Abstract
Hydroxypipecolic acids are bioactive compounds widely distributed in nature and are valuable building blocks for the organic synthesis of pharmaceuticals. We have found a novel hydroxylating enzyme with activity toward l-pipecolic acid (l-Pip) in a filamentous fungus, Fusarium oxysporum c8D. The enzyme l-Pip trans-4-hydroxylase (Pip4H) of F. oxysporum (FoPip4H) belongs to the Fe(II)/α-ketoglutarate-dependent dioxygenase superfamily, catalyzes the regio- and stereoselective hydroxylation of l-Pip, and produces optically pure trans-4-hydroxy-l-pipecolic acid (trans-4-l-HyPip). Amino acid sequence analysis revealed several fungal enzymes homologous with FoPip4H, and five of these also had l-Pip trans-4-hydroxylation activity. In particular, the homologous Pip4H enzyme derived from Aspergillus nidulans FGSC A4 (AnPip4H) had a broader substrate specificity spectrum than other homologues and reacted with the l and d forms of various cyclic and aliphatic amino acids. Using FoPip4H as a biocatalyst, a system for the preparative-scale production of chiral trans-4-l-HyPip was successfully developed. Thus, we report a fungal family of l-Pip hydroxylases and the enzymatic preparation of trans-4-l-HyPip, a bioactive compound and a constituent of secondary metabolites with useful physiological activities.
INTRODUCTION
Hydroxypipecolic acids (HyPips) are naturally occurring six-membered heterocyclic hydroxy amino acids that are widely distributed in nature. For instance, 4-hydroxy-, 5-hydroxy-, and 4,5-dihydroxypipecolic acids have been found in various members of the plant family (1–4). Likewise, 5-hydroxypipecolic acid was found to be present in mammalian brain and blood (5). HyPips are also components of some peptide antibiotics, terpenoids, and alkaloids, for example, trans-3-hydroxy-l-pipecolic acid (trans-3-l-HyPip) in GE81112 (6), cis-3-hydroxy-l-pipecolic acid (cis-3-l-HyPip) in tetrazomine (7), trans-4-hydroxy-l-pipecolic acid (trans-4-l-HyPip) in ulleungamides (8) and damipipecolin (9), and cis-4-hydroxy-l-pipecolic acid (cis-4-l-HyPip) in halichonadins (10).
HyPips have also been used as valuable building blocks for organic synthesis in various industries, such as the pharmaceutical industry. For example, cis-4-l-HyPip is a component of palinavir, an HIV protease inhibitor (11, 12), and cis-5-hydroxy-l-pipecolic acid (cis-5-l-HyPip) is a precursor for the synthesis of the β-lactamase inhibitor MK-7655 (13). Thus, the development of processes for the industrial synthesis of HyPips may be of great significance.
The asymmetric biosynthesis of several l-HyPips was demonstrated by regio- and stereoselective hydroxylation of l-pipecolic acid (l-Pip) with enzymes belonging to the Fe(II)/α-ketoglutarate (αKG)-dependent dioxygenase (Fe/αKG-DO) superfamily. Three Fe/αKG-DOs, l-proline cis-3-hydroxylase (cis-Pro3H) of Streptomyces sp. strain TH1 (14), l-proline trans-4-hydroxylase (trans-Pro4H) of Dactylosporangium sp. strain RH1 (15), and l-proline cis-4-hydroxylase (cis-Pro4H) of Sinorhizobium meliloti (16), were found to possess hydroxylating activities toward l-Pip as well as l-proline and were utilized for the bioconversion of l-Pip into l-HyPip isomers. cis-Pro3H converted l-Pip into cis-3-l-HyPip with a yield of 17%, cis-Pro4H produced 33% cis-3-l-HyPip and 33% cis-5-l-HyPip from l-Pip in parallel, and trans-Pro4H converted l-Pip into trans-5-l-hydroxypipecolic acid (trans-5-l-HyPip) with a yield of 68% (17).
Thus, the enzymatic hydroxylation of l-Pip was achieved as a side reaction of l-proline hydroxylase. To date, specific l-Pip-hydroxylating enzymes have not been reported, despite the fact that HyPips are important metabolites found in plants and mammals. In the present study, we identified and characterized l-Pip hydroxylases broadly conserved in some filamentous fungi. We found that the newly discovered l-Pip hydroxylases were particularly suitable biocatalysts for the asymmetric hydroxylation of cyclic amino acids, such as pipecolic acids and prolines. Moreover, the widespread distribution of genes encoding l-Pip hydroxylases indicates some important physiological roles of fungal secondary metabolites related to trans-4-l-HyPip.
MATERIALS AND METHODS
Strains and culture.
Microorganisms isolated from soil and plant samples were cultured in GP medium comprised of 0.15% (wt/vol) KH2PO4, 0.05% (wt/vol) K2HPO4, 0.1% (wt/vol) NH4Cl, 0.03% (wt/vol) MgSO4·7H2O, 0.01% (wt/vol) yeast extract, 0.025% (wt/vol) glucose, and 0.1% (wt/vol) l-Pip (pH 7.0) and grown at 28°C with shaking. Escherichia coli JM109 and Rosetta2(DE3) (Novagen, WI, USA) were used as host strains for the overexpression of the recombinant enzymes and cultured at 28°C in LB medium, comprised of 1% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, and 1% (wt/vol) NaCl, with the addition of appropriate antibiotics.
Conditions for amino acid analysis by HPLC.
Amino acids were derivatized using an AccQ-Tag derivatization kit (Waters, MA, USA) according to the manufacturer's instructions and analyzed with an Alliance 2695 high-performance liquid chromatography (HPLC) system (Waters). An XBridge C18 column (particle size, 5 μm; 2.1 by 150 mm; Waters) was used for separation at 40°C. The mobile phases were 10 mM ammonium acetate at pH 5.0 (eluent A) and methanol (eluent B), and the flow rate of the eluent was 0.3 ml/min. The eluent gradients were 0 to 1% (vol/vol) eluent B for 0 to 0.5 min, 1 to 5% eluent B for 0.5 to 18 min, 5 to 9% eluent B for 18 to 19 min, 9 to 17% eluent B for 19 to 29.5 min, 17 to 60% eluent B for 29.5 to 40 min, and 60% eluent B for 40 to 43 min. The AccQ-Tag derivatives were detected with a fluorescence detector (excitation, 250 nm; emission, 395 nm). Electrospray ionization (ESI) mass spectrometry (MS) was carried out using an LCMS-2010A liquid chromatography (LC)-MS system (Shimadzu, Kyoto, Japan). MS conditions were as follows: block temperature, 200°C; curved desolvation line (CDL) temperature, 250°C; detector voltage, 1.5 kV; and nebulizing gas flow, 1.5 liters/min.
Conditions for succinate analysis by ion chromatography (IC).
Succinate was quantitatively analyzed by use of a Prominence HPLC system (Shimadzu) equipped with a Shim-pack IC-SA2 column (150 by 4.6 mm; Shimadzu) at 50°C. The sample was isocratically eluted in the mobile phase, consisting of 1.8 mM Na2CO3 and 1.7 mM NaHCO3, at a flow rate of 1.0 ml/min. The succinate concentration was determined with a suppressed conductivity detector.
Assay of l-Pip hydroxylase activity.
The l-Pip hydroxylase activity in the samples, such as wet cell pellets of the microorganisms, their cell lysates, and protein solutions, was assayed. The standard reaction conditions were as follows: the reaction mixture consisted of 10 mM l-Pip, 15 mM αKG, 0.5 mM FeSO4·7H2O, 5 mM ascorbate, and 1 mM dithiothreitol (DTT) in 50 mM MES (morpholineethanesulfonic acid) buffer (pH 6.5), and the reaction was carried out at 20°C for 10 min with shaking at 300 rpm. After the reaction, hydroxylation activity was determined by measurement of the amount of succinate produced by succinate analysis and/or the amino acid content of the samples by amino acid analysis. In order to determine the catalytic properties of l-Pip hydroxylases, 1 mg/ml of purified enzyme was used for the reaction. The effects of pH on hydroxylation activity were examined by varying the reaction pH between pH 3.0 and 10.0. pH stability was examined by measuring the residual activity after incubation in each buffer for 1 h. The effects of temperature on hydroxylation activity were examined by varying the reaction temperature between 10°C and 70°C. The temperature stability was examined by measuring the residual activity after incubation at each temperature for 1 h.
For substrate specificity analysis, various kinds of substrates were used instead of l-Pip. To determine the kinetic parameters, the reaction was carried out at the optimum pH and temperature for each enzyme and with substrate concentrations that varied from 0.25 to 10 mM. Apparent Km and Vmax values were determined by use of Lineweaver-Burk plots of the data. Enzyme activity is expressed in micromoles per minute per milligram (units per milligram).
Microbial screening for hydroxylation activity of l-Pip.
About 100 kinds of soil and plant samples were used as the source of microorganisms. A small amount of untreated sample was added into a test tube containing 5 ml isolation medium, comprised of 0.15% (wt/vol) KH2PO4, 0.05% (wt/vol) K2HPO4, 0.1% (wt/vol) NH4Cl, 0.03% (wt/vol) MgSO4·7H2O, 0.01% (wt/vol) yeast extract, and 0.1% (wt/vol) l-Pip (pH 7.0), and incubated at 28°C with shaking for a few days. Fifty microliters of the culture broth was transferred into 5 ml of fresh isolation medium and further cultivated for a few days. Then, the culture was streaked onto an agar plate with isolation medium and cultivated at 28°C for a few days. After cultivation, more than 2,500 colonies appeared, and these colonies were isolated as l-Pip-assimilating microorganisms. These microorganisms were cultured in GP medium and grown at 28°C with shaking for a few days. Microbial cells were harvested by centrifugation at 3,000 rpm for 10 min. The wet cell pellet was applied to the l-Pip hydroxylase activity assay under the standard reaction conditions, except that a reaction time of 16 h was used.
Microbial conversion of l-Pip.
Fusarium oxysporum c8D was cultured in GP medium at 28°C with shaking for a few days. The wet cell pellet was reacted with l-Pip under the standard reaction conditions, except that a reaction time of 16 h was used. In order to identify the reaction products, the solution remaining after the reaction was centrifuged, and the supernatant was adjusted to pH 11 using 1 M KOH and applied to negative ion-exchange resin (Dowex 2 × 8; Dow Chemical, Midland, USA) that had previously been equilibrated with distilled water. The reaction product of l-Pip was eluted using 3% (wt/vol) KCl and freeze-dried. The dried powder was dissolved in distilled water and separated using a Prominence HPLC system (Shimadzu) equipped with a TSKgel Amide-80 column (7.8 mm by 300 mm; Tosoh, Tokyo, Japan) at 40°C. The mobile phase was 1.5 mM ammonium acetate (pH 5.0) in 85% acetonitrile, and the flow rate was 0.6 ml/min. Amino acids were detected by determination of the UV absorbance at 210 nm. The eluate containing the isolated amino acids was freeze-dried and dissolved in D2O. Proton and carbon nuclear magnetic resonances (1H and 13C NMRs, respectively) were recorded on an Avance 500 spectrometer (Bruker, Billerica, MA, USA).
Measurement of l-Pip hydroxylation activity using F. oxysporum c8D cell lysate.
F. oxysporum c8D was cultivated in a medium comprised of 0.15% (wt/vol) KH2PO4, 0.05% (wt/vol) K2HPO4, 0.1% (wt/vol) NH4Cl, 0.03% (wt/vol) MgSO4·7H2O, 0.01% (wt/vol) yeast extract, and 0.025% (wt/vol) glucose (pH 7.0) and grown at 28°C with shaking. In order to induce l-Pip hydroxylation activity, 0.1% (wt/vol) l-Pip was added to the medium. At several time points during cultivation, F. oxysporum c8D cells were collected by centrifugation and disrupted with glass beads at 4°C using a multibead shocker (Yasui Kikai, Osaka, Japan). The cell lysate was centrifuged at 12,000 × g for 30 min, and the resulting supernatant was applied to the l-Pip hydroxylase activity assay. To test the effects of different cofactors on activity, FeSO4·7H2O, αKG, or ascorbate was eliminated from the reaction mixture.
Purification of l-Pip trans-4-hydroxylase (Pip4H) from F. oxysporum c8D cells (FoPip4H).
F. oxysporum c8D was cultivated for 40 h in 500 ml of GP medium. The cell suspension was centrifuged; the cell pellet (2.5 g [wet weight] of cells) was resuspended in 25 ml of MD buffer, consisting of 40 mM MES buffer (pH 6.5), 1 mM DTT, and Complete protease inhibitor cocktail (Roche Diagnostic, Basel, Switzerland) at the manufacturer's recommended concentration; and the cell pellet was disrupted with glass beads at 4°C using a multibead shocker (Yasui Kikai). The cell lysate was centrifuged at 12,000 × g for 30 min, and the resulting supernatant was dialyzed against MD buffer. The protein solution was applied into a Toyopearl CM-650M column (3.5 by 150 mm; Tosoh, Tokyo, Japan) that had been equilibrated with MD buffer and eluted with MD buffer containing 1 M NaCl. The fractions showing l-Pip-hydroxylating activities were collected and dialyzed against MD buffer. The proteins were sequentially separated with a Toyopearl DEAE-650M column (3.5 by 150 mm; Tosoh) and a Mono Q column (5 by 50 mm; GE Healthcare Biosciences, Uppsala, Sweden) as described above. The active fractions were collected and applied onto a Phenyl Superose column (GE Healthcare Biosciences) that had been equilibrated with MD buffer containing 1.8 M (NH4)2SO4 and eluted using a linear gradient of 1.8 to 0 M (NH4)2SO4 in MD buffer. The active fractions were then concentrated using an Ultracal-10K membrane (Millipore, MA, USA) and separated using a Superdex 200 column (GE Healthcare) in MD buffer containing 150 mM NaCl.
Amino acid sequencing of FoPip4H.
The purified FoPip4H enzyme was freeze-dried and dissolved in 25 μl of 50 mM Tris buffer (pH 9.0) containing 8 M urea, and the solution was incubated at 37°C for 1 h. Then, 25 μl of 50 mM Tris buffer (pH 9.0) and 6 nmol of lysyl endopeptidase were added to the solution and the mixture was incubated at 30°C for 6 h. The resulting peptides were separated with a Prominence HPLC system (Shimadzu) equipped with a Jupiter C18 column (250 by 4.6 mm; Phenomenex, CA, USA) at 35°C, using 10 mM trifluoroacetic acid in H2O (eluent A) and 10 mM trifluoroacetic acid in acetonitrile (eluent B) as the mobile phases. A linear gradient (10 to 45% eluent B over 20 min) was carried out at a flow rate of 1.0 ml/min. The eluate was monitored by determination of the UV absorbance at 210 nm, and the fractions containing the peptides were collected. The N-terminal amino acid sequences of the peptides were determined by automated Edman degradation with a PSQQ-30 protein sequencer (Shimadzu).
Construction of strains for expression of l-Pip hydroxylases.
The FoPip4H gene was amplified by PCR using the following primers: AAGGATCCATGGCCGCCCTCAACGCAGA and CCAAGCTTTTAAGCCTTAGCTTCAGCAG. PCR was carried out with Tks Gflex DNA polymerase (TaKaRa Bio, Shiga, Japan) under the following conditions: 30 cycles of 10 s at 98°C, 15 s at 56°C, and 40 s at 68°C. For the expression of N-terminally 6×His-tagged proteins, the PCR product was digested with BamHI and HindIII endonucleases and cloned into the expression vector pQE80L (Qiagen, CA, USA), which had previously been digested with the same endonucleases. The resultant plasmid, pQE-Fopip4H, was transformed into E. coli JM109.
Codon-optimized genes encoding the proteins of the following filamentous fungi homologous to FoPip4H were synthesized and ligated into the pJexpress411 vector by DNA2.0 Bioengineering Solutions (CA, USA): Colletotrichum gloeosporioides Nara gc5 (CgPip4H; GenBank accession no. XP_007276452), Aspergillus oryzae RIB40 (AoPip4H; GenBank accession no. XP_001827566), Talaromyces marneffei ATCC 18224 (PMAA_075210; GenBank accession no. XP_002146999), Aspergillus flavus NRRL3357 (AFLA_066710; GenBank accession no. XP_002380230), Penicillium rubens Wisconsin 54-1255 (PrPip4H; GenBank accession no. XP_002558179), Fusarium graminearum PH-1 (FgPip4H; GenBank accession no. XP_011322545), and Aspergillus nidulans FGSC A4 (AnPip4H; GenBank accession no. XP_659994). The resultant plasmids were designated pJ-Cgpip4H, pJ-Aopip4H, pJ-PMAA_075210, pJ-AFLA_066710, pJ-Prpip4H, pJ-Fgpip4H, and pJ-Anpip4H, respectively, and transformed into E. coli Rosetta2 (DE3).
Expression and purification of recombinant Pip4H enzymes.
Each of the E. coli transformants expressing the Pip4H enzymes was cultured at 28°C in LB medium, comprised of 1% (wt/vol) tryptone, 0.5% (wt/vol) yeast extract, and 1% (wt/vol) NaCl, with the addition of appropriate antibiotics. At an optical density at 600 nm (OD600) of 1.0, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM, and the cultures were incubated for 16 h at 28°C with shaking at 300 rpm. The cell suspension (250 ml) was centrifuged at 3,000 rpm for 10 min, and the cell pellet was suspended in 10 ml of binding buffer containing 20 mM Tris-HCl buffer (pH 7.4), 0.5 M NaCl, 20 mM imidazole, and 1 mM DTT and disrupted for 1 h by sonication with an Insonator 201R sonicator (Kubota, Osaka, Japan). The lysate was centrifuged at 12,000 × g for 30 min, and the supernatant was filtered through a 0.45-μm-pore-size Millex syringe-driven filter unit (Millipore). The protein solution was applied to an Ni-Sepharose column (His Trap HP 5 ml; GE Healthcare Bioscience) that had previously been equilibrated with binding buffer. The column was washed with binding buffer, and the proteins were eluted with a linear gradient of 0.02 to 0.50 M imidazole in binding buffer. The fractions containing recombinant Pip4H enzyme were pooled, concentrated by ultrafiltration, and used as purified enzyme.
Preparative-scale production of trans-4-l-HyPip by bioconversion with FoPip4H-expressing E. coli cells.
For the production of trans-4-l-HyPip from l-Pip, the reaction was performed in an 80-ml mixture composed of 100 mM l-Pip, 150 mM αKG, 1 mM FeSO4·7H2O, 10 mM tris(2-carboxyethyl)phosphine (TCEP), 10 mM trisodium citrate, 6 g (wet weight) of cells of E. coli JM109/pQE-Fopip4H, and 50 mM bis-Tris buffer (pH 6.5) at 15°C for 6 h with vigorous shaking. The reaction mixture was centrifuged, and the supernatant was adjusted to pH 2.0 with 1 M HCl, applied to positive ion-exchange resin (Dowex 50W × 8; Dow Chemical) which had been equilibrated with distilled water, and eluted with 1 M NH3 solution. The fractions containing the oxidized amino acid products were passed through negative ion-exchange resin (Dowex 2 × 8; Dow Chemical) which had been equilibrated with distilled water and eluted with 1 M HCl. The fractions containing the products were combined and freeze-dried.
Nucleotide sequence accession number.
The FoPip4H gene has been submitted to GenBank and may be found under accession no. LC076551.
RESULTS
l-Pip hydroxylation activities of filamentous fungi isolated from soil.
We isolated more than 2,500 microorganisms assimilating l-Pip from soil and plant samples. These microorganisms, including bacteria, actinomycetes, and filamentous fungi, were screened for hydroxylation activity toward l-Pip. In reaction mixtures containing wet cells, two filamentous fungal isolates in particular, c7D and c8D, produced the same l-Pip-derived reaction product, as observed by amino acid analysis. The molecular mass of this product was greater than that of l-Pip by 16 Da, which indicated that the hydroxylation reaction was catalyzed in the reaction mixtures containing these fungal isolates. According to the morphological characteristics and taxonomic identification obtained using the internal transcribed sequence-5.8S ribosomal DNA sequence by the identification services of TechnoSuruga Laboratory Co., Ltd. (Shizuoka, Japan), strain c7D was identified to be Fusarium merismoides and strain c8D was identified to be Fusarium oxysporum. Because of its higher activity on amino acid analysis, F. oxysporum c8D was chosen for further investigation.
Structural analysis of the product of l-Pip hydroxylation by F. oxysporum c8D.
The hydroxylated product of l-Pip in the reaction mixture with F. oxysporum c8D cells was purified and subjected to structural analysis (see Fig. S1 in the supplemental material). The chemical shifts in NMR analysis were as follows: 1H NMR (500 MHz, D2O) δ 1.82 to 1.95 (3H, m), 2.19 (1H, d, J = 14.6 Hz), 3.27 (2H, m), 3.88 (1H, dd, J = 11.5, 3.1 Hz), 4.20 (1H, s). 13C NMR (500 MHz, D2O) δ 28.18, 32.96, 38.72, 54.14, 62.03, 174.52. On the basis of the information from these NMR spectra and published data from a previous study (18), the reaction product of l-Pip was determined to be trans-4-hydroxy-l-pipecolic acid (trans-4-l-HyPip).
l-Pip-hydroxylating activity of F. oxysporum c8D.
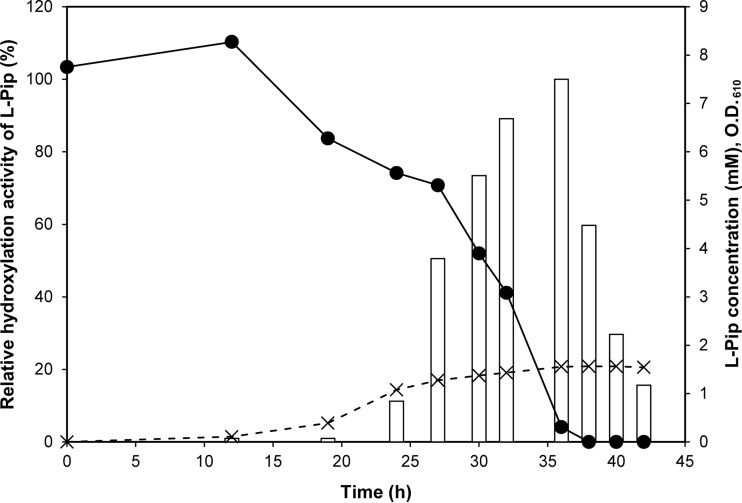
The hydroxylating activity for the production of trans-4-l-HyPip from l-Pip was measured using the lysate of F. oxysporum c8D cells collected at various points during cell cultivation. F. oxysporum c8D cells cultured without l-Pip showed no activity throughout the growth period. When l-Pip was added to the culture medium, strong hydroxylating activity was observed during late log phase and stationary phase; the activity then gradually diminished after l-Pip depletion (Fig. 1). These results indicate that the l-Pip-hydroxylating activity of F. oxysporum c8D is inducible and strictly regulated by the presence of l-Pip during cultivation. In contrast to the effect of l-Pip, no inductive effect of l-Pro on the activity of F. oxysporum c8D was observed. Moreover, the addition of ferrous iron, αKG, and ascorbate to the reaction mixture was required for the maximum activity of the F. oxysporum c8D cell lysate. Since similar observations have been made for other amino acid hydroxylases belonging to the Fe/αKG-DO superfamily (19–21), these results suggest that the enzyme responsible for l-Pip-hydroxylating activity belongs to the Fe/αKG-DO superfamily.
FIG 1.
l-Pip hydroxylation activity in F. oxysporum c8D cells cultivated with l-Pip. Lysates of F. oxysporum c8D cells, collected at several time points during cultivation in medium containing l-Pip, were used for the l-Pip hydroxylation reaction. The relative activity for l-Pip hydroxylation in the cell lysates (white bars), the concentration of l-Pip in the medium (closed circles), and cell densities at an OD610 (multiplication signs) are shown.
Identification of an l-Pip trans-4-hydroxylase from F. oxysporum c8D.
An l-Pip trans-4-hydroxylase, FoPip4H, was purified from F. oxysporum c8D cells by five steps of column chromatography (Table 1) and had an approximate molecular mass of 40 kDa, as determined by SDS-PAGE analysis (data not shown). Although the N terminus of FoPip4H was blocked from Edman degradation, its internal amino acid sequences were successfully determined to be MLAK, TRDIK, and GYEKGLGPLKTRDIKETVD. A BLAST search of these sequences revealed that they correspond to the amino acid sequence for a protein of unknown function, FOXB_08233 (GenBank accession no. EGU81245.1), of F. oxysporum Fo5176. The genome region encoding FoPip4H was sequenced in its entirety and found to include one exon without introns. The full-length amino acid sequences of FoPip4H and FOXB_08233 were identical. The amino acid sequence of FoPip4H also contained the motifs required for ferrous iron binding (His1-XAA-Asp/Glu-XAAn-His2) and αKG binding (Arg) that are conserved in the Fe/αKG-DO superfamily (22), providing further evidence that FoPip4H is a member of the Fe/αKG-DO superfamily. FoPip4H was heterologously expressed in E. coli and purified for use in the enzyme assay, where it was confirmed to have hydroxylating activity toward l-Pip and produce trans-4-l-HyPip (Fig. 2A).
TABLE 1.
Purification of l-Pip-hydroxylating enzyme from F. oxysporum c8D
| Step | Total activity (U) | Total protein (mg) | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|
| Cell lysate | 1.26 | 45 | 0.028 | 100 |
| Toyopearl CM-650M | 0.50 | 9.1 | 0.055 | 40 |
| Toyopearl DEAE-650M | 0.33 | 5.1 | 0.065 | 26 |
| Mono Q | 0.30 | 1.9 | 0.16 | 24 |
| Phenyl Superose | 0.067 | 0.32 | 0.21 | 5.3 |
| Superdex 200 | 0.0022 | 0.0030 | 0.74 | 0.18 |
FIG 2.
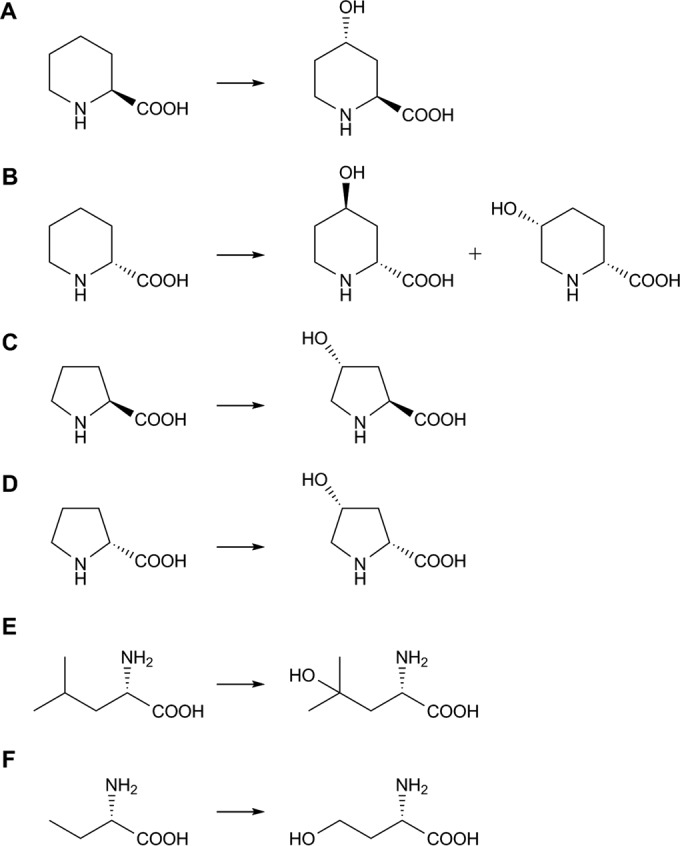
Hydroxylating reactions catalyzed by Pip4Hs. (A) Hydroxylation of l-Pip into trans-4-l-HyPip by FoPip4H and AnPip4H; (B) hydroxylation of d-Pip into trans-4-d-HyPip and cis-5-d-HyPip by AnPip4H; (C) hydroxylation of l-Pro into trans-4-l-HyPro by FoPip4H and AnPip4H; (D) hydroxylation of d-Pro into cis-4-d-HyPro by AnPip4H; (E) hydroxylation of l-Leu into 4-hydroxy-l-Leu by FoPip4H; (F) hydroxylation of l-AABA into l-homoserine by FoPip4H.
Phylogenetic analysis of proteins homologous to FoPip4H.
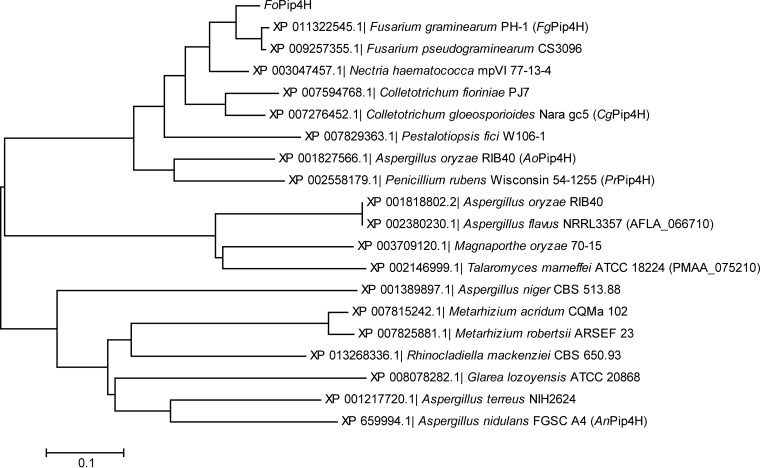
Proteins homologous to FoPip4H were obtained by use of a search of the amino acid sequence of FoPip4H against the sequences in protein reference sequence databases with the BLASTP program. All proteins sharing over 40% identity with FoPip4H were from filamentous fungi and were subjected to phylogenetic analysis using the neighbor-joining method (Fig. 3). The proteins homologous to FoPip4H were widely distributed among several classes of filamentous ascomycetes, such as Sordariomycetes (Fusarium, Nectria, Colletotrichum, Pestalotiopsis, Magnaporthe, Metarhizium), Eurotiomycetes (Aspergillus, Penicillium, Talaromyces, Rhinocladiella), and Leotiomycetes (Glarea genus). Furthermore, all had the conserved amino acid motifs of the Fe/αKG-DO superfamily. This would suggest that these homologous proteins may have hydroxylation activities similar to the hydroxylation activity of FoPip4H and play an important common role in filamentous ascomycetes.
FIG 3.
Phylogenetic tree of amino acid sequences of proteins homologous to FoPip4H. The proteins sharing over 40% identity with FoPip4H from filamentous fungi were subjected to phylogenetic analysis using the neighbor-joining method.
l-Pip hydroxylation activity of proteins homologous to FoPip4H.
Among the 20 proteins shown in Fig. 3, 7 diverse homologues of FoPip4H were selected to be assayed for hydroxylation activity toward l-Pip: FgPip4H, CgPip4H, AoPip4H, PrPip4H, AFLA_066710, AnPip4H, and PMAA_075210. Four of the homologues, FgPip4H, CgPip4H, AoPip4H, and PrPip4H, which had a high degree of similarity to FoPip4H (with over 67% identity), catalyzed the hydroxylation of l-Pip and produced trans-4-l-HyPip like FoPip4H (Table 2). On the other hand, the two proteins with lower degrees of homology, AFLA_066710 and PMAA_075210 (with 44% and 41% identities to FoPip4H, respectively), showed no activity toward l-Pip, even though the successful expression of these proteins was confirmed. Interestingly, AnPip4H, which also had a low level of homology with FoPip4H (42% identity), was found to possess hydroxylation activity and produce trans-4-l-HyPip. Thus, FoPip4H and AnPip4H were used for further characterization of the enzyme activity. The optimal pH and temperature for l-Pip hydroxylation activity were pH 7.0 and 15°C, respectively, for FoPip4H and pH 6.5 and 37°C, respectively, for AnPip4H. FoPip4H maintained over 80% of its maximum activity after 1 h of incubation at pH 5 to 7 and a temperature of 4 to 30°C, and AnPip4H maintained over 80% of its maximum activity after 1 h of incubation at pH 6 to 8 and a temperature of 4 to 40°C.
TABLE 2.
Proteins homologous to FoPip4H and their l-Pip-hydroxylating activities
| Enzyme | Origin | % identity to FoPip4H | l-Pip-hydroxylating activitya |
|---|---|---|---|
| FoPip4H | Fusarium oxysporum c8D | 100 | + |
| FgPip4H | Fusarium graminearum PH-1 | 92 | + |
| CgPip4H | Colletotrichum gloeosporioides Nara gc5 | 81 | + |
| AoPip4H | Aspergillus oryzae RIB40 | 71 | + |
| PrPip4H | Penicillium rubens Wisconsin 54-1255 | 67 | + |
| AFLA_066710 | Aspergillus flavus NRRL3357 | 44 | − |
| AnPip4H | Aspergillus nidulans FGSC A4 | 42 | + |
| PMAA_075210 | Talaromyces marneffei ATCC 18224 | 41 | − |
trans-4-l-HyPip production was detected (+) or not detected (−) by amino acid analysis.
Asymmetric hydroxylation of amino acids with FoPip4H.
Purified recombinant FoPip4H was used to analyze substrate specificity (Table 3). Proteinogenic l-amino acids, nonproteinogenic l-amino acids, d-amino acids, amino acid derivatives, and piperidine derivatives were used as the substrates. Enzymatic activities were confirmed by both amino acid analysis and succinate analysis. In the reaction mixture containing FoPip4H, succinate production and amino acid conversion were detected when l-Pro or cis-5-l-HyPip was used as a substrate, as well as when l-Pip was used as a substrate. Although succinate formation was not detected in reaction mixtures containing l-leucine (l-Leu) or l-2-aminobutyrate (l-AABA) due to the detection limit of the succinate analysis used in this study, a single peak of a newly produced amino acid was observed by amino acid analysis, as was the case with l-Pip, l-Pro, or cis-5-l-HyPip. The molecular masses of all these amino acid products were greater than those of their original substrates by 16 Da, indicating that FoPip4H catalyzed the hydroxylation of these substrates (see Fig. S2 in the supplemental material). In the amino acid analysis, the retention times and molecular masses of the products from l-Pro, l-Leu, and l-AABA corresponded to those of authentic samples of trans-4-hydroxy-l-Pro (trans-4-l-HyPro), 4-hydroxy-l-Leu, and l-homoserine, respectively (see Fig. S2D, F, and G in the supplemental material); however, we were unable to identify the hydroxylated product of cis-5-l-HyPip without some authentic samples, such as dihydroxylated compounds of l-Pip (see Fig. S2C in the supplemental material). We determined that FoPip4H introduced a hydroxyl group onto the carbon at the 4 position of l-amino acids, such as l-Pip, l-Pro, l-Leu, and l-AABA (Fig. 2A, C, E, and F). Specific activities and Km values were determined to be 1.6 U/mg and 1.5 mM, respectively, for l-Pip and 0.55 U/mg and 3.1 mM, respectively, for l-Pro. These results indicate that, compared to l-Pro, l-Pip was the preferred substrate for FoPip4H.
TABLE 3.
Substrate specificity analysis and kinetic parameters of FoPip4H and AnPip4Hb
| Substrate |
FoPip4H |
AnPip4H |
||
|---|---|---|---|---|
| Sp act (U/mg) | Km (mM) | Sp act (U/mg) | Km (mM) | |
| l-Pip | 1.6 ± 0.1 | 1.5 ± 0.1 | 0.76 ± 0.01 | 1.4 ± 0.0 |
| d-Pip | ND | 0.18 ± 0.01a | ||
| l-Pro | 0.55 ± 0.03 | 3.1 ± 0.2 | 0.37 ± 0.02 | 1.4 ± 0.1 |
| d-Pro | ND | 0.044 ± 0.008 | 3.8 ± 0.7 | |
| l-Leu | tr | ND | ||
| l-AABA | tr | ND | ||
| cis-5-l-HyPip | 0.50 ± 0.05a | 0.28 ± 0.05a | ||
Enzyme activities were analyzed by succinate analysis.
All measurements were performed at least three times. ND, not detected; tr, trace activity detected only in the amino acid analysis.
Asymmetric hydroxylation of amino acids with AnPip4H.
Substrate specificity analysis of AnPip4H was performed in a manner similar to that described above (Table 3). AnPip4H reacted with l-Pip, d-pipecolic acid (d-Pip), l-Pro, d-proline (d-Pro), and cis-5-l-HyPip. On the basis of the results of amino acid analysis (see Fig. S2 in the supplemental material), the reaction products were all hydroxylated compounds corresponding to the original substrates, and trans-4-l-HyPip, trans-4-l-HyPro, and cis-4-d-HyPro were identified to be products from l-Pip, l-Pro, and d-Pro, respectively (see Fig. S2A, D, and E in the supplemental material). Interestingly, two kinds of hydroxylated compounds were formed when d-Pip was used as a substrate, and they were identified to be trans-4-hydroxy-d-pipecolic acid and cis-5-hydroxy-d-pipecolic acid by the correspondence of the retention times with those of their enantiomeric counterparts, trans-4-l-HyPip, and cis-5-l-HyPip, respectively (see Fig. S2B in the supplemental material). Likewise, although a single peak of an hydroxylated compound was detected in the reaction mixture containing cis-5-l-HyPip, the structure has not yet been identified (see Fig. S2C in the supplemental material). We determined that AnPip4H introduced a hydroxyl group onto the carbon at the 4 or 5 position of amino acids such as l-Pip, d-Pip, l-Pro, d-Pro, and l-Leu (Fig. 2A to D). Specific activities and Km values were determined to be 0.76 U/mg and 1.4 mM, respectively, for l-Pip; 0.37 U/mg and 1.4 mM, respectively, for l-Pro; and 0.04 U/mg and 3.8 mM, respectively, for d-Pro. These results demonstrated that, compared to all substrates used here, l-Pip was the principal substrate for AnPip4H, similar to the finding for FoPip4H.
Preparative-scale production of trans-4-l-HyPip.
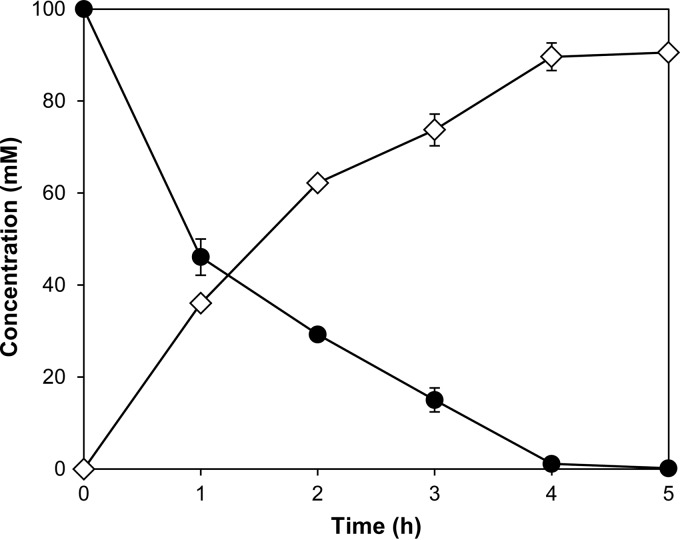
The biotransformation of l-Pip to obtain trans-4-l-HyPip was carried out using E. coli cells expressing FoPip4H as the biocatalyst. As shown in Fig. 4, 100 mM l-Pip was linearly converted into trans-4-l-HyPip and was completely consumed within 5 h. As a result, trans-4-l-HyPip was successfully produced in the reaction mixture with a conversion ratio of 91%. Therefore, a total of 23 g of l-Pip was used as a substrate for the biotransformation, and the resultant trans-4-l-HyPip was purified by ion-exchange chromatography. Consequently, 18 g of trans-4-l-HyPip was obtained, and its final production yield was 81%. According to the 1H NMR spectrum, the compound was confirmed to consist of trans-4-l-HyPip with a >99% diastereomeric excess. Thus, the asymmetric hydroxylation of l-Pip was achieved in the biotransformation by taking advantage of the high regio- and stereoselectivity of FoPip4H.
FIG 4.
Preparative-scale production of trans-4-l-HyPip using FoPip4H. The hydroxylation of l-Pip (closed circles) into trans-4-l-HyPip (open diamonds) with FoPip4H-expressing E. coli cells is shown. All measurements were performed at least three times.
DISCUSSION
Along with l-proline hydroxylases, many Fe/αKG-DOs are known to hydroxylate free amino acids, such as l-asparagine, l-lysine, l-arginine, l-Leu, and l-isoleucine (23–26). Until recently, all Fe/αKG-DOs had been found in bacteria; however, an l-proline hydroxylase was identified to be the first Fe/αKG-DO from a fungus, Glarea lozoyensis (27). Here, we have reported six novel fungal Fe/αKG-DOs, Pip4Hs, which had hydroxylation activities toward l-Pip and were widely distributed among several classes of filamentous ascomycetes. Although both FoPip4H and AnPip4H had catalytic properties similar to those of bacterial proline hydroxylases with respect to kinetic parameters and optimal reaction conditions (14, 16), their amino acid sequences showed no homology to those of known bacterial proline hydroxylases. More importantly, FoPip4H and AnPip4H preferentially reacted with the substrate l-Pip rather than l-Pro, in contrast to bacterial proline hydroxylases, which favor l-Pro over l-Pip (17, 28). Furthermore, we found that FoPip4H is an inducible enzyme strictly regulated by l-Pip but not by l-Pro (Fig. 1). Additionally, we obtained preliminary results by substrate specificity analysis of all Pip4Hs (see Table S1 in the supplemental material). According to those results, CgPip4H showed hydroxylation activity toward both l-Pip and l-Pro, as was the case with FoPip4H and AnPip4H, but FgPip4H, AoPip4H, and PrPip4H did not react with l-Pro. Taken together, these results suggest that Pip4Hs primarily catalyze the hydroxylation of l-Pip.
Under natural circumstances, it is presumed that trans-4-l-HyPip is formed as a secondary metabolite of filamentous ascomycetes possessing Pip4Hs. Here we showed that F. oxysporum c8D produced trans-4-l-HyPip in the culture medium using the substrate of Pip4Hs, l-Pip. Now then, l-Pip is produced by microorganisms, plants, and animals and is therefore readily available for use by fungi (29). Alternatively, l-Pip might be synthesized by these fungi themselves. Indeed, the gene set encoding key enzymes for l-Pip biosynthesis, l-α-aminoadipate aminotransferase, l-aminoadipate-semialdehyde dehydrogenase, and pyrroline-5-carboxylate reductase (30), is found in full in the genome sequences of the ascomycetes listed in Table 2. Thus, it is not unexpected that F. oxysporum c8D and other fungi might produce trans-4-l-HyPip as a metabolite, though its role in fungi is still unknown.
As shown in Fig. 4, the preparative-scale production of optically pure trans-4-l-HyPip was possible using FoPip4H as the biocatalyst. Some reports have indicated that trans-4-l-HyPip has physiological functions. In Acacia maidenii, significant osmotic adjustment was caused by increases in trans-4-l-HyPip levels (31). It was also reported to be a relatively selective agonist of an N-methyl-d-aspartate receptor subtype prevalently found in the mouse cortex (32). Additionally, trans-4-l-HyPip has antidiabetic and antioxidative effects in mice (33) and stimulated glucose uptake and glucose transporter-4 translocation from an intracellular location to the cell surface in skeletal muscle cells (34). Moreover, trans-4-l-HyPip has been found to be present as a component of various bioactive compounds. For example damipipecolin, a bromopyrrole alkaloid isolated from the sponge Axinella damicornis, displayed a modulating effect on the serotonin receptor (9); halichonadin K, a dimeric sesquiterpenoid isolated from a marine sponge (Halichondria sp.), had cytotoxicity against human epidermoid carcinoma KB cells (10); and ulleungamide A, a cyclic depsipeptide isolated from a Streptomyces sp., displayed growth-inhibitory activity against some bacteria (8). Thus, there may be an increasing demand for trans-4-l-HyPip, and our production method is promising for its high efficiency and regio- and stereoselectivity.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by a grant-in-aid for scientific research (no. 24688010 to M. Hibi) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03764-15.
REFERENCES
- 1.Binns WW, Blunden G, Woods DL. 1968. Distribution of leucoanthocyanidinsm phenolic glycosides and amino acids in leaves of Salix spp. Phytochemistry 7:1577–1581. doi: 10.1016/S0031-9422(00)88609-4. [DOI] [Google Scholar]
- 2.Clark-Lewis JW, Mortimer PI. 1959. Occurrence of 4-hydroxypipecolic acid in Acacia species. Nature 184:1234–1235. doi: 10.1038/1841234b0. [DOI] [PubMed] [Google Scholar]
- 3.Romeo JT, Swain LA, Bleecker AB. 1983. cis-4-Hydroxypipecolic acid and 2,4-cis-4,5-trans-4,5-dihydroxypipecolic acid from Calliandra. Phytochemistry 22:1615–1617. doi: 10.1016/0031-9422(83)80098-3. [DOI] [Google Scholar]
- 4.Hatanaka SI, Kaneko S. 1977. cis-5-Hydroxy-l-pipecolic acid from Morus alba and Lathyrus japonicus. Phytochemistry 16:1041–1042. doi: 10.1016/S0031-9422(00)86718-7. [DOI] [Google Scholar]
- 5.Miyata T, Okano Y, Nagatatanoue J, Ijimamiyamura S, Iwamura H, Takahama K, Hitoshi T. 1987. Identification and quantification of 5-hydroxypipecolic acid and 4-hydroxyproline in mammalian brain and blood by selected ion monitoring. Anal Biochem 163:303–308. doi: 10.1016/0003-2697(87)90228-4. [DOI] [PubMed] [Google Scholar]
- 6.Brandi L, Lazzarini A, Cavaletti L, Abbondi M, Corti E, Ciciliato I, Gastaldo L, Marazzi A, Feroggio M, Fabbretti A, Maio A, Colombo L, Donadio S, Marinelli F, Losi D, Gualerzi CO, Selva E. 2006. Novel tetrapeptide inhibitors of bacterial protein synthesis produced by a Streptomyces sp. Biochemistry 45:3692–3702. doi: 10.1021/bi052540k. [DOI] [PubMed] [Google Scholar]
- 7.Sato T, Hirayama F, Saito T, Kaniwa H. 1991. A new alkaloid antibiotic tetrazomine. Structure determination. J Antibiot (Tokyo) 44:1367–1370. doi: 10.7164/antibiotics.44.1367. [DOI] [PubMed] [Google Scholar]
- 8.Son S, Ko SK, Jang M, Lee JK, Ryoo IJ, Lee JS, Lee KH, Soung NK, Oh H, Hong YS, Kim BY, Jang JH, Ahn JS. 2015. Ulleungamides A and B, modified α,β-dehydropipecolic acid containing cyclic depsipeptides from Streptomyces sp. KCB13F003. Org Lett 17:4046–4049. doi: 10.1021/acs.orglett.5b01969. [DOI] [PubMed] [Google Scholar]
- 9.Aiello A, Fattorusso E, Giordano A, Menna M, Muller WE, Perovic-Ottstadt S, Schroder HC. 2007. Damipipecolin and damituricin, novel bioactive bromopyrrole alkaloids from the Mediterranean sponge Axinella damicornis. Bioorg Med Chem 15:5877–5887. doi: 10.1016/j.bmc.2007.05.074. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka N, Suto S, Ishiyama H, Kubota T, Yamano A, Shiro M, Fromont J, Kobayashi J. 2012. Halichonadins K and L, new dimeric sesquiterpenoids from a sponge Halichondria sp. Org Lett 14:3498–3501. doi: 10.1021/ol3014705. [DOI] [PubMed] [Google Scholar]
- 11.Lamarre D, Croteau G, Wardrop E, Bourgon L, Thibeault D, Clouette C, Vaillancourt M, Cohen E, Pargellis C, Yoakim C, Anderson P. 1997. Antiviral properties of palinavir, a potent inhibitor of the human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother 41:965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulieu PL, Lavallee P, Abraham A, Anderson PC, Boucher C, Bousquet Y, Duceppe JS, Gillard J, Gorys V, Grand-Maitre C, Grenier L, Guindon Y, Guse I, Plamondon L, Soucy F, Valois S, Wernic D, Yoakim C. 1997. Practical, stereoselective synthesis of palinavir, a potent HIV protease inhibitor. J Org Chem 62:3440–3448. doi: 10.1021/jo9702655. [DOI] [Google Scholar]
- 13.Miller SP, Zhong YL, Liu Z, Simeone M, Yasuda N, Limanto J, Chen Z, Lynch J, Capodanno V. 2014. Practical and cost-effective manufacturing route for the synthesis of a β-lactamase inhibitor. Org Lett 16:174–177. doi: 10.1021/ol4031606. [DOI] [PubMed] [Google Scholar]
- 14.Mori H, Shibasaki T, Yano K, Ozaki A. 1997. Purification and cloning of a proline 3-hydroxylase, a novel enzyme which hydroxylates free l-proline to cis-3-hydroxy-l-proline. J Bacteriol 179:5677–5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shibasaki T, Mori H, Chiba S, Ozaki A. 1999. Microbial proline 4-hydroxylase screening and gene cloning. Appl Environ Microbiol 65:4028–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara R, Kino K. 2009. Characterization of novel 2-oxoglutarate dependent dioxygenases converting l-proline to cis-4-hydroxy-l-proline. Biochem Biophys Res Commun 379:882–886. doi: 10.1016/j.bbrc.2008.12.158. [DOI] [PubMed] [Google Scholar]
- 17.Klein C, Huttel W. 2011. A simple procedure for selective hydroxylation of l-proline and l-pipecolic acid with recombinantly expressed proline hydroxylases. Adv Synth Catal 353:1375–1383. doi: 10.1002/adsc.201000863. [DOI] [Google Scholar]
- 18.Lloyd RC, Smith MEB, Brick D, Taylor SJC, Chaplin DA, McCague R. 2002. Chemoenzymatic synthesis of the four diastereoisomers of 4-hydroxypipecolic acid from N-acetyl-(R,S)-allylglycine: chiral scaffolds for drug discovery. Org Process Res Dev 6:762–766. doi: 10.1021/op020017x. [DOI] [Google Scholar]
- 19.Hibi M, Kawashima T, Kasahara T, Sokolov PM, Smirnov SV, Kodera T, Sugiyama M, Shimizu S, Yokozeki K, Ogawa J. 2012. A novel Fe(II)/α-ketoglutarate-dependent dioxygenase from Burkholderia ambifaria has β-hydroxylating activity of N-succinyl l-leucine. Lett Appl Microbiol 55:414–419. doi: 10.1111/j.1472-765X.2012.03308.x. [DOI] [PubMed] [Google Scholar]
- 20.Hibi M, Kawashima T, Sokolov PM, Smirnov SV, Kodera T, Sugiyama M, Shimizu S, Yokozeki K, Ogawa J. 2013. l-Leucine 5-hydroxylase of Nostoc punctiforme is a novel type of Fe(II)/α-ketoglutarate-dependent dioxygenase that is useful as a biocatalyst. Appl Microbiol Biotechnol 97:2467–2472. doi: 10.1007/s00253-012-4136-7. [DOI] [PubMed] [Google Scholar]
- 21.Hibi M, Kawashima T, Kodera T, Smirnov SV, Sokolov PM, Sugiyama M, Shimizu S, Yokozeki K, Ogawa J. 2011. Characterization of Bacillus thuringiensis l-isoleucine dioxygenase for production of useful amino acids. Appl Environ Microbiol 77:6926–6930. doi: 10.1128/AEM.05035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausinger RP. 2004. Fe(II)/α-ketoglutarate-dependent hydroxylases and related enzymes. Crit Rev Biochem Mol Biol 39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 23.Ju JH, Ozanick SG, Shen B, Thomas MG. 2004. Conversion of (2S)-arginine to (2S,3R)-capreomycidine by VioC and VioD from the viomycin biosynthetic pathway of Streptomyces sp. strain ATCC11861. Chembiochem 5:1281–1285. doi: 10.1002/cbic.200400136. [DOI] [PubMed] [Google Scholar]
- 24.Baud D, Saaidi PL, Monfleur A, Harari M, Cuccaro J, Fossey A, Besnard M, Debard A, Mariage A, Pellouin V, Petit JL, Salanoubat M, Weissenbach J, de Berardinis V, Zaparucha A. 2014. Synthesis of mono- and dihydroxylated amino acids with new α-ketoglutarate-dependent dioxygenases: biocatalytic oxidation of C-H bonds. ChemCatChem 6:3012–3017. doi: 10.1002/cctc.201402498. [DOI] [Google Scholar]
- 25.Hibi M, Ogawa J. 2014. Characteristics and biotechnology applications of aliphatic amino acid hydroxylases belonging to the Fe(II)/α-ketoglutarate-dependent dioxygenase superfamily. Appl Microbiol Biotechnol 98:3869–3876. doi: 10.1007/s00253-014-5620-z. [DOI] [PubMed] [Google Scholar]
- 26.Strieker M, Kopp F, Mahlert C, Essen LO, Marahiel MA. 2007. Mechanistic and structural basis of stereospecific Cβ-hydroxylation in calcium-dependent antibiotic, a daptomycin-type lipopeptide. ACS Chem Biol 2:187–196. doi: 10.1021/cb700012y. [DOI] [PubMed] [Google Scholar]
- 27.Houwaart S, Youssar L, Huttel W. 2014. Pneumocandin biosynthesis: involvement of a trans-selective proline hydroxylase. Chembiochem 15:2365–2369. doi: 10.1002/cbic.201402175. [DOI] [PubMed] [Google Scholar]
- 28.Shibasaki T, Sakurai W, Hasegawa A, Uosaki Y, Mori H, Yoshida M, Ozaki A. 1999. Substrate selectivities of proline hydroxylases. Tetrahedron Lett 40:5227–5230. doi: 10.1016/S0040-4039(99)00944-2. [DOI] [Google Scholar]
- 29.Vranova V, Lojkova L, Rejsek K, Formanek P. 2013. Significance of the natural occurrence of l- versus d-pipecolic acid. Chirality 25:823–831. doi: 10.1002/chir.22237. [DOI] [PubMed] [Google Scholar]
- 30.He M. 2006. Pipecolic acid in microbes: biosynthetic routes and enzymes. J Ind Microbiol Biotechnol 33:401–407. doi: 10.1007/s10295-006-0078-3. [DOI] [PubMed] [Google Scholar]
- 31.Warren CR, Aranda I, Cano FJ. 2011. Responses to water stress of gas exchange and metabolites in Eucalyptus and Acacia spp. Plant Cell Environ 34:1609–1629. doi: 10.1111/j.1365-3040.2011.02357.x. [DOI] [PubMed] [Google Scholar]
- 32.Moroni F, Galli A, Mannaioni G, Carla V, Cozzi A, Mori F, Marinozzi M, Pellicciari R. 1995. NMDA receptor heterogeneity in mammalian tissues: focus on two agonists, (2S,3R,4S) cyclopropylglutamate and the sulfate ester of 4-hydroxy-(S)-pipecolic acid. Naunyn Schmiedebergs Arch Pharmacol 351:371–376. [DOI] [PubMed] [Google Scholar]
- 33.Singh AB, Khaliq T, Chaturvedi JP, Narender T, Srivastava AK. 2012. Anti-diabetic and anti-oxidative effects of 4-hydroxypipecolic acid in C57BL/KsJ-db/db mice. Hum Exp Toxicol 31:57–65. doi: 10.1177/0960327111407227. [DOI] [PubMed] [Google Scholar]
- 34.Naresh G, Jaiswal N, Sukanya P, Srivastava AK, Tamrakar AK, Narender T. 2012. Glucose uptake stimulatory effect of 4-hydroxypipecolic acid by increased GLUT 4 translocation in skeletal muscle cells. Bioorg Med Chem Lett 22:5648–5651. doi: 10.1016/j.bmcl.2012.06.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.