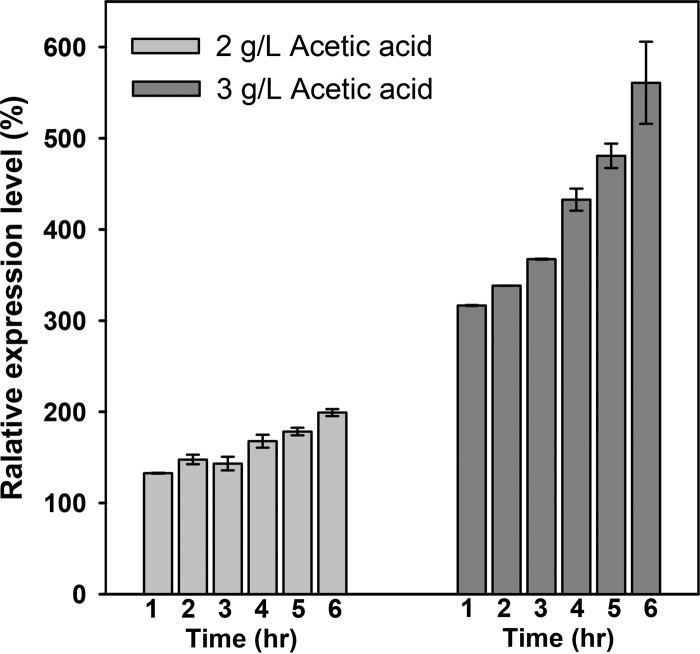
The positive effect brought by overexpression of WHI2 motivated us to examine how the endogenous expression of Whi2 would change in response to acetic acid. The S. cerevisiae strains with GFP fusion proteins (no. 95702; Life Technologies) related to each target were employed to examine protein expression levels. A detailed description of the methods and data analysis was provided in Materials and Methods. The relative expression levels of Whi2 in the presence of acetic acid (2 g/liter or 3 g/liter) versus those under the control condition without acetic acid over time are shown in Fig. 3. Under the condition with 2 g/liter acetic acid (pH 4.0), expression of Whi2 was induced, and the expression level increased during the exposure time period (Fig. 3). At the higher concentration of acetic acid (3 g/liter), the Whi2 protein had substantially increased expression levels. The longer the exposure time was, the higher the expression level was (Fig. 3), and the expression level of Whi2 with 6 h of incubation with acetic acid (3 g/liter) was 550% of that without acetic acid. The results showed that acetic acid activated the endogenous expression of Whi2 in S. cerevisiae.
FIG 3.
Effects of acetic acid on the expression of Whi2. Yeast strains with GFP fusion proteins in exponential phase of growth were incubated in SCD medium amended with acetic acid (2 g/liter or 3 g/liter [pH 4.0]) for 6 h. The protein expression level of Pgk1 was used as the reference for normalization. The normalized protein level in cells without acetic acid stress was taken as 100%. The results are the means from triplicate experiments; error bars represent standard deviations.
To further confirm the expression level of the WHI2 gene product, we determined the transcriptional changes of WHI2 in response to acetic acid stress through reverse transcription-quantitative PCR. The transcriptional level of WHI2 in the wild-type strain under the condition of 2 g/liter acetic acid was (2.84 ± 0.07)-fold of that under the control condition without acetic acid. The transcriptional analysis together with the protein expression results confirmed that endogenous expression of the WHI2 gene was induced by acetic acid stress.
Hypersensitivity to acetic acid in a deficient mutant of Whi2.
To evaluate the importance of Whi2 in yeast resistance to acetic acid, we characterized the susceptibility of the whi2Δ mutant to acetic acid stress compared to that of the wild type. The cell viability under high acetic acid stress was quantified as a function of time in the presence of 4 g/liter acetic acid (pH 4.0) (Fig. 4). From the viability data from 1 h to 7 h, the cell death rates were calculated based on first-order kinetics. The death rate for the whi2Δ mutant strain was determined to be 0.143 ± 0.042 h−1, which was nearly 3 times higher than that of the wild-type strain (death rate, 0.047 ± 0.012 h−1). These results suggested that Whi2 might play a critical role in the yeast response to acetic acid stress.
FIG 4.
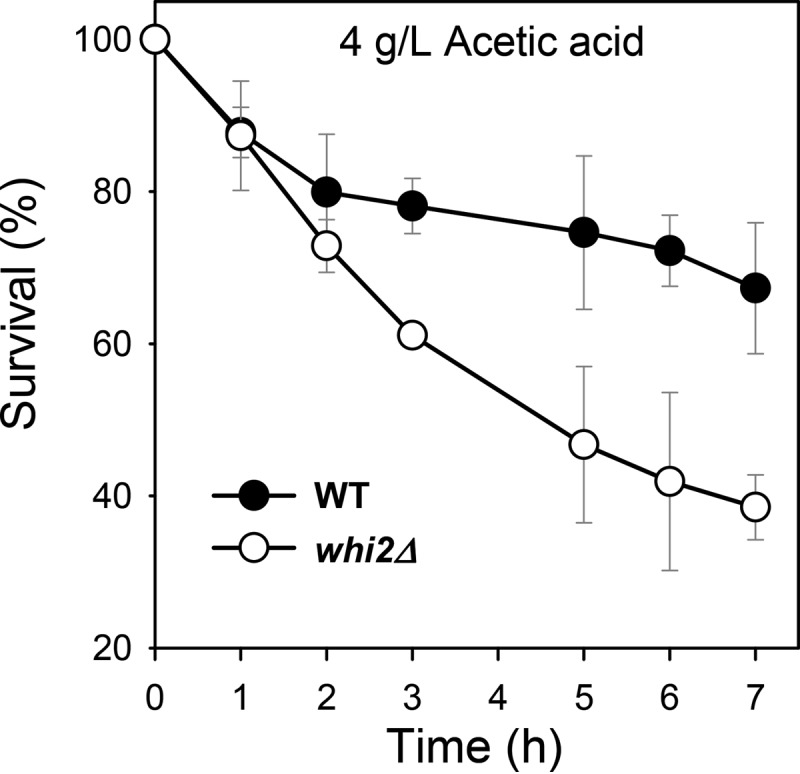
Stress susceptibility of the whi2Δ mutant versus that of the wild-type strain S. cerevisiae BY4741 to acetic acid. Cells in exponential growth phase in minimal medium were treated with acetic acid (4 g/liter [pH 4.0]), and the CFU were counted as a function of time. The results are the means from triplicate experiments; error bars represent standard deviations.
Whi2 and its binding partner Psr1 were both involved in eliciting acetic acid resistance.
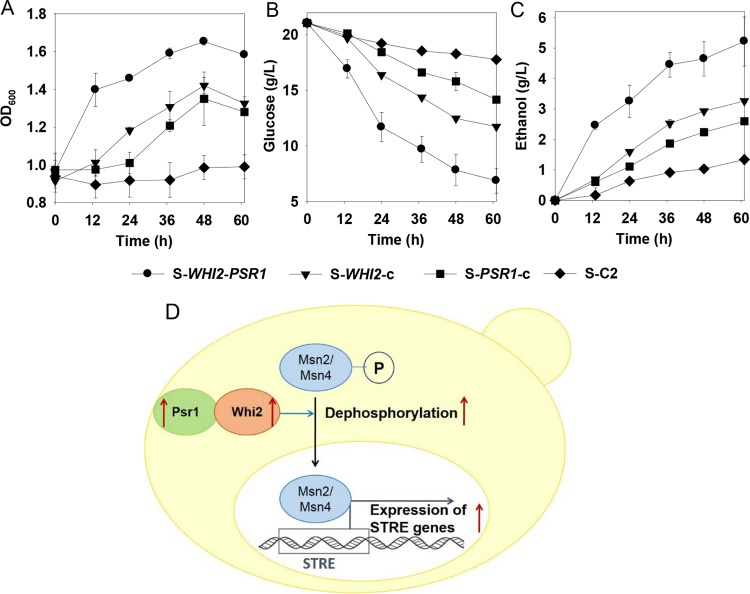
It has been suggested in the existing literature that Whi2 can interact with the plasma membrane phosphatase Psr1 as the binding partner for full activation of the stress response in S. cerevisiae (49, 54). Thus, we hypothesized that Whi2 might function in combination with Psr1 to induce acetic acid resistance. To test the hypothesis, we investigated the effect of perturbation of the PSR1 gene on acetic acid tolerance. We constructed a new group of recombinant yeast strains that overexpressed WHI2, PSR1, or WHI2 plus PSR1 or contained a control plasmid (i.e., the strains S-WHI2-c, S-PSR1-c, S-WHI2-PSR1, and S-C2, respectively, listed in Table 1). The batch fermentation results showed that the strain S-WHI2-PSR1 had the highest cell growth rate (Fig. 5A), glucose consumption rate (Fig. 5B), and ethanol productivity (Fig. 5C) among the four strains. The strain S-PSR1-c showed fermentation performance comparable to that of the strain S-WHI2-c. Noticeably, overexpression of WHI2 and PSR1 simultaneously resulted in substantially higher acetic acid resistance than overexpression of each gene individually (Fig. 5A to C). Such a synergistic effect indicated that the pathway involving both Whi2 and Psr1 might contribute to eliciting endogenous acetic acid resistance in S. cerevisiae. A possible mechanism for Whi2/Psr1 involving the stress response to acetic acid is proposed in Fig. 5D and is addressed in more depth in Discussion.
FIG 5.
Improved acetic acid resistance by overexpression of WHI2 and/or PSR1 gene targets. Cell growth (A), glucose consumption (B), and ethanol production (C) were determined during fermentation in SC medium containing glucose (20 g/liter) and acetic acid (2.5 g/liter [pH 4.0]). The results are the means from duplicate experiments; error bars indicating standard deviations are not visible when smaller than the symbol size. (D) Proposed mechanism involving Whi2 and Psr1 in eliciting the acetic acid stress response in S. cerevisiae.
Improved fermentation by the engineered acetic acid-resistant strain.
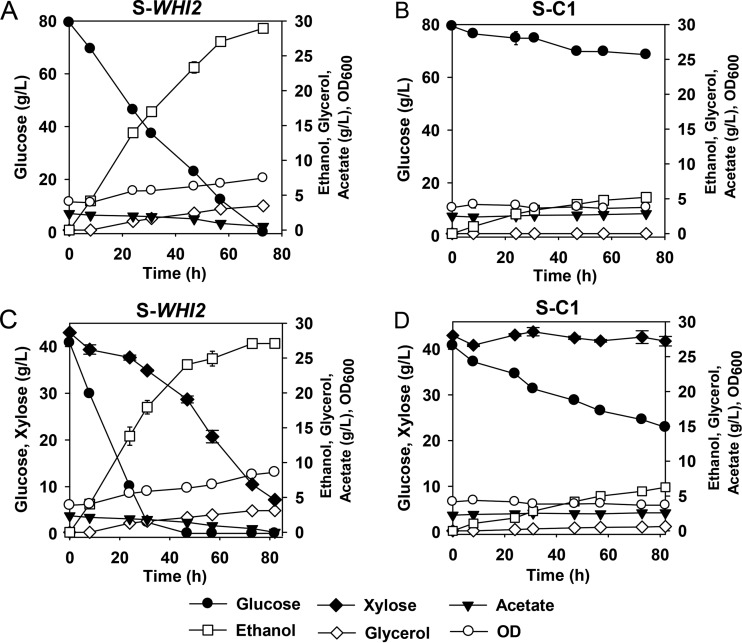
The WHI2-overxpressing strain S-WHI2 was further characterized under industrially relevant conditions, including fermentation of a high glucose concentration (80 g/liter) or of mixed sugars (40 g/liter glucose plus 40 g/liter xylose) with acetic acid (2.5 g/liter [pH 4.0]), and fermentation with real cellulosic hydrolysates. As expected from the results of earlier experiments, the strain S-WHI2 grew better and had much faster sugar fermentation under conditions with a high glucose concentration (Fig. 6A) or a glucose-plus-xylose mixture (Fig. 6B) with acetic acid stress than the control strain. The control strain did not have observable cell growth and consumed only a small portion of glucose under both conditions (10 g/liter and 18 g/liter, respectively). In the fermentation by S-WHI2, the acetic acid concentration did not decrease substantially until the glucose concentration became low after 24 h (Fig. 6A) or almost depleted in a mixed-sugar fermentation (Fig. 6C). A possible explanation is that acetic acid was consumed as an alternative substrate due to alleviated carbon catabolite repression as glucose was fermented in the yeast. In addition, the results indicated that the positive impact by WHI2 overexpression might not be attributed to acetic acid consumption, although the reduced acetic acid concentration during the later phase of sugar fermentation might contribute to detoxification of the medium. The improved sugar fermentation was also demonstrated in corn stover hydrolysates, where fermentation by the control strain was severely inhibited (see Fig. S3 in the supplemental material).
FIG 6.
Improved fermentation by the strain S-WHI2 compared to that of the control strain S-C1 in minimal medium containing 80 g/liter glucose (A and B) and 40 g/liter glucose plus 40 g/liter xylose (C and D), both with acetic acid (2.5 g/liter [pH 4.0]). The results are the means from duplicate experiments; error bars indicating standard deviations are not visible when smaller than the symbol size.
DISCUSSION
Improving the resistance of S. cerevisiae to acetic acid, a major fermentation inhibitor, is highly desirable and important for achieving efficient and cost-effective biofuel production from lignocellulosic biomass (5, 30). Prior research efforts have significantly advanced the understanding of the toxic effects of acetic acid and the stress response in S. cerevisiae (5, 6, 22, 26). However, engineering of yeast strains with superior acetic acid resistance remains a challenge due to the limited information regarding which genes can be effective perturbation targets. The present study applied an inverse metabolic engineering approach and identified a novel gene overexpression target, WHI2, that substantially improved the acetic acid resistance in S. cerevisiae. Results from the characterization experiments, including induced expression of Whi2 under acetic acid stress, increased the susceptibility of the whi2Δ mutant versus that of the wild type, and the synergistic effects of coexpressing Whi2 and its binding partner Psr1 clearly suggested that Whi2 could play an important role in eliciting endogenous acetic acid resistance in S. cerevisiae. The engineered strain developed herein demonstrated improved glucose and/or xylose fermentation in the presence of toxic levels of acetic acid. Results from this study contribute to the breeding of yeast strains with superior acetic acid resistance for achievement of efficient and cost-effective biofuel production from lignocellulosic biomass.
The remarkable improvement in acetic acid resistance by overexpression of the WHI2 gene was observed in different strain backgrounds in our study, suggesting that Whi2 could provide a protective effect against acetic acid stress in S. cerevisiae. Whi2 is a 55-kDa cytoplasmatic globular scaffold protein in S. cerevisiae (38); it is involved in coordinating cell proliferation and nutrient-dependent cell cycle arrest (45–47). For example, whi2Δ mutant cells failed to cease cell division with nutrient depletion (45, 55) and had much smaller cell sizes than the wild-type cells (56). Deletion of WHI2 resulted in cells that failed to accumulate storage carbohydrate compounds and had reduced resistance to environmental stress, such as heat shock (46, 57). This study found that the whi2Δ mutant had significantly lower cell viability than the wild type in the presence of acetic acid stress, which is consistent with a previous observation that deletion of WHI2 in S. cerevisiae led to increased cell susceptibility to propionic acid and acetic acid (22).
While overexpression of WHI2 substantially enhanced acetic acid resistance in the yeast, it was noted that the induction level of endogenous WHI2 expression by 2 g/liter acetic acid was relatively low. Similarly, the HAA1 gene was reported in a previous study to be an overexpression target for improving acetic acid resistance in S. cerevisiae (58), but the induction of HAA1 expression in the wild-type strain under acetic acid stress (6 g/liter acetic acid) was only 2.1-fold of that in the absence of acetic acid (25). These findings suggested that an effective gene overexpression target for improving yeast acetic acid resistance does not necessarily have high endogenous expression in response to acetic acid. The relatively low induction level of WHI2 expression with acetic acid stress could also be a reason why it was not reported in previous transcriptomic analysis, which generally focused on analyzing genes with substantially different expression levels. Furthermore, the exposure time to acetic acid stress had a considerable impact on the induction level of Whi2 expression. The short timing of the response in previous transcriptomic analysis (e.g., sampling after only 30 min of exposure) (26) might be another reason why the gene WHI2 was not identified. Further work is needed to elucidate the mechanism through which Whi2 is involved in the stress response to acetic acid.
The synergistic effect by overexpression of WHI2 and PSR1 together on enhancement of acetic acid resistance provided new insight to possible endogenous stress response mechanisms to acetic acid in S. cerevisiae. Psr1 is a plasma membrane phosphatase which was first identified to be required for the sodium osmostress response (54). It was found that Whi2 can bind Psr1 and form a functional complex to interact with Msn2/Msn4 for full activation of gene expressions controlled by stress response elements (STREs) (49, 59). The transcription factors Msn2/Msn4 were regarded to be localized in the cytoplasm in the phosphorylated form under nonstress conditions and to be dephosphorylated and translocated into the nucleus under stress conditions, where they can activate STRE-mediated gene expression (60). Genetic studies on the function of Whi2/Psr1 showed that whi2 or psr1 null mutant strains had hyperphosphorylation of Msn2/Msn4 and reduced transcription of STRE-controlled genes (49). As such, a possible mechanism to explain the observed synergistic effect of overexpression of WHI2 and PSR1 in improving yeast acetic acid resistance is illustrated in Fig. 5D. Since Whi2 can bind the phosphatase Psr1 to mediate dephosphorylation of Msn2/Msn4, increases in the expression levels of both WHI2 and PSR1 might facilitate the formation of the Whi2-Psr1 functional complex to a larger extent than overexpression of either target alone and thus greatly enhance the dephosphorylation of the phosphorylated forms of Msn2/Msn4. The increased level of dephosphorylated Msn2/Msn4 might induce the expression of stress response genes related to acetic acid tolerance and consequently improve the resistance phenotype. The hypothesized mechanism was supported by the observation that the improvement in acetic acid tolerance brought by overexpression of WHI2 in the msn2Δ mutant strain (i.e., S-msn2Δ-WHI2 versus S-msn2Δ-C1) was smaller than that in the wild-type strain (i.e., S-WHI2 versus S-C1) (see Fig. S4 in the supplemental material). Deletion of MSN2 did not completely eliminate the gain in acetic acid tolerance by overexpression of WHI2, probably because the activity of Msn4 (the Msn2 homolog) still remained. Our ongoing work is focused on the transcriptomic and metabolomic analysis of the engineered resistant strain versus the control strain to further determine the molecular mechanisms for improved acetic acid resistance in yeast.
It is worth mentioning that overexpression of WHI2 had an effect on improving acetic acid resistance superior to that of overexpression of HAA1 or MSN2 (see Fig. S5 in the supplemental material). Haa1 and Msn2 were identified as two important transcription factors that regulate genes associated with acetic acid responses in previous genome-wide analysis studies (21, 26). In particular, Haa1 regulates approximately 80% of the genes induced in acetic acid-stressed cells (26), and the effect of overexpression of HAA1 on acetic acid tolerance was reported previously (58). However, overexpression of HAA1 had a considerably smaller positive effect than overexpression of WHI2 on improving acetic acid resistance in the yeast (see Fig. S5 in the supplemental material). On the other hand, although the transcriptional activator Msn2 (and its homolog Msn4) was found to regulate many genes induced by weak acids (22, 23, 26), overexpression of MSN2 did not result in observable improvement in acetic acid resistance. It was also noted before that activation of the Msn2p/Msn4p regulon did not necessarily result in enhanced resistance to weak acids (22, 23). Based on our proposed mechanism, a possible explanation for no observable improvement in the strain overexpressing MSN2 is that increasing MSN2 transcription alone does not necessarily mean a higher level of dephosphorylated Msn2 and its appropriate localization to the nucleus for activating the STRE-mediated gene expression. All of these observations suggested the need for identifying effective gene targets through methods that directly select resistant yeast strains with traceable genetic perturbations. Successful application of inverse metabolic engineering in the present study and some previous works (34, 61–64) demonstrated the effectiveness of this approach in discovering novel gene perturbation targets for improving the desirable target phenotypes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the University of Pittsburgh and the Mascaro Center for Sustainable Innovation.
We thank Yong-Su Jin for providing the strains SR8, SR8-trp, and SR8-4. We thank Dan Schell from National Renewable Energy Laboratory (NREL) for providing the corn stover hydrolysates.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03718-15.
REFERENCES
- 1.Hahn-Hägerdal B, Galbe M, Gorwa-Grauslund M-F, Lidén G, Zacchi G. 2006. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol 24:549–556. doi: 10.1016/j.tibtech.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 2.Somerville C, Youngs H, Taylor C, Davis SC, Long SP. 2008. Feedstocks for lignocellulosic biofuels. Science 329:790–792. doi: 10.1126/science.1189268. [DOI] [PubMed] [Google Scholar]
- 3.Tilman D, Socolow R, Foley JA, Hill J, Larson E, Lynd L, Pacala S, Reilly J, Searchinger T, Somerville C. 2009. Beneficial biofuels—the food, energy, and environment trilemma. Science 325:270–271. doi: 10.1126/science.1177970. [DOI] [PubMed] [Google Scholar]
- 4.Jeffries TW. 1985. Emerging technology for fermenting d-xylose. Trends Biotechnol 3:208–212. doi: 10.1016/0167-7799(85)90048-4. [DOI] [Google Scholar]
- 5.Jönsson LJ, Alriksson B, Nilvebrant N-O. 2013. Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6:16–25. doi: 10.1186/1754-6834-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmqvist E, Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. I. Inhibition and detoxification. Bioresour Technol 74:17–24. [Google Scholar]
- 7.Carroll A, Somerville C. 2009. Cellulosic biofuels. Annu Rev Plant Biol 60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 8.Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu Rev Plant Biol 61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- 9.Klinke HB, Thomsen AB, Ahring BK. 2004. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- 10.Palmqvist E, Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. II. Inhibitors and mechanisms of inhibition. Bioresour Technol 74:25. [Google Scholar]
- 11.Maiorella B, Blanch HW, Wilke CR. 1983. By-product inhibition effects on ethanolic fermentation by Saccharomyces cerevisiae. Biotechnol Bioeng 25:103–121. doi: 10.1002/bit.260250109. [DOI] [PubMed] [Google Scholar]
- 12.Phowchinda O, Deliadupuy ML, Strehaiano P. 1995. Effects of acetic acid on growth and fermentative activity of Saccharomyces cerevisiae. Biotechnol Lett 17:237–242. doi: 10.1007/BF00127996. [DOI] [Google Scholar]
- 13.Pampulha ME, Loureiro-Dias MC. 2000. Energetics of the effect of acetic acid on growth of Saccharomyces cerevisiae. FEMS Microbiol Lett 184:69–72. doi: 10.1111/j.1574-6968.2000.tb08992.x. [DOI] [PubMed] [Google Scholar]
- 14.Hong K-K, Nielsen J. 2012. Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell Mol Life Sci 69:2671–2690. doi: 10.1007/s00018-012-0945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krivoruchko A, Siewers V, Nielsen J. 2011. Opportunities for yeast metabolic engineering: lessons from synthetic biology. Biotechnol J 6:262–268. doi: 10.1002/biot.201000308. [DOI] [PubMed] [Google Scholar]
- 16.Russell JB. 1992. Another explanation for the toxicity of fermentation acids at low pH: anion accumulation versus uncoupling. J Appl Bacteriol 73:363–370. doi: 10.1111/j.1365-2672.1992.tb04990.x. [DOI] [Google Scholar]
- 17.Narendranath NV, Thomas KC, Ingledew WM. 2001. Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J Ind Microbiol Biotechnol 26:171–177. doi: 10.1038/sj.jim.7000090. [DOI] [PubMed] [Google Scholar]
- 18.Graves T, Narendranath NV, Dawson K, Power R. 2006. Effect of pH and lactic or acetic acid on ethanol productivity by Saccharomyces cerevisiae in corn mash. J Ind Microbiol Biotechnol 33:469–474. doi: 10.1007/s10295-006-0091-6. [DOI] [PubMed] [Google Scholar]
- 19.Weber C, Farwick A, Benisch F, Brat D, Dietz H, Subtil T, Boles E. 2010. Trends and challenges in the microbial production of lignocellulosic bioalcohol fuels. Appl Microbiol Biotechnol 87:1303–1315. doi: 10.1007/s00253-010-2707-z. [DOI] [PubMed] [Google Scholar]
- 20.Vilela-Moura A, Schuller D, Mendes-Faia A, Silva RD, Chaves SR, Sousa MJ, Corte-Real M. 2011. The impact of acetate metabolism on yeast fermentative performance and wine quality: reduction of volatile acidity of grape musts and wines. Appl Microbiol Biotechnol 89:271–280. doi: 10.1007/s00253-010-2898-3. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes AR, Mira NP, Vargas RC, Canelhas I, Sá-Correia I. 2005. Saccharomyces cerevisiae adaptation to weak acids involves the transcription factor Haa1p and Haa1p-regulated genes. Biochem Biophys Res Commun 337:95–103. doi: 10.1016/j.bbrc.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Mira NP, Palma M, Guerreiro JF, Sá-Correia I. 2010. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79–79. doi: 10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mira NP, Teixeira MC, Sá-Correia I. 2010. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 14:525–540. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piper P, Calderon CO, Hatzixanthis K, Mollapour M. 2001. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology 147:2635–2646. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]
- 25.Haitani Y, Tanaka K, Yamamoto M, Nakamura T, Ando A, Ogawa J, Shima J. 2012. Identification of an acetate-tolerant strain of Saccharomyces cerevisiae and characterization by gene expression analysis. J Biosci Bioeng 114:648–651. doi: 10.1016/j.jbiosc.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Mira NP, Becker JD, Sá-Correia I. 2010. Genomic expression program involving the Haa1p-regulon in Saccharomyces cerevisiae response to acetic acid. OMICS 14:587–610. doi: 10.1089/omi.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller G, Ray E, Brown PO, Winge DR. 2001. Haa1, a protein homologous to the copper-regulated transcription factor Ace1, is a novel transcriptional activator. J Biol Chem 276:38697–38702. doi: 10.1074/jbc.M107131200. [DOI] [PubMed] [Google Scholar]
- 28.Swinnen S, Fernández-Niño M, González-Ramos D, van Maris AJA, Nevoigt E. 2014. The fraction of cells that resume growth after acetic acid addition is a strain-dependent parameter of acetic acid tolerance in Saccharomyces cerevisiae. FEMS Yeast Res 14:642–653. doi: 10.1111/1567-1364.12151. [DOI] [PubMed] [Google Scholar]
- 29.Fernández-Niño M, Marquina M, Swinnen S, Rodríguez-Porrata B, Nevoigt E, Ariño J. 2015. The cytosolic pH of individual Saccharomyces cerevisiae cells is a key factor in acetic acid tolerance. Appl Environ Microbiol 81:7813–7821. doi: 10.1128/AEM.02313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ling H, Teo W, Chen B, Leong SSJ, Chang MW. 2014. Microbial tolerance engineering toward biochemical production: from lignocellulose to products. Curr Opin Biotechnol 29:99–106. doi: 10.1016/j.copbio.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Kim S-R, Skerker JM, Kang W, Lesmana AL, Wei N, Arkin AP, Jin Y-S. 2013. Rational and evolutionary engineering approaches uncover a small set of genetic changes efficient for rapid xylose fermentation in Saccharomyces cerevisiae. PLoS One 8:e57048. doi: 10.1371/journal.pone.0057048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. 2013. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang GZ, Kong II, Kim H, Liu JL, Cate JHD, Jin Y-S. 2014. Construction of a quadruple auxotrophic mutant of an industrial polyploidy Saccharomyces cerevisiae using RNA-guided Cas9 nuclease. Appl Environ Microbiol 80:7694−7701. doi: 10.1128/AEM.02310-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee KS, Hong ME, Jung SC, Ha SJ, Yu BJ, Koo HM, Park SM, Seo JH, Kweon DH, Park JC, Jin Y-S. 2011. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng 108:621–631. doi: 10.1002/bit.22988. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119–122. doi: 10.1016/0378-1119(92)90454-W. [DOI] [PubMed] [Google Scholar]
- 37.Gietz RD, Schiestl RH. 2007. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 38.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. 2003. Global analysis of protein localization in budding yeast. Nature 425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 39.Gou N, Onnis-Hayden A, Gu AZ. 2010. Mechanistic toxicity assessment of nanomaterials by whole-cell-array stress genes expression analysis. Environ Sci Technol 44:5964–5970. doi: 10.1021/es100679f. [DOI] [PubMed] [Google Scholar]
- 40.O'Connor STF, Lan J, North M, Loguinov A, Zhang L, Smith MT, Gu AZ, Vulpe C. 2012. Genome-wide functional and stress response profiling reveals toxic mechanism and genes required for tolerance to benzo[a]pyrene in S. cerevisiae. Front Genet 3:316–316. doi: 10.3389/fgene.2012.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Eliasson A, Camilla C, Fredrik W, Hahn-Hägerdal B. 2000. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol 66:3381–3386. doi: 10.1128/AEM.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei N, Quarterman J, Kim SR, Cate JH, Jin Y-S. 2013. Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat Commun 4:2580. doi: 10.1038/ncomms3580. [DOI] [PubMed] [Google Scholar]
- 44.Casey E, Sedlak M, Ho NW, Mosier NS. 2010. Effect of acetic acid and pH on the cofermentation of glucose and xylose to ethanol by a genetically engineered strain of Saccharomyces cerevisiae. FEMS Yeast Res 10:385–393. doi: 10.1111/j.1567-1364.2010.00623.x. [DOI] [PubMed] [Google Scholar]
- 45.Rahman DJ, Sudbery P, Kelyl S, Marison I. 1988. The effect of dissolved oxygen concentration on the growth physiology of Saccharomyces cerevisiae whi2 mutants. J Gen Microbiol 134:2241–2248. [DOI] [PubMed] [Google Scholar]
- 46.Saul D, Walton E, Sudbery P, Carter B. 1985. Saccharomyces cerevisiae whi2 mutants in stationary phase retain the properties of exponentially growing cells. J Gen Microbiol 131:2245–2251. [Google Scholar]
- 47.Sudbery PE, Goodey AR, Carter BL. 1980. Genes which control cell proliferation in the yeast Saccharomyces cerevisiae. Nature 288:401–404. doi: 10.1038/288401a0. [DOI] [PubMed] [Google Scholar]
- 48.Mountain HA, Sudbery PE. 1990. The relationship of growth rate and catabolite repression with whi2 expression and cell size in Saccharomyces cerevisiae. J Gen Microbiol 136:733–737. doi: 10.1099/00221287-136-4-733. [DOI] [PubMed] [Google Scholar]
- 49.Kaida D, Yashiroda H, Toh-e A, Kikuchi Y. 2002. Yeast Whi2 and Psr1-phosphatase form a complex and regulate STRE-mediated gene expression. Genes Cells 7:543–552. doi: 10.1046/j.1365-2443.2002.00538.x. [DOI] [PubMed] [Google Scholar]
- 50.Leadsham JE, Miller K, Ayscough KR, Colombo S, Martegani E, Sudbery P, Gourlay CW. 2009. Whi2p links nutritional sensing to actin-dependent Ras-cAMP-PKA regulation and apoptosis in yeast. J Cell Sci 122:706–715. doi: 10.1242/jcs.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mendl N, Occhipinti A, Mueller M, Wild P, Dikic I, Reichert AS. 2011. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J Cell Sci 124:1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- 52.Pampulha ME, Loureirodias MC. 1989. Combined effect of acetic-acid, pH and ethanol on intracellular pH of fermenting yeast. Appl Microbiol Biotechnol 31:547–550. [Google Scholar]
- 53.Larsson S, Palmqvist E, Hahn-Hägerdal B, Tengborg C, Stenberg K, Zacchi G, Nilvebrant NO. 1999. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb Technol 24:151–159. doi: 10.1016/S0141-0229(98)00101-X. [DOI] [Google Scholar]
- 54.Siniossoglou S, Hurt EC, Pelham HR. 2000. Psr1p/Psr2p, two plasma membrane phosphatases with an essential DXDX (T/V) motif required for sodium stress response in yeast. J Biol Chem 275:19352–19360. doi: 10.1074/jbc.M001314200. [DOI] [PubMed] [Google Scholar]
- 55.Kelly DE, Trevethick J, Mountain H, Sudbery PE. 1988. Transcript characterisation, gene disruption and nucleotide sequence of the Saccharomyces cerevisiae WHI2 gene. Gene 66:205–213. doi: 10.1016/0378-1119(88)90357-5. [DOI] [PubMed] [Google Scholar]
- 56.Auberson LCM, von Stockar U. 1992. The role of the limited respiratory capacity in the Saccharomyces cerevisiae whi2 mutation. J Biotechnol 23:35–53. doi: 10.1016/0168-1656(92)90098-T. [DOI] [Google Scholar]
- 57.Mountain HA, Sudbery PE. 1990. Regulation of the Saccharomyces cerevisiae WHI2 gene. J Gen Microbiol 136:727–732. doi: 10.1099/00221287-136-4-727. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka K, Ishii Y, Ogawa J, Shima J. 2012. Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the HAA1 gene, encoding a transcriptional activator. Appl Environ Microbiol 78:8161–8163. doi: 10.1128/AEM.02356-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, Hao T, Rual J-F, Dricot A, Vazquez A, Murray RR, Simon C, Tardivo L, Tam S, Svrzikapa N, Fan C, de Smet A-S, Motyl A, Hudson ME, Park J, Xin X, Cusick ME, Moore T, Boone C, Snyder M, Roth FP, Barabási A-L, Tavernier J, Hill DE, Vidal M. 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beck T, Hall MN. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 61.Hong M-E, Lee K-S, Yu BJ, Sung Y-J, Park SM, Koo HM, Kweon D-H, Park JC, Jin Y-S. 2010. Identification of gene targets eliciting improved alcohol tolerance in Saccharomyces cerevisiae through inverse metabolic engineering. J Biotechnol 149:52–59. doi: 10.1016/j.jbiotec.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 62.Kim HJ, Turner TL, Jin Y-S. 2013. Combinatorial genetic perturbation to refine metabolic circuits for producing biofuels and biochemicals. Biotechnol Adv 31:976–985. doi: 10.1016/j.biotechadv.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 63.Santos CNS, Stephanopoulos G. 2008. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol 12:168–176. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 64.Tyo KE, Zhou H, Stephanopoulos GN. 2006. High-throughput screen for poly-3-hydroxybutyrate in Escherichia coli and Synechocystis sp. strain PCC6803. Appl Environ Microbiol 72:3412–3417. doi: 10.1128/AEM.72.5.3412-3417.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 66.Nikawa J, Tsukagoshi Y, Yamashita S. 1991. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J Biol Chem 266:11184–11191. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.