Abstract
Background: Specific single nucleotide polymorphisms (SNPs) in the fatty acid desaturase (FADS) gene affect the activity and efficiency of enzymes that are responsible for the conversion of polyunsaturated fatty acids (PUFAs) into their long-chain active form. A high prevalence of SNPs that are associated with slow PUFA conversion has been described in Hispanic populations.
Objective: We assessed the heterogeneity of the effect of prenatal supplementation with docosahexaenoic acid (DHA) on birth weight across selected FADS SNPs in a sample of Mexican women and their offspring.
Design: We obtained information on the maternal genotype from stored blood samples of 654 women who received supplementation with 400 mg DHA/d or a placebo from weeks 18 to 22 of gestation through delivery as part of a randomized controlled trial conducted in Cuernavaca, Mexico. We selected 4 tag SNPs (rs174455, rs174556, rs174602, and rs498793) in the FADS region for analysis. We used an ANOVA to test for the heterogeneity of the effect on birth weight across each of the 4 SNPs.
Results: The mean ± SD birth weight was 3210 ± 470 g, and the weight-for-age z score (WAZ) was −0.24 ± 1.00. There were no intention-to-treat differences in birth weights. We showed significant heterogeneity by SNP rs174602 (P = 0.02); offspring of carriers of alleles TT and TC in the intervention group were heavier than those in the placebo group (WAZ: −0.13 ± 0.14 and −0.20 ± 0.08 compared with −0.55 ± 0.15 and −0.39 ± 0.09, respectively); there were no significant differences in offspring of rs174602 CC homozygotes (WAZ: −0.26 ± 0.09 in the intervention group compared with −0.04 ± 0.09 in the placebo group). We showed no significant heterogeneity across the other 3 FADS SNPs.
Conclusion: Differential responses to prenatal DHA supplementation on the basis of the genetic makeup of target populations could explain the mixed evidence of the impact of DHA supplementation on birth weight. This trial was registered at clinicaltrials.gov as NCT00646360.
Keywords: birth weight, DHA, FADS, long-chain PUFAs, prenatal supplementation
INTRODUCTION
The n–3 long-chain PUFA (LC-PUFA)8 DHA is essential for neurodevelopment and regulates gene expression especially during early development through effects on stem cell proliferation (1). DHA has been suggested to play an important role in fetal growth by increasing the duration of gestation (2, 3) or by increasing blood concentrations of insulin-like growth factor I (4).
Maternal PUFA dietary intake and metabolism determine the availability of essential LC-PUFAs for the offspring during the first months of life (5); hence, an adequate maternal DHA status during pregnancy is an important determinant of offspring development (6). However, results from well-designed randomized controlled trials (RCTs) of prenatal supplementation with DHA have yielded heterogeneous or null effects on birth weight (3). The role of fatty acid desaturase (FADS) genes that modulate the conversion of n–3 and n–6 essential fatty acids into their LC-PUFA derivatives (7) could explain this heterogeneity (6, 8–10). The conversion from dietary precursors to LC-PUFAs [including the biologically active DHA and arachidonic acid (AA)] proceeds via a series of desaturations and elongations that are performed by specific enzymatic complexes in which n–6 and n–3 PUFAs compete for conversion (Figure 1). δ-5 Desaturase and δ-6 desaturase are coded in the fatty acid FADS1, FADS2, and, potentially, FADS 3 gene regions (7). Single nucleotide polymorphisms (SNPs) in these genes have been identified as important determinants of an LC-PUFA plasma status through the regulation of this dehydrogenase complex (11–14). Moreover, a recent study identified SNPs in these FADS genes as determinants of adult height and weight in Greenlandic Inuit, which suggested that important adaptations to a diet extremely high in n–3 LC-PUFAs are mediated by these FADS SNPs (15). Previously, important geographical variations in the prevalence of FADS genotypes that are potentially related to genetic adaptation have been described (16, 17). For example, Hispanics (17) and Native American populations have a greater proportion of carriers of the genotype associated with the decreased activity of the δ-desaturase complex (97% compared with 20–50% in Europe and Asia) (16).
FIGURE 1.
Long-chain PUFA metabolism. Conversion from EPA to DHA requires elongation, desaturation (catalyzed by δ-6 desaturase), and β oxidation. Adapted from reference 11 with permission.
On the basis of the important genetic variation in different populations and the lack of evidence from diverse populations of the effect of FADS SNPs on LC-PUFA metabolism (16, 17), there has been a call to include genetic assessments in cohort and intervention studies from different settings (10); such assessments might contribute to explain the heterogeneous results across studies. In the current analysis, we address this gap by describing the distribution of 15 FADS SNPs and assessing if these SNPs modified the effect of prenatal DHA supplementation on birth weight.
METHODS
Sample description
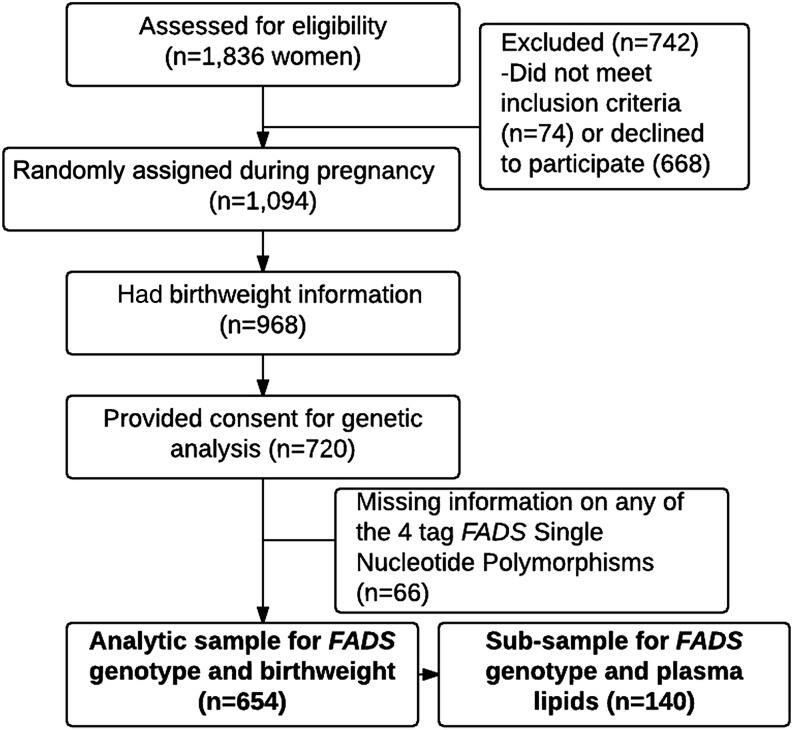
The Prenatal Omega-3 Supplementation on Child Growth and Development study is a double-blind RCT (clinicaltrials.gov; NCT00646360) that was conducted in Mexico from 2004 to 2006 in which 1094 women were randomly assigned, at 18–22 wk of gestation, to receive 400 mg preformed DHA/d or a placebo through delivery. Eligible women were between 18 and 35 y of age and planned to deliver at the Mexican Institute for Social Security General Hospital in Cuernavaca, to breastfeed for ≥3 mo, and to live in the area for ≥2 y after delivery (18). Blood samples of all participating women were stored and, when technology became available, consent to use these samples for genetic testing was requested from participants; of the 980 women who were still participating in the study at delivery, 654 women provided written informed consent for genetic testing (Figure 2) and had valid information for key SNPs.
FIGURE 2.
Sample selection from the Prenatal Omega-3 Supplementation on Child Growth and Development original trial to the analytic sample for this study. FADS, fatty acid desaturase.
The study was conducted according to the guidelines of the Declaration of Helsinki. The Emory University Institutional Review Board and the Mexican National Public Health Institute (INSP) ethics committee approved all procedures involving human subjects.
Birth weight-for-age z score
Anthropometric measurements were obtained from hospital records ≤24 h after delivery. Birth weight was measured to the nearest 10 g with the use of a pediatric scale. Gestational age at birth in days was determined on the basis of the date of birth, maternal recall of the date of the last menstrual period at recruitment, and a maturity evaluation of Capurro made by the pediatrician. Birth weight was converted into z scores relative to WHO Child Growth Standards (19, 20) with the use of an SAS macro (SAS 9.2; SAS Institute Inc.).
Collection and storage of blood samples
Fasting venous blood was obtained at the Mexican Institute of Social Security General Hospital I in Cuernavaca with the use of a evacuated tube system. Samples were centrifuged at 2500 × g for 3 min at 4°C. Plasma, buffy coat, and red blood cells were separated and stored at INSP laboratories at −70°C. Plasma was stored in 200-μL aliquots, whereas erythrocytes were stored in cryotubes after being isolated and washed 3 times with saline solution (0.89%). Samples were kept at INSP laboratories before being transported to the University of Munich for a genetic analysis.
Genotyping
A genetic analysis was conducted at the Helmholtz Center, Munich. DNA was extracted from stored buffy coat with the use of a High Pure PCR Template Preparation Kit (Roche). A total of 5 μL DNA was subjected to polymerase chain reaction amplification followed by the genotyping procedure with the use of the MassARRAY system and iPLEX chemistry as suggested by the manufacturer (Sequenom) and previously described in detail (11). Results of the genetic analysis were entered and cleaned at the University of Munich, and the data sets that contained information on 15 FADS1, FADS2, and FADS3 SNPs (rs174556, rs174455, rs174561, rs174570, rs174574, rs174575, rs2727271, rs174576, rs174578, rs174579, rs174602, rs498793, rs174448, rs174449, and rs174558) were sent to Emory University in encrypted files. These 15 SNPs were selected on the basis of previous evidence of their effects on LC-PUFA metabolism (6, 11, 14, 21, 22). We included additional SNPs located in the FADS3 gene because evidence has suggested that they might also play a role in LC-PUFA metabolism (23).
Determination of plasma fatty acids
A subsample of 140 women with genetic information also had information on plasma concentrations of LC-PUFA at baseline. Samples from these women were randomly selected for the LC-PUFA analysis and analyzed as part of the original study in 2004–2005. Determinations of total fatty acids in plasma were conducted at the INSP laboratories. Total fat from plasma was extracted with a chloroform:methanol mixture (2:1). The yield was determined with the use of gravimetry and was expressed as g/100 g serum. Fatty acids were derivatized with the use of boron trifluoride and were extracted with pure hexane. The extracts were injected into a gas chromatograph (Hewlett-Packard Model 5890 Series II; Hewlett-Packard) with the use of a 100-m length × 0.25-mm internal diameter Supelco SP 2560 column (Supelco). Chromatographic peaks were identified with the use of reference standards for 37 fatty acids (Supelco).
Dietary intake
Maternal dietary intake was assessed with the use of a 110-item food-frequency questionnaire that was specifically designed to include important PUFA sources (24).
Other demographic characteristics
Maternal age was calculated by subtracting the birth date from the date of the interview; women were asked the number of years they had attended school, and socioeconomic status was calculated with the use of principal components analysis on a list of assets ascertained by interview (18).
Statistical analysis
Baseline characteristics of the included subsample were compared with those of the rest of the birth cohort with the use of t tests for continuous variables and chi-square tests for categorical variables. The normality of the concentrations of DHA and AA was assessed with the use of tests for normality by applying PROC UNIVARIATE procedure in SAS 9.2 software (SAS Institute Inc.). Baseline characteristics that differed by intervention groups were included in the models as covariates. P < 0.05 was considered significant.
Distribution of 15 FADS SNPs
Allele frequencies, Hardy-Weinberg equilibrium (HWE), and tests on linkage disequilibrium (LD) were calculated with the use of JLIN (Java LINkage disequilibrium plotter, version 1.6.0; University of Western Australia, 2006) (25). Fisher’s exact test was used to test for HWE. LD was tested by using the likelihood ratio test of allelic associations and pairwise Lewtontin’s D′ squared correlations. JLIN was also used to show LD blocks graphically (Figure 3). A single SNP (or tag SNP) was randomly selected from each cluster of SNPs with an LD > 0.8 to represent the clustering of the 15 SNPs that were originally analyzed.
FIGURE 3.
LD (D′) plot of 15 FADS SNPs. The value 1.0 denotes perfect LD between the SNP pair or 100% power. FADS, fatty acid desaturase; LD, linkage disequilibrium; r^2, r2 power to detect LD; SNP, single nucleotide polymorphism.
Heterogeneity of impact of prenatal DHA on weight-for-age z score by FADS SNPs
We categorized each allele on the basis of the prevalence in this population as previously described (6, 13, 14) where zero denoted a homozygote minor, one denoted a heterozygote, and 2 denoted a homozygote major. We conducted a multivariate linear regression with birth weight as the outcome and tested for a multiplicative interaction between the intervention (zero denoted the control, and one denoted prenatal DHA) and allele categories. We used an ANOVA to assess differences in weight-for-age z scores (WAZs) across intervention groups and FADS SNP genotypes. Analyses were first performed separately for each of the 4 candidate SNPs. The models were then adjusted for all other SNPs and relevant covariates (those which varied between groups or by SNP). Significant confounders (P < 0.05) were included in the final models. We also conducted a mediation analysis by evaluating models with and without controlling for gestational age.
To account for a potentially increased risk of type 1 error because of multiple testing, we used the Bonferroni correction (26). We tested 4 different SNPs and one outcome (WAZ), and Bonferroni-corrected P values <0.013 were considered significant. The sample of n = 654 allowed us to look an effect size of the interaction term as little as 0.3 with >85% power [as calculated with the use of Quanto Power and Sample Size software (version 1.2.4; University of South Carolina, 2009) (27)] for minor allele frequencies (MAFs) as small as 0.2.
Association between FADS SNPs and plasma concentrations of AA and DHA
A linear regression analysis of the additive association between each of the 4 candidate SNPs and DHA and AA plasma concentrations was conducted with the heterozygote with the slowest expected rate of conversion as the reference. A multivariate logistic regression was conducted with all 4 SNPs in the model.
RESULTS
Descriptive characteristics of the 654 women and offspring with FADS genetic information by supplementation group are shown in Table 1. The mean age was 26 y, and the mean height was 155 cm. Mean dietary intakes of n–6 linoleic acid and AA were 19.2 and 0.15 g/d, respectively, and intakes of α-linolenic acid and DHA were 1.7 and 0.08 g/d, respectively. A comparison of the analytic sample to the rest of the birth cohort (n = 326) showed small differences in dietary intakes and sociodemographic characteristics whereby women with genetic information were slightly older (26.5 compared with 25.8 y, respectively), a smaller proportion was primagravid (34% compared with 44%, respectively), and a smaller proportion gave birth to a girl (45% compared with 52%, respectively) (all P < 0.05) (Table 1)
TABLE 1.
Maternal characteristics at baseline and offspring characteristics at birth for women who were participating in the POSGRAD trial with (n = 654) and without (n = 326) genetic information1
| Analytic sample (n = 654) |
Sample missing information (n = 326) |
|||
| Control (n = 318) | DHA (n = 336) | Control (n = 172) | DHA (n = 154) | |
| Maternal characteristics | ||||
| Age,2 y | 26.4 ± 4.73 | 26.5 ± 5.0 | 25.8 ± 4.3 | 25.8 ± 4.3 |
| Socioeconomic status score | 0.06 ± 1.00 | 0.01 ± 0.99 | −0.04 ± 1.04 | 0.10 ± 0.95 |
| Schooling, y | 12.0 ± 3.55 | 11.8 ± 3.45 | 11.9 ± 3.63 | 12.3 ± 3.58 |
| Raven intelligence test score | 41.2 ± 9.41 | 40.6 ± 9.17 | 41.0 ± 8.95 | 41.2 ± 8.43 |
| Height, m | 155 ± 5.58 | 155 ± 5.73 | 156 ± 5.71 | 155 ± 5.96 |
| BMI, kg/m2 | 26.5 ± 4.37 | 26.0 ± 4.16 | 25.8 ± 4.04 | 25.6 ± 4.23 |
| First pregnancy,2 % | 36.2 | 33.3 | 43.6 | 46.8 |
| Dietary intake, g/d | ||||
| n–3 Fatty acids | 1.81 ± 1.14 | 1.85 ± 1.04 | 1.82 ± 0.87 | 1.71 ± 0.94 |
| ALA | 1.69 ± 1.08 | 1.74 ± 1.00 | 1.71 ± 0.83 | 1.59 ± 0.94 |
| DHA | 0.08 ± 0.09 | 0.08 ± 0.07 | 0.08 ± 0.07 | 0.08 ± 0.10 |
| n–6 Fatty acids | 19.3 ± 10.0 | 19.7 ± 9.15 | 18.9 ± 7.79 | 18.9 ± 8.79 |
| LA | 19.2 ± 9.94 | 19.6 ± 9.11 | 18.8 ± 7.78 | 18.8 ± 8.77 |
| AA | 0.15 ± 0.11 | 0.15 ± 0.09 | 0.14 ± 0.06 | 0.15 ± 0.07 |
| Offspring characteristics at birth | ||||
| Girls,2 % | 45.3 | 44.9 | 52.3 | 51.3 |
| Gestational age, wk | 39.0 ± 1.79 | 39.0 ± 1.83 | 39.2 ± 1.96 | 39.0 ± 1.98 |
| Length, cm | 50.4 ± 2.45 | 50.4 ± 2.45 | 50.3 ± 2.30 | 50.0 ± 3.18 |
| Weight, g | 3190 ± 460 | 3210 ± 460 | 3190 ± 520 | 3172 ± 470 |
| Head circumference, cm | 34.2 ± 1.84 | 34.4 ± 1.50 | 34.1 ± 2.01 | 34.2 ± 1.71 |
Chi-square tests, t tests, and an ANOVA were used to test differences between groups. There were no significant differences between control and intervention (DHA) groups for any of the characteristics included in the table. AA, arachidonic acid; ALA, α-linolenic acid; LA, linoleic acid; POSGRAD, Prenatal Omega-3 Supplementation on Child Growth and Development.
Analytic and missing samples were significantly different, P < 0.05.
Mean ± SD (all such values).
Distribution of FADS SNPs
The distribution of 15 FADS SNPs in the study population is presented in Table 2. There were no major violations of the HWE for most of the SNPs (P = 0.005–0.7). The 3 SNPs with a HWE P value <0.05 (rs174449, rs174575, and rs174579) were excluded from additional analyses. Four SNP clusters resulted from the map of LD (Figure 3), and SNPs rs174455, rs174556, rs174602, and rs498793 were selected as tag SNPs representing the FADS1, FADS2, and FADS3 gene clusters. These 4 tag SNPs were included for the assessment of the effect modification of prenatal DHA on birth weight and for the subanalysis of plasma concentrations of AA and DHA.
TABLE 2.
FADS genotype information of 654 women who were participating in the POSGRAD trial1
| Homozygote minor | Heterozygote | Homozygote major | Minor allele frequency | HWE P | |
| FADS1 | |||||
| rs174548 | G (5.1) | GC (36.4) | C (58.6) | 0.22 | 0.05 |
| rs174556 | G (5.2) | GA (38.4) | A (56.4) | 0.24 | 0.19 |
| rs174561 | A (5.2) | GA (38.2) | G (56.7) | 0.24 | 0.19 |
| FADS2 | |||||
| rs174570 | C (7.0) | CT (40.7) | T (52.3) | 0.27 | 0.47 |
| rs174574 | C (4.0) | CA (32.9) | A (62.8) | 0.20 | 0.70* |
| rs174575 | G (14.1) | GC (52.5) | C (33.3) | 0.40 | 0.04 |
| rs174576 | C (3.9) | CA (34.6) | A (61.4) | 0.21 | 0.47 |
| rs174578 | T (4.1) | TA (35.0) | A (61.0) | 0.21 | 0.40 |
| rs174579 | T (11.3) | CT (52.4) | C (36.3) | 0.38 | <0.01** |
| rs174602 | T (14.7) | TC (44.8) | C (40.4) | 0.37 | 0.49 |
| rs498793 | T (13.8) | TC (43.3) | C (42.9) | 0.35 | 0.14 |
| rs2727271 | T (15.1) | AT (48.1) | A (36.7) | 0.40 | 0.80 |
| FADS3 | |||||
| rs174448 | T (7.2) | TC (44.5) | C (48.3) | 0.29 | 0.07 |
| rs174449* | T (6.6) | CT (44.3) | C (48.6) | 0.28 | 0.03 |
| rs174455 | T (4.9) | TC (35.5) | C (59.6) | 0.22 | 0.49 |
Values in parentheses are percentages. *P <0.05, **P <0.01. FADS, fatty acid desaturase; HWE, Hardy-Weinberg equilibrium; POSGRAD, Prenatal Omega-3 Supplementation on Child Growth and Development.
Heterogeneity of the impact of prenatal DHA on birth WAZs across FADS SNPs
In the sample as a whole, the mean ± SD birth weight was 3210 ± 470 g, and the WAZ was −0.24 ± 1.00 with no differences by treatment group (18). We showed a significant interaction by SNP rs174602 (P < 0.01). Offspring of carriers of alleles TT and TC in the intervention group were heavier than those in the placebo group (WAZ: −0.13 ± 0.14 and −0.20 ± 0.08 compared with −0.55 ± 0.15 and −0.39 ± 0.09, respectively); there were no significant differences in the offspring of rs174602 CC homozygotes (WAZ: −0.26 ± 0.09 in the intervention group compared with −0.04 ± 0.09 in the placebo group) (Figure 4). These effects remained significant after adjustment for gestational age although there was evidence of some attenuation (WAZ: −0.18 ± 0.14 and −0.22 ± 0.08 in the DHA group compared with −0.48 ± 0.15 and −0.40 ± 0.09 in the placebo group for TT and TC, respectively; P = 0.03). We showed no significant effect modification by the other 3 FADS SNPs.
FIGURE 4.
Mean ± SEM birth WAZs relative to the WHO Growth Standards of 654 children whose mothers participated in the Prenatal Omega-3 Supplementation on Child Growth and Development trial by 4 tag FADS SNPs and prenatal DHA supplementation. *P-interaction < 0.05 after Bonferroni correction for multiple comparisons. FADS, fatty acid desaturase; SNP, single nucleotide polymorphism; WAZ, weight-for-age z score.
Association between FADS SNPs and plasma concentration of AA and DHA
Plasma concentrations of AA and DHA were available for 140 women, who were similar to the FADS SNP sample in terms of key demographic characteristics (data not shown). In unadjusted single-SNP models, rs174455 (0.66 ± 0.1 mg/dL), rs174556 (0.79 ± 0.1 mg/dL), and rs174602 (0.28 ± 9.1 mg/dL) were positively associated with AA plasma concentrations (P < 0.05), but only rs174556 was still associated with AA after controlling for the other SNPs. For DHA, the only positive association was with rs174556 both in the unadjusted and adjusted models, and it was inversely associated with rs174602 (P < 0.05) (Table 3). The 4 tag SNPs (rs174455, rs174556, rs174602, and rs498793), when included together in the regression analysis, explained 24% and 11% of the variability in plasma concentrations of AA and DHA, respectively.
TABLE 3.
Associations between 4 selected tag FADS SNPs and plasma concentrations of AA and DHA in a randomly selected subsample of 140 women who were participating in the POSGRAD trial1
| AA, g/100 g |
DHA, g/100 g |
|||
| SNPs | Unadjusted2 | Adjusted3 | Unadjusted2 | Adjusted3 |
| rs174455 | 0.67 ± 0.12*** | 0.28 ± 0.19 | 0.12 ± 0.04 | 0.09 ± 0.07 |
| rs174556 | 0.76 ± 0.12*** | 0.69 ± 0.20*** | 0.12 ± 0.04* | 0.14 ± 0.07* |
| rs174602 | 0.28 ± 0.12* | −0.20 ± 0.13 | −0.00 ± 0.04 | −0.12 ± 0.05** |
| rs498793 | −0.03 ± 0.12 | −0.12 ± 0.11 | −0.04 ± 0.04 | −0.06 ± 0.04 |
Values are means ± SDs. Subsample of 140 women was randomly selected and was comparable to the study population and the sample included in the current study. β Coefficients were the results of linear regression models and were interpreted as the changes in plasma fatty acid concentration (mg/dL) per increase in the number of minor alleles (defined as those with the lowest prevalence in this population). *P < 0.05, **P < 0.01, ***P < 0.001. AA, arachidonic acid; FADS, fatty acid desaturase; POSGRAD, Prenatal Omega-3 Supplementation on Child Growth and Development; SNP, single nucleotide polymorphism.
Models represent the linear association of each single SNP and plasma concentrations of AA or DHA independently of the effect of the other 3 tag SNPs.
Models represent the linear association of each SNP and plasma concentrations of AA or DHA adjusted for the other 3 tag SNPs (that were included as covariates).
DISCUSSION
In this sample of Mexican women, the distribution of FADS SNPs was consistent with that previously described in other Hispanic populations in whom FADS SNPs that were associated with a less-efficient conversion of dietary PUFAs into their active forms were particularly prevalent (16, 17). We showed the heterogeneity of the effect of prenatal supplementation with DHA on birth weight. In carriers of the minor allele of FADS2 SNP rs174602, women who received prenatal DHA gave birth to significantly heavier offspring than those of women who received the placebo; this difference was not observed in carriers of the major allele. This interaction was only partially explained by an increase in the duration of pregnancy; hence, the specific biological mechanism behind this effect should be further explored.
Results from the fatty acid analysis may contribute to a better understanding of this interaction. The nature and direction of the associations with the plasma LC-PUFAs AA and DHA varied depending on the gene location of each SNP. FADS1 SNP rs174556 was positively associated with plasma concentrations of AA and DHA, whereas FADS2 SNP rs174602 was inversely associated with DHA after controlling for the effect of the other tag SNPs. This result contributed to the evidence that carriers of the minor allele of SNP rs174602 in our sample were at greater risk of DHA deficiency. A role for FADS2 SNPs on DHA metabolism was expected because the enzyme δ-5 desaturase, which is coded in this gene, catalyzes an important step in the conversion from n–3 EPA into DHA that is not in the AA pathway (11, 28) (Table 1). Previous studies of FADS SNPs and plasma concentrations of LC-PUFAs, which have mostly been conducted in European cohorts, have reported strong associations between FADS SNPs and plasma or erythrocyte concentrations of AA, but the association with DHA has been less clear (9, 10, 11, 21, 22, 28), which is a pattern that is consistent with our results. In a prenatal supplementation trial in the United States (29), women who were homozygous for the minor allele of FADS1 SNP rs174533 had lower red blood cell concentrations of both AA and DHA at the start of the intervention, and after supplementation with 600 mg preformed DHA/d during the last 2 trimesters of pregnancy, the intervention increased the DHA concentration and decreased the AA:DHA ratio only in carriers of the minor allele. Although we did not analyze SNP rs174533, and information on the LD of this SNP with other FADS SNPS was not provided (26), the results provided additional support for a selective beneficial effect of prenatal DHA supplementation in carriers of some minor FADS SNPs alleles.
A potential limitation of this analysis was that a proportion of the women in the original study did not provide consent for the genetic testing; however, there were no major differences between these women and the women included in the current study. Similarly, information on plasma concentrations of LC-PUFAs at baseline was only available for a subsample of 140 women. The demographic characteristics of this more-restricted sub-sample were similar to those of the full study population. In addition, we adjusted for variables that were significantly different between groups (maternal age, parity, and offspring sex). Another consideration was that total fatty acids were measured in plasma and not in erythrocytes because this was the method available in Mexico at the time of the intervention. Plasma total LC-PUFA concentrations have been widely used as a proxy for the tissue fatty acid composition and are considered an effective method in adults (30) but have the limitation that they may be affected by fasting status. In our study, all samples were collected in the morning after overnight fasting. Finally, we showed a significant interaction only with SNP rs174602, which was the one with the highest MAF and, hence, the one with the highest power. It is possible that interactions by other SNPs were not detected because of the lack of power. However, the sample size available was enough to detect interactions as small as 0.3 for MAFs as small as 0.2, which is what was observed for SNPs rs174556 and rs174455.
The high prevalence of alleles associated with the slow n–6 LC-PUFA conversion in our sample and other Latin American populations suggested an evolutionary advantage of alleles that have been associated with the slow conversion of n–6 PUFAs into AA within a context where n–6 linoleic acid is highly abundant (16). However, this genetic panorama raised concerns of greater risk of DHA deficiency in this study population because food sources are scarce (31) and a slow conversion of n–3 PUFAs would further exacerbate this deficiency. Our results that showed the heaviest birth weights in the offspring of major allele homozygotes of SNP 174602 together with the selective impact of prenatal DHA in carriers of the minor allele suggest that this SNP located in the FADS2 gene plays a key role in the adaptation to a diet that is abundant in n–6 PUFAs and poor in n–3. A recent study in Greenlandic Inuit also identified SNP 174602 as showing strong signatures of adaptation to a diet that is high in n–3 PUFAs (15), which provided support of a potentially relevant role of this SNP that induces an intron modification on PUFA metabolism and potentially on long-term growth (15).
Important strengths of this study were as follows: the data collection and laboratory-analysis protocols were standardized, validated, and conducted by trained personnel within a clinical setting; participants of this study were part of a population with high dietary intake of n–6 fatty acids as well as a high prevalence of FADS genotypes reportedly associated with the slow conversion of LC-PUFAs; and we were able to test our hypothesis within an RCT with high-quality information on birth outcomes and a wide range of maternal and offspring sociodemographic characteristics. To our knowledge, this is the first study to report a role of the FADS genotype in the modification of the impact of prenatal DHA supplementation on birth weight.
In conclusion, this study highlights the importance of incorporating current advances in the field of genetics into the design of future nutrition interventions and in the analysis of existing RCTs. In particular, the maternal FADS genotype appears to play a key role in essential LC-PUFA metabolism and availability for the offspring and, consequently, on birth outcomes. Future research should further study the biological mechanisms behind the heterogeneous associations of AA and DHA with different FADS SNPs and haplotypes and assess if the effect modification observed in the current study is consistent across populations with different diets and genetic makeups. Similarly, it will be interesting to study if the impact of prenatal DHA supplementation on birth weight in carriers of the minor allele in our study translates into long-term child growth and development outcomes. Differential responses to prenatal DHA supplementation trials on the basis of the genetic makeup of target populations and PUFA dietary intake could explain the mixed evidence of the impact of DHA supplementation on birth weight.
Acknowledgments
The authors’ responsibilities were as follows—IG-C: drafting and editing of the manuscript; IG-C, PR, ADS, RGF, JARD, AB-V, HD, IR, SV, and RM: collection, analysis, and interpretation of the data; IG-C, PR, ADS, JARD, BK, and UR: concept and study design; and all authors: reading and approval of the submission of the final manuscript and responsibility for the reported research and the manuscript. None of the authors reported a conflict of interest related to the study.
Footnotes
Abbreviations used: AA, arachidonic acid; FADS, fatty acid desaturase; HWE, Hardy-Weinberg equilibrium; INSP, Mexican National Public Health Institute; LC-PUFA, long-chain PUFA; LD, linkage disequilibrium; MAF, minor allele frequency; RCT, randomized controlled trial; SNP, single nucleotide polymorphism; WAZ, weight-for-age z score.
REFERENCES
- 1.Martini I, Di Domenico EG, Scala R, Caruso F, Ferreri C, Ubaldi FM, Lenzi A, Valensise H. Optimization of the viability of stem cells derived from umbilical cord blood after maternal supplementation with DHA during the second or third trimester of pregnancy: study protocol for a randomized controlled trial. Trials 2014;15:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koletzko B, Lien E, Agostoni C, Bohles H, Campoy C, Cetin I, Decsi T, Dudenhausen JW, Dupont C, Forsyth S, et al. The roles of long-chain polyunsaturated fatty acids in pregnancy, lactation and infancy: review of current knowledge and consensus recommendations. J Perinat Med 2008;36:5–14. [DOI] [PubMed] [Google Scholar]
- 3.Campoy C, Escolano-Margarit MV, Anjos T, Szajewska H, Uauy R. Omega 3 fatty acids on child growth, visual acuity and neurodevelopment. Br J Nutr 2012;107:S85–106. [DOI] [PubMed] [Google Scholar]
- 4.Damsgaard CT, Molgaard C, Matthiessen J, Gyldenlove SN, Lauritzen L. The effects of n-3 long-chain polyunsaturated fatty acids on bone formation and growth factors in adolescent boys. Pediatr Res 2012;71:713–9. [DOI] [PubMed] [Google Scholar]
- 5.Innis SM. Essential fatty acids in growth and development. Prog Lipid Res 1991;30:39–103. [DOI] [PubMed] [Google Scholar]
- 6.Koletzko B, Demmelmair H, Schaeffer L, Illig T, Heinrich J. Genetically determined variation in polyunsaturated fatty acid metabolism may result in different dietary requirements. Nestle Nutr Workshop Ser Pediatr Program 2008;62:35–44; discussion 44–9. doi:. [DOI] [PubMed] [Google Scholar]
- 7.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics 2000;66:175–83. [DOI] [PubMed] [Google Scholar]
- 8.Glaser C, Lattka E, Rzehak P, Steer C, Koletzko B. Genetic variation in polyunsaturated fatty acid metabolism and its potential relevance for human development and health. Matern Child Nutr 2011;7(Suppl 2):27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lattka E, Illig T, Heinrich J, Koletzko B. FADS gene cluster polymorphisms: important modulators of fatty acid levels and their impact on atopic diseases. J Nutrigenet Nutrigenomics 2009;2:119–28. [DOI] [PubMed] [Google Scholar]
- 10.Lattka E, Illig T, Heinrich J, Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr 2010;29:277–87. [DOI] [PubMed] [Google Scholar]
- 11.Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, Demmelmair H, Illig T, Koletzko B, Heinrich J. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet 2006;15:1745–56. [DOI] [PubMed] [Google Scholar]
- 12.Guerra A, Demmelmair H, Toschke AM, Koletzko B. Three-year tracking of fatty acid composition of plasma phospholipids in healthy children. Ann Nutr Metab 2007;51:433–8. [DOI] [PubMed] [Google Scholar]
- 13.Rzehak P, Heinrich J, Klopp N, Schaeffer L, Hoff S, Wolfram G, Illig T, Linseisen J. Evidence for an association between genetic variants of the fatty acid desaturase 1 fatty acid desaturase 2 (FADS1 FADS2) gene cluster and the fatty acid composition of erythrocyte membranes. Br J Nutr 2009;101:20–6. [DOI] [PubMed] [Google Scholar]
- 14.Rzehak P, Thijs C, Standl M, Mommers M, Glaser C, Jansen E, Klopp N, Koppelman GH, Singmann P, Postma DS, et al. Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS One 2010;5:e13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 2015;349:1343–7. [DOI] [PubMed] [Google Scholar]
- 16.Ameur A, Enroth S, Johansson A, Zaboli G, Igl W, Johansson AC, Rivas MA, Daly MJ, Schmitz G, Hicks AA, et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet 2012;90:809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–96. [DOI] [PubMed] [Google Scholar]
- 18.Ramakrishnan U, Stein AD, Parra-Cabrera S, Wang M, Imhoff-Kunsch B, Juarez-Marquez S, Rivera J, Martorell R. Effects of docosahexaenoic acid supplementation during pregnancy on gestational age and size at birth: randomized, double-blind, placebo-controlled trial in Mexico. Food Nutr Bull 2010;31(2 Suppl):S108–16. [DOI] [PubMed] [Google Scholar]
- 19.Borghi E, de Onis M, Garza C, Van den Broeck J, Frongillo EA, Grummer-Strawn L, Van Buuren S, Pan H, Molinari L, Martorell R, et al. Construction of the World Health Organization child growth standards: selection of methods for attained growth curves. Stat Med 2006;25:247–65. [DOI] [PubMed] [Google Scholar]
- 20.WHO Multicentre Growth Reference Study Group. WHO Child Growth Standards based on length/height, weight and age. Acta Paediatr Suppl 2006;450:76–85. [DOI] [PubMed] [Google Scholar]
- 21.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol 2010;21:64–9. [DOI] [PubMed] [Google Scholar]
- 22.Lattka E, Koletzko B, Zeilinger S, Hibbeln JR, Klopp N, Ring SM, Steer CD. Umbilical cord PUFA are determined by maternal and child fatty acid desaturase (FADS) genetic variants in the Avon Longitudinal Study of Parents and Children (ALSPAC). Br J Nutr 2013;109:1196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanchard H, Legrand P, Pedrono F. Fatty acid desaturase 3 (Fads3) is a singular member of the Fads cluster. Biochimie 2011;93:87–90. [DOI] [PubMed] [Google Scholar]
- 24.Parra-Cabrera S, Stein AD, Wang M, Martorell R, Rivera J, Ramakrishnan U. Dietary intakes of polyunsaturated fatty acids among pregnant Mexican women. Matern Child Nutr 2011;7:140–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter KW, McCaskie PA, Palmer LJ. JLIN: a java based linkage disequilibrium plotter. BMC Bioinformatics 2006;7:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong RA. When to use the Bonferroni correction. Ophthalmic Physiol Opt 2014;34:502–8. [DOI] [PubMed] [Google Scholar]
- 27.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med 2002;21:35–50. [DOI] [PubMed] [Google Scholar]
- 28.Voruganti VS, Higgins PB, Ebbesson SO, Kennish J, Goring HH, Haack K, Laston S, Drigalenko E, Wenger CR, Harris WS, et al. Variants in CPT1A, FADS1, and FADS2 are associated with higher levels of estimated plasma and erythrocyte delta-5 desaturases in Alaskan Eskimos. Front Genet 2012;3:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholtz SA, Kerling EH, Shaddy DJ, Li S, Thodosoff JM, Colombo J, Carlson SE. Docosahexaenoic acid (DHA) supplementation in pregnancy differentially modulates arachidonic acid and DHA status across FADS genotypes in pregnancy. Prostaglandins Leukot Essent Fatty Acids 2015;94:29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fekete K, Marosvolgyi T, Jakobik V, Decsi T. Methods of assessment of n-3 long-chain polyunsaturated fatty acid status in humans: a systematic review. Am J Clin Nutr 2009;89:2070S–84S. [DOI] [PubMed] [Google Scholar]
- 31.Parra-Cabrera S, Moreno-Macias H, Mendez-Ramirez I, Schnaas L, Romieu I. Maternal dietary omega fatty acid intake and auditory brainstem-evoked potentials in Mexican infants born at term: cluster analysis. Early Hum Dev 2008;84:51–7. [DOI] [PubMed] [Google Scholar]