Abstract
Yesso scallop-pathogenic Vibrio splendidus strain JZ6 was found to have the highest virulence at 10°C, while its pathogenicity was significantly reduced with increased temperature and completely incapacitated at 28°C. In the present study, comparative transcriptome analyses of JZ6 and another nonpathogenic V. splendidus strain, TZ19, were conducted at two crucial culture temperatures (10°C and 28°C) in order to determine the possible mechanism of temperature regulation of virulence. Comparisons among four libraries, constructed from JZ6 and TZ19 cultured at 10°C and 28°C (designated JZ6_10, JZ6_28, TZ19_10, and TZ19_28), revealed that 241 genes were possibly related to the increased virulence of JZ6 at 10°C. There were 10 genes, including 2 encoding Flp pilus assembly proteins (FlhG and VS_2437), 6 encoding proteins of the “Vibrio cholerae pathogenic cycle” (ToxS, CqsA, CqsS, RpoS, HapR, and Vsm), and 2 encoding proteins in the Sec-dependent pathway (SecE and FtsY), that were significantly upregulated in JZ6_10 (P < 0.05) compared to those in JZ6_28, TZ19_10, and TZ19_28, which were supposed to be responsible for adhesion, quorum sensing, virulence, and protein secretion of V. splendidus. When cultured at 10°C, JZ6 cells were larger and tended to aggregate more than those cultured at 28°C. The virulence factor (extracellular metalloprotease) was also found to be highly expressed in the extracellular product (ECP) of JZ6 at 10°C, and this ECP exhibited obvious cytotoxicity to oyster primary hemocytes, A549 cells, and L929 cells. These results indicated that low temperatures (10°C) could enhance adhesion, activate the quorum sensing systems, upregulate virulence factor synthesis and secretion, and, lastly, increase the pathogenicity of JZ6.
INTRODUCTION
The Gram-negative pathogenic bacterium Vibrio splendidus is a ubiquitous and representative species of the Vibrio genus, a causal agent of vibriosis, which causes high rates of mortality in aquaculture animals, including turbot, scallop, clam, and oyster (1–5). Like most Vibrio strains, V. splendidus is an opportunistic pathogen that causes mortality of animals in an optimum environment, whereas it usually acts as a “normal bacterium” in the host or environment under adverse conditions (6–8). Therefore, environmental factors are important regulators of the pathogenicity of V. splendidus.
In previous studies, most opportunistic pathogenic Vibrio bacteria, such as V. splendidus, V. cholerae, V. parahaemolyticus, V. alginolyticus, and V. harveyi, have been reported to cause high rates of mortality of cultured animals in the summer and enter a viable-but-nonculturable (VBNC) state when they are exposed to temperatures below 10°C (6, 8–11). Hence, temperature has been considered one of the crucial environmental factors that can regulate the metabolic process, growth, adhesion, and even pathogenicity of bacteria by influencing their gene expression (12–14). V. splendidus strain JZ6 was previously isolated and identified as a pathogenic agent for Yesso scallop (Patinopecten yessoensis) in low-temperature environments (5). This strain survived in cold environments and exhibited the highest pathogenicity at a temperature of 10°C. Its virulence was significantly reduced with increased temperature and even completely incapacitated at 28°C. This low-temperature adaptation indicated that a special environmental regulation mechanism might exist in V. splendidus strain JZ6.
The mechanism of regulation of gene expression is always involved in complex and specific biological processes composed of a series of functionally related molecules (15), and conventional investigation of single genes is not sufficient to discover the complete regulation and cross talk of these genes. Nowadays, the high-throughput transcriptome sequencing (RNA-seq) technique has dramatically accelerated the analytical capacity and has been widely used in constructing global networks of gene expression (16). In the past decades, there were several comparative transcriptome analyses focused on temperature-dependent genes in some important environmental bacteria, including Escherichia coli (12), Tropheryma whipplei (13), Pseudomonas aeruginosa (14), Streptococcus thermophilus (17), and Bacillus cereus (18). The expression patterns of virulence-related genes in Listeria monocytogenes, Salmonella enterica, Xanthomonas oryzae, and V. cholerae have also been revealed through transcriptome analysis (19–22).
The pathogenicity of bacteria is determined and regulated by a complicated network composed of major virulence factors, transcription factors, transport systems, and protein secretion and intrusion systems (23–25). Some of the virulence systems, such as quorum sensing and adhesion systems, have been confirmed to be regulated by temperature (14, 26–28). Unfortunately, the regulation mechanism for the adaptability and pathogenicity of opportunistic bacteria with changes in temperature is still unclear according to previous studies.
In the present study, the transcripts of JZ6 and another nonpathogenic V. splendidus strain, TZ19, were sequenced and compared at two crucial culture temperatures (10°C and 28°C) in order to identify the major temperature-dependent genes and their specific expression profiles closely associated with pathogenicity at 10°C and further reveal the special regulation mechanism in V. splendidus JZ6 at low temperatures.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Pathogenic V. splendidus strain JZ6 and nonpathogenic strain TZ19 for Yesso scallop (P. yessoensis) were employed in the present study (5). They were cultivated on Zobell 2216E agar at 18°C for 24 h, and the individual bacterial colonies were then subcultured in Trypticase soy broth (TSB) with 20‰ salinities at 10°C or 28°C and with shaking at 170 rpm for 12 h. After cultivation, bacterial cell numbers were adjusted to the same order of magnitude by absorption spectrophotometry and microscopic counting for the following experiments.
RNA extraction.
Total RNA was isolated essentially as described previously (29). After being cultured at different temperatures (10°C or 28°C), the same numbers of V. splendidus JZ6 and TZ19 cells were harvested by centrifugation at 12,000 × g for 1 min. Total cellular RNA was extracted by using TRIzol reagent (Invitrogen), and DNA contamination was ruled out with a Mega Clear kit (Ambion, Life Technologies). mRNA was purified by using the Microb Express bacterial mRNA enrichment kit (Ambion, Life Technologies) to remove rRNA according to the manufacturer's protocol. The purified mRNA was quantified by using a NanoDrop 2000 spectrophotometer (Thermo Scientific) and checked for integrity with an Agilent 2100 bioanalyzer (Agilent Technologies).
Library preparation.
The single-end fragment library was constructed according to the SOLiD Total RNA-seq kit protocol (Life Technologies) with 100 ng mRNA, as previously described (30). First, RiboMinus RNA was fragmented by RNase III and purified by using the RiboMinus concentration module (Invitrogen). The RNA fragments were linked with the adaptor by using hybridization master mix, and reverse transcription was performed in a subsequent procedure. The purified cDNA was size selected after DNA electrophoresis with NovexTBE-urea gel (Invitrogen) at 180 V for 20 min and then purified as the amplification template. PCRs were performed at 95°C for 5 min and then at 95°C for 30 s, 62°C for 30 s, and 72°C for 30 s for 15 cycles in a thermal cycler. All of the components used for amplification were obtained from the SOLiD Total RNA-seq kit. The yield and size distribution of PCR products were checked by using an Agilent 2100 bioanalyzer. Emulsion PCR and bead enrichment were performed by using the SOLiD EZ Bead system (Life Technologies). Workflow analysis (WFA) was first used to verify the quality and density of the template beads. The enriched beads for each sample were then deposited onto a sequencing slide. Finally, four libraries of strains JZ6 and TZ19 cultured at 10°C and 28°C (designated JZ6_10, JZ6_28, TZ19_10, and TZ19_28) were sequenced on the SOLiD 4 platform, and color space reads were outputted.
Bioinformatics analysis.
The full-length genomic sequences of V. splendidus LGP32 were downloaded from the NCBI (http://www.ncbi.nlm.nih.gov/genome/933?project_id=59353) and served as a reference for alignment analysis. Read alignment was performed by using LifeScope software (Life Technologies) with default parameters. The classic mapping strategy “seed and extend” was adopted, with “25.2.0:20” as a mapping scheme (for the 50-base reads, the seed might be 25 bases long with up to two mismatches allowed, and the start site of the seed could be 0 or 20). Cufflinks was used to assemble transcripts, estimate their abundances, and identify differentially expressed genes between the treatment and control groups. The overall situation of RNA-seq was analyzed by using an R package of CummeRbund. The cutoff value for upregulated and downregulated genes between two samples was defined as >2-fold.
For gene ontology (GO) analysis, the protein sequences of V. splendidus were aligned by a local blastp search of the nonredundant (nr) database of the NCBI with an E value of <0.001. The alignment results were parsed for assigning GO terms by using Blast2GO software. The GO distributions and enrichment analysis of differentially expressed genes were implemented by using one-tailed Fisher's exact test with a P value/false-discovery rate (FDR) of <0.001 (31). Significantly overenriched GO terms were identified from the test set. The pathways composed of the differentially expressed genes were retrieved from the KEGG database by using a built-in function of Blast2GO.
Quantitative real-time PCR analysis.
The expression levels of six differentially expressed genes participating in pathogenic processes were validated by quantitative real-time PCR (qRT-PCR). The templates for qRT-PCR were prepared with independently isolated RNA samples from three biological replicates, and first-strand cDNA synthesis was carried out based on Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT) usage information (Promega). Specific primers were designed according to the corresponding sequences in the genome of V. splendidus LGP32 (Table 1), and the comparative threshold cycle (CT) method (2−ΔΔCT method) was used to analyze expression levels as described previously (32). Two 16S rRNA gene primers for V. splendidus, 16S rtf and 16S rtr (Table 1), were used as an internal control to verify successful transcription and to calibrate the cDNA template for the corresponding samples. qRT-PCR was performed by using an Applied Biosystems 7500 instrument (Life Technologies), and the collected data were analyzed with 7500 system SDS software. The assay was conducted with a volume of 20 μl consisting of 10 μl of 2× SYBR Premix Ex Taq II (TaKaRa), 0.8 μl of each forward and reverse primer (10 μmol liter−1), 0.4 μl of 50× ROX reference dye, 2 μl of DNA extract (10 ng μl−1), and 6 μl of nuclease-free water. The reaction was performed in triplicate at 95°C for 30 s, with 40 cycles of primer annealing at 95°C for 5 s and primer extension at 60°C for 31 s. Dissociation curve analysis of amplification products was performed at the end of each PCR to confirm that only one PCR product was amplified and detected. All data were given in terms of relative mRNA levels expressed as means ± standard errors (SE) (n = 4). The results were subjected to one-way analysis of variance (ANOVA) followed by an unpaired, two-tailed t test, and the differences were considered significant at a P value of <0.05.
TABLE 1.
Primers used in the present study
| Target and primer | Sequence (5′–3′) |
|---|---|
| 16S rRNA | |
| 16S rtf | TCGTGTYGTGARATGTTGGGT |
| 16S rtr | CCACCTTCCTCCRGTTTRTCA |
| Vsm | |
| vsmrtf | AAGTCGCCCAAGTGGTGTATCT |
| vsmrtr | CGATGGGAAAGCTAGGGAAGT |
| CqsA | |
| cqsArtf | GCACTTGGTATTGGGTAAA |
| cqsArtr | TTGATGGGCGTCTTGGAT |
| CqsS | |
| cqsSrtf | CTTGTGGGTTGGATGGGCT |
| cqsSrtr | CAAAACAGACGCACGGCTAA |
| ToxS | |
| toxSrtf | CTATTTACTGATTTCGCCAACGG |
| toxSrtr | TGCGTCGGCTTTGCTCTG |
| FlhG | |
| flhGrtf | CGGAGCAATGAGTGAGGAGTA |
| flhGrtr | GAGCCATTGTCACTTTCATCAC |
| RpoS | |
| rpoSrtf | GAGCGTGGCTTCCGTTTC |
| rpoSrtr | CTCTTGCGGTGCGTAGGT |
Morphological analysis of V. splendidus at different temperatures.
V. splendidus strains JZ6 and TZ19 were cultured in TSB medium with 20‰ salinities at both 10°C and 28°C for 12 h (until the optical density [OD] reached 0.5). Bacterial cultures were centrifuged at 4,000 × g for 4 min, resuspended with 100 μl sterilized normal saline, and then washed twice with sterile saline to eliminate interference from the medium. The morphological structures of JZ6 and TZ19 were observed both by scanning electron microscopy (SEM) at a ×10,000 magnification and by using a fluorescence-activated cell sorter (FACS) flow cytometer (Becton Dickinson) with cell size (forward light scatter [FSC]) and complexity (side light scatter [SSC]) measurements.
SDS-PAGE and Western blot analysis of extracellular products of V. splendidus.
Extracellular products (ECPs) of V. splendidus strains JZ6 and TZ19 were obtained according to procedures described previously by Balebona et al. (33). Strains JZ6 and TZ19 were inoculated in TSB medium with 20‰ salinities at 18°C for 12 h, and 200 μl of culture of each bacterial strain was spread onto a Zobell 2216E agar plate overlaid with a sterile cellophane sheet and incubated at 10°C and 28°C for 24 h. Bacterial cells were harvested with sterile saline, and the cell suspensions were centrifuged at 12,000 × g at 4°C for 10 min. The supernatants were filtered through 0.22-μm membrane filters and used as the crude ECPs (designated PJZ6_10, PJZ6_28, PTZ19_10, and PTZ19_28). The total protein contents of the ECPs were measured according to the bicinchoninic acid (BCA) method (34).
These four different ECP samples were used at a concentration of 20 μg μl−1, separated electrophoretically by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and visualized with Coomassie bright blue R250. The expression of metalloprotease (Vsm) in the ECP samples was detected by Western blotting using a monoclonal antibody to JZ6 Vsm (designed by Abmart Inc.). The ECP samples were electrophoretically transferred onto a nitrocellulose transfer membrane (Millipore). After being blocked with a 5% skim milk powder solution, the membrane was first incubated with an anti-Vsm solution (1:1,000, vol/vol) at room temperature for 1 h, washed with Tris-buffered saline–Tween (TBST) (10 mmol liter−1 Tris-HCl [pH 8.0], 100 mmol liter−1 NaCl, and 0.05% [wt/vol] Tween 20), and subsequently incubated with a 1:2,000 (vol/vol) dilution of horseradish peroxidase-conjugated anti-mouse IgG (Life Technologies) at room temperature for 1 h. After repeated washing, the membrane was incubated with SuperSignalWest Pico (Thermo Scientific) and exposed to film. Mouse preimmune serum was used as a negative control, and a recombinant Vsm protein was used as a positive control.
Cytotoxicity test of V. splendidus ECPs.
The toxicities of ECPs were tested by using cells, including oyster primary hemocytes, a human lung cancer cell line (A549), and a mouse fibroblast cell line (L929). Oyster primary hemocytes were grown at 18°C by using Leibovitz's L-15 medium (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (Gibco, Life Technologies), while the commercial cell lines A594 and L929 (Keygen BioTECH) were cultured at 37°C with 5% CO2 by using Dulbecco's modified Eagle medium (DMEM) and RPMI 1640 medium (Gibco, Life Technologies) with 10% fetal bovine serum, respectively, according to the manufacturer's instructions.
Different cells were divided into six groups and cultured as monolayers in 96-well plates (Coast; Corning) at a concentration of 104 cells per well with 100 μl of medium. In the experimental groups, 5 μl (10 μg) of four different ECP samples, PJZ6_10, PJZ6_28, PTZ19_10, and PTZ19_28, was added to each well. Normal cells and cells inoculated with sterile saline were used as blanks and negative controls. The effects of ECPs on cell monolayers were observed 3, 6, and 24 h after stimulation, and the viability of A549 and L929 cells was calculated by using Cell Counting kit 8 (Beyotime) according to the manufacturer's instructions.
Microarray data accession number.
The raw sequencing reads were submitted to the NCBI Sequence Read Archive under accession no. SRP035223.
RESULTS
Global transcriptional profiles of V. splendidus strains JZ6 and TZ19 at 10°C and 28°C.
There were 14.69 million to 17.43 million reads obtained after sequencing of four libraries, and these reads were aligned to the LGP32 genome sequence (NCBI GenBank accession no. NC_010717.1). The global transcriptional profiles of V. splendidus JZ6 and TZ19 at 10°C and 28°C were obtained by data normalization and statistical analysis (see Fig. S1 in the supplemental material). There were 775 genes in strain JZ6 (comparison of JZ6_10/JZ6_28) and 612 genes in strain TZ19 (comparison of TZ19_10/TZ19_28) expressed differently in a temperature-dependent manner, and there were 974 genes with different expression between JZ6 and TZ19 at 10°C (comparison of JZ6_10/TZ19_10). The complete lists of all differentially expressed genes are given in Tables S1 to S3 in the supplemental material.
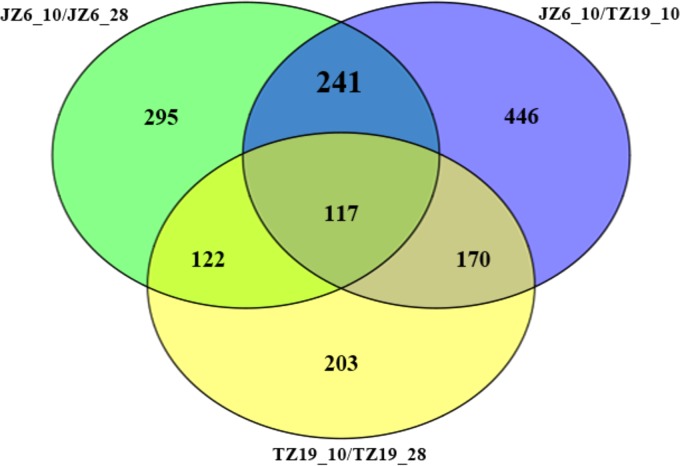
A Venn diagram of differentially expressed genes was plotted with three comparisons of transcriptional profiles (Fig. 1), including JZ6 at 10°C and 28°C (JZ6_10/JZ6_28), TZ19 at 10°C and 28°C (TZ19_10/TZ19_28), and JZ6 and TZ19 at 10°C (JZ6_10/TZ19_10). In total, 358 differentially expressed genes were obtained in the comparisons between groups JZ6_10 and JZ6_28 (temperature difference group) and between groups JZ6_10 and TZ19_10 (pathogenicity difference group). Among them, 117 genes were collectively observed in the three groups of JZ6_10/JZ6_28, JZ6_10/TZ19_10, and TZ19_10/TZ19_28, which represented common genes responding to changes of environmental temperature in strains JZ6 and TZ19.
FIG 1.
Grouping of differentially expressed genes among the three comparison groups, JZ6_10/JZ6_28, TZ19_10/TZ19_28, and JZ6_10/TZ19_10.
Gene ontology and KEGG analysis of specific temperature-regulated genes.
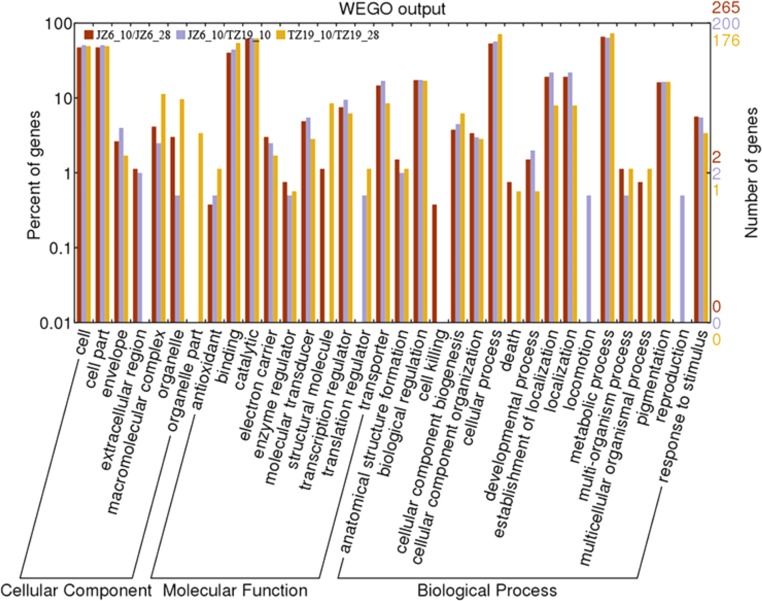
Differentially expressed genes in the three comparison groups were classified by using the Web Gene Ontology Annotation Plot (WEGO) assignment (Fig. 2), and they were classified into three main categories (cellular component, molecular function, and biological process) and subdivided into 34 functional groups. In the WEGO classifications, differentially expressed genes were dominantly clustered into common life processes such as “cell,” “cell part,” “binding,” “catalytic,” “cellular process,” and “metabolic process” and environmental adaptation processes such as “extracellular region,” “transcription regulator,” and “response to stimulus.”
FIG 2.
Distribution analysis of differentially expressed genes in JZ6_10/JZ6_28, TZ19_10/TZ19_28, and JZ6_10/TZ19_10 comparison groups with histogram presentation of gene ontology classifications.
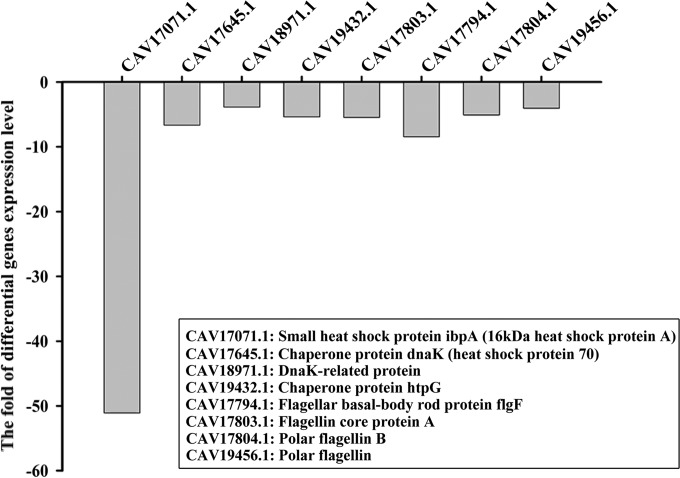
For 358 differentially expressed genes, 213 upregulated and 145 downregulated genes were identified as specific molecules associated with temperature regulation in JZ6. These genes were mapped to the KEGG pathway with reference to V. splendidus LGP32. The upregulated genes in JZ6 at 10°C were mainly clustered into the “two-component system,” “prokaryote-type ABC transporter,” and “Vibrio cholerae pathogenic cycle” pathways (Table 2; see also Fig. S2 to S4 in the supplemental material), while the genes encoding heat shock-related (ibpA, dnaK, and htpG, etc.) and flagellum-assembling (flgF, flagellin core protein A, and polar flagellin B, etc.) proteins were upregulated in JZ6 at 28°C (Fig. 3).
TABLE 2.
Some of the upregulated genes in sample JZ6_10
| Category and GenBank accession no. | Gene product descriptiona | Expression levelb |
Fold change in expression level | |
|---|---|---|---|---|
| JZ6_10 | JZ6_28 | |||
| Two-component systems | ||||
| CAV19495.1 | Nitrogen regulatory protein P-II, GlnB | 4,301.29 | 1,906.74 | 2.25583 |
| CAV18204.1 | Chaperone protein, TorD | 1,094.38 | 400.163 | 2.73484 |
| CAV19181.1 | Cytochrome c-type protein, TorC | 664.605 | 228.382 | 2.91006 |
| CAV18208.1 | Transcriptional regulatory protein, TorR | 1,278.88 | 389.225 | 3.28571 |
| CAV17936.1 | Alkaline phosphatase, PhoA | 259.999 | 52.2893 | 4.97232 |
| CAV27738.1 | Putative two-component sensor | 226.494 | 40.7308 | 5.56076 |
| CAV17573.1 | DNA-binding response regulator, PhoB | 583.169 | 41.2896 | 14.1239 |
| Prokaryote-type ABC transporters | ||||
| CAV25568.1 | Substrate-binding protein precursor | 266.587 | 132.424 | 2.01313 |
| CAV26093.1 | Substrate-binding protein precursor | 460.395 | 220.856 | 2.08459 |
| CAV26059.1 | Transmembrane and ATP-binding protein | 136.434 | 62.1462 | 2.19537 |
| CAV18564.1 | Transmembrane protein | 131.833 | 47.6521 | 2.76657 |
| CAV26801.1 | Substrate-binding protein precursor | 120.826 | 41.7848 | 2.89163 |
| CAV18669.1 | Permease protein | 188.82 | 56.4247 | 3.34641 |
| CAV25570.1 | Transmembrane protein | 199.363 | 58.8266 | 3.38899 |
| CAV26807.1 | Transmembrane protein | 139.94 | 40.7586 | 3.43339 |
| CAV27507.1 | Substrate-binding protein precursor | 107.275 | 27.0638 | 3.96378 |
| CAV25612.1 | Transmembrane protein | 52.2,864 | 10.0918 | 5.18108 |
| CAV26058.1 | Transmembrane and ATP-binding protein | 316.893 | 59.2151 | 5.35156 |
| CAV25872.1 | Substrate-binding protein precursor | 91.3,874 | 16.7836 | 5.44504 |
| CAV17882.1 | Substrate-binding protein precursor | 151.885 | 5.60258 | 27.1098 |
| Bacterial secretion systems | ||||
| CAV20266.1 | Preprotein translocase subunit, SecE | 11,835.6 | 4,540.29 | 2.60679 |
| CAV20299.1 | Cell division protein, FtsY | 817.819 | 232.308 | 3.52041 |
| Pilus assembly proteins | ||||
| CAV19597.1 | Flp pilus assembly protein, FlhG | 189.81 | 10.3781 | 18.2895 |
| CAV19596.1 | Flp pilus assembly protein, VS_2437 | 154.652 | 18.4313 | 8.39073 |
| Vibrio cholerae pathogenic cycle | ||||
| CAV19699.1 | HTH-type transcriptional regulator, HapR | 860.788 | 343.366 | 2.50691 |
| CAV18909.1 | Aminotransferase, CqsA | 1,141.75 | 446.578 | 2.55667 |
| CAV19434.1 | Transmembrane regulatory protein, ToxS | 358.616 | 104.648 | 3.42688 |
| CAV18910.1 | Sensor histidine kinase, CqsS | 67.1,794 | 14.0415 | 4.78435 |
| CAV26646.1 | RpoS-like sigma factor, RpoS | 728.066 | 40.7599 | 17.8623 |
| CAV18407.1 | Extracellular zinc metalloprotease, Vsm | 2,521.28 | 37.0821 | 67.9918 |
HTH, helix-turn-helix.
Expression levels are fragments per kilobase of transcript per million mapped reads (FPKM).
FIG 3.
Partial list of key genes involved in the heat shock response and motility of JZ6 upregulated at 28°C.
Set of upregulated genes related to pathogenicity and virulence of JZ6 at 10°C.
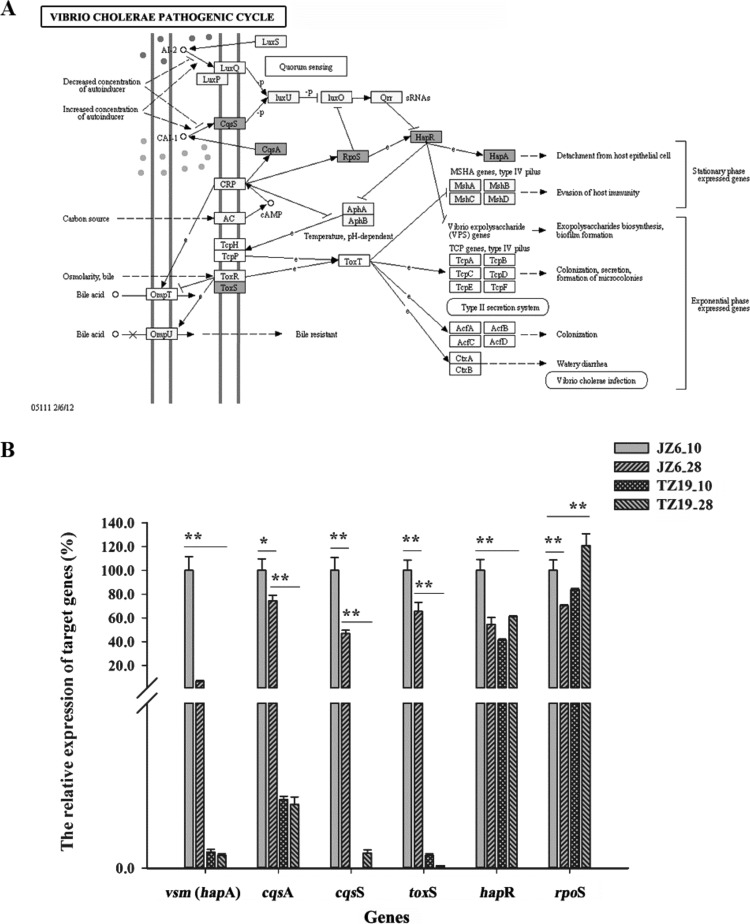
Comprehensive functional and KEGG analyses of the upregulated genes revealed that there were 10 pivotal genes involved in adhesion, protein secretion, and virulence of V. splendidus, including 2 genes (secE and ftsY) in the Sec-dependent pathway, 2 genes (flhG and VS_2437) for Flp pilus assembly, and 6 genes (toxS, cqsA, cqsS, rpoS, hapR, and vsm) in the Vibrio cholerae pathogenic cycle.
SecE (GenBank accession no. CAV20266.1) and FtsY (accession no. CAV20299.1) were essential proteins in the Sec-dependent pathway, and their expression levels were increased 2.61- and 3.58-fold, respectively, in JZ6 at 10°C in comparison with those at 28°C. FlhG (accession no. CAV19597.1), an antiactivator of flagellar biosynthesis, and the pilus assembly protein VS_2437 (accession no. CAV19596.1) were also upregulated 18.29-fold and 8.39-fold, respectively.
The expression levels of six genes in the Vibrio cholerae pathogenic cycle pathway, including genes encoding one transmembrane regulatory protein, ToxS (3.43-fold); two temperature-dependent transcriptional regulators, CqsA (2.56-fold) and CqsS (4.78-fold); one RNA polymerase nonessential primary-like sigma factor, RpoS (17.86-fold); one hemagglutinin (HA)/protease regulatory protein, HapR (2.51-fold); and one extracellular zinc metalloprotease/hemagglutinin, Vsm (67.99-fold) (Fig. 4A), were higher in JZ6 when it was cultured at 10°C. The expression levels of these six genes were further verified by qRT-PCR (Fig. 4B). Their relative expression levels in JZ6_10 were all significantly higher (P < 0.05) than those in JZ6_28, TZ19_10, and TZ19_28, while the expression levels of the vsm, cqsS, toxS, and hapR genes were significantly higher in JZ6_10 (P < 0.01). The expression level of the rpoS gene in JZ6 at 10°C was significantly higher (P < 0.01) than those in JZ6_28 and TZ19_10 but lower than that in TZ19_28.
FIG 4.
Expression pattern of virulence-related genes involved in the Vibrio cholerae pathogenic cycle. (A) KEGG pathway diagram of virulence-related genes in the Vibrio cholerae pathogenic cycle (generated using the KEGG PATHWAY database [Kanehisa Laboratories]). Genes highlighted in gray are upregulated in JZ6 at 10°C. cAMP, cyclic AMP; MSHA, mannose-sensitive hemagglutinin; TCP, toxin-coregulated pilin. (B) Detection of relative expression levels of six upregulated genes of the Vibrio cholerae pathogenic cycle by qRT-PCR.
ECPs and morphological differences in JZ6 at 10°C and 28°C.
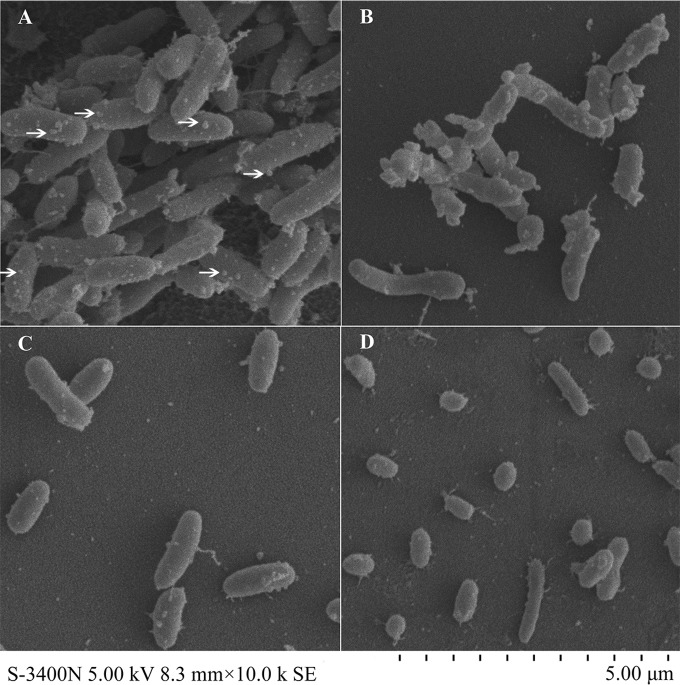
The morphological features of JZ6 and TZ19 were observed, with notable differences between those at 10°C and at 28°C as determined by SEM. Both JZ6 and TZ19 cells cultured at 10°C were larger than when they were grown at 28°C (Fig. 5A and C). There were some “appendages” distributed on the cell surface of JZ6 cells cultured at 10°C, which constituted more complicated structures than those of cells at 28°C. In addition, cells of JZ6 at 10°C were more prone to form aggregations than cells at 28°C (Fig. 5A and B). Conversely, the outer surface of TZ19 cells was smoother than that of JZ6 cells at both 10°C and 28°C, and no aggregation was observed (Fig. 5C and D).
FIG 5.
Scanning electron microscope photographs of V. splendidus strains JZ6 and TZ19 cultured at 10°C and 28°C. (A) JZ6 at 10°C. (B) JZ6 at 28°C. (C) TZ19 at 10°C. (D) TZ19 at 28°C. (Magnification, ×10,000.)
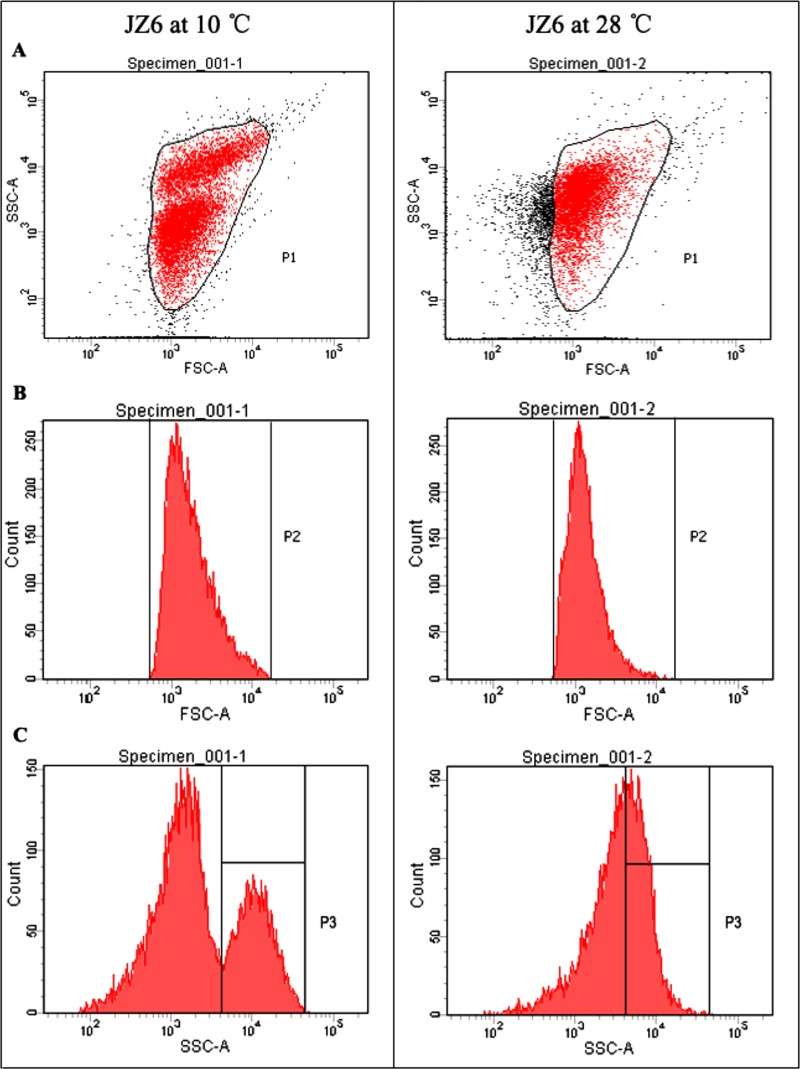
The size and complexity of JZ6 cells at different temperatures were also determined by FACS flow cytometry. The major population of bacterial cells was clearly identified as the peak 1 (P1) area by size and complexity analyses, which covered almost all cells (92.4%) of JZ6 cultured at 10°C (Fig. 6). Only 74% of JZ6 cells at 28°C fell into this area, and other cells outside were smaller and less complex than JZ6 cells at 10°C by both SSC and FSC analyses. JZ6 cells were obviously clustered into two groups when they were cultured at 10°C compared with those at 28°C (Fig. 6A and B). The more intuitive result could also be observed by single-SSC analysis (Fig. 6C), and the cells in this special group occupied 32.3% of the population of JZ6 cells at 10°C.
FIG 6.
FACS flow cytometry analysis of the size and complexity of JZ6 cells at 10°C and 28°C. (A) Scatter diagram for FSC and SSC analyses. (B) Peak diagram for single-FSC analysis. (C) Peak diagram for single-SSC analysis.
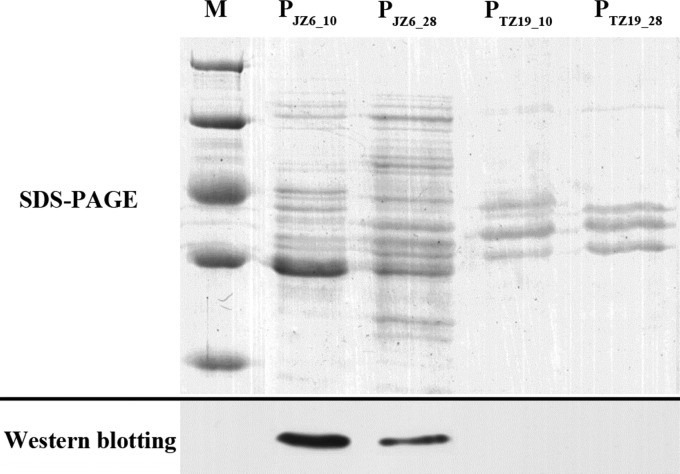
SDS-PAGE analysis revealed differences among the ECP samples (PJZ6_10, PJZ6_28, PTZ19_10, and PTZ19_28) (Fig. 7). The band patterns were different in samples PJZ6_10 and PJZ6_28, and a very thick band was observed in PJZ6_10 but not in PJZ6_28. There were fewer bands in both PTZ19_10 and PTZ19_28 than in PJZ6_10 and PJZ6_28. A distinct band was observed in sample PJZ6_10 by Western blot analysis using the Vsm monoclonal antibody, which had the same molecular weight as that of the rough band observed by SDS-PAGE. No obvious band was detected for the other three ECP samples.
FIG 7.
ECPs of JZ6 and TZ19 cultured at 10°C and 28°C analyzed by SDS-PAGE and Western blotting. Lane M, protein molecular mass standards (in kilodaltons). PJZ6_10, ECP of JZ6 cultured at 10°C; PJZ6_28, ECP of JZ6 cultured at 28°C; PTZ19_10, ECP of TZ19 cultured at 10°C; PTZ19_28, ECP of TZ19 cultured at 28°C. Western blotting detected the extracellular metalloprotease (Vsm) of V. splendidus with polyclonal antiserum (rat anti-Vsm) diluted 1:1,000.
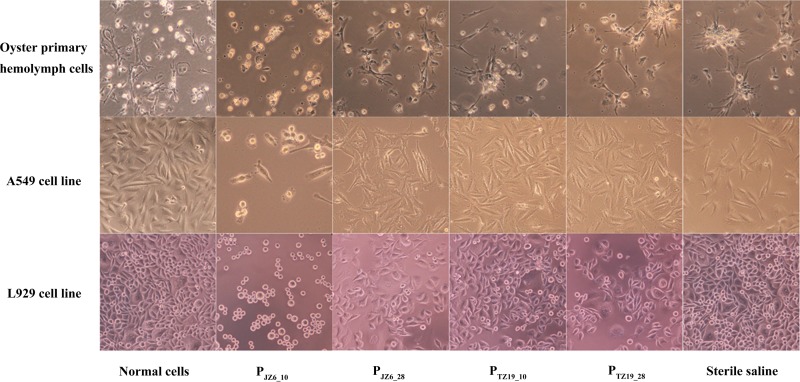
Cytotoxicity of JZ6 ECPs at 10°C.
The ECPs of JZ6 at 10°C (PJZ6_10) exhibited obvious cytotoxicity to oyster primary hemocytes, A549 cells, and L929 cells. After incubation with PJZ6_10, the cell shape became round, shrunken, and detached from 3 h to 24 h. The monolayers of all three cell types were completely destroyed, and only small spheroid textures were observed at 24 h after treatment with PJZ6_10 (Fig. 8). There were no significant changes in cell structures and morphological characteristics for normal cells in the blank, negative-control, or other treatment groups.
FIG 8.
Cytotoxicity of ECPs from different bacterial samples to oyster primary hemolymph cells, human lung cancer cells (A549), and mouse fibroblast cells (L929).
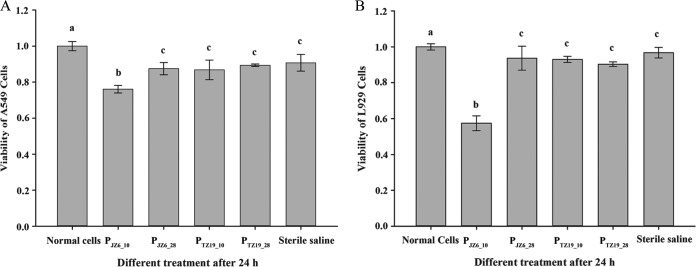
In the Cell Counting kit 8 assay, the viability values for A549 cells were significantly lower (P < 0.01) in the PJZ6_10 treatment group than in the other groups after incubation for 24 h (Fig. 9A). The value for the blank group was higher (P < 0.05) than those for the PJZ6_28, PTZ19_10, PTZ19_28, and sterile saline groups, while no significant differences were observed among the latter four groups. The same result was also obtained for L292 cells in different treatment groups (Fig. 9B).
FIG 9.
Viability analysis of cultured cells treated with ECP samples PJZ6_10, PJZ6_28, PTZ19_10, and PTZ19_28 by using Cell Counting kit 8. (A) Viability of the A549 cell line. (B) Viability of the L929 cell line.
DISCUSSION
V. splendidus strain JZ6 was a pathogenic agent isolated from lesions of diseased Yesso scallops in a low-temperature environment (5). It shared 100% 16S rRNA gene sequence similarity with TZ19, a nonpathogenic V. splendidus isolate from moribund Yesso scallops at the same time in winter (5). Yesso scallops usually grow at an optimal temperature of ∼4°C to 8°C, and juvenile scallops are transferred from the hatching place to a cultured environment in winter (35). Compared with other reported pathogenic V. splendidus strains, JZ6 causes massive mortality of scallops at 10°C, and its virulence was weakened with increased temperature. Hence, environmental temperature was suspected to be one of the key regulators of the virulence of V. splendidus.
In the present study, changes of gene expression in pathogenic strain JZ6 and nonpathogenic strain TZ19 of V. splendidus cultured at 10°C and 28°C were investigated by transcriptome analysis. There were 775 differentially expressed genes in JZ6_10/JZ6_28, 612 differentially expressed genes in TZ19_10/TZ19_28, and 974 differentially expressed genes in JZ6_10/TZ19_10. Interestingly, in total, 388 upregulated genes and 586 downregulated genes were found in JZ6_10 compared with those in TZ19_10, indicating that genes of different V. splendidus strains were expressed differently even though they inhabited the same ecological niche. It has been reported that environmental factors such as temperature, pH, and salinity play major roles in shaping the adaptation of microorganisms to various environments (14, 18, 36). The differentially expressed genes identified in the present study provided new insight to explain the mechanisms underlying the adaptation of V. splendidus at low temperatures and the regulation of virulence as well.
There were 422 genes upregulated and 351 genes downregulated in JZ6_10 compared to those in JZ6_28. After GO distribution analysis, no specific enrichment was found in the upregulated genes from JZ6_10 compared with the reference, indicating that the gene expression and physiological status of strain JZ6 cultured at 10°C might be similar to those under normal conditions. The 351 downregulated genes in JZ6_10 (or, rather, upregulated genes in JZ6_28) were assigned to 25 biological processes by GO distribution analysis, mainly including bacterium-type flagellum, cell projection, and cell motility (see Fig. S5 in the supplemental material). For these biological processes, some representative genes, such as those encoding the small heat shock protein IbpA, heat shock protein 70 (chaperone protein DnaK), DnaK-related protein, the chaperone protein HtpG, the flagellar basal body rod protein FlgF, flagellin core protein A, and polar flagellin B, were involved in the heat stress response and the flagellar assembly process, indicating that these genes might be of benefit for protecting JZ6 from high temperatures and escape from adverse environments (37–39). These results could partly explain the adaptation of JZ6 to low temperatures and its survival in high-temperature environments.
In the Venn diagram of differentially expressed genes in three comparative groups (JZ6_10/JZ6_28, TZ19_10/TZ19_28, and JZ6_10/TZ19_10), there were 241 differentially expressed genes closely related to the high pathogenicity of JZ6 at low temperatures. Most of these genes were involved in adhesion, protein secretion, and virulence of bacteria. In general, the pilus and flagellum are both important proteinaceous structures on the surface of Gram-negative bacteria. The pilus is a critical virulence factor in mediating the attachment and invasion of bacteria into host cells, while the flagellum is mainly responsible for cell motility. It has been reported that some pilus proteins could negatively regulate the expression of flagellar proteins and the process of flagellar biosynthesis (40–42). In Vibrio species, FlhG is a typical pilus assembly protein with a function as an antiactivator for flagellar biosynthesis, and its high expression would impact the synthesis of the flagellum (43). The upregulation of FlhG and downregulation of flagellar proteins in JZ6_10 in the present study were exactly consistent with data from previous reports. Furthermore, another pilus assembly protein, VS_2437, annotated as a protein required for T-pilus biogenesis and virulence to host cells (44, 45), was also upregulated in JZ6_10. The difference in expression between pilus and flagellum assembly proteins suggested that the ability for adhesion was improved and that the motility of strain JZ6 was reduced at 10°C, which might facilitate invasion of the host by strain JZ6_10.
The Vibrio cholerae pathogenic cycle is a classic pathway for the expression of virulence factors in pathogenic Vibrio spp., which was closely associated with the quorum sensing, infection, and secretion systems (46). In the present study, six homologous proteins (HapR, CqsA, CqsS, ToxS, Vsm, and RpoS) involved in the Vibrio cholerae pathogenic cycle were found to have upregulated expression in JZ6_10. The hemagglutinin/protease regulatory protein (HapR) is a master regulator of quorum sensing that operates social behaviors of bacteria, such as alternation between individual and group behaviors and secretion of virulence factors at high cell densities (47–51). When HapR was highly expressed, some autoinducers accumulated to trigger the signaling properties of CqsA-CqsS pairs in the quorum sensing system (52, 53). In the present study, the expression levels of HapR, CqsA, and CqsS in JZ6 were higher at 10°C than at 28°C, indicating that the virulence of strain JZ6 was heightened by the activation of the quorum sensing system at low temperatures, and the activation of quorum sensing was also observed with the aggregation of bacterial cells in JZ6 at 10°C by both SEM and FACS analyses. In addition, as a regulatory protein for the Zn-dependent metalloprotease HA/protease in V. cholerae, HapR could promote the expression of HA/protease by cooperation with the general stress response regulator RpoS (46). In V. splendidus, a metalloprotease (designated Vsm, the homolog of HA) was confirmed as the major determinant of toxicity of ECPs (54, 55), the expression of which was increased 67.99-fold with high expression levels of HapR (2.51-fold) and RpoS (17.86-fold) in strain JZ6 at low temperatures in the present study. Although there was no further information about the regulation effects of HapR and RpoS on Vsm expression in V. splendidus, it was supposed that a pathogenic cycle pathway similar to the Vibrio cholerae pathogenic cycle might be activated in JZ6 at low temperatures, and the upregulation of these genes might contribute to the ability of bacteria with the highest virulence to invade hosts.
For Gram-negative bacteria, secretion systems are vital for pathogenic bacteria to complete their infection. Among them, the general secretory pathway (Sec) is the first secretion system discovered in bacteria (56), which accomplishes the transport and secretion of proteins across the cytoplasmic membrane to the bacterial periplasm and outer membrane by the signal recognition particle (SRP) (57). The SRP-protein complex in the cell cytoplasm is first captured by the SRP receptor (named FtsY) and then delivered to the SecYEG complex and secreted out of the cell (58, 59). In the SecYEG complex, SecE was considered an essential molecule to maintain the stability of this structure, and its high expression could also promote the secretion of proteins (60, 61). In the present study, the expressions of FtsY and SecE were upregulated in JZ6 when it was cultured at 10°C, as determined by transcriptome analysis. Hence, it was suspected that the increased expression of these genes in the Sec-dependent pathway could be the essential mechanism for JZ6 to enhance the transport and secretion of Vsm at low temperatures.
Vsm has been found to have toxicity to mollusk and mouse fibroblastic cell lines (54, 55). In the present study, the high expression level of Vsm in strain JZ6 at 10°C was also confirmed by Western blotting, which was consistent with the results of transcriptome analysis and qRT-PCR analysis, and the cytotoxicity of JZ6 ECPs to oyster primary hemolymph cells, A549 cells, and L929 cells was further examined to determine the association of the high expression level of Vsm with virulence of JZ6. Compared with PTZ19_10, PTZ19_28, and PJZ6_28, ECP sample PJZ6_10 manifested strong cytotoxicity to all tested cells, which suggested a positive correlation of pathogenicity with the expression of Vsm. All these results indicated that the specific transcriptional pattern and the high expression levels of genes in virulence-associated biological processes could regulate the synthesis and secretion of Vsm at 10°C, which endowed JZ6 with high pathogenicity to aquaculture animals at low temperatures.
Conclusion.
In total, 358 differentially expressed genes were identified by comparing the transcripts of pathogenic strain JZ6 and nonpathogenic strain TZ19 at 10°C and 28°C, which were considered to be closely related to the high pathogenicity of JZ6 at low temperatures. Ten of these molecules, FlhG, VS_2437, ToxS, CqsA, CqsS, RpoS, HapR, Vsm, SecE, and FtsY, were identified as key molecules in adhesion, quorum sensing, virulence, and protein secretion systems of V. splendidus. The high expression levels of these genes could enhance adhesion, activation of the quorum sensing system, and upregulation of the synthesis of virulence factors and their secretion and, lastly, increase the pathogenicity of JZ6 at 10°C.
Supplementary Material
ACKNOWLEDGMENTS
We thank all laboratory members for the technical advice and helpful discussion.
This research was supported by a grant (grant no. 31302224) from the National Science Foundation of China, the Modern Agro-Industry Technology Research System (CARS-48).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03486-15.
REFERENCES
- 1.Reichelt JL, Baumann P, Baumann L. 1976. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridisation. Arch Microbiol 110:101–120. doi: 10.1007/BF00416975. [DOI] [PubMed] [Google Scholar]
- 2.Lacoste A, Jalabert F, Malham S, Cueff A, Gélébart F, Cordevant C, Lange M, Poulet SA. 2001. A Vibrio splendidus strain is associated with summer mortality of juvenile oysters Crassostrea gigas in the Bay of Morlaix (North Brittany, France). Dis Aquat Organ 46:139–145. doi: 10.3354/dao046139. [DOI] [PubMed] [Google Scholar]
- 3.Thomson R, Macpherson HL, Riaza A, Birkbeck TH. 2005. Vibrio splendidus biotype 1 as a cause of mortalities in hatchery-reared larval turbot, Scophthalmus maximus (L). J Appl Microbiol 99:243–250. doi: 10.1111/j.1365-2672.2005.02602.x. [DOI] [PubMed] [Google Scholar]
- 4.Mateo DR, Siah A, Araya MT, Berthe FC, Johnson GR, Greenwood SJ. 2009. Differential in vivo response of soft-shell clam hemocytes against two strains of Vibrio splendidus: changes in cell structure, numbers and adherence. J Invertebr Pathol 102:50–56. doi: 10.1016/j.jip.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Liu R, Qiu L, Yu Z, Zi J, Yue F, Wang L, Zhang H, Teng W, Liu X, Song L. 2013. Identification and characterisation of pathogenic Vibrio splendidus from Yesso scallop (Patinopecten yessoensis) cultured in a low temperature environment. J Invertebr Pathol 114:144–150. doi: 10.1016/j.jip.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Du M, Chen JX, Zhang XH, Li AJ, Li Y. 2007. Characterization and resuscitation of viable but nonculturable Vibrio alginolyticus VIB283. Arch Microbiol 188:283–288. doi: 10.1007/s00203-007-0246-5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang XH. 2007. Marine microbiology. China Ocean University Press, Qingdao, China. [Google Scholar]
- 8.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 9.Gómez-León J, Villamil L, Lemos ML, Novoa B, Figueras A. 2005. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl Environ Microbiol 71:98–104. doi: 10.1128/AEM.71.1.98-104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L. 2009. Two pathogens of greenshell mussel larvae, Perna canaliculus: Vibrio splendidus and a V. coralliilyticus/neptunius-like isolate. J Fish Dis 32:499–507. doi: 10.1111/j.1365-2761.2009.01006.x. [DOI] [PubMed] [Google Scholar]
- 11.Jones JL, Lüdeke CH, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. 2012. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol 50:2343–2352. doi: 10.1128/JCM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polissi A, De Laurentis W, Zangrossi S, Briani F, Longhi V, Pesole G, Dehò G. 2003. Changes in Escherichia coli transcriptome during acclimatization at low temperature. Res Microbiol 154:573–580. doi: 10.1016/S0923-2508(03)00167-0. [DOI] [PubMed] [Google Scholar]
- 13.Crapoulet N, Barbry P, Raoult D, Renesto P. 2006. Global transcriptome analysis of Tropheryma whipplei in response to temperature stresses. J Bacteriol 188:5228–5239. doi: 10.1128/JB.00507-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu DQ, Li Y, Xu Y. 2012. Comparative analysis of temperature-dependent transcriptome of Pseudomonas aeruginosa strains from rhizosphere and human habitats. Appl Microbiol Biotechnol 96:1007–1019. doi: 10.1007/s00253-012-4466-5. [DOI] [PubMed] [Google Scholar]
- 15.Cho S, Cho Y, Lee S, Kim J, Yum H, Kim SC, Cho BK. 2013. Current challenges in bacterial transcriptomics. Genomics Inform 11:76–82. doi: 10.5808/GI.2013.11.2.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sorek R, Cossart P. 2010. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11:9–16. doi: 10.1038/nrg2695. [DOI] [PubMed] [Google Scholar]
- 17.Li JS, Bi YT, Dong C, Yang JF, Liang WD. 2011. Transcriptome analysis of adaptive heat shock response of Streptococcus thermophilus. PLoS One 6:e25777. doi: 10.1371/journal.pone.0025777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mols M, Mastwijk H, Nierop Groot M, Abee T. 2013. Physiological and transcriptional response of Bacillus cereus treated with low-temperature nitrogen gas plasma. J Appl Microbiol 115:689–702. doi: 10.1111/jam.12278. [DOI] [PubMed] [Google Scholar]
- 19.Xu Q, Dziejman M, Mekalanos JJ. 2003. Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci U S A 100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 21.Perkins TT, Davies MR, Klemm EJ, Rowley G, Wileman T, James K, Keane T, Maskell D, Hinton JC, Dougan G, Kingsley RA. 2013. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol Microbiol 87:526–538. doi: 10.1111/mmi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, Du Z, Huang L, Vera Cruz C, Zhou Y, Li Z. 2013. Comparative transcriptome profiling reveals different expression patterns in Xanthomonas oryzae pv. oryzae strains with putative virulence-relevant genes. PLoS One 8:e64267. doi: 10.1371/journal.pone.0064267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceccarelli D, Hasan NA, Huq A, Colwell RR. 2013. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front Cell Infect Microbiol 3:97. doi: 10.3389/fcimb.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson CN. 2013. Fitness factors in vibrios: a mini-review. Microb Ecol 65:826–851. doi: 10.1007/s00248-012-0168-x. [DOI] [PubMed] [Google Scholar]
- 25.Wilson JW. 2014. Infectious complications in cancer patients: bacterial pathogens. Cancer Treat Res 161:91–128. doi: 10.1007/978-3-319-04220-6_3. [DOI] [PubMed] [Google Scholar]
- 26.Antunes LC, Schaefer AL, Ferreira RB, Qin N, Stevens AM, Ruby EG, Greenberg EP. 2007. Transcriptome analysis of the Vibrio fischeri LuxR-LuxI regulon. J Bacteriol 189:8387–8391. doi: 10.1128/JB.00736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elgaml A, Higaki K, Miyoshi S. 2014. Effects of temperature, growth phase and luxO-disruption on regulation systems of toxin production in Vibrio vulnificus strain L-180, a human clinical isolate. World J Microbiol Biotechnol 30:681–691. doi: 10.1007/s11274-013-1501-3. [DOI] [PubMed] [Google Scholar]
- 28.Ushijima B, Videau P, Burger AH, Shore-Maggio A, Runyon CM, Sudek M, Aeby GS, Callahan SM. 2014. Vibrio coralliilyticus strain OCN008 is an etiological agent of acute Montipora white syndrome. Appl Environ Microbiol 80:2102–2109. doi: 10.1128/AEM.03463-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yue F, Shi X, Zhou Z, Wang L, Wang M, Yang J, Qiu L, Song L. 2013. The expression of immune-related genes during the ontogenesis of scallop Chlamys farreri and their response to bacterial challenge. Fish Shellfish Immunol 34:855–864. doi: 10.1016/j.fsi.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Sun X, Zhou Z, Zhang T, Yi Q, Liu R, Wang M, Song L. 2014. The promotion of cytoskeleton integration and redox in the haemocyte of shrimp Litopenaeus vannamei after the successive stimulation of recombinant VP28. Dev Comp Immunol 45:123–132. doi: 10.1016/j.dci.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y. 2010. Discovering the false discovery rate. J R Stat Soc B 72:405–416. doi: 10.1111/j.1467-9868.2010.00746.x. [DOI] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Balebona MC, Andreu MJ, Bordas MA, Zorrilla I, Moriñigo MA, Borrego JJ. 1998. Pathogenicity of Vibrio alginolyticus for cultured gilt-head sea bream (Sparus aurata L.). Appl Environ Microbiol 64:4269–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal Biochem 150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 35.Gardner P, IEC International. 2001. Economic potential of sea ranching and enhancement of selected shellfish species in Canada (section IV scallop culture in Japan). Office of the Commissioner for Aquaculture Development, Department of Fisheries and Oceans, Ottawa, Ontario, Canada. [Google Scholar]
- 36.Kato S, Kosaka T, Watanabe K. 2008. Comparative transcriptome analysis of responses of Methanothermobacter thermautotrophicus to different environmental stimuli. Environ Microbiol 10:893–905. doi: 10.1111/j.1462-2920.2007.01508.x. [DOI] [PubMed] [Google Scholar]
- 37.Inoue Y, Baker MA, Fukuoka H, Takahashi H, Berry RM, Ishijima A. 2013. Temperature dependences of torque generation and membrane voltage in the bacterial flagellar motor. Biophys J 105:2801–2810. doi: 10.1016/j.bpj.2013.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu S, Kojima S, Homma M. 2013. Structure, gene regulation and environmental response of flagella in Vibrio. Front Microbiol 4:410. doi: 10.3389/fmicb.2013.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L, Huang L, Su Y, Qin Y, Kong W, Ma Y, Xu X, Lin M, Zheng J, Yan Q. 2015. Involvement of the flagellar assembly pathway in Vibrio alginolyticus adhesion under environmental stresses. Front Cell Infect Microbiol 5:59. doi: 10.3389/fcimb.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo AW, Moonens K, Remaut H. 2013. Chemical attenuation of pilus function and assembly in Gram-negative bacteria. Curr Opin Microbiol 16:85–92. doi: 10.1016/j.mib.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 41.Patenge N, Fiedler T, Kreikemeyer B. 2013. Common regulators of virulence in streptococci. Curr Top Microbiol Immunol 368:111–153. doi: 10.1007/82_2012_295. [DOI] [PubMed] [Google Scholar]
- 42.Volkan E, Kalas V, Pinkner JS, Dodson KW, Henderson NS, Pham T, Waksman G, Delcour AH, Thanassi DG, Hultgren SJ. 2013. Molecular basis of usher pore gating in Escherichia coli pilus biogenesis. Proc Natl Acad Sci U S A 110:20741–20716. doi: 10.1073/pnas.1320528110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Correa NE, Peng F, Klose KE. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol 187:6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward JE Jr, Dale EM, Christie PJ, Nester EW, Binns AN. 1990. Complementation analysis of Agrobacterium tumefaciens Ti plasmid virB genes by use of a vir promoter expression vector: virB9, virB10, and virB11 are essential virulence genes. J Bacteriol 172:5187–5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ripoll-Rozada J, Zunzunegui S, de la Cruz F, Arechaga I, Cabezón E. 2013. Functional interactions of VirB11 traffic ATPases with VirB4 and VirD4 molecular motors in type IV secretion systems. J Bacteriol 195:4195–4201. doi: 10.1128/JB.00437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klose KE, Mekalanos JJ. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol 28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 47.Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu Rev Microbiol 55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 48.Rutherford ST, van Kessel JC, Shao Y, Bassler BL. 2011. AphA and LuxR/HapR reciprocally control quorum sensing in vibrios. Genes Dev 25:397–408. doi: 10.1101/gad.2015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H, Wu JH, Ayala JC, Benitez JA, Silva AJ. 2011. Interplay among cyclic diguanylate, HapR, and the general stress response regulator (RpoS) in the regulation of Vibrio cholerae hemagglutinin/protease. J Bacteriol 193:6529–6538. doi: 10.1128/JB.05166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Kessel JC, Rutherford ST, Shao Y, Utria AF, Bassler BL. 2013. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J Bacteriol 195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rutherford ST, Bassler BL. 2012. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng WL, Perez LJ, Wei Y, Kraml C, Semmelhack MF, Bassler BL. 2011. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol Microbiol 79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ke X, Miller LC, Ng WL, Bassler BL. 2014. CqsA-CqsS quorum-sensing signal-receptor specificity in Photobacterium angustum. Mol Microbiol 91:821–833. doi: 10.1111/mmi.12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl Environ Microbiol 73:777–784. doi: 10.1128/AEM.02147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Binesse J, Delsert C, Saulnier D, Champomier-Vergès MC, Zagorec M, Munier-Lehmann H, Mazel D, Le Roux F. 2008. Metalloprotease Vsm is the major determinant of toxicity for extracellular products of Vibrio splendidus. Appl Environ Microbiol 74:7108–7117. doi: 10.1128/AEM.01261-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rusch SL, Kendall DA. 2007. Interactions that drive Sec-dependent bacterial protein transport. Biochemistry 46:9665–9673. doi: 10.1021/bi7010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kudva R, Denks K, Kuhn P, Vogt A, Müller M, Koch HG. 2013. Protein translocation across the inner membrane of Gram-negative bacteria: the Sec and Tat dependent protein transport pathways. Res Microbiol 164:505–534. doi: 10.1016/j.resmic.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 58.Bernstein HD, Hyndman JB. 2001. Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J Bacteriol 183:2187–2197. doi: 10.1128/JB.183.7.2187-2197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park E, Rapoport TA. 2012. Bacterial protein translocation requires only one copy of the SecY complex in vivo. J Cell Biol 198:881–893. doi: 10.1083/jcb.201205140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gumbart J, Schulten K. 2007. Structural determinants of lateral gate opening in the protein translocon. Biochemistry 46:11147–11157. doi: 10.1021/bi700835d. [DOI] [PubMed] [Google Scholar]
- 61.Beckwith J. 2013. The Sec-dependent pathway. Res Microbiol 164:497–504. doi: 10.1016/j.resmic.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.