Abstract
Members of the marine Roseobacter clade are major participants in global carbon and sulfur cycles. While roseobacters are well represented in cultures, several abundant pelagic lineages, including SAG-O19, DC5-80-3, and NAC11-7, remain largely uncultivated and show evidence of genome streamlining. Here, we analyzed the partial genomes of three single cells affiliated with CHAB-I-5, another abundant but exclusively uncultivated Roseobacter lineage. Members of this lineage encode several metabolic potentials that are absent in streamlined genomes. Examples are quorum sensing and type VI secretion systems, which enable them to effectively interact with host and other bacteria. Further analysis of the CHAB-I-5 single-cell amplified genomes (SAGs) predicted that this lineage comprises members with relatively large genomes (4.1 to 4.4 Mbp) and a high fraction of noncoding DNA (10 to 12%), which is similar to what is observed in many cultured, nonstreamlined Roseobacter lineages. The four uncultured lineages, while exhibiting highly variable geographic distributions, together represent >60% of the global pelagic roseobacters. They are consistently enriched in genes encoding the capabilities of light harvesting, oxidation of “energy-rich” reduced sulfur compounds and methylated amines, uptake and catabolism of various carbohydrates and osmolytes, and consumption of abundant exudates from phytoplankton. These traits may define the global prevalence of the four lineages among marine bacterioplankton.
INTRODUCTION
Members of the marine Roseobacter clade play a prominent role in global carbon and sulfur cycles (1). They constitute a monophyletic Alphaproteobacteria lineage with a maximum divergence of 11% in their 16S rRNA gene sequences (1). Roseobacters are abundant in the pelagic oceans, representing up to 20% of the bacterial cells in coastal waters and 3 to 5% in open ocean waters (2–4). They also dominate the microbial communities associated with a variety of marine seaweeds and animals and often act as probiotics or pathogens and thus are an important component in marine conservation (5). Because of their large genome sizes, versatile metabolic pathways, and regulatory circuits, roseobacters have been considered classical patch-associated bacteria that take advantage of transient microscale organic matter and nutrient hot spots occurring in seawater (6–9), in contrast to the free-living bacteria with streamlined metabolic and regulatory capabilities and growing under low organic matter and nutrient concentrations typical of bulk seawater (9–11).
Recent analyses of >3,000 high-quality Roseobacter reads from the Global Ocean Sampling (GOS) (12) metagenomes based on an assembly-free bioinformatics pipeline showed that the uncultivated roseobacters do not fit the patch-associated model (13). Several signature genes of free-living bacteria (e.g., sec [secretion system]) in contrast to those of patch-associated bacteria (e.g., antibiotic production, chemotaxis, and cell surface modification genes) are statistically enriched in the wild and cultured roseobacters, respectively (13). In addition, roseobacters in GOS show a lower percentage of noncoding DNA and a smaller estimated genome size than do those in cultures (5, 11, 14). These genomic features are consistent with the hypothesis that some uncultivated planktonic roseobacters conform to the free-living ecological paradigm. Another major characteristic is that oceanic roseobacters display a bimodal distribution of G+C content, with a major peak centered at 42% and a secondary peak at 54%, which differs from the unimodal distribution of cultured roseobacters, which peaks at 62% (5, 14).
These distinct traits of wild roseobacters are nicely captured in three single-cell amplified genomes (SAGs) comprising a monophyletic Roseobacter lineage, SAG-O19. They consistently have streamlined (2.6 to 3.5 Mbp) and G+C-poor (39 to 40%) genomes, a reduced percentage of noncoding DNA, and a gene repertoire matching that of the free-living model (14). These features are also evident in another two Roseobacter lineages, NAC11-7 (15–17) and DC5-80-3 (or RCA, or Planktomarina temperata in ARB SILVA) (17). They are among the most abundant bacteria in some ocean regions such as Monterey Bay (18) and the North Sea (4), respectively, but remain largely uncultivated. Indeed, the only cultured strain, HTCC2255 of the lineage NAC11-7, was lost soon after isolation, and no other closely related strains have been isolated since (15). Likewise, members of the lineage DC5-80-3 are rarely cultured, and the cultures resist to being transferred through the traditional streak-plating method (19).
CHAB-I-5 is another major pelagic Roseobacter lineage that comprises ∼6% of all bacterioplankton cells and ∼20% of the roseobacters in some surface ocean waters (1). Despite its ecological relevance, this lineage remains uncultivated and has not been subjected to any genomic and metagenomic analysis. Here we utilized single-cell genomics to compare CHAB-I-5 to other roseobacters and to gain understanding about its ecological niche.
MATERIALS AND METHODS
Sampling, single-cell sorting, genome sequencing, and assembly.
Water samples for single-cell analyses were collected from the Gulf of Maine (43°50′39.87″N, 69°38′27.49″W) on 16 September 2009, the South Atlantic gyre (12°29′41.4″N, 4°59′55.2″W) on 1 December 2007, and the North Pacific gyre (22°45′N, 158°00′W) on 9 September 2009, cryopreserved with 6% glycine betaine, and stored at −80°C, as previously described (11). SAGs of bacterioplankton cells were generated and identified using fluorescence-activated cell sorting, multiple displacement amplification, and sequencing of the 16S rRNA gene at the Bigelow Laboratory Single Cell Genomics Center (SCGC; Boothbay Harbor, ME, USA), following previously described protocols (11, 20).
Five SAGs, including three affiliated with the CHAB-I-5 lineage and two with the SAG-O19 lineage, were selected for genome sequencing and de novo assembly at SCGC. Standard SCGC protocols were applied. Briefly, between 11.4 and 14.8 million paired-end reads (2 × 150 bp) per SAG were generated using NextSeq 500 (Illumina, San Diego, CA, USA). The reads were quality trimmed with Trimmomatic v0.30 (21), digitally normalized with kmernorm (http://sourceforge.net/projects/kmernorm/), and assembled with SPAdes version 3.1.0 (22). Only contigs with a length greater than 2,000 bp were retained.
Phylogenomic tree.
Genomes of three single cells of the CHAB-I-5 lineage, three single cells of the SAG-O19 lineage, two cultured strains of the DC5-80-3 lineage, one cultured strain and one single cell of the NAC11-7 lineage, 53 cultured strains of various Roseobacter lineages, and two outgroup species (Rhodovulum sp. PH10, Ahrensia sp. 13) were used for phylogenomic tree construction. At the time of this analysis, two additional single cells (AAA015-L03, AAA160-J18) of the SAG-O19 lineage were not available. Orthologous gene families were identified using the GET_HOMOLOGUES package (23), which reimplements the algorithm of the OrthoMCL software (24) but is easier to access. A total of 78 single-copy shared gene families were chosen for phylogenomic analysis, each of which requires the presence of at least three CHAB-I-5, two SAG-O19, two DC5-80-3, two NAC11-7 strains, 49 other roseobacters, and the two outgroup species. Members in each gene family were aligned at the amino acid sequence level using the MAFFT software (25), and the columns with gaps were deleted. The trimmed alignments were concatenated to comprise a superalignment with 22,712 sites.
The concatenated amino acid sequence was recoded into the following six Dayhoff groups (26): (i) cysteine; (ii) alanine, serine, threonine, proline, and glycine; (iii) asparagine, aspartic acid, glutamic acid, and glutamine; (iv) histidine, arginine, and lysine; (v) methionine, isoleucine, leucine, and valine; and (vi) phenylalanine, tyrosine, and tryptophan. The phylogenomic tree was built on these recoded data using the P4 Bayesian phylogenetic software (27). The procedure for phylogenetic model selection and tree construction follows a recent Roseobacter study (14). The only difference is that the present study selected a nonstationary model of NDCH(8) + NDRH(3), that is, a model with 8 composition vectors and 3 general time-reversible (GTR) rate matrices, while the previous study employed NDCH(8) + NDRH(2).
Metabolic pathway.
Analyses of metabolic genes focused on the four largely uncultivated lineages and six cultured pelagic strains (Phaeobacter sp. Y4I, Ruegeria pomeroyi DSS-3, Oceanicola batsensis HTCC2597, Roseobacter sp. GAI101, Oceanicola granulosus HTCC2516, Jannaschia sp. CCS1). The online Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) (28) was used to analyze the metabolic pathways. The BlastKOALA (KEGG orthology and links annotation [http://www.kegg.jp/blastkoala/]), a new genome annotation server accomplished by the SSEARCH program, was implemented for the computational assignments of a K number to each protein (29). Proteins assigned with a valid K number were mapped to KEGG pathways using the “Reconstruct Pathway” function provided by KEGG Mapper (http://www.kegg.jp/kegg/tool/map_pathway.html).
The identification of ecologically relevant metabolic genes was also facilitated by taking advantage of the model strain Ruegeria pomeroyi DSS-3, in which an updated annotation became recently available (30), as well as another four Roseobacter strains (Dinoroseobacter shibae DFL 12, Ruegeria sp. TM1040, Rhodobacteraceae bacterium KLH11, and Phaeobacter inhibens DSM 17395), in which genes encoding several key metabolic pathways have been experimentally identified. Orthologous genes families among all these genomes were identified using the GET_HOMOLOGUES package (23). The well-annotated genes served as anchors to search for functional genes in the four largely uncultivated lineages and the six cultured pelagic strains. When a gene of interest was missing in a certain lineage from the orthologous table, a BLAST (31) search against all predicted proteins in this lineage was used to confirm its absence.
Proportion of the four largely uncultivated lineages in the Roseobacter communities.
A custom reference database was created by combining the NCBI microbial RefSeq database (32) and the predicted protein sequences of the lineages CHAB-I-5, DC5-80-3, NAC11-7, and SAG-O19. Redundant data were removed. Query metagenome data sets were collected from the iMicrobe database (http://imicrobe.us/), the NCBI SRA archive, and links provided in related publications. All the metagenomic reads were subjected to quality trimming using the PRINSEQ software (33) for removal of ambiguities (Ns), duplicates, and low-quality and low-complexity reads. Qualified metagenomic reads were searched against the custom reference database using the RAPsearch2 software (34) with an E value cutoff of 1e−3. To avoid any bias in counting the number of reads hitting to the largely uncultivated lineages as a result of missing genes in SAGs, we focused on conserved genes shared by all Roseobacter lineages. Using GET_HOMOLOGUES (23), 1,206 orthologous protein families were identified, each of which had at least one member from each of the four largely uncultivated lineages and one member from the remaining roseobacters. Only reads with a best hit affiliated to any of these orthologous protein families were counted. Accordingly, the relative abundance of a certain lineage was approximate to the proportion of the best hits to its members in these orthologous families.
Proportion of the Roseobacter clade in the bacterioplankton communities from global oceans.
We implemented a three-step pipeline to evaluate the relative abundance of the Roseobacter clade in the bacterioplankton communities via the 16S rRNA gene profile of global metagenomic data sets (see Table S2 in the supplemental material). Initially, metagenomic reads in each data set were scanned for ribosomal small subunit (SSU) sequences that belong to Bacteria and Archaea using the Metaxa2 software (35). Metaxa2 identifies partial rRNA sequences from huge sequencing data sets containing reads as short as 100 bp with a very low false-positive rate and outperforms other single sequence repeat (SSR) classification tools in common use (35). Given that a read may span the boundaries of genes, reads identified as bacterioplankton SSUs were subjected to a BLAST search against a combined 16S rRNA gene database consisting of GreenGenes (gg_13_5) (36), an RDP classifier training set (version 15) (37), and SILVA release 111 (38), to exclude potential non-SSU regions. After the BLAST search, only DNA that was aligned to the genes in these databases with a length no smaller than 100 bp was retained. The extracted DNA fragments were pooled with 114 Roseobacter 16S rRNA gene sequences retrieved from the NCBI RefSeq database, and operational taxonomic units (OTUs) were generated using CD-HIT (39) with a percentage of identity cutoff of 0.89. This cutoff was set because roseobacters differ by up to 11% in their 16S rRNA gene sequences (5, 7). Other cutoffs (0.97, 0.95, 0.9, and 0.85) were also tried, but only under the criterion of 0.89 did all the RefSeq Roseobacter 16S rRNA gene sequences fall into one cluster. Consequently, metagenomic fragments grouped with this cluster were regarded as roseobacters. Next, the relative abundance of roseobacters in the bacterioplankton communities in each metagenomic data set was approximated to the ratio of the base counts of all Roseobacter 16S rRNA genes to the base counts of all bacterioplankton 16S rRNA genes.
Proportion of Roseobacter lineages in marine aerobic anoxygenic phototrophic communities.
The GOS metagenomic reads (12) were screened through three steps to identify valid PufM orthologs. Initially, the hidden Markov model (HMM) profile of the photosynthetic reaction center protein family (Photo_RC; accession no. PF00124) in Pfam (40) was queried against the six-frame translations of the metagenomic reads for significant matches using hmmsearch (HMMER v3.1b2) with the profile-specific trusted cutoffs (41). The peptides retained through this process are homologous to either PufM, PufL, PsbA, or PsbD, since the Photo_RC family was constructed on conserved domains of these four subfamilies. To separate PufM orthologs out of this mix, the retained peptides were searched for the best hit against a combined database, including 465 PufM, 491 PufL, 5,424 PsbA, and 1,828 PsbD nonredundant proteins retrieved from the InterPro database (42) using BLASTP (31) with an E value cutoff of 1e−5. To exclude potential paralogs, another round of BLASTP search with an E value cutoff of 1e−5 was performed on the selected peptides against the custom reference database built earlier (RefSeq + SAGs). Eventually, only peptides whose best hit was a PufM protein with an identity of >30% and an alignment length of >100 amino acids (aa) were kept. This procedure yielded 354 PufM partial sequences.
Next, these PufM sequences were clustered based on their pairwise identities by using BLASTclust (31) with the parameters “-S 80 -L 0.8,” and 65 representative sequences from the clusters and 25 RefSeq PufM sequences were combined to construct a phylogeny. Multiple-sequence alignment was built using MAFFT (25) and trimmed using trimAl (43), and a maximum likelihood tree was built using RAxML v8.1.22 (44) with the Protgammalg model.
Genomic characteristics.
To obtain a distribution of G+C content for the cultured Roseobacter cells, we chopped their genome sequences into 800-bp fragments, approximate to the average length of GOS reads, at a step of 1 bp. A number of reads, calculated by the formula , were randomly sampled to simulate an average sequencing depth of 10×, where Gleni represents the base count of genome i and read_len was set to 800. For the distribution of the G+C content of roseobacters in metagenomes, the 5,608 GOS Roseobacter reads sampled by the dN pipeline (13) were used. The percentage of noncoding DNA and the genome size were estimated according to the method described in a previous Roseobacter study (14).
Sigma factors were identified using hmmsearch (HMMER v3.1b2) (41) against Alphaproteobacteria proteomes with an E value cutoff of 1e−5 based on representative sigma factor SFam HMMs (45). Genomes analyzed include the CHAB-I-5, SAG-O19, DC5-80-3, and NAC11-7 lineages, other roseobacters (n = 116), and other Alphaproteobacteria (n = 500) from the NCBI RefSeq database, each having a unique species name (32). The results were cross-validated by mapping to clusters of orthologous groups (COGs) using RPS-BLAST (31) with an E value cutoff of 1e−5. Another 20 SAR11 and 7 SAR116 proteomes were used as a control to show the consistency between our estimates and results from the previous study (10).
RESULTS AND DISCUSSION
Genome characteristics and evolutionary position of the CHAB-I-5 lineage.
Genome sequencing of three CHAB-I-5 SAGs, AAA076-A02, AAA076-M18, and AA076-I17 (see Fig. S1 in the supplemental material), led to a total assembly size of 1.89, 2.32, and 3.14 Mbp, respectively (Table 1; see also Table S1 in the supplemental material). The 16S rRNA genes of the three SAGs share 99.74% to 99.87% sequence identity and therefore can be considered members of the same, operationally defined species (46). The genomic G+C content of these assemblies is 49.5 to 49.7%. We estimated that complete genome sizes of the analyzed cells are between 4.1 and 4.4 Mbp (see Fig. S2 in the supplemental material), which translates to genome recoveries of 46%, 53%, and 77%, respectively, for these single cells. In addition, two new single cells (AAA015-L03, AAA160-J18) of the lineage SAG-O19 (see Fig. S1 in the supplemental material) were sequenced, and their genomic characteristics are consistent with the three previously reported single cells of this lineage (Table 1; see also Table S1 in the supplemental material). These data suggest that members of lineage CHAB-I-5 have larger genomes than those of members of lineages SAG-O19, DC5-80-3, and NAC11-7 (Table 1).
TABLE 1.
Overview of the genome characteristics of the lineages CHAB-I-5, SAG-O19, NAC11-7, and DC5-80-3
| SAG ID (SCGC) or isolated strain name | Clade | Geographical source | No. of assembled nucleotides (Mbp) | No. of contigs | No. of coding sequences identified | G+C (%) | Predicted genome size (Mbp) | Reference or source | Genome recovery method |
|---|---|---|---|---|---|---|---|---|---|
| AAA076-I17 | CHAB-I-5 | Gulf of Maine | 3.14 | 98 | 3,390 | 49.5 | 4.07 | This study | SAG |
| AAA076-M18 | CHAB-I-5 | Gulf of Maine | 2.32 | 87 | 2,512 | 49.6 | 4.36 | This study | SAG |
| AAA076-A02 | CHAB-I-5 | Gulf of Maine | 1.89 | 62 | 2,044 | 49.7 | 4.10 | This study | SAG |
| AAA015-O19 | SAG-O19 | South Atlantic gyre | 1.70 | 159 | 1,780 | 38.5 | 3.10 | 11 | SAG |
| AAA300-J04 | SAG-O19 | North Pacific gyre | 0.62 | 77 | 650 | 39.1 | 2.65 | 11 | SAG |
| AAA298-K06 | SAG-O19 | North Pacific gyre | 1.70 | 231 | 1,771 | 39.9 | 3.50 | 11 | SAG |
| AAA015-L03 | SAG-O19 | South Atlantic gyre | 1.18 | 78 | 1,195 | 39.6 | 3.40 | This study | SAG |
| AAA160-J18 | SAG-O19 | Gulf of Maine | 2.21 | 47 | 2,293 | 40.5 | 2.97 | This study | SAG |
| HTCC2255 | NAC11-7 | Oregon coast | 2.29 | 14 | 2,174 | 36.8 | 2.55 | GenBank | Culture |
| AAA076-C03 | NAC11-7 | Gulf of Maine | 2.00 | 107 | 1,925 | 36.7 | 2.64 | 11 | SAG |
| RCA23 | DC5-80-3 | North Sea | 3.29 | 1 | 3,091 | 53.6 | 3.29 | 17 | Culture |
| LE17 | DC5-80-3 | California coast | 2.97 | 212 | 2,891 | 54.4 | 3.06 | 82 | Culture |
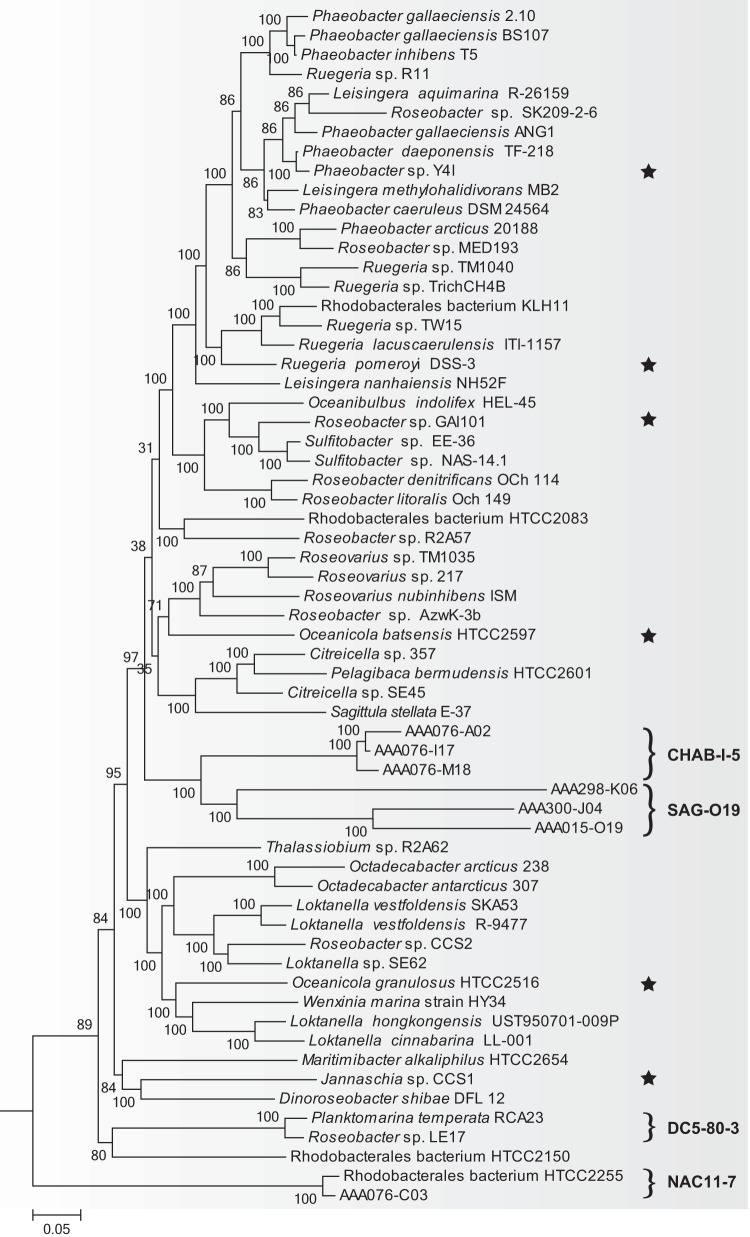
In a previous concatenation-based maximum likelihood phylogenetic analysis using RAxML (44), it was shown that the addition of SAG-O19 leads to a distortion of the Roseobacter phylogeny, including a considerable difference in the branch length of two strains with identical 16S rRNA gene sequences and the reordering of several previously well-established phylogenetic groups (14, 47). These issues were well resolved using a composition-heterogeneous phylogenetic model implemented in the P4 Bayesian software (27). In the present study, the same model resolved all major Roseobacter phylogenetic clusters and their branching orders congruent with the reported phylogeny (14), and interestingly, it placed CHAB-I-5 next to SAG-O19, which together constitute an exclusively uncultivated, monophyletic clade (Fig. 1). The other two, largely uncultivated lineages, NAC11-7 and DC5-80-3, are part of the basal lineages of the Roseobacter clade (Fig. 1), which is again consistent with other studies (5, 17).
FIG 1.
Bayesian phylogenomic tree of the Roseobacter clade (shaded) using a composition-heterogeneous model in the P4 software package based on a concatenation of 22,712 amino acid sites over 78 single-copy orthologous genes. The scale bar indicates the number of substitutions per site. The value near each internal branch is the posterior probability (after multiplying by 100) for that branch, with values lower than 75 not shown. The tree is rooted using two genomes from sister clades, and the outgroup lineages are not shown. The four largely uncultivated lineages (CHAB-I-5, SAG-O19, DC5-80-3, and NAC11-7) are specified, and the six cultured pelagic strains are marked with a star.
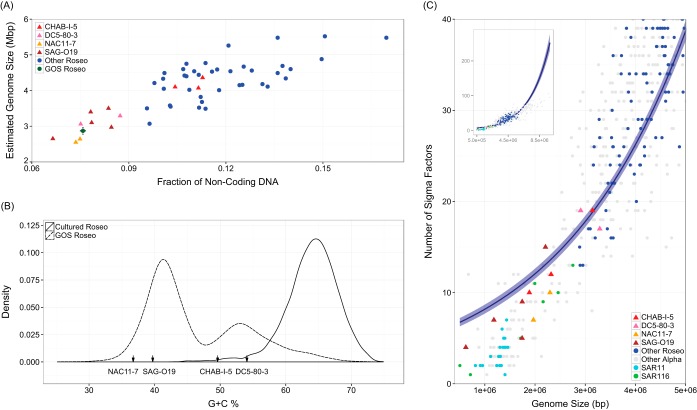
Genome streamlining of marine planktonic bacteria may be an important cause of their resistance to cultivation (10, 11). It is thus interesting to test whether members of the exclusively uncultivated CHAB-I-5 lineage are also under streamlining. Streamlined genomes are often manifested by reduced genome sizes, a decreasing percentage of noncoding DNA, depletion of G/C bases, and a low number of sigma factors for global regulation (10, 11, 14). While these features are evident in lineages SAG-O19, NAC11-7, and DC5-80-3, they are not shown in the SAGs of CHAB-I-5 (Fig. 2).
FIG 2.
Genomic characteristics of the four largely uncultivated Roseobacter lineages (CHAB-I-5, SAG-O19, DC5-80-3, and NAC11-7), cultured roseobacters, and GOS metagenomic Roseobacter reads. (A) Scatter plot of the percentage of noncoding DNA and the estimated genome size. (B) Frequency distribution of the G+C content of GOS Roseobacter reads and cultured roseobacters. G+C contents of the four major lineages are indicated by arrows along the x axis. (C) Number of sigma factors plotted against the length of assembled genomes for the four major lineages, as well as another 116 roseobacters, 20 SAR11, 7 SAR116, and 500 other Alphaproteobacteria genomes. The embedded panel displays the overall trend of the regression line, whereas the main graph provides a closeup view of the genome size between 0 to 5 Mbp in length. Data points from the four major lineages are symbolized with triangles and others with round dots. The data were fitted to a negative binomial generalized linear model with a log link, and the lighter blue shades confine the 95% confidence intervals. All four panels are colored with the same strategy: red for CHAB-I-5, pink for DC5-80-3, yellow for NAC11-7, brown for SAG-O19, green for GOS Roseobacter reads, blue for other roseobacters, cyan for SAR11, lime for SAR116, and gray for other members of the Alphaproteobacteria.
Global distribution of CHAB-I-5 and other uncultivated Roseobacter lineages.
Analysis of the 16S rRNA gene sequences showed that the Roseobacter clade represents a highly variable fraction (1 to 40%) of the surface ocean bacterioplankton (bacteria and archaea) communities, with an average of >5%. In general, roseobacters rarely exceed 10% of the tropical/subtropical bacterioplankton cells, and nearly all samples with a high Roseobacter abundance (>10%) are from temperate and polar oceans (see Fig. S3 in the supplemental material). Next, we estimated the relative abundance of the four largely uncultivated lineages (CHAB-I-5, SAG-O19, NAC11-7, and DC5-80-3) within the pelagic Roseobacter communities. This analysis used a functional gene binning approach to assign reads from multiple shotgun metagenomic data sets with a total of ∼55 Gbp (after quality trimming using PRINSEQ) sampled from global oceans (see Fig. S4 and Table S2 in the supplemental material). These four lineages together represent 61% of free-living roseobacters in tropical (between 23.5°S and 23.5°N), 76% in temperate (23.5°N to 66.5°N and 23.5°S to 66.5°S), and 56% in polar (>66.5°N and >66.5°S) waters.
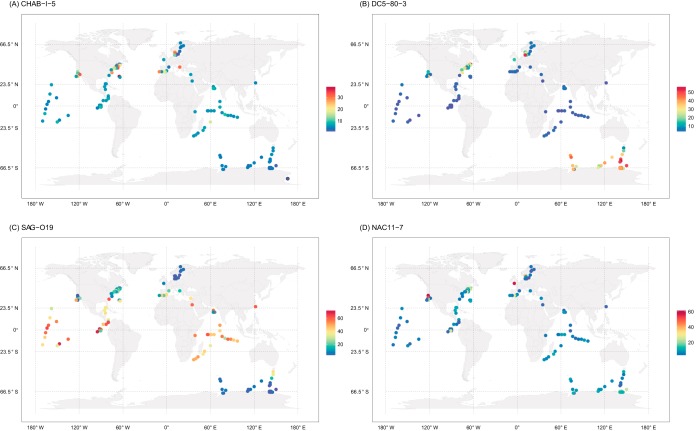
These four largely uncultivated lineages often distinctly dominate over the Roseobacter communities in different ocean regions (Fig. 3). The Western English Channel (48) and Monterey Bay, for instance, each are dominated by the single lineage NAC11-7 (each ∼60% of all roseobacters) (Fig. 3D; see also Table S3 in the supplemental material) and each contain the most abundant roseobacters of all other sampled oceans (18% and 25% of all prokaryotic cells, respectively) (see Fig. S3 in the supplemental material). In contrast, in all sampled stations from tropical/subtropical oceans, the Roseobacter communities are overrepresented by the SAG-O19 lineage (each 40 to 60% of all roseobacters) (see Table S3 in the supplemental material), and in samples from the Southern Ocean the lineage DC5-80-3 dominates (∼40%) (see Table S3 in the supplemental material), the latter consistent with previous studies based on the analysis of the 16S rRNA gene sequences (49–51). In several other oceans, the Roseobacter communities appear to be dominated by multiple uncultivated lineages. Two examples are the Baltic Sea and North Atlantic sampled during spring blooms, in which the Roseobacter communities are dominated by DC5-80-3 (22%) and NAC11-7 (34%) and by CHAB-I-5 (19%) and SAG-O19 (21%), respectively (see Table S3 in the supplemental material). This trend continues in the Roseobacter communities in the Gulf of Maine (52) and a transect between Southern California Bight and California Current sampled at a seasonal upwelling event (53), where all four lineages represent a significant proportion (see Table S3 in the supplemental material).
FIG 3.
Geographic distribution of the metagenomic samples and the relative abundance of CHAB-I-5 (A), DC5-80-3 (B), SAG-O19 (C), and NAC11-7 (D) at each location. The equator and boundaries of the main climate zones (tropic, temperate, polar) are marked in dashed lines. Each solid dot represents at least one metagenomic sample observed within the ±0.5° latitude/longitude range represented by the center of the dot and is colored according to the relative abundance of a particular lineage in the Roseobacter community. Colors are assigned in distinct steps in each panel, according to the different scales of the estimated population proportion. Only metagenomic samples with at least 50 reads (within the 1,206 gene families) mapped to the Roseobacter clade are displayed. Maps were created using the R package rworldmap (81).
Pooling all samples according to the temperature zone, we identified an opposite trend of the relative abundance of lineages SAG-O19 and DC5-80-3, in which the former decreases but the latter increases from tropical to polar waters (Fig. 3B and C). The major jumps of these two lineages both are at the transition from tropical to temperate waters, with the former lineage decreasing from 46% to 13% and the latter increasing from 3% to 23% (Table 2). In the face of global warming, the geographic ranges of these two lineages are expected to expand and contract with the rising temperature, respectively. These global scale patterns, however, are less prominent in CHAB-I-5 and NAC11-7. The CHAB-I-5 lineage, for instance, is largely distributed in selected temperate oceans in the Northern hemisphere (Fig. 3A). Likewise, while the NAC11-7 lineage is highly successful in selected temperate oceans, making up to 87% of all roseobacters, it accounts for <10% of the free-living roseobacters in 70% of the global ocean samples (Fig. 3D).
TABLE 2.
Percentages of the lineages CHAB-I-5, SAG-O19, NAC11-7, and DC5-80-3 in the free-living and particle-associated Roseobacter communities at different temperature zones
| Living style; geographical zonea | Total no. of roseobacter reads | % of lineage in community |
|||
|---|---|---|---|---|---|
| CHAB-I-5 | DC5-80-3 | SAG-O19 | NAC11-7 | ||
| FL; tropical | 348,245 | 6.7 | 3.2 | 45.6 | 5.2 |
| FL; temperate | 1,002,370 | 9.2 | 23.1 | 12.9 | 30.3 |
| FL; polar | 184,498 | 3.3 | 35.0 | 3.6 | 14.6 |
| PA; tropical | 3,331 | 10.8 | 2.8 | 28.7 | 4.2 |
| PA; temperate | 366,145 | 10.7 | 33.3 | 4.6 | 12.5 |
| PA; polar | 276,186 | 2.4 | 26.5 | 1.9 | 8.3 |
FL, free-living; PA, particle associated. Geographical zones: tropical, between 23.5°S and 23.5°N latitude; temperate, from 23.5°N and 66.5°N and from 23.5°S to 66.5°S; polar, from 66.5°N to the North Pole and from 66.5°S to the South Pole.
It was somehow unexpected that these free-living lineages are also successful in the particle-associated communities, as they represent 40 to 60% of particle-associated roseobacters from various temperature zones (Table 2). If one half of the pelagic roseobacters are associated with particles and the other half are free-living (13), marine particles may be an important ecological niche to support these four lineages. In particular, the single DC5-80-3 lineage accounts for 33% and 26% of the particle-associated Roseobacter communities in temperate and polar waters, respectively, and the CHAB-I-5, SAG-O19, and NAC11-7 lineages also make a sizable contribution in tropical-temperate (11% each), tropical (29%), and temperate (12%) oceans, respectively (Table 2). However, these results need to be interpreted with caution. The large volumes of seawater filtered for metagenomic analyses inevitably led to clogging, which makes it unavoidable that some free-living bacteria are caught in the particle-associated fraction. Hence, these observations need validations from future studies.
The findings of high relative abundance of the four largely uncultivated lineages are remarkable if one considers that the 16S rRNA gene diverges only by 1% within lineages CHAB-I-5 and NAC11-7 (1), 2% within lineage DC5-80-3 (4), and up to 5% within lineage SAG-O19, falling well within nominal delineations of species or genus (46), compared to the described cultured roseobacters that have already reached ∼200 species over ∼70 genera (54). A previous study partitioned the cultured roseobacters into two lifestyle groups, those from pelagic oceans and those from associations with other organisms, surfaces, or sediments (17). To understand the ecological success of the four largely uncultivated lineages, we selected six cultured and phylogenetically diverse (Fig. 1) Roseobacter strains from the pelagic group and contrasted the genomic sequences of the uncultivated lineages with those of the six cultured strains with a focus on metabolic potential for energy, carbon, and nutrient acquisition. One caveat is that the lineages CHAB-I-5 and SAG-O19 are represented only by partial genomes, and thus the absence of any metabolic pathway requires additional evidence in future studies.
Energy acquisition.
Many successful heterotrophic marine bacterioplankton utilize sunlight to power their activity in otherwise energy-deficient waters (photoheterotrophy) mainly through bacteriochlorophyll a (BChl-a) or proteorhodopsin (PR) (55, 56), and those possessing BChl-a are called aerobic anoxygenic phototrophs (AAPs) (57). AAPs and PR-based photoheterotrophs make 1 to 30% and 15 to 70% of noncyanobacterial bacterioplankton, respectively, in various oceans (58). While the latter are based on a single PR gene, the former require a photosynthesis gene cluster (PGC) that is composed of genes coding for a photosynthetic reaction center, light harvesting complexes, BChl, carotenoids, and assembly factors (59). Previous studies showed that the representative strain HTCC2255 of lineage NAC11-7 is the only known Roseobacter that uses PR to harvest light energy (5) and that the type strain Planktomarina temperata RCA23 of lineage DC5-80-3 is an AAP (17). We identified a complete and partial PGC in the SAG-O19 lineage (found in AAA298-K06) and in the CHAB-I-5 lineage, respectively (see Fig. S5 in the supplemental material). For the latter, the puf operon (pufQALMC) encoding the photosynthetic reaction center, the puh operon (puhABCE) for assembling the reaction center, the bchCXYZ and bchIDO operons for BChl synthesis, the crtAIBKCDEF operon for carotenoid synthesis, and a few other associated genes were identified (see Fig. S5 in the supplemental material). If the PGC is classified based on the rearrangement of its component operons (60), the PGCs in CHAB-I-5 and SAG-O19 fall into type I, which differs from type II in DC5-80-3 (see Fig. S5 in the supplemental material). Apparently, this classification of Roseobacter AAPs is not consistent with the phylogeny of these strains (Fig. 1), suggestive of a convoluted evolutionary history of the PGCs in roseobacters.
Since CHAB-I-5, SAG-O19, and DC5-80-3 are among the most abundant Roseobacter lineages and are potential AAPs, we further estimated their relative abundance in the oceanic AAP community. In general, roseobacters account for 64% of oceanic AAP community based on the relative abundance of the signature gene pufM in the global ocean metagenomic data sets (both free-living and particle-associated) (Table 3; see also Table S2 in the supplemental material), suggesting that roseobacters are the most abundant AAPs. Interestingly, these three largely uncultivated lineages together represent 72% of the oceanic Roseobacter AAPs, though they account for only 45% of all roseobacters. This overrepresentation of oceanic Roseobacter AAP is most evident in lineages SAG-O19 (41% of the oceanic Roseobacter AAPs versus 14% of all roseobacters) and CHAB-I-5 (14% versus 7%). In contrast, lineage DC5-80-3 is underrepresented in the Roseobacter AAP community (17% versus 20%), suggesting that not all members of this particular lineage are AAPs.
TABLE 3.
Number of pufM genes found in the metagenomic datasets listed in Table S2 in the supplemental material and distribution among largely uncultivated lineages at different temperature zones
| Temp zonea | Total no. of Roseobacter pufM genes | % of pufM genes in lineage: |
||
|---|---|---|---|---|
| CHAB-I-5 | DC5-80-3 | SAG-O19 | ||
| Tropical | 251 | 5.2 | 10.4 | 53.8 |
| Temperate | 189 | 21.2 | 29.6 | 32.3 |
| Polar | 43 | 32.6 | 0 | 2.3 |
Geographical zones: tropical, between 23.5°S and 23.5°N latitude; temperate, from 23.5°N and 66.5°N and from 23.5°S to 66.5°S; polar, from 66.5°N to the North Pole and from 66.5°S to the South Pole.
Energy production through nonobligate chemolithotrophy is a common feature of roseobacters. Oxidation of carbon monoxide (CO), for instance, has been demonstrated in a number of Roseobacter strains (8, 61). Two types of carbon monoxide dehydrogenase (CODH) often cooccur in roseobacters, and the presence of type I is a requirement for CO oxidation (61). Type I is present in lineages SAG-O19 and DC5-80-3 but missing in lineage NAC11-7. In the case of CHAB-I-5, only type II (with the presence of a signature motif, AYRGAGR, in CoxL) was identified, and more genomic data are needed to identify type I. This differential presence of potential CO oxidizers was similarly observed in the cultured pelagic roseobacters (Table 4). In contrast, a nearly complete gene cluster for the oxidation of sulfide or thiosulfate (soxRSVWXYZABCDEGH), a more productive process for energy generation (62), is present in all uncultivated lineages, in contrast to its occurrence in only one-half of the six cultured pelagic strains (Table 4).
TABLE 4.
Survey of select genes and metabolic pathways in pelagic Roseobacter representatives
| Functional category | Gene and/or pathway | CHAB-I-5 | SAG-O19 | DC5-80-3 | NAC11-7 | CCS1 | HTCC2597 | HTCC2516 | Y4I | GAI101 | DSS-3 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Photoheterotrophy | AAP, aerobic anoxygenic photoheterotroph | • | • | • | • | ||||||
| Proteorhodopsin | • | ||||||||||
| Other energy generation pathways | sox, sulfur oxidation | • | • | • | • | • | • | • | |||
| cox type I, CO oxidation | • | • | • | • | • | ||||||
| napA, dissimilatory nitrate reductase | • | • | • | ||||||||
| narG, dissimilatory nitrate reductase | |||||||||||
| nirK, dissimilatory nitrite reductase | • | ||||||||||
| nirS, dissimilatory nitrite reductase | • | ||||||||||
| nosZ, nitrous oxide reductase | |||||||||||
| norB, nitric oxide reductase | • | ||||||||||
| TTT | • | • | • | • | • | • | • | • | • | ||
| DHPS catabolism | hpsNOP | • | • | • | • | • | • | • | |||
| hpsKLM | • | • | • | • | |||||||
| suyA suyB | • | ||||||||||
| comE comD xsc | • | • | • | • | • | ||||||
| cuyA | • | ||||||||||
| ferA, feruloyl-coenzyme A synthetase | • | • | • | • | • | • | • | • | • | ||
| One-carbon metabolism | fsdD, C1 compound catabolism | • | • | ||||||||
| fae, C1 compound catabolism | |||||||||||
| Tmm, methylated amine catabolism | • | • | • | • | • | ||||||
| tmoXVW, methylated amine uptake | • | • | • | • | |||||||
| tdm, methylated amine catabolism | • | • | • | • | • | ||||||
| gmaS, methylated amine catabolism | • | • | • | • | • | • | • | ||||
| mgsABC, methylated amine catabolism | • | • | • | • | • | • | • | ||||
| betABC, choline catabolism | • | • | • | • | • | • | • | • | • | • | |
| fhs, choline catabolism | • | • | • | • | • | • | • | • | • | • | |
| dmdA, DMSP catabolism | • | • | • | • | • | • | • | • | |||
| dddW, DMSP catabolism | • | ||||||||||
| dddQ, DMSP catabolism | • | • | • | • | • | ||||||
| dddD, DMSP catabolism | • | ||||||||||
| dddP, DMSP catabolism | • | • | • | • | • | • | • | • | |||
| tauABC, tpa, xsc, pta, ald, taurine catabolism | • | • | • | • | • | ||||||
| Organismal interaction | T4SS | • | |||||||||
| T6SS | • | • | • | • | • | ||||||
| pepA, M42 glutamyl aminopeptidase | • | • | • | • | • | ||||||
| GTA | • | • | • | • | • | • | |||||
| luxRI quorum sensing | • | • | • | • | • | • | |||||
| igiBC, indigoidine synthesis | • | • | |||||||||
| tdaABCDEF, TDA synthesis |
Carbon and nutrient metabolism.
A systematic survey in the genome sequences of the transporter systems for alpha-glucoside (aglEFGK), rhamnose (rhaPSQT), fructose (frcABC), ribose (rbsABC), sorbitol/mannitol (smoEFG), d-xylose (xylFGH), and glycerol (glpVPQST), among others, showed that the potential for uptake of carbohydrates is more prevalent in the uncultivated lineages than in the cultured pelagic members (see Table S4 in the supplemental material). This is not the case in the potential uptake of organic acids, in which the tripartite ATP-independent periplasmic (TRAP) transporter (dctPQM) and the tripartite tricarboxylate transporter (TTT) system (tctABC) are more evenly distributed among these Roseobacter lineages (see Table S4 in the supplemental material). In addition, we surveyed various metabolic pathways for nitrogen, phosphorus, and vitamin uptake and metabolism (see Table S4 in the supplemental material). Since similar analyses were conducted in previous studies (7, 17), these observations and interpretations are included in Supplemental results in the supplemental material. Notably, key genes (hpsNOP) responsible for converting 2,3-dihydroxypropane-1-sulfonate (DHPS), recently identified as a key currency mediating the oceanic Roseobacter-diatom interaction (63), to (R)-sulfolactate are present in the four dominant lineages but missing in three of the six cultured pelagic isolates (Table 4). Following this initial oxidation step, three routes are known to carry out desulfonation via a different set of genes (suyAB, comDE/xsc, or cuyA) (63), and the CHAB-I-5 lineage possesses a distinct route from the other three largely uncultivated lineages (Table 4).
Next, we discuss in detail the genetic potential of one-carbon (C1) metabolism. The substrates for microbial C1 metabolism can be either C1 compounds (e.g., methanol and formaldehyde) or methylated compounds, the latter including osmolytes (e.g., taurine, dimethylsulfoniopropionate [DMSP], glycine betaine [GBT], choline, trimethylamine oxide [TMAO]) and their catabolic products, such as various methylated amines (MAs) (e.g., monomethylamine [MMA], dimethylamine [DMA], trimethylamine [TMA]). These compounds are produced in large amounts by all cellular organisms in the ocean (64, 65). Catabolic genes for C1 compounds, such as those encoding the NAD-dependent formate dehydrogenase (fsdD) and the formaldehyde-activating protein (fae), are rare in all the roseobacters analyzed here (Table 4), suggesting that roseobacters in general are not competitive in the utilization of C1 compounds. But this is not the case for the utilization of many methylated compounds. For instance, the complete genetic pathway (tmm, tmoXVW, tdm, gmaS, mgsABC) for the uptake and catabolism of MAs (62, 64) is consistently present in operon structures in the four uncultivated lineages but missing in a majority of the cultured strains (Table 4). The MAs are often utilized as C and energy source by roseobacters (e.g., Roseovarius sp. 217, Roseovarius sp. 216, Roseovarius mucosus DFL-24, Leisingera aquimarina LMG24366) (66, 67), though they may serve as N sources as well (68). Catabolic potentials for several other abundant methylated compounds (DMSP, taurine, and choline) are equally present among cultured and uncultivated Roseobacter lineages (Table 4).
Genomic features mediating bacterium-bacterium and bacterium-host interactions.
The type VI secretion system (T6SS) allows bacteria to efficiently target competitors by injection of antibacterial toxins (69). It was also demonstrated to be important in the bacterium-host interactions, as the deletion of T6SS reduces the ability of Agrobacterium tumefaciens to cause gall disease of its host plant (70) and attenuate virulence of Vibrio cholerae in animal hosts (71). Its key role in interaction with the host was recently hypothesized in roseobacters, where T6SS is overrepresented in roseobacters isolated from the accessory nidamental gland of squid, compared to those from other environments (72). A few key genes of T6SS (e.g., vasK, vasF, hcp, vgrG, vasG) were found in four of the six cultured pelagic strains and in the lineage CHAB-I-5 but are completely missing in the other three largely uncultivated lineages (Table 4). This is strong evidence that members of CHAB-I-5 colonize detrital particles or eukaryotic organisms. A more familiar secretion system in marine bacteria, similarly considered a fundamental component of the infection machinery (73), is the type IV secretion system (T4SS) (5, 74). Interestingly, this system (virB4, virD4) is present only in HTCC2597 and missing from all dominant lineages.
The quorum sensing (QS) system that uses acylated homoserine lactones has been shown to regulate motility, bacterium-host interaction, antibiotic production, polysaccharide production, and biofilm formation in diverse members of Proteobacteria (75) and is thus considered another key feature of patch-associated bacteria that is absent from free-living marine bacteria (76). While five of the six cultured pelagic strains carry this QS system, only lineage CHAB-I-5 possesses it among the four largely uncultivated lineages. The luxRI homologous genes encoding this QS system are orthologous to those in Phaeobacter sp. Y4I, which are involved in regulating the biosynthesis of indigoidine, an antibiotic shown to be critical to inhibit the colonization of Vibrio fischeri on surfaces (77). A few important genes (igiBC) involved in indigoidine production were identified only in CHAB-I-5 among the uncultivated lineages, but a key gene (igiD) encoding nonribosomal peptide synthetase (NRPS) was not found in the partial genomes of CHAB-I-5. To further this point, we followed a previous phylogenetic analysis of NRPS and polyketide synthase (PKS) responsible for secondary metabolite production (78) and did not find genes homologous to any of the four types of NRPS/PKS in the uncultivated Roseobacter lineages (see Fig. S6 in the supplemental material).
Concluding remarks.
All surveyed single cells and cultured strains of lineages CHAB-I-5, SAG-O19, DC5-80-3, and NAC11-7 have the potential to harvest light for increased biomass yields (55) or survive under stressful conditions (79). In contrast, only one of the six analyzed genomes of the less abundant pelagic Roseobacter lineages encodes these genetic potentials. Genes encoding other energy-producing processes, such as the oxidation of “energy-rich” reduced-sulfur compounds and trimethylamine (62), are similarly more common in the four mostly uncultured lineages. Moreover, these uncultivated lineages are more versatile in the uptake and catabolism of various carbohydrate compounds, methylated osmolytes, and key currencies underlying the trophic interactions between bacteria and phytoplankton. A few metabolic pathways that are completely missing from them but are more frequent in the cultured pelagic strains include nitrogen respiratory genes, siderophore uptake, and vitamin B7 and B1 synthesis (see Table S4 in the supplemental material). The complete absence of gene transfer agent (GTA) in the four dominant lineages and its consistent presence in the cultured pelagic roseobacters are remarkable but in agreement with the low frequency of GTA in marine metagenomes (80), and this suggests a limited role of GTA in Roseobacter adaptation to pelagic environments. An alternative explanation for the absence of these metabolic potentials in uncultivated lineages is that these genes could be part of the missing nucleotides that were not sequenced in SAGs.
While there is compelling evidence showing that members of lineages SAG-O19, DC5-80-3, and NAC11-7 are under genome streamlining (14, 16, 17), analyses of various genomic traits of the CHAB-I-5 roseobacters, including estimated genome size, percentage of noncoding DNA, and number of sigma factors, argue against streamlining in this lineage. Identification of a few genes involved in bacterium-bacterium and bacterium-host interactions (e.g., quorum sensing, antibiotic indigoidine synthesis, type VI secretion) that are typically missing in all streamlined genomes provides further evidence that members of CHAB-I-5 are adapted to a particle- and eukaryote-associated lifestyle (9).
The next question is whether we are missing more ecologically relevant but uncultivated lineages of the Roseobacter clade. The CHAB-I-5, SAG-O19, DC5-80-3, and NAC11-7 lineages together represent ∼65% of roseobacters in the pelagic oceans. The majority of the remaining 35% oceanic roseobacters may also not be a bona fide reflection of the cultured ones, as evidently shown by a limited overlap in the distribution of the G+C content between oceanic and cultured roseobacters (Fig. 2B). Furthermore, in 153 of the 391 samples in which there are at least 50 reads affiliated with roseobacters, these four lineages account for <50% of all roseobacters. These samples are predominantly from the Antarctic and the Baltic Sea. In particular, in most samples from marine-derived Antarctic lakes (e.g., Organic Lake, Ace Lake) and low-salinity waters of the Baltic Sea, these four lineages make only <15% and <20% in most of the local Roseobacter communities, respectively. Even in many typical oceanic waters, like the ALOHA station of the North Pacific subtropical gyre, these four lineages represent only one-half of the Roseobacter community. These data strongly suggest that we are still missing abundant lineages of the Roseobacter clade and that further sampling efforts will add new insights into Roseobacter diversity.
Supplementary Material
Funding Statement
Hong Kong Research Grants Council (RGC) Early Career Scheme provided funding to Haiwei Luo under grant number 24101015. The Chinese University of Hong Kong provided funding to Haiwei Luo under grant numbers 4930062 and 4053105. National Natural Science Foundation of China (NSFC) provided funding to Haiwei Luo and Yao Zhang under grant numbers 41576141 and 41422603, respectively, and the U.S. National Science Foundation provided funding to Ramunas Stepanauskas under grant numbers OCE-1232982 and OCE-1335810.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03678-15.
REFERENCES
- 1.Buchan A, Gonzalez JM, Moran MA. 2005. Overview of the marine Roseobacter lineage. Appl Environ Microbiol 71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran MA, Belas R, Schell MA, Gonzalez JM, Sun F, Sun S, Binder BJ, Edmonds J, Ye W, Orcutt B, Howard EC, Meile C, Palefsky W, Goesmann A, Ren Q, Paulsen I, Ulrich LE, Thompson LS, Saunders E, Buchan A. 2007. Ecological genomics of marine roseobacters. Appl Environ Microbiol 73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wemheuer B, Wemheuer F, Hollensteiner J, Meyer F-D, Voget S, Daniel R. 2015. The green impact: bacterioplankton response toward a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front Microbiol 6:805. doi: 10.3389/fmicb.2015.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giebel H-A, Kalhoefer D, Lemke A, Thole S, Gahl-Janssen R, Simon M, Brinkhoff T. 2011. Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J 5:8–19. doi: 10.1038/ismej.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo H, Moran MA. 2014. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78:573–587. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polz MF, Hunt DE, Preheim SP, Weinreich DM. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos Trans R Soc Lond B Biol Sci 361:2009–2021. doi: 10.1098/rstb.2006.1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, Rinta-Kanto JM, Sharma S, Sun S, Varaljay V, Vila-Costa M, Westrich JR, Moran MA. 2010. Genome characteristics of a generalist marine bacterial lineage. ISME J 4:784–798. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 8.Moran MA, Buchan A, Gonzalez JM, Heidelberg JF, Whitman WB, Kiene RP, Henriksen JR, King GM, Belas R, Fuqua C, Brinkac L, Lewis M, Johri S, Weaver B, Pai G, Eisen JA, Rahe E, Sheldon WM, Ye W, Miller TR, Carlton J, Rasko DA, Paulsen IT, Ren Q, Daugherty SC, Deboy RT, Dodson RJ, Durkin AS, Madupu R, Nelson WC, Sullivan SA, Rosovitz MJ, Haft DH, Selengut J, Ward N. 2004. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432:910–913. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 9.Luo H, Moran MA. 2015. How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends Microbiol 23:577–584. doi: 10.1016/j.tim.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Giovannoni SJ, Cameron Thrash J, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J 8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, Luo H, Wright JJ, Landry ZC, Hanson NW, Thompson BP, Poulton NJ, Schwientek P, Acinas SG, Giovannoni SJ, Moran MA, Hallam SJ, Cavicchioli R, Woyke T, Stepanauskas R. 2013. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci U S A 110:11463–11468. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y-H, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077.17355176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo H, Löytynoja A, Moran MA. 2012. Genome content of uncultivated marine Roseobacters in the surface ocean. Environ Microbiol 14:41–51. doi: 10.1111/j.1462-2920.2011.02528.x. [DOI] [PubMed] [Google Scholar]
- 14.Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA. 2014. Evolutionary analysis of a streamlined lineage of surface ocean Roseobacters. ISME J 8:1428–1439. doi: 10.1038/ismej.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA. 2014. Comparing effective population sizes of dominant marine alphaproteobacteria lineages. Environ Microbiol Rep 6:167–172. doi: 10.1111/1758-2229.12129. [DOI] [PubMed] [Google Scholar]
- 16.Luo H, Csűros M, Hughes AL, Moran MA. 2013. Evolution of divergent life history strategies in marine Alphaproteobacteria. mBio 4:e00373-13. doi: 10.1128/mBio.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voget S, Wemheuer B, Brinkhoff T, Vollmers J, Dietrich S, Giebel H-A, Beardsley C, Sardemann C, Bakenhus I, Billerbeck S, Daniel R, Simon M. 2015. Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J 9:371–384. doi: 10.1038/ismej.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ottesen EA, Marin R, Preston CM, Young CR, Ryan JP, Scholin CA, DeLong EF. 2011. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J 5:1881–1895. doi: 10.1038/ismej.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giebel H-A, Kalhoefer D, Gahl-Janssen R, Choo Y-J, Lee K, Cho J-C, Tindall BJ, Rhiel E, Beardsley C, Aydogmus ÖO, Voget S, Daniel R, Simon M, Brinkhoff T. 2013. Planktomarina temperata gen. nov., sp. nov., belonging to the globally distributed RCA cluster of the marine Roseobacter clade, isolated from the German Wadden Sea. Int J Syst Evol Microbiol 63:4207–4217. doi: 10.1099/ijs.0.053249-0. [DOI] [PubMed] [Google Scholar]
- 20.Stepanauskas R, Sieracki ME. 2007. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci U S A 104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger AM, Lohse M, Usadel B. 1 April 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katoh K, Kuma K-I, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Martin Embley T. 2004. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature 432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 27.Foster PG. 2004. Modeling compositional heterogeneity. Syst Biol 53:485–495. doi: 10.1080/10635150490445779. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivers AR, Smith CB, Moran MA. 2014. An updated genome annotation for the model marine bacterium Ruegeria pomeroyi DSS-3. Stand Genomic Sci 9:11. doi: 10.1186/1944-3277-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruitt KD, Tatusova T, Klimke W, Maglott DR. 2009. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res 37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye Y, Choi JH, Tang H. 2011. RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinformatics 12:159. doi: 10.1186/1471-2105-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bengtsson-Palme J, Hartmann M, Eriksson KM, Pal C, Thorell K, Larsson DG, Nilsson RH. 2015. metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol Ecol Resour 15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 36.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maidak BL, Olsen GJ, Larsen N, Overbeek R, McCaughey MJ, Woese CR. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res 25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. 2006. Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res 37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharpton TJ, Jospin G, Wu DY, Langille MGI, Pollard KS, Eisen JA. 2012. Sifting through genomes with iterative-sequence clustering produces a large, phylogenetically diverse protein-family resource. BMC Bioinformatics 13:264. doi: 10.1186/1471-2105-13-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yarza P, Yilmaz P, Pruesse E, Glockner FO, Ludwig W, Schleifer K-H, Whitman WB, Euzeby J, Amann R, Rossello-Mora R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 47.Luo H. 2015. The use of evolutionary approaches to understand single cell genomes. Front Microbiol 6:174. doi: 10.3389/fmicb.2015.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilbert JA, Field D, Swift P, Thomas S, Cummings D, Temperton B, Weynberg K, Huse S, Hughes M, Joint I, Somerfield PJ, Mühling M. 2010. The taxonomic and functional diversity of microbes at a temperate coastal site: a ‘multi-omic’ study of seasonal and diel temporal variation. PLoS One 5:e15545. doi: 10.1371/journal.pone.0015545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selje N, Simon M, Brinkhoff T. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445–448. doi: 10.1038/nature02272. [DOI] [PubMed] [Google Scholar]
- 50.Giebel H-A, Brinkhoff T, Zwisler W, Selje N, Simon M. 2009. Distribution of Roseobacter RCA and SAR11 lineages and distinct bacterial communities from the subtropics to the Southern Ocean. Environ Microbiol 11:2164–2178. doi: 10.1111/j.1462-2920.2009.01942.x. [DOI] [PubMed] [Google Scholar]
- 51.Landa M, Blain S, Christaki U, Monchy S, Obernosterer I. 2015. Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J 10:39–50. doi: 10.1038/ismej.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tully BJ, Nelson WC, Heidelberg JF. 2012. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol 14:254–267. doi: 10.1111/j.1462-2920.2011.02628.x. [DOI] [PubMed] [Google Scholar]
- 53.Zeigler Allen L, Allen EE, Badger JH, McCrow JP, Paulsen IT, Elbourne LDH, Thiagarajan M, Rusch DB, Nealson KH, Williamson SJ, Venter JC, Allen AE. 2012. Influence of nutrients and currents on the genomic composition of microbes across an upwelling mosaic. ISME J 6:1403–1414. doi: 10.1038/ismej.2011.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pujalte MJ, Lucena T, Ruvira MA, Arahal DR, Macián MC. 2014. The family Rhodobacteraceae. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Germany. doi: 10.1007/978-3-642-30197-1_377. [DOI] [Google Scholar]
- 55.Koblížek M. 2015. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol Rev 39:854–70. doi: 10.1093/femsre/fuv032. [DOI] [PubMed] [Google Scholar]
- 56.DeLong EF, Béjà O. 2010. The light-driven proton pump proteorhodopsin enhances bacterial survival during tough times. PLoS Biol 8:e1000359. doi: 10.1371/journal.pbio.1000359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yurkov VV, Beatty JT. 1998. Aerobic anoxygenic phototrophic bacteria. Microbiol Mol Biol Rev 62:695–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirchman DL, Hanson TE. 2013. Bioenergetics of photoheterotrophic bacteria in the oceans. Environ Microbiol Rep 5:188–199. doi: 10.1111/j.1758-2229.2012.00367.x. [DOI] [PubMed] [Google Scholar]
- 59.Swingley WD, Sadekar S, Mastrian SD, Matthies HJ, Hao J, Ramos H, Acharya CR, Conrad AL, Taylor HL, Dejesa LC, Shah MK, O'Huallachain ME, Lince MT, Blankenship RE, Beatty JT, Touchman JW. 2007. The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism. J Bacteriol 189:683–690. doi: 10.1128/JB.01390-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng Q, Zhang R, Koblížek M, Boldareva EN, Yurkov V, Yan S, Jiao N. 2011. Diverse arrangement of photosynthetic gene clusters in aerobic anoxygenic phototrophic bacteria. PLoS One 6:e25050. doi: 10.1371/journal.pone.0025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cunliffe M. 2011. Correlating carbon monoxide oxidation with cox genes in the abundant marine Roseobacter clade. ISME J 5:685–691. doi: 10.1038/ismej.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lidbury, IDEA, Murrell JC, Chen Y. 2015. Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J 9:760–769. doi: 10.1038/ismej.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, Dearth SP, Van Mooy BAS, Campagna SR, Kujawinski EB, Armbrust EV, Moran MA. 2015. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci U S A 112:453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lidbury I, Murrell JC, Chen Y. 2014. Trimethylamine N-oxide metabolism by abundant marine heterotrophic bacteria. Proc Natl Acad Sci U S A 111:2710–2715. doi: 10.1073/pnas.1317834111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lidbury I, Kimberley G, Scanlan DJ, Murrell JC, Chen Y. 2015. Comparative genomics and mutagenesis analyses of choline metabolism in the marine Roseobacter clade. Environ Microbiol 17:5048–62. doi: 10.1111/1462-2920.12943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schäfer H, McDonald IR, Nightingale PD, Murrell JC. 2005. Evidence for the presence of a CmuA methyltransferase pathway in novel marine methyl halide-oxidizing bacteria. Environ Microbiol 7:839–852. doi: 10.1111/j.1462-2920.2005.00757.x. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y. 2012. Comparative genomics of methylated amine utilization by marine Roseobacter clade bacteria and development of functional gene markers (tmm, gmaS). Environ Microbiol 14:2308–2322. doi: 10.1111/j.1462-2920.2012.02765.x. [DOI] [PubMed] [Google Scholar]
- 68.Chen Y, McAleer KL, Murrell JC. 2010. Monomethylamine as a nitrogen source for a nonmethylotrophic bacterium, Agrobacterium tumefaciens. Appl Environ Microbiol 76:4102–4104. doi: 10.1128/AEM.00469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coulthurst SJ. 2013. The type VI secretion system—a widespread and versatile cell targeting system. Res Microbiol 164:640–654. doi: 10.1016/j.resmic.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 70.Wu H-Y, Chung P-C, Shih H-W, Wen S-R, Lai E-M. 2008. Secretome analysis uncovers an Hcp-family protein secreted via a type VI secretion system in Agrobacterium tumefaciens. J Bacteriol 190:2841–2850. doi: 10.1128/JB.01775-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collins AJ, Fullmer MS, Gogarten JP, Nyholm SV. 2015. Comparative genomics of Roseobacter clade bacteria isolated from the accessory nidamental gland of Euprymna scolopes. Front Microbiol 6:123. doi: 10.3389/fmicb.2015.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de la Cuesta-Zuluaga JJ, Sánchez-Jiménez MM, Martínez-Garro J, Olivera-Angel M. 2013. Identification of the virB operon genes encoding the type IV secretion system, in Colombian Brucella canis isolates. Vet Microbiol 163:196–199. doi: 10.1016/j.vetmic.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 74.Persson OP, Pinhassi J, Riemann L, Marklund B-I, Rhen M, Normark S, González JM, Hagström Å. 2009. High abundance of virulence gene homologues in marine bacteria. Environ Microbiol 11:1348–1357. doi: 10.1111/j.1462-2920.2008.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 76.Kirchman DL. 2016. Growth rates of microbes in the oceans. Annu Rev Mar Sci 8:285–309. doi: 10.1146/annurev-marine-122414-033938. [DOI] [PubMed] [Google Scholar]
- 77.Cude WN, Mooney J, Tavanaei AA, Hadden MK, Frank AM, Gulvik CA, May AL, Buchan A. 2012. Production of the antimicrobial secondary metabolite indigoidine contributes to competitive surface colonization by the marine Roseobacter Phaeobacter sp. strain Y4I. Appl Environ Microbiol 78:4771–4780. doi: 10.1128/AEM.00297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martens T, Gram L, Grossart H-P, Kessler D, Müller R, Simon M, Wenzel S, Brinkhoff T. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb Ecol 54:31–42. doi: 10.1007/s00248-006-9165-2. [DOI] [PubMed] [Google Scholar]
- 79.Fuhrman JA, Schwalbach MS, Stingl U. 2008. Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol 6:488–494. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- 80.Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. 2008. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol 74:2933–2939. doi: 10.1128/AEM.02129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.South A. 2011. rworldmap: a new R package for mapping global data. R J 3:35–43. [Google Scholar]
- 82.Mayali X, Franks PJS, Azam F. 2008. Cultivation and ecosystem role of a marine Roseobacter clade-affiliated cluster bacterium. Appl Environ Microbiol 74:2595–2603. doi: 10.1128/AEM.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.