Abstract
Chronic exposure to airborne fungi has been associated with different respiratory symptoms and pathologies in occupational populations, such as grain workers. However, the homogeneity in the fungal species composition of these bioaerosols on a large geographical scale and the different drivers that shape these fungal communities remain unclear. In this study, the diversity of fungi in grain dust and in the aerosols released during harvesting was determined across 96 sites at a geographical scale of 560 km2 along an elevation gradient of 500 m by tag-encoded 454 pyrosequencing of the internal transcribed spacer (ITS) sequences. Associations between the structure of fungal communities in the grain dust and different abiotic (farming system, soil characteristics, and geographic and climatic parameters) and biotic (wheat cultivar and previous crop culture) factors were explored. These analyses revealed a strong relationship between the airborne and grain dust fungal communities and showed the presence of allergenic and mycotoxigenic species in most samples, which highlights the potential contribution of these fungal species to work-related respiratory symptoms of grain workers. The farming system was the major driver of the alpha and beta phylogenetic diversity values of fungal communities. In addition, elevation and soil CaCO3 concentrations shaped the alpha diversity, whereas wheat cultivar, cropping history, and the number of freezing days per year shaped the taxonomic beta diversity of these communities.
INTRODUCTION
Occupational exposure to grain dust is associated with both acute and chronic effects on the respiratory tract. The main reported respiratory effects of exposure to grain dust are asthma and acute asthma-like symptoms (1), reduced lung volume, and symptoms evoking chronic bronchitis (2, 3). However, the etiology of these effects is not completely understood, mainly because of the complexity and variety of potentially causative agents within grain dust. Indeed, grain dust is a generic name for the dust generated by handling different species as well as cultivars of cereal crops. Grain dust is of heterogeneous composition, consisting of the variable microbial flora associated with multiple plants, fragments of wheat plants and seeds generated by their abrasion when handled, fragments of insects and mites, particles of inorganic matter, and pesticides (4). Although several of these components might affect respiratory health by acting through different mechanisms, the adverse health effects of an exposure to high levels of airborne fungi (2 × 109 spores/m3 in grain workers with organic dust toxic syndrome; 1 × 108 to 4 × 108 spores/m3 in farmers with hypersensitivity pneumonitis [HP]; 3 × 106 to 1 × 107 spores/m3 in nonsymptomatic individuals) (5) direct the research on the etiology of these respiratory diseases toward the nature and the diversity of these biological agents within and between samples. Indeed, although several fungal species were identified as causes of HP (6) or asthma (7) in other occupational populations, limited information on the fungal species identified as causes of these pathologies and on the frequency of these species in the aerosols inhaled by the grain workers is available. Thus, evaluating the homogeneity of exposure to harmful fungi during grain and straw handling as well as identifying the abiotic and biotic factors that might shape the structure of the fungal community in grain dust will allow for the targeting of measures to prevent respiratory impairment in grain workers.
Wheat is a major crop in Western Europe. For this reason, the study of its associated fungal communities, particularly of its fungal pathogens, is of significant economic and health interest. Previous studies of wheat-associated fungal communities revealed a large number of operational taxonomical units (OTUs) that are dominated by a few basidiomycete yeasts (e.g., Cryptococcus and Sporobolomyces roseus), filamentous saprobes (e.g., Alternaria, Cladosporium, and Epicoccum), and plant pathogens (Fusarium) (8, 9). Among these plant pathogens, the Fusarium species complex, which is responsible for Fusarium head blight (FHB) (10), produces a range of secondary metabolites in the infected host plant that are harmful to human health when they are ingested and potentially when they are inhaled (11). Moreover, Alternaria, Cladosporium, and Epicoccum are well known for their allergenic properties (7). Although the drivers of wheat phytopathogen infection, such as climatic factors, wheat cultivar, and cropping history, have been intensively studied (12), little is known about the factors that shape the structure of entire fungal communities (e.g., pesticides) (13), and even less is known about fungal diversity in airborne dust generated during wheat harvesting. Thus, currently the homogeneity of the exposure of grain workers to fungal aerosols from one field to another during their work is difficult to estimate. There is a need to identify the factors that shape the fungal community composition that is aerosolized from wheat dust to better characterize the exposure of grain workers.
The structures of fungal communities in aboveground ecosystems can be shaped by various drivers. One known selective filter is the plant. Major differences in aboveground fungal community composition have been noticed between plant species (14) and genotypes (15, 16). Another selective filter is the environment. Gradients of elevation (17), temperature, and rainfall (18) as well as variations in soil nutrient content (19) have all been associated with changes in aboveground fungal assemblage. In temperate regions, even seasonal thermal fluctuations have been shown to affect the dynamics of plant pathogens (20, 21). In landscapes disturbed by agricultural practice, an additional selective filter acts on aboveground fungal communities: the fungicide treatments (13, 22). However, no studies have simultaneously investigated how farming system, environmental variables, and plant cultivars affect the composition of fungal communities along ecological gradients and what are the consequences of these eventual changes on fungal particles aerosolized during plant handling. Thus, identifying the determinants of the fungal community structure will help us propose efficient preventive measures for the harvesters who are exposed to it.
The aims of this study were the following: (i) to identify the abiotic (e.g., climate and geographic parameters) and biotic (e.g., plant cultivar and farming system) drivers that might shape the fungal community structure in grain dust; (ii) to determine the similarity between the fungal community structure in grain dust and that in the aerosols released during harvesting; and (iii) to identify the allergenic and mycotoxigenic species to which grain workers are usually exposed during the harvesting activities. We used 454 sequencing of amplicons from the fungal internal transcribed spacer 1 (ITS1) of the ribosomal DNA to determine the composition of fungal communities associated with wheat grain dust and with aerosols collected during harvest from farms that are managed organically, extensively, and conventionally. Then, we used a combination of multivariate statistical approaches, including model selection procedures (23, 24) and multivariate analyses (25, 26), which have been used for decades to study variation among communities of macroorganisms and, in recent years, among communities of microorganisms (27, 28), to study assemblages of fungal species.
MATERIALS AND METHODS
Study site and sampling design.
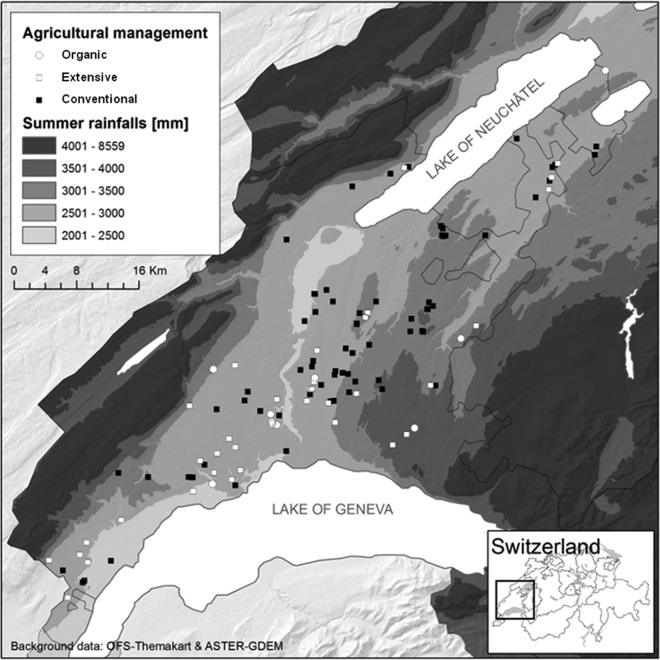
This study was conducted in the grain-growing region of Vaud in Switzerland, which provides approximately 25% of Switzerland's internal consumption of wheat and covers approximately 560 km2 (Fig. 1). In this region, the elevation ranges from 300 to 800 m, and the annual mean air temperature and precipitation are quite variable (Fig. 1). Interestingly, in this geographic region there is no correlation between the elevation and the number of freezing days although the elevation and the annual or summer precipitation are positively correlated (see Fig. S1 in the supplemental material). Wheat is produced in one of the following farming systems: conventional, extensive, or organic farming. The differences among these three farming systems are described in Table 1. Note that no fungicides are applied on the fields in the extensive and organic farming systems and that all fields were lowed prior to seeding. In the conventional farming system, the fungicide applications occurred at BBCH (Biologische Bundesanstalt, Bundessortenamt, and chemical industry) wheat growth stages 30 to 32 and 45 to 51. Detailed information on the fungicide applications in each field is provided in Table S1 in the supplemental material. The grain is ready to be harvested when it loses its moisture at the end of the ripening stage. During this last stage, the plant undergoes senescence and finally dies.
FIG 1.
Study area in the grain-growing region, the canton of Vaud, Switzerland. The dots correspond to the sampled wheat fields. Each type of dot corresponds to a wheat field that was managed by a distinct farming system: white circles represent organic farming, white squares represent extensive farming, and black squares represent conventional farming. The background represents the summer rainfall level. Elevation and rainfall were strongly correlated.
TABLE 1.
Description of the farming systems used to cultivate wheat in the study area
| Farming system | Tillage | Fertilizer type | Herbicide | Insecticide | Fungicide | Plant growth regulator | N input type | Soil pH correction |
|---|---|---|---|---|---|---|---|---|
| Organic | Yes | Organic | No | No | No | No | Organic | No |
| Extensive | Yes | Petrochemical | Yes | No | No | No | Mineral | No |
| Conventional | Yes | Petrochemical | Yes | Yes | Yes | Yes | Mineral | Yes |
Samples were collected from 98 fields that were cultured with winter wheat (67 conventional, 24 extensive, and 7 organic fields) (Fig. 1), 3 with triticale (2 conventional fields and 1 organic field), and 2 with spelt wheat (both, organic fields) at the full ripening stage (BBCH stages 95 to 100) during the harvesting process between 15 July and 7 August 2010. The fields were chosen randomly and stratified by the elevation gradient (29). Overall, the samples represent the diversity of farming practices of the canton of Vaud with respect to wheat cultivars, wheat species (winter wheat, spelt wheat, and triticale), soil types, fertilization, and fungicide treatments.
The sampling consisted of collecting aerosols, grains, and soil from each field site during the harvesting process. Aerosols were collected on a gelatin filter at a flow rate of 2.0 liters min−1 for 70 min on average (minimum, 11 min; maximum, 400 min) (see Table S2 in the supplemental material) using a pocket pump (MSA Escort Elf [Mine Safety Appliance Company, Pittsburgh, PA, USA] or SKC pocket pump 210-1002 [SKC, Inc., USA]) and clear styrene cassettes with three sections (25-mm diameter; SKC, Inc.). The pump was placed on the harvester 3 m above the soil surface. The temperature, barometric pressure, and relative air humidity were measured at each sampling site with a thermo-hygrometer and barometer (PCE-THB 40; PCE Group Iberica). Wind intensity was estimated as absent, low, middle, or strong (see Table S2). A 1-kg sample of harvested grain was collected from the bin. Ten soil cores were sampled from the organo-mineral horizon (10 cm deep) every 20 m. The gelatin filters and grain and soil samples were kept at 4°C during transportation back to the lab. Once in the lab, the aerosol samples (the gelatin filters) were stored at 4°C for 12 h, and the grain samples were frozen at −20°C until DNA analyses could be performed. The soil samples from each field were processed immediately.
Collection of climatic variables and farming practices.
Two groups of environmental variables were used in this study: climatic and soil variables (see Table S3 in the supplemental material). Climatic variables and wind speed and direction were recorded by the Swiss network of meteorological stations and interpolated using a digital elevation model at 25-m resolution (30, 31). The location of the weather stations, monthly mean precipitation, and mean temperature in June, July, and August 2010 are illustrated in Fig. S2 in the supplemental material. The data can be found at the Swiss office of meteorology and climatology, MeteoSwiss (http://www.meteoswiss.admin.ch/). For describing the average monthly time courses of temperature and precipitation and the number of freezing days across Vaud, all of the stations that delivered data during the period from 1961 to 2010 covering the entire studied region were selected.
Farming practices, including the wheat cultivar, the cropping history over the last 5 years, tillage, fertilization, and fungicide treatments (Table 1), were recorded directly from each farmer according to a standardized questionnaire. Data on the incidence of wheat diseases in 2010 in the wheat fields of Vaud, such as FHB (percentage of symptomatic heads), powdery mildew, rust, septoria, or eyespot (percentage of leaves), came from a cereal disease monitoring program coordinated by the phytosanitary office of the canton of Vaud (A. Zimmermann, personal communication). The severity of FHB was estimated by the visual inspection of 200 grains from each field after harvesting.
Generation of soil chemical variables.
The soil chemical variables were determined from the 10 soil cores sampled from each field that were pooled to smooth out the intrafield variation. The soil samples were dried in an oven at 40°C for 48 h, ground, and sieved through a 2-mm-pore-size sieve. The pH was measured with an automatic system in a solution containing 20 g of dry soil and 50 ml of water. The total nitrogen (Ntot) and carbon (Ctot) contents were extracted with sulfuric acid according to the Kjeldahl method. The organic carbon content (Corg) was estimated by removing the amount of mineral carbon that was derived by calcimetric analyses. The total phosphorus (Ptot) and available phosphorus (Pa) contents were determined using the ignition method developed by Saunders and Williams (32), which has been shown to be fairly reliable for weakly weathered soils (33). All of the soil analyses were performed by Sol-Conseil (Nyon, Switzerland).
DNA extraction, PCR, and 454 pyrosequencing.
Wheat grain dust was obtained from 100 g of grain by shaking the grain with 50 ml of 0.9% NaCl for 1 min and by centrifuging the liquid phase for 30 min at 8,500 × g. The gelatin filters were shaken at room temperature for 15 min in 6 ml of 0.9% NaCl before centrifugation under conditions identical to those used for grain dust. The recovered pellet from both types of samples was mechanically disrupted with a Tissue Lyser (Qiagen) in the first buffer of a FastDNA Spin Kit for Soil (MP Biomedicals, Switzerland). Then, the total DNA was extracted according to the manufacturer's instructions.
The internal transcribed spacer 1 (ITS1) region was amplified on a Biometra PCR thermal cycler using the forward primer ITS1F (34) and the reverse primer ITS2 (35). The 454 A adapter and a 10-bp molecular identification (MID) tag were appended to the 5′ end of the ITS1F primer to allow for sample multiplexing as described in Pellissier et al. (28). The 454 B adapter sequence was added at the 5′ end of the ITS2 primer. PCR was performed in 50-μl reaction mixtures with 50 ng of DNA, 0.2 μM each primer, 200 μl of each nucleotide, 1 U of peqGOLD Taq DNA Polymerase (PeqLab, Switzerland), 1× PCR buffer S, and 1× Enhancer solution P. The PCR conditions were as follows: denaturation at 94°C for 3 min, followed by 25 amplification cycles of 30 s at 94°C, 35 s at 50°C, and 90 s at 72°C, with a 7-min final extension at 72°C. The PCR products were visualized using gel electrophoresis. Ninety-six grain dust samples (68 from conventional, 20 from extensive, and 8 from organic farming) and 76 airborne samples (52 from conventional, 16 from extensive, and 8 from organic farming) were successfully amplified. The amplicons were cleaned using a QIAquick PCR Purification kit (Qiagen, Switzerland), quantified using a Quant-iT PicoGreen dsDNA Assay kit (Life Technologies, Switzerland), and pooled in equimolar concentrations. The sequencing was performed on 5/16 of a plate on a GS FLX Titanium sequencer (Life Sciences/Roche Applied Biosystems, Switzerland) by Microsynth, Switzerland.
Bioinformatics analyses.
The raw reads were sorted by MID sequence. Sequences with no mismatch within the MID and the ITS2 primer sequence were retrieved and then denoised with a Denoiser 0.851 (36). The highly variable ITS1 region was extracted with an ITS extractor program (37). The sequences shorter than 100 bp were removed. The cleaned sequences were grouped into operational taxonomic units (OTUs) using the UCLUST algorithm (38) implemented in the pick_otus function of QIIME (39) at a 97% sequence similarity cutoff. The OTUs were given taxonomic assignment in QIIME based on a previously published sequence database (28) modified for QIIME compatibility. When the taxonomy assigned was ambiguous, DBC454, a clustering tool which allows the identification of sequences not correctly assigned in GenBank, was used to resolve the ambiguity (40).
Only nonsingleton OTUs were retained for further analysis. Nonphylogenetic diversity estimates using Chao1 and observed species numbers for alpha diversity and Bray-Curtis for beta diversity calculations were carried out. Alpha diversity matrices based on observed species at the sample level were used to produce rarefaction curves using EstimateS, version 9.1.0 (41, 42). Good's estimator of coverage was calculated in QIIME.
Alpha diversity of fungal assemblages.
Two measures of alpha diversity, the taxonomic and the phylogenetic alpha diversity, were calculated and correlated with the biotic, abiotic, and management variables mentioned in Table S3 in the supplemental material. Each of these two measures accounts for the abundance of the nonsingleton OTUs assigned as fungi. The Shannon diversity index was used as a measure of taxonomic alpha diversity. The net relatedness index (NRI) implemented in the Picante, version 1.6, package in R (43), together with a tree of the OTUs generated on the basis of known phylogenetic relationships between taxonomically assigned OTUs (44), was used as a measure of the phylogenetic alpha diversity. The NRI is a standardized effect size (SES) of the observed mean pairwise phylogenetic distance (MPD) of all of the species in a community. Null models were built by reshuffling the tip label on the fungal phylogeny in a way that preserved the important property over the study area (e.g., species prevalence and species richness).
The correlations between the taxonomic diversity (Shannon) and phylogenetic (NRI) alpha diversities and different abiotic, biotic, and management variables were assessed by a multiple ordinary least-squares (OLS) regression model that included all of these variables. Based on the full OLS regression model, information-theoretic-based model selection was undertaken by comparing all of the competing models, including all of the possible combinations of variables. To do so, we used the Akaike information criterion (AIC) to quantify the support for each model (23). The selection of the minimum adequate model (MAM) was based on the lowest AIC value. From all of the subset models, we calculated the Akaike weight (w), which can be interpreted as the probability that a specific model is the best (23). We then estimated the relative importance of each predictor in explaining the fungal species richness by summing the Akaike weight values across all of the models that included the predictor variable (23, 24, 45). These summed Akaike weights (weighted AIC, or WAIC) ranged from 0 to 1, thereby providing a means for ranking the predictor variables in terms of information content. These analyses were performed using the package MuMIn in R (23).
Beta diversity of fungal assemblages.
As a measure of taxonomic beta diversity, we calculated the pairwise variation in composition using the Jaccard dissimilarity index with the vegdist function implemented in the Vegan R package (46). To determine the fungal phylogenetic beta diversity, the mean pairwise distance was calculated using the comdist function in the Picante, version 1.6, package (43). The standardized effect size of the mean phylogenetic distance across communities was computed in the same way used for the NRI by calculating a standardized effect size, as described by Swenson (47) and Pellissier et al. (48). Finally, the two beta diversities, taxonomic and phylogenetic, were correlated with the variation in abiotic and biotic variables and with the differences in farming systems (Table 2). The significance was tested using Mantel tests and 9,999 permutations.
TABLE 2.
Factors influencing the fungal alpha diversity described by the Shannon index or phylogenetic diversity (NRI)a
| Diversity measure and factor (unit) | R2 | Estimate | P valueb | WAIC |
|---|---|---|---|---|
| Shannon index (R2 = 0.31) | ||||
| CaCO3 (mg kg−1) | 0.13 | −0.3225 | ** | 1 |
| Elevation (m) | 0.04 | 0.6272 | ** | 0.8 |
| Annual rainfall (mm) | 0.03 | −0.6056 | * | 0.74 |
| No. of freezing days/year | 0.03 | −0.2477 | * | 0.51 |
| Corg (mg kg−1) | 0.02 | 0.1529 | 0.4 | |
| pH | 0.03 | 0.2443 | 0.22 | |
| Pa (mg kg−1) | 0.06 | −0.2367 | 0.15 | |
| K (mg kg−1) | 0.02 | 0.1844 | 0.07 | |
| Phylogenetic diversity (R2 = 0.32) | ||||
| Extensive vs organic system | 0.27 | −0.2682 | ** | 1 |
| Extensive vs conventional system | 0.2719 | ** | ||
| Annual rainfall (mm) | 0.07 | −0.1953 | * | 0.54 |
The table shows the coefficient of determination (R2) and the standardized regression coefficients estimated from ordinary least-squares multiple regression (OLS) of the minimum adequate model (MAM) with the lowest AIC. The relative importance of each predictor variable was assessed using the summed Akaike weights (WAIC).
*, P < 0.05; **, P < 0.01.
Comparison between grain and airborne fungal communities.
To estimate the similarity between grain dust and aerosol fungal communities, the prevalence of nonsingleton OTUs in the grain dust and aerosols was determined. Because the number of successfully sequenced samples was uneven between the grain and airborne samples, only the fields from which both the grain dust and airborne fungal communities were obtained were retained for further analysis. A coinertia analysis was run using the function coinertia implemented in the ade4 R package (49) to test for the shared structures between the grain dust and the aerosol fungal compositions from each field. The significance of the correlation was tested using a randomization test implemented in the Vegan R package. The null hypothesis was that species are present in the air according to their prevalences on the grain (i.e., rare species on the plant are found rarely in the air, whereas frequent species on the plant are found frequently in the air). This relationship should be close to a 1:1 relationship. A Wilcoxon test was performed to assess differences in the number of OTUs between the different farming systems. Linear regressions using the “lm” function in R were employed to calculate the relationship between the relative abundances of Fusarium OTUs and the average score of FHB severity on grain.
Nucleotide sequence accession numbers.
The raw sequence data are available at the NCBI under BioProject accession number PRJNA240301, and the representative OTUs have been deposited in GenBank under accession numbers KT886526 to KT886841.
RESULTS
Diversity of grain and aerosol fungal communities.
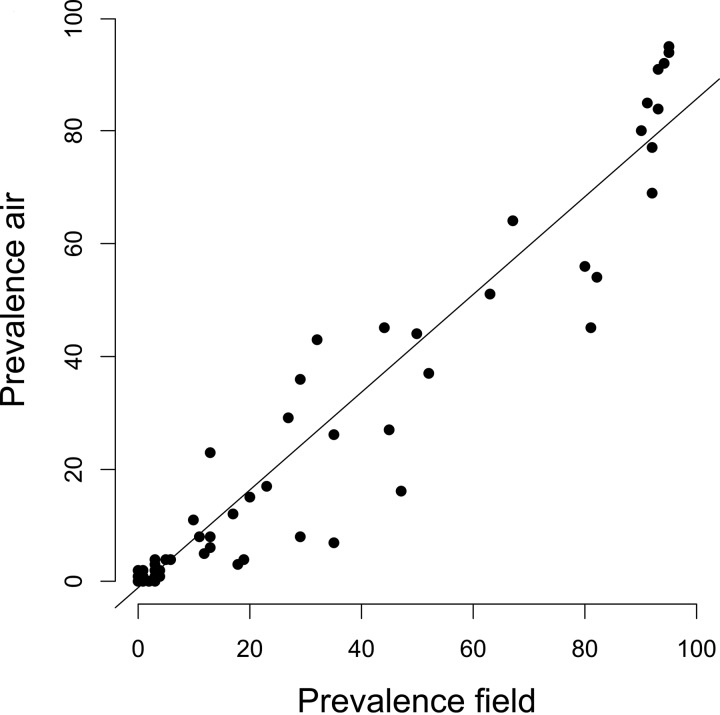
We assembled an extensive spatial data set of the fungi present in 96 grain dust samples and 76 aerosols released during the harvesting process of the 96 fields. These fields were spread across a region of 560 km2 and along an elevation gradient of 500 m (Fig. 1). Twenty-two different winter wheat cultivars, the triticale, and the spelt wheat were represented. After quality filtering, denoising, the removal of chimera and short sequences, and the extraction of the ITS1 variable region, a total of 115,777 sequences were retained. The average read length was 281 bp. The average number of sequences in each grain dust sample was 933 ± 390, and the average number of sequences in each airborne sample was 708 ± 274 (means ± standard deviations [SD]). Rarefaction and species accumulation curves indicated that the sequencing depth was sufficient to cover the fungal diversity. Good's coverage indicated that we recovered most of the expected OTUs for all of the air and grain dust samples (median ± SD for grain samples, 95.5% ± 1.1%; median for aerosol samples, 95.5% ± 1.9%) (see Table S4 in the supplemental material). The number of OTUs detected in the grain dust samples was significantly higher than that within the aerosol samples, independent of the farming system (median numbers of OTUs that were found in the grain samples were 30.5, 30.0, and 32.0, and in the aerosol samples the median numbers of OTUS were 19.0, 20.5, and 22.0 in the fields that were managed following organic, extensive, or conventional protocols, respectively; Wilcoxon test, W = 2,533; P < 0.0001 for the Shannon overall diversity indices between the grain dust and aerosol samples) (see Table S4). Despite this difference, the fungal communities associated with the grain and those found in the associated aerosols shared significant assemblage structures, as shown by the coinertia analysis (coinertia coefficient [RV] = 0.17; P < 0.0001). Moreover, the abundance of the fungal OTUs in the grain dust was highly correlated with that in aerosols (R2 = 0.99, P < 0.0001) (Fig. 2). Thus, the abundant OTUs in the grain dust samples were also abundant in the aerosol samples, whereas the OTUs that were scarcely represented in the grain dust were also rare in the aerosol samples.
FIG 2.
Correlation between the OTU prevalence in the grain and aerosols that were released during the harvesting of the corresponding fields. All of the OTUs appear to be aerosolized in the same proportion as their prevalence on the grain.
Taxonomic composition of wheat-associated fungal community.
In total, 319 OTUs were obtained, of which 197 OTUs were identified to the species or genus level as fungi (see Table S5 in the supplemental material). The majority of the remaining OTUs corresponded to fungal species that were not isolated by culture, namely, uncultured fungi in GenBank (116 OTUs), with only one being identified as a nonfungus, a plant species (Pleuropterus multiflorus). However, for five OTUs, we found no similarity with any GenBank sequence. Concerning the representativeness of these OTUs in the data set, 43 OTUs were singletons, 75 OTUs were doubletons, and 78 OTUs were represented by more than 20 sequences. The abundant OTUs were more easily assigned than the rare ones. Therefore, only 37% of the OTUs represented by 2 to 19 sequences could be assigned, whereas 78.5% of those with more than 20 sequences were identified at the genus or species level.
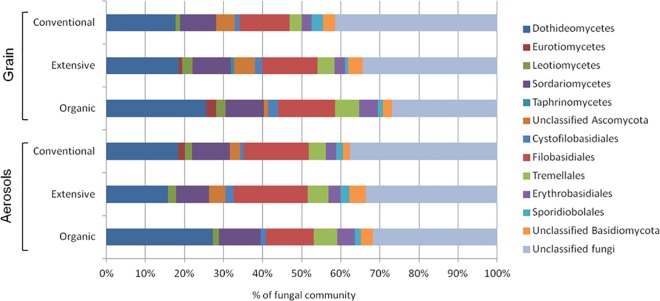
Overall, pyrosequencing revealed 37 different fungal genera that were assigned to the phylum Ascomycota (59% of the OTUs) or Basidiomycota (41% of the OTUs). Of the 37 fungal genera, 30 corresponded to 13 different orders of Ascomycota, and 7 corresponded to five orders of Basidiomycota, encompassing primarily phytopathogenic (44%) and saprobe (40%) fungal species (see Table S5 in the supplemental material). However, two fungal classes, the Tremellomycetes and the Dothideomycetes, contributed ∼60% of the taxon richness. The most frequent species belonged to Pleosporales, Capnodiales, Hypocreales, Filobasidiales, and Erythrobasidiales (Fig. 3). Among them, several allergenic species, members of the genera Epicoccum, Alternaria, and Cladosporium, were present in almost all of the grain dust and aerosol samples. Toxigenic species (members of Fusarium) were also frequently detected in more than 81% of the grain dust samples and in 43 to 61% of the aerosol samples (Table 3; see also Table S6 in the supplemental material). The relative abundance of the most frequent Fusarium species in the grain dust and aerosol samples was found to significantly correlate with the severity of FHB disease (correlation coefficient of 0.029 and P value of 0.000 for grain dust and correlation coefficient of 0.386 and P value of 0.000 for aerosol). The low frequency and abundance of the other wheat pathogens Blumeria graminis, Puccinia spp., Mycosphaerella graminicola, Phaeosphaeria avenaria f. sp. triticae, Phaeosphaeria nodorum, and Pyrenophora tritici-repentis in the 454 data set was confirmed by the low incidence of the corresponding diseases in wheat in the year of the sampling (unpublished data).
FIG 3.
Fungal community composition in wheat grain dust and in aerosols at the order level depending on the farming systems: conventional, extensive, and organic. The data are represented as the percentage of OTUs affiliated to one particular order that was detected in the samples taken from each farming system.
TABLE 3.
The relative abundances of the most frequent OTUs in the grain and aerosol samples, depending on the farming system
| Taxonomic assignmenta | GenBank accession no. | Frequency (%) by sample type and farm system |
Mean relative abundance (%) by sample type and farm system |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grain sample |
Aerosol sample |
Grain sample |
Aerosol sample |
||||||||||
| Conventional | Extensive | Organic | Conventional | Extensive | Organic | Conventional | Extensive | Organic | Conventional | Extensive | Organic | ||
| Epicoccum nigrum | KT886532 | 100 | 100 | 100 | 100 | 100 | 100 | 56.4 | 57.3 | 54.2 | 60.8 | 60.3 | 63.2 |
| Passalora robiniae | KT886526 | 100 | 100 | 100 | 100 | 100 | 100 | 14.9 | 12.8 | 20.6 | 20.5 | 17.3 | 20.1 |
| Alternaria ethzedia | KT886531 | 100 | 100 | 100 | 100 | 100 | 100 | 8.5 | 8.2 | 3.6 | 5.5 | 4.8 | 4.3 |
| Cryptococcus victoriae | KT886536 | 100 | 100 | 100 | 98 | 94 | 100 | 3.3 | 1.8 | 2.3 | 3.4 | 2.7 | 2 |
| Cladosporium cladosporioides | KT886527 | 99 | 100 | 100 | 98 | 100 | 100 | 2.4 | 2.4 | 4 | 1.7 | 2 | 3.1 |
| Didymella exitialis | KT886541 | 97 | 100 | 100 | 83 | 100 | 100 | 1.8 | 2.6 | 1.2 | 1.1 | 1.7 | 1 |
| Cryptococcus sp. | KT886538 | 91 | 100 | 100 | 64 | 83 | 50 | 0.7 | 0.8 | 0.5 | 0.5 | 1 | 0.3 |
| Alternaria alternata | KT886534 | 94 | 96 | 88 | 85 | 83 | 100 | 1.4 | 1 | 1.5 | 0.7 | 0.6 | 1.8 |
| Alternaria infectoria | KT886761 | 91 | 89 | 88 | 53 | 61 | 63 | 0.8 | 0.8 | 0.5 | 0.4 | 0.3 | 0.2 |
| Fusarium graminearum | KT886533 | 90 | 89 | 63 | 51 | 61 | 0 | 2.2 | 3.9 | 0.9 | 0.6 | 0.4 | 0 |
| Mycosphaerella graminicola | KT886549 | 75 | 89 | 25 | 47 | 94 | 25 | 0.4 | 1.5 | 0.4 | 0.3 | 1.3 | 0.1 |
| Fusarium poae | KT886562 | 91 | 81 | 75 | 43 | 50 | 0 | 0.9 | 0.5 | 0.4 | 0.2 | 0.2 | 0 |
| Gibberella sp. | KT886557 | 84 | 78 | 63 | 34 | 56 | 25 | 1.1 | 0.5 | 0.5 | 0.3 | 0.2 | 0 |
| Sporobolomyces roseus | KT886529 | 90 | 74 | 100 | 81 | 67 | 75 | 0.7 | 0.6 | 1.9 | 0.7 | 1.5 | 0.7 |
| Aureobasidium pullulans | KT886530 | 59 | 74 | 88 | 42 | 61 | 25 | 0.5 | 1 | 1.8 | 0.1 | 0.2 | 0 |
| Microdochium nivale | KT886558 | 68 | 70 | 38 | 64 | 89 | 50 | 0.4 | 0.7 | 0.1 | 0.4 | 1.2 | 0.1 |
| Cryptococcus wieringae | KT886535 | 48 | 63 | 88 | 42 | 50 | 63 | 0.4 | 0.2 | 1.1 | 0.2 | 0.7 | 0.3 |
In bold are the known allergenic or toxigenic species.
The majority of the OTUs assigned as fungi were shared between grain and aerosol samples (73% of nonsingleton OTUs; 96.8% of the OTUs were represented by at least 20 reads) (Table 3; see also Table S6 in the supplemental material). Fifty nonsingleton OTUs were detected only in grain samples, and seven OTUs were isolated only from aerosol samples (see Table S6).
Alpha diversity of the fungal communities.
We considered only the 197 assigned OTUs to calculate the taxonomic and phylogenetic diversities. These two diversity indexes were found to correlate significantly with different abiotic and biotic interfering variables at the local scale (alpha diversity) or among habitats (beta diversity).
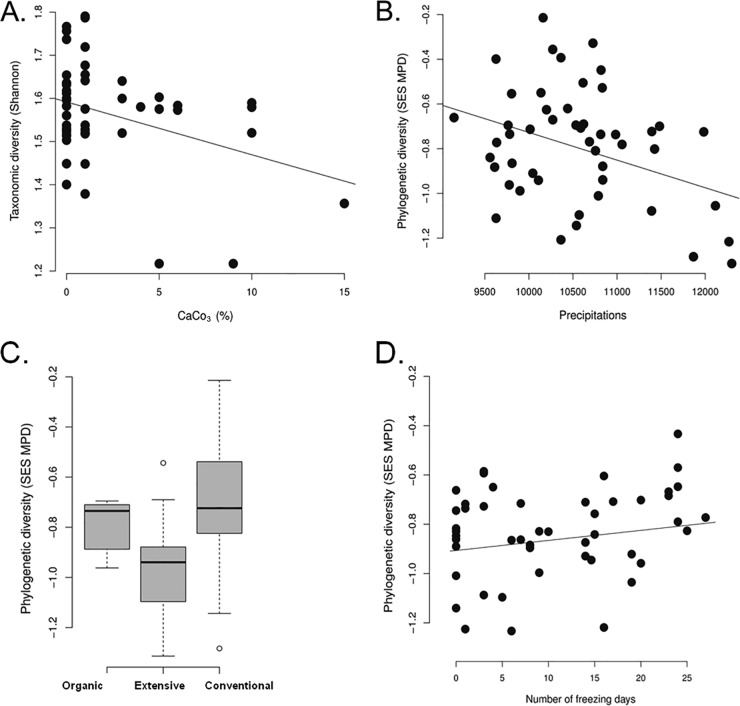
The Shannon alpha diversity of fungal communities was found to be negatively related to one geographic variable (elevation), two climatic variables (annual rainfall and the number of freezing days per year), and one soil variable (the amount of CaCO3). In contrast, the phylogenetic alpha diversity of fungal communities was significantly associated only with the farming systems and the annual rainfall (Table 2 and Fig. 4). Thus, the diversity slightly increased in the samples that originated from organic farming essentially because of an increase in the proportion of the saprobe and epiphyte species, such as Aureobasidium pullulans.
FIG 4.
Examples of correlations between facets of fungal diversity and major predictors described in Tables 2 and 4. (A) The negative correlation between soil CaCO3 and alpha taxonomic diversity. (B) The negative correlation between annual precipitation (mm) and alpha phylogenetic diversity. (C) Differences between the three farming systems in the beta phylogenetic diversity of wheat-related fungi. (D) Positive correlation between the beta phylogenetic diversity and the number of freezing days.
Beta diversity of the fungal communities.
The taxonomic beta diversity of the fungal communities differed between the wheat cultivars, depended on the cropping history, and was found to be significantly correlated with summer rainfall, the number of freezing days/year, and elevation (Table 4). The phylogenetic composition was significantly related only to the farming system and to the number of freezing days per year (Table 4 and Fig. 4). However, although the frequencies of some allergenic species such as Aureobasidium pullulans differed between the farming systems, no significant difference in the frequencies of other allergenic (Alternaria, Cladosporium, and Epicoccum) and mycotoxigenic (Fusarium) species depending on the farming system was found (Table 3).
TABLE 4.
Results of the Mantel test correlating fungal taxonomic (Jaccard) and phylogenetic (MPD) beta diversity and various predictor variables
| Diversity measure and factor (unit) | Mantel r | P valuea |
|---|---|---|
| Taxonomic diversity | ||
| No. of freezing days/yr | 0.23 | *** |
| Wheat cultivar | 0.23 | *** |
| Cropping history | 0.21 | *** |
| Summer rainfall (mm) | 0.11 | * |
| Elevation (m) | 0.07 | * |
| pH | 0.07 | 0.07 |
| Geographic distance | 0.06 | 0.1 |
| CaCO3 (mg kg−1) | 0.05 | 0.19 |
| Mg (mg kg−1) | 0.04 | 0.19 |
| Pa (mg kg−1) | 0.03 | 0.3 |
| Ctot/Ptot ratio | 0.03 | 0.3 |
| Ctot/Ntot ratio | 0.03 | 0.31 |
| Ntot/Ptot ratio | 0.03 | 0.34 |
| Ntot (mg kg−1) | 0.02 | 0.4 |
| Corg (%) | 0.02 | 0.38 |
| K (mg kg−1) | 0.02 | 0.41 |
| Temp (°C) | 0.02 | 0.31 |
| Annual rainfall (mm) | 0.02 | 0.31 |
| Farming system | 0.02 | 0.44 |
| Ptot (mg kg−1) | 0.001 | 0.51 |
| Phylogenetic diversity | ||
| Farming system | 0.17 | ** |
| No. of freezing days/yr | 0.16 | * |
| Ntot | 0.12 | 0.05 |
| CaCO3 (mg kg−1) | 0.11 | 0.06 |
| Ctot/Ptot ratio | 0.11 | 0.07 |
| Corg (%) | 0.11 | 0.06 |
| Ntot/Ptot ratio | 0.08 | 0.1 |
| Annual rainfall (mm) | 0.08 | 0.09 |
| K (mg kg−1) | 0.06 | 0.19 |
| Summer rainfall (mm) | 0.05 | 0.15 |
| Elevation (m) | 0.04 | 0.21 |
| Geographic distance | 0.04 | 0.22 |
| Ptot | 0.04 | 0.2 |
| Cropping history | 0.04 | 0.26 |
| Ctot/Ntot ratio | 0.03 | 0.24 |
| pH | 0.02 | 0.39 |
| Wheat cultivar | 0.01 | 0.29 |
| Mg (mg kg−1) | 0.008 | 0.52 |
| Temp (°C) | 0.006 | 0.49 |
| Pa (mg kg−1) | 0.001 | 0.5 |
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
Note that in the study area, elevation was positively related to summer precipitation (R2 = 0.78) but not to the number of freezing days. Thus, summer precipitation, and not elevation, is likely to be the most proximal variable affecting fungal communities.
DISCUSSION
In this study including 96 grain and 76 aerosols samples, we characterized the mycobiome to which the grain workers are exposed during wheat harvesting, and we simultaneously examined how farming system, environmental variables, and plant cultivars affect the composition of fungal communities along ecological gradients. Our results show a relatively homogenous composition of fungal communities in wheat grain dust across the 96 fields despite the large geographic scale (560 km2). The strong relationship found between the airborne and grain dust fungal communities suggests that grain workers are exposed to a relatively homogenous composition of fungal species during wheat harvesting. However, some variation in the wheat-related fungal community composition and diversity emerged. The farming system was identified as the major driver of the alpha and beta phylogenetic diversity of fungi although cultivar, cropping history, and the number of freezing days per year were the major factors shaping the taxonomic beta diversity of fungal communities. In particular, the frequency of some allergenic species such as Aureobasidium pullulans was found to be different between the farming systems. However, no significant differences in the frequency of other allergenic (Alternaria, Cladosporium, and Epicoccum) or mycotoxigenic species between farming systems or cultivars were found. Only the degree of severity of FHB diseases might play a role in the level of Fusarium particles in the air.
The decrease in fungal community similarity with increasing geographic distance is one of the most discussed aspects in fungal community ecology. Our results showed that the fungal community in grain dust was relatively homogenous at distances as large as hundreds of kilometers. In concordance with this, Sapkota et al. (16) found that crop genotype at the species level and fungicide treatment had a much stronger effect than geographic distance on wheat fungal communities. The homogeneity of the wheat-associated fungi in grain dust has a direct impact on the fungal particles aerosolized during the harvesting of wheat. Indeed, the mechanical process of harvesting breaks the fungal mycelia that were present on the wheat plants into small fragments, forcefully releases fungal spores from foliage, grain, and straw, and creates a cloud of fungal fragments and spores in the air. Thus, independently of their spore size (Table 2), particles of fungi growing on the plant might be mechanically projected into the air. The overall high frequency of some species compared to that of others may actually result from differences in fungal strategies for colonizing wheat. After human perturbations of the domesticated plant ecosystem, the regional fungal species pool may consist of a restricted number of ubiquitous species that can cope with the frequent temporal variability in agricultural management.
Although the variation in fungal community diversity is low, this variation was sufficient to allow detection of the major factors that shape the taxonomic and phylogenetic diversity of the fungal community. The major selective filter on the phylogenetic diversity of the aboveground fungal community is the farming system. Indeed, the fungal phylogenetic diversity has been found to be statistically higher in organic than in extensive farming but lower in extensive farming than in a conventional system. Whereas conventional and extensive farming systems share the same wheat varieties, the difference in fungal phylogenetic diversity values between the two farming systems indicates that the local fungal communities of conventional fields contain fungal species that are more distantly related to one another than those contained in extensive fields. This finding suggests that closely related species compete more strongly or are under more distinct environmental constraints in the conventional farming system than in the extensive farming system (this concept is reviewed by Cavender-Bares et al. [50]). The major difference between these two farming systems that is expected to affect fungal community structure is the application of fungicides. Previous studies have presented evidence of the effects of fungicide treatment on fungal community structure (13, 16). Thus, we suggest that fungicide application might be one of the factors that differentiate the fungal community compositions between the extensive and conventional farming systems. Even more differences between the conventional and extensive systems are expected in conventional farms with a more intensive use of fungicides. Indeed, the reduced use of fungicides in the conventional farms the year of sampling is explained by the low disease pressure (A. Zimmermann, personal communication). Nevertheless, because the correlations between farming systems and fungal community phylogenetic diversity are only partial and because only some cultivars are shared between the extensive and conventional systems and hardly any between the organic system and the two others, we cannot exclude the role of wheat cultivars in shaping the diversity of the associated fungal community. A controlled experimental design would allow us to better disentangle the relative effect of each factor.
Although wheat cultivars and species were not identified in this study to act on the phylogenetic diversity of the fungal community, they were determined to be the major selective filter on the beta taxonomic diversity of this community. These results confirmed the major role played by plant genotype on the structure of aboveground fungal communities, as previously proposed for grain fungal communities (16) and for wheat fungal phytopathogens (51). Thus, the selection of wheat cultivars for their resistance to different wheat diseases affects the infection of wheat not only by the fungal phytopathogens but also by the other members of the fungal community. In addition, our results provide evidence that cropping history might influence both the diversity of fungal phytopathogens (51, 52) and the overall diversity of the fungal community. These findings are similar to those previously published by Nicolaisen et al. (9).
Climatic factors such as summer precipitation and the number of freezing days per year have also been identified in the present study to shape taxonomic fungal community diversity. Weather conditions during the postanthesis period have previously been shown to affect the composition of Fusarium communities in winter wheat grain, and rainfall during the flowering stage is a major determinant of grain infection by Fusarium (12, 51). Our results illustrate the role of climate on the overall taxonomic diversity of fungi. Thus, by favoring Fusarium infection, summer rainfall indirectly reduces overall fungal diversity. Moreover, the winter climatic conditions have been shown to affect pathogen survival depending on host and pathogen life histories. Thus, fungal disease prevalence might decrease in years with more days of exposure to subzero temperature (e.g., for wheat, the Puccinia striiformis system [53], and for plantago, the Podosphaera system [20]). Although our results cannot directly show the impact of the number of freezing days per year on pathogen survival, they nonetheless suggest such an impact by the decrease in species diversity with the number of freezing days. Concerning the impact of soil parameters, only CaCO3 was shown to impact the fungal community structure, which suggests a limited exchange between soil and plant-associated fungi or a limited indirect effect on plant-associated fungi through the effect of soil characteristics on the plant.
The similarity of the fungal communities in grain dust and in the associated aerosols is an important finding for the occupational exposure sciences. Thus, grain dust samples can be used in place of air samples to predict the fungal species present in aerosol during harvesting. This information could be useful for future epidemiological studies on respiratory health in grain workers. The frequent detection of several allergenic (genera Epicoccum, Alternaria, and Cladosporium) and mycotoxigenic (genus Fusarium) species highlights the species that might be monitored in such epidemiological studies. Nevertheless, particular attention should be given to members of the genus Fusarium which are known to produce highly toxigenic secondary metabolites for human health, such as deoxynivalenol, nivalenol, and fusarenon X, and whose relative abundance in aerosols furthermore increases with the severity of FHB in grain. These allergenic and mycotoxigenic species were frequently detected not only in the aerosol samples of the present study in Switzerland (∼50% or more samples) but also in Danish (9) and Argentinian (8) wheat grain samples. The consistency of our results with those obtained in these previous high-throughput sequencing (9) and culture-based studies (8) underlines the potential exposure of grain workers to allergenic and toxigenic fungal particles. Their frequent presence in aerosols calls for further research on their aggregate effect on the respiratory health of grain workers.
In conclusion, this study provides the most in-depth characterization to date of airborne fungi associated with a single fungal source (wheat harvest) along a large geographic gradient. Our multivariate analyses demonstrated relationships between fungal assemblages and environmental conditions that were similar to those classically observed for macroorganisms (48, 54, 55). Our results demonstrated that fungal diversity in the aerosols generated during this activity is quite homogenous in fungal composition in spite of geographic distance. The taxonomic composition of airborne fungi is highly correlated with that of fungi in grain dust. Both allergenic and mycotoxigenic fungal species, which were previously detected in grain, have been shown in this study to also be frequent in these aerosols. This is an important finding for occupational health scientists who seek to understand the etiology of respiratory diseases described in grain worker populations. In addition, our results answer ecological questions about the natural and human factors that drive the composition of aboveground fungal communities. They elucidated the effects of plant cultivar, climate, and farming systems on the diversity of the grain dust fungal community.
Supplementary Material
ACKNOWLEDGMENTS
We greatly appreciate the support of all of the grain workers who participated in this study. We also thank the two reviewers for their constructive comments, and we thank Laure Bernardet, Briseis Castella, Baptiste Jimenez, and Jeremie Projer for assistance in sample collection in the field, Celine Pattaroni for assistance in sample processing in the lab, and Horatio Herrera and Pierre-Alain Porchet for advice about air sampling.
This work was supported by the Swiss National Foundation grant PMPDP3-129027, an interdisciplinary grant from the Faculty of Biology and Medicine of the University of Lausanne (Switzerland), and Agence Nationale de Sécurité Sanitaire de l'Alimentation, de l'Environnement et du Travail (ANSES, France) grant 2011/1/087 to H.N.-H.
We declare that we have no conflicts of interest.
H.N.-H., A.O., F.M., and S.K. designed the study; H.N.-H. collected the data; D.S.-B. and G.M. processed the samples in the lab; L.P., A.H.H., and H.N.-H. conducted the analysis; H.N.-H. and L.P. wrote the manuscript, and the other authors contributed.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03336-15.
REFERENCES
- 1.Rask-Andersen A. 2011. Asthma increase among farmers: a 12-year follow-up. Ups J Med Sci 116:60–71. doi: 10.3109/03009734.2010.503287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pahwa P, Senthilselvan A, McDuffie HH, Dosman JA. 2003. Longitudinal decline in lung function measurements among Saskatchewan grain workers. Can Respir J 10:135–141. [DOI] [PubMed] [Google Scholar]
- 3.Jouneau S, Boche A, Brinchault G, Fekete K, Guillot S, Bayat S, Desrues B. 2012. On-site screening of farming-induced chronic obstructive pulmonary disease with the use of an electronic mini-spirometer: results of a pilot study in Brittany, France. Int Arch Occup Environ Health 85:623–630. doi: 10.1007/s00420-011-0708-6. [DOI] [PubMed] [Google Scholar]
- 4.Moira CYM, Enarson DA, Kennedy SM. 1992. The impact of grain dust on respiratory health. Am Rev Respir Dis 145:476–487. doi: 10.1164/ajrccm/145.2_Pt_1.476. [DOI] [PubMed] [Google Scholar]
- 5.Eduard W. 2009. Fungal spores: a critical review of the toxicological and epidemiological evidence as a basis for occupational exposure limit setting. Crit Rev Toxicol 39:799–864. doi: 10.3109/10408440903307333. [DOI] [PubMed] [Google Scholar]
- 6.Selman M, Lacasse Y, Pardo A, Cormier Y. 2010. Hypersensitivity pneumonitis caused by fungi. Proc Am Thorac Soc 7:229–236. doi: 10.1513/pats.200906-041AL. [DOI] [PubMed] [Google Scholar]
- 7.Denning DW, O'Driscoll BR, Hogaboam CM, Bowyer P, Niven RM. 2006. The link between fungi and severe asthma: a summary of the evidence. Eur Respir J 27:615–626. doi: 10.1183/09031936.06.00074705. [DOI] [PubMed] [Google Scholar]
- 8.Larran S, Perelló A, Simón M, Moreno V. 2007. The endophytic fungi from wheat (Triticum aestivum L.). World J Microbiol Biotechnol 23:565–572. doi: 10.1007/s11274-006-9266-6. [DOI] [Google Scholar]
- 9.Nicolaisen M, Justesen AF, Knorr K, Wang J, Pinnschmidt HO. 2014. Fungal communities in wheat grain show significant co-existence patterns among species. Fungal Ecol 11:145–153. doi: 10.1016/j.funeco.2014.06.002. [DOI] [Google Scholar]
- 10.Scherm B, Balmas V, Spanu F, Pani G, Delogu G, Pasquali M, Migheli Q. 2013. Fusarium culmorum: causal agent of foot and root rot and head blight on wheat. Mol Plant Pathol 14:323–341. doi: 10.1111/mpp.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vacher G, Ciarlo E, Savova-Bianchi D, Le Roy D, Hantier G, Niculita-Hirzel H, Roger T. 2015. Innate immune sensing of Fusarium culmorum by mouse dendritic cells. J Toxicol Environ Health A 78:871–885. doi: 10.1080/15287394.2015.1051201. [DOI] [PubMed] [Google Scholar]
- 12.Czaban J, Wroblewska B, Sulek A, Mikos M, Boguszewska E, Podolska G, Nieróbca A. 2015. Colonisation of winter wheat grain by Fusarium spp. and mycotoxin content as dependent on a wheat variety, crop rotation, a crop management system and weather conditions. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32:874–910. doi: 10.1080/19440049.2015.1019939. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson I, Friberg H, Steinberg C, Persson P. 2014. Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS One 9:e111786. doi: 10.1371/journal.pone.0111786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meiser A, Balint M, Schmitt I. 2014. Meta-analysis of deep-sequenced fungal communities indicates limited taxon sharing between studies and the presence of biogeographic patterns. New Phytol 201:623–635. doi: 10.1111/nph.12532. [DOI] [PubMed] [Google Scholar]
- 15.Bazzicalupo AL, Bálint M, Schmitt I. 2013. Comparison of ITS1 and ITS2 rDNA in 454 sequencing of hyperdiverse fungal communities. Fungal Ecol 6:102–109. doi: 10.1016/j.funeco.2012.09.003. [DOI] [Google Scholar]
- 16.Sapkota R, Knorr K, Jorgensen LN, O'Hanlon KA, Nicolaisen M. 2015. Host genotype is an important determinant of the cereal phyllosphere mycobiome. New Phytol 207:1134–1144. doi: 10.1111/nph.13418. [DOI] [PubMed] [Google Scholar]
- 17.Cordier T, Robin C, Capdevielle X, Fabreguettes O, Desprez-Loustau ML, Vacher C. 2012. The composition of phyllosphere fungal assemblages of European beech (Fagus sylvatica) varies significantly along an elevation gradient. New Phytol 196:510–519. doi: 10.1111/j.1469-8137.2012.04284.x. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerman NB, Vitousek PM. 2012. Fungal endophyte communities reflect environmental structuring across a Hawaiian landscape. Proc Natl Acad Sci U S A 109:13022–13027. doi: 10.1073/pnas.1209872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eschen R, Hunt S, Mykura C, Gange AC, Sutton BC. 2010. The foliar endophytic fungal community composition in Cirsium arvense is affected by mycorrhizal colonization and soil nutrient content. Fungal Biol 114:991–998. doi: 10.1016/j.funbio.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Penczykowski RM, Walker E, Soubeyrand S, Laine AL. 2015. Linking winter conditions to regional disease dynamics in a wild plant-pathogen metapopulation. New Phytol 205:1142–1152. doi: 10.1111/nph.13145. [DOI] [PubMed] [Google Scholar]
- 21.Suffert F, Ravigne V, Sache I. 2015. Seasonal changes drive short-term selection for fitness traits in the wheat pathogen Zymoseptoria tritici. Appl Environ Microbiol 81:6367–6379. doi: 10.1128/AEM.00529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulas C, Petsoulas C, Rousidou K, Perruchon C, Karas P, Karpouzas DG. 2013. Effects of systemic pesticides imidacloprid and metalaxyl on the phyllosphere of pepper plants. Biomed Res Int 2013:969750. doi: 10.1155/2013/969750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach, 2nd ed Springer, New York, NY. [Google Scholar]
- 24.Johnson JB, Omland KS. 2004. Model selection in ecology and evolution. Trends Ecol Evol 19:101–108. doi: 10.1016/j.tree.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 25.ter Braak CJF. 1986. Canonical correspondence analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67:1167–1179. doi: 10.2307/1938672. [DOI] [Google Scholar]
- 26.Lepš J, Šmilauer P. 2003. Multivariate analysis of ecological data using CANOCO. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 27.Buttigieg PL, Ramette A. 2014. A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol 90:543–550. doi: 10.1111/1574-6941.12437. [DOI] [PubMed] [Google Scholar]
- 28.Pellissier L, Niculita-Hirzel H, Dubuis A, Pagni M, Guex N, Ndiribe C, Salamin N, Xenarios I, Goudet J, Sanders IR, Guisan A. 2014. Soil fungal communities of grasslands are environmentally structured at a regional scale in the Alps. Mol Ecol 23:4274–4290. doi: 10.1111/mec.12854. [DOI] [PubMed] [Google Scholar]
- 29.Hirzel A, Guisan A. 2002. Which is the optimal sampling strategy for habitat suitability modelling. Ecol Modell 157:331–341. doi: 10.1016/S0304-3800(02)00203-X. [DOI] [Google Scholar]
- 30.Zimmermann NE, Kienast F. 1999. Predictive mapping of alpine grasslands in Switzerland: species versus community approach. J Veg Sci 10:469–482. doi: 10.2307/3237182. [DOI] [Google Scholar]
- 31.Zimmermann NE, Edwards TC, Moisen GG, Frescino TS, Blackard JA. 2007. Remote sensing-based predictors improve distribution models of rare, early successional and broadleaf tree species in Utah. J Appl Ecol 44:1057–1067. doi: 10.1111/j.1365-2664.2007.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saunders WMH, Williams EG. 1955. Observations on the determination of total organic phosphorus in soils. J Soil Sci 6:254–267. doi: 10.1111/j.1365-2389.1955.tb00849.x. [DOI] [Google Scholar]
- 33.Williams JDH, Syers JK, Walker TW, Rex RW. 1970. A comparison of methods for the determination of soil organic phosphorus. Soil Sci 110:13–18. doi: 10.1097/00010694-197007000-00003. [DOI] [Google Scholar]
- 34.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol 2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 35.White T, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis M, Gelfand D, Sninsky J, White T (ed), PCR protocols: a guide to methods and applications. Academic Press, New York, NY. [Google Scholar]
- 36.Reeder J, Knight R. 2010. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods 7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson RH, Veldre V, Hartmann M, Unterseher M, Amend A, Bergsten J, Kristiansson E, Ryberg M, Jumpponen A, Abarenkov K. 2010. An open source software package for automated extraction of ITS1 and ITS2 from fungal ITS sequences for use in high-throughput community assays and molecular ecology. Fungal Ecol 3:284–287. doi: 10.1016/j.funeco.2010.05.002. [DOI] [Google Scholar]
- 38.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagni M, Niculita-Hirzel H, Pellissier L, Dubuis A, Xenarios I, Guisan A, Sanders IR, Goudet J, Guex N. 2013. Density-based hierarchical clustering of pyro-sequences on a large scale—the case of fungal ITS1. Bioinformatics 29:1268–1274. doi: 10.1093/bioinformatics/btt149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao A, Chazdon RL, Colwell RK, Shen T-J. 2005. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol Lett 8:148–159. doi: 10.1111/j.1461-0248.2004.00707.x. [DOI] [Google Scholar]
- 42.Colwell RK, Chao A, Gotelli NJ, Lin S-Y, Mao CX, Chazdon RL, Longino JT. 2012. Models and estimators linking individual-based and sample-based rarefaction, extrapolation, and comparison of assemblages. J Plant Ecol 5:3–21. doi: 10.1093/jpe/rtr044. [DOI] [Google Scholar]
- 43.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. 2010. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 44.Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, Huhndorf S, James T, Kirk PM, Lücking R, Thorsten Lumbsch H, Lutzoni F, Matheny PB, McLaughlin DJ, Powell MJ, Redhead S, Schoch CL, Spatafora JW, Stalpers JA, Vilgalys R, Aime MC, Aptroot A, Bauer R, Begerow D, Benny GL, Castlebury LA, Crous PW, Dai YC, Gams W, Geiser DM, Griffith GW, Gueidan C, Hawksworth DL, Hestmark G, Hosaka K, Humber RA, Hyde KD, Ironside JE, Kõljalg U, Kurtzman CP, Larsson KH, Lichtwardt R, Longcore J, Miadlikowska J, Miller A, Moncalvo JM, Mozley-Standridge S, Oberwinkler F, Parmasto E, et al. 2007. A higher-level phylogenetic classification of the fungi. Mycol Res 111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Burnham KP, Anderson DR. 2004. Multimodel inference: understanding AIC and BIC in model selection. Sociol Methods Res 33:261–304. doi: 10.1177/0049124104268644. [DOI] [Google Scholar]
- 46.Oksanen J, Kindt R, Legendre P, O'Hara B, Stevens MHH, Oksanen MJ. 2007. Vegan: community ecology package, version 1.15-1. http://vegan.r-forge.r-project.org/.
- 47.Swenson NG. 2012. Phylogenetic analyses of ecological communities using DNA barcode data. Methods Mol Biol 858:409–419. doi: 10.1007/978-1-61779-591-6_20. [DOI] [PubMed] [Google Scholar]
- 48.Pellissier L, Ndiribe C, Dubuis A, Pradervand JN, Salamin N, Guisan A, Rasmann S. 2013. Turnover of plant lineages shapes herbivore phylogenetic beta diversity along ecological gradients. Ecol Lett 16:600–608. doi: 10.1111/ele.12083. [DOI] [PubMed] [Google Scholar]
- 49.Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. [Google Scholar]
- 50.Cavender-Bares J, Kozak KH, Fine PV, Kembel SW. 2009. The merging of community ecology and phylogenetic biology. Ecol Lett 12:693–715. doi: 10.1111/j.1461-0248.2009.01314.x. [DOI] [PubMed] [Google Scholar]
- 51.Bernhoft A, Torp M, Clasen PE, Loes AK, Kristoffersen AB. 2012. Influence of agronomic and climatic factors on Fusarium infestation and mycotoxin contamination of cereals in Norway. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 29:1129–1140. doi: 10.1080/19440049.2012.672476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogelgsang S, Hecker A, Musa T, Dorn B, Forrer H-R. 2011. On-farm experiments over 5 years in a grain maize/winter wheat rotation: effect of maize residue treatments on Fusarium graminearum infection and deoxynivalenol contamination in wheat. Mycotoxin Res 27:81–96. doi: 10.1007/s12550-010-0079-y. [DOI] [PubMed] [Google Scholar]
- 53.Sharma-Poudyal D, Chen X, Rupp RA. 2014. Potential oversummering and overwintering regions for the wheat stripe rust pathogen in the contiguous United States. Int J Biometeorol 58:987–997. doi: 10.1007/s00484-013-0683-6. [DOI] [PubMed] [Google Scholar]
- 54.Klimek S, Hofmann M, Isselstein J. 2007. Plant species richness and composition in managed grasslands: the relative importance of field management and environmental factors. Biol Conserv 134:559–570. doi: 10.1016/j.biocon.2006.09.007. [DOI] [Google Scholar]
- 55.Schaffers AP, Raemakers IP, Sýkora KV, Ter Braak CJ. 2008. Arthropod assemblages are best predicted by plant species composition. Ecology 89:782–794. doi: 10.1890/07-0361.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.