Abstract
The ability of certain oral biofilm bacteria to moderate pH through arginine metabolism by the arginine deiminase system (ADS) is a deterrent to the development of dental caries. Here, we characterize a novel Streptococcus strain, designated strain A12, isolated from supragingival dental plaque of a caries-free individual. A12 not only expressed the ADS pathway at high levels under a variety of conditions but also effectively inhibited growth and two intercellular signaling pathways of the dental caries pathogen Streptococcus mutans. A12 produced copious amounts of H2O2 via the pyruvate oxidase enzyme that were sufficient to arrest the growth of S. mutans. A12 also produced a protease similar to challisin (Sgc) of Streptococcus gordonii that was able to block the competence-stimulating peptide (CSP)–ComDE signaling system, which is essential for bacteriocin production by S. mutans. Wild-type A12, but not an sgc mutant derivative, could protect the sensitive indicator strain Streptococcus sanguinis SK150 from killing by the bacteriocins of S. mutans. A12, but not S. gordonii, could also block the XIP (comX-inducing peptide) signaling pathway, which is the proximal regulator of genetic competence in S. mutans, but Sgc was not required for this activity. The complete genome sequence of A12 was determined, and phylogenomic analyses compared A12 to streptococcal reference genomes. A12 was most similar to Streptococcus australis and Streptococcus parasanguinis but sufficiently different that it may represent a new species. A12-like organisms may play crucial roles in the promotion of stable, health-associated oral biofilm communities by moderating plaque pH and interfering with the growth and virulence of caries pathogens.
INTRODUCTION
Dental caries remains an enormous health problem worldwide (1–4). The current view of the initiation and progression of dental caries is one of a dynamic process in which the normal balance between the complex environmental factors that control demineralization and remineralization of the tooth is disrupted in a way that favors the loss of tooth mineral. Demineralization is driven primarily by microbial fermentation of carbohydrates to organic acids, which can lower the pH of oral biofilms to the extent that there is significant dissolution of tooth mineral. Alkalization of oral biofilms occurs during fasting periods and is attributable to salivary clearance of acids, buffering of oral biofilms by host-derived and microbially derived components, and the production of basic compounds by microbial metabolism of amino acids, polyamines, urea, and other compounds (5–10). Development or worsening of a carious lesion occurs when acid-mediated enamel demineralization predominates and is associated with changes in the composition and biochemical activities of the associated microflora. Evidence is now accumulating from a variety of sources that supports that alkali generation, particularly in the form of ammonia, plays a major role in pH homeostasis in oral biofilms and inhibits the initiation and progression of dental caries (5, 10–15).
Arginine is an amino acid found in a wide variety of foods and is relatively abundant in saliva and other oral secretions (16). In the oral cavity, arginine and peptides that contain arginine are primarily catabolized by the arginine deiminase (AD) system (ADS), which is present in several bacterial species that constitute a major proportion of the oral microbiome (5, 17). The ADS consists minimally of three enzymes that convert one molecule of arginine to one molecule of ornithine and carbon dioxide and two molecules of ammonia, with the concomitant production of ATP from ADP. The ADS is believed to contribute to the inhibition of dental caries in a variety of ways. The ammonia generated elevates the biofilm pH, which prevents the loss of tooth mineral and promotes remineralization. The elevation in pH creates an environment that is less favorable for the outgrowth of cariogenic bacteria and more favorable for the persistence of health-associated taxa. There are also bioenergetic benefits to the bacteria that have this system: the ATP generated by arginine catabolism can be used for growth and maintenance by the organism, while neutralization of the cytoplasm by ammonia enhances the ΔpH component of the proton motive force (5, 11, 18, 19). It is now becoming apparent that arginine may impact oral biofilms and their pathogenic potential in other ways (20, 21).
Multiple studies have established an association of arginine levels and ADS activity with caries status in human subjects. For example, the levels of free arginine in the parotid saliva of caries-free (CF) adults were significantly higher than those found in age- and sex-matched controls who had a history of dental decay (16). A number of early studies showed that bacterial metabolism of arginine, but not other common amino acids, by salivary bacteria could elicit a rapid rise in pH (12, 14, 18, 22). Studies that compared the capacities of plaque and saliva samples from human subjects to hydrolyze arginine via the ADS found that caries-active (CA) subjects or tooth sites had substantially lower ADS activity than did similar samples from CF individuals, adding further support showing that arginine metabolism may be a deterrent to dental caries (18). Studies with subjects who consumed mints or used dentifrices containing an arginine-bicarbonate formulation showed that arginine was effective at inhibiting the initiation and progression of dental caries (13, 18, 23–25). More recently, large-scale randomized clinical trials revealed that dentifrices containing 1.5% arginine plus an insoluble calcium compound and 1,450 ppm F provided significantly greater protection against the initiation and progression of caries than did dentifrices containing 1,450 ppm F alone (23, 25). Furthermore, the use of a fluoride-free toothpaste containing 1.5% arginine was associated with increases in the ADS activity of dental plaque from CA individuals and induced changes in the composition of their plaque microflora such that the microbiomes of CA subjects began to more closely resemble those of CF subjects (18), compared to a control group using the same oral hygiene regimen but with a fluoride-containing dentifrice. Thus, a substantial amount of data now supports that arginine and the ADS can have a profoundly positive impact on dental health and oral biofilm ecology.
Another major factor shaping the composition and virulence of dental biofilms is antagonism between commensal and cariogenic bacteria (26). One of the primary ecological advantages for caries pathogens is acid tolerance (aciduricity). Highly acidogenic and aciduric caries pathogens, such as Streptococcus mutans, Lactobacillus spp., Scardovia spp., Bifidobacteria spp., and certain other taxa (6, 27, 28), rapidly ferment dietary carbohydrates to pH values that are far lower than those that permit the growth of health-associated commensal bacteria (27, 29–31). While the antagonistic effects of acidification of oral biofilms are a major driver of changes in the composition and biochemical activities of dental biofilms during caries development, there are more sophisticated antagonistic strategies used by caries pathogens to gain a competitive advantage over health-associated commensals, including the production of bacteriocins. Of relevance here, the human dental caries pathogen Streptococcus mutans produces a variety of lantibiotic and nonlantibiotic bacteriocins (often termed “mutacins”) that are active against a spectrum of bacteria, some of which are associated with oral health. Mutacin production is dominantly controlled by the ComDE two-component system, which is orthologous to BlpRH of Streptococcus pneumoniae (32, 33). CSP (competence-stimulating peptide) (34), an 18-amino-acid (aa) peptide derived from secretion and processing of the ComC protein, induces mutacin expression through ComDE. Notably, CSP can also be a potent activator of genetic competence (the ability to take up exogenous DNA) under certain growth conditions (35, 36). The concurrent activation of mutacins to lyse neighboring species and activation of competence to take up exogenous DNA have been proposed to be a strategy used by S. mutans to increase nutrient availability and enhance its genetic diversity (2, 37).
Activation of competence in S. mutans occurs transiently and requires the induction of the gene for the alternative sigma factor ComX. Notably, the ComDE-CSP circuit does not directly activate comX expression. Instead, the Rgg-like transcriptional regulator ComR is the proximal regulator of comX expression, and ComR activity is enhanced by binding to the 7-aa comX-inducing peptide (XIP). The ComRS regulatory pathway includes processing of the 17-aa ComS to XIP, which appears extracellularly, followed by active transport of XIP into the cell by the Opp oligopeptide permease and formation of a ComR-XIP complex that binds near the comX promoter to activate transcription (35, 38, 39). Higher levels of XIP can also trigger the activation of the bacteriocin genes of S. mutans, at least in part because ComX can enhance the transcription of the comE gene (40, 41). It is also known that CSP is able to activate comX only in complex medium containing peptides and only in a subpopulation of cells (bimodally), although CSP-dependent activation of mutacin genes occurs unimodally (across the population) in complex medium or in peptide-free chemically defined medium. In contrast, XIP is able to activate comX only in defined medium, presumably because peptides in complex medium compete with XIP for internalization by Opp.
Commensal bacteria also have various antagonistic strategies that they deploy against S. mutans and other cariogenic species. As noted above, neutralization of the environment by arginine or urea metabolism (5, 6, 14) diminishes the competitive advantage that caries pathogens gain at low pH. Thus, alkali generation by the ADS can be thought of as a nonspecific defense against caries pathogens, which do not compete effectively with many commensals at neutral pH values. Streptococcus sanguinis, S. gordonii, and various other oral streptococcal species secrete substantial amounts (micromolar) of hydrogen peroxide (H2O2), which is a potent antagonist of the growth of S. mutans and other oral pathogens. Oxidase enzymes that use various substrates can yield H2O2, but the pyruvate oxidase enzyme (Pox) encoded by spxB has been shown to be a dominant source of H2O2 for certain commensal streptococci (42). In addition to antagonism of caries pathogens through neutralization of the environment and H2O2 generation, S. gordonii can interfere with peptide signaling by S. mutans through the production of a proteinase termed challisin, which appears to degrade CSP to inhibit bacteriocin production and competence development by S. mutans (43).
Recently, in an effort to better understand the basis for the significantly higher ADS activities of oral biofilms obtained from CF subjects than of those obtained from CA subjects, our laboratory isolated a panel of ADS-positive (ADS+) strains from supragingival plaque (44), including strains of S. sanguinis S. gordonii, S. parasanguinis, S. intermedius, S. australis, and S. cristatus, and conducted an analysis of the ability of these isolates to express the ADS under a variety of conditions known to repress or induce the genes for the ADS (5, 19, 44–46). Substantial variability in constitutive and condition-specific arginolytic capacities was noted between and within taxa of commensal streptococci. During characterization of these strains, one strain in particular, a streptococcal strain designated strain A12, was found to be especially effective at expressing high ADS activity (44), and a preliminary analysis revealed a potent capacity to interfere with the growth of S. mutans. The purpose of this study was to begin to explore the microbiological, molecular, and genomic basis for the desirable properties of Streptococcus clinical isolate A12.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
The bacterial strains and plasmids used in this study are shown in Table 1 (36, 44, 45, 47, 48). A12 was isolated from supragingival plaque of a CF subject. Escherichia coli DH5α was grown in Luria broth (49). A12, S. mutans UA159, and derivatives of these strains were routinely grown in brain heart infusion (BHI) (3.7%) broth, on BHI agar plates (Difco), or in chemically defined FMC medium (50). Multiple medium formulations were developed empirically to examine the effects of carbohydrate source and nutrient availability on interspecies interactions: (i) TY (3% tryptone, 0.5% yeast extract) agar plates were supplemented with 25 mM galactose or 25 mM glucose, (ii) a more dilute version of TY (TY-D) (1% tryptone, 0.2% yeast extract) agar plates supplemented with 25 mM galactose, or (iii) half-strength BHI broth (1.85% BHI). For the selection of antibiotic-resistant colonies after genetic transformation or for enumeration of bacteria from mixed cultures, erythromycin (300 μg ml−1 for E. coli or 12 μg ml−1 for S. mutans), kanamycin (50 μg ml−1 for E. coli or 1 mg ml−1 for S. mutans), or spectinomycin (50 μg ml−1 for E. coli or 1 mg ml−1 for S. mutans) was added to the medium. Unless noted otherwise, cultures were grown overnight in BHI medium, with antibiotics when necessary, at 37°C in a 5% CO2 aerobic atmosphere.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source and/or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH5α | Laboratory stock | |
| S. mutans UA159 | S. mutans wild-type reference strain | Laboratory stock (ATCC 700610) |
| S. gordonii DL1 | S. gordonii wild-type reference strain | Laboratory stock |
| S. sanguinis SK150 | S. sanguinis wild-type reference strain | Laboratory stock |
| A12 | Isolated from supragingival dental plaque of caries-free subject | Clinical strain, 44 |
| SAB358 | S. mutans UA159::PcomX-lacZ; Kmr | 36 |
| SAB249 | S. mutans UA159::PcipB-lacZ; Kmr | This study |
| S. mutans pBGS | S. mutans UA159::pBGS; Spr | This study |
| S. gordonii ΔarcR | S. gordonii DL1 ΔarcR; Kmr | 45 |
| S. gordonii Δsgc | S. gordonii DL1 Δsgc; Emr | This study |
| S. gordonii ΔspxB | Streptococcus gordonii DL1 ΔspxB; Kmr | This study |
| S. gordonii Δsgc ΔspxB | Streptococcus gordonii DL1 Δsgc ΔspxB; Emr Kmr | This study |
| A12 ΔarcR | A12 ΔarcR; Emr | This study |
| A12 Δsgc | A12 Δsgc; Emr | This study |
| A12 ΔspxB | A12 ΔspxB; Kmr | This study |
| A12 Δsgc ΔspxB | A12 Δsgc ΔspxB; Emr Kmr | This study |
| Plasmids | ||
| pVA838 | Escherichia coli-Streptococcus shuttle vector; Emr | |
| pJL184 | Escherichia coli-Streptococcus shuttle vector; Kmr | 47 |
| pDL278 | Escherichia coli-Streptococcus shuttle vector; Spr | 48 |
| pBGS | Escherichia coli-Streptococcus shuttle vector; Spr |
In A12, arcR, sgc, and spxB are the homologs of the arcR, sgc, and spxB genes of S. gordonii, respectively. Spr, spectinomycin resistance.
Synthetic CSP (sCSP), corresponding to the 21-aa peptide (34, 35), was synthesized by the Interdisciplinary Center for Biotechnology Research (ICBR) facility at the University of Florida, and its purity was confirmed by high-performance liquid chromatography (HPLC). Synthetic XIP (sXIP), corresponding to residues 11 to 17 of ComS (amino acid sequence of GLDWWSL), was synthesized and purified to 96% homogeneity by NeoBioSci (Cambridge, MA, USA). Lyophilized sXIP was reconstituted in 99.7% dimethyl sulfoxide (DMSO) to a final concentration of 2 mM and stored in 40-μl aliquots at −20°C.
Construction of mutant strains and reporter gene fusions.
Mutant strains of A12 were constructed by double-crossover recombination using linear DNA engineered to replace the gene of interest with an antibiotic resistance gene. Briefly, using a colony of A12 as the template, 0.5-kbp fragments upstream and downstream of the genes of interest were amplified by PCR using primers (Table 2) based on the genome sequence of A12 (see below). Antibiotic resistance markers were amplified by PCR from pJL184 (kanamycin resistance [Kmr]) or pVA838 (erythromycin resistant [Emr]). Each PCR product was gel purified, and 100 ng of each fragment was fused by using PCR, as described previously (51). PCR products were excised from an agarose gel, purified, and used to transform A12 by using a modification of a method originally developed for Streptococcus australis (52). Briefly, a single colony was inoculated into Todd-Hewitt broth (THB) containing 10% heat-inactivated horse serum and incubated for 16 to 18 h. Cells were diluted 1:40 into THB supplemented with 10% horse serum and incubated until the optical density at 600 nm (OD600) reached 0.05 to 0.08. PCR products to be used for gene inactivation (0.5 μg) were added to the cultures, and incubation was continued for 3 h. Plasmid DNA (pDL278) (48) was used as a positive control to ensure that cells were competent for transformation. Cultures were plated onto BHI agar plates containing the desired antibiotics. Transformants were verified by PCR and DNA sequencing, including sequencing of the regions used for recombination to confirm that no unwanted mutations had been introduced into flanking genes. Mutant derivatives of S. gordonii DL1 were constructed by using similar methods (51).
TABLE 2.
Primers used in this study
| Primer | Sequencea | Application |
|---|---|---|
| A12-arcR-5′-F | 5′-TTTCACACCCGAGCTCTTAC-3′ | A12 ΔarcR; Emr |
| A12-arcR-5′-R-SOE | 5′-ACTCCTTCTGGTGGCGACTTTCAATCTTATTC-3′ | A12 ΔarcR; Emr |
| A12-arcR-Erm-F-SOE | 5′-GTCGCCACCAGAAGGAGTGATTACATGAACAAA-3′ | A12 ΔarcR; Emr |
| A12-arcR-Erm-R-SOE | 5′-GAGGCAGACCCCTTTAGTAACGTGTAACTTT-3′ | A12 ΔarcR; Emr |
| A12-arcR-3′-F-SOE | 5′-ACTAAAGGGGTCTGCCTCATCATCTGTGAA-3′ | A12 ΔarcR; Emr |
| A12-arcR-3′-R | 5′-CTCTCCATTGATGAACAGCAA-3′ | A12 ΔarcR; Emr |
| A12-spxB-5′-F | 5′-GAATTGGCGCAAGGGATGATAA-3′ | A12 ΔspxB; Kmr |
| A12-spxB-5′-R-SOE | 5′-TTTCTACAGGATAGTGTCTACGCCCCATGTT-3′ | A12 ΔspxB; Kmr |
| A12-spxB-Kan-F-SOE | 5′-GACACTATCCTGTAGAAAAGAGGAAGGAA-3′ | A12 ΔspxB; Kmr |
| A12-spxB-Kan-R-SOE | 5′-TGGTACAAGGAGTATGGACAGTTGCGGATGTA-3′ | A12 ΔspxB; Kmr |
| A12-spxB-3′-F-SOE | 5′-TCCATACTCCTTGTACCATTCCGTCTCTTC TT-3′ | A12 ΔspxB; Kmr |
| A12-spxB-3′-R | 5′-CCACTTACTAGCAGGGATTCGGTT-3′ | A12 ΔspxB; Kmr |
| A12-sgc-5′-F | 5′-CCATTTGGCCAAGGAGAGTTGTT-3′ | A12 Δsgc; Emr |
| A12-sgc-5′-R-SOE | 5′-CACTCCTTCGAACAGATCCTAATAGCACCGA-3′ | A12 Δsgc; Emr |
| A12-sgc-Erm-F-SOE | 5′-GATCTGTTCGAAGGAGTGATTACATGAACAA-3′ | A12 Δsgc; Emr |
| A12-sgc-Erm-R-SOE | 5′-CAGGATTAGCCCTTTAGTAACGTGTAACTTT-3′ | A12 Δsgc; Emr |
| A12-sgc-3′-F-SOE | 5′-ACTAAAGGGCTAATCCTGGTGCTAGTCAA-3′ | A12 Δsgc; Emr |
| A12-sgc-3′-R | 5′-CCTGGAGAAGAACTATACGTAA-3′ | A12 Δsgc; Emr |
| cipB5′ | 5′-TCATGGATTGAGCTCAAAAAGTAAT-3′ | PcipB::lacZ reporter strain |
| cipB3′ | 5′-TGTATTCATGGATCCAATACCCCTT-3′ | PcipB::lacZ reporter strain |
The underlined sequences were included in primers to facilitate PCR ligation of DNA fragments used for mutant construction.
To construct a strain of S. mutans with a lacZ gene fused to the promoter of the cipB bacteriocin gene (PcipB::lacZ; strain SAB249), the promoter region of cipB was amplified using primers (Table 2) that incorporated SacI and BamHI sites (53) and cloned into a pMC340B-based lacZ reporter vector (54), which carries a Kmr gene. The vector has sequence homology to the mtlA and phnA genes flanking the cloning sites, allowing for integration of the PcipB-lacZ construct in a single copy into the S. mutans chromosome through double-crossover homologous recombination. The PcipB-lacZ construct was transformed into the wild-type strain, and the correct integration and sequence of the gene fusion were verified by PCR and DNA sequencing. To create a marked strain for coculture experiments, S. mutans was transformed with pBGS, a spectinomycin-resistant (Spr) derivative of pBGK in which the Kmr marker was replaced with a Spr marker. Transformation of S. mutans UA159 with pBGS allowed for stable integration of the Spr marker into the gtfA gene (55), a nonessential gene that does not adversely affect growth or competitive fitness of S. mutans, to create S. mutans strain UA159::pBGS.
ADS activity.
ADS activity was measured by monitoring citrulline production from arginine using protocols detailed previously (44). ADS activity was measured in cells grown as follows: cultures of each strain grown overnight were diluted 1:20 into fresh TY medium containing 25 mM galactose, with or without 10 mM arginine, and incubated as described above until the OD600 reached 0.5 to 0.6. The cells were then permeabilized by using a 1/10 volume of toluene-acetone (1:9) prior to determination of their ADS activity. The concentration of protein in the permeabilized cell preparations was measured by using a Pierce (Waltham, MA, USA) bicinchoninic acid (BCA) protein assay kit with bovine serum albumin as the standard. ADS activity levels of bacterial strains were normalized to protein content and defined as nanomoles of citrulline generated × [minutes × (milligrams protein)]−1. Alternatively, cells were cultivated in FMC medium containing arginine in concentrations ranging from 0 to 100 mM. Dilutions, incubation, cell preparation, and ADS assays for cells grown in FMC medium were the same as those used for cells grown in TY medium.
Measurement of H2O2.
Cultures of bacterial strains grown overnight were diluted 1:20 into TY medium plus 25 mM glucose, TY medium plus 25 mM galactose, or TY-D medium plus 25 mM galactose and incubated statically in a 5% CO2 aerobic atmosphere. When the cultures reached an OD600 of 0.3, they were shaken at 200 rpm for 30 min, and the amount of H2O2 present in culture supernatants was measured as previously described (56), with minor modifications: 0.65 ml of the culture supernatant was added to 0.6 ml of a solution containing 2.5 mM 4-amino-antipyrine (4-amino-2,3-dimethyl-1-phenyl-3-pyrazolin-5-one; Sigma) and 0.17 M phenol (Fisher). The reaction was allowed to proceed for 4 min at room temperature, and horseradish peroxidase (Pierce, Thermo Scientific) in 0.2 M potassium phosphate buffer (pH 7.2) was then added to the reaction mixture at a final concentration of 13 mU/ml. After a 20-min incubation at room temperature, the OD510 was measured in a Beckman DU-640 spectrophotometer. A standard curve was generated for each assay mixture with known concentrations of H2O2 (Fisher Scientific).
Assays for H2O2-generating oxidase enzymes.
Cultures grown overnight were centrifuged at 4,000 × g for 10 min and washed twice with 2 ml of phosphate-buffered saline (PBS) (pH 7.3) (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, 1.4 mM KH2PO4). The pelleted cells were resuspended in 2 ml PBS. Aliquots of the cell suspension were used for determining lactate oxidase activity, and the remaining cell suspensions were permeabilized for determination of pyruvate oxidase, l-arginine oxidase, and NADH oxidase activities by mixing the cell suspension with 0.02 volumes of toluene-acetone (1:9, vol/vol) and vortexing the mixture for 2 min. The concentration of protein in the cell preparations was determined by using a Pierce (Waltham, MA, USA) BCA protein assay kit with bovine serum albumin as the standard (57).
Pyruvate oxidase activity was determined by measuring the production of acetyl phosphate (AcP) (57, 58). Briefly, the reaction mixture consisted of 0.45 ml of the permeabilized cell suspension and 0.45 ml of a solution containing 50 mM potassium phosphate buffer (pH 6.0), 10 μM MgCl2, 0.2 μM thiamine pyrophosphate (Sigma), 50 mM potassium pyruvate, and 12 μM flavin adenine dinucleotide (FAD; Sigma). Reaction mixtures lacking permeabilized cells or pyruvate were included as negative controls. The reaction mixtures were incubated at 37°C for 30 min with shaking at a speed of 250 rpm, and the amount of acetyl phosphate generated during the reaction was then measured as follows. An aliquot of cell suspensions (0.3 ml) was preincubated at 37°C for 1 min in a model 111002 heat block (Boekel Scientific, Feasterville, PA, USA). Fifty microliters of a 2 M hydroxylamine hydrochloride solution was added, and the mixture was placed into a heat block at 60°C and incubated for 5 min. Next, 100 μl of development solution consisting of equal volumes of 0.5 M ferric chloride in 5 M HCl and 30% trichloroacetic acid was added, and color was allowed to develop for at least 1 min at room temperature. The samples were centrifuged, and the OD540 was measured. A standard curve was generated with known concentrations of lithium potassium acetyl phosphate (Sigma).
Lactate oxidase, l-arginine oxidase, and NADH oxidase activities were assessed by assaying H2O2 production (57). For the lactate oxidase assay, 0.45 ml of intact cells was added to 0.45 ml of 0.2 M sodium phosphate buffer (pH 7.0) containing 20 mM sodium l-lactate; reaction mixtures without sodium l-lactate were used as negative controls. The mixtures were incubated at 37°C with shaking for 30 min at 250 rpm. For l-arginine oxidase assays, 0.45 ml of permeabilized cell suspensions was added to 0.45 ml of 0.2 M sodium phosphate buffer (pH 7.0) containing 20 mM l-arginine; reaction mixtures without arginine were used as negative controls. The mixtures were incubated at 37°C with shaking for 2 h at 250 rpm. For NADH oxidase assays, 0.45 ml of permeabilized cell suspensions was added to 0.45 ml of 0.2 M sodium phosphate buffer (pH 7.0) containing 13 mM NADH (sodium salt); reaction mixtures without NADH were used as negative controls. The mixtures were incubated at 37°C with shaking for 3 h at 250 rpm, and the concentration of H2O2 in the supernatants was determined.
Competition assays on agar plates.
For competition assays between S. mutans UA159 and strains of A12 or S. gordonii DL1, a culture of each strain grown overnight was adjusted to an OD600 of 0.5 in BHI medium. An aliquot from each culture (6 μl) was inoculated adjacent to the other strain on agar plates as follows: (i) A12 or DL1 was inoculated first, followed by inoculation of S. mutans 24 h later and an additional 24 h of incubation; (ii) S. mutans was inoculated first, followed by inoculation with A12 or DL1 after 24 h plus an additional 24 h of incubation; or (iii) DL1 or A12 and S. mutans were inoculated at the same time, and plates were incubated for a total of 24 h.
Bacteriocin assays.
Cultures of S. mutans UA159::pBGS (55), S. gordonii DL1, A12, and derivatives of these strains grown overnight were centrifuged, washed twice with PBS, resuspended in PBS, and adjusted to an OD600 of 0.5. S. mutans was then mixed with other strains at a ratio of 1:1. Pure cultures or mixed cultures were spotted onto BHI agar plates and grown aerobically or anaerobically. After 24 h of incubation at 37°C, 3 ml of a soft agar overlay (1% BHI agar) containing 107 cells of the indicator strain Streptococcus sanguinis SK150 was poured evenly onto the plate, the agar was allowed to set, and the plates were incubated for 24 h. Zones of inhibition were measured and documented with a digital imager. In some cases, the agar plug from mixed cultures was harvested, serially diluted, and plated onto selective agar to enumerate cells of S. mutans, S. gordonii DL1, strain A12, or derivatives of these strains. CFU were counted after incubation at 37°C for 48 h.
Monitoring of comX and cipB gene promoter activities.
The ability of supernatants from A12 and S. gordonii to interfere with CSP or XIP signaling by S. mutans was assessed by monitoring the expression of genes that have been established to be activated by these peptides (36, 37, 59). Specifically, strains derived from S. mutans UA159 that carried lacZ reporter gene fusions to the promoter regions of cipB, encoding a bacteriocin (PcipB-lacZ), or to comX, encoding the alternative sigma factor required for the development of genetic competence (PcomX-lacZ), were used to monitor the ability of S. mutans to respond to sCSP or sXIP, respectively. Supernatants from cultures of strains grown in BHI medium overnight were obtained after removal of cells by centrifugation, the pH was adjusted to pH 7.0, and the supernatants were filter sterilized. A subset of aliquots of supernatants was kept on ice, while others were heat treated to inactive putative proteases. sCSP was added to supernatants to a final concentration of 2 μM, and the mixtures were incubated for 3 h. S. mutans UA159::PcipB-lacZ that had been grown to an OD600 of 0.12 was then suspended in the supernatants. The cultures were then incubated for 2 h, and S. mutans cells were assayed for β-galactosidase activity. For XIP signaling, which occurs in defined medium, supernatant fluids from cultures of A12 and its derivatives, S. gordonii, or S. mutans UA159 that had been grown overnight in FMC medium were adjusted to pH 7.0, supplemental glucose was added to increase the glucose concentration by 25 mM, and the supernatants were filter sterilized. sXIP was added to heat-treated or untreated supernatants as described above and incubated overnight at 37°C. Cultures of S. mutans UA159::PcomX-lacZ that had been grown in FMC medium to an OD600 of 0.12 were added to supernatants. The cultures were incubated for 2 h, and LacZ assays were performed to detect XIP-dependent activation of the comX promoter. Where noted, A12 was also grown under anaerobic conditions, or the supernatants were treated with catalase to examine the influence of oxygen and/or hydrogen peroxide on the ability of A12 to inhibit peptide signaling.
LacZ (β-galactosidase) activity was measured by using a modification of the Miller protocol (35, 60). Briefly, cells were harvested by centrifugation, washed once with Z buffer (Na-phosphate buffer [pH 7.0], 10 mM KCl, 1 mM MgSO4, 5 mM β-mercaptoethanol), and resuspended in 1.2 ml of Z buffer. A 400-μl aliquot of the sample was vortexed with 20 μl of toluene-acetone (1:9) for 2 min, and the mixture was kept at 37°C. The remainder of the cell suspension was used to measure the OD600. The reaction was initiated by the addition of 80 μl of an ONPG (o-nitrophenyl-β-d-galactopyranoside) solution (4 mg/ml) to the mixture and was terminated by the addition of 400 μl of 1 M Na2CO3 to the mixture. Samples were centrifuged at 15,250 × g for 1 min, and the OD of the supernatant fluid was measured at 420 and 550 nm. β-Galactosidase activity was expressed in Miller units (35, 60).
Whole-genome sequencing and phylogenomic analysis.
Genomic DNA was isolated by using Qiagen Genomic-Tip 20/G columns and the Qiagen Genomic DNA buffer set according to the supplier's protocols. A 10-kb library was made by the ICBR Core at the University of Florida and sequenced by using PacBio RSII SMRT sequencing technology. The Bioinformatics Core at the University of Florida assembled the genome by using SMRT analysis software. The genome was annotated by using the RAST server (61). A comprehensive phylogenetic analysis comparing A12 to 14 other members of the genus Streptococcus (8 species from the S. mitis group, 3 species from the S. sanguinis group, and 3 species from the S. anginosus group) (see Table S1 in the supplemental material) was performed as described previously by Richards et al. (62). Briefly, a Markov cluster algorithm was used to determine the core gene set. Core gene clusters were aligned, and those judged to be recombinant were removed, leaving 374 putatively nonrecombinant gene alignments. Two phylogenetic approaches were taken. In the first approach, a maximum likelihood (ML) phylogeny was generated from a concatenation of the 374 alignments. In the second approach, 374 separate ML phylogenies (gene trees) were constructed, and a consensus phylogeny was constructed.
Nucleotide sequence accession number.
The DNA sequence for Streptococcus sp. A12 has been deposited with NCBI and assigned BioSample acccession number SAMN04287214.
RESULTS
Effects of arginine on ADS induction in A12.
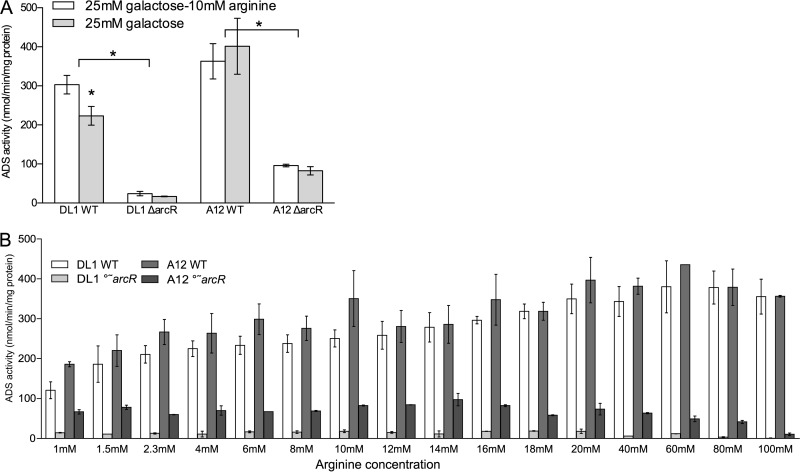
The arginine deiminase (AD) enzyme activity in A12 was higher than that in the highly arginolytic reference strain S. gordonii DL1 in cells grown under conditions that have been shown to be optimal for ADS expression in vitro (TY medium containing 25 mM galactose and 10 mM arginine) (44). Importantly, full induction of AD activity in A12 did not require supplementation of TY medium containing galactose with arginine, whereas supplementation with arginine was necessary for full expression of the ADS in DL1 (Fig. 1A) and many other ADS+ bacteria (5, 19, 44, 45). ArcR is the transcriptional activator of the ADS genes in streptococci, governing ADS gene expression in response to arginine (45). When arcR was inactivated in A12 and S. gordonii DL1, both strains produced much lower AD activity. The S. gordonii DL1 ΔarcR strain expressed only 8% of the AD activity of the parental strain, whereas the A12 ΔarcR strain retained 25% of the AD activity of wild-type A12. TY medium contains 3% tryptone, so the concentration of available arginine would be estimated to be in the 5 to 10 mM range and present primarily in the peptide form. To explore the requirement of arginine for ADS expression in more detail, cells were cultured in chemically defined FMC medium with a modified arginine content (Fig. 1B). In FMC medium lacking arginine, both A12 and DL1 were unable to grow (data not shown). As the arginine levels in FMC were increased, both A12 and DL1 showed increased AD activity, with A12 producing higher AD levels than DL1, especially at lower concentrations of arginine. Both the S. gordonii DL1 ΔarcR and A12 ΔarcR strains had much lower ADS levels than the wild-type strains, as observed with cells cultured in TY medium. However, the S. gordonii DL1 ΔarcR strain had almost no AD activity, whereas the A12 ΔarcR strain retained considerable ADS activity under the same conditions. Collectively, these data reveal that A12 requires less arginine to produce substantial levels of AD activity and that ArcR-independent ADS gene expression may, at least in part, explain the higher constitutional arginolytic activity of A12.
FIG 1.
(A) Arginine deiminase enzyme activity in S. gordonii DL1 and A12 wild-type (WT) and ΔarcR mutant strains grown in TY medium with or without supplemental arginine. (B) Arginine deiminase activity in wild-type and ΔarcR mutant strains of S. gordonii grown in chemically defined FMC medium containing the indicated amounts of arginine. A12 and S. gordonii DL1 were not able to grow in FMC medium unless some arginine was added. In panel A, asterisks indicate statistical significance between the wild-type and arcR mutant strains under both growth conditions using an alpha value of 0.05. In panel B, comparisons are between the arcR mutants of A12 and S. gordonii.
A12 inhibits growth of S. mutans by pyruvate oxidase-dependent H2O2 production.
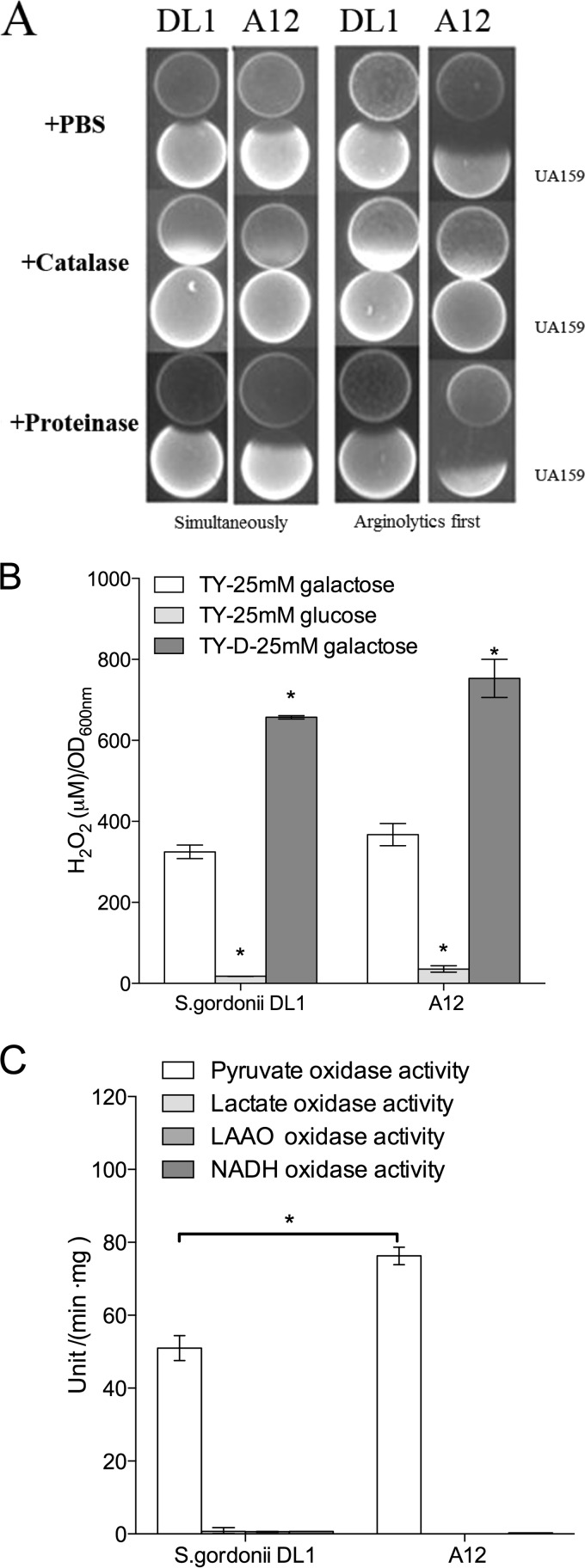
By using a plate-based zone-of-inhibition assay, A12 inhibited the growth of S. mutans UA159 under aerobic conditions more effectively than did DL1 when inoculated prior to or simultaneously with inoculation of S. mutans UA159. H2O2 was clearly a factor in inhibition since the inclusion of catalase in the medium effectively eliminated inhibition (Fig. 2A). Both A12 and DL1 produced substantial quantities of H2O2 (Fig. 2B) and displayed evidence of carbohydrate catabolite repression (CCR) of the activity responsible for H2O2 generation. More specifically, inhibition of S. mutans by A12 was less efficient on medium with glucose than on medium containing galactose, which elicits little or no evidence of CCR (Table 3). The use of a less-rich version of TY medium (TY-D medium) also resulted in enhanced H2O2 production, further linking the inhibitory activity of A12 to nutrient source and availability. Figure 2C shows that pyruvate oxidase (Pox) activity was the dominant source of H2O2 (Fig. 2C), with lactate oxidase, certain l-amino-acid oxidases (LAAOs), and NADH oxidase producing negligible amounts of H2O2 under the conditions tested. Notably, A12 expressed significantly higher Pox enzyme activity than did S. gordonii DL1 (Fig. 2C).
FIG 2.
(A) Plate-based growth inhibition assays of arginolytic strains versus S. mutans UA159. In all cases, cultures grown overnight were collected by centrifugation and resuspended at an OD600 of 0.5 in PBS. In the left two columns, the arginolytic strain S. gordonii DL1 or A12 was spotted onto plates and incubated for 24 h in a 5% CO2 aerobic atmosphere, an equivalent amount of S. mutans UA159 was then spotted adjacent to S. gordonii or A12, and the plates were incubated an additional 24 h. In the two columns on the right, A12 or S. gordonii DL1 was spotted at the same time as S. mutans UA159, and the plates were incubated for a total of 24 h. The addition of catalase to the medium effectively eliminated the inhibition of S. mutans by the commensal, but inclusion of proteinase K did not have the same effect. (B) H2O2 production by S. gordonii or A12 is dependent on carbohydrate source and nutrient availability. Cells were grown under aerobic conditions in the indicated media, and the concentration of H2O2 in the supernatants was measured as described in Materials and Methods. Asterisks indicate statistical significance between growth on glucose and growth on galactose for each organism (alpha = 0.05). (C) Comparison of pyruvate, lactate, l-amino acid, and NADH oxidase enzyme levels in S. gordonii DL1 and A12 (see the text for methodology). Pyruvate oxidase (Pox) was the dominant H2O2-producing oxidase measured in both organisms under the conditions tested, and A12 produced significantly higher levels of Pox enzyme activity than did S. gordonii. Asterisks signify statistical significance between pyruvate oxidase levels in A12 and those in S. gordonii.
TABLE 3.
Zones of inhibition by A12 of S. mutans as a function of growth medium conditionsa
| Strain | Mean zone of inhibition (mm) ± SD |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TY medium + 25 mM galactose |
TY medium + 25 mM glucose |
Half-strength BHI medium |
BHI medium |
TY-D medium + 25 mM galactose |
||||||
| I | II | I | II | I | II | I | II | I | II | |
| DL1 | 0.7 ± 0.0 | 0.7 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.0 | 1.4 ± 0.3 | 0.4 ± 0.1 | 1.3 ± 0.2 | 0.4 ± 0.0 | 1.7 ± 0.2 | 0.9 ± 0.1 |
| A12 | 2.1 ± 0.1 | 0.9 ± 0.1 | 0.9 ± 0.1 | 1.2 ± 0.1 | 1.6 ± 0.1 | 0.5 ± 0.1 | 1.5 ± 0.2 | 0.5 ± 0.1 | 2.6 ± 0.7 | 1.6 ± 0.2 |
Cells were incubated in an aerobic atmosphere. The zone of inhibition was measured in millimeters. I, arginolytic bacteria colonized first; II, arginolytic bacteria and S. mutants UA159 colonized simultaneously.
Modulation of S. mutans bacteriocin production by A12.
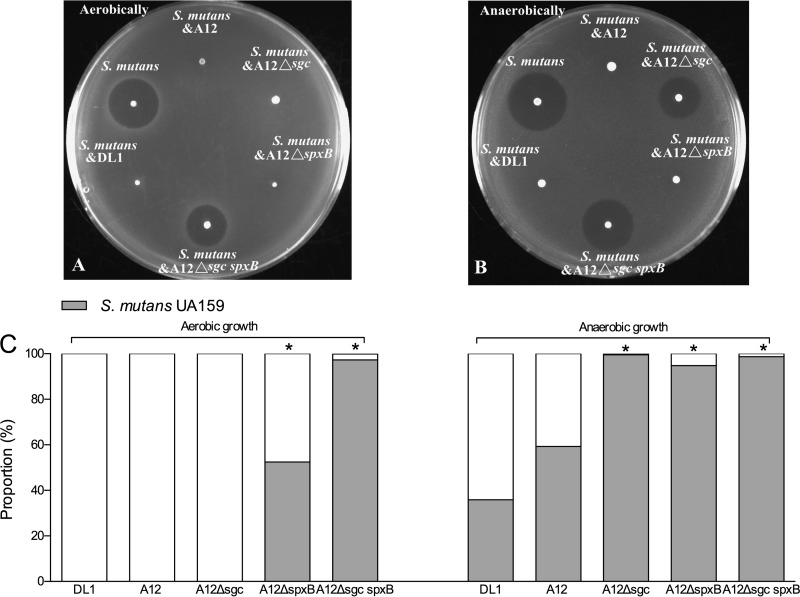
As described in Materials and Methods, a standard agar overlay antagonism assay was modified to examine mechanisms by which A12 may interfere with the growth of S. mutans and whether there were additional beneficial activities associated with A12. In particular, a dual-species cultivation system was created, in which agar plates were coinoculated with A12 and S. mutans and the plates were subsequently overlaid with the indicator strain S. sanguinis SK150, which is sensitive to the mutacins produced by S. mutans UA159 (see Materials and Methods for more detail). This allowed us to (i) examine whether A12 could modulate the capacity of S. mutans to kill sensitive streptococci and (ii) enumerate, by removing and dispersing the bacteria at the inoculation site, the proportions of A12 and S. mutans bacteria remaining on the plates. First, virtually no S. mutans bacteria could be recovered after coinoculation/cocultivation with A12 under aerobic conditions (Fig. 3A and C), whereas S. mutans accounted for nearly 60% of the recovered organisms under anaerobic conditions (Fig. 3B and C), the difference being apparently attributable to H2O2-mediated killing of S. mutans in aerobic cocultures. In fact, when the predicted homolog of the spxB gene of A12 encoding pyruvate oxidase was deleted, the mutant lost the ability to produce H2O2 during aerobic growth (see Fig. S1 in the supplemental material) and to inhibit the growth of S. mutans in the dual-species competition assay (see Fig. S2 in the supplemental material).
FIG 3.
(A and B) Plate-based mutacin assays using S. sanguinis SK150 as the indicator strain. The indicated strains were mixed in equal proportions and spotted onto BHI agar plates, and the plates were incubated overnight under aerobic (A) or anaerobic (B) conditions. Subsequently, 3 ml of a soft BHI agar overlay containing 107 S. sanguinis SK150 bacteria was gently and evenly overlaid onto the plate, and incubation was continued for 24 h prior to measurement of zones of inhibition. Mutants of A12 or S. gordonii lacking the Sgc protease were unable to block mutacin production to protect the indicator strain. (C) An agar plug was obtained from the inoculation spot after the experiment, cells were dispersed, and the proportions of bacteria of S. mutans UA159 and the indicated S. gordonii or A12 strains were enumerated by dilution and plating (see Materials and Methods for more details). Asterisks indicate statistically significant differences in the proportions of S. mutans bacteria cocultivated with the indicated mutant strains compared to cocultivation of wild-type A12 with S. mutans.
The same plate-based assay also revealed that the ability of S. mutans to produce bacteriocins that kill or inhibit the growth of S. sanguinis SK150 could be modulated by A12 and DL1. In particular, when UA159 and A12 were coinoculated, there was no evidence of inhibition of the indicator strain (Fig. 3A and B). While the lack of inhibition of SK150 under aerobic conditions was easily explained by a failure of S. mutans to grow in the presence of A12 or DL1, the lack of inhibition of SK150 was equally as evident under anaerobic conditions. It was reported previously that production of a protease termed challisin, encoded by the sgc gene, by S. gordonii is responsible for degradation of CSP (43), the signal peptide that induces bacteriocin gene expression through the ComDE (32–34) two-component signal transduction system of S. mutans. We were able to identify a gene for a putative challisin-like protease in the genome (see below) of A12 that had 60.4% amino acid sequence identity to S. gordonii challisin. Mutant strains of A12 and DL1 that lacked the predicted sgc homologs were created, as detailed in Materials and Methods. When S. mutans and the A12 Δsgc strain were cocultured under aerobic conditions, there was no killing of the indicator strain, and no S. mutans bacteria were recovered, so the loss of Sgc did not impede the ability of A12 to inhibit S. mutans via H2O2 production. Interestingly, under anaerobic conditions, the proportions of the colony represented by the A12 Δsgc mutant were substantially lower than those represented by the A12 wild-type strain. Similarly, when S. mutans and the A12 ΔspxB strain were grown together aerobically, A12 accounted for almost 50% of the colony but only 5% under anaerobic conditions (Fig. 3C). Notwithstanding, even when A12 represented as little as 5% of the recovered organisms, there was still no killing of the indicator strain (Fig. 3B). Importantly, the A12 Δsgc spxB double mutant strain lost both the ability to inhibit S. mutans through H2O2 production (see Fig. S2 in the supplemental material) and the ability to protect the indicator strain from inhibition. However, again, it was noted that the proportions of S. mutans bacteria were greater during cocultivation with the spxB single mutant than during cocultivation with the spxB sgc double mutant. Therefore, it must be considered that the poorer competition of A12 strains carrying the sgc mutation may indicate that Sgc provides some protection to A12 against antagonistic factors of S. mutans.
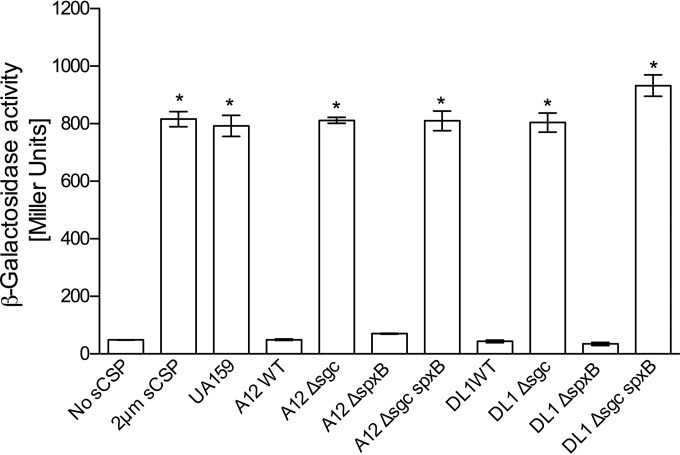
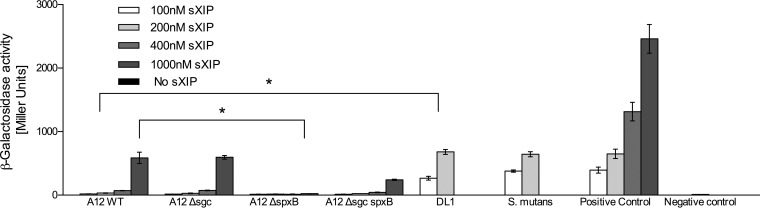
To address in more detail the mechanisms by which A12 might impact the capacity of S. mutans to produce bacteriocins, the ability of supernatants from the wild-type and sgc mutant strains derived from A12 and DL1 to degrade CSP was assessed by monitoring CSP-dependent signaling through ComDE using a cipB-lacZ promoter-reporter fusion. When 2 μM sCSP was incubated for 3 h with supernatants of A12 or S. gordonii DL1, there was almost no induction of LacZ activity (Fig. 4). If the supernatants were boiled for 20 min prior to incubation with sCSP (data not shown), LacZ activity expressed from the cipB promoter was similar to that of the sCSP control or treatment of sCSP with supernatants from S. mutans (Fig. 4). Importantly, supernatants from the Δsgc mutants of A12 and DL1 lost the ability to degrade sCSP (Fig. 4). We also examined the ability of supernatants from spxB mutants of A12 and DL1 to degrade sCSP (Fig. 4) and saw no differences compared to the wild-type strains. However, the A12 Δsgc spxB and DL1 Δsgc spxB double mutants were unable to degrade sCSP.
FIG 4.
Effects of treatment of sCSP with supernatants from cultures of the indicated strains grown overnight. A strain of S. mutans containing a lacZ fusion to the cipB promoter was grown to an OD600 of 0.12 in BHI medium (optimal responsiveness to sCSP). Supernatants from cultures of the indicated strains grown overnight were obtained after centrifugation, adjusted to pH 7.0 (also optimal for sCSP signaling), and filter sterilized. sCSP was added to the supernatants, and the mixtures were incubated for 2 h. The S. mutans cipB-lacZ reporter strain was then resuspended in the indicated supernatants for 2 h prior to measurement of β-galactosidase (LacZ) activity. The Sgc protease was necessary for inhibition of sCSP signaling. The results shown are from a minimum of three biological replicates, each performed in triplicate. Values are averages, and error bars indicate standard deviations. Asterisks indicate statistically significant differences compared with wild-type A12.
A12, but not S. gordonii DL1, inhibits XIP-dependent signaling.
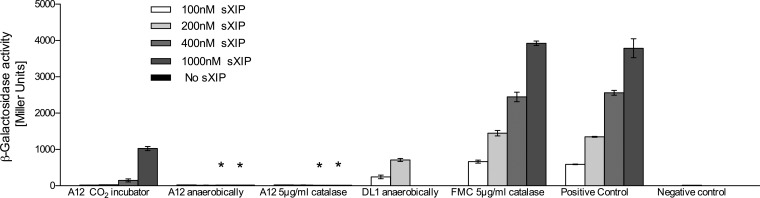
CSP is able to substantially enhance the ability of S. mutans to be transformed with exogenous DNA, but activation of competence by CSP is indirect. The proximal regulator of the comX gene encoding the alternative sigma factor that activates late competence gene expression is ComR, and ComR binding is regulated by the XIP signal peptide. The abilities of supernatants from A12 and DL1 to degrade XIP were compared by using the same methods as those used for CSP, with the exceptions that the experiments were performed with chemically defined FMC medium and a derivative of S. mutans UA159 carrying a fusion of the comX promoter to lacZ was used as the reporter for XIP-dependent activation of gene expression. After overnight treatment of XIP with supernatants from S. gordonii DL1, XIP signaling was as robust as that in the untreated controls, i.e., untreated sXIP in medium alone or sXIP added to supernatants from S. mutans UA159 (Fig. 5). In contrast, exposure of sXIP to supernatants from wild-type A12 eliminated the ability of sXIP to activate comX. If the supernatants of A12 were boiled, XIP activation of comX was similar to that of the sXIP controls (data not shown), indicating that the activity responsible for degradation of or interference with sXIP was heat sensitive. Interestingly, deletion of the sgc gene of A12 did not eliminate the ability of supernatants from A12 to inhibit sXIP signaling, and deletion of spxB resulted in an enhanced ability of supernatants from A12 to inhibit sXIP signaling. The primary difference between the A12 wild-type strain and the A12 ΔspxB strain is that the former can produce substantial quantities of H2O2 and the latter produces almost no H2O2 under the conditions used for these particular experiments. When A12 was grown in either BHI or FMC broth under static conditions in a 5% CO2 aerobic atmosphere, no H2O2 was detected in supernatants of cultures grown overnight (data not shown). To be certain that the effects on XIP were not associated with H2O2, supernatants were pretreated with catalase prior to the addition of XIP (Fig. 6). As noted for the spxB mutant, elimination of H2O2 resulted in an enhancement of the capacity of the supernatants from A12 to block sXIP-dependent activation of comX. Thus, the production or stability of the activity that interferes with XIP signaling may be enhanced under anaerobic conditions, perhaps reflecting an adaptation that could enhance the protection of A12 against S. mutans when A12 is unable to produce H2O2.
FIG 5.
Effects of treatment of XIP with supernatants from cultures of the indicated strains grown overnight. Supernatant fluids from cultures of A12 and its derivatives, S. gordonii, or S. mutans UA159 that had been grown in FMC medium overnight were adjusted to pH 7.0, supplemental glucose was added to increase the glucose concentration by 25 mM, and the supernatants were filter sterilized. sXIP was added to the supernatants, and the mixtures were incubated overnight at 37°C. Cultures of S. mutans UA159::PcomX-lacZ that had been grown in FMC medium to an OD600 of 0.12 were pelleted by centrifugation and resuspended in the supernatants. The cultures were incubated for 2 h, and LacZ assays were performed to detect XIP-dependent activation of the comX promoter. The results shown are from a minimum of three biological replicates, each performed in triplicate. Values are averages, and error bars indicate standard deviations. Data from treatment with 400 or 1,000 nM sXIP for DL1 and S. mutans are not shown because neither supernatant influenced sXIP signaling any differently than the positive control. Asterisks indicate statistical significance in comparisons between wild-type A12 with 200 nM XIP and DL1 with 200 nM XIP and between wild-type A12 with 1,000 XIP and Spx-deficient A12 with 1,000 nM XIP.
FIG 6.
Effects of growth conditions on interference of XIP signaling by supernatants from cultures of the indicated strains grown overnight. All experiments were performed as detailed in the legend of Fig. 5 except that cells were grown in different atmospheres or supernatants were treated with catalase. Supernatants from A12 cells grown anaerobically had an apparently enhanced capacity to interfere with sXIP signaling, and the effects were not impacted by the inclusion of catalase. S. gordonii again was unable to interfere with sXIP signaling, and data for treatment with 400 or 1,000 nM sXIP for DL1 are not shown because there was no difference from the positive control. Catalase itself also had no effect on signaling (FMC 5 μg/ml catalase bars). The results shown are from a minimum of three biological replicates, each performed in triplicate. Values are averages, and error bars indicate standard deviations. Asterisks indicate statistical significance between A12 grown in CO2 and A12 grown anaerobically and between A12 grown in CO2 and A12 grown with catalase treatment.
Whole-genome sequencing and phylogenomic analysis of A12.
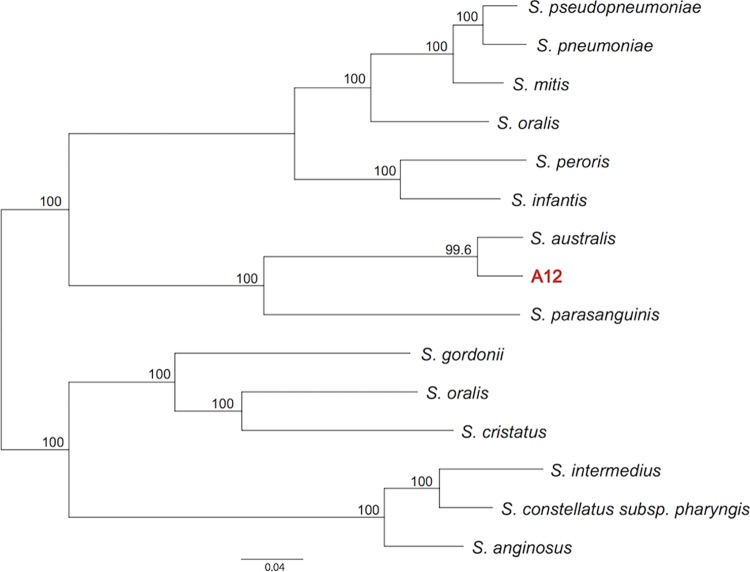
Comparison of the entire 16S sequence of A12 to the 16S sequences of other organisms by BLAST revealed that the 16S rRNA gene was most highly conserved with that of the reference strain for Streptococcus australis (NCBI GenBank accession number NZ_AFUD01000002.1. To more carefully evaluate the relatedness of A12 to other streptococci, the entire genome sequence of A12 was determined by using PacBio sequencing technology to attain 60× coverage. A single 1.8-Mb contig was obtained and annotated (BioSample accession number SAMN04287214), and a comprehensive phylogenomic analysis was conducted according to methodologies detailed previously (62). By using these approaches, it was determined (Fig. 7) that the patristic distance (i.e., divergence between lineages) between A12 and its closest relative, S. australis, was 0.058 (5.8% divergence), whereas the patristic distance between Streptococcus pneumoniae and Streptococcus pseudopneumoniae, two distinct species, was 0.049 (4.9%). Although it will be necessary to isolate and sequence additional A12-like bacteria to propose that A12 represents a new species of Streptococcus, data from phylogenomic analysis suggest that A12 may not be appropriately classified as S. australis or S. parasanguinis. Of note, A12 has a colony morphology distinct from those of other commonly isolated oral streptococcal species, and comparison of the translated sequences of certain genes from A12 with apparent homologs from S. australis or S. parasanguinis revealed substantial degeneracy, which has proven useful in distinguishing A12-like organisms from close relatives in plaque samples. With this knowledge, we have been able to isolate a small number of putative A12-like organisms from clinical samples. All of the isolates showed the expected sequence conservation in a gene of interest, with significant differences from S. australis and S. parasanguinis being notable (data not shown), along with constitutively high ADS activity and potent antagonistic capacity against S. mutans (X. Huang and R. A. Burne, unpublished data).
FIG 7.
Maximum likelihood phylogeny showing relationships among 14 Streptococcus species and A12. Numbers on the branches show the proportions of gene phylogenies that support the grouping.
DISCUSSION
In light of the strong correlation between the absence of dental caries and high dental plaque ADS activity (5, 12, 14, 18), oral bacteria with constitutionally high ADS expression levels may have significant potential for applications in probiotic therapies to prevent and control dental caries. Such strains would be even more desirable if they could antagonize the growth of known caries pathogens. Here, we characterize a novel Streptococcus isolate, designated A12, with the ability to express high ADS activity under conditions that commonly occur in human dental biofilms (44). A12 was also shown to have a particularly potent inhibitory effect on the growth of S. mutans, mainly through pyruvate oxidase-dependent H2O2 production. Moreover, A12 is able to interfere with the two dominant intercellular peptide-based communication pathways of S. mutans, which has the impact of disabling a primary antagonistic strategy (bacteriocins) of S. mutans while rendering S. mutans unable to induce genetic competence, competence being associated with nutritional benefits, genome diversification, acid tolerance, and biofilm formation. The identification of an oral isolate with this combination of characteristics, when coupled with our findings on the tremendous heterogeneity of commensal streptococci (44), highlights the profound gap in our understanding of the phenotypic capacity and plasticity of the oral microbiome as these factors relate to the behaviors of particular taxa and populations. The data also reinforce the importance of complementing current metagenomic approaches that utilize 16S rRNA sequences (37, 44, 62) with more in-depth approaches to characterize the oral microbiota if reliable correlations for the composition and activities of the microbiome and oral health are to be established.
The ADS is widely distributed among prokaryotes, and the primary structures of the enzymes in this pathway have been conserved throughout evolution (63). Most microorganisms studied so far have their ADS genes organized into one cluster. In S. gordonii, which has the most extensively characterized ADS of any oral bacterium, the genes for the three enzymes of the pathway, arginine deiminase (AD) (arcA), ornithine carbamoyltransferase (arcB), and carbamate kinase (arcC), are cotranscribed in an operon with arcD (arginine:ornithine antiporter) and arcT (arginine aminopeptidase). Induction of the ADS genes (arcABCDT) by arginine is mediated by a transcriptional activator encoded by the divergently transcribed arcR gene located immediately downstream of the arcABCDT operon (45). Notably, arcR of S. gordonii is the second gene in a two-gene operon that includes the queA gene, which has been shown in other organisms to catalyze the final step in queosine modification of tRNAs. QueA-deficient S. gordonii mutants show altered ADS expression. An Fnr-like protein (flp) is encoded immediately upstream of arcA and is required for the activation of expression of the arcA promoter under anaerobic conditions (46). Catabolite repression of the operon by preferred carbohydrate sources (CCR), such as glucose, is exerted primarily through CcpA (46) binding to two cis-acting catabolite response elements (CREs) near the arcA promoter (64).
There are some similarities of the ADS operon of A12 with those of its closest relatives, S. australis and S. parasanguinis, and with the well-characterized operon of S. gordonii DL1 (see Fig. S3A in the supplemental material). Most notably, arcABCDT of A12 appear to be cotranscribed, as is the case for arcABCDT of S. gordonii DL1 (45). Also present in A12 are arcR and queA, which are immediately downstream of arcT and transcribed in the opposite direction, as is the case for S. gordonii (63). There is also an apparent homolog of the flp gene upstream of arcA, with Flp of A12 sharing 63% sequence identity (82% similarity) with Flp of S. gordonii (46). The promoter for the S. gordonii arcA gene has been mapped and is located the proper distance from a fairly well-conserved (TAGAAT) −10 sequence but a weaker −35 sequence (see Fig. S3B in the supplemental material). A near-consensus −10 sequence (TAAAAT) was identified in A12, which is located 1 nucleotide (nt) closer to the arcA start codon than the −10 element of S. gordonii. However, the predicted −35 sequence of A12, which is also 1 nt closer to the initiation codon than that of S. gordonii, was more similar to the consensus than the S. gordonii −35 element. Also, similar to S. gordonii and consistent with the fact that both genes appear to require a transcriptional activator (ArcR) for optimal expression, an ArcR binding site was identified upstream of arcA (see Fig. S3B in the supplemental material).
The arc operons of S. gordonii and A12 are sensitive to CCR, although A12 is less so. In particular, in cells grown in the presence of 25 mM glucose, AD activity in S. gordonii DL1 was only ∼15% of that expressed in cells grown on 25 mM galactose, whereas A12 grown on glucose retained >50% of the AD activity of cells grown on 25 mM galactose (44). Catabolite repression of the ADS operon of S. gordonii is predominantly regulated by CcpA, which is predicted to bind to two conserved CREs located in the arcA promoter region. Two sequences in the A12 arcA promoter region with conservation of the derived consensus sequence (TQWNANCGNTNWCA) (64) for binding of the catabolite control protein CcpA were identified at positions −116 to −104 and at positions −34 to −21 in the A12 operon, essentially in the same positions as the CREs in S. gordonii with respect to the arcA start codon. The upstream CcpA binding site in A12 at positions −116 to −104 (AGAAAGCGGTTCAT) has 4 nucleotides that differ from the consensus sequence, whereas the binding site at positions −21 to −34 (TGAAAGCGGTACCA) had 1 nucleotide that differed from the consensus. Overall, then, adherence of the CREs to the consensus sequence was not as high in A12 as that of the CREs in the arcA promoter region of S. gordonii. While the promoter strength and the effectiveness of the putative cis- and trans-acting factors in induction and repression of the ADS of A12 remain to be determined, the constitutionally higher levels of A12 ADS activity may be associated with an inherently stronger promoter and diminished CcpA-dependent CCR.
In a recent communication, we reported that the absolute level of ADS activity expressed by multiple commensal streptococci under ideal conditions can be highly variable across and within species, as can the ability of the system to be repressed by carbohydrate or oxygen or induced by arginine. We posit that there may be an evolutionary basis for this inter- and intraspecies variation that leads to certain isolates, like A12, having higher constitutive ADS gene expression levels and a diminished sensitivity to CCR and other factors. In particular, we propose that A12 and similar isolates may have evolved to rely more heavily on the ADS for protection from the detrimental effects of low pH, whereas low-ADS producers or strains that are more sensitive to repression by environmental inputs, such as high levels of carbohydrate, evolved other acid tolerance strategies, for example, higher proton-extruding ATPase activity, H+-ATPases with lower optimal pH values, diminished membrane proton permeability, or other traits associated with resistance to low pH (5, 29). Should this hypothesis be proven correct, it can then be extrapolated that the latter group of commensals, with lower ADS expression and higher inherent acid tolerance, might tend to be associated with caries activity, whereas the former group would tend to be associated with health. The difficulty, of course, is that these strains cannot be discriminated by comparing 16S sequences. Isolation and characterization of additional A12-like organisms and genomic/phenotypic characterization of these isolates and isolates of other streptococcal species to test this hypothesis are under way.
A variety of oral streptococci that are predominant members of early and mature biofilms are able to generate H2O2, which is thought to have a profound impact on the composition and biochemical activities of the microbiota (26, 42, 65). NADH oxidase enzymes were believed to be a primary source of H2O2 in the oral cavity (66), but more recent studies support that other enzymatic pathways may contribute substantially to endogenously generated H2O2 in oral biofilms. In particular, pyruvate oxidase (Pox), which is encoded by spxB, is a major source of H2O2 produced by A12, S. sanguinis, S. gordonii, and some other arginolytic streptococci. Utilization of pyruvate via Pox under aerobic conditions may have several benefits for those organisms that produce the enzyme as well as having a beneficial impact on biofilm ecology. In particular, Pox directly converts pyruvate and inorganic phosphate to the high-energy intermediate acetyl phosphate (AcP), which can be used for ATP synthesis by acetate kinase (67, 68). AcP also serves as a signal molecule and as a substrate for certain two-component systems (69). Thus, Pox provides bioenergetic benefits while potentially contributing to the optimization of gene expression and physiology through modulation of AcP levels. Pox also liberates CO2, which can be converted to bicarbonate by carbonic anhydrases to buffer biofilms against acidification. It is also notable that under aerobic conditions, Pox shunts pyruvate away from lactate and toward the production of the weaker organic acid acetate, which may further diminish the extent of dental plaque acidification. Streptococcus oligofermentans (56) and some isolates of S. gordonii and S. cristatus (data not shown) can also produce H2O2 from lactic acid via the lactate oxidase (Lox) enzyme (56). LAAOs in these species may also contribute, to a lesser degree, to the production of H2O2 by these organisms (70). However, Lox, LAAO(s), or NADH oxidase does not appear to be a major contributor to H2O2 generation by A12 or S. gordonii DL1 compared to Pox under the conditions tested.
The production of Pox by A12 also appears sensitive to CCR, which is consistent with what has been reported previously for certain other commensal streptococci (70) and with the presence of a CRE in the promoter region for the spxB gene in A12 (TGAAACGTTTTCA) with only one mismatch from the consensus CRE. Clearly, then, there may be conditions in dental biofilms that are not favorable for H2O2-dependent inhibition of S. mutans, such as when oral biofilms become more anaerobic and/or when there is a sufficient amount of dietary carbohydrate ingested by the host to repress the expression of gene products needed for H2O2 production. In this regard, the demonstration that A12 has evolved strategies to subvert the bacteriocin- and competence-activating signal pathways of S. mutans and that these systems remain active under anaerobic conditions and when glucose is abundant (Fig. 3 and 6) (44) may be highly significant for the role of A12 in the antagonism of S. mutans and protection of beneficial organisms.
Mutacin production has been postulated, although not directly demonstrated, to be an essential ecological determinant that enhances the establishment, persistence, and emergence of S. mutans in cariogenic biofilms. Two factors have been shown to dominantly control bacteriocin expression in S. mutans, CSP and exposure to oxygen. CSP, at levels as low as 30 nM, is sufficient to rapidly and robustly induce the lantibiotic and nonlantibiotic bacteriocins of S. mutans as well as an endogenous bacteriocin (CipB) that has been proposed to play a role in altruistic cell death. Growth in air also dramatically induces bacteriocin production by S. mutans, with the gene for the nonlantibiotic mutacin (nlmD) being induced >40-fold in cells exposed to oxygen compared to cells cultured anaerobically (71). Thus, S. mutans, when in early or immature dental biofilms where oxygen and redox levels may be fairly high (66), may activate mutacin expression as a deterrent to competition with those commensals that are better adapted to tolerate growth in the presence of air. Conversely, when biofilms mature and oxygen levels and redox drop significantly, S. mutans can still activate mutacin gene expression through the accumulation of the CSP signal (72). While commensals that are competing with S. mutans may gain an advantage under aerated conditions through H2O2 production, the possession by A12, S. gordonii, and possibly other health-associated oral streptococci of an activity that can degrade CSP may offer protection from mutacins in mature biofilms. There is also a role for CSP in competence and cell lysis, the latter providing extracellular DNA (eDNA) that can stabilize biofilms by serving as part of the extracellular matrix (73). While much remains to be learned about CSP signaling, competence, and programmed cell death as well as how these factors influence plaque ecology, interference with the CSP signaling pathway would be predicted to diminish the ability of S. mutans to persist and emerge as a dominant member of the oral flora.
Phylogenomic studies clearly show that S. mutans is a remarkably diverse species and that this genomic diversity is, in large part, attributable to its highly efficient capacity to acquire genes through lateral gene transfer, mainly due to its ability to become genetically competent (transformable). Conjugation and bacteriophage, on the other, do not appear to be major contributors to genetic diversity in this caries pathogen (62). While CSP can clearly enhance the transformability of S. mutans, the primary control point for the activation of competence genes is through the ComRS-XIP system described above. Interestingly, while isolates of S. mutans show considerable heterogeneity in terms of the CSP-ComDE system, with some strains completely lacking the genes and others having divergent sequences or pseudogenes, the ComRS systems shows a remarkably high level of conservation and is part of the core genome (37). A full appreciation of the contribution of XIP and ComRS to S. mutans colonization, growth in vivo, persistence, or virulence has not yet been realized, but the high degree of evolutionary conservation and the essential role of ComRS in competence development provide significant support for an essential role(s) for this system and for strong evolutionary pressure to maintain a fully functional ComRS-XIP pathway. In this regard, the unusual capacity of A12 to interfere with XIP-dependent signaling may be a particularly beneficial attribute, if for no other reason than DNA may be an important nutrient source in vivo. Clearly, though, our data show that the A12 activity that interferes with XIP signaling is not produced by S. gordonii under the conditions tested and that it is distinct from the Sgc enzyme. Recently, the PepO protease in group A streptococci has been shown to degrade small hydrophobic signal peptides, including XIP. A12 contains a gene with 71% identity and 82% similarity to the PepO protease of Streptococcus pyogenes. We are currently exploring potential candidates for XIP-degrading activity in A12.
Finally, there is the question of whether A12-like bacteria represent a new species of Streptococcus that occupies a niche in oral biofilms that distinguishes them from their closest relatives, S. australis and S. parasanguinis. The phylogenomic analyses utilized in this study revealed that the relatedness of A12 to S. australis is less than that of two established streptococcal species, Streptococcus pneumoniae and S. pseudopneumoniae. While it would be premature to propose A12 as a new species, the distinct colony morphology and ability to find highly conserved genes in what appear to be A12-like organisms that have diverged substantially in sequence from apparent homologs in S. australis or S. parasanguinis should allow us to gather sufficient data to establish whether designation as a new species is warranted. Regardless of the outcome, A12 is an isolate that is both highly arginolytic and able to inhibit growth and intercellular signaling in S. mutans in a way that may promote the stability of health-associated biofilm communities. Our recent communication shows that isolates that, by 16S sequence, are classified as known streptococcal species display a spectrum of arginolytic and antagonistic properties. In contrasting the genome sequences of A12-like organisms and isolates with similar beneficial properties with poorly arginolytic and poorly antagonistic members of the same taxa, it should be possible to develop molecular probes and other tools that can be utilized to ascertain the extent to which biofilms of human subjects are populated with beneficial organisms. Such knowledge could aid the practice of clinical dentistry by augmenting caries risk assessment and improving the ability to evaluate the impact of therapeutic approaches on oral microbiomes.
Supplementary Material
ACKNOWLEDGMENT
We thank Zezhang (Tom) Wen at the LSU Health Sciences Center for constructing pBGS.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03887-15.
REFERENCES
- 1.Selwitz RH, Ismail AI, Pitts NB. 2007. Dental caries. Lancet 369:51–59. doi: 10.1016/S0140-6736(07)60031-2. [DOI] [PubMed] [Google Scholar]
- 2.Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang X, Cheng L, Exterkate RAM, Liu M, Zhou X, Li J, ten Cate JM. 2012. Effect of pH on Galla chinensis extract's stability and anti-caries properties in vitro. Arch Oral Biol 57:1093–1099. doi: 10.1016/j.archoralbio.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Huang X, Guo Q, Ren B, Li Y, Zhou X. 2016. Models in caries research, p 157–173. In Zhou X. (ed), Dental caries. Springer, Berlin, Germany. [Google Scholar]
- 5.Burne RA, Marquis RE. 2000. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett 193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi N, Nyvad B. 2011. The role of bacteria in the caries process: ecological perspectives. J Dent Res 90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 7.Marsh PD. 2003. Are dental diseases examples of ecological catastrophes? Microbiology 149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 8.ten Cate JM. 2006. Biofilms, a new approach to the microbiology of dental plaque. Odontology 94:1–9. doi: 10.1007/s10266-006-0063-3. [DOI] [PubMed] [Google Scholar]
- 9.Huang X, Liu M, Li J, Zhou X, ten Cate JM. 2012. Chemical composition of Galla chinensis extract and the effect of its main component(s) on the prevention of enamel demineralization in vitro. Int J Oral Sci 4:146–151. doi: 10.1038/ijos.2012.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burne RA, Zeng L, Ahn SJ, Palmer SR, Liu Y, Lefebure T, Stanhope MJ, Nascimento MM. 2012. Progress dissecting the oral microbiome in caries and health. Adv Dent Res 24:77–80. doi: 10.1177/0022034512449462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X, Exterkate RAM, ten Cate JM. 2012. Factors associated with alkali production from arginine in dental biofilms. J Dent Res 91:1130–1134. doi: 10.1177/0022034512461652. [DOI] [PubMed] [Google Scholar]
- 12.Nascimento MM, Gordan VV, Garvan CW, Browngardt CM, Burne RA. 2009. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol Immunol 24:89–95. doi: 10.1111/j.1399-302X.2008.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ten Cate JM, Cummins D. 2013. Fluoride toothpaste containing 1.5% arginine and insoluble calcium as a new standard of care in caries prevention. J Clin Dent 24:79–87. [PubMed] [Google Scholar]
- 14.Kleinberg I. 2002. A mixed-bacteria ecological approach to understanding the role of the oral bacteria in dental caries causation: an alternative to Streptococcus mutans and the specific-plaque hypothesis. Crit Rev Oral Biol Med 13:108–125. doi: 10.1177/154411130201300202. [DOI] [PubMed] [Google Scholar]
- 15.Margolis HC, Duckworth JH, Moreno EC. 1988. Composition of pooled resting plaque fluid from caries-free and caries-susceptible individuals. J Dent Res 67:1468–1475. doi: 10.1177/00220345880670120601. [DOI] [PubMed] [Google Scholar]
- 16.Van Wuyckhuyse BC, Perinpanayagam HE, Bevacqua D, Raubertas RF, Billings RJ, Bowen WH, Tabak LA. 1995. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J Dent Res 74:686–690. doi: 10.1177/00220345950740021001. [DOI] [PubMed] [Google Scholar]
- 17.Rogers AH, Zilm PS, Gully NJ, Pfennig AL. 1988. Response of a Streptococcus sanguis strain to arginine-containing peptides. Infect Immun 56:687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nascimento MM, Browngardt C, Xiaohui X, Klepac-Ceraj V, Paster BJ, Burne RA. 2014. The effect of arginine on oral biofilm communities. Mol Oral Microbiol 29:45–54. doi: 10.1111/omi.12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Y, Nascimento MM, Burne RA. 2012. Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int J Oral Sci 4:135–140. doi: 10.1038/ijos.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolderman E, Bettampadi D, Samarian D, Dowd SE, Foxman B, Jakubovics NS, Rickard AH. 2015. l-Arginine destabilizes oral multi-species biofilm communities developed in human saliva. PLoS One 10:e0121835. doi: 10.1371/journal.pone.0121835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakanaka A, Kuboniwa M, Takeuchi H, Hashino E, Amano A. 2015. Arginine-ornithine antiporter ArcD controls arginine metabolism and interspecies biofilm development of Streptococcus gordonii. J Biol Chem 290:21185–21198. doi: 10.1074/jbc.M115.644401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nascimento MM, Burne RA. 2014. Caries prevention by arginine metabolism in oral biofilms: translating science into clinical success. Curr Oral Health Rep 1:79–85. doi: 10.1007/s40496-013-0007-2. [DOI] [Google Scholar]
- 23.Kraivaphan P, Amornchat C, Triratana T, Mateo LR, Ellwood R, Cummins D, DeVizio W, Zhang YP. 2013. Two-year caries clinical study of the efficacy of novel dentifrices containing 1.5% arginine, an insoluble calcium compound and 1,450 ppm fluoride. Caries Res 47:582–590. doi: 10.1159/000353183. [DOI] [PubMed] [Google Scholar]
- 24.Acevedo AM, Montero M, Rojas-Sanchez F, Machado C, Rivera LE, Wolff M, Kleinberg I. 2008. Clinical evaluation of the ability of CaviStat in a mint confection to inhibit the development of dental caries in children. J Clin Dent 19:1–8. [PubMed] [Google Scholar]
- 25.Yin W, Hu DY, Li X, Fan X, Zhang YP, Pretty IA, Mateo LR, Cummins D, Ellwood RP. 2013. The anticaries efficacy of a dentifrice containing 1.5% arginine and 1450 ppm fluoride as sodium monofluorophosphate assessed using quantitative light-induced fluorescence (QLF). J Dent 41:S22–S28. doi: 10.1016/j.jdent.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Kreth J, Merritt J, Qi F, Dong X, Shi W. 2011. Antagonistic, synergistic, and counteroffensive strategies for streptococcal interspecies interactions, p 331–343. In Kolenbrander P. (ed), Oral microbial communities. ASM Press, Washington, DC. [Google Scholar]
- 27.van Houte J. 1994. Role of micro-organisms in caries etiology. J Dent Res 73:672–681. [DOI] [PubMed] [Google Scholar]
- 28.Bowden GH. 1990. Microbiology of root surface caries in humans. J Dent Res 69:1205–1210. doi: 10.1177/00220345900690051701. [DOI] [PubMed] [Google Scholar]
- 29.Burne RA. 1998. Oral streptococci. Products of their environment. J Dent Res 77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- 30.Bradshaw D, Marsh PD. 1998. Analysis of pH-driven disruption of oral microbial communities in vitro. Caries Res 32:456–462. doi: 10.1159/000016487. [DOI] [PubMed] [Google Scholar]
- 31.Van Ruyven FOJ, Lingström P, Van Houte J, Kent R. 2000. Relationship among mutans streptococci, “low-pH” bacteria, and iodophilic polysaccharide-producing bacteria in dental plaque and early enamel caries in humans. J Dent Res 79:778–784. doi: 10.1177/00220345000790021201. [DOI] [PubMed] [Google Scholar]
- 32.Martin B, Quentin Y, Fichant G, Claverys JP. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol 14:339–345. doi: 10.1016/j.tim.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Burne RA, Bessen DE, Broadbent JR, Claverys JP. 2007. The Seventh International Conference on the Genetics of Streptococci, Lactococci, and Enterococci J Bacteriol 189:1209–1218. doi: 10.1128/JB.01363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hossain MS, Biswas I. 2012. An extracellular protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol 194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Q, Ahn SJ, Kaspar J, Zhou X, Burne RA. 2014. Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J Bacteriol 196:227–236. doi: 10.1128/JB.00995-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Son M, Ahn SJ, Guo Q, Burne RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmer SR, Miller JH, Abranches J, Zeng L, Lefebure T, Richards VP, Lemos JA, Stanhope MJ, Burne RA. 2013. Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS One 8:e61358. doi: 10.1371/journal.pone.0061358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaspar J, Ahn SJ, Palmer SR, Choi SC, Stanhope MJ, Burne RA. 2015. A unique open reading frame within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance. Mol Microbiol 96:463–482. doi: 10.1111/mmi.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Son M, Ghoreishi D, Ahn SJ, Burne RA, Hagen SJ. 2015. Sharply tuned pH response of genetic competence regulation in Streptococcus mutans: a microfluidic study of environmental sensitivity of comX. Appl Environ Microbiol 81:5622–5631. doi: 10.1128/AEM.01421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reck M, Tomasch J, Wagner-Döbler I. 2015. The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet 11:e1005353. doi: 10.1371/journal.pgen.1005353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Son M, Shields RC, Ahn S, Burne RA, Hagen SJ. 10 September 2015. Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol Lett doi: 10.1093/femsle/fnv159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kreth J, Merritt J, Shi W, Qi F. 2005. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol 187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B-Y, Kuramitsu HK. 2005. Interactions between oral bacteria: inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl Environ Microbiol 71:354–362. doi: 10.1128/AEM.71.1.354-362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang X, Schulte RM, Burne RA, Nascimento MM. 2015. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res 49:165–176. doi: 10.1159/000365296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong Y, Chen YYM, Snyder JA, Burne RA. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl Environ Microbiol 68:5549–5553. doi: 10.1128/AEM.68.11.5549-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong Y, Chen YYM, Burne RA. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J Bacteriol 186:2511–2514. doi: 10.1128/JB.186.8.2511-2514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santiago B, MacGilvray M, Faustoferri RC, Quivey RG. 2012. The branched-chain amino acid aminotransferase encoded by ilvE is involved in acid tolerance in Streptococcus mutans. J Bacteriol 194:2010–2019. doi: 10.1128/JB.06737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn SJ, Wen ZT, Burne RA. 2006. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun 74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994. Methods for general and molecular bacteriology. ASM Press, Washington, DC. [Google Scholar]
- 50.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect Immun 11:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer SR, Crowley PJ, Oli MW, Ruelf MA, Michalek SM, Brady LJ. 2012. YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology 158:1702–1712. doi: 10.1099/mic.0.059139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warburton PJ, Ciric L, Lerner A, Seville LA, Roberts AP, Mullany P, Allan E. 2013. TetAB(46), a predicted heterodimeric ABC transporter conferring tetracycline resistance in Streptococcus australis isolated from the oral cavity. J Antimicrob Chemother 68:17–22. doi: 10.1093/jac/dks351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ahn SJ, Wen ZT, Burne RA. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J Bacteriol 189:8519–8527. doi: 10.1128/JB.01180-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Zeng L, Burne RA. 2009. AguR is required for induction of the Streptococcus mutans agmatine deiminase system by low pH and agmatine. Appl Environ Microbiol 75:2629–2637. doi: 10.1128/AEM.02145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wen ZT, Burne RA. 2001. Construction of a new integration vector for use in Streptococcus mutans. Plasmid 45:31–36. doi: 10.1006/plas.2000.1498. [DOI] [PubMed] [Google Scholar]
- 56.Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. 2007. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol 63:872–880. doi: 10.1111/j.1365-2958.2006.05546.x. [DOI] [PubMed] [Google Scholar]
- 57.Liu L, Tong H, Dong X. 2012. Function of the pyruvate oxidase-lactate oxidase cascade in interspecies competition between Streptococcus oligofermentans and Streptococcus mutans. Appl Environ Microbiol 78:2120–2127. doi: 10.1128/AEM.07539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fowler ML, Ingram-Smith CJ, Smith KS. 2011. Direct detection of the acetate-forming activity of the enzyme acetate kinase. J Vis Exp 58:e3474. doi: 10.3791/3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perry J, Jones M, Peterson S, Cvitkovitch D, Levesque C. 2009. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol 72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zubay G, Morse DE, Schrenk WJ, Miller JHM. 1972. Detection and isolation of the repressor protein for the tryptophan operon of Escherichia coli. Proc Natl Acad Sci U S A 69:1100–1103. doi: 10.1073/pnas.69.5.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richards VP, Palmer SR, Pavinski Bitar PD, Qin X, Weinstock GM, Highlander SK, Town CD, Burne RA, Stanhope MJ. 2014. Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol Evol 6:741–753. doi: 10.1093/gbe/evu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Dong Y, Chen YYM, Burne RA. 2008. Environmental and growth phase regulation of the Streptococcus gordonii arginine deiminase genes. Appl Environ Microbiol 74:5023–5030. doi: 10.1128/AEM.00556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng L, Dong Y, Burne RA. 2006. Characterization of cis-acting sites controlling arginine deiminase gene expression in Streptococcus gordonii. J Bacteriol 188:941–949. doi: 10.1128/JB.188.3.941-949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng L, Itzek A, Chen Z, Kreth J. 2011. Environmental influences on Streptococcus gordonii competitive hydrogen peroxide production. Appl Environ Microbiol 77:4318–4328. doi: 10.1128/AEM.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marquis R. 1995. Oxygen metabolism, oxidative stress and acid-base physiology of dental plaque biofilms. J Ind Microbiol 15:198–207. doi: 10.1007/BF01569826. [DOI] [PubMed] [Google Scholar]
- 67.Kreth J, Zhang Y, Herzberg MC. 2008. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J Bacteriol 190:4632–4640. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe AJ, Chang DE, Walker JD, Seitz-Partridge JE, Vidaurri MD, Lange CF, Prüß BM, Henk MC, Larkin JC, Conway T. 2003. Evidence that acetyl phosphate functions as a global signal during biofilm development. Mol Microbiol 48:977–988. doi: 10.1046/j.1365-2958.2003.03457.x. [DOI] [PubMed] [Google Scholar]
- 69.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 70.Tong H, Chen W, Shi W, Qi F, Dong X. 2008. SO-LAAO, a novel L-amino acid oxidase that enables Streptococcus oligofermentans to outcompete Streptococcus mutans by generating H2O2 from peptone. J Bacteriol 190:4716–4721. doi: 10.1128/JB.00363-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahn SJ, Ahn SJ, Browngardt CM, Burne RA. 2009. Changes in biochemical and phenotypic properties of Streptococcus mutans during growth with aeration. Appl Environ Microbiol 75:2517–2527. doi: 10.1128/AEM.02367-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li YH, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao S, Klein MI, Heim KP, Fan Y, Bitoun JP, Ahn SJ, Burne RA, Koo H, Brady LJ, Wen ZT. 2014. Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J Bacteriol 196:2355–2366. doi: 10.1128/JB.01493-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.