Abstract
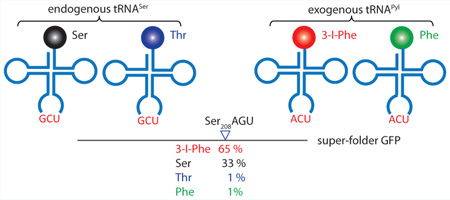
Expansion of the genetic code through engineering the translation machinery has greatly increased the chemical repertoire of the proteome. This has been accomplished mainly by read-through of UAG or UGA stop codons by the noncanonical aminoacyl-tRNA of choice. While stop codon read-through involves competition with the translation release factors, sense codon reassignment entails competition with a large pool of endogenous tRNAs. We used an engineered pyrrolysyl-tRNA synthetase to incorporate 3-iodo-l-phenylalanine (3-I-Phe) at a number of different serine and leucine codons in wild-type Escherichia coli. Quantitative LC-MS/MS measurements of amino acid incorporation yields carried out in a selected reaction monitoring experiment revealed that the 3-I-Phe abundance at the Ser208AGU codon in superfolder GFP was 65 ± 17%. This method also allowed quantification of other amino acids (serine, 33 ± 17%; phenylalanine, 1 ± 1%; threonine, 1 ± 1%) that compete with 3-I-Phe at both the aminoacylation and decoding steps of translation for incorporation at the same codon position. Reassignments of different serine (AGU, AGC, UCG) and leucine (CUG) codons with the matching tRNAPyl anticodon variants were met with varying success, and our findings provide a guideline for the choice of sense codons to be reassigned. Our results indicate that the 3-iodo-l-phenylalanyl-tRNA synthetase (IFRS)/tRNAPyl pair can efficiently outcompete the cellular machinery to reassign select sense codons in wild-type E. coli.
Keywords: genetic code, sense codons reassignment, pyrrolysyl-tRNA synthetase (PylRS), 3-iodo-l-phenylalanyl-tRNA synthetase (IFRS), aminoacyl-tRNA synthetase (aaRS), noncanonical amino acid (ncAA), tRNA competition, quantitative proteomic analysis, selected reaction monitoring (SRM)
Graphical abstract
Expansion of the genetic code through the engineering of aminoacyl-tRNA synthetases (aaRSs) and their corresponding tRNAs has allowed a great number of noncanonical amino acids (ncAAs) to be incorporated into proteins.1 While the engineering of essential components of the translation machinery (tRNAs, aaRSs, elongation factors, ribosomes) is crucial for the development of an orthogonal translation system, ultimately, any orthogonal translation system requires a codon in the mRNA to direct the incorporation of the desired amino acid. Over the past decade, ncAAs have been cotranslationally inserted at stop codons1,2 and quadruplet codons3,4 using engineered aaRSs and ribosomes. However, the former is restricted to three stop codons, and the latter faces frameshifting problems. In contrast, sense codon reassignment avoids the problem of frameshifting, and a large number of sense codons are available for reassignment due to the degeneracy of the genetic code. While sense codon reassignment entails competition with a large pool of endogenous tRNA and is not amenable to read-through analyses, these issues can be bypassed.
Sense codon reassignment requires an orthogonal aaRS/tRNA pair that does not recognize the tRNA anticodon. At the same time, the host aaRS must not recognize the exogenous tRNA; thus, its tRNA substrate recognition mechanism must not include the anticodon. In Escherichia coli, the endogenous aaRSs that recognize their tRNAs in an anticodon-independent manner are seryl-tRNA synthetase (SerRS), leucyl-tRNA synthetase (LeuRS), and alanyl-tRNA synthetase (AlaRS).5 Thus, the serine, leucine, and alanine codons are good targets for reassignment since their corresponding aaRSs would not acylate the exogenous tRNA, which would result in higher incorporation levels of the desired ncAA.
Attempts to reassign sense codons have yielded various levels of success. Utilizing the Saccharomyces cerevisiae phenylalanyl-tRNA synthetase (PheRS) and engineered tRNAPheAAA, the phenylalanine UUU codon was partially reassigned to l-3-(2-naphthyl)alanine (Nal) in E. coli.6 While the authors were able to replace, on average, 4.4 of the 9 Phe codons with Nal in a heterologously expressed murine dihydrofolate reductase, this level of reassignment required the E. coli host strain to be auxotrophic for Phe. Inefficient reassignment can be further explained by the anticodon-dependent recognition of engineered tRNAPhe by PheRS,7 which reverts its ncAA designation back to Phe.
To overcome the limitation of aaRS anticodon recognition in sense codon reassignment, several studies have utilized the Methanosarcina mazei pyrrolysyl-tRNA synthetase (PylRS)/tRNAPyl pair to reassign the rare arginine AGG codon. Pyrrolysine (Pyl) is a rarely used amino acid encoded by the amber UAG codon in methanogenic archaea and Gram-positive bacteria.8 UAG decoding is achieved by the aminoacylation of tRNAPylCUA by PylRS,9 which had biochemically10 and structurally11 been shown not to recognize the tRNAPyl anticodon. Since PylRS will aminoacylate tRNAPyl regardless of its anticodon sequence, mutating the anticodon theoretically would allow the decoding of any codon of choice. This has been demonstrated most successfully via the insertion of ncAAs at ochre UAA,12 opal UGA,13 as well as quadruplet AGGA,14 CUAG, AGUA, and UAGA15 codons. However, initial attempts to reassign the sense Arg CGG codon in Mycoplasma16,17 using M. mazei PylRS/tRNAPyl were unsuccessful, and it has been speculated that successful AGG reassignment was not achieved due to the recognition of the tRNAPylCCG anticodon by the endogenous arginyl-tRNA synthetase, which resulted in only Arg incorporation.
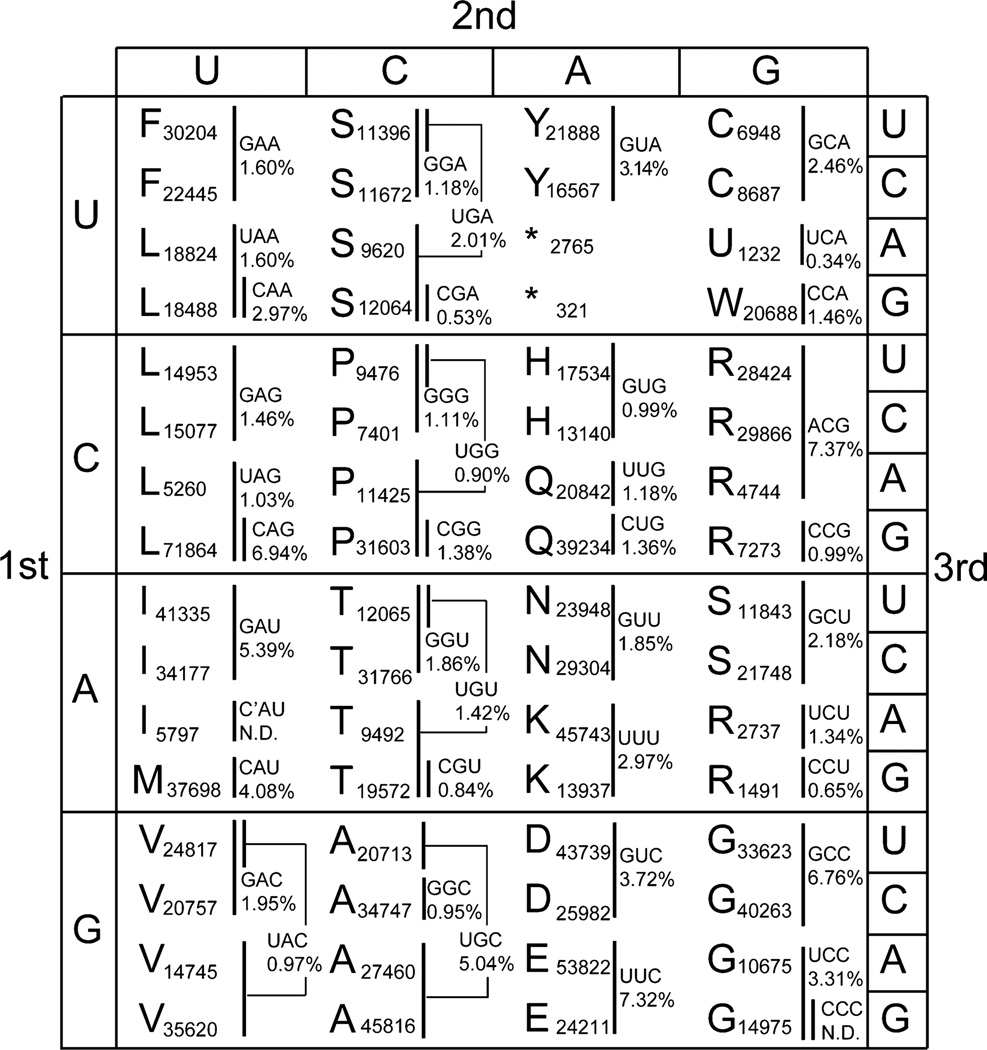
Successful Arg AGG codon reassignment in E. coli was subsequently reported with M. mazei PylRS/tRNAPylCCU18 and Methanocaldococcus jannaschii TyrRS/tRNATyrCCU,19 with efficiencies in the 80–83% range18 and at the quantitative level.19 In a recent study, complete reassignment of the Arg AGG codon in E. coli was achieved by converting all instances of the Arg AGG codon in essential genes to synonymous codons.20 These levels of reassignment required the deletion or downregulation of competing endogenous tRNAArgCCU,18–20 the deletion of genes involved in arginine biosynthesis,19 or providing a high concentration of the ncAA,18 which can result in toxicity. These studies achieved efficient reassignment in large part due to the low codon usage of the rare Arg AGG codon (1491 instances in E. coli MG1655) and the fact that Arg AGG has the smallest pool size of endogenous tRNA isoacceptors (0.65% of total tRNAs, Figure 1). The success of these studies indicated that, in principle, sense codon reassignment could be achieved with an engineered aaRS/tRNA pair. However, the feasibility was demonstrated only for rare codons, and it remains unknown if reassignment can be accomplished for high-frequency codons.
Figure 1.
A genetic code representation that marks the codons decoded by each of the 46 tRNA isoacceptors, whose anticodons and intracellular abundances are stated. In E. coli, the UGA stop codon is reassigned to selenocysteine (U). The decoding patterns and cellular abundances of all 46 endogenous tRNA isoacceptors had previously been reported,26 and we calculated the isoacceptor abundance for each of the 64 codons using this equation: isoacceptor abundance for a codon = (biochemically determined isoacceptor abundance) × (codon usage)/(total codon usage of all codons decoded by isoacceptor). Codon usage values are based on NC_000913.2 (National Center for Biotechnology Information, 1 September 2011) and taken from Lajoie et al.25 N.D. indicates values not determined.
In our efforts to reassign high-frequency codons, we decided to reassign Ser and Leu sense codons since SerRS21 and LeuRS22 recognize their tRNAs in an anticodon-independent fashion. This approach ensured that the exogenous tRNA anticodon would not be recognized by the endogenous aaRS, a phenomenon that had previously been shown to limit codon reassignment of the Arg CGG codon.16,17 By choosing to reassign, for instance, a Ser AGU codon, the endogenous SerRS will not serylate the exogenous tRNAPylACU. Additionally, the strength of the codon–anticodon interaction is affected by the composition of base pairs as well as post-transcriptional tRNA modifications in and around the anticodon.23 An endogenous tRNA decodes a codon through either perfect Watson–Crick base pairing or wobble pairing at the third base position. If a codon is ordinarily decoded by a wobble pairing tRNA isoacceptor, then we can engineer an ncAA-bearing tRNA that provides perfect Watson–Crick base pairing, and this thermodynamic advantage should increase the ncAA incorporation yield.6 For example, decoding of Ser AGU with tRNASerGCU normally involves a wobble G–U base pair, whereas an engineered tRNAPylACU would provide a Watson–Crick A–U base pair at the third base position. Thermodynamic studies of RNA hairpins revealed the Gibbs free energy of melting A–U and G–U base pairs to be 6.3 and 4.1 kcal/mol, respectively.24 Therefore, with tRNAPylACU we are thermodynamically more likely to outcompete the endogenous wobble decoding tRNASerGCU to decode Ser AGU.
If a codon is decoded by a wobble base-pairing tRNA, then reassignment with a complete Watson–Crick base pairing tRNAPyl variant may increase codon-biased incorporation of ncAAs. To test the prediction that an anticodon–codon pair that provides three Watson–Crick base pairs may allow more efficient decoding than a wobble-decoded anticodon–codon pair that provides only two Watson–Crick base pairs, we evaluated the reassignability of three serine and leucine codons with very different properties. The serine AGU codon (11 843 instances in E. coli MG1655) has median usage frequency among the serine codons,25 and its natural tRNA isoacceptor decodes purely via wobble pairing. Hence, this codon presents an excellent scenario for high-yield ncAA incorporation. The Ser UCG codon (12 064 instances in E. coli MG1655) has the highest codon usage in the UCN box and is decoded by two tRNA isoacceptors, including a dedicated tRNA isoacceptor that decodes via standard Watson–Crick pairing. Hence, reassignment of this codon should be more challenging, and we wanted to qualitatively evaluate the possibility of reassignment. The leucine CUG codon (71 864 instances in E. coli MG1655) not only is the most frequently used codon in the E. coli genome25 but also gets decoded by a dedicated tRNALeuCAG that decodes by Watson–Crick base pairing and is the most abundant tRNA in the cell, constituting 6.94% of total tRNA.26 Furthermore, the Leu CUG codon is also decoded by a second tRNALeuUAG isoacceptor that decodes by wobble base pairing (Figure 1). On the basis of these parameters, the Leu CUG codon should be the most difficult to reassign.
To achieve reassignment of the selected codons, we used an engineered PylRS, 3-iodo-l-phenylalanyl-tRNA synthetase (IFRS), that efficiently utilizes 3-iodo-l-phenylalanine (3-I-Phe).27 After a biochemical analysis of IFRS/tRNAPyl binding and aminoacylation kinetics to confirm anticodon-independent aminoacylation of diverse tRNAPyl anticodon variants, we constitutively expressed IFRS/tRNAPyl in wild-type E. coli strain K12 (MG1655) to evaluate in vivo tRNAPyl aminoacylation levels. We confirmed via qualitative LC-MS/MS that 3-I-Phe was incorporated in serine and leucine codons of superfolder green fluorescent protein (sfGFP). At a specific Ser AGU codon in sfGFP, we quantified via selected reaction monitoring (SRM) the incorporation of 3-I-Phe and Ser and, additionally, Phe and Thr, whose incorporations were predicted due to known misactivation by PylRS28 and SerRS,29 respectively. SRM is a highly sensitive mass spectrometric method developed to quantify absolute peptide abundances30 and facilitates the detection of low-frequency mistranslation events.31 With the use of isotopically labeled calibration standards, we can measure the absolute abundances of peptides containing 3-I-Phe, Ser, Phe, and Thr at a targeted Ser codon position. In this study, we demonstrate that by choosing a codon with consideration for anticodon independence and wobble decoding, incorporation can be efficient at a frequent codon in a wild-type E. coli strain.
RESULTS AND DISCUSSION
IFRS Recognizes tRNAPyl Variants Independent of Their Anticodons
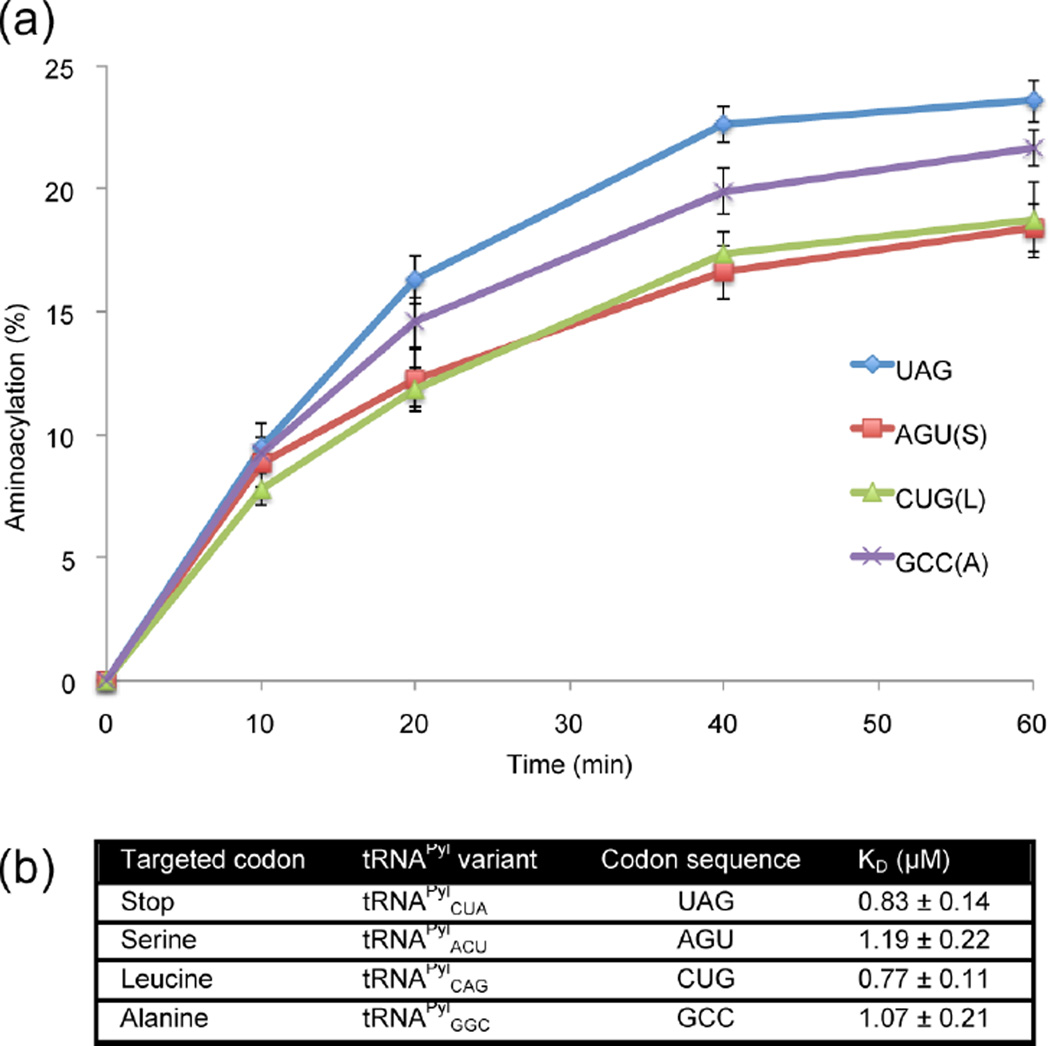
While the anticodon independence of PylRS had previously been demonstrated,10 this had not been shown for the engineered IFRS. To test for the anticodon independence of IFRS activity, we measured the aminoacylation efficiencies and dissociation constants of IFRS for representative tRNAPyl anticodon variants: tRNAPylCUA, tRNAPylACU, tRNAPylCAG, and tRNAPylGGC. We included the tRNAPyl variant decoding Ala GCC due to its diametrically opposite nature, being 3 transversion mutations away from the natural UAG codon. Aminoacylation assays were performed by monitoring the amount of aminoacyl-tRNA product formed over time when IFRS was incubated with 32P 3′-end-labeled tRNAPyl anticodon variants in the presence of 3-I-Phe. The results showed that IFRS aminoacylated the anticodon variants with similar efficiencies, approximating 20% aminoacylation after 60 min (Figure 2a). Next, dissociation constants were determined by incubating IFRS with known concentrations of each tRNAPyl anticodon variant, followed by a filter binding assay to measure the fraction of IFRS bound at each tRNAPyl concentration. These binding assays revealed that binding affinities were similar for all of the tRNAPyl variants, ranging from 0.83 to 1.19 µM (Figure 2b). Since IFRS had similar aminoacylation efficiencies and binding affinities to these tRNAPyl anticodon variants, we concluded that the engineering of PylRS to produce IFRS did not significantly affect its anticodon-independent nature. Overall, these biochemical results confirmed that IFRS is a good candidate for reassignment of Ser and Leu codons.
Figure 2.
Biochemical characterization of the aminoacylation efficiencies and dissociation constants of IFRS for each tRNAPyl anticodon variant. (a) The aminoacylation efficiency of IFRS for each of the anticodon variants was measured via an aminoacylation assay, where 3′-end-labeled tRNAPyl anticodon variants were incubated with IFRS and 3-I-Phe to measure the amount of product formed over 60 min at 37 °C. (b) Binding affinities were determined via filter binding assay, where varying concentrations of IFRS were incubated on ice with 3′-end-labeled tRNAPyl and the fraction of bound IFRS was measured to obtain dissociation constants.
Intracellular Aminoacylation of tRNAPyl Variants
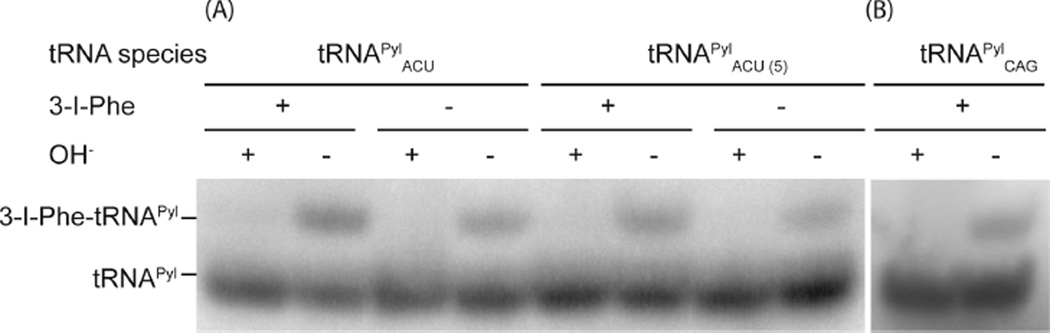
Although the aminoacylation levels of tRNAPyl variants were nearly identical in vitro, this did not necessarily reflect the situation in vivo. To fully characterize IFRS aminoacylation in vivo, IFRS and tRNAPylACU were expressed in E. coli strain MG1655. Previous reports had shown that three32 or six33 copies of a plasmid-borne tRNA gene yielded more tRNA in vivo than one copy. To optimize the copy number, we prepared constructs containing one or five copies of the pyrrolysyl tRNA gene encoding tRNAPylACU, constitutively expressed under the lpp promoter. To evaluate the tRNAPylACU aminoacylation levels in vivo, total RNA was purified and tRNA aminoacylation levels were assessed by acid urea PAGE and northern blot analyses.34 The levels of aminoacylated tRNAPyl were found to be comparable for both the single- and five-copy constructs (Figure 3a); thus, single-copy constructs were used in subsequent experiments. Aminoacylation of tRNAPylACU in the presence and absence of 1 mM 3-I-Phe was 30 and 10%, respectively. Notably, PylRS has been shown to have evolved from PheRS and to misactivate Phe to form Phe-AMP.28 As IFRS was engineered from PylRS, IFRS may similarly misactivate Phe, and we postulate that in the absence of 3-I-Phe IFRS may aminoacylate tRNAPylACU with Phe and cause Phe incorporation at targeted codons. This hypothesis was tested further in the LC-MS/MS experiment.
Figure 3.
Northern blot analyses of intracellular aminoacylation of tRNAPyl isoacceptors. Total RNA was obtained from E. coli cells grown in the presence or absence of 3-I-Phe (1 mM) expressing IFRS and (A) either one or five gene copies of tRNAPylACU or (B) one gene copy of tRNAPylCAG, from the pCAM plasmid. Total RNA was suspended in either 10 mM sodium acetate, pH 4.5 (acylated), or 200 mM Tris, pH 8.0 (deacylated, OH−). Positions of acylated and deacylated tRNAPyl isoacceptors are indicated.
Reassignment of Targeted Ser AGU Codons in sfGFP with 3-I-Phe
To evaluate the success of Ser AGU reassignment, overexpressed sfGFP was purified from E. coli that constitutively expressed IFRS and tRNAPylACU. LC-MS/MS analyses of chymotrypsin/trypsin-digested samples revealed 3-I-Phe incorporation at 1 of 2 targeted AGU sites and 1 of 4 wobble decoded AGC sites (Figures 4, S1, and S3). This decoding pattern is consistent with the natural decoding pattern of the tRNAPylGCU isoacceptor, which decodes both Ser AGU and AGC codons in E. coli.23 Since chymotrypsin cleaves at Phe residues, it may inadvertently cleave 3-I-Phe. To detect all reassignment events including low-occurrence events, we performed a separate digest with Asp-N, an endoproteinase that cleaves at aspartate residues. As predicted, additional 3-I-Phe-containing peptides were observed in Asp-N digested sfGFP (Table S1). Digestion with Asp-N conserves all instances of 3-I-Phe incorporation; thus, subsequent quantitative analyses of reassignment levels were performed with Asp-N instead of chymotrypsin to avoid underreporting 3-I-Phe incorporation levels.
Figure 4.
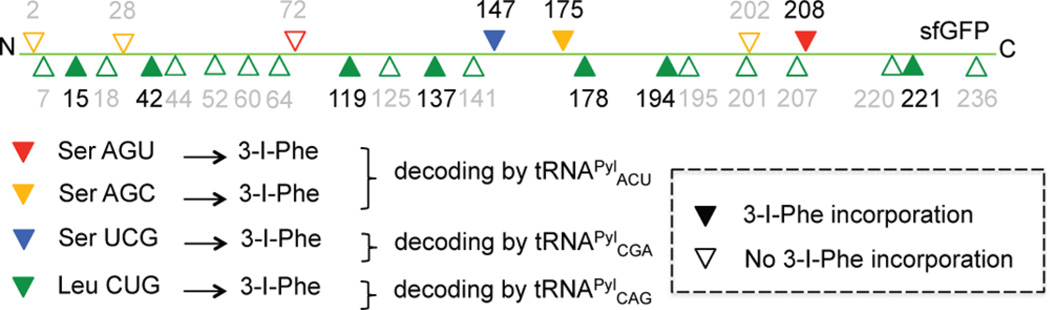
Positions of successful 3-I-Phe incorporation in sfGFP, as detected by LC-MS/MS of chymotrypsin/trypsin-digested samples. E. coli cultures expressing each individual tRNAPyl anticodon variant (tRNAPylACU, tRNAPylCGA, or tRNAPylCAG) were grown in the presence of 1 mM 3-I-Phe, and sfGFP was overexpressed, purified, and chymotrypsin/trypsin-digested, followed by analysis via tandem mass spectrometry. Triangles denote targeted codons; empty triangles indicate the failure of 3-I-Phe incorporation, and filled triangles indicate the successful incorporation of 3-I-Phe.
To further assess the reassignment of Ser AGU codons, we analyzed a single peptide that contained three different serine codons (residues 201–209: LS202TQS205VLS208K). LC-MS/MS revealed that 3-I-Phe was detected only at the targeted Ser208AGU codon and that Ser was detected only at the nontargeted Ser202AGC and Ser205UCU codons (Figure 5). The ability of tRNAPylACU to preferentially decode only the targeted Ser208AGU codon in a sequence of multiple off-target Ser codons further highlights the importance of anticodon–codon pair selection in sense codon reassignment. To identify all amino acids present at the targeted Ser208AGU codon, we searched the LC-MS/MS data for incorporation of each of the 20 canonical amino acids. In addition to 3-iodo-Phe and Ser, we detected the incorporation of Phe and Thr at this position. The observation of Phe incorporation agrees with the prediction that IFRS may have the ability to aminoacylate tRNAPyl with Phe, as suggested from the northern blot analyses, where expressed tRNAPylACU was observed to be aminoacylated in the absence of 3-I-Phe. The presence of Thr was also expected based on the ability of SerRS to misactivate Thr.29
Figure 5.
Tandem mass spectrometric analysis of the LSTQS208VLXK fragment of sfGFP purified from E. coli expressing tRNAPylACU. The notations b0 and y0 denote loss of water from the b and y fragments, respectively. X denotes 3-I-Phe incorporated at the targeted Ser208AGU position. This peptide contains 3 different serine codons, of which only 1 was encoded by AGU (bolded), decoded by tRNAPylACU, and reassigned to 3-I-Phe.
To determine if IFRS was responsible for the in vivo Phe incorporation at Ser AGU codons, cells that constitutively expressed tRNAPylACU were grown in 3-I-Phe, with and without expression of IFRS. Purified sfGFP was digested with Asp-N and analyzed via LC-MS/MS, and searches were performed for Ser → 3IF, Thr, and Phe modifications. In the presence of IFRS, all four amino acids (Ser, Thr, 3-I-Phe, and Phe) were incorporated at Ser AGU codons. In contrast, in the absence of IFRS, only Ser and Thr were incorporated (Table S1). The emergence of Phe incorporation at Ser AGU codons only when IFRS was expressed indicates that the in vivo Phe incorporation at Ser AGU codons could be attributed to IFRS misacylation of tRNAPyl.
Quantification of Amino Acid Incorporation at a Single Ser AGU Site
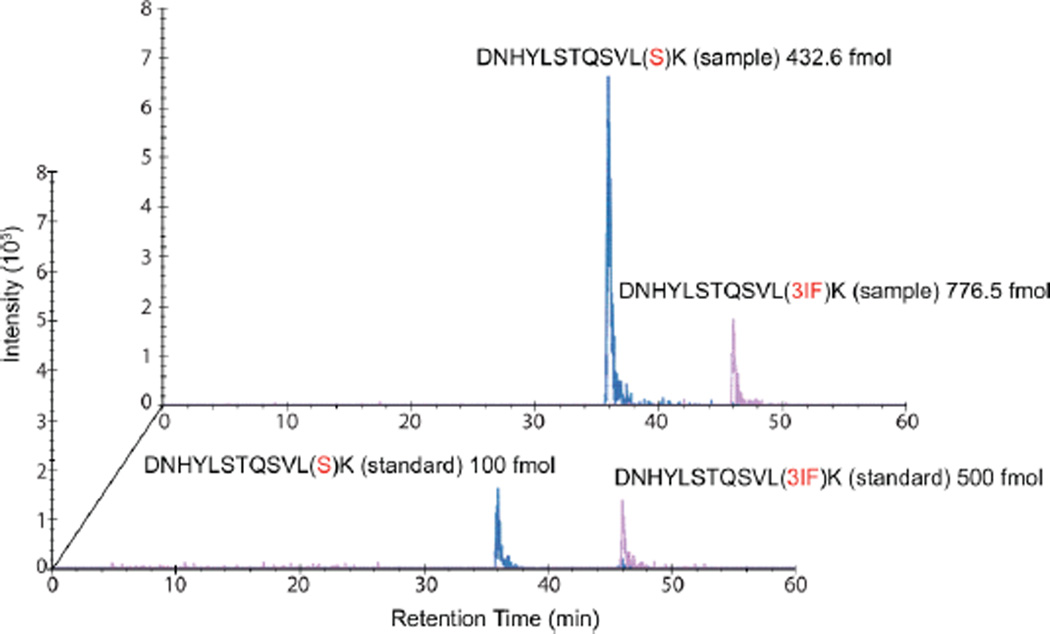
Having demonstrated qualitative incorporation of 3-I-Phe at targeted Ser AGU codons, we next measured the extent of reassignment. The absolute abundances of amino acids incorporated at the Ser208AGU position in Asp-N digested sfGFP were measured by quantitative LC-MS/MS through an SRM experiment.30 Isotopically labeled variants of peptide DNHYLSTQSVLX208K (residues 197–209, with a terminal [13C6;15N2] K label) containing 3-I-Phe, Ser, Phe, or Thr at X208 were chemically synthesized and utilized as internal standards. Importantly, the use of calibration standards allows us to fully account for changes in peptide ionization potential caused by ncAA incorporation. Absolute abundances of each peptide in four biological replicates were measured (Table 1), and signal peaks for the Ser- and 3-I-Phe-containing peptides are shown in the extracted chromatogram (Figure 6). Details of precursor ions derived from the product ions for both biologically derived peptides and isotopically labeled calibration standards (Table S2), as well as values for the lower limit of detection (LLOD) and lower limit of quantification (LLOQ) for each peptide (Table S3), are provided.
Table 1.
Values of 3-I-Phe Incorporation Efficiency and Coefficient of Variancea
| Biological replicate | AA at Ser208AGU | % incorporation | % Coeff. Var. |
| Sample 1 | Serine | 56 | 1 |
| Threonine | 1 | 13 | |
| 3-iodo-Phe | 43 | 5 | |
| Phenylalanine | 0 | 100 | |
| Sample 2 | Serine | 20 | 11 |
| Threonine | 0 | 14 | |
| 3-iodo-Phe | 79 | 19 | |
| Phenylalanine | 1 | 100 | |
| Sample 3 | Serine | 19 | N.D. |
| Threonine | 1 | N.D. | |
| 3-iodo-Phe | 78 | N.D. | |
| Phenylalanine | 2 | N.D. | |
| Sample 4 | Serine | 37 | 1 |
| Threonine | 1 | 43 | |
| 3-iodo-Phe | 60 | 11 | |
| Phenylalanine | 2 | 26 | |
| Average values | AA at Ser208AGU | % incorporation | % St. Dev. |
| Average | Serine | 33 | 17 |
| Threonine | 1 | 1 | |
| 3-iodo-Phe | 65 | 17 | |
| Phenylalanine | 1 | 1 |
Values are from four biological replicates. Exact molar quantities of the Ser, Thr, 3-I-Phe, and Phe-containing peptides in each of the four AspN-digested biological samples were measured. For each heavy-labeled standard, 500 fmol (S, T, P) or 2500 fmol (3-I-Phe) was spiked into 10 µg of digested biological sample. The samples were then brought up in 10 µL of Buffer A, and 2–5 µL of each sample was injected into the instrument. The ratios of each light peptide were multiplied by the amount of internal standard present to obtain the fmol amount of each light peptide per 5 µg of total protein in each sample. N.D. indicates values not determined.
Figure 6.
Extracted ion chromatograms from SRM analyses to quantify 3-I-Phe incorporation at Ser208AGU of sfGFP. The extent of 3-I-Phe incorporation was measured by quantitative mass spectrometric analyses (SRM) to be 65 ± 17% at position Ser208AGU. Signals from known amounts of the isotopically labeled Ser (blue, in foreground; 100 fmol) and 3-I-Phe (pink, in foreground; 500 fmol) peptides are superposed on signals from the digested sample, which contained a heterogeneous mixture of Ser (blue, at back; 432.6 fmol), 3-I-Phe (pink, at back; 776.5 fmol), Thr, and Phe peptides.
The relative abundances of amino acid incorporation at Ser208AGU were 3-I-Phe, 65 ± 17%; Ser, 33 ± 17%; Phe, 1 ± 1%; and Thr, 1 ± 1% (Table 1). The addition of 1 mM 3-I-Phe to the growth media of cells expressing IFRS/tRNAPylACU resulted in remarkably high 3-I-Phe incorporation efficiency of the Ser AGU codon, despite the presence of the competing endogenous tRNASer and the complete Ser biosynthetic pathway. This is in contrast to the previous studies that reassigned the rare Arg AGG codon in Arg auxotrophic strains19 that also included the deletion19 or downregulation18 of the competing endogenous tRNA.
The levels of amino acid incorporation detected here are a result of competition between 3-I-Phe-tRNAPylACU, Phe-tRNAPylACU, Ser-tRNASerGCU, and Thr-tRNASerGCU for decoding Ser AGU. In the absence of any engineering of the E. coli strain, this successful reassignment of the median frequency Ser AGU codon further validates reassigning the Ser AGU codon based on the anticodon-independent nature of SerRS and attests to the merit of using an engineered 3-I-Phe-tRNAPylACU that can outcompete the endogenous wobble decoding tRNASerGCU isoacceptor.
Reassignment of the Most Frequent Codon in the Ser UCN Box
To determine if sense codon reassignment to 3-I-Phe with the IFRS/tRNAPyl pair could be extended to the UCN box, we attempted to reassign Ser UCG (12 064 instances in E. coli MG1655), which has the highest codon usage in the UCN box. Ser UCG is normally decoded by two isoacceptors: a dedicated tRNASerCGA isoacceptor and tRNASerUGA that also decodes UCU and UCA.26 To evaluate the reassignment of targeted and off-target Ser codons, overexpressed sfGFP was purified from E. coli that constitutively expressed IFRS and tRNAPylCGA. After chymotrypsin/trypsin double digestion of the reporter protein sfGFP, LC-MS/MS analyses revealed 3-I-Phe incorporation at 1 of 1 targeted UCG site (Ser147UCG), and only Ser was incorporated at the other 4 nontargeted UCN sites (2 UCA, 1 UCC, and 1 UCU) (Figures 4 and S4). The observed decoding pattern of exogenous tRNAPylCGA is identical to that of the dedicated endogenous tRNASerCGA isoacceptor, and tRNAPylCGA is able to compete with tRNASerCGA to specifically decode only the Ser UCG codon. Since chymotrypsin may inadvertently cleave at 3-I-Phe residues, the LC-MS/MS analyses of sfGFP digested with chymotrypsin/trypsin will reveal 3-I-Phe-containing peptides only if they are abundant. Since we observed 3-I-Phe incorporation at the Ser UCG codon even when sfGFP was digested with chymotrypsin/trypsin, the reassignment efficiency was likely to have been high.
Reassignment of Codons with the Highest Codon Usage
Given the success of Ser UCG reassignment, we moved on to reassigning the Leu CUG codon. The Leu CUG codon is the most frequently used codon in E. coli (71 864 instances in E. coli MG1655) and has the largest pool size of tRNA isoacceptors.26 Furthermore, it has a dedicated tRNALeuCAG that is the most abundant tRNA in the cell26 and decodes via standard Watson–Crick base pairing. We first measured the in vivo aminoacylation of tRNAPylCAG inside cells, which was found to be 10% (Figure 3b). Next, to evaluate the reassignment of targeted Leu CUG codons, overexpressed sfGFP was purified from E. coli grown in the presence of 3-I-Phe (1 mM) that constitutively expressed IFRS and tRNAPylCAG. After chymotrypsin/trypsin double digestion of sfGFP, LC-MS/MS analyses revealed site-specific 3-I-Phe incorporation at 7 of 20 Leu CUG codons (Figures 4 and S5–S11). It was previously shown that the pool of decoding tRNALeuCAG is aminoacylated at a very low level (~2%) in cells grown in minimal media.35,36 Under these growth conditions, overexpressed tRNAPylCAG was 10% charged (Figure 3b), thus allowing for 3-I-Phe-tRNAPylCAG to compete with Leu-tRNALeuCAG to decode Leu CUG codons despite the high-frequency codon usage and large cellular tRNA pool size.
Next, we sought to quantify the extent of 3-I-Phe incorporation at Leu178CUG, whose corresponding peptide had a high peptide quality score. Asp-N digestion was performed on sfGFP purified from E. coli, and SRM analyses were performed. However, reassignment of Leu178CUG to 3-I-Phe could not be measured by SRM, and the low incorporation suggests that the exogenous tRNA was not able to substantially outcompete the large pool of Watson–Crick base pairing tRNA isoacceptors. In addition, analysis of the sequence coverage map (Figures 4 and S3) revealed that not every targeted codon was reassigned. In peptides with two consecutive Leu CUG codons, only one was reassigned (Figures 4, S10, and S11). Incorporation may depend on sequence context, local steric environment, and chemical similarity of the ncAA to the original amino acid.
Conclusions
In sum, we successfully reassigned the Ser AGU codon and quantified the reassignment efficiency through selected reaction monitoring (SRM). The use of calibration standards in SRM permitted accurate quantification of incorporation yield at a specific codon and fully accounted for peptide ionization changes upon ncAA incorporation. We were able to quantify incorporation yields of three amino acids (Ser, Phe, Thr) that compete with the 3-I-Phe reassignment at both the aminoacylation and decoding steps of translation when IFRS/tRNAPyl is expressed in wild-type E. coli. Our study demonstrates that the Ser AGU sense codon can be efficiently reassigned in wild-type E. coli K12, without the need to delete or downregulate the competing endogenous tRNA isoacceptor or use strains deficient in amino acid biosynthesis. By choosing a codon whose tRNA isoacceptor decodes via wobble pairing and whose aaRS has an anticodon-independent substrate recognition mechanism, we can maximize the extent of reassignment. In addition, we provide some evidence suggesting that codons whose tRNAs decode by wobble pairing may be more readily reassigned by tRNAPyl variants that decode by perfect Watson–Crick base pairing. Notably, the efficiency of ncAA incorporation depends on multiple factors. For instance, studies have revealed that inefficient incorporation of ncAAs can be caused by inefficient delivery of noncanonical aminoacyl-tRNA species to the ribosome by the bacterial elongation factor EF-Tu37 and that swapping to use a tRNA body with high EF-Tu affinity has been shown to greatly improve ncAA incorporation.38 Additionally, the thermodynamics of codon–anticodon interactions can also be modulated by post-transcriptional tRNA modifications,23 which provide an additional layer of complexity to the hypothesis that codons decoded by wobble pairing may be more readily reassigned. The contributions of these factors may be further evaluated in subsequent studies.
Future studies can focus on the parameters that affect reassignability and potential applications of sense codon reassignment. Sense codon reassignment in wild-type E. coli results in ambiguous decoding, which can increase microbial fitness under stress conditions.39 Since ncAA incorporation at the active site of an essential protein may alter cell fitness in unexpected ways, each targeted codon will have different parameters that control reassignment success. Sequence context rules that guide incorporation can also be tested via global proteomics analyses to identify sites that consistently get reassigned to the ncAA. The pattern of reassigned codons will change depending on the ncAA used, and the sequence context rules can be tested with the large repertoire of amino acid substrates that PylRS can be selected to utilize.40–43 In addition, new IFRS variants with enhanced catalytic efficiencies can be made to increase the extent of reassignment. Recent genome recoding efforts have also generated an E. coli strain that is free of amber UAG codons,44,45 and the de novo synthesis of a recoded E. coli that has unused or “open” sense codons is within the realm of possibility.25 Given the high level of ncAA incorporation in wild-type E. coli, this engineered pair should provide complete codon reassignment in a genomically recoded organism where competition at the targeted sense codon is reduced.
MATERIALS AND METHODS
Purification of Proteins
His-tagged T7 RNA polymerase and CCA-adding enzyme were individually overexpressed in E. coli BL21 (DE3), which was induced with 0.5 mM IPTG at A600 = 0.4, cultivated at 37 °C for 4 h, and harvested by centrifugation at 5000 rpm for 15 min. The cells were resuspended in buffer containing 50 mM Tris (pH 8.0), 0.3 M sodium chloride, 10% glycerol, and 5 mM imidazole; cells were disrupted by sonication, and the supernatant from a 1 h 12 000 rpm spin step was loaded on an equilibrated Ni-NTA (Qiagen) column, washed (resuspension buffer with 20 mM imidazole), eluted (resuspension buffer with 250 mM imidazole), and dialyzed with buffer containing 50 mM Tris (pH 8.0), 300 mM sodium chloride, and 40% glycerol.
IFRS was prepared by fusion of the Methanosarcina barkeri PylRS N-terminal and M. mazei PylRS C-terminal domains and bore mutations (N311S/C313I) in its amino acid binding pocket obtained from selection for binding of 3-I-Phe.27 IFRS was appended with an N-terminal 6×His tag in plasmid pET15b and overexpressed in E. coli strain BL21 (DE3). Cells overexpressing IFRS were similarly grown, induced, and harvested. Cells were suspended in 50 mM Tris-HCl (pH 7.5), 1 M sodium chloride, 10 mM 2-mercaptoethanol, 10% glycerol, and 10 mM imidazole and then similarly purified as above, with the exception of the elution step (resuspension buffer with 200 mM imidazole). The eluate was dialyzed against buffer containing 50 mM HEPES (pH 7.4), 300 mM sodium chloride, 5 mM MgCl2, 10 mM 2-mercaptoethanol, and 40% glycerol.
Preparation of Radiolabeled M. mazei tRNAPyl Anticodon Variants
The M. mazei tRNAPyl anticodon variants were prepared by site-directed mutagenesis of a pCAM plasmid containing the gene encoding M. mazei tRNAPyl, preceded by an lpp promoter. The pUC18 plasmid containing the gene encoding M. mazei tRNAPyl, preceded by the T7 promoter, was expressed in E. coli XL10-Gold (Agilent). The plasmid was purified, BstNI digested, phenol/chloroform extracted, and transcribed using T7 RNA polymerase. The in vitro transcript generated by runoff transcription was gel purified on a 12% urea polyacrylamide gel. The transcript was incubated at 8 µM with CCA-adding enzyme and 0.5 µCi/µL [α-32P] ATP for 1 h at room temperature in buffer containing 50 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 5 mM dithiothreitol (DTT), and 50 µM sodium pyrophosphate. After phenol/chloroform extraction the sample was passed over a Sephadex G25 Microspin column (Amersham Biosciences) to remove excess ATP.
Biochemical Characterization of the IFRS/tRNAPyl Pair
The aminoacylation assays were carried out for 60 min at 37 °C in aminoacylation buffer containing 100 mM Tris-HCl (pH 7.0), 10 mM MgCl2, 50 mM potassium chloride, and 5 mM DTT, with 1 mM 3-I-Phe (ChemImpex), 2 mM ATP, 500 nM IFRS, and 5 µM 32P 3′-end-labeled transcript. Aliquots (2 µL) of each reaction were quenched on ice with 3 µL of 100 mM sodium citrate (pH 5.0) and then incubated with 0.66 mg/mL nuclease P1 (Sigma) at room temperature for 1 h.46 To separate 3-I-Phe-AMP from AMP, 1 µL of quenched, digested sample was spotted on glass polyethylenimine (PEI) cellulose 20 cm × 20 cm thin-layer chromatography (TLC) plates (EMD) and developed for 75 min in 50 mM sodium acetate and 5% acetic acid. The air-dried plates were exposed on an imaging plate (FujiFilms), scanned on a Molecular Dynamics Storm 860 PhosphorImager, and quantified using ImageQuant software.
Dissociation constants were determined by mixing increasing concentrations of IFRS (50 nM to 10 µM) with 10 nM 32P 3′-end-labeled tRNAPyl in 10 µL of binding buffer containing 100 mM Tris-HCl (pH 7.0), 10 mM MgCl2, 50 mM potassium chloride, and 5 mM DTT. This experiment was performed in triplicate. After a 15 min incubation at room temperature, 7 µL of each sample was spotted onto a 96-well vacuum manifold (Hybri-dot 96; Whatman Biometra) encasing a pre-equilibrated sandwich of an upper nitrocellulose membrane (MF-Membrane Filter; Millipore) and a lower nylon membrane (Hybond-N+; Amersham). The membranes were washed with 200 µL of room-temperature binding buffer, air-dried, and exposed on an imaging plate (FujiFilms), which was scanned on a Molecular Dynamics Storm 860 PhosphorImager. The levels of radiolabeled tRNA on each filter were quantified using Phosphor-Imager and ImageQuant, and the background-corrected ratio of RNAbound to RNAtotal was plotted against the log of IFRS concentration. This semilog plot was fitted to the following equation: [R.E] = ([Rtotal] × [E])/(KD + [E]) using Kaleidagraph v. 3.6 (Synergy Software) (R, RNA; E, enzyme).
Northern Blot Analyses of Intracellular Aminoacyl-tRNA
A 40 mL culture of MG1655 that constitutively expressed tRNAPyl (pCAM or pKD) and IFRS (pCDF) was grown in LB until A600 = 1.2, washed twice, and suspended in 1% glycerol M9 minimal media supplemented with 1 mM 3-I-Phe. Cells were grown for another 6 h, harvested, disrupted in 0.3 M sodium acetate, pH 4.5, and 10 mM Na2EDTA, and split into two tubes. Total RNA was extracted and suspended in either 10 mM sodium acetate, pH 4.5 (acylated), or 200 mM Tris hydrochloride, pH 8.0 (deacylated). 1–5 µg of each sample was loaded onto a 6.5% polyacrylamide gel, pH 5.0, containing 8 M urea. Gels were run at 4 °C at 600 V for 22 h, and the RNA was transferred onto Hybond-N+ membrane (GE Healthcare) with a Trans-Blot SD semidry transfer cell (Bio-Rad) at 400 mA for 40 min. RNA was cross-linked to the membrane with a UVC 500 Ultraviolet Cross-linker (Amersham Biosciences) at 120 000 µJ/cm2. Membranes were prehybridized in ULTRAhyb Ultrasensitive buffer (Ambion) at 42 °C for 30 min, hybridized with 32P 5′-end-labeled DNA oligonucleotides complementary to tRNAPyl at 42 °C for 16 h, and washed with 2× SSC, 0.1% SDS, followed by a wash with 0.1× SSC, 0.1% SDS. Membranes were exposed to an imaging plate (FujiFilms), and plates were scanned on a Molecular Dynamics Storm 860 PhosphorImager.
Reporter Protein sfGFP Overexpression and Purification
A 200 mL culture of MG1655 that constitutively (lpp promoter) expressed tRNAPyl (pCAM) and IFRS (pCDF) was grown in LB with continuous shaking at 200 rpm in a 37 °C incubator. At A600 = 1.2, cells were washed twice and resuspended in 1% glycerol-supplemented M9 minimal media supplemented with 1 mM 3-I-Phe. After shaking for 30 min at 37 °C, 0.2% l-arabinose was added to induce the production of sfGFP (pBAD). Cells were resuspended in 1% glycerol-supplemented minimal media, and sfGFP was induced after a 30 min interval to allow accumulation of intracellular 3-I-Phe-tRNAPyl in an effort to maximize 3-I-Phe incorporation. Cells were grown for another 6 h, collected by centrifugation at 5000 rpm for 15 min, and resuspended in 10 mL of phosphate buffer solution. The cell suspension was subjected to two rounds of sonication (3 min with 6 s pulse and 6 s intervals). After centrifugation at 7000 rpm for 20 min, the supernatant was poured into an equilibrated Ni-NTA column. The column was washed twice with wash buffer containing 50 mM NaH2PO4 (pH 7.4), 300 mM sodium chloride, and 10 mM imidazole. Finally, sfGFP was eluted with elution buffer containing 500 mM NaH2PO4 (pH 7.4), 300 mM sodium chloride, and 300 mM imidazole. The eluate was concentrated using Amicon filters (Millipore), buffer exchanged with 1× PBS to remove traces of imidazole, and run on an SDS gel. A single band was always observed and was excised for LC-MS/MS analyses at the Yale Keck Mass Spectrometry Facility. Digestion was performed either with both chymotrypsin and trypsin or with Asp-N alone.
Asp-N Digestion
Dithiothreitol was added to 10 µg of each sample to a final concentration of 10 mM, the samples were then heated at 55 °C for 20 min and allowed to cool to room temperature. Iodoacetamide was added to a final concentration of 15 mM, and the samples were incubated in the dark at room temperature for a further 20 min to complete alkylation of free sulfhydryl groups. Triethylammonium bicarbonate was added to a final concentration of 50 mM, and 1% Protease max surfactant was added to a final concentration of 0.1%. Next, 0.2 µg of endoproteinase Asp-N (Roche Diagnostics) was added to the samples, which were digested overnight at 37 °C and dried in vacuo. Peptides were reconstituted with 10 µL of mobile phase A (2% acetonitrile/0.1% formic acid) containing internal standard peptides at a concentration of 100 fmol/µL, with the exception of the 3-I-Phe-containing internal standard peptide which was at a concentration of 500 fmol/µL.
Capillary LC Mass Spectrometry
A Thermo Vantage triple quadrupole mass spectrometer (Thermo Scientific) equipped with an ESI spray source was coupled to EASY-nLC system (Thermo Scientific). The nanoflow LC system was configured with a 180 µm i.d. fused silica capillary trap column containing 3 cm of Aqua 5-µm C18 material (Phenomenex) and a self-pack PicoFrit 100 µm analytical column with an 8 µm emitter (New Objective, Woburn, MA) packed to 15 cm with Aqua 3-µm C18 material (Phenomenex). Mobile phase A consisted of 2% acetonitrile/0.1% formic acid, and mobile phase B consisted of 90% acetonitrile/0.1% formic Acid. For each sample, 5 µL was dissolved in mobile phase A and injected through the autosampler onto the trap column. Peptides were then separated using the following linear gradient steps at a flow rate of 300 nL/min: 3% B for 2 min, 3–8% B over 3 min, 8–40% B over 30 min, 40–80% B over 1 min, held at 80% B for 5 min, 80–5% B over 1 min, and held at 5% B for the final 5 min. Eluted peptides were directly electrosprayed into the Vantage triple quadrupole mass spectrometer with the application of a distal 2.3 kV spray voltage and a capillary temperature of 200 °C. SRM was used to monitor for 24 transitions: 3 transitions for the heavy and light versions of the 4 different peptides (Table 1). Peaks were integrated, and peak areas were calculated using Skyline analysis software. Isotopically labeled standard peptides containing the residues of interest (Ser/Thr/3-I-Phe/Phe) at targeted sites were synthesized (Thermo Scientific Pierce Protein Research).
Supplementary Material
Acknowledgments
We thank Anna Chase, Markus Englert, Corwin Miller, Oscar Vargas, and Yane-Shih Wang for insightful discussions and comments. This work was supported by grants from the NIH [GM22854 (to D.S.), 5P30CA045508 (to D.J.P.), and GM049857 (to G.M.C.)].
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.5b00197.
Construct sequences and mass spectrometric data (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 2.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annu. Rev. Biochem. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 3.Hohsaka T, Ashizuka Y, Taira H, Murakami H, Sisido M. Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry. 2001;40:11060–11064. doi: 10.1021/bi0108204. [DOI] [PubMed] [Google Scholar]
- 4.Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: from triplet to quadruplet codes. Angew. Chem., Int. Ed. 2012;51:2288–2297. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- 5.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon I, Kirshenbaum K, Tirrell DA. Breaking the degeneracy of the genetic code. J. Am. Chem. Soc. 2003;125:7512–7513. doi: 10.1021/ja0350076. [DOI] [PubMed] [Google Scholar]
- 7.Pallanck L, Schulman LH. Anticodon-dependent aminoacylation of a noncognate tRNA with isoleucine, valine, and phenylalanine in vivo. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3872–3876. doi: 10.1073/pnas.88.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srinivasan G, James CM, Krzycki JA. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 9.Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Söll D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, Söll D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kavran JM, Gundllapalli S, O’Donoghue P, Englert M, Söll D, Steitz TA. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11268–11273. doi: 10.1073/pnas.0704769104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. A facile system for genetic incorporation of two different noncanonical amino acids into one protein in Escherichia coli. Angew. Chem., Int. Ed. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 13.Odoi KA, Huang Y, Rezenom YH, Liu WR. Nonsense and sense suppression abilities of original and derivative Methanosarcina mazei pyrrolysyl-tRNA synthetase-tRNAPyl pairs in the Escherichia coli BL21(DE3) cell strain. PLoS One. 2013;8:e57035. doi: 10.1371/journal.pone.0057035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Sachdeva A, Cox DJ, Wilf NM, Lang K, Wallace S, Mehl RA, Chin JW. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnakumar R, Ling J. Experimental challenges of sense codon reassignment: an innovative approach to genetic code expansion. FEBS Lett. 2014;588:383–388. doi: 10.1016/j.febslet.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 17.Krishnakumar R, Prat L, Aerni HR, Ling J, Merryman C, Glass JI, Rinehart J, Söll D. Transfer RNA misidentification scrambles sense codon recoding. ChemBioChem. 2013;14:1967–1972. doi: 10.1002/cbic.201300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y, Wang W, Liu WR. Towards reassigning the rare AGG codon in Escherichia coli. ChemBioChem. 2014;15:1750–1754. doi: 10.1002/cbic.201400075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee BS, Shin S, Jeon JY, Jang KS, Lee BY, Choi S, Yoo TH. Incorporation of unnatural amino acids in response to the AGG codon. ACS Chem. Biol. 2015;10:1648–1653. doi: 10.1021/acschembio.5b00230. [DOI] [PubMed] [Google Scholar]
- 20.Mukai T, Yamaguchi A, Ohtake K, Takahashi M, Hayashi A, Iraha F, Kira S, Yanagisawa T, Yokoyama S, Hoshi H, Kobayashi T, Sakamoto K. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015;43:8111–8122. doi: 10.1093/nar/gkv787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Normanly J, Ogden RC, Horvath SJ, Abelson J. Changing the identity of a transfer RNA. Nature. 1986;321:213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- 22.Asahara H, Himeno H, Tamura K, Hasegawa T, Watanabe K, Shimizu M. Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr . J. Mol. Biol. 1993;231:219–229. doi: 10.1006/jmbi.1993.1277. [DOI] [PubMed] [Google Scholar]
- 23.Agris PF. Decoding the genome: a modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meroueh M, Chow CS. Thermodynamics of RNA hairpins containing single internal mismatches. Nucleic Acids Res. 1999;27:1118–1125. doi: 10.1093/nar/27.4.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lajoie MJ, Kosuri S, Mosberg JA, Gregg CJ, Zhang D, Church GM. Probing the limits of genetic recoding in essential genes. Science. 2013;342:361–363. doi: 10.1126/science.1241460. [DOI] [PubMed] [Google Scholar]
- 26.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 27.Guo LT, Wang YS, Nakamura A, Eiler D, Kavran JM, Wong M, Kiessling LL, Steitz TA, O’Donoghue P, Söll D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16724–16729. doi: 10.1073/pnas.1419737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ko JH, Wang YS, Nakamura A, Guo LT, Söll D, Umehara T. Pyrrolysyl-tRNA synthetase variants reveal ancestral aminoacylation function. FEBS Lett. 2013;587:3243–3248. doi: 10.1016/j.febslet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I. Hydrolysis of non-cognate aminoacyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett. 2007;581:5110–5114. doi: 10.1016/j.febslet.2007.09.058. [DOI] [PubMed] [Google Scholar]
- 30.Barnidge DR, Dratz EA, Martin T, Bonilla LE, Moran LB, Lindall A. Absolute quantification of the G protein-coupled receptor rhodopsin by LC/MS/MS using proteolysis product peptides and synthetic peptide standards. Anal. Chem. 2003;75:445–451. doi: 10.1021/ac026154+. [DOI] [PubMed] [Google Scholar]
- 31.Moghal A, Mohler K, Ibba M. Mistranslation of the genetic code. FEBS Lett. 2014;588:4305–4310. doi: 10.1016/j.febslet.2014.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W, Brock A, Chen S, Chen S, Schultz PG. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 33.Ryu Y, Schultz PG. Efficient incorporation of unnatural amino acids into proteins in Escherichia coli. Nat. Methods. 2006;3:263–265. doi: 10.1038/nmeth864. [DOI] [PubMed] [Google Scholar]
- 34.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 35.Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 37.Ieong KW, Pavlov MY, Kwiatkowski M, Forster AC, Ehrenberg M. Inefficient delivery but fast peptide bond formation of unnatural L-aminoacyl-tRNAs in translation. J. Am. Chem. Soc. 2012;134:17955–17962. doi: 10.1021/ja3063524. [DOI] [PubMed] [Google Scholar]
- 38.Ieong KW, Pavlov MY, Kwiatkowski M, Ehrenberg M, Forster AC. A tRNA body with high affinity for EF-Tu hastens ribosomal incorporation of unnatural amino acids. RNA. 2014;20:632–643. doi: 10.1261/rna.042234.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu. Rev. Genet. 2013;47:121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tharp JM, Wang YS, Lee YJ, Yang Y, Liu WR. Genetic incorporation of seven ortho-substituted phenylalanine derivatives. ACS Chem. Biol. 2014;9:884–890. doi: 10.1021/cb400917a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuley A, Wang YS, Fang X, Kurra Y, Rezenom YH, Liu WR. The genetic incorporation of thirteen novel non-canonical amino acids. Chem. Commun. (Cambridge, U. K.) 2014;50:2673–2675. doi: 10.1039/c3cc49068h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang YS, Fang X, Chen HY, Wu B, Wang ZU, Hilty C, Liu WR. Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant. ACS Chem. Biol. 2013;8:405–415. doi: 10.1021/cb300512r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang YS, Russell WK, Wang Z, Wan W, Dodd LE, Pai PJ, Russell DH, Liu WR. The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol. BioSyst. 2011;7:714–717. doi: 10.1039/c0mb00217h. [DOI] [PubMed] [Google Scholar]
- 44.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, Mercer JA, Wang HH, Carr PA, Mosberg JA, Rohland N, Schultz PG, Jacobson JM, Rinehart J, Church GM, Isaacs FJ. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfson AD, Uhlenbeck OC. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl. Acad. Sci. U. S. A. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.