Abstract
Guanine (G) is a target for oxidation by reactive oxygen species in DNA, RNA and the nucleotide pool. Damage to DNA yields products with alternative properties toward DNA processing enzymes compared to the parent nucleotide. A new lesion, 5-carboxamido-5-formamido-2-iminohydantoin (2Ih) bearing a stereocenter in the base, was recently identified from the oxidation of G. DNA polymerase and base excision repair processing of this new lesion has now been evaluated. Single nucleotide insertion opposite (S)-2Ih and (R)-2Ih in the template strand catalyzed by the DNA polymerases Klenow fragment exo−, DPO4, and Hemo KlenTaq demonstrate these lesions to cause point mutations. Specifically, they promote threefold more G•C → C•G transversion mutations than G•C → T•A, and (S)-2Ih was twofold more blocking for polymerase bypass than (R)-2Ih. Both diastereomer lesions were found to be substrates for the DNA glycosylases NEIL1 and Fpg, and poorly excised by endonuclease III (Nth). The activity was independent of the base pair partner. Thermal melting, CD spectroscopy, and density functional theory geometric optimization calculations were conducted to provide insight into these polymerase and DNA glycosylase studies. These results identify that formation of the 2Ih lesions in a cell would be mutagenic in the event that they were not properly repaired.
Graphical Abstract
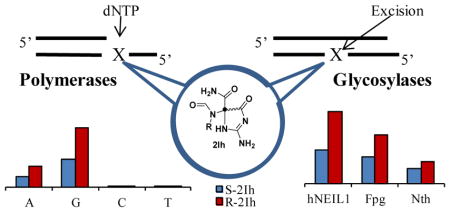
Introduction
DNA damage has been implicated in aging, carcinogenesis, and neurological disorders.1 Reactive oxygen species (ROS) represent one of the key threats that effect damage to the genome yielding strand breaks and base lesions. Base lesions have the ability to stall or cause replicative polymerases to incorporate the incorrect nucleotide that can lead to mutations.2,3 To avoid mutations, biology has a repair system, of which the base excision repair (BER) pathway is one approach to maintain the integrity of the genome.3,4 Mutation profiles observed in a number of cancers have identified critical mutations at guanine nucleotides.5
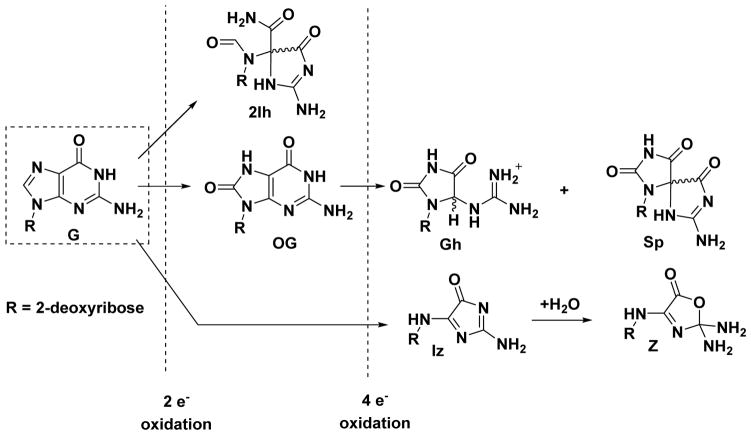
The heterocyclic base of 2’-deoxyguanosine (G) is the most susceptible to oxidative damage because it has the lowest redox potential among the four DNA bases.6 Oxidation of G by two electrons yields 8-oxo-7,8-dihydro-2’-deoxyguanosine (OG).7,8 The four-electron oxidation of G yields two diastereomers of spiroiminodihydantoin-2’-deoxyribonucleoside (Sp), two diastereomers of 5-guanidinohydantoin-2’-deoxyribonucleoside (Gh), and 2,5-diaminoimidazolone-2’-deoxyribonucleoside (Iz) that hydrolyzes to 2,2,4-triamino-2H-oxazol-5-one-2’-deoxyribonucleoside (Z) and other products.7,9–19 Lastly, a second two-electron oxidation product resulting from G oxidation is the diastereomeric pair 5-carboxamido-5-formamido-2-iminohydantoin-2’-deoxyribonucleoside (2Ih, Figure 1).20–28 In eukaryotic and prokaryotic models, many of these lesions have been detected;29–33 however, 2Ih has not yet been the subject of such an investigation.
Figure 1.
Products observed from the oxidation of 2′-deoxyguanosine.
Lesions in templating strands of DNA can induce misinsertions of nucleotides opposite the lesion that are determined by the polymerase active site and the base pairing properties of the lesion. Previous studies have demonstrated that DNA polymerases can synthesize past OG and incorporate dATP or dCTP opposite the lesion, that upon further replication cause G•C → T•A transversion mutations in the absence of repair.34–36 When Sp or Gh are in the template strand, they show preference for incorporation of dATP or dGTP, in which dGTP is more efficiently incorporated opposite Gh than Sp. These polymerase studies identify both lesions as inducing either G•C → C•G or G•C → T•A transversion mutations.34,37–41 Three common classes of DNA polymerases include those that are replicative and replicate the genome, those involved in the DNA damage response whose role is to bypass damaged nucleotides, and PCR polymerases used in sequencing applications. Ideally, mutations are avoided by the action of multiple repair enzymes that remove the damage before polymerase bypass can occur.
The base excision repair (BER) pathway is one mechanism responsible for removal of damaged DNA nucleotides. The first step in the BER process employs a DNA glycosylase to recognize and remove the damaged base. The repair process is then completed by the actions of an endonuclease yielding a gap at the damage site, followed by polymerase filling of the gap, and finally a ligase seals the nick to furnish a repaired strand.42 The ability to repair OG has been found in bacterial and mammalian cells.3 In E. coli, MutM (i.e., Fpg) can remove OG from an OG•C base pair, while MutY removes A from an OG•A base pair.3,43–45 In mammalian cells, OGG1 acts on the OG•C lesion while MUTYH acts on the A in an OG•A base pair.46 Endonuclease VIII (Nei) and endonuclease III (Nth) in prokaryotes have the ability to repair oxidized pyrimidines,3,43,44 while the oxidized purines such as Z, Gh, and Sp are substrates for these glycosylases with reduced activity.47,48 A “Nei-like” glycosylase in humans (NEIL1) has been identified and characterized.49 NEIL1, like Fpg and Nei, is a bifunctional repair enzyme (glycosylase/lyase) that catalyzes N-glycosidic bond hydrolysis and β-,δ-elimination reactions to affect strand scission.2,3,42 NEIL1 was originally found to cleave Fapy-G, Fapy-A, thymine glycol (Tg), 5-hydroxycytosine (5-OHC), and 5-hydroxyuracil (5-OHU);50–52 however, the best substrates found so far for NEIL1 are the diastereomers of Gh and Sp. Furthermore, the efficient removal of Sp or Gh is not dependent on the base opposite in dsDNA.47,53–55 Gh and Sp have also been found to be substrates for Fpg.56,57 As new lesions to DNA are found, their ability to be repaired by BER DNA glycosylases should be assessed to determine if the lesion will persist in DNA and cause problems during polymerase activity.
To date, the ability of 2Ih to cause mutations via incorrect polymerase activity or to evade the BER pathway has not been evaluated. In this study, we investigated polymerase activity toward (R)-2Ih or (S)-2Ih in the template strand using the three polymerases Kf exo− (replicative), DPO4 (translesion synthesis), and Hemo KlenTaq (PCR). Next, we investigated the excision of (R)-2Ih and (S)-2Ih lesions with the BER DNA glycosylases Fpg, Endonuclease III (Nth), and human NEIL1. These polymerase studies identify the diastereomers of 2Ih to induce mutations when bypassed by all three polymerases studied, and the 2Ih isomers are substrates for Fpg and NEIL1, but are poorly acted on by Nth.
Experimental
Materials and Methods
All chemicals were obtained from commercially available sources and used without further purification. All oligodeoxynucleotides (ODNs) were synthesized by the DNA/peptide core facility at the University of Utah and then cleaved, deprotected, and purified following standard protocols.58 NiCR was synthesized by a literature protocol by Dr. James G. Muller (University of Utah).59
Synthesis of Oligonucleotides containing 2Ih or Sp
The 2Ih-containing ODNs were synthesized by oxidizing ODNs 1 (18 mer) and 3 (15 mer) when X and Y = G (Table 1), respectively, in the presence of the nickel(II) complex NiCR as catalyst with KHSO5 as oxidant yielding 2Ih at the only G site via a method previously described by our laboratory.22,60 Briefly, 10 μM ODN in 20 mM NaPi buffer (pH 7.4) and 100 mM NaCl was incubated with 5 μM NiCR at 37 °C for 20 min prior to the addition of 100 μM KHSO5, and the reaction was allowed to progress for 30 min. The reaction was quenched by adding 50 mM HEPES (pH 8). The Sp-containing ODNs were used as a standard to compare how polymerases and BER enzymes react with 2Ih-containing ODNS. The Sp-containing ODNs were synthesized by oxidizing ODNs 1 and 3, in which X and Y = OG. To 10 μM ODN in 20 mM NaPi buffer (pH 7.4) and 100 mM NaCl was added 100 μM Na2IrCl6 at 37 °C for 30 min. The 2Ih- and the Sp-containing ODN diastereomers were separated and purified by an analytical ion-exchange HPLC column in which mobile phase A = 10% CH3CN and 90% ddH2O, and B = 1.5 M NaOAc (pH 7) in 10% CH3CN/90% ddH2O running at 1 mL/min while monitoring the absorbance at 260 nm. The absoluteconfigurations for the diastereomers of 2Ih and Sp have previously been determined, as well as their elution order in an ODN on the ion-exchange HPLC column used in the present studies.60,61 These ODNs were further purified by HPLC to obtain > 95% purity before use. The ODNs with 2Ih or Sp were characterized by either ESI-MS for ODNs 1 (G: calcd 5357.5, found 5356.9; (S)-2Ih or (R)-2Ih): calcd 5391.5, found 5391.0 and 5391.1, respectively; a mixture of the Sp diastereomers: calcd 5389.5, found 5389.6) or MALDI-MS for ODNs 3 (G: calcd 4463.8, found 4464.2; mixture of 2Ih diastereomers: calcd 4497.8, found 4498.2; mixture of Sp diastereomers: calcd 4495.8, found 4496.6).
Table 1.
Oligodeoxynucleotide sequences used in the polymerase and repair enzyme studies.
| ODN1 5’-AAX CCA CCT ACA CAC CTC-3’ |
| ODN2 5’-GAG GTG TGT AGG TGG-3’ ( primer ) |
| ODN3 5’-AAT CCA CYA CAC CTC-3’ |
| ODN4 5’-GAG GTG TNG TGG ATT-3’ |
X or Y = G, OG, (S-2Ih, R-2Ih, S-Sp, or R-Sp), N = A, C, G, or T
Thermal denaturation studies (Tm) and CD measurements
ODNs 3 and 4 (Y = G, (S)-2Ih, or (R)-2Ih, and N = A, C, G, or T, Table 1) were annealed in a ratio of 2:1, respectively, at 10 μM duplex in analysis buffer (20 mM NaPi (pH 7.4) and 100 mM NaCl), followed by heating to 90 °C for 5 min and then allowing to cool slowly to room temperature (~3 h). The samples were kept at 4 °C prior to their analysis. The Tm analysis was conducted by monitoring the absorbance at 260 nm while heating the samples from 15 to 80 °C ramped at 1 °C per min and then cooling down to 15 °C via the same rate. The CD spectra for these samples (5 μM) were obtained on a JACSO 815 CD spectrophotometer. The wavelength scanned ranged from 200 to 320 nm while maintaining the temperature at 20 °C.
ODN labeling
For the polymerase studies, ODN 2 (primer, Table 1) was radiolabeled at the 5’ end using T4-polynucleotide kinase and [γ-32P]-ATP at 37 °C for 30 min following literature methods.58 Unreacted [γ-32P]-ATP and enzyme were removed using a Microspin G-25 column (GE Health Sciences) following the manufacturer’s protocol. The primer was annealed to the template ODN 1 (X = (S)-2Ih, (R)-2Ih, (S)-Sp, or (R)-Sp) in 100 μL of buffer solution (10 mM Tris-HCl, 5 mM MgCl2, and 7.5 mM DTT, pH 7.5) with a molar ratio 2:1 template:primer. In the BER studies, ODNs 3 (Y = (S)-2Ih, (R)-2Ih, (S)-Sp, or (R)-Sp) were labeled at the 5’ end following the same procedure prescribed above, and then were annealed with 20% excess complementary strand ODN 4 (N = A, C, G, or T) in annealing buffer (10 mM EDTA, 150 mM NaCl, and 20 mM Tris-HCl at pH 7.6).
Polymerase studies
Single nucleotide insertion or full primer extension reactions were conducted using the DNA polymerases Kf exo−, DPO4, or Hemo KlenTaq. The polymerization was initiated by mixing a solution containing the labeled template/primer ODNs (final concentration = 50 nM), 10 μM of each dNTP (for the single nucleotide insertion studies), or a mixture of the four dNTPs (for the full primer extension studies), and polymerase (0.2 unit of Kf exo−, 0.5 unit of DPO4, or 0.5 unit Hemo KlenTaq). Reaction mixtures were incubated at 37 °C for 30 min, then quenched with 5 μL of termination solution (95% formamide, 0.1% bromophenol blue, and 0.1% xylene cyanol) and heated to 90 °C for 5 min. These samples were applied to a 20% polyacrylamide gel in the presence of 7 M urea, electrophoresed at 45 W for 2 h, and then put on a phosphor screen for 18 h. The screen was imaged by phosphorimager autoradiography, and the band intensities were quantified using ImagQuant software. The % yields were based on the intensity of the extended strand relative to the total intensity of the reaction.
Glycosylase studies
Single-turnover experiments where [Enz] > [DNA] were conducted by mixing 50 nM of labeled ssODN or dsODN and 500 nM enzyme (Fpg, NEIL1, or Nth) in assay buffer (20 mM Tris-HCl, pH 7.5, 10 mM EDTA, 0.1 mg/mL BSA, and 150 mM NaCl) at 37 °C for 20 min. In order to measure the glycosylase activity, these samples were quenched by adding 5 μL of 0.5 N NaOH and heating to 90 °C for 2 min. The samples were analyzed by gel electrophoresis as described for the polymerase assays. The % yields were based on the intensity of the cleaved strand relative to the total intensity of the reaction. The Fpg and Nth were from commercial sources, while NEIL1 was a kind gift from Dr. Sheila David (University of California-Davis).
Density functional theory calculations
All density functional theory (DFT) energy minimization calculations were conducted using the Gaussian 09 software package.62 Each structure was geometrically optimized using the B3LYP functional63,64 and 6–31G basis set,65,66 while implicitly defining the solvent as water with the polarizable continuum model (PCM).67,68 Scans of the dihedral angles provided lowest energy conformations for the 2Ih diastereomers.
Results and Discussion
Synthesis of 2Ih in oligodeoxynucleotides
The modified nucleoside containing 2Ih is not available in the form of a phosphoramidite, nor is there a suitable precursor to this lesion other than guanine. Therefore, the only available synthetic route to 2Ih incorporation into a specific site in an oligodeoxynucleotide is oxidation of an oligomer that contains only one G. This imposes a limitation on the number of sequence contexts that can be studied. Additionally, the yield of 2Ih is ~10% under ideal conditions, requiring HPLC purification before biochemical studies can be performed. Although 2Ih is formed from a variety of reactive oxygen species including hydroxyl radical formed by iron and copper Fenton-like reactions,21,28 we have found that nickel(II)-catalyzed decomposition of monoperoxysulfate is a high yielding method for conversion of G to 2Ih.22 The square-planar coordination compound NiCR, formed by Schiff base condensation of 2,6-diacetylpyridine with 1,5,9-triazanonane, is a convenient catalyst for this reaction.22
Polymerase studies
To investigate how polymerases would process 2Ih in a template strand, a series of standing start and full extension assays were conducted with the replicative polymerase Kf exo− , DNA damage response polymerase DPO4, and the PCR polymerase Hemo KlenTaq. The template ODN 1 (18 mer; X = G, (S)-2Ih, (R)-2Ih, (S)-Sp, or (R)-Sp) was annealed with the 5’-labeled primer ODN 2 (15 mer) for the single nucleotide insertion and full primer extension experiments. In this study, the concentration of dNTPs was initially studied at near physiological concentrations (10 μM) for Kf exo− and DPO4,69 and Hemo KlenTaq was studied with the standard 100 μM dNTP concentrations. However, DPO4 had activity too low to be reliably quantified at physiological dNTPs, and therefore, the dNTP concentrations were increased threefold.
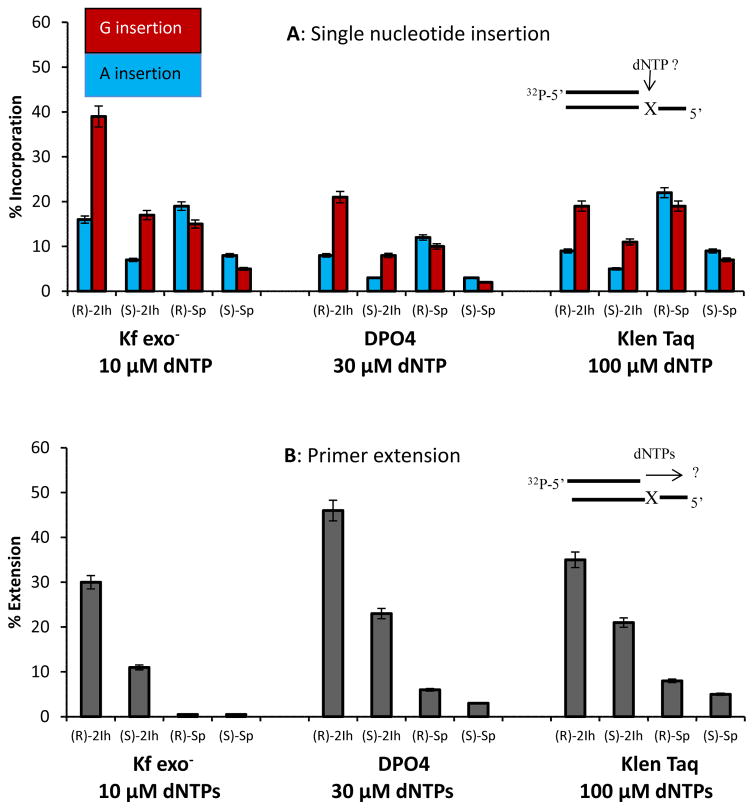
The control experiment with a template G led to the expected result with Kf exo− in which dCTP was inserted opposite; however, when dTTP was the only possible nucleotide, it was inserted in 39% yield, likely due to a feasible G•T wobble base pair (Figure S6). The (R)-2Ih-containing template showed insertion of dGTP (40%) more than dATP (14%); similarly (S)-2Ih-containg templates showed insertion of dGTP (19%) more than dATP (7%, Figure 2A). In these assays, dTTP or dCTP were not observed to be inserted opposite either diastereomer of 2Ih in a yield >1% (Figure S6), and for this reason, only dGTP and dATP insertion data are shown in Figure 2A. Next, all dNTPs were added to the reaction mixture to determine if full primer extension could occur. The control study with a template G showed 95% full extension (Figure S6). When (R)-2Ih was in the template 30% full extension was observed, while a template (S)-2Ih led to only 11% full extension (Figure 2B). These studies with the replicative polymerase Kf exo− demonstrate that when either diastereomer of 2Ih was in the template, dGTP insertion was threefold greater than dATP insertion. Additionally, higher yields of dNTP insertion were observed for the R isomer relative to the S isomer of 2Ih. Lastly, bypass of (R)-2Ih was threefold greater than bypass of (S)-2Ih when all dNTPs were present.
Figure 2.
Comparison of the relative amount of nucleotide insertion opposite (R)-2Ih, (S)-2Ih, (R)-Sp, and (S)-Sp by Kf exo−, DPO4, and Hemo KlenTaq polymerases. (A) Single nucleotide insertion, the dCTP and dTTP insertions were studied but they were not incorporated in a yield >1% and data for these studies are provided in Figures S6, S7, and S8. (B) Full extension of the primer in the presence of all four nucleotides. The error bars represent the standard deviation obtained from three independent reactions.
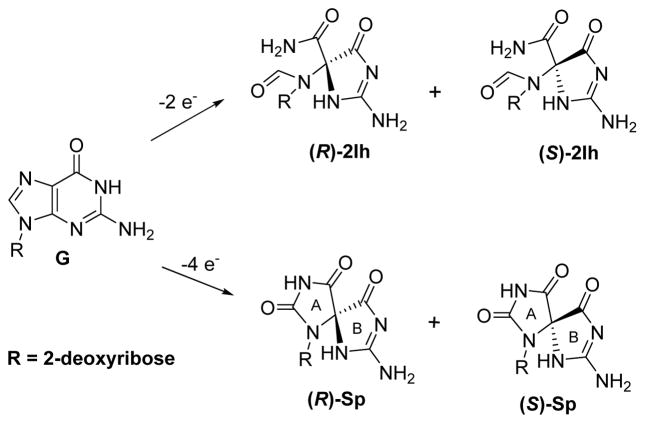
A comparison was then made between the 2Ih data and the diastereomers of Sp in the template strand (Figure 2A). First, it should be noted that the geometric configurations for the R isomers of 2Ih and Sp or S isomers of 2Ih and Sp are essentially the same; the only difference is that in 2Ih, the five-membered ring attached to the sugar is ring opened compared to Sp (Figure 3) and the oxidation state of what was C8 of G is reduced in 2Ih compared to Sp.60,61 When (R)- or (S)-Sp were in the template, dATP (R = 19% and S = 8%) was inserted more often than dGTP (R = 15% and S = 5%). For both template Sp diastereomers, dATP vs. dGTP were selected in nearly equal amounts. Next, when all dNTPs were added to the mixture, primer extension past both Sp diastereomers was not observed with physiologically relevant amounts of dNTPs (Figure 2B). A comparison of 2Ih with Sp identifies the diastereomers of 2Ih to show a much stronger preference for base pairing with G over A (3:1) compared to the ~1:1 preference opposite Sp. Furthermore, 2Ih is more easily bypassed than Sp under identical conditions.
Figure 3.
Comparison of the geometric configurations for the diastereomers of 2Ih and Sp that are products resulting from the oxidation of G.
The lesion bypass polymerase DPO4 was then analyzed for nucleotide insertion opposite 2Ih. Insertion of dGTP opposite (R)-2Ih was observed in a yield of 7%, (S)-2Ih at a 3% yield and no dATP insertion was observed at physiological concentrations of dNTPs (Figure S8). When the dNTP concentrations were increased to 30 μM, the single nucleotide insertion opposite (R)-2Ih and (S)-2Ih yielded more dGTP (R = 25% and S = 9%) than dATP (R = 6% and S = 2%) opposite the lesion (Figure 2A). No insertion of dCTP or dTTP opposite the 2Ih diastereomers was observed with DPO4. When all four nucleotides were added to the reaction mixture, DPO4 was able to provide full extension in a yield of 49% and 23% for (R)-2Ih and (S)-2Ih, respectively (Figure 2B). These results demonstrate that (R)-2Ih is more easily bypassed by DPO4 than (S)-2Ih, and dGTP is inserted nearly five times more than dATP opposite either diastereomer.
Comparisons were then made between DPO4 activities with template 2Ih vs. Sp (Figure 2). Insertions opposite the Sp diastereomers were not observed with 10 μM dNTPs; however, increasing the dNTP concentrations to 30 μM produced insertion opposite the Sp isomers. Specifically, insertion opposite (R)-Sp and (S)-Sp gave dATP (R = 12% and S = 4%) and dGTP (R = 10% and S = 3%) in nearly equal yields. When all four dNTPs were added to the mixture, (R)-Sp was extended by 10% and (S)-Sp was extended by 8%. This observation demonstrates that DPO4 shows similar activity with the Sp diastereomers; in contrast, DPO4 accommodated the R diastereomer of 2Ih better than the S isomer leading to more efficient bypass, and both 2Ih isomers yielded dGTP insertion fivefold more than dATP.
In the last study, the PCR polymerase Hemo KlenTaq was allowed to insert dNTPs opposite a template (R)- or (S)-2Ih diastereomer. At the typical 100 μM dNTP concentration used in PCR amplification, template (R)-2Ih or (S)-2Ih gave more dGTP (R = 17% and S = 11%) insertion than dATP insertion (R = 10% and S = 7%). When all four dNTPs were added to the mixture, the (R)-2Ih was extended with a 34% yield and (S)-2Ih was extended in a 21% yield (Figure 2). These observations demonstrate when 2Ih is present in DNA to be PCR amplified by Hemo KlenTaq, these strands will be poorly extended to full length amplicons, and they will have twofold more G•C → C•G than G•C → T•A transversion mutations. The (R)-2Ih isomer showed less inhibition for Hemo KlenTaq bypass than (S)-2Ih. The (R)-Sp and (S)-Sp diastereomers were shown to yield insertion for dATP (20% and 9%) or dGTP (17% and 7%) by Hemo KlenTaq with full primer extension of 8% and 5% observed for (R)- and (S)-Sp, respectively (Figure 2).
These results illustrate how three polymerases select dNTPs when either of the 2-Ih diastereomers were in the template strand. The incorporation efficiencies were found to be polymerase dependent. All three polymerases studied incorporated dGTP opposite 2Ih more than dATP; in contrast, when Sp was in the template, there was a nearly identical level of insertion of dGTP vs. dATP opposite. Because 2Ih and Sp have similar structures in the B ring (Figure 3), the large contrast in dNTP insertion opposite them was unexpected. Thus, selection of dNTPs opposite 2Ih or Sp must be determined by other features than their similar B-ring structures or similarity in the geometric arrangement of atoms for the diastereomers (Figure 3). Full extension of the primer with either 2Ih diastereomer in the template could be extended with all three polymerases, but not with high efficiency like that observed for a template G. According to this, 2Ih lesions can inhibit the replication by polymerases during in vitro DNA synthesis. These results identify that 2Ih lesions in DNA can predominantly induce G•C → C•G transversion mutations and to a lesser degree cause G•C → T•A transversion mutations. The Sp lesions showed a higher inhibition and cause greater blocking for polymerases compared to 2Ih. The Sp lesions cause nearly equal amounts of G•C → C•G and G•C → T•A transversion mutations, a result consistent with previous studies.37,38
DNA Glycosylase activity toward the diastereomers of 2Ih
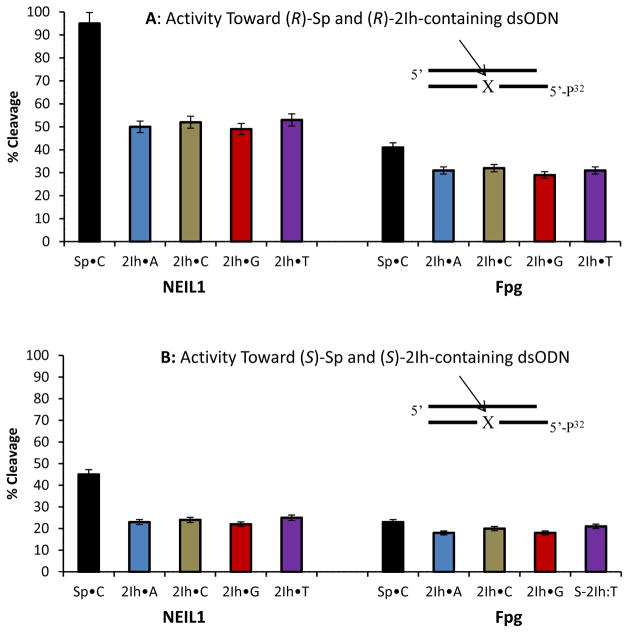
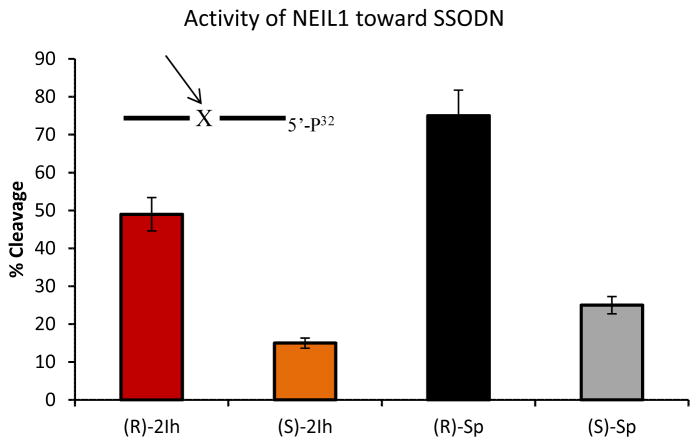
DNA glycosylase activity was measured using ODN 3 (Y = (R)-2Ih, (S)-2Ih, (R)-Sp, and (S)-Sp) for a ssODN context with NEIL1, and ODN 3 (Y = (R)-2Ih, (S)-2Ih, (R)-Sp, and (S)-Sp) annealed with ODN 4 (N = A, C, G, or T) as a dsODN context with the NEIL1, Fpg, or Nth enzymes. Repair activity was analyzed by PAGE after the reactions were run for 20 min and quenched with NaOH (0.2 M) and heat (90 °C for 2 min). NEIL1 successfully cleaved 2Ih-containing dsODNs and showed a 50% yield for (R)-2Ih•C and a 25% yield for (S)-2Ih•C (Figure 4A and B). These reactivities were compared to Sp-containing dsODN that gave > 95% cleavage for (R)-Sp•C and 45% for (S)-Sp•C. From this comparison, the activity of NEIL1 toward 2Ih-containing dsODNs was half that observed with Sp. Therefore, 2Ih is a substrate for NEIL1, but not as good as Sp in this context. Next, NEIL1 activity toward (R)-2Ih and (S)-2Ih-containing dsODNs with different bases (A, C, G, or T) opposite showed the repair efficiency to be independent of the base-pair partner (Figure 4A and B). When Fpg was assayed for cleavage of (R)-2Ih- and (S)-2Ih-containing dsODNs, 35% and 20% product formation was observed, respectively (Figure 4A and B). The cleavage activity of Fpg toward (R)-2Ih and (S)-2Ih was a little lower than observed for (R)-Sp•C (40%) and (S)-Sp•C (25%). When the base pair partners were changed, Fpg showed a similar activity toward (R)-2Ih and (S)-2Ih opposite any of the four nucleotides. The 2Ih lesions were shown to be substrates for Fpg with similar cleavage efficiency as observe for Sp. Finally, Nth was able to cleave the dsODNs with (R)-2Ih (15%), (S)-2Ih (10%), (R)-Sp (16%), (S)-Sp (11%, Figure S15). Compared to Fpg and NEIL1, Nth showed the lowest activity toward 2Ih and Sp lesions, the latter result is consistent with literature reports.47 The DNA glycosylase activity toward 2Ih-containing dsODNs, both the R or S isomers were found to give activity in the order of NEIL1> Fpg> Nth. NEIL1 was able to cleave the 2Ih-containing ssODN to 50% yield for (R)-2Ih and 20% yield for S-2Ih (Figure 5). The Sp-containing ssODNs containing either R or S isomers were better substrates than 2Ih-containing ssODNs, and (R)-Sp cleavage was 70% while 25% cleavage was observed for (S)-Sp.
Figure 4.
Comparison of NEIL1 or Fpg DNA glycosylase activity toward dsODNs containing 2Ih or Sp lesions. (A) Studies when (R)-2Ih was base paired with A, C, G, or T and compared to (R)-Sp•C, and (B) studies when (S)-2Ih base paired with A, C, G, or T was compared to (S)-Sp•C. The error bars represent the standard deviation obtained from three independent reactions.
Figure 5.
Glycosylase activity of NEIL1 toward ssODNs containing (R)-2Ih, (S)-2Ih, (R)-Sp, and (S)-Sp. The error bars represent the standard deviation obtained from three independent reactions.
When DNA lesions cause polymerases to miscode the original message, they cause a mutation if not repaired.3,70 The ability of NEIL1 to initiate repair of the 2Ih lesions in ssODN and dsODNs suggests that cells can defend against them. NEIL1 showed a higher cleavage activity toward Sp in agreement with previous studies.53–55 The 2Ih lesions are viable candidate substrates for cleavage by NEIL1. Fpg was able to cleave both 2Ih and Sp in dsODN with slightly lower efficiency compared to NEIL1 (Figure 4A and B). Again, the Sp lesion cleavage by Fpg was in agreement with previous studies.56,57 The 2Ih lesion is a potential substrate for Fpg to act upon, similarly to other modified purines71 including the initially identified substrates Fapy-G and OG.43,72,73
Nth has shown good activity toward several modified pyrimidine bases,74–78 and very low activity toward modified purines such as Z, Gh, and Sp.47,48 In this study, Nth has the lowest activity among the glycosylases to remove 2Ih and Sp. The results showed that the R isomer of either Sp or 2Ih was always a better substrate for BER glycosylases than the S isomer. The preference of (R)-Sp over (S)-Sp for NEIL1 is not in agreement with what was previously reported in our laboratory.53,54 This difference may indicate that DNA glycosylase activity is dependent on the configuration of lesions as well as the sequence context. The length of the ODN in our previous studies was 30 nucleotides with a context of 5’-CXT-3’ (X = (R)-Sp or (S)-Sp), while the present ODN was 15 nucleotides with the context of 5’-CXA-3’ (X = (R)-Sp or (S)-Sp). Further studies with different sequence contexts and different lengths of ODN would address the importance of sequence length and context for repair activity by NEIL1.
Tm and CD analysis of 2Ih in dsODNs
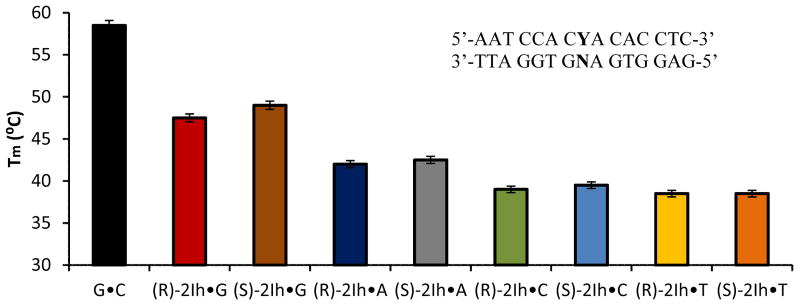
Next, experiments were conducted to address the favorability of dGTP insertion opposite 2Ih in a template strand by a DNA polymerase. As a first test, the halfway points through the melting transition (Tm) for duplexes containing 2Ih and their CD spectra were measured. The duplex ODN comprised of the 15-mer ODN 3 with (R)-2Ih or (S)-2Ih opposite all four canonical nucleotides in ODN 4 (N = A, C, G, or T) allowed Tm and CD spectra of these dsODNs in comparison to a control dsODN containing a G•C base pair. The Tm of ODN with a G•C base pair was 58.5 °C (Figure 6). The Tm of ODNs with (R)-2Ih•G and (S)-2Ih•G base pairs were 11.0 and 9.5 °C less than the G•C base pair, respectively. The (R)-2Ih•A and (S)-2Ih•A base pairs showed a Tm of 16.5 and 16.0 °C lower than the G•C base pair, respectively. The 2Ih•A base pairs gave Tm values 6.5 °C less than the 2Ih•G base pair. The Tm of dsODNs when 2Ih was base paired opposite C or T was relatively lower than the 2Ih•G or 2Ih•A base pairs, and was as follows: (R)-2Ih•C was 39.0 °C, (S)-2Ih•C was 39.5 °C, while (R)-2Ih•T and (S)-2Ih•T were 38.5 °C (Figure 6). From these results, the thermal stability of duplexes containing 2Ih was significantly reduced compared to duplexes with unmodified G. The thermal stability for all duplexes followed a trend decreasing from 2Ih•G > 2Ih•A > 2Ih•C ~ 2Ih•T. These results indicate that 2Ih-containing ODN duplexes with G opposite are the most stable. This adds one line of evidence why dGTP was the preferred nucleotide inserted opposite the 2Ih lesions. Further reasoning for dGTP selection opposite the 2Ih lesions may include hydrogen bonding and base stacking. These properties will be determined when a high resolution structure is solved of a duplex with the 2Ih base pairs.
Figure 6.
Tm analysis data for the 15 mers dsODN, when Y = (R)-2Ih or (S)-2Ih and N = A, C, G, or T. The error bars represent the standard deviation obtained from three independent reactions.
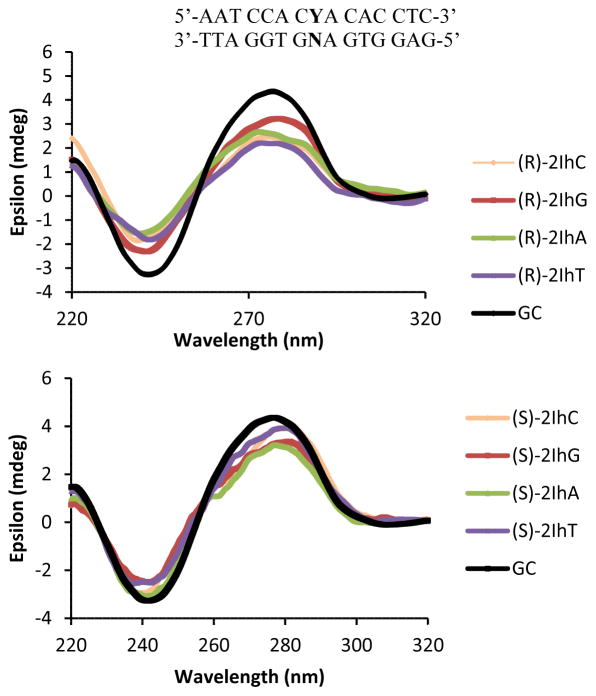
The CD spectra for dsODNs with a G•C base pair as a control and (R)-2Ih or (S)-2Ih when base paired with A, C, G, or T were measured. These CD spectra showed no significant difference in the λmax (277 nm) and λmin (240 nm, Figure 7) between the control duplex and those bearing the 2Ih lesions. These results highlight that 2Ih does not significantly distort the structure of a 15-mer duplex. Previous studies also demonstrated that the Sp diastereomers did not significantly distort the CD spectra for a short duplex.79 These studies support the structural hypothesis that 2Ih and Sp only lead to local disruption of the duplex structure. In the case of Sp, molecular dynamic simulations found that Sp distorted two base pairs 5′ and 3′ to the lesion,80 2Ih most likely has a similar impact on the duplex structure.
Figure 7.
CD analysis data for the 15 mers dsODN, when Y = (R)-2Ih (Top) or (S)-2Ih (Bottom)) and N = A, C, G, T.
Determination of energetically preferred conformations for the 2Ih diastereomers by DFT calculations
Inspection of the 2Ih nucleosides identifies three rotatable bonds that could lead to energetically favorable conformations (Figure 3). A series of DFT calculations were conducted to scan the dihedral angles of these bonds to identify the lowest energy conformations for the two diastereomers. To allow these calculations to finish in a reasonable amount of time, scans of the energy vs. bond angle were conducted for the C1′,N9 bond and the C4,C5 bond when they were locked (Figure S16), while letting the N9,C8 bond to freely rotate during each calculation. The bond angles were scanned in 18° increments for each bond (400 total calculations) in the diastereomers utilizing the B3LYP functional and 6–31G basis set while implicitly defining the solvent as water with the PCM model. Lastly, methyl groups were placed on the 3′ and 5′ alcohols to prevent these groups from acting as H-bond donors. The energy minimization calculations identified distinct conformations, and these lowest energy conformations from the scans were resubmitted to geometric optimization calculations in which all bonds were allowed to freely rotate. These final structures are the ones reported in Figure 8.
Figure 8.
The lowest energy conformations for the 2Ih diastereomers identified by DFT dihedral scan calculations.
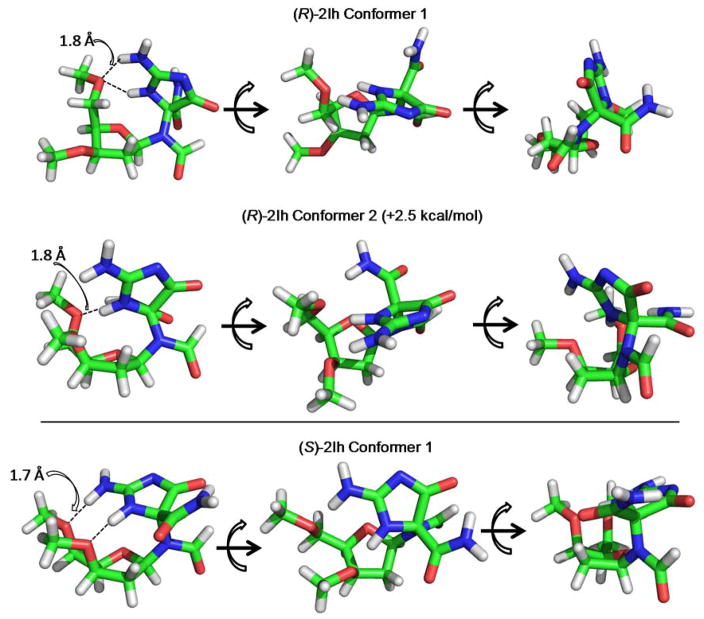
The computational geometric optimizations identified two low energy conformations for (R)-2Ih that differed in energy by 2.5 kcal/mol and one low energy conformation for (S)-2Ih. A few key structural features were identified from these energy minimization calculations. The lowest energy structures were all observed to have H-bonds (distance ~1.6 – 2.0 Å) between the base and sugar, a feature that was not observed in the higher energy structures. Specifically, the lowest energy (R)-2Ih conformer has two H-bonds (N3-O5′, and N2-O5′), while the second lowest energy conformation has one H-bond (N3-O5′, Figure 8). The lowest energy (S)-2Ih conformation has two H-bonds (N3,O3′, and N2,O5′, Figure 8) between the base and sugar. The ability to H-bond between the base and sugar is a feature that has not been observed for the diastereomers of Sp.81 These structures provide initial clues to the stereochemical dependency of the polymerase and BER assays previously described; however, these calculations do not provide a complete understanding of the molecular interactions. Full molecular details of enzyme interactions with 2Ih-containing DNA strands will better be modeled by molecular dynamics, an approach that has been successful in understanding enzyme interactions with Sp-containing DNA substrates.82 The polymerase studies consistently found the R diastereomer of 2Ih to yield twofold greater insertion of dNTPs and bypass relative to the S isomer (Figure 2 and 3). Further, this stereochemical dependency is independent of the polymerase studied suggesting that polymerase activity is dependent on the lesion. The selection of dGTP vs. dATP was nearly the same for the two diastereomers of 2Ih. Polymerase activity on lesion-containing DNA templates will be most efficient if the lesion has complementary H-bonds with an incoming dNTP and the lesion does not distort the polymerase active site preventing the ability to catalyze further polymerization of dNTPs. The 2Ih diastereomers are not planar, and based on the calculations the closed ring is poorly positioned to select a dNTP based on H-bonding potential. Therefore, selection of dGTP or dATP could be due toH-bonds to the amide or formamide groups. Alternatively, if selection is based on dNTP size, slightly larger dGTP would be favored over dATP. Confirmation of nucleotide selection will be best answered when an x-ray crystal structure is solved for 2Ih in the active site of a polymerase. In contrast to the 2Ih results, the Sp diastereomers yielded nearly equal amounts of dATP and dGTP insertion (Figure 2), an observation that identifies dNTP selection is not entirely sterochemically dependent.
Lesion bypass for distorting lesions, such as the 2Ih diastereomers, will show the greatest yield for the isomer that is most accommodating for dNTP insertion after processing of the lesion. On the basis of the DFT scans, the R isomer of 2Ih has a greater degree of flexibility, likely allowing greater bypass. In contrast, the S isomer of 2Ih adopts one conformation that is stabilized by two internal H-bonds leading to a potentially more rigid structure that could be more challenging for polymerases to bypass (Figure S16 and 8). Further, this structure provides a nonplanar base for the next dNTP to π stack with, leading to lower bypass efficiency. In contrast, the Sp diastereomers with two fused rings are more poorly bypassed by polymerases than 2Ih (Figure 2). This observation results from the propeller shape of the Sp base, a feature that has previously been described in the literature.80
Lesion selection and cleavage rate by BER enzymes will be dependent on a number of factors. First, lesion recognition by the DNA glycosylase, and the ability for the active-site nucleophilic lysine residue (Fpg = Lys57 and NEIL = Lys53)83,84 to initiate N-glycosydic bond hydrolysis and strand scission are lesion structure dependent. Again, the DFT calculations identified (R)-2Ih to adopt more conformations than (S)-2Ih. This greater degree of flexibility may allow the R isomer to be more easily processed than the S isomer. This claim is consistent with the experimental results in which (R)-2Ih was more efficiently acted on by Fpg and NEIL1 than (S)-2Ih (Figure 4 and 5). Experiments that evaluate the stereochemical dependency of 2Ih to be repaired and cause mutations in vivo will ultimately determine if these structure based arguments hold in the cell. A similar set of comparative studies have been conducted for the Sp diastereomers that found the in vitro work to provide some explanations for the in vivo studies.37,40
These comparative studies between 2Ih and Sp as substrates for polymerases and BER DNA glycosylase removal have identified properties of 2Ih that might be characteristic of this lesion in the cell. First, the diastereomers of 2Ih will lead to twofold greater G•C → C•G transversion mutations than G•C → T•A; in contrast, the Sp diastereomers gave nearly equal amounts of G•C → C•G and G•C → T•A transversion mutations. Further, the R diastereomer of 2Ih was most easily bypassed by polymerases leading to a greater degree of mutations in the cell. On the basis of these studies, 2Ih will not be as efficiently removed by DNA glycosylases compared to Sp. This feature supports a hypothesis that 2Ih is a highly mutagenic lesion in DNA.
Conclusions
This study demonstrates that the G-oxidation lesion 2Ih when bypassed by DNA polymerases induces misinsertion of dGTP and to a lesser extent dATP during replication in vitro and leads to a significant block for DNA elongation. The insertion of dGTP or dATP opposite 2Ih leads to G•C → C•G and G•C → T•A transversion mutations in the absence of repair. Transversion mutations at G similar to those identified in these studies for 2Ih have been characterized in a number of cancers and diseases.5 These studies also verify that 2Ih is a substrate for certain BER DNA glycosylases. The NEIL1 DNA glycosylase showed activity toward 2Ih in ssODN and dsODNs. Curiously, NEIL1 was able to remove 2Ih in dsODNs regardless of which base was in the opposite strand. Lastly, Fpg and Nth were able to remove 2Ih in dsODNs with lower efficiency compared to NEIL1. Fpg was similar to NEIL1 with removal of 2Ih being independent of the base opposite. These results suggest that polymerases maintain a low degree of activity when 2Ih is in the template strand and DNA glycosylases can initiate the BER process to avoid mutations derived from this lesion.
In comparison to its close analog Sp, 2Ih shows greater differences between the (R) and (S) diastereomers when being processed by either polymerases or BER glycosylases. Additionally, the extent of lesion bypass was significantly greater for both 2Ih diastereomers compared to Sp but particularly high for (R)-2Ih, while the extent of repair was lower for 2Ih compared to Sp. These results suggest that 2-Ih, particularly the (R) diastereomer, could accumulate in the genome, inducing G•C → C•G and, to a lesser extent, G•C → T•A transversion mutations.
The present work paints an initial picture of the potential mutational profile of the 2Ih lesions. We recently showed that 2Ih is formed in vitro from ionizing radiation and Fenton-catalyzed oxidation to a similar extent as OG,21,28 suggesting that it is a significant lesion in vivo. While the OG lesion is very readily bypassed, it also benefits from an efficient repair system for both the OG:C and OG:A base pairs in DNA.3 OG has been widely studied in various contexts in DNA and RNA, and these studies benefit from the synthetic availability of the OG phosphoramidite. Until a better synthetic route to oligomers containing 2Ih in specific sequence contexts is available, further biochemical studies of 2Ih remain challenging.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by a National Institutes of Health grant (R01 CA090689). The oligonucleotides were provided by the DNA/Peptide core facility at the University of Utah that is supported in part by the NCI Cancer Center Support grant (P30 CA042014).
We are appreciative of Dr. Anita M. Orendt (University of Utah) for helpful discussions with the computational work. Computational resources are gratefully acknowledged from the Center for High Performance Computing at the University of Utah.
Abbreviations
- 2Ih
5-carboxamido-5-formamido-2-iminohydantoin
- d
2′-deoxyribose
- BER
base excision repair
- CD
circular dichroism
- dATP
adenosine-2′-deoxynucleoside-5′-triphosphate
- dCTP
cytidine-2′-deoxynucleoside-5′-triphosphate
- DFT
density functional theory
- dGTP
guanosine-2′-deoxynucleoside-5′-triphosphate
- dNTP(s)
2′-deoxynucleoside-5′-triphosphate(s)
- dsODNs
double-strand oligonucleotides
- dTTP
thimidine-2′-deoxynucleoside-5′-triphosphate
- ESI
electrospray ionization
- Fapy•G
2,6-diamino-4-hydroxy-5-formamidopyrimidine
- Fpg
formamidopyrimidine-DNA glycosylase
- G
guanine
- Gh
5-guanidinohydantoin
- Iz
2,5-diaminoimidazolone
- MALDI
matrix-assisted laser desorption ionization
- MS
mass spectrometry
- NEIL1
human endonuclease VIII-like 1
- Nth
endonuclease III
- ODN(s)
oligonucleotide(s)
- OG
8-oxo-7,8-dihydroguanine
- PAGE
polyacrylamide gel electrophoresis
- ROS
reactive oxygen species
- Sp
spiroiminodihydantoin
- ssODNs
single-strand oligonucleotides
- Tm
thermal melting temperature
- Z
2,2,4-triamino-2H-oxazol-5-one
Footnotes
Conflict of Interest
The authors declare no competing financial interests.
Supporting Information
ESI-MS, MALDI-MS, PAGE analysis for the polymerase and BER studies, dihedral scan energies, and coordinates for DFT optimized structures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Beckman KB, Ames BN. Oxidative decay of DNA. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 2.Wallace S, Murphy D, Sweasy J. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer G, Besaratinia A. Mutational spectra of human cancer. Hum Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 7.Angelov D, Spassky A, Berger M, Cadet J. High-intensity UV laser photolysis of DNA and purine 2'-deoxyribonucleosides:_ formation of 8-oxopurine damage and oligonucleotide strand cleavage as revealed by HPLC and gel electrophoresis studies. J Am Chem Soc. 1997;119:11373–11380. [Google Scholar]
- 8.Douki T, Martini R, Ravanat J-L, Turesky RJ, Cadet J. Measurement of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 8-oxo-7,8-dihydroguanine in isolated DNA exposed to gamma radiation in aqueous solution. Carcinogenesis. 1997;18:2385–2391. doi: 10.1093/carcin/18.12.2385. [DOI] [PubMed] [Google Scholar]
- 9.Crean C, Geacintov NE, Shafirovich V. Oxidation of guanine and 8-oxo-7,8-dihydroguanine by carbonate radical anions: insight from oxygen-18 labeling experiments. Ang Chem Int Ed. 2005;44:5057–5060. doi: 10.1002/anie.200500991. [DOI] [PubMed] [Google Scholar]
- 10.Cui L, Ye W, Prestwich EG, Wishnok JS, Taghizadeh K, Dedon PC, Tannenbaum SR. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, singlet oxygen, and γradiation. Chem Res Toxicol. 2013;26:195–202. doi: 10.1021/tx300294d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleming AM, Muller JG, Dlouhy AC, Burrows CJ. Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2'-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing. J Am Chem Soc. 2012;134:15091–15102. doi: 10.1021/ja306077b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gremaud JN, Martin BD, Sugden KD. Influence of substrate complexity on the diastereoselective formation of spiroiminodihydantoin and guanidinohydantoin from chromate oxidation. Chem Res Toxicol. 2010;23:379–385. doi: 10.1021/tx900362r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joffe A, Geacintov NE, Shafirovich V. DNA lesions derived from the site selective oxidation of guanine by carbonate radical anions. Chem Res Toxicol. 2003;16:1528–1538. doi: 10.1021/tx034142t. [DOI] [PubMed] [Google Scholar]
- 14.Luo W, Muller JG, Burrows CJ. The pH-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2001;3:2801–2804. doi: 10.1021/ol0161763. [DOI] [PubMed] [Google Scholar]
- 15.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- 16.Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem Res Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- 17.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin and guanidinohydantoin are the dominant products of 8-oxoguanosine oxidation at low fluxes of peroxynitrite: mechanistic studies with 18O. Chem Res Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 18.Sugden KD, Campo CK, Martin BD. Direct oxidation of guanine and 7,8-dihydro-8-oxoguanine in DNA by a high-valent chromium complex: a possible mechanism for chromate genotoxicity. Chem Res Toxicol. 2001;14:1315–1322. doi: 10.1021/tx010088+. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T, Friesen MD, Ohshima H. Identification of products formed by reaction of 3',5'-di-O-acetyl-2'-deoxyguanosine with hypochlorous acid or a myeloperoxidase H2O2 Cl− System. Chem Res Toxicol. 2003;16:382–389. doi: 10.1021/tx025638y. [DOI] [PubMed] [Google Scholar]
- 20.Banu L, Blagojevic V, Bohme DK. Lead(II)-catalyzed oxidation of guanine in solution studied with electrospray ionization mass spectrometry. J Phys Chem B. 2012;116:11791–11797. doi: 10.1021/jp302720z. [DOI] [PubMed] [Google Scholar]
- 21.Fleming AM, Muller JG, Ji I, Burrows CJ. Characterization of 2'-deoxyguanosine oxidation products observed in the Fenton-like system Cu(II)/H2O2/reductant in nucleoside and oligodeoxynucleotide contexts. Org Biomol Chem. 2011;9:3338–3348. doi: 10.1039/c1ob05112a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghude P, Schallenberger MA, Fleming AM, Muller JG, Burrows CJ. Comparison of transition metal-mediated oxidation reactions of guanine in nucleoside and single-stranded oligodeoxynucleotide contexts. Inorg Chem Acta. 2011;369:240–246. doi: 10.1016/j.ica.2010.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Murthy NN, Telser J, Zakharov LN, Yap GP, Rheingold AL, Karlin KD, Rokita SE. Targeted guanine oxidation by a dinuclear copper(II) complex at single stranded/double stranded DNA junctions. Inorg Chem. 2006;45:7144–7159. doi: 10.1021/ic0605930. [DOI] [PubMed] [Google Scholar]
- 24.Rokhlenko Y, Geacintov NE, Shafirovich V. Lifetimes and reaction pathways of guanine radical cations and neutral guanine radicals in an oligonucleotide in aqueous solutions. J Am Chem Soc. 2012;134:4955–4962. doi: 10.1021/ja212186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomaszewska A, Mourgues S, Guga P, Nawrot B, Pratviel G. A single nuclease-resistant linkage in DNA as a versatile tool for the characterization of DNA lesions: application to the guanine oxidative lesion “G+34” generated by metalloporphyrin/KHSO5 reagent. Chem Res Toxicol. 2012;25:2505–2512. doi: 10.1021/tx300319y. [DOI] [PubMed] [Google Scholar]
- 26.Vialas C, Claparols C, Pratviel G, Meunier B. Guanine oxidation in double-stranded DNA by Mn-TMPyP/KHSO5:_ 5,8-dihydroxy-7,8-dihydroguanine residue as a key precursor of imidazolone and parabanic acid derivatives. J Am Chem Soc. 2000;122:2157–2167. [Google Scholar]
- 27.Ye W, Sangaiah R, Degen DE, Gold A, Jayaraj K, Koshlap KM, Boysen G, Williams J, Tomer KB, Mocanu V, Dicheva N, Parker CE, Schaaper RM, Ball LM. Iminohydantoin lesion induced in DNA by peracids and other epoxidizing oxidants. J Am Chem Soc. 2009;131:6114–6123. doi: 10.1021/ja8090752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alshykhly OR, Fleming AM, Burrows CJ. 5-Carboxamido-5-formamido-2-iminohydantoin, in addition to 8-oxo-7,8-dihydroguanine, is the major product of the iron-Fenton or X-ray radiation-induced oxidation of guanine under aerobic reducing conditions in nucleoside and DNA contexts. J Org Chem. 2015;80:6996–7007. doi: 10.1021/acs.joc.5b00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gedik CM, Collins AF. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 30.Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem Res Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matter B, Malejka-Giganti D, Csallany AS, Tretyakova N. Quantitative analysis of the oxidative DNA lesion, 2,2-diamino-4-(2-deoxy-beta-D-erythro-pentofuranosyl)amino]-5(2H)-oxazolone (oxazolone), in vitro and in vivo by isotope dilution-capillary HPLC-ESI-MS/MS. Nucleic Acids Res. 2006;34:5449–5460. doi: 10.1093/nar/gkl596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pouget J-P, Frelon S, Ravanat J-L, Testard I, Odin F, Cadet J. Formation of modified DNA bases in cells exposed either to gamma radiation or to high-LET particles. Radiat Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 34.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe LG, Guengerich FP. Steady-state and pre-steady-state kinetic analysis of dNTP insertion opposite 8-oxo-7,8-dihydroguanine by Escherichia coli Polymerases I exo− and II exo−. Biochemistry. 1996;35:9840–9849. doi: 10.1021/bi960485x. [DOI] [PubMed] [Google Scholar]
- 36.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 37.Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- 38.Kornyushyna O, Burrows CJ. Effect of the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin on proofreading by Escherichia coli DNA polymerase I (Klenow fragment) in different sequence contexts. Biochemistry. 2003;42:13008–13018. doi: 10.1021/bi0350755. [DOI] [PubMed] [Google Scholar]
- 39.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 40.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- 41.Neeley W, Delaney S, Alekseyev Y, Jarosz D, Delaney J, Walker G, Essigmann J. DNA polymerase V allows bypass of toxic guanine oxidation products in vivo. J Biol Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- 42.Wilson DM, III, Bohr VA. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair. 2007;6:544–559. doi: 10.1016/j.dnarep.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 43.Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- 44.David SS, Williams SD. Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem Rev. 1998;98:1221–1262. doi: 10.1021/cr980321h. [DOI] [PubMed] [Google Scholar]
- 45.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 46.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 47.Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duarte V, Gasparutto D, Jaquinod M, Cadet J. In vitro DNA synthesis opposite oxazolone and repair of this DNA damage using modified oligonucleotides. Nucleic Acids Res. 2000;28:1555–1563. doi: 10.1093/nar/28.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano T, Katafuchi A, Shimizu R, Terato H, Suzuki T, Tauchi H, Makino K, Skorvaga M, Van Houten B, Ide H. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 53.Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair. 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- 57.Krishnamurthy N, Muller JG, Burrows CJ, David SS. Unusual structural features of hydantoin lesions translate into efficient recognition by Escherichia coli Fpg. Biochemistry. 2007;46:9355–9365. doi: 10.1021/bi602459v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fleming AM, Alshykhly O, Zhu J, Muller JG, Burrows CJ. Rates of chemical cleavage of DNA and RNA oligomers containing guanine oxidation products. Chem Res Toxicol. 2015;28:1292–1300. doi: 10.1021/acs.chemrestox.5b00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karn JL, Busch DH. Nickel (II) complexes of the tetradentate macrocycle 2,12-dimethyl-3,7,11,17-tetraazabicyclo (11.3.1) heptadeca-1 (17),2,11,13,15-pentaene. Nature. 1966;211:160–162. [Google Scholar]
- 60.Fleming AM, Alshykhly O, Orendt AM, Burrows CJ. Computational studies of electronic circular dichroism spectra predict absolute configuration assignments for the guanine oxidation product 5-carboxamido-5-formamido-2-iminohydantoin. Tetrahedron Lett. 2015;56:3191–3196. doi: 10.1016/j.tetlet.2014.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fleming AM, Orendt AM, He Y, Zhu J, Dukor RK, Burrows Reconciliation of chemical, enzymatic, spectroscopic and computational data to assign the absolute configuration of the DNA base lesion spiroiminodihydantoin. J Am Chem Soc. 2013;135:18191–18204. doi: 10.1021/ja409254z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frisch GW, et al. Gaussian 09 Revision C01. Gaussian; Wallingford, CT: 2010. [Google Scholar]
- 63.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 64.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 65.Krishnan R, Binkley JS, Seeger R, Pople JA. Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J Chem Phys. 1980;72:650–654. [Google Scholar]
- 66.Frisch MJ, Pople JA, Binkley JS. Self-consistent molecular orbital methods 25. Supplementary functions for Gaussian basis sets. J Chem Phys. 1984;80:3265–3269. [Google Scholar]
- 67.Scalmani G, Frisch MJ. Continuous surface charge polarizable continuum models of solvation. I. General formalism. J Chem Phys. 2010;132:114110. doi: 10.1063/1.3359469. [DOI] [PubMed] [Google Scholar]
- 68.Tomasi J, Mennucci B, Cammi R. Quantum mechanical continuum solvation models. Chem Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 69.Traut T. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 70.Wallace S. Base excision repair: a critical player in many games. DNA Repair. 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J Biol Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- 72.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tchou J, Kasai H, Shibutani S, Chung MH, Laval J, Grollman AP, Nishimura S. 8-oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc Natl Acad Sci U S A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boiteux S, O'Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 75.Wang D, Essigmann JM. Kinetics of oxidized cytosine repair by endonuclease III of Escherichia coli. Biochemistry. 1997;36:8628–8633. doi: 10.1021/bi970341y. [DOI] [PubMed] [Google Scholar]
- 76.Hatahet Z, Kow YW, Purmal AA, Cunningham RP, Wallace SS. New substrates for old enzymes. 5-Hydroxy-2'-deoxycytidine and 5-hydroxy-2'-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2'-deoxyuridine is a substrate for uracil DNA N-glycosylase. J Biol Chem. 1994;269:18814–18820. [PubMed] [Google Scholar]
- 77.Breimer LH, Lindahl T. DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J Biol Chem. 1984;259:5543–5548. [PubMed] [Google Scholar]
- 78.Boorstein RJ, Hilbert TP, Cadet J, Cunningham RP, Teebor GW. UV-induced pyrimidine hydrates in DNA are repaired by bacterial and mammalian DNA glycosylase activities. Biochemistry. 1989;28:6164–6170. doi: 10.1021/bi00441a007. [DOI] [PubMed] [Google Scholar]
- 79.Khutsishvili I, Zhang N, Marky L, Crean C, Patel D, Geacintov N, Shafirovich V. Thermodynamic profiles and nuclear magnetic resonance studies of oligonucleotide duplexes containing single diastereomeric spiroiminodihydantoin lesions. Biochemistry. 2013;52:1354–1363. doi: 10.1021/bi301566v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia L, Shafirovich V, Shapiro R, Geacintov N, Broyde S. Structural and thermodynamic features of spiroiminodihydantoin damaged DNA duplexes. Biochemistry. 2005;44:13342–13353. doi: 10.1021/bi050790v. [DOI] [PubMed] [Google Scholar]
- 81.Jia L, Shafirovich V, Shapiro R, Geacintov N, Broyde S. Spiroiminodihydantoin lesions derived from guanine oxidation: structures, energetics, and functional implications. Biochemistry. 2005;44:6043–6051. doi: 10.1021/bi0473657. [DOI] [PubMed] [Google Scholar]
- 82.Jia L, Shafirovich V, Geacintov NE, Broyde S. Lesion specificity in the base excision repair enzyme hNeil1: Modeling and dynamics studies. Biochemistry. 2007;46:5305–5314. doi: 10.1021/bi062269m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sidorkina OM, Laval J. Role of lysine-57 in the catalytic activities of Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg protein) Nucleic Acids Res. 1998;26:5351–5357. doi: 10.1093/nar/26.23.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: A new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.