Abstract
OBJECTIVE
Exposure to microorganisms has repeatedly been found to influence development of atopic diseases, such as asthma. Innovative techniques have been developed that can comprehensively characterize microbial communities. The objective of this study was to characterize the home microbiota of asthmatic children utilizing 16S rRNA based phylogenetic analysis by microarray.
METHODS
In this cross-sectional study DNA was extracted from home dust and bacterial 16S rRNA genes amplified. Bacterial products were hybridized to the PhyloChip Array and scanned using a GeneArray scanner. The Adonis test was used to determine significant differences in the whole microbiome. Welch’s t-test was used to determine significant abundance differences and genus-level richness differences.
RESULTS
Nineteen homes were included in the analysis (14 asthma, 5 no asthma). 1741 operational taxonomic units (OTUs) were found in at least one sample. Bacterial genus richness did not differ in the homes of asthmatics and non-asthmatics (p=0.09). The microbial profile was significantly different between the two groups (p=0.025). All of the top 12 OTUs with significant abundance differences were increased in homes of asthmatics and belonged to one of the five phyla (p=0.001 to p=7.2 × 10−6). Nearly half of significant abundance differences belonged to the phylum Cyanobacteria or Proteobacteria.
CONCLUSIONS
These results suggest that home dust has a characteristic microbiota which is disturbed in the homes of asthmatics, resulting in a particular abundance of Cyanobacteria and Proteobacteria. Further investigations are needed which utilize high throughput technology to further clarify how home microbial exposures influence human health and disease.
Keywords: Microbiome, Built Environment, Pediatrics, Cyanobacteria, Damp Environments
Introduction
Microbial exposures are thought to be an important influence in the development of atopic diseases, such as asthma, allergic rhinitis, and atopic dermatitis. Previous studies have demonstrated that the risk for asthma development is decreased when a child is in a rich microbial environment in early life, particularly after early life exposure to livestock and pets.1–3 This observation was clarified by Ege, et. al. who found that children living on farms were exposed to a higher diversity of microbes than suburban children, an observation which was also found to explain the inverse relationship between growing up on a farm and asthma.4 Unfortunately, very little is known about the microbial exposure in low income, urban homes which are more likely to have moderate to severe physical problems that lead to increased dampness, including cracked foundations, leaky roofs, and inadequate guttering.5 Further, previous studies that have sought to characterize the microbial exposures in these unique environments have utilized techniques that are limited in their ability to fully characterize microbial communities.
Demographic differences exist in asthma prevalence in the United States with children from low- income families and non-hispanic black children disproportionately affected by this epidemic.6,7 Although it likely contributes to the increased susceptibility to asthma, genetic variation alone does not fully explain this disparity.3 As many low-income and non-hispanic black children are raised within cities, the unique exposures to which these children are subject in early life is of considerable interest for their possible contribution to asthma. The home environment is of particular concern as this is the environment in which children spend a substantial part of their day and in which they have relatively little control. Howell, et. al. previously found 25% of children living in deteriorated housing have been diagnosed with asthma compared to 8% in other housing.8 This fact leads to the hypothesis that if the exact exposures can be identified that trigger the onset of asthma in children, targeted remediation of the home environment could lead to a decrease in the overall burden of asthma.
In this study, we aimed to characterize the bacterial exposures in low income, urban homes of asthmatics and compare these exposures to that of homes of non-asthmatics from a similar demographic. In order to accomplish this aim, 16S rRNA based phylogenetic analysis was used. We hypothesized that the overall microbiome would differ between the homes of asthmatics and non-asthmatics and certain bacterial species could be identified that are found in a higher abundance in the homes of non-asthmatics.
Methods
Study design
This cross-sectional pilot study utilized data and dust samples collected as part of the Kansas City Safe and Healthy Homes program (KCHHP) which aimed to determine the influence of home remediation on asthma severity. The primary aim of the current study being reported was to compare the dust in homes of smokers and nonsmokers enrolled in the KCHHP; while, the secondary aim was to compare dust in homes of asthmatics and the homes of those without asthma from the same population as stated previously. Both the KCHHP and this study were approved by the local Institutional Review Board.
Study Population
Volunteers were recruited from the greater Kansas City area between October 2008 and November 2011 as previously described.9 Briefly, interested volunteers responded to advertisements by contacting the study coordinator directly by phone. Inclusion criteria for participation in the KCHHP were families with a child that has been diagnosed with asthma, chronic respiratory symptoms, chronic allergy symptoms, or other chronic symptoms affected by a home environment; were living in the Kansas City area; were staying at the same home at least four nights per week; had lived in the same home for the past six months; planned to live in the same home for the next 12 months; and were from families with a total family income less than 80% of the Kansas City median family income. Family income from the previous year was verified prior to enrollment.
Eligible families attended one clinic visit where written informed permission by a parent or guardian was obtained. Assent was obtained when age appropriate. A detailed questionnaire was completed which included a review of systems as well as past medical, family, social, and environmental histories. Asthma diagnosis was determined by parent report.
A subset of homes was selected for dust analysis by microarray by the following methods. First, homes with any type of pet were excluded from analysis as homes with dogs may protect against the development of asthma and allergic disease.10 Second, homes were included if dust samples were available. The remaining homes were divided into groups based on smoking status (no smokers lived in the home, smokers live in the home but do not smoke in the home, smokers live and smoke in the home) as smoking status has been shown to influence the microbiome in at least one study.11 As an excess of samples were available in the no smoking group, homes were randomly selected by an investigator who only had access to a study number.
Dust Collection
Dust collection protocols have been described previously.12,13 Briefly, dust was either collected by a special vacuum nozzle developed at Children’s Mercy Hospital and transported in dust collection sample bags (X-cell 100, Midwest filtration of Cincinnati, Ohio) to the Children’s Mercy Pediatric Immunology Laboratory or removed from the home vacuum and placed into the dust collection sample bag. Samples collected by the environmental hygienist were taken from 9 square feet of carpeting in either the family room or the child’s bedroom. Samples were filtered through a mesh screen (Thermo Fisher Scientific, Waltham, MA) and stored at −20°C until DNA extraction.
DNA Isolation and PhyloChip sample processing
DNA was isolated from the collected dust utilizing the PowerMax DNA isolation kit (Mobio, Carlsbad, Ca) according to manufacturer instructions. The purified DNA was resuspended in 5mL of RNase-free water and the concentration determined using a Nanodrop spectrophotometer. Extracted DNA was stored at −20°C until shipped to the Second Genome company for further analysis.
The bacterial 16S rRNA genes were amplified using the degenerate forward primer: 27F.1 5’-AGRGTTTGATCMTGGCTCAG-3’and the non degenerate reverse primer: 1492R.jgi 5’-GGTTACCTTGTTACGACTT-3’. The PCR products were concentrated using a solid-phase reversible immobilization method for the purification of PCR products and quantified by electrophoresis using an Agilent 2100 Bioanalyzer. PhyloChip Control Mix was added to each amplified product. Thirty-five cycles of bacterial 16S rRNA gene PCR amplification was performed. Labeled bacterial products were fragmented, biotin labeled, and hybridized to the PhyloChip Array, version G3 (Second Genome, San Bruno, CA). PhyloChip arrays were washed, stained, and scanned using a GeneArray scanner (Affymetrix). Each scan was captured using standard Affymetrix software (GeneChip Microarray Analysis Suite). Hybridization values, the fluorescence intensity, for each taxon were calculated as a trimmed average, with maximum and minimum values removed before averaging.
Statistics and Bioinformatics
PhyloChip data analyses, including data preprocessing and reduction, construction of sample-to-sample distance functions, ordination, clustering, and classification methods and phylogenetic tree construction and visualization, have been previously described.14 Briefly, bacterial profiles from homes were compared to determine a Bray-Curtis Dissimilarity Index. The Adonis test was used to determine significant differences in the whole microbiome. We calculated that with 5 homes in each group, we would be able to detect a Bray-Curtis difference of 0.1 between each sample with 80% power (α=0.05). Principal coordinate analysis (PCoA) and average-neighbor hierarchical clustering (HC-AN) was performed based on Bray-Curtis distance to plot the relationships between samples. Welch’s t-test was used to determine significant abundance differences between samples of individual OTUs as well as genus-level richness differences. Randomization tests were used to confirm that noted differences were unlikely due to chance alone. χ2 analysis was used in order to determine a significant difference in DNA retrieval from homes. All analyses were performed on PhyCA-Stats™ analysis software or an Excel spreadsheet. A p-value < 0.05 was considered statistically significant.
Results
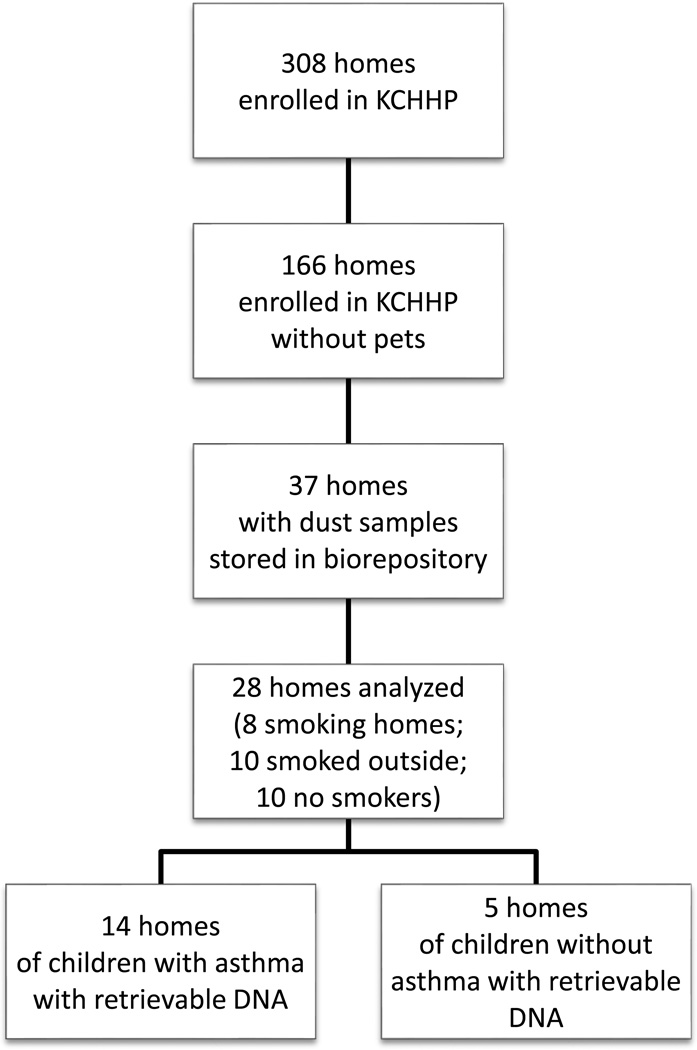
Three hundred eight homes were enrolled in the KCHHP as previously reported. Of those, 166 homes did not have a pet living in the home. Thirty-seven homes were then identified that had a dust sample stored in the KCHHP biorepository. Eight homes had a smoker living in the home who smoked in the home; ten homes had a smoker living in the home that did not smoke in the home; and nineteen homes did not have a smoker living in the home. All 8 homes of smokers living in the home who smoked in the home and all 10 homes that had a smoker living in the home that did not smoke in the home were included. Of the final 19 available samples (all from homes of non-smokers), 10 were selected for analysis based on budgetary constraints. In total, 28 homes were selected for analysis, and 6 homes were selected to have duplicate samples analyzed. Twenty-two of these homes had a child with asthma living in it. The remaining 6 homes had a child living in it that did not have asthma. Of the 36 samples processed, 23 yielded usable amounts of DNA which included 19 distinct homes (Figure 1; 14 asthma, 5 no asthma). Characteristics of each group can be found in Table 1. Homes with smokers who smoked in the home were more likely to have an insignificant amount of bacterial DNA, and therefore, the primary aim of this study could not be completed (p<0.05).
Figure 1. Study Enrollment.
Table 1.
Characteristics of compared groups
| Non-asthmatics n=5 n (%) |
Asthmatics n=14 n (%) |
|
|---|---|---|
| Demographics | ||
| Gender | ||
| Female | 4 (80%) | 4 (29%) |
| Age (mean ± SD) | 2.4 (0.9) | 4.8 (3.1) |
| Race/ethnicity | ||
| African-American | 3 (60%) | 6 (43%) |
| Hispanic | 1 (20%) | 3 (21%) |
| Caucasian | 1 (20%) | 2 (14%) |
| Other | 0 | 3 (21%) |
| Family history of asthma | 3 (60%) | 6 (43%) |
| Socio-economic status | ||
| <80% but >50% of MFI in KC | 2 (40%) | 3 (21%) |
| <50% of MFI in KC | 3 (60%) | 11 (79%) |
| Exposed to SHS inside home | 0 | 3 (21%) |
| Lives with a smoker1 | 3 (60%) | 8 (73%) |
SD, standard deviation; MFI, median family income; KC, Kansas City
Smoker does not live inside the home
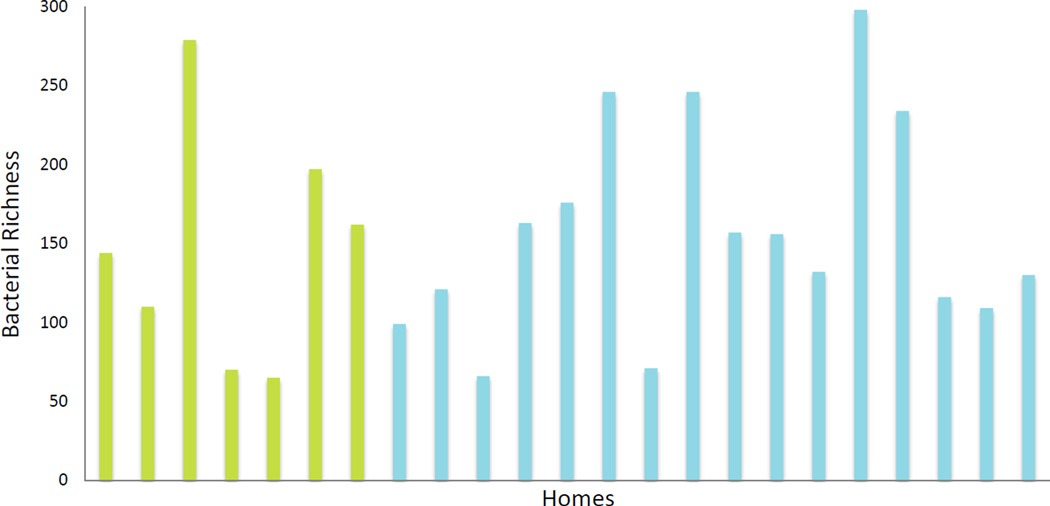
Of over 59,000 operational taxonomic units (OTUs) included on the PhyloChip, 1,741 unique OTUs were identified from at least one sample. Firmicutes, Proteobacteria, and Bacteroidetes were the three most prominent phyla found. The top 9 identified bacterial families represented, on average, 32% of the OTUs from each sample: Lachnospiraceae, Pseudomonadaceae, Rikenellaceaell, Rhodospirillaceae, Ruminococcaceae, Prevotellaceae, Flexibacteraceae, Flavobacteriaceae, and Anaerolinaceae. Bacterial genus richness ranged from 65 to 298. No significant difference in genus-level richness was found between the asthma homes and control homes (Figure 2, p=0.09).
Figure 2. Genus level richness is not different between homes of asthmatics and homes of controls.
Bacterial richness ranged from 65 to 298. No significant difference in genus-level richness exists between homes of asthmatics (blue) and healthy controls (green, p=0.0923).
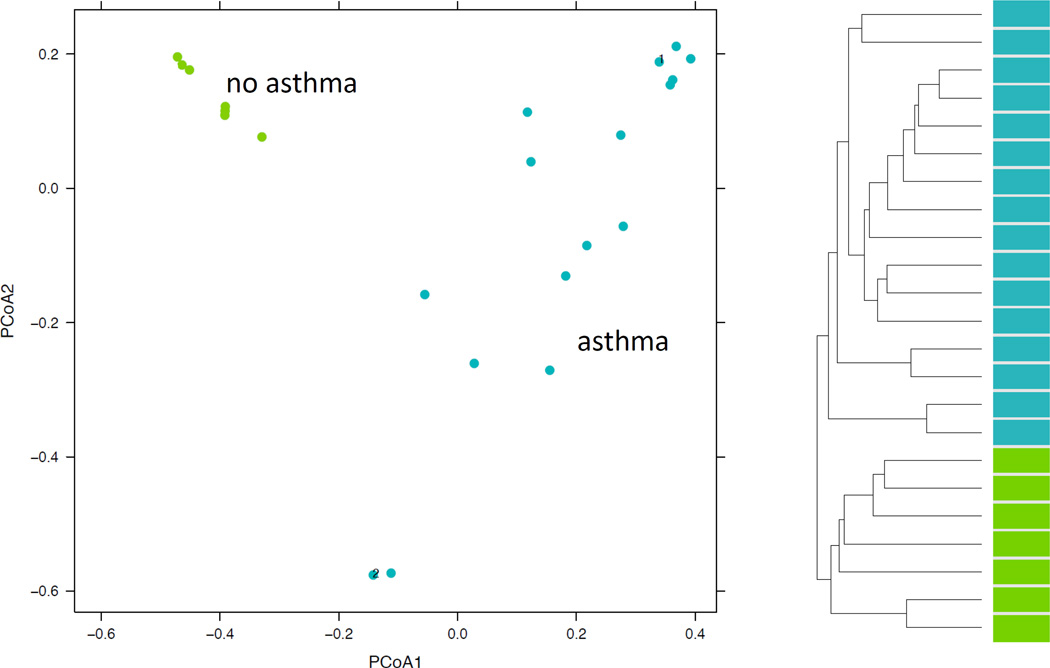
Direct comparison of the dust microbiota from homes of asthmatics with that of controls revealed a significant difference in the overall microbial profile (p=0.025). Bacterial communities were structured according to asthma status (Figure 3). On principal coordinate analysis, the microbiome from the dust of homes of non-asthmatics clustered into a distinct group; while, homes of asthmatics were separated but did not distinctly cluster. Hierarchical clustering showed that samples cluster distinctly into two groups based on asthma status. Repeat samples which were taken from the same home were in close distance to each other but not overlying demonstrating some intra-home variation.
Figure 3. Phylochip analysis of 16S rRNA reveals that the microbiome of homes of asthmatics is varied compared to the consistent microbiome of homes of controls.
A. Communities are clustered using principal coordinates analysis. Each point corresponds to a community colored according to asthma status. Axis 1: 24% of variation; Axis 2: 13% of variation B. Hierarchical clustering demonstrates a distinct clustering of the microbiomes of dust from the homes of asthmatics (blue) and controls (green).
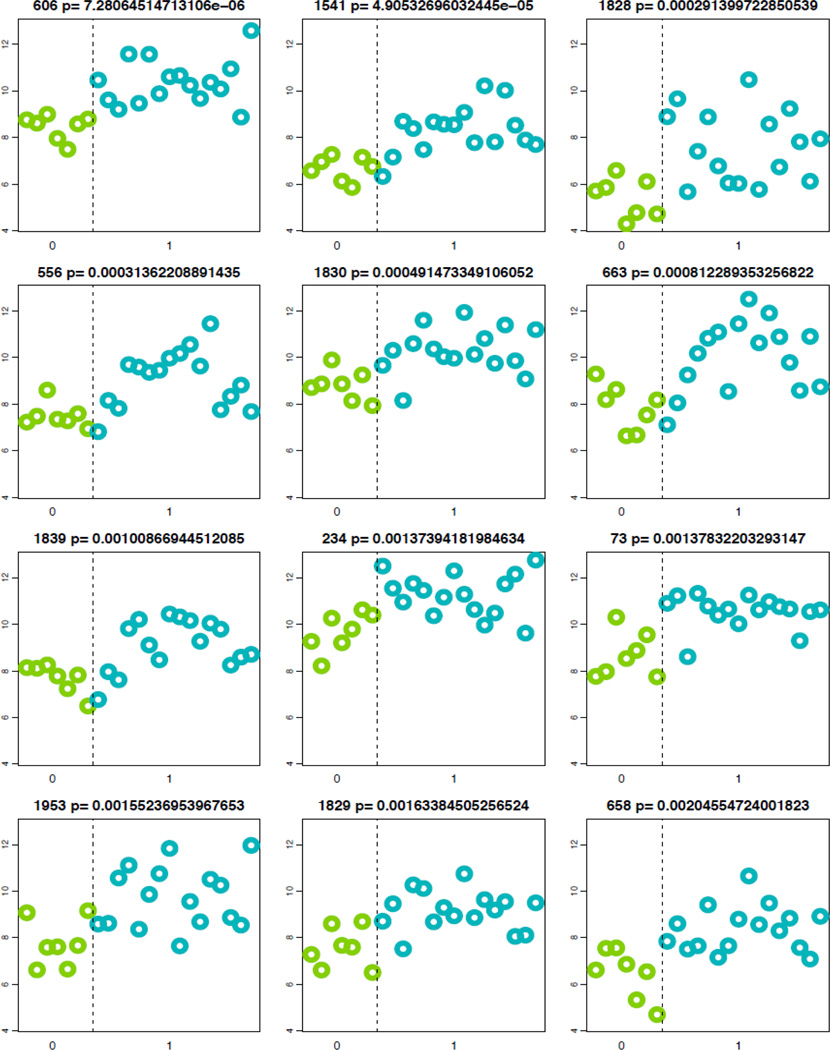
When taxa abundance was compared between the samples from homes of asthmatics and the homes of non-asthmatics, each of the top 12 OTU’s belonged to one of the five phyla Cyanobacteria, Firumicutes, Actinobacteria, Proteobacteria, and Bacteroidetes and all were increased in abundance in the dust from the homes of asthmatics relative to the dust from homes of non-asthmatics (Figure 4, Table 2, p=7 × 10−6 to p=0.002). Seven of the top 12 OTUs belonged to the phylum Cyanobacteria. In all, 182 OTUs within 71 families were found to have significantly different abundances between homes of asthmatics when compared to that of non-asthmatics (p<0.05). These OTUs were not restricted to one phylum but nearly half were from the phyla Proteobacteria or Cyanobacteria (Figure 5). Of the 182 OTUs identified with abundance differences, 179 OTU’s was found to have an increased abundance in the homes of asthmatics. One OTU within the family Lachnospiraceae (Phylum: Firmicutes) and 2 OTUs within the family Enterobacteriaceae (Phylum: Proteobacteria) were increased in homes of non-asthmatics.
Figure 4. OTUs Differing in Abundance Between Homes of Asthmatics and Non-Asthmatics.
All of the top 12 OTUs with significant abundance differences were increased in abundance in the dust from homes of asthmatics (blue). Each plot represents one OTU and each point represents one sample.
Table 2.
OTUs differing in abundance between homes of asthmatics and non-asthmatics.
| Taxa ID | Phylum | Class | Order | Family | Genus | species |
|---|---|---|---|---|---|---|
| 606 | Cyanobacteria | Oscillatoriophycideae | Chroococcales | Xenococcaceae | Chroococcidiopsis | unclassified |
| 1541 | Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium | paraputrificum |
| 1828 | Cyanobacteria | Chloroplast | Rhodophyta | unclassified | unclassified | unclassified |
| 556 | Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | unclassified |
| 1830 | Cyanobacteria | Chloroplast | Stramenopiles | unclassified | unclassified | unclassified |
| 663 | Firmicutes | Bacilli | Lactobacillales | Carnobacteriaceae | Trichococcus | collinsii |
| 1839 | Proteobacteria | Betaproteobacteria | Burkholderiales | Comamonadaceae | unclassified | unclassified |
| 234 | Cyanobacteria | Oscillatoriophycideae | Chroococcales | Xenococcaceae | Chroococcidiopsis | unclassified |
| 73 | Cyanobacteria | Chloroplast | Stramenopiles | unclassified | unclassified | unclassified |
| 1953 | Cyanobacteria | Nostocophycideae | Nostocales | Rivulariaceae | Calothrix | unclassified |
| 1829 | Cyanobacteria | Chloroplast | Stramenopiles | unclassified | unclassified | unclassified |
| 658 | Bacteroidetes | Sphingobacteria | Sphingobacteriales | Sphingobacteriaceae | unclassified | unclassified |
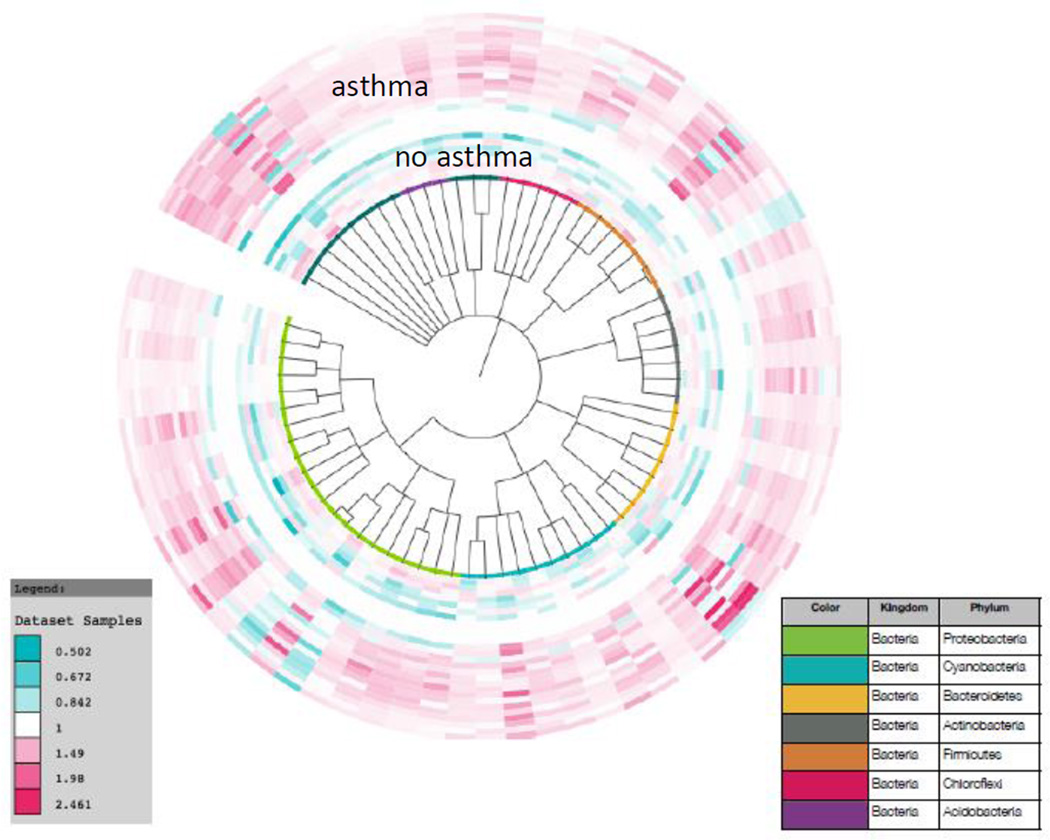
Figure 5. Phylogenetic tree of significantly different abundances of OTUs in homes of asthmatic children (outer rings) and homes of non-asthmatic children (inner rings).
From the 1741 OTUs found in each home, 182 OTUs from 72 families were found to be increased in the homes of asthmatic (p<0.05).
Discussion
Several observations were made about the home microbiome in this study. First, bacterial DNA could not be isolated from homes of smokers using the stated methods. In the remaining homes, 1741 distinct OTUs could be identified in house dust which demonstrates a much higher diversity of bacterial exposures than previously measured. Further, this study found that the microbiota in homes of asthmatic children did not differ in genus level richness from that of children without asthma. When the whole microbiome was compared between the two groups, the dust from the homes of non-asthmatic children clustered strongly together demonstrating a characteristic microbiome, while that from homes of asthmatic children was dissimilar with no distinct clustering.
When the differences were further examined, 179 OTUs were found to be in increased abundance in the homes of asthmatic children while only 3 OTUs could be found that were in increased abundance in the homes of non-asthmatic children. All the top 12 OTUs that were found in higher abundance in the asthmatic homes belonged to one of the five phyla; while, 7 of these belonged to the phyla Cyanobacteria. Of all the OTUs found to have significant abundance differences, nearly all were increased in the homes of asthmatics and nearly half of these were from the phyla Proteobacteria and Cyanobacteria. The 3 OTUs that were found in higher abundance in the homes of non-asthmatic children were from the families Lachnospiraceae and Enterobacteriaceae.
These results differ from that of one previous study which demonstrated a decrease in bacterial diversity in the homes of children with asthma and numerous studies that have correlated more abundant levels of bacterial markers in homes with asthma.4,15–17 Our results found no difference in genus richness between the two groups; rather, they demonstrate different bacteria to be in abundance in the homes of asthmatic children. One possible explanation for the difference is the unique populations. While the study by Ege et al analyzed dust from homes in farming environments and suburban areas in Europe, the dust analyzed in this study was taken from homes in a Midwestern, urban core. Numerous other factors may also be influencing the results of both the study by Ege et al as well as this leading to disparate results, including temperature and humidity at the time of sampling, the season of sampling, and the type of dwelling sampled. Regression models for 16s sequencing have recently been developed and will allow for analysis of the multiple covariates in future studies.18,19 Finally, this difference may also be explained by the limited number of homes in this study, ass this study was powered to determine the difference in the whole microbiome and not genus-level richness.
Previous work has demonstrated that the microbiome in the lung of asthmatics had a striking increase in Proteobacteria species.11 Our work likewise identified Proteobacteria species as having a higher abundance in the homes of asthmatics. This suggests that that home environmental exposure to Proteobacteria could lead to lung colonization in a susceptible host. The consequence of Proteobacteria in the lung, however, is poorly understood and further investigation is needed to confirm that the home microbiome influences microbial colonization in the respiratory tract and the clinical consequence of this potential influence. In addition both this study, as well as the study by Hilty et al, failed to isolate bacterial DNA from samples isolated from smokers. Although no explanation for this observation exists in the literature, current techniques may fail to liberate DNA from the cigarette tar; however, this theory needs to be tested in controlled studies.
The clinical implication of exposure to cyanobacteria or “blue-green algae”, the bacteria found in particular abundance in the homes of asthmatics, is also poorly understood. Considerable disagreement has occurred over the proper classification of cyanobacteria, as OTUs from this phylum have characteristics of both Gram-negative and Gram-positive organisms.20 Notably, however, cyanobacteria does contain lipopolysaccharide (LPS) or endotoxin which has shown to be both protective and pathogenic in regards to asthma depending on timing of exposure, the presence of absence of preexisting disease, and genetic polymorphisms.21 Interestingly, the LPS found in cyanobacteria has certain structural alterations from the LPS of E. coli, Salmonella, and other Gram-negative, gut-dwelling bacteria, specifically in the bioactive lipid A moiety.20 These alterations are suspected to provide cyanobacteria not only with LPS-antagonistic properties but also the ability to reduce the activity of glutathione S-transferase, an enzyme linked to asthma and wheeze.22–25 This proposed mechanism may also provide explanation to the previous observations that water-damaged homes are associated with asthma and/or wheeze as the optimal conditions for cyanobacteria growth is in damp environments.26–30 Finally, Bernstein et al recently found that up to 29% of a population with chronic rhinitis were sensitized to cyanobacteria, implicating cyanobacteria as an unrecognized sensitizing allergen.31
The strength of this study is the utilization of high throughput technology to study bacterial exposures in homes of children similar to another recent study which utilized 18S sequencing to fully characterize fungal exposures in homes.12 This newer technology allows for comprehensive characterization of the home microbial communities and provides an exciting basis for further study in this area. This study is, however, subject to several limitations. Since it is a cross-sectional analysis, this study is hypothesis generating and causality of asthma cannot be attributed to the identified bacteria. A large scale cohort utilizing similar high throughput techniques is needed to clarify the observed associations. In addition, this was a pilot study, and the sample size is small. Wider analysis may reveal other important differences in microbial populations not recognized in this study. Further, a larger sample size in future studies will allow for regression modeling and inclusion of several important covariates. Finally, we acknowledge that the control group in this study may not be representative of a healthy population due to the inclusion criteria of the KCHHP which recruited children with chronic diseases that may be influenced by the home environment such as chronic rhinitis.
Conclusions
Through the use of high-throughput techniques, this study has shown that the microbial diversity in homes is greater than previously imagined. In addition, although the genus level richness did not differ is our study, two phyla, Cyanobacteria and Proteobacteria, were found to be in significantly higher abundance in the homes of asthmatics compared to non-asthmatic children. The wide application of the methodology used in this study and similar high throughput techniques are likely to result in improved characterization of home microbial exposures and a better understanding of the relationship between these exposures and human health and disease.12,32
Acknowledgments
We thank Edward Ellerbeck for his thoughtful review of this manuscript and the families that have participated in the KCHHP who have selflessly volunteered to allow us into their homes and have provided us with intimate details of their lives with the hope that future generations may become less burdened with allergic disease.
Footnotes
Declaration of Interest:
The authors do not have any relevant conflicts of interest. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- 1.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA. 2002;288:963–972. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- 2.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10:861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 3.Eder W, Ege MJ, von Mutius E. The asthma epidemic. N Engl J Med. 2006;355:2226–2235. doi: 10.1056/NEJMra054308. [DOI] [PubMed] [Google Scholar]
- 4.Ege MJ, Mayer M, Normand AC, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 5.Krieger J, Higgins DL. Housing and health: time again for public health action. Am J Public Health. 2002;92:758–768. doi: 10.2105/ajph.92.5.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease C, Prevention. Vital signs: asthma prevalence, disease characteristics, and self-management education: United States, 2001–2009. MMWR Morbidity and mortality weekly report. 2011;60:547–552. [PubMed] [Google Scholar]
- 7.Weiss KB, Gergen PJ, Crain EF. Inner-city asthma. The epidemiology of an emerging US public health concern. Chest. 1992;101:362S–367S. doi: 10.1378/chest.101.6.362s. [DOI] [PubMed] [Google Scholar]
- 8.Howell E, Harris LE, Popkin SJ. The health status of HOPE VI public housing residents. Journal of health care for the poor and underserved. 2005;16:273–285. doi: 10.1353/hpu.2005.0036. [DOI] [PubMed] [Google Scholar]
- 9.Ciaccio CE, DiDonna AC, Kennedy K, Barnes CS, Portnoy JM, Rosenwasser LJ. Association of tobacco smoke exposure and atopic sensitization. Ann Allergy Asthma Immunol. 2013;111:387–390. doi: 10.1016/j.anai.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimura KE, Demoor T, Rauch M, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111:805–810. doi: 10.1073/pnas.1310750111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rittenour WR, Ciaccio CE, Barnes CS, et al. Internal transcribed spacer rRNA gene sequencing analysis of fungal diversity in Kansas City indoor environments. Environmental science Processes & impacts. 2014;16:33–43. doi: 10.1039/c3em00441d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnes CS, Kennedy K, Gard L, et al. The impact of home cleaning on quality of life for homes with asthmatic children. Allergy Asthma Proc. 2008;29:197–204. doi: 10.2500/aap.2008.29.3099. [DOI] [PubMed] [Google Scholar]
- 14.Noval Rivas M, Burton OT, Wise P, et al. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201–212. doi: 10.1016/j.jaci.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gehring U, Strikwold M, Schram-Bijkerk D, et al. Asthma and allergic symptoms in relation to house dust endotoxin: Phase Two of the International Study on Asthma and Allergies in Childhood (ISAAC II) Clin Exp Allergy. 2008;38:1911–1920. doi: 10.1111/j.1365-2222.2008.03087.x. [DOI] [PubMed] [Google Scholar]
- 16.Douwes J, van Strien R, Doekes G, et al. Does early indoor microbial exposure reduce the risk of asthma? The Prevention and Incidence of Asthma and Mite Allergy birth cohort study. J Allergy Clin Immunol. 2006;117:1067–1073. doi: 10.1016/j.jaci.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Sordillo JE, Hoffman EB, Celedon JC, Litonjua AA, Milton DK, Gold DR. Multiple microbial exposures in the home may protect against asthma or allergy in childhood. Clin Exp Allergy. 2010;40:902–910. doi: 10.1111/j.1365-2222.2010.03509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Li H. Variable Selection for Sparse Dirichlet-Multinomial Regression with an Application to Microbiome Data Analysis. The annals of applied statistics. 2013;7 doi: 10.1214/12-AOAS592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia F, Chen J, Fung WK, Li H. A logistic normal multinomial regression model for microbiome compositional data analysis. Biometrics. 2013;69:1053–1063. doi: 10.1111/biom.12079. [DOI] [PubMed] [Google Scholar]
- 20.Stewart I, Schluter PJ, Shaw GR. Cyanobacterial lipopolysaccharides and human health - a review. Environmental health : a global access science source. 2006;5:7. doi: 10.1186/1476-069X-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams LK, Ownby DR, Maliarik MJ, Johnson CC. The role of endotoxin and its receptors in allergic disease. Ann Allergy Asthma Immunol. 2005;94:323–332. doi: 10.1016/S1081-1206(10)60983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabesch M, Hoefler C, Carr D, Leupold W, Weiland SK, von Mutius E. Glutathione S transferase deficiency and passive smoking increase childhood asthma. Thorax. 2004;59:569–573. doi: 10.1136/thx.2003.016667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fryer AA, Bianco A, Hepple M, Jones PW, Strange RC, Spiteri MA. Polymorphism at the glutathione S-transferase GSTP1 locus. A new marker for bronchial hyperresponsiveness and asthma. Am J Respir Crit Care Med. 2000;161:1437–1442. doi: 10.1164/ajrccm.161.5.9903006. [DOI] [PubMed] [Google Scholar]
- 24.Spiteri MA, Bianco A, Strange RC, Fryer AA. Polymorphisms at the glutathione S-transferase, GSTP1 locus: a novel mechanism for susceptibility and development of atopic airway inflammation. Allergy. 2000;55(Suppl 61):15–20. doi: 10.1034/j.1398-9995.2000.00502.x. [DOI] [PubMed] [Google Scholar]
- 25.Mapp CE, Fryer AA, De Marzo N, et al. Glutathione S-transferase GSTP1 is a susceptibility gene for occupational asthma induced by isocyanates. J Allergy Clin Immunol. 2002;109:867–872. doi: 10.1067/mai.2002.123234. [DOI] [PubMed] [Google Scholar]
- 26.Zock JP, Jarvis D, Luczynska C, Sunyer J, Burney P European Community Respiratory Health S. Housing characteristics, reported mold exposure, and asthma in the European Community Respiratory Health Survey. J Allergy Clin Immunol. 2002;110:285–292. doi: 10.1067/mai.2002.126383. [DOI] [PubMed] [Google Scholar]
- 27.Nicolai T, Illi S, von Mutius E. Effect of dampness at home in childhood on bronchial hyperreactivity in adolescence. Thorax. 1998;53:1035–1040. doi: 10.1136/thx.53.12.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taskinen T, Hyvarinen A, Meklin T, Husman T, Nevalainen A, Korppi M. Asthma and respiratory infections in school children with special reference to moisture and mold problems in the school. Acta Paediatr. 1999;88:1373–1379. doi: 10.1080/080352599750030112. [DOI] [PubMed] [Google Scholar]
- 29.Jedrychowski W, Flak E. Separate and combined effects of the outdoor and indoor air quality on chronic respiratory symptoms adjusted for allergy among preadolescent children. International journal of occupational medicine and environmental health. 1998;11:19–35. [PubMed] [Google Scholar]
- 30.Slezak JA, Persky VW, Kviz FJ, Ramakrishnan V, Byers C. Asthma prevalence and risk factors in selected Head Start sites in Chicago. J Asthma. 1998;35:203–212. doi: 10.3109/02770909809068208. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein JA, Ghosh D, Levin LS, et al. Cyanobacteria: an unrecognized ubiquitous sensitizing allergen? Allergy Asthma Proc. 2011;32:106–110. doi: 10.2500/aap.2011.32.3434. [DOI] [PubMed] [Google Scholar]
- 32.Vesper S, Barnes C, Ciaccio CE, et al. Higher Environmental Relative Moldiness Index (ERMI) values measured in homes of asthmatic children in Boston, Kansas City, and San Diego. J Asthma. 2013;50:155–161. doi: 10.3109/02770903.2012.740122. [DOI] [PMC free article] [PubMed] [Google Scholar]