Abstract
Psychrophilic enzymes play crucial roles in cold adaptation of microbes and provide useful models for studies of protein evolution, folding, and dynamic properties. We examined the crystal structure (2.2-Å resolution) of the psychrophilic β-glucosidase BglU, a member of the glycosyl hydrolase 1 (GH1) enzyme family found in the cold-adapted bacterium Micrococcus antarcticus. Structural comparison and sequence alignment between BglU and its mesophilic and thermophilic counterpart enzymes (BglB and GlyTn, respectively) revealed two notable features distinct to BglU: (i) a unique long-loop L3 (35 versus 7 amino acids in others) involved in substrate binding and (ii) a unique amino acid, His299 (Tyr in others), involved in the stabilization of an ordered water molecule chain. Shortening of loop L3 to 25 amino acids reduced low-temperature catalytic activity, substrate-binding ability, the optimal temperature, and the melting temperature (Tm). Mutation of His299 to Tyr increased the optimal temperature, the Tm, and the catalytic activity. Conversely, mutation of Tyr301 to His in BglB caused a reduction in catalytic activity, thermostability, and the optimal temperature (45 to 35°C). Loop L3 shortening and H299Y substitution jointly restored enzyme activity to the level of BglU, but at moderate temperatures. Our findings indicate that loop L3 controls the level of catalytic activity at low temperatures, residue His299 is responsible for thermolability (particularly heat lability of the active center), and long-loop L3 and His299 are jointly responsible for the psychrophilic properties. The described structural basis for the cold adaptedness of BglU will be helpful for structure-based engineering of new cold-adapted enzymes and for the production of mutants useful in a variety of industrial processes at different temperatures.
INTRODUCTION
The β-glucosidases (BGs; β-d-glucoside glycohydrolases, EC 3.2.1.21) are a widely occurring group of enzymes that cleave the carbohydrate moiety of short-chain oligosaccharides (two to six degrees of polymerization), alkyl, and aryl β-glucosides (1, 2). BGs have been classified into 135 glycoside hydrolase (GH) families, based on amino acid sequence similarity. GHs can also be classified into two major types, retaining and inverting enzymes, according to changes in anomeric configuration during hydrolytic reactions. Early studies of BGs were focused on their role in degrading the plant polymers cellulose and xylan (3, 4). More recently, these enzymes have gained commercial significance because of their biotechnological application in processes related to preparation of plant-based foods, e.g., conversion of phytoestrogen glucosides in fruits and vegetables to aglycone moieties, detoxification of cassava, and degradation of bitter compounds in citrus fruit juices and unripe olives (5, 6). Many mesophilic and thermophilic BGs have been cloned, purified, and characterized during the past 2 decades (5, 7–10). In contrast to mesophilic or thermophilic homologues, BGs from organisms adapted to cold environments have potential application in fields that include food processing to prevent spoilage or changes of nutrient content and taste, detergents, textiles, and bioremediation of polluted soils and wastewaters at low to moderate temperatures (11–13).
To date, studies indicate that cold-adapted enzymes typically display a high catalytic constant (kcat), low optimal temperature, and high degree of thermolability, particularly of active center (14–16). A cold-adapted BG of the GH1 family, termed BglU, was previously cloned from the DNA library of Micrococcus antarcticus, a psychrophilic spherical bacterium found in Antarctica (17). BglU displays typical properties of cold-adapted enzymes, including high catalytic efficiency at low temperatures and relative instability at high temperatures. Its optimal temperature is 25°C, and its activity decreases greatly within 30 min at higher temperatures (18).
Many studies and hypotheses since 2000 have addressed the structural basis of cold adaptation in psychrophilic proteins (19–22). However, structural data for psychrophilic enzymes remain quite limited, and no such data are available for cold-adapted BGs. To clarify the mechanism of cold adaptation of BglU, we mapped its crystal structure at 2.2-Å resolution. This is the first structural description of a cold-adapted BG of the GH1 family. We identified the structural basis and specific amino acid residues responsible for the psychrophilic properties of BglU by (i) comparison of its structure and amino acid sequence to those of mesophilic and thermophilic enzymes with which it shares >40% amino acid identity, e.g., BglB from Paenibacillus polymyxa (23) and GlyTn from Thermus nonproteolyticus Hg102 (24), and (ii) mutagenesis studies. Two key features, long-loop L3 and residue His299, were found to be essential for cold adaptation, and coordination between them is the basis for psychrophilicity of the enzyme. Our findings help elucidate the principles that underlie relationships between protein architecture and cold adaptation properties of psychrophilic GH1 family BGs and will have practical application in structure-based engineering of cold-adapted enzymes and production of mutants useful for low-temperature industrial processes.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Escherichia coli BL21(DE3) was used for production of His-tagged recombinant BglU from expression vectors. Escherichia coli XL1-Blue was used for the construction and selection of mutations. All E. coli strains were grown at 37°C, with vigorous shaking in LB medium, unless stated otherwise. Plasmid pET28-a(+) was used for expression of bglU gene and its mutants (18).
Shortened-loop L3 mutagenesis by fusion PCR.
Two mutants were constructed by this procedure: delta-L3 (10 amino acids deleted [315 to 324], PQPVVPTDSP) and delta-L3/H299Y. For construction of loop L3 mutants, fusion PCR was performed as described by Walker et al. (25) and Yu et al. (26), using BglU as the template and the primer pairs listed in Table 1. In brief, three rounds of PCR were conducted, with primers P1 and P2 for round 1, P3 and P4 for round 2, and P1 and P4 for round 3. Rounds 1 and 2 were performed using Pfu polymerase under the following conditions: a 1-min denaturation at 95°C, followed by 30 cycles of a 1-min denaturation at 95°C, a 1-min annealing at 60°C, and a 1-min extension at 68°C. Two chimeric fragments were obtained, with ∼30-bp overlapping region. The fragments were gel purified, mixed, and amplified (fusion reaction) using Pfu polymerase under the following conditions: 2 min of denaturation at 95°C, followed by eight cycles consisting of a 30-s denaturation at 95°C, a 5-min annealing at 60°C, and a 1-min extension at 72°C. Primers 1 and 4 were added, and PCR was performed as follows: 2 min of denaturation at 95°C, followed by 30 cycles of a 30-s denaturation at 95°C, a 30-s annealing at 55°C, and a 1-min extension at 72°C.
TABLE 1.
Primers used for mutations
| Mutation and primer | Sequence (5′-3′)a |
|---|---|
| delta-L3 | |
| P1 | CGCGGATCCATGATGAACCACTTATC |
| P2 | GCCGGGAAGGTCACATACTCGCTGCCGACAAACTGGCCGG CGGGCAGCGGAT |
| P3 | ACGTTTCAGGCCATCCGCTGCCCGCCGGCCAGTTTGTCGGCAGCGAGTATGT |
| P4 | CCCAAGCTTTTATGCCATTGTACGG |
| H299Y | |
| Pf | CTTGGGCGTGAACTACTACCACGATGACAACG |
| Pr | CGTTGTCATCGTGGTAGTAGTTCACGCCCAAG |
| delta-L3/H229Y | All of the above |
Restriction sites are indicated by underlining.
Site-directed mutagenesis.
BglU was used as the template for all cloning and mutagenesis procedures. Plasmid pET28-a(+) was used for expression procedures. The primers used are listed in Table 1. Homology searches for protein sequences were performed using the BLAST and FASTA programs. Mutant genes were prepared by site-directed mutagenesis with PCR amplification. PCR products were sequenced to confirm the identity of the desired mutations by the primer-walking technique using an automated sequencer (model ABI 3730xl; Applied Biosystems, USA). Sequence information for the mutant enzymes was analyzed using the DNAman program (v5.1; Lynnon Biosoft).
Two overlapping complementary oligonucleotides, Pf and Pr, for each mutation were designed, containing the corresponding nucleotide changes. Each plasmid was amplified by using 1 U of Pfu polymerase in a linear extension reaction by PCR under the following conditions: 1-min denaturation at 95°C, followed by 18 cycles consisting of a 1-min denaturation at 95°C, a 1-min annealing at 60°C, and a 5-min extension at 68°C. The reaction products were cooled to 20°C and then treated with DpnI for 1 h at 37°C to hydrolyze the methylated parental plasmid strands. Then, 1 μl of DpnI-treated DNA was used to transform E. coli XL1-Blue.
De novo synthesis of BglB and GlyTn gene sequences.
Gene sequences of BglB (PDB 2Z1S), GlyTn (PDB 1NP2), and their mutants were de novo synthesized, and the sequences were confirmed by Sangon Biotech Co. (Shanghai, China).
Expression and purification of recombinant BglU and its mutants.
The recombinant plasmids were transformed into E. coli BL21(DE3) and cultured at 37°C in LB medium containing 50 μg/ml kanamycin. IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mmol liter−1 when the optical density at 600 nm reached 0.7, and the culture was continued for another 6 h at 16°C. Cells were harvested by centrifugation, washed twice with 50 mmol liter−1 phosphate buffer (pH 6.5), suspended in the same buffer, and disrupted by sonication. Insoluble debris was removed by centrifugation (10,000 × g, 4°C, 20 min; Sigma rotor 12159-H). The supernatant was applied to Ni-NTA agarose (Novagen) and purified according to a standard protocol (Ni-NTA His-Bind resin kit; Novagen). Eluates were pooled, concentrated by using an Amicon Ultra-15 (Millipore), and stored at −20°C.
Enzyme assay and kinetic parameters.
The BG activity was assayed by measuring the increase in absorbance at 400 nm resulting from the release of p-nitrophenol from p-nitrophenyl-β-glucoside (pNPG) (27). The reaction mixture (0.5 ml) contained 0.2 μg of enzyme except the mutant delta-L3 (20 μg), and 50 mmol liter−1 phosphate buffer (pH 6.5) with 5 mmol liter−1 pNPG (final concentration). The mixture was incubated for 5 min, and the reaction was stopped by the addition of 1 ml of 1 mol liter−1 Na2CO3. One unit (U) of BG activity was defined as the amount of enzyme that caused release of 1 μmol of p-nitrophenol per min. The values of the kinetic constants Km and Vmax of the Michaelis-Menten equation were determined by nonlinear regression plot using the SPSS19.0 software program. The kinetic constant kcat and the catalytic efficiency ratio kcat/Km were determined based on the Km and Vmax values.
The optimal temperature and thermal stability were analyzed as described previously (18). The melting temperature (Tm) was determined by differential scanning fluorimetry (DSF). In brief, DSF was performed using ∼0.2 mg of protein/ml in phosphate-buffered saline added with 5% glycerol with SYPRO Orange (Invitrogen, catalog no. S6650; final concentration, 5×). The mixture was monitored in hard-shell 96-well skirted PCR plates (Bio-Rad, catalog no. HSP-9601) using a 40-μl volume and a Bio-Rad CFX96 RT-PCR system (excitation wavelength, 450 to 490 nm; emission wavelength, 560 to 590 nm).
Crystallization and data collection.
Crystallization was performed at 4°C using sitting-drop vapor diffusion with a drop ratio of 1 μl of 20-mg/ml protein solution in 50 mmol liter−1 Tris-HCl and 500 mmol liter−1 NaCl to 1 μl of reservoir solution. Crystals were grown by the hanging-drop vapor diffusion method. BglU crystals were obtained by mixing 1 μl of reservoir solution (0.1 mol liter−1 Tris-HCl, 0.2 mol liter−1 MgCl2, and 30% [vol/vol] PEG4000). Crystals were flash-cooled in a nitrogen stream at 100K. X-ray diffraction data (2.2-Å resolution) for BglU were collected on an R-Axis IV++ image plate detector using a Rigaku rotating anode X-ray generator.
Structure determination and refinement.
Data sets were processed using the Mosflm program (28) and then scaled and reduced using the SCALA program of the CCP4 suite (29). Initial phases for BglU were obtained by the molecular replacement method using the Phaser program of the CCP4 suite. The crystal structure of Paenibacillus polymyxa β-glucosidase B (BglB; PDB ID 2Z1S) (23) was used as a search model. Model building and refinement were conducted using the COOT (30), CNS (31), and PHENIX (32) programs. Several cycles of refinement were used to obtain acceptable root mean square deviation (RMSD) and Rfree values. Data collection and refinement statistics are presented in Table 2. Stereochemical properties were evaluated using the PROCHECK program (33). Figures were prepared using the PyMOL program (34).
TABLE 2.
Data collection and refinement statistics
| Parameter | Value(s) for parameter |
|---|---|
| Space group | C2221a |
| Unit cell parameters | |
| a, b, c (Å) | 57.28, 84.67, 184.77 |
| α, β, γ | 90, 90, 90 |
| Resolution (Å) | 61.55–2.20 |
| No. of unique reflections | 22,807 |
| Completeness (%) | 97.9 (87.4) |
| Rmerge (%) | 3.9 (7.3) |
| Mean I/σ〈I〉 | 32.6 (14.2) |
| Redundancy | 6.4 (3.5) |
| Rwork/Rfree (%) | 16.13/19.97 |
| No. of molecules | |
| Protein | 3,714 |
| Water | 442 |
| Tris | 1 |
| B factor (Å2) | |
| Mean | 13.70 |
| Protein | 12.95 |
| Water | 20.02 |
| Tris | 12.96 |
| Ramachandran plot (%) | |
| Favored | 96.8 |
| Allowed | 3.2 |
| Disallowed | 0 |
| RMSDb | |
| Bond length (Å) | 0.0073 |
| Bond angle (°) | 1.070 |
Numbers in parentheses indicate values in the highest-resolution shell.
RMSD, root mean square deviation.
Protein Data Bank accession number and mutant gene accession numbers.
Coordinates and structure factors for BglU were deposited in the Protein Data Bank (http://www.wwpdb.org) under accession number 3W53. Sequences of mutant genes delta-L3, H299Y, and delta-L3/H229Y were deposited in GenBank database under accession numbers KU198288, KU198286, and KU198287, respectively.
RESULTS
Psychrophilic β-glucosidase BglU adopts a TIM barrel fold structure, as do its mesophilic and thermophilic counterparts.
The crystal structure of BglU, a psychrophilic BG from Micrococcus antarcticus, was determined by the molecular replacement method using the structure of BglB (23) as a search model and refinement at 2.2-Å resolution with R factor 0.161 and Rfree = 0.199 (Table 2). The final model contains 3,714 protein atoms, 1 Tris molecule, and 442 water molecules. BglU exists as a monomer in the asymmetric unit, in accordance with its monomeric state in solution. The overall structure of BglU (Fig. 1) adopts a typical (α/β)8 “TIM barrel fold” (named after triosephosphate isomerase), as also observed in other GH1 family members (23, 24).
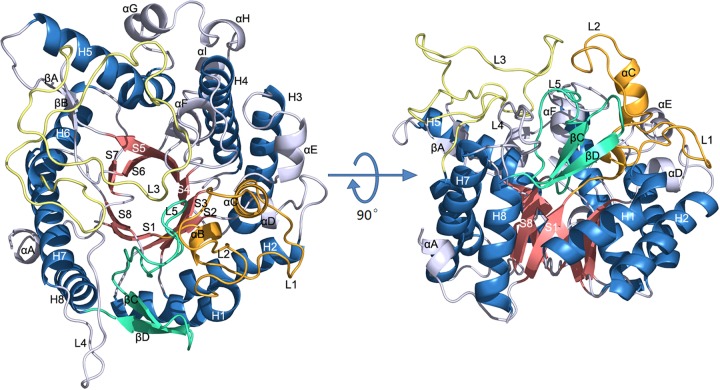
FIG 1.
Ribbon representation of the overall structure of BglU. The α-helices and β-strands of the TIM barrel are shown by salmon and sky blue. Loops L1, L2, αB, and αC are orange. L5, βC, and βD are green/cyan. L3 is yellow. L4 and other residues are gray.
BglU, the first psychrophilic BG ever reported, shows similarities in sequence and structure to other GH1 family enzymes, including its mesophilic and thermophilic counterparts. Sequence alignments showed that BglU shows 49% identity with mesophilic BglB (PDB ID 2Z1S) (23) and 42% identity with thermophilic GlyTn (PDB ID 1NP2) (24) (Fig. 2). Structure superposition (Fig. 3) showed that the structure of BglU is similar to those of BglB and GlyTn, with Ca atom RMSD values of 1.5 and 1.2 Å, respectively.
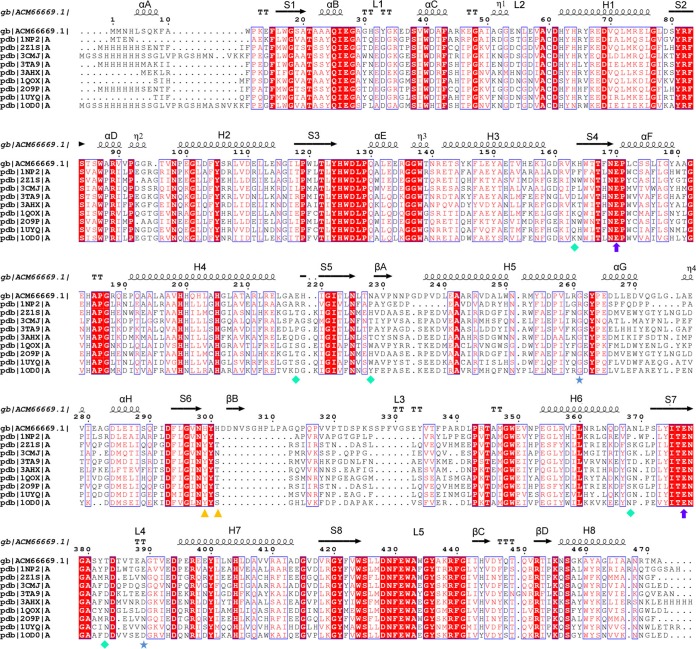
FIG 2.
Sequence alignment of BglU and its mesophilic (2Z1S) and thermophilic (1NP2) homologues in the GH1 family. Sequence alignment was performed using the ClustalW (v2.0) and ESPript programs. Conserved sequences are indicated by boxes, and similar sequences are indicated by colored background. α-Helix, β-sheet, random coil, and beta turn are labeled α, β, η, and T, respectively. Secondary structures and their designations are shown. β-Strands and α-helices are represented by arrows and coils, respectively. Residues Glu170 and Glu377 are indicated by purple circles; His299 and His301 are indicated by yellow triangles; Lys163, Glu218, Asn228, Ala368, and Thr383 are indicated by cyan diamonds; Gly261 and Ala389 are indicated by blue stars.
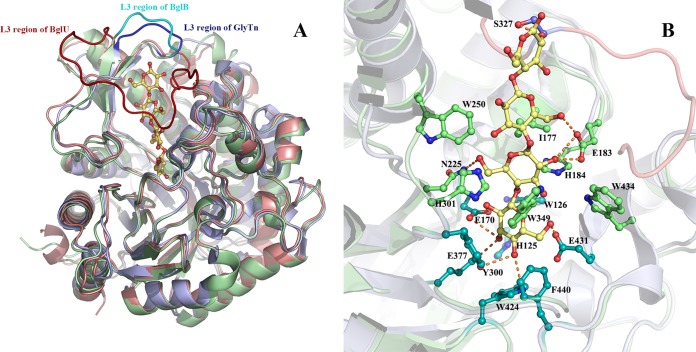
FIG 3.
(A) Structure superposition of BglU on the thermophilic counterpart GlyTn (A chain) and the mesophilic counterpart BglB. BglU, GlyTn, and BglB are colored in salmon, light blue, and pale green, whereas their loop L3 is shown in red, blue, and cyan, respectively. The cellotetraose molecule modeled into the substrate-binding pocket is shown as sticks. (B) Detail of substrate-binding pocket with cellotetraose. Residues His125, Trp126, Glu170, Tyr300, Glu377, Trp424, and Glu431, Phe440 (indicated in cyan) interact mainly with the first nonretaining monosaccharide group, whereas residues Glu183 (also with the third monosaccharide group), His184, His301, Ile177, Asn225, Trp250, Trp349, and Trp434 (indicated in green) interact mainly with the second nonretaining monosaccharide. Residues from loop L3, which is indicated in violet but with residue Ser327 emphasized in steel blue, interact with the third and fourth nonretaining monosaccharides. The figures were prepared using the PyMOL program.
Substrate-binding pocket.
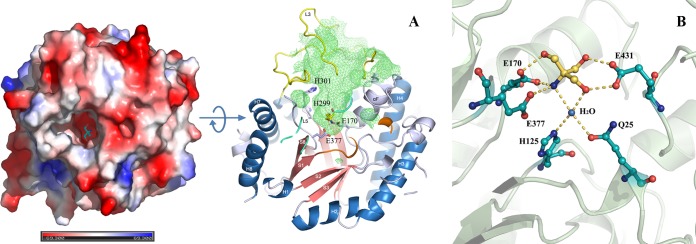
A large, deep pocket is present on the surface of BglU (Fig. 4) and has a depth of 20 Å, an inner surface area of 1,088 Å2, and a volume of 1,858 Å3 as calculated by CASTp. Glu170 and Glu377, the two residues directly related to enzyme activity, are located at the bottom, indicating that this is the substrate-binding pocket of the enzyme. The pocket is at the top of the TIM barrel core and is surrounded mainly by the residues of loops L1, L2, L3, L4, and L5, helix αF, and strand βB. One Tris molecule is located at the bottom of the pocket. Residue Glu431 forms two hydrogen bonds with two hydroxyl groups of Tris via its carboxyl group. Residue Glu170 forms one hydrogen bond with the third hydroxyl group of Tris via its carboxyl group. Residues Glu170 and Glu377 form two hydrogen bonds with the amino group of Tris via their carboxyl groups. One water molecule is involved in the protein-Tris interaction and is bonded by a hydrogen bond network. Jeng et al. (35) reported that the active site of TrBgl2 and NkBgl is also occupied by a Tris molecule. The Tris molecule is presumed to mimic the substrate, as reported previously for other GH1 family members (23, 24), because both are rich in hydroxyl groups.
FIG 4.
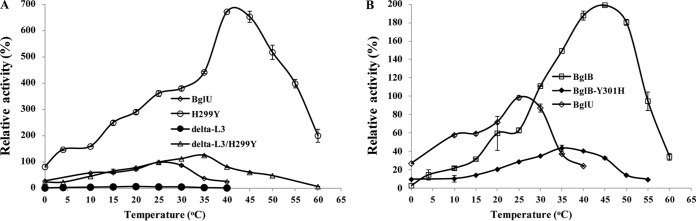
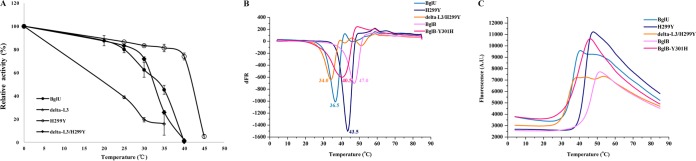
Comparison of optimal temperatures of BglU and its mutants. (A) BglU, H299Y, delta-L3, and delta-L3/H299Y. (B) BglU, BglB, and BglB-Y301H. The maximal activity (385.6 U/mg) of BglU was defined as 100%. The experiments were performed in triplicate or quadruplicate.
The 20-Å depth of the substrate-binding pocket of BglU is consistent with the length of a tetrasaccharide molecule. A cellotetraose molecule was modeled into the substrate-binding pocket by homologue modeling, using the BglB-cellotetraose complex structure as a template. In this model, the conserved residues Glu377, Glu170, Glu431, Trp424, Phe440, Trp126, His125, and Tyr300 interact mainly with the first nonretaining monosaccharide. Residues Ile177, Glu183, His184, Asn255, His301, Trp349, Try250, and Trp434 interact mainly with the second nonretaining monosaccharide. Residues from loop L3 (see below) interact with the third and fourth nonretaining monosaccharides, such as Ser327. Most residues involved in recognition of the first and second monosaccharides are conserved (Fig. 3B).
Loop L3 and His299 constitute the major difference between BglU and its mesophilic and thermophilic counterparts in terms of both structure and sequence.
Sequence alignment and structural comparison between BglU and its mesophilic and thermophilic counterparts (BglB and GlyTn) revealed that the long-loop L3 and His299 constitute the major difference in terms of both sequence and structure (Fig. 2 and 3A). Loop L3 of BglU is 28 amino acid residues longer than that of BglB or GlyTn. This long-loop L3 has a unique structure and surrounds the entrance to the active site. Because BglB and GlyTn do not have this long loop, their substrate-binding pockets are more accessible than that of BglU. The long-loop L3 increases the solvent-accessible area, mobility, and flexibility of the BglU structure and is evidently an important factor contributing to substrate recognition and low-temperature activity of the enzyme.
Loop L3 can be divided into two parts: an N-terminal nonconserved part (residues 307 to 342) and a C-terminal conserved part (residues 343 to 349). The N-terminal part is not conserved among BglU, BglB, and GlyTn. It forms an O-ring-like structural motif and exclusively comprises the entrance of the substrate-binding pocket in BglU. It is the primary feature accounting for the difference of loop 3 between BglU and other GH1 enzymes. This part is 35 residues long in BglU, but it is only 7 residues long in BglB and GlyTn. In the modeled BglU-cellotetraose complex structure, the N-terminal part contributes to interactions with the third and fourth nonretaining monosaccharides via van der Waals interaction. The corresponding segments in BglB, GlyTn, and other GH1 members have similar (∼7 amino acids) lengths and structures but totally different amino acid compositions.
The C-terminal part of loop L3 is conserved in GH1 enzymes and has sequence motif PXTXX[M/I]GW. This part mainly interacts with the second monosaccharide via van der Waals interaction, e.g., the side chain of residue Trp349.
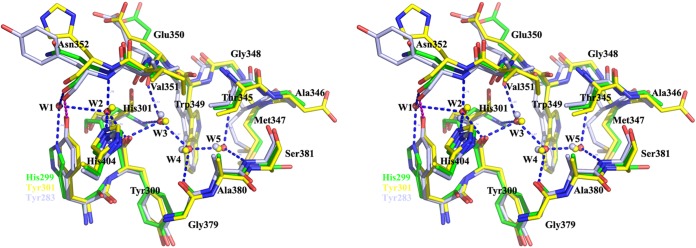
The side chain of His299 forms a hydrogen bond with a water molecule, termed W1, which is accessible to the solvent. An ordered chain consisting of water molecules W1, W2, W3, W4, and W5 was found to be present in a tunnel extending from the protein surface to the catalytic site. Residues located around this tunnel, including His299, Tyr300, His301, Asn350, Val351, Gly379, Ala380, and Ser381, form a complicated hydrogen bond network with atoms in the water molecule chain and help stabilize the chain (see Fig. 7).
FIG 7.
Structure superposition of the ordered water molecule chains, as well as residues His299 and His301, of BglU (green), BglB (yellow), and GlyTn (gray).
Similar water molecule chains have been observed in BglB and GlyTn. In BglB, the chain is evidently involved in substrate binding (24). Structure superpositions revealed that the chain is conserved except that His299 and adjunct water molecule W1 in BglU are structurally replaced by a Tyr residue in the other two enzymes, i.e., Tyr301 in BglB and Tyr283 in GlyTn. Other residues involved in water molecule chain binding are identical. Hydrogen bonds other than those formed by His299 and W1 in BglU are identical in the three enzymes. We hypothesized that replacement of this Tyr residue in mesophilic BglB and thermophilic GlyTn with His in psychrophilic BglU is related to the cold adaptedness of BglU.
Loop L3 and His299 play key roles in cold adaptation of BglU.
To clarify the role of loop L3 and residue His299 in cold adaptedness, we constructed a deletion mutant of loop L3, a site-directed mutant of H299Y, and a loop L3 deletion/His299Y substitution double mutant. As a reference, an Y301H mutant of BglB was constructed to clarify the role of residue Tyr301 in BglB (corresponding to His299 in BglU).
BglU mutant delta-L3 (25 amino acids), in comparison to BglU, showed a lower catalytic activity (kcat = 0.46 × 103 s−1 versus 6.7 × 103 s−1), substrate-binding ability (Km = 19.39 mmol liter−1 versus 5.9 mmol liter−1), Tm (<25°C versus 36.5°C; the Tm of delta-L3 was too unstable to determine [see Fig. 6B]), and optimal temperature (20°C versus 25°C) (Fig. 5A and Table 3). Catalytic activity was undetectable (<1%) when loop L3 was further shortened to <25 amino acids (including substitution by the L3 of GlyTn) and in H299Y double mutants (data not shown). These mutants were therefore not studied further. These findings indicate that shortening of loop L3 in BglU significantly reduced catalytic activity, substrate-binding ability, thermostability, and optimal temperature.
FIG 6.
Substrate-binding pocket of BglU. (A) Electronic potential surface of BglU (left) showing the large, deep cavity for substrate binding, and a section of BglU showing the inner surface (right, green grid) of the substrate-binding pocket. Residues Glu170, His299, His301, and Glu377 and a Tris molecule in the pocket are indicated by sticks (right). (B) Closer inspection of the binding mode of the Tris molecule, in yellow, located at the substrate-binding pocket. Residues Gln25, His125, Glu170, Glu377, and Glu431 interacting with the Tris molecule are indicated by sticks.
FIG 5.
Relative activity at various temperatures of BglU and its mutants (A), the Tm value from the derivative of the fluorescent signal (B), and the fluorescence adsorption of BglU, BglB, and their mutants (C). The calculated Tm values were 36.5°C for BglU, 43.5°C for H299Y, 34°C for delta-L3/H299Y, 47°C for BglB, and 40.5°C for BglB-Y301H. BglU and its mutant enzymes were maintained at various temperatures as shown for 40 min. For each protein, activity at 0 min was defined as 100%. The Tm was determined by differential scanning fluorimetry (DSF). The experiments were performed in triplicate or quadruplicate.
TABLE 3.
Kinetic parameters for BglU and its mutantsa
| Enzyme or mutant | Mean ± SEM |
|||
|---|---|---|---|---|
| Km (mmol/liter) | Vmax × 104 (μmol min−1 mg−1) | kcat × 103 (s−1) | kcat/Km × 106 (liter mol−1 s−1) | |
| BglU | 5.9 ± 0.57 | 0.843 ± 0.1 | 6.7 ± 0.7 | 1.14 ± 0.13 |
| H229Y | 3.99 ± 0.14 | 2.83 ± 0.04 | 22.64 ± 0.07 | 5.7 ± 1.00 |
| delta-L3 | 19.39 ± 1.27 | 0.058 ± 0.01 | 0.46 ± 0.09 | 0.0238 ± 0.002 |
| delta-L3/H299Y | 5.2 ± 0.6 | 0.909 ± 0.19 | 7.27 ± 1.5 | 1.48 ± 0.19 |
Kinetic parameters were determined using pNPG as the substrate at 25°C. The experiments were performed in triplicate.
In comparison to H299Y, delta-L3/H299Y had a lower catalytic activity (kcat = 1.48 × 103 s−1 versus 5.70 × 103 s−1), Tm (34.0°C versus 43.5°C), and optimal temperature (35°C versus 45°C) and a slightly lower substrate-binding ability (Km = 5.2 mmol liter−1 versus 4.0 mmol liter−1). Low-temperature activity (i.e., high activity at low temperatures) was lost in delta-L3/H299Y but was retained in H299Y (Fig. 5A). Taken together, our findings indicate that (i) loop L3 is essential for low-temperature catalytic activity of BglU and relative thermostability at low temperatures and that (ii) loop L3 is essential for substrate-binding ability; however, the loss of such ability resulting from loop L3 deletion is restored by mutation of His299 to Tyr.
Compared to BglU, H299Y had a higher Tm (43.5°C versus 36.5°C) (Fig. 6), optimal temperature (45°C versus 25°C) (Fig. 5A), and catalytic activity (kcat = 22.64 × 103 s−1 versus 6.7 × 103 s−1) and a slightly higher substrate-binding ability (Km = 3.99 mmol liter−1 versus 5.9 mmol liter−1). In comparison to delta-L3, delta-L3/H299Y had a higher Tm (35°C versus <25°C) (Fig. 6), optimal temperature (35°C versus 20°C), substrate-binding ability (Km = 5.2 mmol liter−1 versus 19.39 mmol liter−1), and catalytic activity (kcat = 7.72 × 103 s−1 versus 0.46 × 103 s−1). These findings indicate that the mutation of His299 to Tyr results in significant increases of thermostability, optimal temperature, substrate-binding ability, and catalytic activity.
This concept is supported by findings for mutation of corresponding residue Tyr301 to His in mesophilic BglB. Compared to BglB, the BglB-Y301H mutant had reduced Tm (40.5°C versus 47°C) (Fig. 5B and C) (36, 37) and optimal temperature (35°C versus 45°C) (Fig. 4B). BglB-Y301H displayed no low-temperature activity because of the absence of long-loop L3.
DISCUSSION
BglU, the first reported psychrophilic BG, shows typical features of cold adaptation: high activity at low temperature, a low optimal temperature, low thermostability, and a large difference between optimal temperature (25°C) and Tm (36.5°C) (38). Like other psychrophilic enzymes, BglU is inactivated by temperature prior to structure unfolding. This difference indicates that the active site and catalytic intermediates of BglU have strong heat lability. A long-loop L3, ∼28 amino acids longer than loops described for other BGs, is a unique structure in BglU. It increases the solvent-accessible area, mobility, and flexibility of the BglU structure. When long-loop L3 was shortened to medium length (25 amino acids, in mutant delta-L3), and at the even shorter length (7 amino acids) in mesophilic (BglB) and thermophilic (GlyTn) counterpart enzymes, the catalytic activity was greatly reduced. The shorter the length, the lower the observed activity.
The residue His299 is unique to BglU. In comparison with BglU, H299Y had significantly higher catalytic activity, optimal temperature, and thermostability (Fig. 5A, Fig. 6A to C). Similar trends were seen in comparisons between delta-L3/H299Y and delta-L3. Large differences between Tm and optimal temperature were not seen for delta-L3/H299Y or H299Y. These mutants have properties of mesophilic or psychrotrophilic rather than psychrophilic enzymes, indicating that the mutation of His299 to Tyr may stabilize the active site and the catalytic intermediate. In view of the fact that His299 is involved in stabilizing an ordered water molecule chain accessible to the substrate-binding pocket, the properties of the mutants suggest that His299 and the chain play key roles in the catalytic reaction. His299 and the chain contribute to the low optimal temperature and thermolability of BglU and, in particular, to the greater heat lability of the active center than the overall structure.
The structure of BglU includes an ordered water molecule chain comprised of five water molecules, termed W1 to W5 in sequence from the protein surface to the active site. The side chains of His299 and His301 interact via hydrogen bonds with the outer and inner molecules W1 and W3, respectively, to stabilize the chain (Fig. 7). This hydrogen-bonded network also involves the catalytic residues Glu170 and Glu377. In BglB and GlyTn, His299 of BglU is substituted by Tyr (Tyr297 and Tyr283, respectively). These Tyr residues interact directly with the main chain at the site corresponding to Asn352 of BglU, preventing the existence of W1 and blocking the pathway of W2 to W5 accessible to the protein surface. In brief, His299 and water molecule W1 of BglU are structurally substituted by Tyr297 of BglB and Tyr283 of GlyTn.
We presume that Tyr299 in H299Y similarly prevents the existence of W1 and interacts directly with the main chain at site Asn352, as the corresponding residue does in BglB and GlyTn (Fig. 7). According to our proposed model, this interaction stabilizes the conformation of segment 347 to 352, which is located at the bottom of the substrate-binding pocket and interacts directly with the substrate. Tyr299 stabilizes the conformation of His301 via the water molecule chain and the hydrogen-bonded network. Segment 347-352 and residue His301 are involved in catalysis and prefer a stable conformation, accounting for the fact that optimal temperature and thermostability of H299Y are higher than those of BglU. The conserved water molecule chain is essential for transferring protons from the solvent to the active site. His299, located on the protein surface, initiates the proton transfer and is involved in subsequent catalytic reaction. A proton is transferred from His299 to W1, W2, and W3 in turn, to His301, and then to the substrate. It is easier to deprotonate a Tyr than a His residue at position 299, accounting for the fact that enzyme activity of H299Y is higher than that of BglU. Differing deprotonating velocities of His and Tyr residues at different pHs may explain the observed differences in optimal pH between BglU and its mutants. The functional pH range of H299Y is wider than that of BglU (data not shown).
Why does M. antarcticus utilize a psychrophilic BG, BglU? This bacterium is found in the Antarctic region and has presumably become adapted to survive in a permanently cold environment. The optimal temperature for growth of M. antarcticus is 16.8°C, and growth is possible even at 0°C (17). BglU is an essential enzyme in cellular metabolism and is necessary (in combination with other enzymes) to maintain sufficient activity for growth at 0 to 16°C. In view of the environmental temperatures typically encountered by M. antarcticus, BglU presumably maintains significant enzyme activity below 10°C. None of the mesophilic or thermophilic BGs so far described is capable of efficient catalytic activity at temperatures below 10°C. The present findings, as explained above, indicate that both long-loop L3 and Tyr299 promote low-temperature activity in GH1 enzymes. The lowest enzyme activity was seen for the mutant delta-L3, which lacks both long-loop L3 and Tyr299. Activity was higher when either long-loop L3 (BglU) or Tyr299 (delta-L3/H299Y) was present. The highest activity was observed for H299Y, which has both long-loop L3 and Tyr299. At a moderate temperature, the activity of delta-L3/H299Y became similar to that of BglU, but there was no low-temperature activity; i.e., delta-L3/H299Y had the properties of a mesophilic enzyme. The low-temperature activity of H299Y was significantly higher than that of delta-L3/H299Y. These findings all indicate that low-temperature activity in these BGs is controlled by long-loop L3.
Aside from its high activity at low temperature, BglU shows typical thermolabile properties similar to those of other cold-adapted enzymes. Thermostability and optimal temperature were increased significantly for all the H299Y mutants, suggesting that His299 controls thermolability in psychrophilic BGs. Cold adaptation properties result from the combined effects of various sequences and secondary structures in psychrophilic enzymes. Cross talk between loop L3 and residue Tyr (His299) contributes to the diversity of enzyme activity, optimal temperature, and thermostability. Loop L3 of a certain length, and Tyr, can substitute for each other to maintain a certain level of enzyme activity, but different strategies using various lengths of loop L3 or Tyr299 allow for adaptation to different temperatures. The results of our mutagenesis assay indicate that a long, flexible loop L3 can readily change its construction with low active energy, allowing efficient substrate binding. Cold-adapted bacteria such as M. antarcticus therefore tend to utilize a combination of long-loop L3 structure and His299 to retain high activity at low temperatures and become inactive immediately to avoid wasting substrates when temperatures are not suitable for cell growth (i.e., >25°C). Mesophilic and thermophilic BGs such as BglB and GlyTn display no (or very low) activity at low temperature because of the absence of long-loop L3 and are not suitable for survival of cold-adapted bacteria. Tyr299 requires more active energy for efficient enzyme activity, and the rigid structure of short-loop L3 promotes thermostability in BGs. Mesophilic and thermophilic bacteria therefore tend to utilize a combination of Tyr299 and very short (7 amino acids) loop L3 for survival in their higher-temperature environments.
Why do microbes living in cold environments not utilize the cold-resistant combination of long-loop L3 and Tyr299 to facilitate low-temperature activity? The combination of long-loop L3 and His299 presumably resulted from evolutionary selection in M. antarcticus during the process of adaptation to the permanently cold environment in Antarctica. There was no need to retain high activity at higher temperatures. Both long-loop L3 and His299 are flexible, and this flexibility increases the overall thermolability of BglU; i.e., the enzyme can be more easily degraded to save energy and food when the environment becomes unsuitable for cell growth. His299 is less efficient than Tyr for degradation of substrates by BGs, but the loss of such capability from utilizing His299 is compensated by long-loop L3. The stability and high activity of long-loop L3 and Tyr at high temperatures would lead to wastage of food and energy in the case of microbes for which cell growth is limited at high temperature. Increased understanding of these molecular mechanisms will help us construct “super GH1 enzymes” with optimal combinations of amino acid residues and structures to achieve higher enzyme activity and proper thermostable in laboratory and industrial applications.
In summary, the present findings demonstrate that long-loop L3 and residue His299 are jointly responsible for the cold-adapted (psychrophilic) properties of BglU and are essential for proton transfer during the enzyme reaction. Each of the structures and residue replacements involved in the psychrophilic properties of BglU has its own specific effect on temperature-dependent enzyme activity or stability. Many previous studies have demonstrated an inherent trade-off between the rigidity necessary for enzyme stability and the flexibility required for enzyme activity. Our results suggest that thermostability can be improved at no cost to low-temperature activity and vice versa. High stability is not necessarily incompatible with high catalytic activity at low temperatures. There is no strict inverse relationship between stability and low-temperature activity (39). The fact that enzymes displaying both properties are not found in nature most likely reflects the effects of evolutionary processes rather than any intrinsic physical-chemical limitations on protein structure and function.
The present findings will facilitate the design of novel enzymes useful in a variety of industrial processes (e.g., food production and sewage treatment) at different temperatures. The structural basis for the cold adaptedness of BglU described here will be helpful for structure-based engineering of new cold-adapted enzymes and for the production of mutants useful in such industrial processes.
ACKNOWLEDGMENTS
We are grateful to S. Anderson for English editing of the manuscript.
This study was supported by grants from the National Nature Science Foundation of China (30970102 and 31270792) and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCS2-YW-G-055-01).
REFERENCES
- 1.Chuenchor W, Pengthaisong S, Robinson RC, Yuvaniyama J, Oonanant W, Bevan DR, Esen A, Chen CJ, Opassiri R, Svasti J, Cairns JR. 2008. Structural insights into rice BGlu1 β-glucosidase oligosaccharide hydrolysis and transglycosylation. J Mol Biol 377:1200–1215. doi: 10.1016/j.jmb.2008.01.076. [DOI] [PubMed] [Google Scholar]
- 2.Hong MR, Kim YS, Park CS, Lee JK, Kim YS, Oh DK. 2009. Characterization of a recombinant β-glucosidase from the thermophilic bacterium Caldicellulosiruptor saccharolyticus. J Biosci Bioeng 108:36–40. doi: 10.1016/j.jbiosc.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Faure D. 2002. The family-3 glycoside hydrolases: from housekeeping functions to host-microbe interactions. Appl Environ Microbiol 68:1485–1490. doi: 10.1128/AEM.68.4.1485-1490.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flannelly DF, Aoki TG, Aristilde L. 2015. Short-time dynamics of pH-dependent conformation and substrate binding in the active site of beta-glucosidases: a computational study. J Struct Biol 191:352–364. doi: 10.1016/j.jsb.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia Y, Mishra S, Bisaria VS. 2002. Microbial β-glucosidases: cloning, properties, and applications. Crit Rev Biotechnol 22:375–407. doi: 10.1080/07388550290789568. [DOI] [PubMed] [Google Scholar]
- 6.Zhou L, Li S, Zhang T, Mu W, Jiang B. 2015. Properties of a novel polydatin-β-d-glucosidase from Aspergillus niger SK34.002 and its application in enzymatic preparation of resveratrol. J Sci Food Agric doi: 10.1002/jsfa.7465. [DOI] [PubMed] [Google Scholar]
- 7.Hong J, Tamaki H, Kumagai H. 2007. Cloning and functional expression of thermostable β-glucosidase gene from Thermoascus aurantiacus. Appl Microbiol Biotechnol 73:1331–1339. doi: 10.1007/s00253-006-0618-9. [DOI] [PubMed] [Google Scholar]
- 8.Wallecha A, Mishra S. 2003. Purification and characterization of two β-glucosidases from a thermo-tolerant yeast Pichia etchellsii. Biochim Biophys Acta 1649:74–84. doi: 10.1016/S1570-9639(03)00163-8. [DOI] [PubMed] [Google Scholar]
- 9.Faved MR, Buthe A, Rashid MH, Wang P. 2016. Cost-efficient entrapment of β-glucosidase in nanoscale latex and silicone polymeric thin films for use as stable biocatalysts. Food Chem 190:1078–1085. doi: 10.1016/j.foodchem.2015.06.040. [DOI] [PubMed] [Google Scholar]
- 10.Tsai CT, Meyer AS. 2014. Enzymatic cellulose hydrolysis: enzyme reusability and visualization of β-glucosidase immobilized in calcium alginate. Molecules 19:19390–19406. doi: 10.3390/molecules191219390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeom SJ, Kim BN, Kim YS, Oh DK. 2012. Hydrolysis of isoflavone glycosides by a thermostable β-glucosidase from Pyrococcus furiosus. J Agric Food Chem 60:1535–1541. doi: 10.1021/jf204432g. [DOI] [PubMed] [Google Scholar]
- 12.Choo DW, Kurihara T, Suzuki T, Soda K, Esaki N. 1998. A cold-adapted lipase of an Alaskan psychrotroph, Pseudomonas sp. strain B11-1: gene cloning and enzyme purification and characterization. Appl Environ Microbiol 64:486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerday C, Aittaleb M, Bentahir M, Chessa JP, Claverie P, Collins T, D'Amico S, Dumont J, Garsoux G, Georlette D, Hoyoux A, Lonhienne T, Meuwis MA, Feller G. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol 18:103–107. doi: 10.1016/S0167-7799(99)01413-4. [DOI] [PubMed] [Google Scholar]
- 14.D'Amico S, Claverie P, Collins T, Georlette D, Gratia E, Hoyoux A, Meuwis MA, Feller G, Gerday C. 2002. Molecular basis of cold adaptation. Philos Trans R Soc London 357:917–925. doi: 10.1098/rstb.2002.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feller G, Gerday C. 1997. Psychrophilic enzymes: molecular basis of cold adaptation. Cell Mol Life Sci 53:830–841. doi: 10.1007/s000180050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bialkowska AM, Cieslinski H, Nowakowska KM, Kur J, Turkiewicz M. 2009. A new β-galactosidase with a low temperature optimum isolated from the Antarctic Arthrobacter sp. 20B: gene cloning, purification, and characterization. Arch Microbiol 191:825–835. doi: 10.1007/s00203-009-0509-4. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Xu Y, Ma Y, Zhou P. 2000. Characterization of Micrococcus antarcticus sp. nov., a psychrophilic bacterium from Antarctica. Int J Syst Evol Microbiol 50:715–719. doi: 10.1099/00207713-50-2-715. [DOI] [PubMed] [Google Scholar]
- 18.Fan HX, Miao LL, Liu Y, Liu HC, Liu ZP. 2011. Gene cloning and characterization of a cold-adapted β-glucosidase belonging to glycosyl hydrolase family 1 from a psychrotolerant bacterium Micrococcus antarcticus. Enzyme Microb Technol 49:94–99. doi: 10.1016/j.enzmictec.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Wintrode PL, Miyazaki K, Arnold FH. 2000. Cold adaptation of a mesophilic subtilisin-like protease by laboratory evolution. J Biol Chem 275:31635–31640. doi: 10.1074/jbc.M004503200. [DOI] [PubMed] [Google Scholar]
- 20.Nakazawa H, Okada K, Onodera T, Ogasawara W, Okada H, Morikawa Y. 2009. Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl Microbiol Biotechnol 83:649–657. doi: 10.1007/s00253-009-1901-3. [DOI] [PubMed] [Google Scholar]
- 21.Romero PA, Arnold FH. 2009. Exploring protein fitness landscapes by directed evolution. Nat Rev 10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almog O, Gonzalez A, Godin N, de Leeuw M, Mekel MJ, Klein D, Braun S, Shoham G, Walter RL. 2009. The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state. Proteins 74:489–496. doi: 10.1002/prot.22175. [DOI] [PubMed] [Google Scholar]
- 23.Isorna P, Polaina J, Lorena LG, Canada FJ, Gonzalez B, Julia SA. 2007. Crystal structures of Paenibacillus polymyxa β-glucosidase B complexes reveal the molecular basis of substrate specificity and give new insights into the catalytic machinery of family I glycosidases. J Mol Biol 371:1204–1218. doi: 10.1016/j.jmb.2007.05.082. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, He X, Yang S, An X, Chang W, Liang D. 2003. Structural basis for thermostability of β-glycosidase from the thermophilic eubacterium Thermus nonproteolyticus HG102. J Bacteriol 185:4248–4255. doi: 10.1128/JB.185.14.4248-4255.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker A, Taylor J, Rowe D, Summers D. 2008. A method for generating sticky-end PCR products which facilitates unidirectional cloning and the one-step assembly of complex DNA constructs. Plasmid 59:155–162. doi: 10.1016/j.plasmid.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Yu JH, Hamari Z, Han KH, Seo JA, Yazmid RD, Scazzocchio C. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Badhan AK, Chadha BS, Kaur J, Saini HS, Bhat MK. 2007. Production of multiple xylanolytic and cellulolytic enzymes by thermophilic fungus Myceliophthora sp. IMI 387099. Bioresour Technol 98:504–510. doi: 10.1016/j.biortech.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 28.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. [DOI] [PubMed] [Google Scholar]
- 30.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:8. [Google Scholar]
- 34.Delano WL. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA. [Google Scholar]
- 35.Jeng WY, Wang NC, Lin MH, Lin CT, Liaw YC, Chang WJ, Liu CL, Liang PH, Wang AH. 2011. Structural and functional analysis of three β-glucosidases from bacterium Clostridium cellulovorans, fungus Trichoderma reesei and termite Neotermes koshunensis. J Struct Biol 173:46–56. doi: 10.1016/j.jsb.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Adlakha N, Sawant S, Anil A, Lali A, Yazdani SS. 2012. Specific fusion of β-1,4-endoglucanase and β-1,4-glucosidase enhances cellulolytic activity and helps in channeling of intermediates. Appl Environ Microbiol 78:7447–7454. doi: 10.1128/AEM.01386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menandro CC, Georgina GR, Marinana P, Gerardo PH, Rafael AZ. 2011. Thermal denaturation of β-glucosidase B from Paenibacillus polymyxa proceeds through a lumry-eyring mechanism. Protein J 30:318–323. doi: 10.1007/s10930-011-9334-0. [DOI] [PubMed] [Google Scholar]
- 38.Feller C, Gerday G. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat Rev Microbiol 1:200–208. doi: 10.1038/nrmicro773. [DOI] [PubMed] [Google Scholar]
- 39.Wolterink-van Loo S, Siemerink MA, Perrakis G, Kaper T, Kengen SW, van der Oost J. 2009. Improving low-temperature activity of Sulfolobus acidocaldarius 2-keto-3-deoxygluconate aldolase. Archaea 2:233–239. doi: 10.1155/2009/194186. [DOI] [PMC free article] [PubMed] [Google Scholar]