Abstract
Objective
To characterize meningococcal strains isolated from five cases of meningococcal disease (MD) associated with an outbreak in Trancoso – BA, occurred in October 2009. All cases, with the exception of a 39-year-old male, attended a dance party with approximately 1000 youngsters in a rural site.
Materials and methods
The epidemiological investigation was conducted by the Epidemiological Surveillance Service of Bahia State. Meningococcal strains were characterized at Adolfo Lutz Institute, the Brazilian National Reference Laboratory for Bacterial Meningitis by conventional techniques (serotype, serosubtype and antimicrobial susceptibility test) and by molecular methods (Pulsed-field gel electrophoresis – PFGE and Multilocus Sequence Typing – MLST).
Results
The PFGE showed 2 closely related restriction profiles, designated as PFGE types A and A1, having 92% relatedness to each other. MLST characterization showed both A and A1 clones were ST-3780, which belongs to the ST-103 complex. All isolates displayed the phenotype C:23:P1.5 and were susceptible to all antibiotics tested.
Conclusions
This is the first reported MD outbreak associated with serogroup C ST-103 complex in Brazil, as well as the party and illicit drug-use associated outbreak.
Keywords: Neisseria meningitidis, Outbreak, Serogroup C
Introduction
Meningococcal disease (MD) remains a major worldwide health problem, with most disease globally being caused by serogroups A, B, C, W135 and Y.1 In recent years, MD in Brazil has been caused primarily by serogroup B and C strains. During the past decade, the proportion of cases caused by serogroup B strains declined from 74.5% in 2000 to 25.7% in 2008, corresponding to a decline from 0.6 to 0.1 per 100,000 inhabitants in the average annual incidence of serogroup B MD. At the same time, serogroup C MD has been steadily increasing in several Brazilian states, from 22.4% to 67.7%. The annual incidence of serogroup C MD increased from 0.2 cases in 2000 to 0.41 cases per 100,000 habitants in 2008.2 N. meningitidis serogroup C:23:P1.14–6, belonging to the ST-103 complex, was first described in São Paulo State in 19893 and by 2009 accounted for 75.4% of MD cases. In addition, since 2004 serogroup C ST-103 complex strains have also emerged as the main cause of MD outbreaks in Brazil.
In October 2009, the local authorities of Porto Seguro, a coastal town in Bahia State reported a cluster of meningococcal serogroup C cases associated with a party (http://www.saude.ba.gov.br/arquivos/Nota%20Meningite%20em%20Porto%20Seguro%20(04-11-2009.pdf). To further investigate the outbreak, we characterized the outbreak-associated isolates using molecular methods such as pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST).
Material and methods
Epidemiological data and bacterial isolates
The epidemiological investigation was conducted by the Epidemiological Surveillance Service of Bahia, State and the data are presented in Table 1. Nine cases, 6 (67%) of which were fatal, were reported to public health authorities in Porto Seguro, Bahia between October 21 and 26, 2009. The outbreak occurred in Trancoso, a small coastal town of 4538 inhabitants. There were 7 male and 2 female patients, and all were between the ages of 14 and 39 years. All cases, with the exception of a 39-year-old male, attended a dance party with approximately 1000 youngsters on October 18 in a rural site in Trancoso. Alcohol consumption, cigarette smoking and illicit drug use were reported during the outbreak investigation. People who had close contact with patients, such as a family and classroom colleagues received rifampin chemoprophylaxis. The MD cases were laboratory confirmed, either by isolation of N. meningitidis serogroup C in blood or cerebrospinal fluid (5/9, 55.5%), or by the detection of serogroup C meningococcal polysaccharide in cerebrospinal fluid by latex agglutination (4/9; 44.5%). All meningococcal isolates were confirmed and serogrouped at Adolfo Lutz Institute, the Brazilian National Reference Laboratory for Bacterial Meningitis, using conventional microbiologic methods.4–6
Table 1.
Summary of cases, meningococcal disease outbreak, Trancoso, Brazil.
| Case number | Specimen ID | Age (y)/gender | Link with party | Date of onset (Y Mon DD) | Clinical presentation | Outcome |
|---|---|---|---|---|---|---|
| 1 | No | 23/M | Yes | 2009 Oct 21 | Headache, vomiting, fever, rash, meningismus | Death |
| 2 | No | 15/M | Yes | 2009 Oct 21 | Headache, vomiting, fever, rash | Death |
| 3 | N.665/09 | 20/F | Yes | 2009 Oct 23 | Headache, vomiting, fever, rash, meningismus, myalgia | Death |
| 4 | No | 17/M | Yes | 2009 Oct 23 | Headache, vomiting, fever, rash, meningismus, convulsion | Death |
| 5 | N.664/09 | 25/F | Yes | 2009 Oct 23 | Headache, vomiting, meningismus | Death |
| 6 | N.686/09 | 22/M | Yes | 2009 Oct 23 | Headache, vomiting, fever, rash, meningismus | Survived |
| 7 | N.663/09 | 22/M | Yes | 2009 Oct 23 | Headache, vomiting, fever, rash | Survived |
| 8 | No | 14/M | Yes | 2009 Oct 23 | Headache, vomiting, meningismus | Survived |
| 9 | N.684/09 | 39/M | No | 2009 Oct 28 | Headache, vomiting, fever, rash, meningismus | Death |
ID, identification.
Pulsed-field gel eletrophoresis (PFGE)
For PFGE, genomic DNA was prepared in agarose plugs and digested with the restriction enzyme Nhe I7 and DNA digested were separated by electrophoresis in agarose 1% w/v gels as described by Popovic et al.7 and restriction profiles were analyzed according to the criteria of Tenover et al.8 and using Bionumerics (Applied Maths, St-Martens-Latem, Belgium). The isolates showing similarity >85% were considered to be closely related.
Serological typing
Serotyping and serosubtyping were performed by dot blotting using whole cell suspensions as previously described9 using a panel of 18 PorB and 15 PorA murine MAbs specific for the variable regions, respectively.3
Multilocus sequence typing (MLST)
MLST was performed according to the methods of Maiden et al.10 Primers, determination of a sequence alleles, and designation of sequence types are described on the MLST website (http://neisseria.org/nm/typing/mlst).
Antimicrobial susceptibility testing
Isolates were tested for susceptibility to penicillin, ampicillin, ceftriaxone, ciprofloxacin, chloramphenicol and rifampin using the broth microdilution procedure established by the Clinical and Laboratory Standards Institute (CLSI).11 The susceptibility/resistance breakpoints were those recommended by the European Monitoring Group on Meningococci.12
Results
Based on the 9 cases, the MD incidence was 198 per 100,000 Trancoso inhabitants and, considering the 8 party-associated cases separately, the incidence among party participants was 800 per 100,000. Among the 5 N. meningitidis isolates, there were 2 closely related restriction profiles: one (4 isolates) was designated PFGE type A and was considered to be the outbreak strain, and the remaining isolate was designated as PFGE type A1; PFGE types A and A1 had 92% relatedness to each other (Fig. 1). Both types were shown by MLST to be ST-3780, which belongs to the ST-103 complex. All the isolates displayed the phenotype C:23:P1.5. All strains were susceptible to all antibiotics tested (Table 2).
Fig. 1.
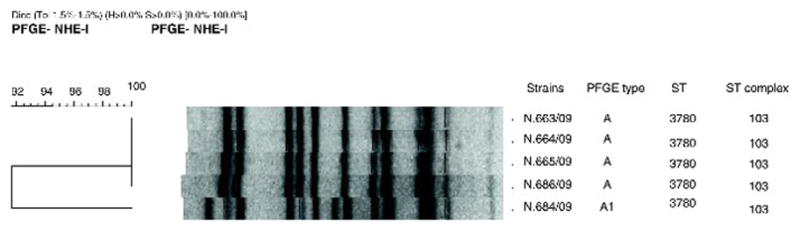
Dendrogram showing PFGE patterns of NheI-digested genomic DNA of the meningococcal strains isolated. The strain identification, PFGE type, sequence type, sequence type complex and phenotype are indicated.
Table 2.
Susceptibility to six different antimicrobials of Neisseria meningitidis strains isolated from Porto Seguro – Bahia cases.
| Specimen ID | MIC (μg/mL)
|
|||||
|---|---|---|---|---|---|---|
| CIP | CLO | CRO | PEN | AMP | RIF | |
| N.665/09 | 0.007 (S) | 1.00 (S) | 0.0003 (S) | 0.015 (S) | 0.06 (S) | 0.250 (S) |
| N.664/09 | 0.003 (S) | 1.00 (S) | 0.0003 (S) | 0.015 (S) | 0.06 (S) | 0.125 (S) |
| N.686/09 | 0.003 (S) | 1.00 (S) | 0.0007 (S) | 0.015 (S) | 0.06 (S) | 0.250 (S) |
| N.663/09 | 0.007 (S) | 1.00 (S) | 0.0003 (S) | 0.030 (S) | 0.06 (S) | 0.125 (S) |
| N.684/09 | 0.003 (S) | 1.00 (S) | 0.0007 (S) | 0.015 (S) | 0.06 (S) | 0.125 (S) |
MIC breakpoints to CIP: Susceptible, ≤0.03 μg/mL; Resistant, ≥0.5 μg/mL; CLO: Susceptible, ≤2 μg/mL; Resistant, ≥8.0 μg/mL; CRO: Susceptible, ≤0.12 μg/mL; PEN: Susceptible, ≤0.06 μg/mL; Resistant, ≥1.0; AMP: Susceptible, ≤0.12 μg/mL; Resistant, ≥2.0 μg/mL; RIF: Susceptible, ≤0.25 μg/mL; Resistant, ≥2.0 μg/mL.
MIC, minimum inhibitory concentration; CIP, ciprofloxacin; CLO, choramphenicol; CRO, ceftriaxone; PEN, penicillin; AMP, ampicillin; RIF, rifampicin; (S), susceptible.
Discussion
N. meningitidis serogroup C strains isolated from areas where the disease is endemic, or from outbreaks, have usually belonged to the ST-11 or ST-8 complex.13 N. meningitidis serogroup C isolates belonging to the ST-11 complex have been responsible for numerous epidemics and outbreaks in the United States, Canada, and Europe, since the early 1990s. These hyperinvasive strains have predominantly affected adolescents and young adults, and have required frequent, massive public health investigations and interventions.14,15 In contrast, most serogroup C sporadic disease and outbreaks in Brazil since 2000 have been caused by strains belonging to the ST-103 complex,3 which differs from ST-11 at all 7 MLST loci, indicating that they are highly unrelated to each other (www.pubmlst.org). This is in contrast to ST-8 N. meningitidis which caused substantial serogroup C disease in Brazil during in the 1990s.3 ST-8 has been previously shown by whole genome sequencing to be closely related to ST-11, with which it shares 3 MLST loci16 (www.pubmlst.org).
The origin of the ST-103 complex is unknown. In addition, the impact of serogroup C conjugate vaccine, which was introduced into the routine childhood immunization schedule in 2010, on pharyngeal carriage caused by ST-103 strains is also not known. Efforts are needed to continue to monitor its spread and define its virulence in order to understand the evolution of this clone in Brazil.
The ST-103 complex, with most strains displaying the 23:P1. 14-6 phenotype,3 have been present in Brazil since 1989 and were associated with the serogroup C epidemic in the 2000. Although ST-103 is relatively uncommon globally as a cause invasive disease, it has recently been reported in Poland (phenotype C:NT:P1.3,6).17
Although most of the N. meningitidis serogroup C, ST-103 complex strains isolated in Brazil have displayed the 23:P1.14-6 phenotype, in the present outbreak, all of the tested strains displayed the 23:P1.5 phenotype. This change in antigenic profile likely occurred through horizontal gene transfer and would have contributed to both the generation and the spread of novel antigenic variants of the protein.18 Antigenic shift within ST-11 serogroup C strains in association with increase in MD incidence has been described in the United States.19
PFGE has been successfully applied to determine the relatedness of bacterial isolates suspected of being part of an outbreak.8 PFGE has the advantage of being able to identify subclones of the same clone20 and has been systematically used to identify N. meningitidis outbreaks.7,21–23 In this study, we identified 2 PFGE patterns among strains belonging to the same clone (ST-103 complex) and observed excellent correlation between the epidemiological data and PFGE results: all the 4 strains isolated from the cases linked with the party had indistinguishable PFGE, consistent with an outbreak; in addition, PFGE was able to differentiate the outbreak strain from the case that was not linked to the party.
Overcrowding facilitates the spread of meningococcal carriage and, in susceptible individuals infected with a virulent strain, subsequent disease. Our data suggest that the party could have provided an opportunity for the dissemination of N. meningitidis. Four MD cases linked to the party, including 3 survivors and 1 who died, were users of illicit drugs or used cocaine at the party. The short period between the party and the onset of symptoms, in addition to genetic similarity of the strains, allow us to hypothesize that the cocaine inhalation associated with the party could have predisposed to invasive disease through injury to the respiratory mucosa.24 An outbreak of MD involving illicit drugs use was reported in New York, with cocaine use reported as the most common risk factor for MD.25 MD has also been associated with other factors that can promote a breakdown in host defense mechanisms or damage to the upper respiratory tract, such as influenza and others acute respiratory diseases.21,26,27
Other social behaviors associated with MD include alcohol consumption, cigarette smoking, and attendance at nightclubs, discotheques, and bars.22,23,28 Other crowded environments, such as the Hajj pilgrimage, educational and military institutions have also been associated with outbreaks, indicating that crowding also facilitates transmission.29,30
In the outbreak described here, all the party cases occurred within 5 days of the event, similar to an outbreak of serogroup C disease associated with nightclub attendance in Australia.21
The combination of epidemiologic data and molecular characterization were essential for understanding this outbreak. To our knowledge, this is the first reported MD outbreak associated with the serogroup C ST-103 complex and the first reported party and illicit drug use-associated outbreak in Brazil.
Acknowledgments
We thank Joana D’Arc dos Reis, the coordinator of Public Health Laboratories, Ministry of Health from 2008 to 2010; Marta Galhardo, Conceição Zanelato and Maria Vaneide de Paiva for serogrouping and antimicrobial susceptibility testing the isolates; Samanta Cristine Grassi Almeida for assistance with Bionumerics software. This work was supported in part by a career development award to Lee H Harrison, National Institute of Allergy and Infectious Diseases (K24 AI52788); and by a Fogarty International Center Global Infectious Diseases Research Training Program grant, National Institutes of Health, to the University of Pittsburgh (D43TW006592).
Footnotes
Conflict of interest
The authors declare no conflicts of interest related to this study.
References
- 1.Tzeng YL, Stephens DS. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2000;2:687–700. doi: 10.1016/s1286-4579(00)00356-7. [DOI] [PubMed] [Google Scholar]
- 2.De Lemos APS, Gorla MCO, Galhardo M, Zanelato MC, Paiva MV, Brandileone MCC. Rapid changing in trends of meningococcal disease in Brazil from 2000 to 2008. Proceedings of the 6th World Congress of the World Society for Pediatric Infectious Diseases. 2009:806. [Google Scholar]
- 3.De Lemos AP, Yara TY, Gorla MCO, de Paiva MV, De Souza AL, Goncalves MIC, et al. Clonal distribution of invasive Neisseria meningitidis serogroup C strains circulating from 1976 to 2005 in greater São Paulo, Brazil. J Clin Microbiol. 2007;45:1266–73. doi: 10.1128/JCM.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alkmin MGA, Shimizu SH, Landgraf IM, Gaspari EN, Melles CEA. Production and immunochemical characterization of Neisseria meningitidis group B antiserum for the diagnosis of purulent meningitis. Braz J Med Biol Res. 1994;27:1627–34. [PubMed] [Google Scholar]
- 5.Popovic T, Ajello GW, Facklam RR. Laboratory Methods for the Diagnosis of Meningitis caused by Neisseria meningitidis, Streptococcus pneumonia, and Haemophilus influenzae. Atlanta, GA: Centers for Disease Control, World Health Organization; 1998. [Google Scholar]
- 6.Winn WCJ, Allen SD, Janda WM, Koneman EM, Procop GW, Schreckenberger PC. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 6. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 566–622. [Google Scholar]
- 7.Popovic T, Schmink S, Rosenstein NA, Ajello GW, Reeves MW, Plikaytis B, et al. Evaluation of pulsed-field gel electrophoresis in epidemiological investigations of meningococcal disease outbreaks caused by Neisseria meningitidis serogroup C. J Clin Microbiol. 2001;39:75–85. doi: 10.1128/JCM.39.1.75-85.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wedege E, Hoiby EA, Rosenqvist E, Froholm LO. Serotyping and subtyping of Neisseria meningitidis isolates by co-agglutination, dot-blotting and ELISA. J Med Microbiol. 1990;31:195–201. doi: 10.1099/00222615-31-3-195. [DOI] [PubMed] [Google Scholar]
- 10.Maiden MC, Bygraves JA, Feil E, Morelli G, Russel JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–5. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Wayne: Clinical and Laboratory Standards Institute; 2006. pp. M7–M17. [Google Scholar]
- 12.Vázquez JA. Resistance testing of meningococci: the recommendations of the European Monitoring Group on Meningococci. FEMS Microbiol Rev. 2007;31:97–100. doi: 10.1111/j.1574-6976.2006.00050-x. [DOI] [PubMed] [Google Scholar]
- 13.Maiden MCJ. Population structure of Neisseria meningitidis. In: Ferreirós C, Criado MT, Vasquez JA, editors. Emerging Strategies in the Fight Against Meningitis: Molecular and Cellular Aspects. Wymondham: Horizon Scientific Press; 2002. pp. 150–169. [Google Scholar]
- 14.Caugant DA. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS. 1998;106:505–25. [PubMed] [Google Scholar]
- 15.Whalen CM, Hockin JC, Ryan JA, Ashton FE. The changing epidemiology of invasive meningococcal disease in Canada, 1985 through 1992. JAMA. 1995;273:390–4. [PubMed] [Google Scholar]
- 16.Budroni S, Siena E, Hotopp JC, Seib KL, Serruto D, Nofroni C, et al. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci USA. 2011;15:4494–9. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadlubowski M, Wasko I, Kalrowicz A, Hryniewicz W. Invasive meningococcal disease at a military base in Warsaw, January 2007. Euro Surveill. 2007;12 doi: 10.2807/esw.12.09.03147-en. Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=3147. [DOI] [PubMed] [Google Scholar]
- 18.Feavers IM, Heath AB, Bygraves JA, Maiden MC. Role of horizontal genetic exchange in the antigenic variation of the class 1 outer membrane protein of Neisseria meningitidis. Mol Microbiol. 1992;6:489–95. doi: 10.1111/j.1365-2958.1992.tb01493.x. [DOI] [PubMed] [Google Scholar]
- 19.Harrison LH, Jolley KA, Shutt KA, Marsh JW, O’Leary M, Thomson Sanza L, et al. Antigenic shift and the incidence of meningococcal disease. J Infect Dis. 2006;193:1266–74. doi: 10.1086/501371. [DOI] [PubMed] [Google Scholar]
- 20.Strathdee CA, Tyler SD, Ryan JA, Johnson WM, Ashton FE. Genomic fingerprinting of Neisseria meningitidis associated with group C meningococcal disease in Canada. J Clin Microbiol. 1993;31:2506–8. doi: 10.1128/jcm.31.9.2506-2508.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison LH, Armstrong CW, Jenkins RS, Harmon MW, Ajello GW, Miller GB, et al. A cluster of meningococcal disease on a school bus following epidemic influenza. Arch Intern Med. 1991;151:1005–9. [PubMed] [Google Scholar]
- 22.Imrey PB, Jackson LA, Ludwinski PH, England AC, Fella GA, Fox BC, et al. Meningococcal carriage, alcohol consumption and campus bar patronage in a serogroup C meningococcal disease outbreak. J Clin Microbiol. 1995;33:3133–7. doi: 10.1128/jcm.33.12.3133-3137.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jelfs J, Jalaludin B, Munro R, Patel M, Kerr M, Daley D, et al. A cluster of meningococcal disease in western Sydney, Australia initially associated with a nightclub. Epidemiol Infect. 1998;120:263–70. doi: 10.1017/s0950268898008681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perper JA, Van Thiel DH. Respiratory complications of cocaine abuse. Recent Dev Alcohol. 1992;10:363–77. doi: 10.1007/978-1-4899-1648-8_18. [DOI] [PubMed] [Google Scholar]
- 25.Weiss D, Stern EJ, Zimmerman C, Bregman B, Yeung A, Das D, et al. Epidemiologic investigation and targed vaccination initiative in response to an outbreak of meningococcal disease among illicit drug users in Brooklyn, New York. Clin Infect Dis. 2009;48:894–901. doi: 10.1086/597257. [DOI] [PubMed] [Google Scholar]
- 26.Young LS, LaForce FM, Head JJ, Feeley JC, Bennett JV. A simultaneous outbreak of meningococcal and influenza infections. N Engl J Med. 1972;287:5–9. doi: 10.1056/NEJM197207062870102. [DOI] [PubMed] [Google Scholar]
- 27.Moore PS, Hierholzer J, De Witt W, Goun K, Djore D, Lippeveld T, et al. Respiratory viruses and mycoplasma a cofactors for epidemic group A meningococcal meningitis. JAMA. 1990;264:1271–5. [PubMed] [Google Scholar]
- 28.Finn R, Groves C, Coe M, Pass M, Harrison LH. Cluster of serogroup C meningococcal disease associated with attendance at a party. South Med J. 2001;94:1192–4. [PubMed] [Google Scholar]
- 29.Almog R, Gdalevich M, Lev B, Wiener M, Ashkenazi S, Block C. First recorded outbreaks of meningococcal disease in the Israel defence force: three clusters due to serogroup C and the emergence of resistance to rifampicin. Infection. 1994;22:69–71. doi: 10.1007/BF01739006. [DOI] [PubMed] [Google Scholar]
- 30.Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, et al. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet. 2000;356:2159. doi: 10.1016/S0140-6736. [DOI] [PubMed] [Google Scholar]


