Abstract
The pandemic of hospital-acquired infections caused by methicillin-resistant Staphylococcus aureus (MRSA) has declined, but the evolution of strains with enhanced virulence and toxins and the increase of community-associated infections are still a threat. In previous studies, 107 MRSA bacteria applied as simulated droplet contamination were killed on copper and brass surfaces within 90 min. However, contamination of surfaces is often via finger tips and dries rapidly, and it may be overlooked by cleaning regimes (unlike visible droplets). In this new study, a 5-log reduction of a hardy epidemic strain of MRSA (epidemic methicillin-resistant S. aureus 16 [EMRSA-16]) was observed following 10 min of contact with copper, and a 4-log reduction was observed on copper nickel and cartridge brass alloys in 15 min. A methicillin-sensitive S. aureus (MSSA) strain from an osteomyelitis patient was killed on copper surfaces in 15 min, and 4-log and 3-log reductions occurred within 20 min of contact with copper nickel and cartridge brass, respectively. Bacterial respiration was compromised on copper surfaces, and superoxide was generated as part of the killing mechanism. In addition, destruction of genomic DNA occurs on copper and brass surfaces, allaying concerns about horizontal gene transfer and copper resistance. Incorporation of copper alloy biocidal surfaces may help to reduce the spread of this dangerous pathogen.
INTRODUCTION
Intrinsic penicillin resistance and acquisition of resistance to methicillin in the 1980s by Staphylococcus aureus led to a pandemic of infections worldwide. Initially, the majority of infections were contracted in health care environments, but incorporation of measures to control the spread, including preadmission screening, decolonization, improved disinfection, and antibiotic treatment, have stemmed the tide (1). The increased use of antibiotics required for the epidemic of infections caused by Gram-positive pathogens has allowed the evolution of multidrug-resistant Gram-negative pathogens, effectively transforming some commensal gut bacteria into potential killers. However, new strains of S. aureus that have acquired further virulence factors and toxins or that have adapted to a specific environment, for example, an increased ability to cause bacteremia (2), are still a considerable threat. There is now widespread community-associated methicillin-resistant S. aureus (CA-MRSA), and infections can spread within households, day care centers, and schools (3). In addition, Giuffre et al. observed an increasing incidence of MRSA in neonates (4). The ability of some strains of MRSA to revert to methicillin-susceptible isolates, particularly in skin and soft tissue infections, has been observed (5).
Colonization with MRSA increases the risk of MRSA infection, particularly following illness, surgical procedures, and treatment with immunosuppressive drugs. Colonization and/or infection may also occur from touching contaminated surfaces. In the community, a recent study observed that 58% and 82% of surfaces in 19 fire stations in Washington were positive for MRSA and methicillin-sensitive S. aureus (MSSA), respectively (6), and 37% of fire service professionals had MRSA requiring medical attention. Otter and French observed that 8% of sites tested in the London public transport system had MSSA contamination, but no MRSA was detected (7). The use of biocidal surfaces may play a role in preventing infection transmission from contaminated surfaces when combined with stringent cleaning regimes. Laboratory studies have suggested that copper surfaces may be effective against a range of bacteria, fungi, and viruses (8–11) and that the irreversible pathogen nucleic acid destruction observed may allay fears of biocide resistance (12). A previous study observed that strains of MRSA were killed on copper surfaces in simulated wet-droplet contamination (13). However, surface contamination is often the result of fingertip touch, which dries rapidly. In this study, we investigated the abilities of several copper alloys to kill MRSA and MSSA and the mechanism of bacterial death on surfaces contaminated by simulated droplet and fingertip touch.
MATERIALS AND METHODS
Bacterial strains.
Methicillin-resistant Staphylococcus aureus NCTC 13143 (epidemic methicillin-resistant S. aureus 16 [EMRSA-16]) was supplied by Public Health England, United Kingdom. Methicillin-sensitive Staphylococcus aureus ATCC 49230 (CDC587), which was originally isolated from a patient with chronic osteomyelitis, was supplied by the American Type Culture Collection, United States.
Metal surfaces.
Metal samples were provided by the Copper Development Association (Table 1) and were cut into coupons (10 by 10 by 0.5 mm) that were cleaned and sterilized as described previously (14).
TABLE 1.
Composition of metals used for the study
| Metal type | UNSa no. | Composition (%) of: |
|||||
|---|---|---|---|---|---|---|---|
| Cu | Zn | Sn | Ni | Fe | Cr | ||
| Copper | C11000 | 100 | |||||
| Copper nickel 10 | C70600 | 89–90 | 10 | <1 | |||
| Cartridge brass | C26000 | 70 | 30 | ||||
| Muntz metal | C28000 | 60 | 40 | ||||
| Stainless steel 18/8 | S30400 | 8 | 74 | 18 | |||
UNS, unified numbering system.
Bacterial survival on surfaces assessed by culture and in situ detection of respiring cells.
For simulated fingertip touch contamination, aliquots of exponentially growing cultures of the two species in tryptone soy broth (TSB) were centrifuged to pellet the cells and resuspended in fresh growth medium to give approximately 107 CFU/μl. One microliter was spread over the surface of a coupon to allow rapid drying within seconds and was incubated for various amounts of time. This size of the inoculum represents a heavy bioburden and permits measuring an endpoint for inactivation. Cells were removed by vortexing them in phosphate-buffered saline (PBS) with glass beads as described previously (14) (there was no significant difference if chelators were present to neutralize copper ions), dilutions were spread onto tryptone soy agar (TSA) plates that were incubated at 37°C for up to 48 h, and numbers of CFU per coupon were calculated. Replicate coupons were done for each time point. Simulated droplet contamination was done by inoculating the same number of cells in 20 μl per coupon. Cells were also inoculated in the presence of the chelators EDTA (20 mM) and bathocuproinedisulfonic acid (BCS; 20 mM) to chelate Cu(II) and Cu(I) ions, respectively. Reactive oxygen species (ROS) quenchers d-mannitol (20 mM), 4,5-dihydroxy-1,3-benzenedisulfonic acid (Tiron), and superoxide dismutase (SOD; 500 U/ml) were used to remove hydroxyl and superoxide radicals, respectively; 500 U/ml catalase and 10% (wt/vol) sucrose were also used to decompose hydrogen peroxide and to investigate osmotic stress, respectively.
Respiring bacterial cells produce electrons that reduce the redox dye CTC (5-cyano-2,3-ditolyl tetrazolium chloride) to insoluble formazan, which can be visualized as a red fluorescent stain using epifluorescence microscopy. SYTO 9 is a membrane-permeable dye that binds to intact double-stranded DNA and emits green fluorescence. The stain will not bind to degraded nucleic acid. Metal coupons were inoculated as described for culture assessment and were stained in situ as described previously (14). Briefly, cells were dually stained with 5 mM CTC and 5 μM SYTO 9 for the final 90 and 30 min of the tested 2-h contact time, respectively. Bacterial cells were observed in situ for actively respiring cells (red) and for all cells with intact DNA, live or dead (green). Cells were also inoculated in the presence of chelators or ROS quenchers as described for culture. Stainless steel was used as a control surface throughout.
In situ DNA destruction of MRSA (simulated fingertip touch contamination).
Bacterial cells in 1 ml of overnight culture were pelleted and washed in PBS. The cells were stained with 10 μM SYTO 9 for 30 min at room temperature. The stained cells were washed to remove excess stain and resuspended in 50 μl fresh culture medium; 1 μl was applied to metal coupons, and fluorescence microscopy images were recorded every minute for 20 min.
DNA fragmentation.
The protocol was originally described by Fernandez et al. (15) and has been adapted in our laboratory (14). Briefly, approximately 107 bacterial cells that had been exposed to copper, stainless steel, cartridge brass, or Muntz metal for 2 h were treated with 40 U lysostaphin for 15 min at 37°C. The cells were then encased in low-melting-point agarose on a slide previously coated with standard agarose. Following membrane permeabilization, the slides were dried and stained with SYBR gold (Invitrogen, United Kingdom), which detects single- and double-stranded DNA, for 5 min at room temperature in the dark. Cells were observed with epifluorescence microscopy for the presence of DNA loops representing the intact genome or dispersed fragments.
Statistical analysis.
Data are expressed as means ± standard errors of the means (SEM) and are from multiple independent experiments. Statistical analyses and graphical representations were performed using GraphPad Prism version 6.
RESULTS AND DISCUSSION
Staphylococcus aureus can persist on surfaces for many months and is easily transferred to hands contacting surfaces (reviewed by Kramer et al. [16]). This represents a significant infection risk, especially in the health care environment and often via the hands of health care workers. In addition, a recent small study in Egypt observed that 100% of mobile phones belonging to patients and health care workers were contaminated with bacterial pathogens, and MRSA was detected in >50%, which may result in transferring pathogens out of the health care environment into the community (17).
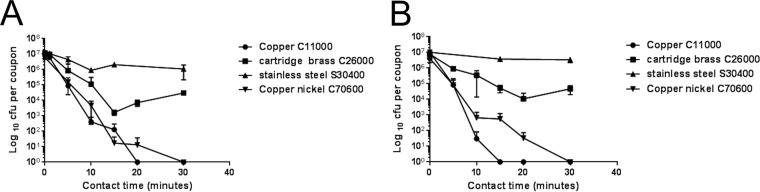
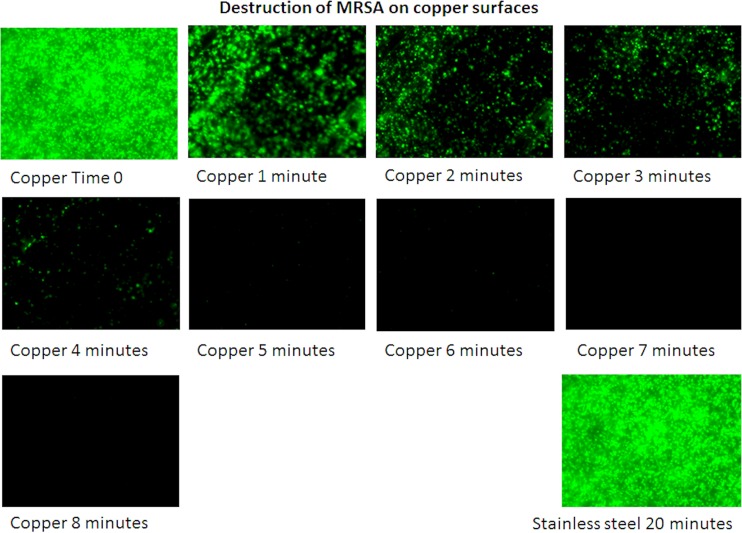
A previous study (13) suggested that strains of MRSA applied as a wet droplet (107 bacteria in 20 μl/cm2) were killed on copper surfaces in 45 to 90 min of contact, with EMRSA-16 being the most resilient. In this new study, we have shown that the same inoculum size of EMRSA-16 applied in a low volume that dries in seconds, to simulate fingertip contamination, was killed in 20 min, and almost a 5 log-reduction occurred within the first 10 min (Fig. 1A). Fingertip touch will also contaminate the surface with biomolecules transferred from the skin surface, so the cells were inoculated in bacteriological medium, which contains a complex mixture of proteins, salts, and sugars. Following 30 min of contact, there was a 2-log reduction in viable cells on cartridge brass (70% copper) and a complete kill on copper nickel 10 (90% copper), which has been shown previously to have excellent antiviral activity. The results were similar for MSSA, except complete killing on copper occurred within 15 min (Fig. 1B). A previous study observed that exposure of MRSA to copper inhibits respiration and compromises DNA integrity (18). Analysis of the bacterial DNA in situ on copper surfaces in the new study suggested that rapid destruction occurs over the first 5 min of contact, which does not occur on stainless steel (Fig. 2) and is equivalent to an approximately 2-log reduction in viable cells observed in the culture results (Fig. 1). This suggests that the development of copper resistance is unlikely and that the problems that have arisen with using some biocides and with concomitant antibiotic resistance (19) should not occur with the use of antimicrobial copper surfaces.
FIG 1.
Rapid death of an epidemic strain of methicillin-resistant Staphylococcus aureus (EMRSA) (A) and methicillin-susceptible Staphylococcus aureus (MSSA) (B) fingertip contamination on copper. Approximately 107 CFU in 1 μl was applied to 1-cm2 test surfaces and was immediately spread over the surface. Bacteria were removed after various contact times at 21°C as described in the text, and they were assessed for viability by growth on agar. Results are expressed as CFU per coupon (± SEM) and are the results of multiple experiments.
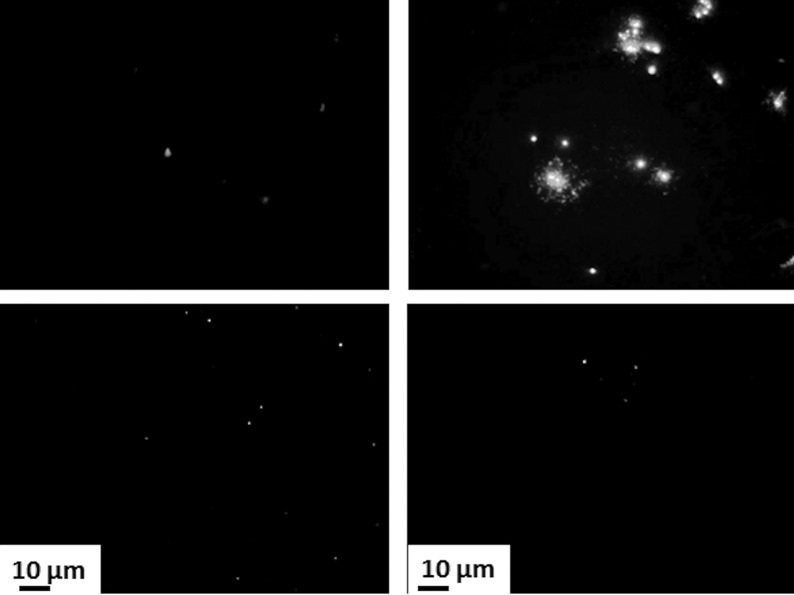
FIG 2.
Destruction of the DNA of an epidemic strain of methicillin-resistant Staphylococcus aureus (EMRSA) on copper surfaces. Approximately 107 bacterial cells were prestained with SYTO 9, which intercalates into intact DNA, and were applied to copper surfaces. Cells with intact DNA appear green using epifluorescence microscopy. Images were recorded every minute (at different fields of view to eliminate the bleaching effect of the excitation light source). A rapid reduction in cells with intact DNA occurred on copper surfaces following the first 5 min of contact. In contrast, there was no reduction in the staining of cells exposed to stainless steel for the experiment duration.
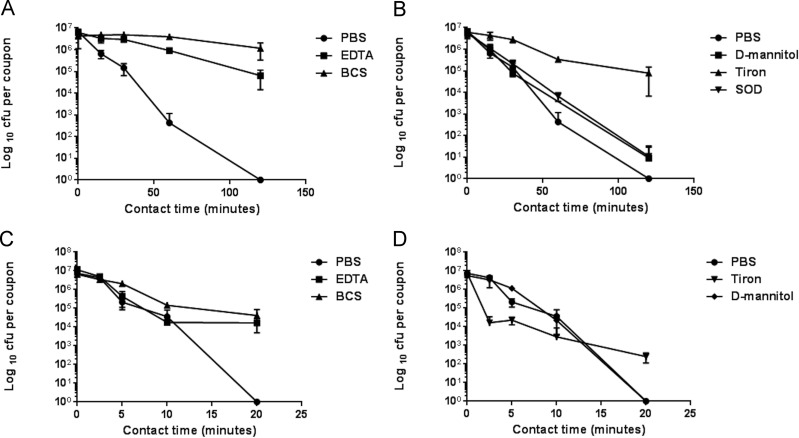
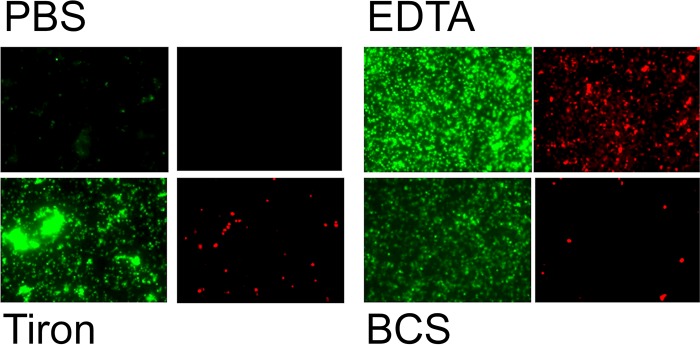
Further investigations into the mechanism of copper killing of MRSA were done for the two types of surface contamination. Chelators of Cu(I) and Cu(II) offered significant protection for EMRSA-16 on copper surfaces (Fig. 3A and C), suggesting that these moieties are directly or indirectly responsible for bacterial death. Generation of free radicals has been suggested to be involved in the copper kill mechanism, and the quenching potential of superoxide generation with membrane-permeable Tiron gave significant protection on copper surfaces for droplet contamination (Fig. 3B); however, the addition of superoxide dismutase (SOD) to dismutate superoxide was not as effective. This may be because the enzyme activity is reduced at the neutral pH used in the test procedure or that the large protein was unable to cross the cell membrane to dismutate intracellular superoxide. Tiron was protective for simulated dry-touch contamination but only after the initial 10 min of contact; thereafter, there were still log-3 survivors on copper after 20 min (Fig. 3D). d-Mannitol displayed a minimal protective effect, suggesting that hydroxyl radicals were not the primary instigators of bacterial death on copper (Fig. 3B and D). It has been shown that generation of highly toxic hydroxyl radicals via Fenton reaction between Cu(I) and hydrogen peroxide combined with direct copper ion action led to the rapid death of Gram-negative bacteria on copper surfaces (20). The results from this new study suggest that reactive oxygen species (ROS) are involved in bacterial killing on copper but not via Fenton chemistry. This was further supported when removal of copper ionic species (by chelators) and superoxide (by quenchers) allowed the bacterial cells to continue respiring and DNA to remain intact (Fig. 4), but addition of d-mannitol to quench hydroxyl radicals and catalase to decompose hydrogen peroxide was not protective. Further evidence for the DNA damage can be observed using the genomic DNA fragmentation assay. Bacteria exposed to stainless steel display intact loops of genomic DNA emanating from and surrounding each lysed bacterial cell, but in cells previously exposed to copper and copper alloys, the DNA has degraded to small fragments that are too small to be visualized (Fig. 5). The loss of genomic DNA was commensurate with cell death.
FIG 3.
Determination of the role of copper ions and reactive oxygen species (ROS) in the rapid death of an epidemic strain of methicillin-resistant Staphylococcus aureus (EMRSA) on copper surfaces. Approximately 107 cells were applied to copper surfaces in 20 μl to simulate wet-droplet (A, B) or dry-touch (C, D) contamination as described in the text. If cells were inoculated in the presence of the chelators BCS and EDTA to chelate Cu(I) and Cu(II), respectively, a protective effect was seen in the droplet and dry-touch scenarios compared to that in inoculation in PBS (A, C). Inoculation in the presence of Tiron to quench superoxide generation also had a significant protective effect, especially in the droplet contamination (B), although superoxide dismutase (SOD) protection was much lower. d-Mannitol, which quenches hydroxyl radicals, had a small protective effect in simulated droplet contamination (B).
FIG 4.
Protection of bacterial DNA and respiration with chelators and reactive oxygen species quenchers in an epidemic strain of methicillin-resistant Staphylococcus aureus (EMRSA) exposed to copper surfaces (simulated droplet contamination). Approximately 107 cells were applied to copper surfaces in 20 μl of PBS or PBS supplemented with chelators or ROS quenchers. During 2 h of contact time at 21°C, the cells were dually stained in situ with CTC and SYTO 9 to detect actively respiring cells (fluoresce red) and total cells with intact DNA (fluoresce green), respectively, as described in the text. Cells inoculated in PBS did not stain with either stain, suggesting that cells were not respiring and DNA had disintegrated. However, when EDTA, BCS, and Tiron were present, the bacterial DNA was protected and cells respired; this suggests that Cu(II), Cu(I), and superoxide generation, respectively, are required for the killing mechanism on copper. No protective effect was observed on copper surfaces with d-mannitol, catalase, or sucrose (not shown). Cells exposed to stainless steel surfaces in PBS or with any supplements displayed intact DNA and active respiration (not shown).
FIG 5.
Exposure to copper and brass surfaces affects the DNA of an epidemic strain of methicillin-resistant Staphylococcus aureus (EMRSA) (simulated droplet contamination). The DNA fragmentation assay allows the DNA integrity of individual bacterial cells to be observed. Bacteria were exposed to copper, brass, and stainless steel surfaces for 2 h at 21°C (±1°C), immersed in agarose, permeabilized, and stained as described in the text. On steel, individual cells with intact DNA loops protruding though the permeabilized membrane can be seen. There is virtually no stained DNA in cells exposed to copper and brasses, suggesting that extensive disintegration of the DNA has occurred. (Top left) Copper C11000; (top right) stainless steel S30400; (bottom left) brass C28000; (bottom right) brass C26000.
These results are comparable to the mechanism of copper toxicity that was observed by our laboratory for other Gram-positive bacteria, i.e., pathogenic enterococci (21). This supports our previous conclusions that the mechanism of bacterial death on copper surfaces is multifaceted and involves a combination of direct copper ion attack of bacterial structural and metabolic biomolecules and suicidal generation of ROS. Although Gram-positive and Gram-negative bacteria die on copper surfaces, the targets vary and rapid breakdown of nucleic acid is observed in the former.
The tide of MRSA infections observed in the 1980s has been reduced by implementation of many interventions, including screening regimes, isolation, decolonization, antibiotic stewardship, and disinfection, and has been superseded by multidrug-resistant Enterobacteriaceae. We must guard against complacency, especially with the increase in Staphylococcus aureus community infections and the evolution of strains with increased virulence. This study is the first to show very rapid killing of fingertip contamination of MRSA and MSSA on copper alloys, and the authors propose that incorporation of copper alloy surfaces may help to reduce the transmission of MRSA and MSSA from contaminated surfaces. The real-life bacterial bioburden is considerably less than that tested here, suggesting that kill times may be even faster, and a hospital trial has already shown that incorporation of just 6 copper surfaces in intensive care units significantly reduced MRSA colonization and health care-acquired infection over a 12-month period (22, 23). Further trials are now urgently needed to determine if the wealth of data from laboratory studies showing high efficacy of copper alloys to kill or inactivate a large range of microbial pathogens can be extrapolated to real-life environments to reduce the number of infections contracted from touching contaminated surfaces.
ACKNOWLEDGMENTS
The Copper Development Association (CDA), New York, and the International Copper Association, New York, provided funding for this research. CDA personnel were not involved in the development, design, or execution of the experiments in this study.
Funding Statement
The Copper Development Association (CDA), New York, provided funding to Sarah L. Warnes and C. William Keevil. The Copper Development Association (CDA), New York, was not involved in the work or in the writing of the paper.
REFERENCES
- 1.Otto M. 2012. MRSA virulence and spread. Cell Microbiol 14:1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgeworth JD, Yadegarfar G, Pathak S, Batra R, Cockfield JD, Wyncoll D, Beale R, Lindsay JA. 2007. An outbreak in an intensive care unit of a strain of methicillin-resistant Staphylococcus aureus sequence type 239 associated with an increased rate of vascular access device-related bacteremia. Clin Infect Dis 44:493–501. doi: 10.1086/511034. [DOI] [PubMed] [Google Scholar]
- 3.Knox J, Uhlemann AC, Lowy FD. 2015. Staphylococcus aureus infections: transmission within households and the community. Trends Microbiol 23:437–444. doi: 10.1016/j.tim.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giuffre M, Amodio E, Bonura C, Geraci DM, Saporito L, Ortolano R, Corsello G, Mammina C. 2015. Methicillin-resistant Staphylococcus aureus nasal colonization in a level III neonatal intensive care unit: incidence and risk factors. Am J Infect Control 43:476–481. doi: 10.1016/j.ajic.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 5.Patel AB, Hill E, Simpson EL, Hanifin JM. 2013. Reversion of methicillin-resistant Staphylococcus aureus skin infections to methicillin-susceptible isolates. JAMA Dermatol 149:1167–1171. doi: 10.1001/jamadermatol.2013.4909. [DOI] [PubMed] [Google Scholar]
- 6.Roberts MC, No DB. 2014. Environment surface sampling in 33 Washington State fire stations for methicillin-resistant and methicillin-susceptible Staphylococcus aureus. Am J Infect Control 42:591–596. doi: 10.1016/j.ajic.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 7.Otter JA, French GL. 2009. Bacterial contamination on touch surfaces in the public transport system and in public areas of a hospital in London. Lett Appl Microbiol 49:803–805. doi: 10.1111/j.1472-765X.2009.02728.x. [DOI] [PubMed] [Google Scholar]
- 8.Wilks SA, Michels H, Keevil CW. 2005. The survival of Escherichia coli O157 on a range of metal surfaces. Int J Food Microbiol 105:445–454. doi: 10.1016/j.ijfoodmicro.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Weaver L, Michels HT, Keevil CW. 2008. Survival of Clostridium difficile on copper and steel: futuristic options for hospital hygiene. J Hosp Infect 68:145–151. doi: 10.1016/j.jhin.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Noyce JO, Michels H, Keevil CW. 2007. Inactivation of influenza A virus on copper versus stainless steel surfaces. Appl Environ Microbiol 73:2748–2750. doi: 10.1128/AEM.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnes SL, Keevil CW. 2013. Inactivation of norovirus on dry copper alloy surfaces. PLoS One 8:e75017. doi: 10.1371/journal.pone.0075017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warnes SL, Highmore CJ, Keevil CW. 2012. Horizontal transfer of antibiotic resistance genes on abiotic touch surfaces: implications for public health. mBio 3:e00489-12. doi: 10.1128/mBio.00489-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Noyce JO, Michels H, Keevil CW. 2006. Potential use of copper surfaces to reduce survival of epidemic meticillin-resistant Staphylococcus aureus in the healthcare environment. J Hosp Infect 63:289–297. doi: 10.1016/j.jhin.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Warnes SL, Green SM, Michels HT, Keevil CW. 2010. Biocidal efficacy of copper alloys against pathogenic enterococci involves degradation of genomic and plasmid DNAs. Appl Environ Microbiol 76:5390–5401. doi: 10.1128/AEM.03050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez JL, Cartelle M, Muriel L, Santiso R, Tamayo M, Goyanes V, Gosalvez J, Bou G. 2008. DNA fragmentation in microorganisms assessed in situ. Appl Environ Microbiol 74:5925–5933. doi: 10.1128/AEM.00318-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6:130. doi: 10.1186/1471-2334-6-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selim HS, Abaza AF. 2015. Microbial contamination of mobile phones in a health care setting in Alexandria, Egypt. GMS Hyg Infect Control 10:Doc03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver L, Noyce JO, Michels HT, Keevil CW. 2010. Potential action of copper surfaces on meticillin-resistant Staphylococcus aureus. J Appl Microbiol 109:2200–2205. doi: 10.1111/j.1365-2672.2010.04852.x. [DOI] [PubMed] [Google Scholar]
- 19.Webber MA, Whitehead RN, Mount M, Loman NJ, Pallen MJ, Piddock LJ. 2015. Parallel evolutionary pathways to antibiotic resistance selected by biocide exposure. J Antimicrob Chemother 70:2241–2248. doi: 10.1093/jac/dkv109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol 14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 21.Warnes SL, Keevil CW. 2011. Mechanism of copper surface toxicity in vancomycin-resistant enterococci following wet or dry surface contact. Appl Environ Microbiol 77:6049–6059. doi: 10.1128/AEM.00597-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michels HT, Keevil CW, Salgado CD, Schmidt MG. 2015. From laboratory research to a clinical trial: copper alloy surfaces kill bacteria and reduce hospital-acquired infections. HERD 9:64–79. doi: 10.1177/1937586715592650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salgado CD, Sepkowitz KA, John JF, Cantey JR, Attaway HH, Freeman KD, Sharpe PA, Michels HT, Schmidt MG. 2013. Copper surfaces reduce the rate of healthcare-acquired infections in the intensive care unit. Infect Control Hosp Epidemiol 34:479–486. doi: 10.1086/670207. [DOI] [PubMed] [Google Scholar]