SUMMARY
Dihydrolipoyl acetyltransferase (E2) is the central component of pyruvate dehydrogenase complex (PDC), which converts pyruvate to acetyl-CoA. Structural comparison by cryo-electron microscopy (cryo-EM) of the human full-length and truncated E2 (tE2) cores revealed flexible linkers emanating from the edges of trimers of the internal catalytic domains. Using the secondary structure constraints revealed in our 8 Å cryo-EM reconstruction and the prokaryotic tE2 atomic structure as a template, we derived a pseudo atomic model of human tE2. The active sites are conserved between prokaryotic tE2 and human tE2. However, marked structural differences are apparent in the hairpin domain and in the N-terminal helix connected to the flexible linker. These permutations away from the catalytic center likely impart structures needed to integrate a second component into the inner core and provide a sturdy base for the linker that holds the pyruvate dehydrogenase for access by the E2-bound regulatory kinase/phosphatase components in humans.
INTRODUCTION
Pyruvate dehydrogenase complex (PDC) is one of the largest multienzyme, supramolecular machines found in both prokaryotes and eukaryotes (Reed, 1974; Patel and Roche, 1990). PDC belongs to the family of enzymes that catalyzes the oxidative decarboxylation of α-keto acids. By converting pyruvate to acetyl-coenzyme A (CoA), PDC allows glucose carbons to be used as an oxidative energy source or in fatty acid biosynthesis. This catalysis is a key reaction that occurs at the junction of glycolysis and the citric acid cycle. Because this conversion leads to a net depletion of glucose carbons in mammals, mammalian PDC plays an important gatekeeper role in the irreversible consumption of carbohydrates (Randle, 1986), and its activity is tightly regulated by a set of dedicated kinases and phosphatases (Randle and Priestman, 1996; Roche et al., 2001).
Human PDC contains six major components: pyruvate dehydrogenase (E1), dihydrolipoyl acetyltransferase (E2), dihydrolipoyl dehydrogenase (E3), E3-binding protein (E3BP), and two dedicated regulatory components—pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatase (PDP) (Roche et al., 2001, 2003; Kato et al., 2007). E1, E2, and E3 are found in species ranging from prokaryotes to higher organisms, and E3BP is found in most eukaryotes. E2 plays a central role by acting as a scaffold in organizing and connecting the components participating in the sequential reactions of the complex (Yang et al., 1997). In humans, tissue-specific isoforms of PDK and PDP provide valuable targets for therapeutic intervention of diabetes, heart ischemia, and cancer (Kato et al., 2007; Roche and Hiromasa, 2007).
The outer globular domains of human E2 consist of two lipoyl domains (L1 and L2) and a small E1-binding domain, connected by alanine- and proline-rich hinge regions that are 20–30 residues in length (Figure 1A) (Thekkumkara et al., 1988). The composition of lipoyl domains and the roles of the binding domains vary among E2 from different sources (Perham, 2000). For instance, the number of lipoyl domains per E2 subunit can be one (yeast, chloroplast, or Bacillus stearothermophilus PDC), two (mammalian, plant mitochondria, and Streptococcus faecalis PDC), or three (Escherichia coli or Azotobacter vinelandii E2). The small binding domain of E2 in eukaryotes and humans binds only to E1, whereas the structurally related E2 domains from bacteria can bind to either E1 or E3. The C-terminal half of E2, which is also called the inner catalytic (IC) domain, shares less than 35% sequence identity in prokaryotic PDC and human PDC. Human E3BP is composed of three linker-connected domains in a configuration similar to that of human E2 (Figure 1A), and its IC domain has ~50% sequence identity with human E2 (Harris et al., 1997). The outer linker regions are responsible for the delivery of the lipoyl domains, through a “swinging arm” active-site coupling mechanism (Reed and Hackert, 1990), to the E1, E2, and E3 components, in which the prosthetic group covalently bound to the lipoyl domain extends into active-site channels (Roche and Cox, 1996). Every E2 trimer has three active-site channels located at the three inter-E2 interfaces. These channels span the distance from the outer surface (lipoyl group entry) to the interior of the dodecahedron (CoA entry) and serve as conduits through which acetyl groups pass (Mattevi et al., 1992).
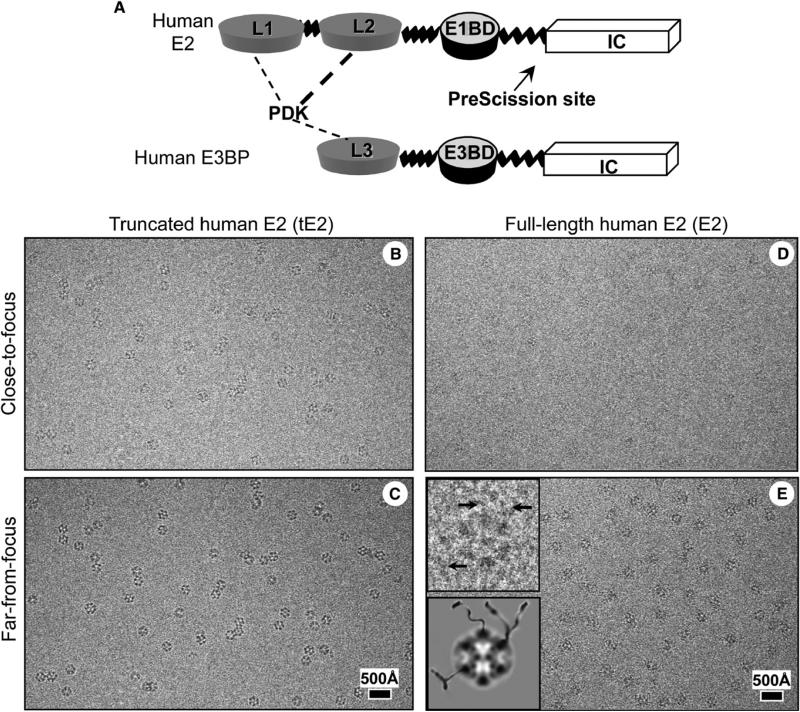
Figure 1. Schematic Diagram of Human E2 and E3BP and Cryo-EM of the Human PDC Core.
(A) Schematic diagram of full-length E2 and E3BP molecules. PDK has a stronger binding affinity to L2 of E2 (indicated by a thick, dotted line) than to L1 of E2 and L3 of E3BP (indicated by thin, dotted lines). The arrow points to the PreScisssion cleavage site of E2.
(B–E) Focal pairs of cryo-EM images of (B and C) recombinant human tE2 and (D and E) full-length E2 cores are shown. The underfocus values for close-to-focus and far-from-focus images, respectively, were 0.9 and 3.0 mm for tE2 and 1.2 and 3.3 μm for E2. Insets in (E) show one enlarged view of the E2 particle and its cartoon illustration, revealing peripheral densities (arrows) tethered to cores.
The three-dimensional (3D) structures of prokaryotic, yeast, and bovine PDCs and their subcomplexes have been studied by X-ray crystallography (Mattevi et al., 1992; Mande et al., 1996), NMR (Kalia et al., 1993), and cryo-electron microscopy (cryo-EM) (Wagenknecht et al., 1991; Stoops et al., 1992, 1997; Zhou et al., 2001a; Milne et al., 2002; Borgnia et al., 2004). These structural analyses have revealed significant evolutionary diversity in structure and subunit composition that imparts unique regulatory properties in spite of the conserved enzymatic activities of PDCs (Patel and Roche, 1990). The E. coli and A. vinelandii PDCs, for example, are built around an octahedral core of E2 chains to which multiple copies of E1 and E3 are bound. In contrast, B. stearothermophilus, yeast, and mammalian E2 complexes form a pentagonal dodecahedron with a large internal cavity. Most notably, the structure of the truncated E2 (tE2) C-terminal half (i.e., the IC domain) from A. vinelandii has been determined to atomic resolution (2.6 Å) by X-ray crystallography (Mattevi et al., 1992). Atomic models of B. stearothermophilus and Enterococcus faecalis tE2 molecules have also been constructed by homology modeling and fitting to X-ray diffraction data at 4.4 Å resolution (Izard et al., 1999).
Functional human PDC complexes and subcomplexes can be obtained in solution by incubating individually expressed proteins (Yang et al., 1997). By introducing a PreScission protease site in the full-length human E2, we were able to obtain a limited amount of soluble human tE2 cores by chemical cleavage of the recombinant full-length E2 (Hiromasa et al., 2004). However, human and other eukaryotic E2 or tE2 cores have eluded X-ray crystallographic efforts. The intrinsic flexibility of full-length human E2 may have prevented the formation of crystals suitable for X-ray diffraction analyses. Attempts to grow crystals of human tE2 have also been fruitless due to difficulties in obtaining an adequate quantity of soluble human tE2 cores either by directly expressing tE2 or by chemically cleaving full-length E2. Consequently, our understanding of the human E2 structure has remained limited.
In this study, we used cryo-EM single-particle reconstructions to obtain the 3D structures of the full-length human E2 and tE2 cores. Comparison of these structures established that human E2 is organized as a pentagonal dodecahedron of 20 E2 trimers with a stiff inner linker emanating from the edge of each subunit of the trimer. Secondary structure elements resolved in our 8 Å resolution cryo-EM map were used as constraints to build a pseudo atomic model of human tE2, which reveals a highly conserved catalytic center and key structural differences in the peripheral domains. These structural differences are consistent with the unique requirements of the human E2, compared with its prokaryotic homologs, to serve its highly regulated functional roles. Based on these observations, we have proposed a mechanistic model to account for the catalytic and unique regulatory activities of human PDC.
RESULTS
3D Structures of the Human PDC E2 and tE2 Cores
We imaged the E2 cores assembled with recombinantly expressed full-length human E2 and N-terminal tE2 (Figure 1A) subunits that contained only the IC domain. Cryo-EM images of tE2 (Figures 1B and 1C) and full-length E2 (Figures 1D and 1E) cores showed cage-like structures with the characteristic pentagonal dodecahedral shape. The contrast of the E2 images was consistently lower than that of the tE2 images, presumably because thicker ice was necessary to fully embed the larger cores containing the full-length E2 subunits. Upon closer examination of the E2 images, we identified densities that extended ~50–80 Å from the dodecahedral cores that appeared to be tethered to the cores (insets in Figure 1E), and this finding is similar to what has been previously seen in 2D images of bovine PDC cores (Wagenknecht et al., 1991).
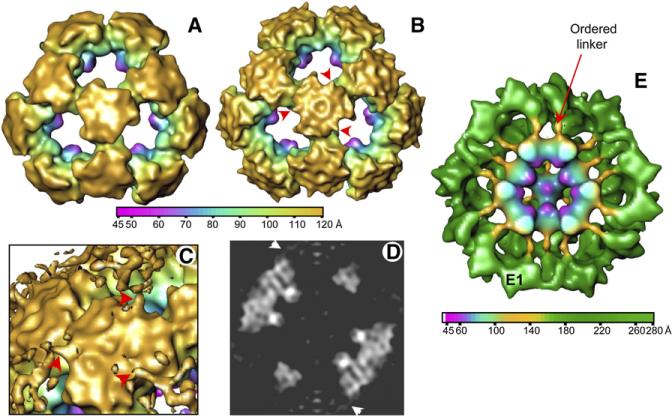
As summarized in Table 1, we determined the 3D structures of human tE2 and E2 cores by cryo-EM single-particle reconstruction to an effective resolution of 8.8 Å and 15 Å, respectively. The lower resolution of the full-length E2 structure is probably due to the greater molecular flexibility and the lower contrast of its cryo-EM images resulting from thicker ice. The 3D reconstructions of tE2 (Figure 2A) and full-length E2 (Figure 2B) cores filtered to the same resolution appear to be very similar when displayed as shaded surface representations at the same density threshold (one standard deviation above the mean). They have the shape of a dodecahedron cage consisting of 60 E2 subunits arranged as 20 trimers. In particular, the E2 IC trimer densities are identical and superimposable in their internal regions. However, the resolved E2 structure also appears to contain some extra densities (indicated by arrowheads in Figure 2B) that were not present in the tE2 structure. When the density contour level was reduced, these additional extensions became more apparent, as shown in the close-up (Figure 2C) and central slice (Figure 2D) of the full-length E2 reconstructions. We attribute these features, which emanate from the periphery of the IC trimers, to ordered segments of the linker between the IC domain and the E1-binding domain. This linker region was visualized in the cryo-EM reconstruction of the intact PDC purified from bovine kidney tissues (Figure 2E). The extra feature on each IC domain seen in Figures 2C and 2D represents only a partially ordered portion of the linker. The flexible portion of the linker (Figure 2C) was further stabilized by binding E1 (Zhou et al., 2001b). In addition to the linker densities emanating from the edge of the E2 IC trimer that are only visible in the full-length E2 structure, there are some minor differences in the outer surface of the trimer of the tE2 and E2 structures. Interference from the flexibly positioned outer domains of E2 may account for the more convoluted appearance of the surface of the E2 inner core (Figure 2B).
Table 1.
Summary of Data Processing of the Human E2 and tE2 Cores
| tE2 |
E2 |
|||
|---|---|---|---|---|
| Far-from-Focus | Close-to-Focus | Far-from-Focus | Close-to-Focus | |
| CCD frames used | 197 | 197 | 100 | 100 |
| Boxed-out particles | 13,199 | 13,002 | 5,169 | 5,054 |
| Particles processed | 9,167 | 2,432 | 4,080 | 1,391 |
| Defocus range (μm) | 2.7–4.2 | 0.6–2.1 | 3.0–4.1 | 0.9–2.0 |
| B factor range (Å2) | NAa | 138–235 | NAa | 154–240 |
| Resolution (Å) | NAa | 8.8 | NAa | 15 |
Not available.
Figure 2. 3D Structures of Human PDC Cores.
(A and B) Outer view of a shaded surface representation of the (A) human tE2 and (B) full-length E2 cores, reconstructed to 15 Å resolution. Maps are color coded according to particle radius. The E2 core contains additional features (arrowheads) that are not present in the tE2 structure.
(C) Close-up view of the region surrounding a trimer shown at a lower-density threshold.
(D) Central slice through the reconstruction in (B), shown as a gray-level display. Arrowheads in (C) and (D) indicate the same features as in (B).
(E) A 25 Å resolution reconstruction of intact PDC isolated from bovine tissues. The linker densities emanate from the E2 core, at the same locations as revealed in the human E2 core structure (see [B]–[D]). The linker extensions are more clearly visible in the bovine PDC structure than in the human E2 core structure because the extensions are likely stabilized by binding of E1 (Zhou et al., 2001a).
E2 and tE2 can vary in size and conformation, which may limit the achievable resolution of the reconstruction (Zhou et al., 2001a; Kong et al., 2003; Borgnia et al., 2004). The dynamic variation in the structure of the tE2 has been documented (Zhou et al., 2001a; Kong et al., 2003); human tE2 and the inner core of full-length E2 exhibit a similar flexibility and variation in particle diameter. To obtain higher resolution, our final structures of tE2 and E2 were reconstructed only from those particles within a 2% size difference of the most probable size group, as classified by an iterative size classification program, sizeDiff (Zhou et al., 2001a). As expected, the resolutions of both the tE2 and E2 structures improved when the amount of size variations among the particles included in the 3D reconstruction decreased. In addition, extra densities were resolved as definite features in the E2 structure (discussed below) when only particles with inner cores within the 0.99–1.01 size group were used.
Human tE2 Structure at Subnanometer Resolution
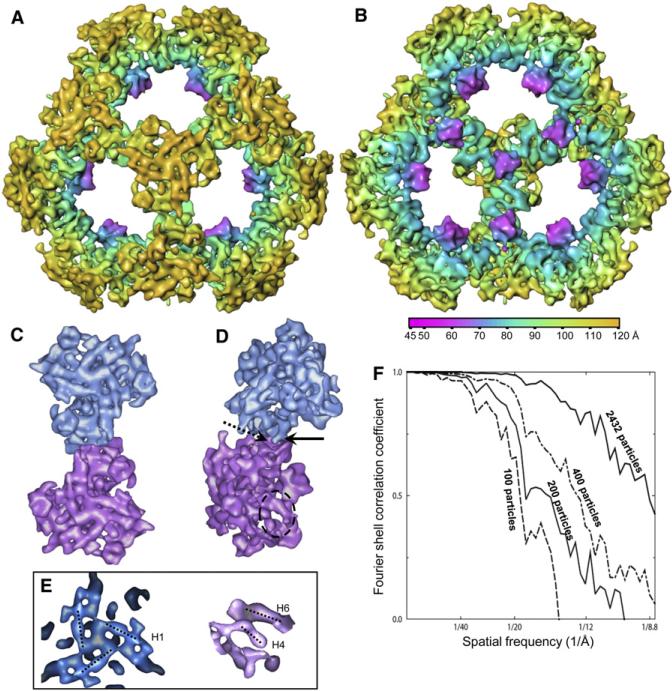
Because positions of the lipoyl domains and linker regions are variable within the full-length E2 structure, we focused on the tE2 core for our higher-resolution analyses (Figure 3; Movie S1, see the Supplemental Data available with this article online). The effective resolution of our 3D structure increased as the number of particles in our data set increased (Figure 3F), and it reached 8.8 Å for the final data set of 2432 particles. At this resolution, secondary structure elements of the core, including α helices (cylindrical rods in Figure 3A) and β sheets (globular shapes in Figure 3B), are clearly visible. Seven α helices could be identified in each tE2 subunit; three of these (H1, H4, and H6) are indicated by dotted lines in Figure 3E. Closer examination of the individual tE2 trimers revealed points at which interactions between adjacent trimers could be seen (Figures 3C and 3D). In contrast to the intensive, interdigitated associations among the three monomers within each trimer, the contact between adjacent trimers is very limited. A short, rod-like structure (arrow in Figure 3D), which is connected to the C-terminal end of helix H6 by a turn, interacts with its neighboring two-fold-axis-related trimer (areas of interaction are indicated by arrows).
Figure 3. 3D Reconstruction of Human tE2 at 8.8 Å Resolution.
(A and B) Shaded, radially colored surface representation of (A) outside and (B) inside views of the human tE2 core along a three-fold axis. Secondary structure elements (cylindrical helices and globular β sheets) are discernable.
(C and D) (C) Top and (D) side views of two adjacent tE2 trimers extracted from the 3D reconstruction of the tE2 core, viewed along a two-fold axis. The solid and dashed arrows in (D) indicate the “ball” and “socket,” respectively, of the characteristic interaction between two adjacent trimers. The dashed oval marks a region with two helices, which is blown up in the right panel of (E).
(E) Close-ups of the 3D map of human tE2, showing some rod-shaped helix densities. Left: top view of three H1 helices surrounding a three-fold axis. Right: side view of the circled area in (D) showing H4 and H6 helices (see helix numbering and details in Figures 4 and 6).
(F) Fourier shell correlation coefficient showing improvement in resolution as the number of particles increases.
Cryo-EM-Constrained Homology Model of Human tE2
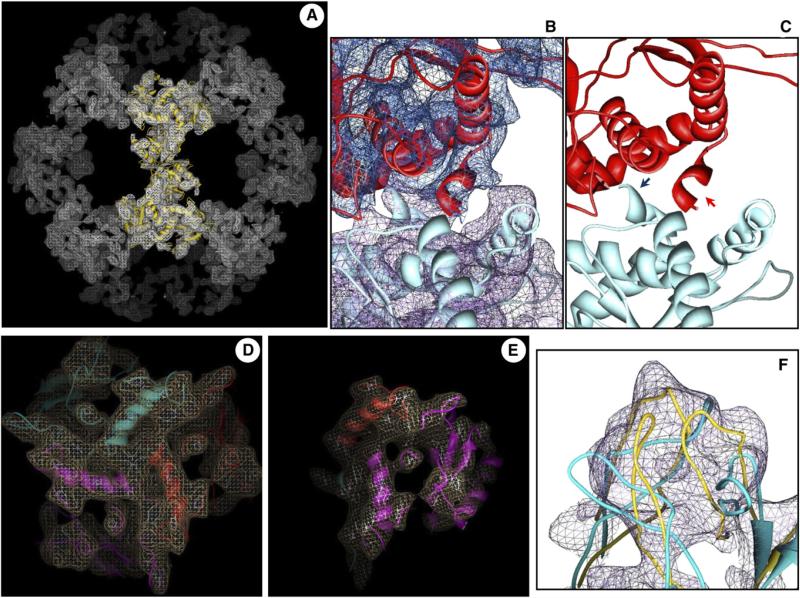
The IC domains of human E2 and prokaryotic E2 (A. vinelandii and B. stearothermophilus) share ~32% of their amino acid sequences (Figure 4). We used the SWISS-MODEL protein modeling server to generate a homology model of human tE2 from the known 2.6 Å resolution tE2 structure of A. vinelandii (Mattevi et al., 1992). The homology model was then refined by using the 8 Å cryo-EM 3D map. Without this refinement, the homology model (e.g., blue ribbon in Figure 5F) contained errors in the positioning of the secondary structure elements. The disposition or fold of the secondary structure elements in our final energy minimized model of the human tE2 core (e.g., ribbons in Figures 5A–5E and gold ribbon in Figure 5F) is more accurately determined, as indicated by its excellent agreement with experimentally resolved helices and β sheets in the cryo-EM map.
Figure 4. Sequence Homology between Human and Bacterial E2s.
Alignment of human, Bacillus stearothermophilus (B. ste.), and A. vinelandii (A. vin.) tE2 sequences. Identical residues are bold and capitalized; similar residues are capitalized only. Helices (H) are indicated by shading; β sheets (S) are indicated by dotted boxes. The prokaryotic tE2s have two short β strands between H3 and S2 that are not present in human tE2, and two other β strands (S6 and S7) are shorter in human than in prokaryotic tE2.
Figure 5. Cryo-EM Density-Restrained Atomic Modeling of Human tE2.
(A) Wire-frame representation of a high-resolution cryo-EM density map from Figure 3, superimposed with our cryo-EM-derived pseudo atomic model (gold ribbon) (also see Movie S1).
(B and C) Close-up views of two neighboring tE2 monomers (in different colors), shown either (B) with or (C) without the cryo-EM density superimposed. Arrows in (C) indicate the C-terminal helix H7 that is involved in the interaction between two E2 monomers.
(D and E) Close-up views of two representative regions of a single E2 trimer, showing agreement of the disposition of secondary structure elements between the cryo-EM densities (wire-frame) and the pseudo atomic model (ribbon). Different colors represent structure elements from different tE2 monomers.
(F) Close-up view of the internal hairpin loop region of tE2, showing the cryo-EM density (wire-frame), the unrefined homology model (blue ribbon), and the refined, cryo-EM-restrained model (gold ribbon).
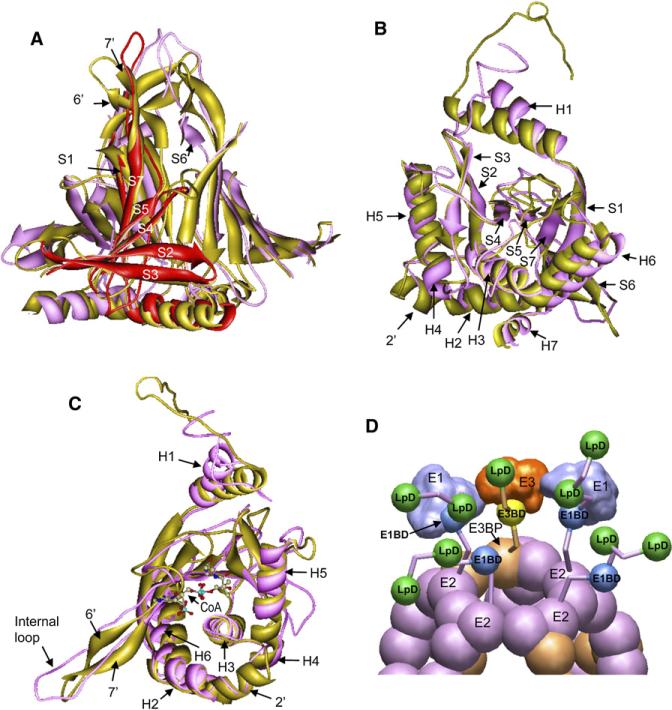
A detailed comparison of our pseudo atomic model of human tE2 with the atomic model of A. vinelandii tE2 is shown in Figures 6A–6C (also see Movie S2). As expected, the structures at the active sites are highly conserved, and there are superimposable helices and β sheets at or near the active sites. For example, the secondary structure elements form the active sites in A. vinelandii tE2 (Mattevi et al., 1992), in which CoA is shown by stick-and-ball models, and are fully conserved in the human tE2 structure (Figure 6C). Similar to A. vinelandii tE2, the active-site pocket in human tE2 is formed by participation of α helices H3–H5 and β sheets composed of strands S2–S3 (Figures 6A–6C). Ser480 and His534 from the neighboring tE2 domain are conserved residues that are responsible for acetyltransferase activity. The secondary structure elements of the lipoyl group-binding site are also preserved. An internal β barrel formed by joining β strands from trimer subunits (Figure 6A) is almost identical to its A. vinelandii tE2 counterpart, suggesting that the key building block of tE2 is preserved from prokaryotes to humans. In addition, the “spring” and “ball” described in the “ball-and-socket” model (Zhou et al., 2001a) are also similar in human and prokaryotic tE2. This structure, which connects each IC domain of a trimer to the IC domain in its neighboring trimer along the two-fold axis, is collectively formed by C-terminal helices H6 and H7 in human tE2. The C-terminal hydrophobic “ball” (Leu561-Leu560) joins a 310 helix, helix H7, next to a Pro-containing turn that functions as an elbow in connecting to helix H6. This entire segment is proposed to act as a cantilever with “spring-like” flexibility that is facilitated by the rotation of the “ball” sheathed in a hydrophobic “socket” that is provided by the neighboring two-fold-symmetric subunit (Zhou et al., 2001a).
Figure 6. Comparison of Human and Prokaryotic tE2 Structures and Mechanistic Model of Human PDC Function.
(A–C) The human (purple and red) and A. vinelandii (gold) (Mattevi et al., 1992) tE2 structures are superimposed and are shown in three approximately orthogonal views. The helices (H) and β strands (S) are numbered consecutively from the N to C termini. (A) The highly conserved β barrel region of the tE2 trimers. For clarity, only the internal β barrel center region of the trimer is shown, and this region consists of β strands and a flanking helix from each of the three tE2 subunits. One subunit of human tE2 is colored red. (B and C) Orthogonal views of the superposition of the human (purple) and A. vinelandii (gold) tE2 monomer showing the most significant structural differences located in the N-terminal loop and its connected helix (H1), and the internal hairpin loop. In (C), coenzyme A (CoA) is shown as a ball-and-stick model to highlight the location of the conserved active sites in the two models (Mattevi et al., 1992).
(D) Proposed model of human PDC structural organization and mechanism for catalysis and regulation (see details in Movie S3). Six E3BP dimmers (brown) substitute for 12 E2 subunits (purple) to form the E2•E3BP core as described (Hiromasa et al., 2004). Around each pentagonal opening, one E3 and two E1 molecules interact to form a shell connected to the E2•E3BP core through 50 Å linkers. The catalytic intermediates and regulatory enzymes are transferred through the “swinging arm” and “hand-over-hand” walking mechanisms. Abbreviations: E1BD, E1-binding domain; E3BD, E3-binding domain; LpD, lipoyl domain.
There are two major regions of difference between the human and prokaryotic tE2 structures (Figures 6B and 6C). First, there is a difference in the orientation of the associated H1 helix at the N terminus of the tE2 molecule. In the A. vinelandii tE2 structure, the extended N-terminal loop projecting from the H1 helix folds back to the middle of the trimer. Instead, the 14 residue H1 helix in human tE2 (residues 343–356) rotates clockwise away from the five-fold axis by ~20°, which places H1 closer to its neighboring subunit than the 16 residue H1 helix in A. vinelandii tE2 (residues 417–432) (Figures 5D, 5E, 6B, and 6C). The marked difference in the N-terminal helix is important for forming a firm base for the departure of the linker region from this structure, which is observed with full-sized E2 (see Discussion).
The other major difference concerns the internal domain extending into the central space of the dodecahedron. In A. vinelandii, this domain exists as a two-stranded β sheet (Figures 6A, and 6C, labeled as 6′ and 7′) with a 2 residue turn at its tip, whereas, in human tE2, it stretches farther as an extended loop (Figures 5F and 6C). Inside the tE2 trimer, each tE2 subunit has a cascading set of β strands aligned to form one-third of an inner β sheet barrel (Figure 6A). The human tE2 end loop or its corresponding A. vinelandii tE2 end β sheet forms the internal extension of this β sheet barrel. In the prokaryotic tE2 trimer structure, the β hairpin bends away from the central axis of the extension tip. In contrast, the longer human tE2 loops align straight down the axis of the extension tip with an inward bend (Figures 4, 6A, and 6C), forming a more compact and more extended inner tip than the prokaryotic tE2 structure.
Aside from these major differences, there are some minor structural differences worth noting. For example, a kink divides helix H2 into two parts in A. vinelandii tE2, whereas, in the human tE2 structure, the C-terminal portion of helix H2 exists as a coil (labeled as 2′ in Figure 6B). Similarly, the long helix H4 in A. vinelandii tE2 corresponds to two shorter helices joined by a kink in the human tE2 structure (labeled as H4 and H5).
DISCUSSION
Cryo-EM-Constrained Atomic Modeling
Cryo-EM single-particle reconstruction has advanced in recent years such that density maps can be resolved beyond the subnanometer resolution level, thereby revealing the topological arrangement, or the folds, of secondary structure elements within protein subunits (e.g., Böttcher et al., 1997; Zhou et al., 2000, 2003; Cheng et al., 2004; Ludtke et al., 2004). Although these structures represent a significant technological advancement in cryo-EM, their value was significantly reduced in the absence of atomic coordinate information of their structural elements, such as an atomic model. Cryo-EM images can now be recorded near atomic resolution by using state-of-the-art instruments, but 3D single-particle reconstructions at atomic resolution are still lacking, largely because of the exponential increase in the amount of image data that need to be processed accurately for higher-resolution structure determination. In the favorable situation in which the structures of homologous or at least remotely homologous molecules have been determined to atomic resolution by the complementary techniques of NMR and X-ray crystallography, as is the case in this study, a pseudo atomic model can be built by integrating the subnanometer resolution cryo-EM density map with the results from widely available comparative modeling techniques (Topf et al., 2006). This approach represents an attractive alternative to de novo atomic modeling that would require ~3 Å resolution structure determination, which remains technically challenging via cryo-EM single-particle reconstruction today.
Comparison of the Human, Yeast, and Prokaryotic PDC Structures
We have used cryo-EM to obtain structures of the human PDC E2 and tE2 cores. To our knowledge, this represents the first study of any full-length E2 in which a segment of 3D structure has been resolved beyond the inner core. Key findings in this study are that the active sites within the IC domain are highly conserved between the human and prokaryotic tE2 molecules, but that other exterior regions have very different structures (Figures 6A–6C), which have relevant functional implications.
There are two structural requirements for the IC domain and its adjacent linker region of mammalian E2 that do not need to be met in prokaryotic and yeast E2. The first is binding of the IC domain of E3BP to the IC domain of E2. Small-angle X-ray scattering and analytical ultracentrifuge studies find that the E2 60-mer is larger than E2•E3BP, and that the E2 structure binds more E1 (Hiromasa et al., 2004). This seems to require the E2-related IC domain of E3BP to be integrated into the inner core. In yeast PDC the E3BP molecule has been proposed to bind near the internal tips of the cone-shaped trimers within the pentagonal openings of the E2 core (Stoops et al., 1997). Consistent with the observation in yeast PDC, the internal hairpin β strands that form the inner tips of prokaryotic tE2 become extended loops in the human molecule, which is probably related to the requirement for human E2 to bind to the C-terminal domain of E3BP (Rahmatullah et al., 1989b). In fact, the C-terminal domains of human E2 and E3BP are 49% similar, which is in marked contrast to the dissimilarities in the IC domain sequences of yeast E2 and E3BP. As we mentioned above, human E3BP is believed to integrate into the E2 core by replacing some E2 subunits during the E2•E3BP assembly process. Therefore, it seems that the internal structural difference between human, yeast, and prokaryotic E2s may relate to specific structural requirements involved in E2•E3BP assembly.
The second unique feature not required in prokaryotic E2 is the need to support PDK and PDP in acting efficiently on their E1 substrate. In humans, the enzymatic function of E2 is highly regulated by the control of four PDK isoforms and two PDP isoforms (Huang et al., 1998; Roche et al., 2001, 2003). Decreasing the activity of PDC by phosphorylation is a critical step in preventing carbohydrate depletion. In fact, inhibiting the activity of specific PDK isoforms is an active drug target with therapeutic potential for treating diabetes, heart ischemia, and cancer (Roche and Hiromasa, 2007). Based on E2 and E2•E3BP maximally binding 60 and 48 E1, respectively (Hiromasa et al., 2004), it would appear that two β subunits of the E1-α2β2 tetramer bind one E1-binding domain of E2 (Figure 1A), as observed with related E1-α2β2 (Aevarsson et al., 1999, 2000). The cluster of three phosphorylation sites of E1 are at the active-site channel in a sunken interface formed between the α and β subunits (Ciszak et al., 2001). PDK and PDP bind to the L2 domain (Figure 1A) (Liu et al., 1995; Chen et al., 1996; Roche et al., 2003) that is connected by a 30 amino acid linker region to the E1-binding domain. With this organization, PDK and PDP need to be able to move into the space under the large α subunits to access the serine residues that undergo phosphorylation and dephosphorylation. This suggests that the linker regions connecting IC to the E1-binding domain in mammalian E2 must maintain substantial separation. Particularly important would be the stiffness at the beginning of the linker region at the IC domain upon E1 binding. Indeed, our findings uncover structural differences in human E2 that are consistent with meeting these unique requirements.
The most striking difference between our human structure and the X-ray structure of prokaryotic tE2 is located near the region connected to the linkers. In particular, the N-terminal H1 helices have different orientations (Figure 6). Interestingly, a recently determined atomic structure of the dihydrolipoamide acyltransferase (E2b) component of the mammalian branched-chain α-ketoacid dehydrogenase complex, which has a cubic architecture, also shows a disposition of the H1 helix similar to that revealed in our human tE2 structure (Kato et al., 2006). The N-terminal segment appears to be forming a foundation for the stable beginning of the linker segment exterior to the tE2 core. It seems likely that this is needed to support the stiff linker extension arising from just above this location in E2. We propose that this stable base/stiff linker region supports an arrangement of the bound E1s that is more conducive to the regulation of the E1 activity by the kinases and phosphatases. The E2-bound E1s form an outer shell in the PDC ~50 Å above the core (Figure 2) (Zhou et al., 2001b). The E1-binding domain forms the pivot about which the swinging linker arms holding the L1 and L2 lipoyl domains move. The first and continuous role is for the lipoyl domains to shuttle the intermediates of catalysis between the three catalytic centers, which are equally disposed and located ~50 Å from the pivot (Zhou et al., 2001b) (Figure 6D). In vivo, there are 20–30 E1 molecules bound to each core of the mammalian PDC, and our cryo-electron tomography studies show that they self associate to form patch-like E1 clusters (not shown). The active-site region of E1 is in a sunken cleft between E1's α and β subunits; this is also the region that is phosphorylated (Ciszak et al., 2003). The second role of the lipoyl domains, particularly the L2 domain, is to shuttle PDK and PDP1 to this active-site region to carry out the kinase and phosphatase reactions. This clearly requires extensive space for the L2-bound 92 kDa kinase dimers and 150 kDa PDP1 to access the E1 active site region located below the larger and more exterior α subunts, while E1 is held by the E1-binding domain of E2 via its interior β subunits at the most interior position (Rahmatullah et al., 1989a, 1990; Zhou et al., 2001b).
Proposed Model of Human PDC Functions and Regulation
Based on our observation and previous data regarding the E2•E3BP complex (Hiromasa et al., 2004), we proposed a working model that accounts for the catalytic and unique regulatory mechanism of mammalian PDC (Figure 6D; also see Movie S3). In this model, there are five flexible E2 or E3BP N termini (i.e., one from E3BP and four from E2 subunits) organized around each five-fold opening of the core. Among them, one E3BP and two of the four E2 subunits are likely to be bound by E3 and E1 molecules, respectively. The E3 molecule interacts with E1s to form the outer shell, and these E1s or E3s are connected to the IC domains of the E2 or E3BP through the internal linkers. Although some lipoyl domains shuttle the catalytic intermediates between active sites by way of the “swinging arm” mechanism with the E1- or E3-binding domain as a pivot (Zhou et al., 2001b), the lipoyl domains in the free E2 molecules may rotate about multiple pivots, including the one near the edge of the E2 core extended to the linker that connects the E1-binding domain. We believe that the latter mode is the key to the efficient delivery of the regulatory enzymes PDK and PDP (shown as red balls in the Movie S3) around each PDC (Turkan et al., 2004). A PDK dimer (1–2 copies per PDC) is proposed to be transferred by a “hand-over-hand” mechanism between different lipoyl domains by alternately holding on to one or two lipoyl domains (Liu et al., 1995; Hiromasa and Roche, 2003; Roche et al., 2003). This nondissociative movement would be aided by the linker-region-supported “swinging arm” mechanism by either using the E1-binding domain as a pivot (transfer within the pentagonal opening) or by using the multiple pivots of the non-E1-bound/unconstrained N terminus of E2 (transfer between different pentagonal openings) as described above.
EXPERIMENTAL PROCEDURES
Enzyme Preparation and Functional Assay
Human E2 and tE2 samples were prepared as described previously (see Supplemental Data in Hiromasa et al., 2004). Briefly, E2 was expressed in E. coli at 37°C to mid-log phase and was purified at 4°C. Harvested cells were disrupted by ultrasonication, and particulate material was removed by centrifugation at 10,000 × g for 20 min. PEG 8000 (50% wt/vol) was added dropwise to a final concentration of 8% (vol/vol), and the precipitated protein was removed by centrifugation. An additional 8% volume of PEG was added, and the precipitate was collected by centrifugation. The 8%–16% PEG precipitate was dissolved in 15 ml 50 mM potassium phosphate buffer (pH 7.2) containing 0.5 mM EDTA, 0.2 M ammonium chloride, 1 mM β-mecaptoethanol, and 0.1% Pluronics F68. This fraction was treated with DNase I and was run through a Sephacryl S-400 HR column (3.0 × 120 cm). Fractions from the column were assayed for E2 activity and analyzed by SDS-PAGE; those containing high E2 activity were combined. E2 was pelleted by ultracentrifugation of the supernatant in a Beckman type 50.1 Ti angle rotor at 35,000 rpm for 4 hr at 4°C. The resuspended pellets were further clarified by centrifugation, and the resulting supernatant was subsequently applied to a Sephacryl S-400 HR column (2.0 × 105 cm). Fractions (1.1 ml/tube) were monitored according to their absorbance at 260 and 280 nm, their E2 activity, as well as their SDS-PAGE profile.
Human tE2 was prepared from scE2, which contains a PreScission site in the third linker region (see Supplemental Data in Hiromasa et al., 2004). scE2 was purified as described above. Treatment of scE2 with the PreScission protease (Amersham Biosciences) removed the N-terminal 319 amino acids. The resulting tE2 was purified by gel filtration with a Sephacryl S-300 HR column. The assembly of the recombinant molecules into fully functional, pentagonal, dodecahedral cores was confirmed by a previously described method and by analytical ultracentrifugation (Yang et al., 1997; Hiromasa et al., 2004).
Cryo-EM
Cryo-EM of the human E2 and tE2 cores was carried out by following standard procedures. Briefly, a 3 μl purified sample in a solution containing 0.8 mg/ml E2 or 0.2 mg/ml tE2 was applied to Quantifoil grids with 2 μm diameter holes (Quantifoil Micro Tools GmbH, Germany) at 4°C. The grids were blotted with filter paper and quickly plunged into liquid-nitrogen-cooled ethane so that the E2 or tE2 molecules were suspended in vitreous ice. Focal-pair image frames were acquired by using a JEOL 2010F electron microscope with a field emission gun operated at 200 kV. This microscope was equipped with a Gatan Model 626 cryostage and a US4000 4k × 4k CCD camera. Digital cryo-EM images were recorded at an effective magnification of 69,250× on the CCD, and this value corresponds to an effective pixel size of 2.17 Å/pixel and a specimen dosage of ~12 electrons/Å2/micrograph. Focal-pair CCD images were recorded and preprocessed for translation and rotation correction by using the JAMES semiautomatic data collection program (Booth et al., 2004). The electron beam was underfocused, and there was a 2 μm difference between the close-to-focus (~1 μm under focus) and far-from-focus (~3 μm under focus) images. The actual defocus value of each micrograph was determined by using incoherently averaged Fourier transforms of selected particle images (Zhou et al., 1996) with the ctfit program (Ludtke et al., 1999).
Computer Image Processing and Reconstruction
Data processing statistics for the 3D reconstructions are summarized in Table 1. Orientation determination and 3D reconstruction were performed by using the IMIRS software package (Liang et al., 2002; Zhou and Chiu, 2003) on multiprocessor MS Windows XP computer workstations from Dell. The orientations were first estimated from the particle images in the far-from-focus micro-graphs and were refined to ~30 Å resolution. These orientation parameters were then further refined by using the particles in the close-to-focus micro-graphs. During each step, the particles were classified according to size to eliminate those particles of larger or smaller diameters. At first, the “average” reconstructions from all particles were used as templates. Each particle image was then classified into different size groups by intervals of 1%. The 0.99–1.01 size group particles were used for the new reconstructions, which were refined and used as templates for the next round of size classifications. The above-described process was repeated until all of the particles fell within the 0.99–1.01 size group. The correction of contrast transfer function was performed as described previously (Zhou and Chiu, 2003), except that varied B factors were used in the current studies (Table 1). The resolution of the reconstructions was calculated by using the criterion of Fourier shell correlation coefficient being greater than 0.5.
Homology Modeling and Cryo-EM Structure-Restrained Refinement
We used the CLUSTALW program (http://www.ebi.ac.uk/clustalw/) to determine the optimal sequence alignment between human tE2, A. vinelandii tE2 (Mattevi et al., 1992), and B. stearothermophilus tE2 (Izard et al., 1999). We submitted this alignment, as well as the atomic structure of A. vinelandii tE2 (PDB code: 1EAA), to the SWISS-MODEL automated protein modeling server (http://www.expasy.org/swissmod/) (Guex and Peitsch, 1997) to obtain a homology model of human tE2. Although the structures of B. stearothermophilus and E. faecalis tE2 cores are also available, we did not use either of them as a template because they are unrefined models based on low-resolution (4.4 Å) diffraction data and their backbones are superimposable with that of the A. vinelandii tE2 structure (Izard et al., 1999).
The preliminary homology model of human tE2 was then aligned by using the constraints derived from our 8 Å cryo-EM density map by maximizing the agreement of secondary structure elements, including helices, β sheets, and loops, by using the Chimera package (Pettersen et al., 2004). Briefly, we first adjusted helices in the homology model by using the interactive segment adjustment tools in Chimera to better fit the cryo-EM density based on visual inspection. The criterion is to enclose as many secondary structure elements (helices or β sheets) as possible in the envelope of our density map. The programs Foldhunter (Jiang et al., 2001) and Situs (Wriggers et al., 1999) were also used to validate the best fit of the enclosed secondary structure elements. This model is subjected to energy minimization with the SWISS-MODEL server, leading to a new and improved trial model. The above-described steps are reiterated until the root-mean-square deviation (rmsd) of two consecutive trial models is less than 2.5 Å and there is no further improvement in the correlation between the cryo-EM density map and the model.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported in part by American Heart Association grant 0240216N (to Z.H.Z.), and by National Institutes of Health (NIH) grants GM071940 (to Z.H.Z.) and DK18320 (to T.E.R.), as well as by Kansas Agriculture Experiment Station Contribution 05-139-J (to T.E.R.). H.T. was supported in part by the Gulf Coast Consortia of the Keck Center for Computational and Structural Biology. We acknowledge the use of the cryo-EM facility at the National Center for Macromolecular Imaging (supported by NIH grant P41RR02250 to Wah Chiu). We thank Lester Reed for helpful discussions and Pierrette Lo for editorial assistance.
Footnotes
Accession Numbers
The cryo-EM density maps and atomic coordinates have been deposited in the Protein Data Bank and the European Bioinformatics Institute under accession codes 3B8K and EMD-1448, respectively.
Supplemental Data
Supplemental Data include three movies illustrating the human tE2 core structure and a mechanistic model of human PDC catalysis and regulation, and are available at http://www.structure.org/cgi/content/full/16/1/104/DC1/.
REFERENCES
- Aevarsson A, Seger K, Turley S, Sokatch JR, Hol WG. Crystal structure of 2-oxoisovalerate dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes. Nat. Struct. Biol. 1999;6:785–792. doi: 10.1038/11563. [DOI] [PubMed] [Google Scholar]
- Aevarsson A, Chuang JL, Wynn RM, Turley S, Chuang DT, Hol WG. Crystal structure of human branched-chain α-ketoacid dehydrogenase and the molecular basis of multienzyme complex deficiency in maple syrup urine disease. Structure. 2000;8:277–291. doi: 10.1016/s0969-2126(00)00105-2. [DOI] [PubMed] [Google Scholar]
- Booth CR, Jiang W, Baker ML, Zhou ZH, Ludtke SJ, Chiu W. A 9 Å single particle reconstruction from CCD captured images on a 200 kV electron cryomicroscope. J. Struct. Biol. 2004;147:116–127. doi: 10.1016/j.jsb.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Borgnia MJ, Shi D, Zhang P, Milne JL. Visualization of α-helical features in a density map constructed using 9 molecular images of the 1.8 MDa icosahedral core of pyruvate dehydrogenase. J. Struct. Biol. 2004;147:136–145. doi: 10.1016/j.jsb.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- Chen G, Wang L, Liu S, Chuang C, Roche TE. Activated function of the pyruvate dehydrogenase phosphatase through Ca2+-facilitated binding to the inner lipoyl domain of the dihydrolipoyl acetyltransferase. J. Biol. Chem. 1996;271:28064–28070. doi: 10.1074/jbc.271.45.28064. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- Ciszak E, Korotchkina LG, Hong YS, Joachimiak A, Patel MS. Crystallization and initial X-ray diffraction analysis of human pyruvate dehydrogenase. Acta Crystallogr. D Biol. Crystallogr. 2001;57:465–468. doi: 10.1107/s0907444901000427. [DOI] [PubMed] [Google Scholar]
- Ciszak EM, Korotchkina LG, Dominiak PM, Sidhu S, Patel MS. Structural basis for flip-flop action of thiamin pyrophosphate-dependent enzymes revealed by human pyruvate dehydrogenase. J. Biol. Chem. 2003;278:21240–21246. doi: 10.1074/jbc.M300339200. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Harris RA, Bowker-Kinley MM, Wu P, Jeng J, Popov KM. Dihydrolipoamide dehydrogenase-binding protein of the human pyruvate dehydrogenase complex. DNA-derived amino acid sequence, expression, and reconstitution of the pyruvate dehydrogenase complex. J. Biol. Chem. 1997;272:19746–19751. doi: 10.1074/jbc.272.32.19746. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Roche TE. Facilitated interaction between the pyruvate dehydrogenase kinase isoform 2 and the dihydrolipoyl acetyltransferase. J. Biol. Chem. 2003;278:33681–33693. doi: 10.1074/jbc.M212733200. [DOI] [PubMed] [Google Scholar]
- Hiromasa Y, Fujisawa T, Aso Y, Roche TE. Organization of the cores of the mammalian pyruvate dehydrogenase complex formed by E2 and E2 plus the E3-binding protein and their capacities to bind the E1 and E3 components. J. Biol. Chem. 2004;279:6921–6933. doi: 10.1074/jbc.M308172200. [DOI] [PubMed] [Google Scholar]
- Huang B, Gudi R, Wu P, Harris RA, Hamilton J, Popov KM. Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J. Biol. Chem. 1998;273:17680–17688. doi: 10.1074/jbc.273.28.17680. [DOI] [PubMed] [Google Scholar]
- Izard T, Aevarsson A, Allen MD, Westphal AH, Perham RN, de Kok A, Hol WG. Principles of quasi-equivalence and Euclidean geometry govern the assembly of cubic and dodecahedral cores of pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA. 1999;96:1240–1245. doi: 10.1073/pnas.96.4.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Baker ML, Ludtke SJ, Chiu W. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 2001;308:1033–1044. doi: 10.1006/jmbi.2001.4633. [DOI] [PubMed] [Google Scholar]
- Kalia YN, Brocklehurst SM, Hipps DS, Appella E, Sakaguchi K, Perham RN. The high-resolution structure of the peripheral subunit-binding domain of dihydrolipoamide acetyltransferase from the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. J. Mol. Biol. 1993;230:323–341. doi: 10.1006/jmbi.1993.1145. [DOI] [PubMed] [Google Scholar]
- Kato M, Wynn RM, Chuang JL, Brautigam CA, Custorio M, Chuang DT. A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain α-ketoacid dehydrogenase complex. EMBO J. 2006;25:5983–5994. doi: 10.1038/sj.emboj.7601444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Li J, Chuang JL, Chuang DT. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Ming D, Wu Y, Stoops JK, Zhou ZH, Ma J. Conformational flexibility of pyruvate dehydrogenase complexes: a computational analysis by quantized elastic deformational model. J. Mol. Biol. 2003;330:129–135. doi: 10.1016/s0022-2836(03)00555-2. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ke EY, Zhou ZH. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J. Struct. Biol. 2002;137:292–304. doi: 10.1016/s1047-8477(02)00014-x. [DOI] [PubMed] [Google Scholar]
- Liu S, Baker JC, Roche TE. Binding of the pyruvate dehydrogenase kinase to recombinant constructs containing the inner lipoyl domain of the dihydrolipoyl acetyltransferase component. J. Biol. Chem. 1995;270:793–800. doi: 10.1074/jbc.270.2.793. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semi-automated software for high resolution single particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 Å resolution by single particle electron cryomicroscopy. Structure. 2004;12:1129–1136. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Mande SS, Sarfaty S, Allen MD, Perham RN, Hol WG. Protein-protein interactions in the pyruvate dehydrogenase multienzyme complex: dihydrolipoamide dehydrogenase complexed with the binding domain of dihydrolipoamide acetyltransferase. Structure. 1996;4:277–286. doi: 10.1016/s0969-2126(96)00032-9. [DOI] [PubMed] [Google Scholar]
- Mattevi A, Obmolova G, Schulze E, Kalk KH, Westphal AH, de Kok A, Hol WG. Atomic structure of the cubic core of the pyruvate dehydrogenase multienzyme complex. Science. 1992;255:1544–1550. doi: 10.1126/science.1549782. [DOI] [PubMed] [Google Scholar]
- Milne JL, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu X, Brooks BR, Perham RN, Henderson R, Subramaniam S. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rahmatullah M, Gopalakrishnan S, Andrews PC, Chang CL, Radke GA, Roche TE. Subunit associations in the mammalian pyruvate dehydrogenase complex. Structure and role of protein X and the pyruvate dehydrogenase component binding domain of the dihydrolipoyl transacetylase component. J. Biol. Chem. 1989a;264:2221–2227. [PubMed] [Google Scholar]
- Rahmatullah M, Gopalakrishnan S, Radke GA, Roche TE. Domain structures of the dihydrolipoyl transacetylase and the protein X com ponents of mammalian pyruvate dehydrogenase complex. Selective cleavage by protease Arg C. J. Biol. Chem. 1989b;264:1245–1251. [PubMed] [Google Scholar]
- Rahmatullah M, Radke GA, Andrews PC, Roche TE. Changes in the core of the mammalian-pyruvate dehydrogenase complex upon selective removal of the lipoyl domain from the transacetylase component but not from the protein X component. J. Biol. Chem. 1990;265:14512–14517. [PubMed] [Google Scholar]
- Randle PJ. Fuel selection in animals. Biochem. Soc. Trans. 1986;14:799–806. doi: 10.1042/bst0140799. [DOI] [PubMed] [Google Scholar]
- Randle PJ, Priestman DA. Shorter term and longer term regulation of pyruvate dehydrogenase kinases. In: Patel MS, Roche TE, Harris RA, editors. α-Keto Acid Dehydrogenase Complexes. Birkhauser Verlag; Basel, Switzerland: 1996. pp. 151–161. [Google Scholar]
- Reed LJ. Multienzyme complexes. Acc. Chem. Res. 1974;7:40–46. [Google Scholar]
- Reed LJ, Hackert ML. Structure-function relationships in dihydrolipoamide acyltransferases. J. Biol. Chem. 1990;265:8971–8974. [PubMed] [Google Scholar]
- Roche TE, Cox DJ. Multifunctional 2-Oxo acid dehydrogenase complexes. In: Agius L, Sherratt HSA, editors. Channeling in Intermediary Metabolism. Portland Press Ltd.; London: 1996. pp. 115–132. [Google Scholar]
- Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell. Mol. Life Sci. 2007;64:830–849. doi: 10.1007/s00018-007-6380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche TE, Baker JC, Yan X, Hiromasa Y, Gong X, Peng T, Dong J, Turkan A, Kasten SA. Distinct regulatory properties of pyruvate dehydrogenase kinase and phosphatase isoforms. Prog. Nucleic Acid Res. Mol. Biol. 2001;70:33–75. doi: 10.1016/s0079-6603(01)70013-x. [DOI] [PubMed] [Google Scholar]
- Roche TE, Hiromasa Y, Turkan A, Gong X, Peng T, Yan X, Kasten SA, Bao H, Dong J. Central organization of mamalian pyruvate dehydrogenase (PD) complex and lipoyl domain-mediated function and control of PD kinases and phosphatase. In: Jordan F, Patel MS, editors. Thiamine: Catalytic Mechanisms and Role in Normal and Disease States. Marcel Decker; New York: 2003. pp. 363–386. [Google Scholar]
- Stoops JK, Baker TS, Schroeter JP, Kolodziej SJ, Niu XD, Reed LJ. Three-dimensional structure of the truncated core of the Saccharomyces cerevisiae pyruvate dehydrogenase complex determined from negative stain and cryoelectron microscopy images. J. Biol. Chem. 1992;267:24769–24775. [PMC free article] [PubMed] [Google Scholar]
- Stoops JK, Cheng RH, Yazdi MA, Maeng CY, Schroeter JP, Klueppelberg U, Kolodziej SJ, Baker TS, Reed LJ. On the unique structural organization of the Saccharomyces cerevisiae pyruvate dehydrogenase complex. J. Biol. Chem. 1997;272:5757–5764. doi: 10.1074/jbc.272.9.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thekkumkara TJ, Ho L, Wexler ID, Pons G, Liu TC, Patel MS. Nucleotide sequence of a cDNA for the dihydrolipoamide acetyltransferase component of human pyruvate dehydrogenase complex. FEBS Lett. 1988;240:45–48. doi: 10.1016/0014-5793(88)80337-5. [DOI] [PubMed] [Google Scholar]
- Topf M, Baker ML, Marti-Renom MA, Chiu W, Sali A. Refinement of protein structures by iterative comparative modeling and CryoEM density fitting. J. Mol. Biol. 2006;357:1655–1668. doi: 10.1016/j.jmb.2006.01.062. [DOI] [PubMed] [Google Scholar]
- Turkan A, Hiromasa Y, Roche TE. Formation of a complex of the catalytic subunit of pyruvate dehydrogenase phosphatase isoform 1 (PDP1c) and the L2 domain forms a Ca2+ binding site and captures PDP1c as a monomer. Biochemistry. 2004;43:15073–15085. doi: 10.1021/bi048901y. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, Grassucci R, Radke GA, Roche TE. Cryoelectron microscopy of mammalian pyruvate dehydrogenase complex. J. Biol. Chem. 1991;266:24650–24656. [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Yang D, Song J, Wagenknecht T, Roche TE. Assembly and full functionality of recombinantly expressed dihydrolipoyl acetyltransferase component of the human pyruvate dehydrogenase complex. J. Biol. Chem. 1997;272:6361–6369. doi: 10.1074/jbc.272.10.6361. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Chiu W. Determination of icosahedral virus structures by electron cryomicroscopy at subnanometer resolution. Adv. Protein Chem. 2003;64:93–124. doi: 10.1016/s0065-3233(03)01003-9. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Hardt S, Wang B, Sherman MB, Jakana J, Chiu W. CTF determination of images of ice-embedded single particles using a graphics interface. J. Struct. Biol. 1996;116:216–222. doi: 10.1006/jsbi.1996.0033. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Dougherty M, Jakana J, He J, Rixon FJ, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Liao W, Cheng RH, Lawson JE, McCarthy DB, Reed LJ, Stoops JK. Direct evidence for the size and conformational variability of the pyruvate dehydrogenase complex revealed by three-dimensional electron microscopy. J. Biol. Chem. 2001a;276:21704–21713. doi: 10.1074/jbc.M101765200. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, McCarthy DB, O'Connor CM, Reed LJ, Stoops JK. The remarkable structural and functional organization of the eukary otic pyruvate dehydrogenase complexes. Proc. Natl. Acad. Sci. USA. 2001b;98:14802–14807. doi: 10.1073/pnas.011597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Zhang H, Jakana J, Lu X-Y, Zhang J-Q. Cytoplasmic polyhedrosis virus structure at 8 Å by electron cryomicroscopy: structural basis of capsid stability and mRNA processing regulation. Structure. 2003;11:651–663. doi: 10.1016/s0969-2126(03)00091-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.